Abstract
mRNA applications have undergone unprecedented applications—from vaccination to cell therapy. Natural killer (NK) cells are recognized to have a significant potential in immunotherapy. NK-based cell therapy has drawn attention as allogenic graft with a minimal graft-versus-host risk leading to easier off-the-shelf production. NK cells can be engineered with either viral vectors or electroporation, involving high costs, risks, and toxicity, emphasizing the need for alternative way as mRNA technology. We successfully developed, screened, and optimized novel lipid-based platforms based on imidazole lipids. Formulations are produced by microfluidic mixing and exhibit a size of approximately 100 nm with a polydispersity index of less than 0.2. They are able to transfect NK-92 cells, KHYG-1 cells, and primary NK cells with high efficiency without cytotoxicity, while Lipofectamine Messenger Max and D-Lin-MC3 lipid nanoparticle-based formulations do not. Moreover, the translation of non-modified mRNA was higher and more stable in time compared with a modified one. Remarkably, the delivery of therapeutically relevant interleukin 2 mRNA resulted in extended viability together with preserved activation markers and cytotoxic ability of both NK cell lines and primary NK cells. Altogether, our platforms feature all prerequisites needed for the successful deployment of NK-based therapeutic strategies.
Keywords: MT: Delivery Strategies, natural killer, messenger RNA, lipid nanoparticles, lipoplexes, interleukin-2, LNP, NK cell therapy, IL-2, non-modified mRNA, NK-92 cells, KHYG-1 cells
Graphical abstract

Pichon and colleagues developed imidazole lipids-based mRNA platforms very efficient to transfect both NK-92 cells, KHYG-1 cells and primary NK cells without cytotoxicity. They succeeded to replace IL-2 protein by IL-2 mRNA transfection and obtained NK cells with extended viability with preserved biomarkers and full functionalities to kill target cells.
Introduction
Immunotherapy represents an unprecedented hope for the treatment of cancer. This approach mainly targets CD8+ cytotoxic T lymphocytes by optimizing their anti-tumor functions. Nevertheless, a lasting clinical response is still only observed in a minority of patients. Activating natural killer (NK) cells is another therapeutic modality that holds promise due to their unique ability to recognize and eliminate target cells.1 NK cells are gaining renewed prominence as a strong candidate for the development of novel immunotherapeutic strategies.
NK cells are a subset of immune cells that play a critical role in innate immunity, especially in the host's defense against viral infections and cancers. They can kill various altered cells, including virus-infected and cancer cells, without inducing toxicity to healthy cells.2 Unlike CD8+ T cells, NK cells have the advantage of not relying on antigen specificity. Indeed, they are characterized by the lack of clonally distributed receptors on their surface such as the B cell receptor of B cells and T cell receptor of T cells and their ability to lyse damaged cells without prior immunization.3
NK cell activation is triggered by a combination of activating and inhibitory receptors that recognize specific ligands on target cells. It relies on a mechanism of sensing the absence of major histocompatibility complex class I together with the activation of receptors and co-receptors associated with diseased cells.4 The distinction between a healthy cell and a diseased cell is made possible by a panel of surface receptors capable of integrating danger signals. Activated NK cells release cytotoxic granules containing perforin and granzymes, which induce direct apoptosis of target cells.5 Moreover, they can also produce a variety of cytokines and chemokines, including interferon-gamma (IFN-γ) and tumor necrosis factor-alpha (TNF-α), which can further enhance the immune response by the recruitment of other immune cell types, such as macrophages, dendritic cells, and T cells.6,7
NK cells have a low risk of inducing autoimmunity or cytokine release syndrome. These features make them particularly attractive and approaches that trigger and/or reconstitute NK cell function and proliferation are of great interest. Finding a specific delivery system that strikes a balance between safety, efficiency, and ease of use of NK cells engineering represents a powerful contender for future immunotherapies.8,9
To date, NK engineering is mostly performed with lentiviral vectors10,11 and electroporation,12 which have well-known limitations as biosecurity and cytotoxicity, respectively. Therefore, the use of technology associated with a non-viral nanomedicine platform seems to be an interesting alternative. The use of mRNA technology, which does not require nuclear import, is attractive to express any proteins of interest in NK cells. mRNA delivery in NK cells by lipid-based nanoparticles remains an under-developed field.13 To our knowledge, few studies related to mRNA delivery in NK cells have been published. The first one is based on the use of cationic polymer termed charge altering releasable transporters,14 while others were performed with lipid nanoparticles (LNPs), for a proof-of-concept transfection with a reporter mRNA.15,16
Interleukin 2 (IL-2), a 15.5-kDa cytokine, that plays a critical role in the activation, cytolytic activity, and expansion of NK cells. Mainly produced by T cells,17 IL-2 signals through an intermediate receptors complex composed of IL-2Rβ and IL-2Rγ chains, which are expressed on the surface of NK cells.18 Binding of IL-2 to its receptor leads to the activation of various intracellular signalling pathways, including the Janus kinase-signal transducer and activator of the transcription pathway, resulting in the proliferation and NK cell activation.19 IL-2 has been shown to enhance the cytotoxic activity of NK cells against cancer cells and viral-infected cells by increasing the expression of activating receptors and the release of cytotoxic granules and by enhancing the production of other cytokines.20,21
IL-2 administration was the first immunotherapy to be approved 30 years ago.22 Therapies based on in vivo infusion of IL-2 at high doses showed significant toxicities caused mainly by the activation of immunosuppressive regulatory T cells (Tregs). However, in adoptive cell transfer therapies, low doses of IL-2 in combination with Treg-depleting reagent are nowadays used in pre-clinical settings to decrease toxicity and maintain the efficacity of transferred cytotoxic cells.23 Finding safer and more effective ways to use IL-2 for NK cell therapy has become a priority. For this purpose, different strategies have been developed, including the delivery of plasmid DNA encoding for IL-2 gene or the engineering of IL-2 protein itself.22,24 More recently, engineering a membrane-bound IL-2 on the surface of NK cells was proposed as an alternative to IL-2 infusion.25
The present study showcases the development of novel, stable, scalable, and homogeneous microfluidic lipid-based formulations: namely, liposomes and LNPs. These formulations have proven to be efficient in delivering mRNA into both NK cell lines and primary NK cells, demonstrating their potential as promising tools for future gene therapy and immunotherapy treatments. We produced (1) lipoplexes (Lx), called imidazole cationic delivery-Lx (iCD-Lx), with liposomes made of a cationic lipid (CL) bearing imidazolium and an ionizable lipid (IL) bearing imidazole moiety and (2) LNP, called imidazole delivery-LNP (iD-LNP) made of the same ionizable imidazole lipid with a mixture of helper lipids. Those formulations were efficient to transfect both NK-92 and KHYG-1, two well-characterized human NK cell lines, and human primary NK cells. The delivery of IL-2 mRNA by these formulations sustained the NK cell proliferation, as did IL-2 recombinant protein. Kinetics of mRNA expression, the impact of mRNA modifications on transfection efficiency, and the cytotoxic activity of NK cells transfected with IL-2 mRNA were investigated. Overall, our data indicate the great potential of these formulations for mRNA transfection in NK cell lines and primary NK cells.
Results
Two delivery systems based on imidazole-functionalized lipids were produced with the microfluidic system (Figure 1A). iCD-Lx was made of an ionizable lipid with an imidazole function combined with CL-bearing imidazolium and different helper lipids. iD-LNP was made of the same ionizable imidazole lipid and a mixture of helper lipids (Figure 1B).
Figure 1.
Development of lipid-based systems for mRNA delivery in NK cells
(A) Microfluidic formulation process of LNPs and liposomes, with handmade addition of mRNA to liposome leading to lipoplexes. (B) Schematic representation of lipoplex and LNP composition.
Optimization of iCD-Lx
Different formulations were first produced by varying the percentage of ionizable lipid and Cationic lipid (CL), as well as cholesterol and polyethylene glycol (PEG) lipid analogues. The physicochemical parameters of the produced formulations show that they had an average size ranging from 60 to 130 nm and a low value of polydispersity index (PDI) ≤0.2 (Table S1). As expected, the global cationic charge varies with the amount of CL with a maximum of approximately 55 mV for the Lx made with 50% CL.
The first set of transfections was done in NK-92 cells using unmodified nucleoside (UNR) mRNA bearing an ARCA cap and encoding eGFP. Figure 2A shows that the transfection efficiency varied from approximately 20% eGFP+ cells with 10% molar ratio of CL, up to approximately 75% eGFP+ cells with 20% of CL and a 2-fold mean of fluorescence intensity (MFI) value ranging from 400 to 800. Increasing the amount of CL above 20% molar ratio resulted in a decrease in both the percentage of transfected cells and the translation efficiency (MFI) without any drastic enhancement of cell toxicity. In fact, the highest toxicity was approximately 20% obtained with the formulation made with 10% CL.
Figure 2.
Lipoplexes transfection efficiency
NK-92 cells were incubated at 37°C overnight with the indicated formulation containing eGFP mRNA. After 24 h, the transfection efficiency was measured by flow cytometry. Full bars, % of transfected cells; hatched bars, MFI; grey bars, % of dead cells. (A) Impact on transfection of the CL at different molar percentages (10%, 20%, 30%, 40%, and 50% for CL10, CL20, CL30, CL40, and CL50, respectively). (B) impact on transfection of cholesterol type and DMPE-PEG in liposomes containing 20% CL. iCD-Lx, cholesterol and DMPE-PEG; iCD-Lx (DMG-PEG), cholesterol and DMG-PEG; iCD-Lx (Sito), β-sitosterol and DMPE-PEG; iCD-Lx (DMG-PEG/Sito), β-sitosterol and DMG-PEG. (C) Impact on transfection of lipoplexes made with 20% CL at various N/P ratios [1/1, 2/1, and 3/1]. (D) Stability of liposomes (iCD-Lip) and (E) Lx (iCD-Lx) stored at 4°C. Data are presented as mean ± SD, ∗p < 0.03, ∗∗p < 0.002, ∗∗∗p < 0.001; ns, not significant, in comparison with iCD-Lx.
When the transfection was performed with formulations made with 20% CL and 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (DMPE-PEG) (iCD-Lx-DMPE-PEG) instead of 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (DMG-PEG) (iCD-Lx-DMG-PEG), the percentage of transfected cells decreased by approximately 50% without impacting drastically the MFI although the toxicity increased from 5% to 10% (Figure 2B). Exchanging cholesterol (iCD-Lx) to β-sitosterol (iCD-Lx-β-sitosterol [Sito]) in the formulations did not affect the transfection potency, but having Sito in combination with DMG-PEG (iCD-Lx-DMG-PEG/Sito) tends to accentuate the decrease of transfection efficiency in terms of eGFP+ cells (Figure 2B). Overall, Lx formed with liposomes comprising 20% CL, 40% ionizable lipid, cholesterol, and DMPE-PEG gave the best results in terms of percentage of transfected cells (approximately 75%) and mRNA translation (MFI of 800). The impact of the nitrogen/phosphate (N/P) ratio on mRNA complexation and on transfection efficiency was assessed. An N/P ratio of 1 was enough to fully condense mRNA (Figure S1A). At this ratio, the percentage of transfected cells was maximum (Figure 2C). Note that an N/P ratio of 3 results in a decrease in transfection efficiency (approximately 30%) associated with an increase in toxicity (approximately 20%).
Liposomes (iCD-Lip) and Lx (iCD-Lx) were stable over 8 weeks of storage at 4°C in PBS as assessed by size and PDI measurements (Figures 2D and 2E), with no significant differences in terms of transfection efficiency for iCD-Lx stored at 4°C compared with freshly prepared (Figure S1D). Moreover, when kept at 37°C iCD-Lx were relatively stable in terms of size up to 10 h, either in the absence or in the presence of serum (Figure S1B). The impact of cell density and mRNA dose was checked. The optimal condition for NK-92 and KHYG-1 cells transfection was 0.5 μg mRNA with a cell density of 4 × 105 cells/mL (Figure S1C).
Optimization of iD-LNP
As for Lx, we produced different LNP made with either the imidazole ionizable lipid or Dlin-MC3-DMA for benchmark, combined with different mixtures of helper lipids at an N/P of 10 or 20 (Table S2). All LNPs had a size that ranged between 80 and 100 nm with a high homogeneity (PDI <0.2), and a mRNA encapsulation efficiency of approximately 90%. Transfection data obtained with eGFP MNR (5-MoU) cap1 showed that, in contrast with our ionizable lipid-LNP, MC3-LNPs were not effective to transfect NK-92 cells after overnight incubation in a serum-free medium (Figure 3A). The best LNP was iD-LNP 19 made with 50% of our ionizable lipid, 39% Sito, 10% Dioleoylphosphatidylethanolamine (DOPE), and 1% DMG-PEG and N/P 20. This formulation, further named iD-LNP, was able to transfect approximately 60% of NK-92 cells with the highest MFI. The size of this LNP is stable over 8 weeks at 4°C, with only a slight PDI increase from 0.1 to 0.2 (Figure 3B). When incubated at 37°C in the absence or presence of serum, no significant change in size and PDI occurred up to 10 h (Figure 3C).
Figure 3.
Screening of LNPs for NK-92 cell transfection
(A) Transfection efficiency of various LNPs made with 0.5 μg of MNR (5-MoU) eGFP mRNA on NK-92 cells upon overnight incubation. (B) Size and PDI stability over weeks of storage of iD-LNPs in PBS at 4°C (n = 3 measurements). (C) Stability at 37°C of iD-LNPs in PBS or PBS with 10% serum. Data are presented as mean ± SD.
Morphology and concentration of iCD-Lip and iD-LNP formulations
The morphology of the best liposome and LNP were assessed by cryoelectron microscopy (cryo-EM). Images of iCD-Lip revealed multilamellar structures as expected for a liposome known to be constituted of a lipid bilayer surrounding an aqueous core. Bleb structures comprising a pocket connected to a denser inner core were observed for iD-LNP (Figure S2A). This structure was distinct from solid core nanoparticles often described for LNPs.26 VideoDrop technology based on interferometric light imaging was also used to measure the particles concentration in Lx and in LNP preparations (Figure S2B). Those formulations were prepared at different final lipids concentrations (LNPs: 5 mg/mL; iCD-Lip: 13.41 mg/mL). Measurement of their concentration after mixing revealed a similar order of magnitude for both, with approximately 1012 particles/mL. However, LNPs are made at an N/P of 20 and Lx are prepared at an N/P of 1. Since the transfection was performed with 0.5 μg mRNA, the resulting calculation of particle numbers in the medium of transfection is different: 1.93 × 1010 for LNP vs. 1.7 × 109 for iCD-Lx.
Intracellular distribution and cell uptake studies
Intracellular distribution and uptake studies were performed to gain more knowledge on how our formulations were able to transfect NK-92 cells. For this purpose, mRNA was labeled with fluorescein (fluorescence isothiocyanate [FITC]-mRNA), while liposomes and LNP were labelled by incorporating 0.25% molar ratio of lipid lissamine-rhodamine-B PE. Note that the use of those labelled molecules did not induce significant modifications of the physico-chemical features of both liposomes and LNPs (data not shown). As a control, cells were also incubated with LFM Lx made with FITC-mRNA. Confocal microscopy images of cells incubated for 30 min and 4 h at 37°C with either iCD-Lx or iD-LNP are presented in Figures S3A-a and S3A-b. Both iCD-Lx and iD-LNP were found inside and on the plasma membrane mostly as yellow punctate spots resulting from the merge of red (lipid) and green (mRNA) fluorescence. The number of spots appeared higher with iCD-Lx than with iD-LNP. Concerning iCD-Lx, images obtained after 30 min and 4 h were quite different (Figure S3A-a). After 30 min, iCD-Lx were scattered as yellow spots inside the cell suggesting the colocalization of mRNA and liposome. Upon 4 h incubation, iCD-Lx were mostly localized close to or in the plasma membrane. While no significant differences were observed for iD-LNP after 30 min and 4 h (Figure S3A-b). Strikingly, no labelled spots were observed with LFM Lx after 4 h incubation, which presumably accounts for its inefficiency to transfect those cells (Figure S3A-c).
Knowing that the state of nucleosides modification has an impact on the intracellular fate of mRNA, we conducted uptake experiments with Lx and LNP made with UNR or MNR (5-MoU) mRNA labelled with FITC. A high amount of iCD-Lx was observed on the surface of the cells both after 2 h and 4 h of incubation. No notable difference was found between those made with non-modified and those with modified mRNA. The surface associated-fluorescence intensity of cells incubated with iD-LNP was at least 8- to 6-fold less compared with that of iCD-Lx at 2 h or 4 h of incubation, respectively (Figures S3B-a and S3B-b). This was in line with the higher zeta potential of Lx compared with LNP favoring their binding to the cell surface via electrostatic interactions. The intracellular amount of iCD-Lx measured after 2 h was low and it almost doubled after 4 h. By contrast, the associated intracellular fluorescence of cells incubated with iD-LNP was equal to that bound on the cell surface (Figures S3B-c–S3B-d). As for iCD-Lx, no significant difference was observed between LNPs made with non-modified versus modified mRNA. To assess the presence of mRNA in acidic compartments (endosomes and lysosomes), cells were treated with monensin to neutralize the intracellular environment. It is known that FITC-labelled molecules will have a quenched fluorescence intensity in acidic compartments that can be released by monensin treatment. The more acidic the environment in which mRNA complexes are found is, the higher the increase in fluorescence intensity will be. No increase of the fluorescence intensity was observed (value close to 1) for iCD-Lx both after 2 h and 4 h of incubation, suggesting the presence of FITC-mRNA in a neutral environment as the cytosol (Figure S3). A monensin-increased value of 1.5 for iD-LNP was observed after 4 h, suggesting that some of these particles migrated to late endosomes and/or lysosomes. Those results were in line with their localization close to the plasma membrane for iCD-Lx and inside the cells around the nucleus for LNP observed by confocal microscopy.
Impact of mRNA modifications on the transfection efficiency
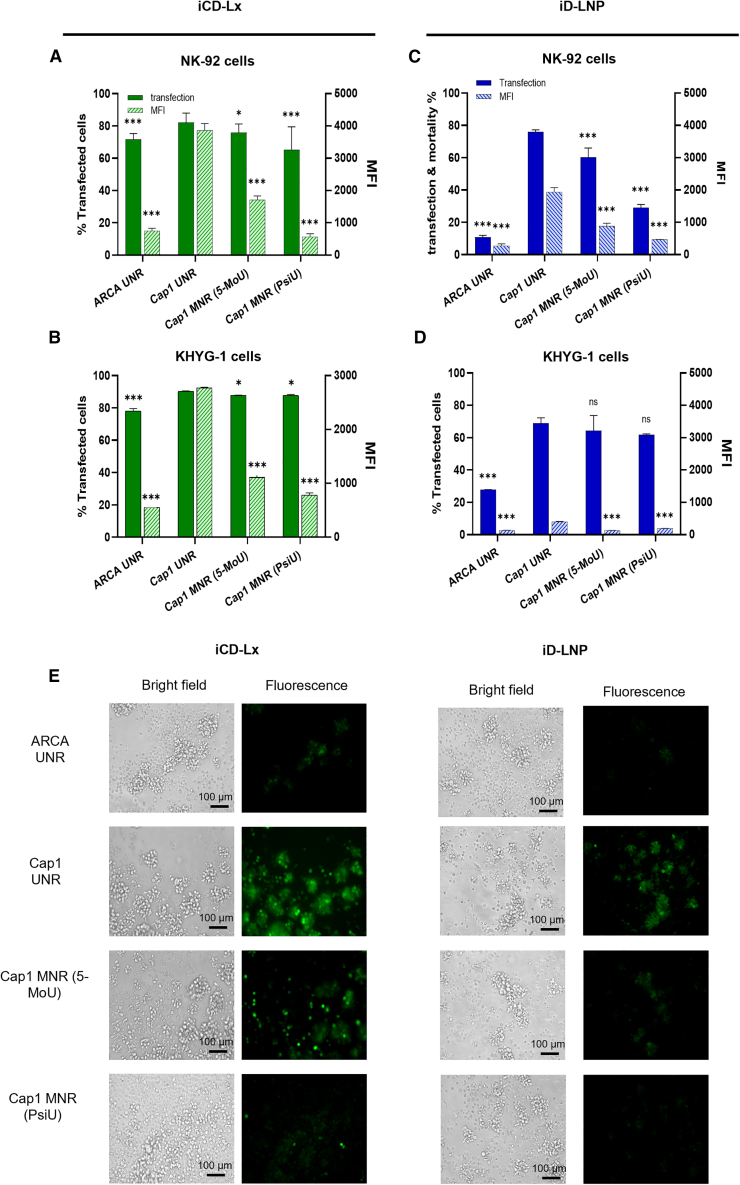
To examine further how NK-92 and KHYG-1 cells respond to mRNA modifications, transfections were performed with four different types of eGFP mRNA: ARCA UNR (non-modified, bearing anti-reverse cap analog), UNR cap1 (non-modified, bearing cap-1), MNR 5-MoU (modified with 5′ moU nucleoside bearing cap-1), and MNR PsiU cap1 (modified with N1-methyl pseudo-uridine, cap-1) (Figure 4). For iCD-Lx, the type of cap used had a drastic impact on the translation efficiency which corresponds to the MFI. The translation efficiency obtained with ARCA UNR was lower compared with cap1 UNR but the percentage of transfected cells was similar in both conditions (Figures 4A and 4B). For iD-LNP, both parameters were affected, ARCA UNR led to the lowest percentage of transfected cells and MFI in both NK-92 and KHYG-1 cells with regard to other cap1 mRNAs (Figures 4C and 4D). Surprisingly, the incorporation of modified nucleosides (5-MoU and PsiU) both in iCD-Lx and iD-LNP had a negative impact on the translation efficiency as shown by the lower values of MFI while the percentage of transfected cells remained the same. iCD-Lx made with cap1 UNR transfected 80% of NK-92 cells with an MFI of 4,000 and 90% of KHYG-1 cells with an MFI of 3,000. It is worth noting that the translation efficiency of iCD-Lx was higher than that obtained with iD-LNP, independent of mRNA type. Fluorescence microscopy observations of transfected cells showed no significant difference in the morphology of NK-92 cells transfected with either iD-Lx or iD-LNP made with different types of mRNA (Figure 4E).
Figure 4.
Impact of mRNA modifications on the transfection efficiency
Cells were transfected overnight with ARCA UNR, cap1 UNR, MNR (5-MoU) or MNR (PsiU) mRNA encoding eGFP formulated as Lx or LNPs. (A) NK-92 cells and (B) KHYG-1 cells transfected with iCD-Lx. (C) NK-92 cells and (D) KHYG-1 cells were transfected with iD-LNPs. After 24 h, the transfection efficiency was measured by flow cytometry. Full bars, percentage of eGFP+ cells; hatched bars, MFI. (E) Fluorescence microscopy observations of NK-92 cells transfected with iCD-Lx or iD-LNP made with indicated mRNA types. Data are presented as mean ± SD, ∗p < 0.03, ∗∗p < 0.002, ∗∗∗p < 0.001; ns, not significant, in comparison with cap-1 UNR.
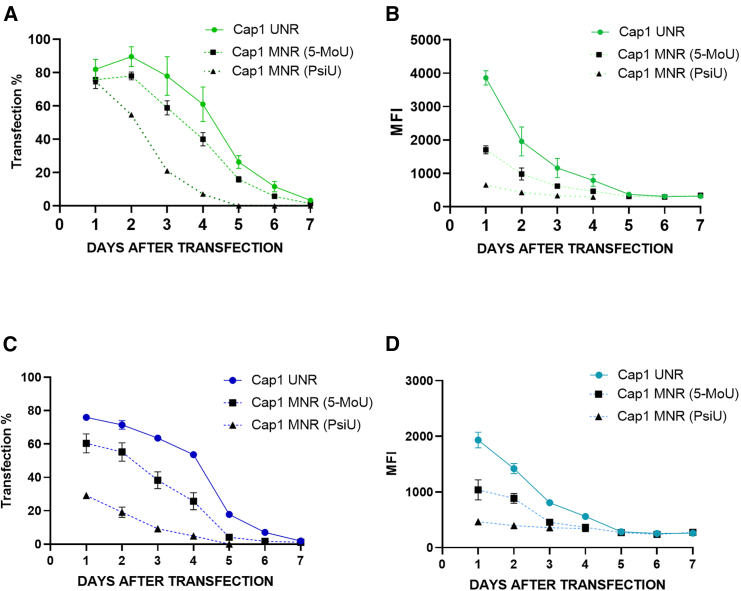
Next, we determined the kinetic of mRNA expression in NK-92 cells upon transfection with either iCD-Lx or iD-LNP (Figure 5). With both formulations, the transfection efficiency (% transfected cells) plateaued for 3–4 days, but the eGFP expression continuously decreased during the following days. Moreover, no differences were observed regarding the use of modified or non-modified nucleotides; UNR mRNA being the highest compared with 5-MoU and PsiU mRNA in terms of eGFP+ cells. Of interest, eGFP translation was already efficient after 6 h transfection with iCD-Lx and iD-LNP (Figure S4).
Figure 5.
Kinetics of mRNA expression
NK-92 cells were transfected overnight with UNR, MNR (5-MoU) or MNR (PsiU) cap-1 eGFP-mRNA formulated as (A and B) iCD-Lx or (C and D) iD-LNP. (A and C) Percentage of transfected cells and (B and D) MFI measured by flow cytometry every day for 1 week. Values are means ± SD of three independent measurements.
Imidazole lipids-based formulations performance compared with commercial and gold standard mRNA delivery systems
The efficacy of iCD-Lx and iD-LNP for eGFP mRNA transfection of NK-92 cells and KHYG-1 cells was first compared with that of commercially available reagents, such as ScreenFect, mRNA-Fect, and Lipofectamine Messenger Max (LFM) (Figures 6A and 6B). None of the commercial reagents tested was significantly efficient to transfect NK-92 cells. ScreenFect and LFM were able to transfect only around 10% of KHYG-1 cells. In contrast, iD-LNP and iCD-Lx allowed approximately 70% and approximately 90% of transfection of both cell lines, respectively. In terms of MFI, which reflects the translation activity, iCD-Lx outperformed iD-LNP, especially on KHYG-1 cells. Compared with Lx, mRNA with LNP was less translated. It was at least 2-fold (4,000 vs. 2,000) to 5-fold less MFI value (2,000 vs. 500) in NK-92 and KHYG-1 cells, respectively. Interestingly, the toxicity was very low (approximately 10%). Then, we compared the impact of the imidazole ionizable lipid with SM-102, ALC-0315, and Dlin-MC3, gold standard ionizable lipids for LNP formulations. For the sake of comparison, the different LNP formulations were made with exactly the same helper lipids composition (Figure 6C), which could be different from their clinically available composition.27 All LNPs exhibited properties (size, PDI, and encapsulation efficiency) very close to our iD-LNP. The latter gave the best transfection efficiency in terms of the percentage of transfected cells and mRNA translation (IL > ALC0315 > SM102 > MC3) on NK-92 cells (Figure 6D). We have also compared the efficiency of our formulations with LNPs made with SM102 ionizable lipid, cholesterol, DSPC, and DMG PEG2K at a 50:38.5:10:1.5 ratio that has been reported to be efficient for NK cells by Douka et al.16 As shown in Figure S5, iCD-Lx is at least as potent as SM102-based LNPs to transfect KHYG-1 cells. While iD-LNP has a comparable efficiency in terms of the percentage of transfected cells, its translation efficiency is lower, as shown by MF1 values (Figure S5B). Our data revealed as well that our formulations have a faster kinetic of transfection. As soon as 5 h after transfection, 25% and 50% of cells were transfected with iCD-Lx and iD-LNP, respectively, while it required 10 h for LNP SM102 to reach this value. However, the efficiency of LNP SM102 and iD-lNP became similar at 20 h after transfection. Overall, these results showcase that our lipid-based platforms (both iCD-Lx and iD-LNP) are promising tools for NK cell mRNA engineering, even when compared with a benchmark LNP made of SM102. We obtained a relatively good transfection efficiency of Jurkat cells used as a gold standard of T cell lines (Figure 6E). Those data suggest that our ionizable lipid could be a versatile ionizable lipid with broad applications.
Figure 6.
Transfection efficiency of iCD-Lx or iD-LNP compared with commercial mRNA transfecting reagents
NK-92 cells (A) and KHYG-1 cells (B) were transfected with Screenfect, mRNAfect, LFM, iCD-Lx, or iD-LNP containing 0.5 μg cap1 UNR eGFP mRNA. Comparison between IL and other ionizable lipids. (C) Table showing the size, PDI, encapsulation efficiency, and concentration of LNPs made with exactly the same lipid composition except for the ionizable lipid (Imidazole-based), SM102, ALC-0315 and D-lin-MC3). (D) NK-92 cells were transfected overnight with indicated LNP encapsulating cap1 UNR eGFP. (E) Jurkat cells were transfected with either iCD-Lx or iD-LNP made of different ionizable lipids. Full blue bars, transfected cells (%); hatched blue bars, MFI; hatched black bars, cell mortality (%). Data are presented as mean ± SD, ∗p < 0.03, ∗∗p < 0.002, ∗∗∗p < 0.001; ns, not significant, in comparison with iD-LNPs.
IL-2 mRNA transfection supports survival and proliferation of NK-92 cells, as does the recombinant IL-2 protein
NK-92 cells require IL-2 supplementation for their survival and proliferation. We investigated whether IL-2 mRNA transfection could replace supplementation with recombinant protein. Cells were first washed and deprived of IL-2 recombinant protein before transfection with either iCD-Lx or iD-LNP containing cap1 UNR IL-2 mRNA. In the absence of IL-2, approximately 100% of cells died after 3 days in culture (Figures 7A–7C). When recombinant IL-2 protein was added in the medium, the cells survived up to 6 days. The same viability was observed with cells transfected with IL-2 mRNA LNP (Figure 7B). Interestingly, IL-2 mRNA transfection with iCD-Lx tended to extend the cell viability up to 8 days (Figure 7A). The production of IL-2 protein by transfected cells was validated by western blots performed with both the cell lysate and the cell medium (Figure S6A). Starvation of IL-2 protein triggered a decrease in cell size and granularity that was not observed in the presence of IL-2 added as IL-2 recombinant protein or produced by IL-2 mRNA transfection (Figure S6B). The proliferation of transfected cells was demonstrated from the cell cycle analysis. Cells were arrested in G0/G1 phase when cultured in IL-2-free medium, while they were in S and G2/M phase upon IL-2 mRNA transfection, as well as when cultured in a medium containing IL-2 protein (Figure S6C). Next, we were wondering if we could make a co-delivery of IL-2 mRNA with another mRNA such as eGFP mRNA (chosen for its easy readout). NK-92 cells were transfected with iCD-Lx complexed with an equal amount (0.25 μg each) of UNR eGFP mRNA and IL-2 mRNA (Figure 7C). The co-delivery also allowed the expression of eGFP protein up to 6 days (Figure S7). When cells were transfected with 0.25 μg IL-2 mRNA, the cells were viable up to 6 days compared with 8 days obtained with 0.5 μg IL-2 mRNA (Figures 7B vs. 7C).
Figure 7.
Impact of IL-2 mRNA transfection on cell viability and cytotoxicity activity
NK-92 cells were either incubated with or without IL-2 protein or transfected with cap1 UNR IL-2 mRNA formulated as iCD-Lx (A) or iD-LNP (B). (C) Cells were incubated with iCD-Lx containing 0.25 μg cap1 UNR eGFP and 0.25 μg IL-2 mRNA. The viability of the cells was measured every day by flow cytometry and expressed as a percentage relative to the total cells. Statistical analysis was done by comparison to IL-2 protein values. (D and E) NK-92 and K562 cells were co-cultured at diverse effector/target ratios for 4 h. The cytotoxicity was evaluated by measuring the expression of caspase 3/7 in K562 cells after NK-92 cells transfection with iD-LNP (D) and iCD-Lx (E). All data are represented by means ± SEM; n = 3 for each experiment. Paired Student t tests were used for statistical analysis.
IL-2 mRNA transfection preserves NK-92 cells functionality
Next, we examined NK-92 cell functionality after IL-2 mRNA transfection. We checked NK-92 cell functional activity in co-culture with K562 cancer cells known to be sensitive to NK cell killing. The addition of either IL-2 protein, IL-2 mRNA iD-LNP or IL-2 mRNA iCD-Lx allowed an increased response in the expression of caspase 3/7 (a cytotoxicity marker) of supplemented NK-92 cells compared with starved cells (Figures 7E and 7F). We then checked the phenotype of NK-92 cells after iD-LNP transfection. As expected, IL-2 starvation led to a decrease in CD69 expression that was not observed with IL-2-supplemented or IL-2 mRNA-transfected NK-92 cells. The expression of NK activation receptors such as NKG2D or NKP30 is essential for NK cell activation and cytotoxicity. Interestingly, the expression of NKG2D and NKP30 was maintained when IL-2 was available compared with starved cells (Figures S8A and S8B). Moreover, during the incubation with K562 cells, the production of TNF-α was maintained in IL-2 transfected cells at a level similar to that obtained with IL-2 recombinant protein, while IL-2 starvation led to a significant decrease (Figure S8C). Altogether, our results show that the supplementation of IL-2 by mRNA transfection is comparable to the use of recombinant protein, validating the potency of our formulation.
Assessing transfection potency on primary NK cells
Finally, we wanted to study those new formulations in primary NK cells. NK cells from different donors were isolated and expanded. As observed for NK-92 cells, the transfection with iCD-Lx of eGFP mRNA allowed the transfection of more than 80% of primary NK cells, while iD-LNP allowed a lower efficacy at around approximately 50% (Figure 8A) and did not affect cell viability (Figure 8B). We then compared the phenotype and functions of primary NK cells when starved or supplemented with either IL-2 protein or after IL-2 transfection by iCD-Lx. As observed previously for NK-92 cells (Figure S8), IL-2-deprived primary NK cells express less CD69 activation marker, NKG2D, and NKP30 compared with conditions with IL-2 protein supplementation or tIL2 mRNA transfection (Figures 8C–8E). Indeed, the proliferation of primary NK cells was comparable with either IL-2 protein supplementation or IL-2 mRNA transfection and it is higher compared with that of starved cells (Figure 8F). Last, we checked the primary NK cell killing activity in co-culture with K562 cancers cells. The addition of IL-2 either as a protein or mRNA transfected allowed a slight increase in the expression of TNF-α for each donor (Figure 8G), but it triggered a greater cytotoxicity response compared with starved cells by looking at caspase 3/7 activity from K562 cells (Figure 8H). Nevertheless, it is important to emphasize the remarkable efficacy of iCD-Lx to transfect the notoriously difficult primary NK cells.
Figure 8.
Transfection efficiency of primary human NK cells with iCD-Lx IL-2 mRNA
Primary human NK cells were obtained from at least three different donors. (A) The cells were transfected with iCD-Lx and iD-LNP containing eGFP mRNA. After 24 h the number of eGFP+ cells was determined by flow cytometry. (B) Viability of primary NK cells 48 h after eGFP mRNA transfection with iCD-Lx and iD-LNP measured by flow cytometry using live/dead dye and expressed as a percent relative to the total cells. (C–E) CD69, NKP30 and NKG2D expressions measured by flow cytometry after 48h of transfection with IL-2 mRNA iCD-Lx, IL-2 protein addition or starvation. (F) Evaluation of primary NK cells proliferation 48 h after transfection with IL-2 mRNA iCD-Lx, IL-2 protein addition or starvation. (G and H) primary NK cells and K562 cells were co-cultured for 4 h at various Effector/Target cell ratios. (G) The production of TNF-α in primary NK cells measured by flow cytometry. (H) Cytotoxicity measured by the expression of caspase 3/7 in K562 cells by flow cytometry. All data are represented by means ± SEM; n = 3 for each experiment. Each condition represents a donor. Paired Student t tests were used for statistical analysis.
Discussion
Today, viral vectors and electroporation are considered as state-of-the-art methodologies for NK cell engineering. Despite their efficiency, they have well-known limitations due to the use of viral components and the high cost of production for viral vectors, as well as the high toxicity for the electroporation. Therefore, there is a crucial need for the development of new transfection modalities. Recent advances in non-viral nanomedicine platforms for nucleic acids delivery have prompted the use of mRNA technology to address this technical shortcoming. Herein, we described the development of Lx (iCD-Lx) and LNP (iD-LNP) that are highly efficient for mRNA delivery in NK cells. Both formulations share the same ionizable lipid bearing an imidazole head group. Lx contain an additional CL with an imidazolium head group. While imidazolium exhibits a positive charge at a neutral pH, allowing an interaction with a negatively charged nucleic acid, the imidazole head group with a pKa in the range of 5.5–6.2,28 is protonated at a pH of <7 (typically encountered in endosomes during nanoparticle trafficking), inducing fusion and/or permeation of negatively charged endosomal membrane.29 The imidazole group can be found in histidine, nucleic acids, and some natural compounds, and it has been demonstrated to have favorable features in many therapeutic strategies.30 The heterocyclic structure of imidazole can be interesting for LNP-mRNA delivery, as it was found to activate the STING pathway for adjuvant properties31 or to simply enhance the condensation of mRNA through π interactions.32 Using the same parameters of the microfluidic system, we produced LNP and liposomes with an average size around 100 nm and a PDI of <0.2, which falls within the range of most other nanoparticles.33 Moreover, our iCD-Lx and iD-LNP showed a good stability in terms of size and PDI within at least 8 weeks when stored at 4°C in PBS. The most efficient Lx was made with liposomes comprising 20% CL and 40% ionizable lipid in combination with 38.5% cholesterol and 1.5% DMPE-PEG. When liposomes were made with higher amount of CL, iCD-Lx had a lower transfection efficiency, which is not due to high toxicity, as usually reported.34 The reduced transfection efficiency could result from a stronger interaction between mRNA and liposomes that likely impacts the structure and/or the availability of mRNA for translation, as we described previously.35
Data from optimization experiments highlight how important the composition of helper lipids is in the shaping of a formulation’s efficiency. The transfection efficiency of iD-LNP containing cholesterol (Chol)/DSPC/DMPE-PEG was improved by changing cholesterol for Sito, while the same change in liposomes for iCD-Lx did not improve it. It is widely known that cholesterol in lipidic formulations has the capacity to impact the membrane’s fluidity. This improved transfection efficiency of LNPs brought by Sito agrees with the study reported by Patel et al.,36,37 showing that this replacement has led to a 4,000-fold higher transfection efficiency of their LNP. This positive impact was explained by the induction of some edge imperfections in the organization of lipids within LNP, which could improve intracellular uptake and endosome destabilization during endosomal escape. Similarly, the presence of DMG-PEG rather than DMPE-PEG impacted negatively iCD-Lx transfection, while it improved that of LNP. The main difference between the two PEG lipids is in their head, being glycerol for DMG-PEG and ethanolamine for DMPE-PEG. Those results are also in line with previous reports suggesting that the tailored structure of DMG-PEG can improve its dissociation from LNP enhancing the transfection efficiency.38,39
An important parameter to consider is the charge ratio (N/P) between the ionizable or CL and mRNA. While most of the N/P ratio for LNP focuses between 6 and 10 in the literature, our LNP is used at an N/P ratio of 20, which did not induce high toxicity and exhibit a greater transfection potency than other counterparts with N/P 10. Of note, LNPs with low N/P ratio are often related to vaccination purpose that remains to be studied in the case of ex vivo cell engineering.
Considering Lx, increasing the N/P ratio from 1 to 3 led to a decrease in transfection efficiency (Figure 2). As stated, too much condensation of mRNA by cationic liposomes (as shown by loss of signal on gel shift assay) can cause a denaturation of the mRNA structure, which leads to a lower translation efficiency.35 Interestingly, while LNPs are often described as solid core nanoparticles,40 ours exhibit structures referred as blebs that have been reported by others, and for which the formation is not yet well understood. It is worth mentioning that such bleb structures have also been observed in LNPs containing another type of imidazole lipid and produced with a buffer concentration in a similar range than ours.32 Bleb formation could depend on either the acidic buffer concentration used to solubilize mRNA or the type of the ionizable lipid.26,32,40 In contrast with LNPs, mRNA is sandwiched by lipid bilayers in Lx. Imidazolium Lx exposes positive charges on their surface, which likely favor the interaction with the cell plasma membrane. LNPs are neutral nanoparticles, the ionizable lipid being inside the particles does not interact with the plasma membrane. Those differences in structures and composition can explain the distinction between observed cellular interactions and internalization of those two types of mRNA particles in the uptake study with NK-92 cells (Figure S2). As expected, the cell surface binding of iCD-Lx was higher than iD-LNP, because of their cationic charge due to the CL. Despite a higher binding of iCD-Lx compared with iD-LNP, the internalized payload is equivalent for both delivery systems.
These data suggest that the amount of internalized particles does not dictate the transfection efficiency. Monensin effect on the fluorescence intensity of cells incubated with iD-LNP and iCD-Lx revealed that the former was likely located in acidic compartments while the latter was in neutral compartments (caveolae) or in the cytosol.41 The presence of iD-LNP in acidic compartments can be associated with a lower and/or slower escape potency and thus less mRNA availability in the cytosol. This result could be linked with their lower transfection efficiency compared with Lx.
Interestingly, our results indicate that the transfection with cap1 UNR mRNA in both iD-LNP and iCD-Lx results in a better efficiency in comparison to the transfection made with cap1 MNR (5-MoU) and (PsiU) as well as ARCA UNR on KHYG-1 and NK-92 cells as well as primary NK cells. Cap1-bearing mRNAs are well known to reduce innate sensors activation compared with mRNA bearing cap0 or its analog as ARCA.42,43 Unexpectedly, when the transfection was performed with modified mRNA bearing cap1, a reduced translation of eGFP mRNA occurred while the percentage of transfected cells remained the same.42,44 The impact of nucleosides modification depends on both the mRNA sequence and/or the lipid composition and/or the cell type.45 In fact, studies conducted with non-modified uridine mRNA in cationic liposomes for chimeric antigen receptor (CAR)-T cells therapy, showed the potency of such non-modified mRNA in therapeutics.46,47 If the incorporation of modified nucleotides offers advantages such as reduced immunogenicity and increased stability, as published by Karikó et al.,48 a recent study published by Mulroney et al.49 unveils a potential drawback. The introduction of N1-methylpseudouridine into mRNA sequences has been demonstrated to induce +1 ribosomal frameshifting. This phenomenon disrupts the fidelity of protein translation, potentially leading to the production of aberrant polypeptides. These abnormal proteins may lack functionality or even exert detrimental effects within the cell. More investigation is required to understand this negative effect on the translation efficiency, and it is ongoing in the lab.
The successful replacement of IL-2 recombinant protein by IL-2 mRNA transfection with iD-LNP and iCD-Lx confirms both the efficiency and the biocompatibility of those formulations. Interestingly, we observed that cell viability was improved following iCD-Lx transfection compared with recombinant protein supplementation, while it was similar following iD-LNP IL-2 mRNA transfection. This strongly suggests that the amount of IL-2 produced after Lx transfection was higher compared with that obtained using LNP. The intracellular presence of IL-2 mRNA can be of interest as the drawbacks of IL-2 protein for many therapeutic approaches are known.50 It is worth noticing that the concomitant delivery of two mRNA encoding different proteins, in the same formulation, is indeed possible and efficient. This would expand the range of therapeutic use such as the delivery of another mRNA coding for ILs (IL-2 + IL-15) or in combination with CAR mRNA for next generation therapy.51,52
More importantly, our data demonstrate that the transfection of NK cells with IL-2 mRNA with both delivery systems does not alter the phenotype of the cell lineage or primary human, with regard to surface markers and cytotoxicity capacity. The use of IL-2 mRNA in this study represents a proof of concept, opening new avenues towards non-virally genetically engineered NK cells. Indeed, the use of viruses is the most common safety hurdle to succeed in reaching the clinical phase. Other cytokines or important signalling checkpoints could be expressed as well to optimize NK cells functions. Further efforts could even lead to the use of our formulations for the CRISPR-Cas9 technology, opening the way to the possibility to modify cells permanently.
Since our Lx and LNP were stable in the presence of serum, their efficiency to target NK cells in vivo deserves to be investigated as it has been shown for T cells.53 Work is in progress to test Lx or LNP formulations in mice. Altogether, our results pave the way for mRNA engineering of NK cells with lipid-based platforms.
Materials and methods
mRNA
The pT7-GFP plasmid was linearized with either SpeI or NotI, for mRNAs production with the mMessage mMachine T7 kit (Ambion) according to the manufacturer’s instructions. IVT UNR mRNAs, containing ARCA, were isolated with phenol/chloroform and isopropanol precipitation. Purity, size and integrity of mRNAs were checked by UV absorbance (NanoDrop One, Thermo Fisher Scientific), denaturing agarose gel electrophoresis, and capillary electrophoresis (Agilent). UNR mRNA EGFP and modified nucleoside mRNA (MNR) (5-MoU) (5-Methoxyuridine) eGFP, both containing CleanCap, were purchased from Tebu-bio (Tebubio). MNR (psiU) (N1-Methylpseudouridine) EGFP and UNR IL-2 mRNAs, containing cap-1, were purchased from OZ Biosciences.
Lipids
DOPE, Chol, Sito, DMG-PEG, DMPE-PEG, and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(Lissamine rhodamine B sulfonyl) were purchased from Avanti Polar Lipid and stocked in ethanol at −20°C. Ionizable imidazole lipid and cationic imidazolium lipid were synthetized internally,35 SM-102, ALC-0315 were obtained from Cayman Therapeutics and D-Lin-MC3 from Cliniscience.
Microfluidic preparation of liposomes and LNP
Microfluidic production of liposomes and LNPs was achieved using either the NanoAssemblr Spark or Ignite platforms (Precision Nanosystems Inc). Depending on the formulation, lipids dissolved in ethanol were mixed with aqueous phase—either PBS (pH 7.4) or mRNA in citrate buffer (50 mM, pH 4)—to obtain liposomes and LNP, respectively. Microfluidic mixing was undertaken at a flow rate ratio of 3:1 (aqueous: organic phase) and at a total flow rate of 10 mL/min (for liposomes) and 4 mL/min (for LNPs). Liposomes were prepared at a 21.6 mM final concentration. Then, liposomes were further dialyzed against PBS for at least 6 h at 4°C, while buffer exchange for LNP was performed with Amicon Ultra-15 Centrifugal Filters of 10 kDa MWCO (Millipore Sigma). The size (hydrodynamic diameter) and zeta potential were determined by dynamic light scattering and electrophoretic mobility, respectively, using the SZ-100 nanoparticle analyzer (Horiba).
Complexation of mRNA with liposomes was analyzed by gel shift assay. Briefly, mRNA and mRNA complexed with liposomes were loaded onto 0.6% agarose gel containing 0.02% of Ribogreen (Thermo Fisher Scientific). Encapsulation efficiency of mRNA in LNP and Lx was determined by using the Ribogreen assay. Briefly, in a black 96-well plate, LNP, as well as mRNA concentration standards, were added in Tris-EDTA (TE) buffer without and with Triton X-100. This was followed by the addition of Ribogreen, which emits fluorescence upon binding to RNA measured with a fluorescent plate reader (λexcitation: 485 nm, λemission: 528 nm). The percentage of mRNA encapsulation was calculated from the ratio of average fluorescence in TE buffer and average fluorescence in TE-Triton buffer. Encapsulated mRNA concentration was calculated from the percentage of encapsulation and the total mRNA concentration (calculated using mRNA standards).
Cryo-EM
A Vitrobot Mark IV (Thermo Fischer Scientific) was used for cryo-grid preparation of iD-LNP and iCD-Lip. The temperature inside the chamber was set to 4°C, humidity was maintained at 100%, blotting time was set to 4 s, and blotting force to 0. Two types of grids were used: Lacey/Carbon (300 Mesh Cu 25/Bx) and UltrAuFoil R 1.2/1.3 (Au, 200 Mesh; Quantifoil Micro Tools GmbH). The grids were treated with Air (Solarus plama cleaner, Gatan) for 15 s immediately before sample application. The liposome sample (3 μL) was deposited onto the grid and after the blot step, the grids were plunge-frozen into liquid ethane and cooled by liquid nitrogen. Specimens were maintained at −170°C using a cryo-holder. Cryo-EM micrographs were recorded on a Glacios electron microscope (Thermo Fischer Scientific) operated at 200 keV. CryoEM images were collected at 79,000× magnification with an exposure time of 2.41 s and a total dose of 644.30 e/nm2 (pixel size 0.16 nm, defocus −3 to −7 μm).
Cells and cell cultures
NK-92 (CLR-2407) and KHYG-1 (ACC 725) cell lines were purchased from the American Type Culture Collection (ATCC) and DSMZ, respectively. Jurkat cells (TIB-152) were purchased from ATCC. Jurkat cells were cultured in RPMI-1640 with 10% fetal bovine serum (FBS). KHYG-1 cells were cultured in RPMI-1640 with 10% FBS and NK-92 cells in the complete medium MyeloCult H5100 (STEMCELL Technologies) supplemented with 10 mM hydrocortisone (STEMCELL Technologies), and both were supplemented with 100 units/mL IL-2 (Miltenyi). Cells were maintained every two to three days (either by addition or replacement of medium). When replacing media, cells were seeded at a concentration of 2–3 × 105 viable cells/mL. Cell culture was performed at 37°C in a humidified atmosphere containing 5% CO2. All cells were mycoplasma-free, as they were regularly tested with MycoAlert Mycoplasma Detection Kit (Lonza).
Primary NK cell cultures
Human NK cells were isolated from peripheral blood mononuclear cells obtained by centrifugation on density gradient of whole blood from healthy volunteers provided by the local Blood Bank (Etablissement Francais du Sang). Negative selection of NK was performed on EasySep Human NK Cell Enrichment Kit system (StemCell Technologies). We cultured 2 x 106 sorted NK cells with 100 Gy-irradiated EBV-LCL cells in a 1:20 ratio, in 20 mL of RPMI supplemented with 10% fetal bovine, 1× GlutaMAX (both from Gibco; Thermo Fisher Scientific), and 500 UI/mL IL-2 (Proleukin).
Preparation of mRNA-Lx and LNP
Lx were prepared by mixing liposomes with eGFP mRNA in N/P ratio of 1/1 (if not stated otherwise) in PBS and gently mixed. After 15 min incubation at room temperature, Lx were added to the cells. LNP were diluted in OptiMEM (Gibco) to reach the desired mRNA quantity and added to the cells. Complexation of mRNA with Lipofectamine, Screenfect, and mRNAfect was performed according to the manufacturer’s instructions.
Transfection of Jurkat, NK-92, and KHYG-1 cells
The a day of transfection, cells were counted (with trypan blue to discriminate dead cells) and were plated in a NuncTM 24-well non-treated plate (Thermo Fisher Scientific) in 400 μL Opti-MEM (Gibco) with 50 U/mL IL-2 (unless noted differently). Lx and LNP were added to the cells to get a final volume of 500 μL. Cells were incubated at 37°C in a humidified atmosphere containing 5% CO2. Typical transfection was performed for 24h. For the kinetic experiments requiring transfection longer than 24 h, 300 μL of complete medium was added to the cells. For primary NK cells, 100 μL of transfecting solution were gently added to the cells in 400 μL serum-free media, then kept in the incubator, 37°C, 5% CO2. After 4 h, FBS was added to the well to obtain the same concentration as in culture medium (20% here). Cells can be split the next day using the usual culture medium.
For eGFP transfection measurement, cells were collected in round-bottom polystyrene tubes and then centrifuged at 1,500 rpm for 5 min. Supernatant was removed and 500 μL PBS was added to each tube. Propidium iodide (PI) (0.01 mg/mL final concentration) was used to detected dead cells. Then the percentage of transfected cells (%), the mean of the fluorescence intensity (MFI) and mortality (PI + cells) were measured by flow cytometry (BD LSR Fortessa X20 Flow Cytometry Analyzer, BD Biosciences).
For kinetic experiments performed on KHYG-1 cells, the transfection efficiency and time course of eGFP mRNA translation were automatically monitored in a real-time using Incucyte S3 system (Sartorius) followed by subsequent cell by cell analysis.
Confocal microscopy observations
mRNA was labelled with fluorescein by using the Label IT Nucleic acid labelling kit (Mirus Bio) and manufacturer instructions. Liposomes and LNPs were labelled by incorporation of 0.25% (molar ratio deducted from cholesterol or β-sitosterol) of lissamine-rhodamine phosphatidylethanolamine. After different incubation time points, cells were washed with PBS, fixed with 2% paraformaldehyde for 20 min, and washed twice with PBS. Then, cells were resuspended in 80 μL and samples of 10 μL were mixed with 10 μL of Vectashield (Vector Laboratories) on a microscope slide before analysis with Zeiss LSM 980 Airyscan 2 confocal microscope.
Uptake experiments
Lx and LNPs were made with FITC-mRNA and uptake assay was performed as already described.54 Briefly, 4.0 × 105 cells were plated in a 24-well plate and were then incubated for different times with Lx or LNP in a serum-free media. Afterwards, cells were carefully washed with cold PBS to remove medium and complexes without removing nanoparticles attached to the cells. The cells were harvested and the cell suspensions were then transferred to round-bottomed polystyrene tubes and kept on ice. For intracellular fluorescence measurement, cells were treated with Trypan blue before analysis to quench cell surface fluorescence. For monensin treatment, aliquots of cells were deposited into new tubes and monensin (0.0354 mg/mL in PBS) was added and cells were kept for 30 min on ice. The fluorescence intensity of the cells was then measured by flow cytometry with the BD LSR Fortessa Flow Cytometry Analyzer.
Cell-cycle analysis
Cells from 24-well plate were pooled (to have ≥1 million cells), washed with PBS and fixed with cold absolute ethanol for 15 min at −20°C. After washing with PBS, they were treated with RNase A (final concentration 50 μg/μL), and finally stained with PI (final concentration of 50 μg/μL) for at least 30 min at 4°C. DNA content was determined by BD FACSCalibur. Data analysis was performed using FlowJo software.
Western blot analysis
To determine IL-2 expression, proteins were isolated 24 h from cells with CCLR lysis reagent (Promega) and from the medium which was concentrated with Amicon Ultra-0.5 Centrifugal Filter Unit (10 kDa MWCO). Proteins separated by size with SDS-PAGE were then transferred to ethanol activated-PVDF membrane (Merck Millipore). Afterwards, incubation in 5% low-fat milk dissolved in TBST was performed for 1 h at room temperature to block the membrane. This was followed by incubation at 4°C overnight with primary antibodies (IL-2 (D7A5) Rabbit #12239, Cell Signaling) diluted either in 5% BSA or 5% milk. The next day, the membrane was washed and incubated for 1 h with a secondary antibody (Goat anti-Rabbit IgG [horseradish peroxidase conjugate] ADI-SAB-300-J, Enzo Lifescience) diluted in 5% low-fat milk. Signals on membranes were detected with Clarity Western ECL Substrate (BIO-RAD) according to the manufacturer’s instructions, followed by a chemiluminescence analysis with ChemiDoc (BIO-RAD).
Cytotoxicity assay
Standard 4 h cytotoxicity assays were performed as described elsewhere.11 The measurement of apoptotic cells was performed using the CellEvent Caspase-3/7 Green Flow Cytometry Assay Kit (catalog #: C10427; Thermo Fisher Scientific) following the manufacturer’s instructions.
Cytokine assay
For the measurement of IFN-γ, TNF production, NK cells (E) were either co-cultured with K562 cells (T) at increasing effector/target (E/T) ratios or stimulated with PMA (10 ng/mL) and ionomycin (1 μg/mL). Cells were incubated with monensin and brefeldin A (BD GolgiPlug and GolgiStop) in a complete medium for 4 h at 37°C. The cells were subjected to surface staining (anti CD56, #130-113-313, Milteny Biotech) and intracellular staining (anti-TNFα: #130-120-063, Milteny Biotech; anti-IFNy: #130-119-577, Milteny Biotech) was performed by using the FoxP3/Transcription Factor Staining Buffer Set (eBioscience).
Flow cytometry and cell sorting
Single-cell suspensions were stained with the appropriate monoclonal antibody in PBS containing 2% FBS. LSRII, Fortessa, (BD Biosciences) were used for cell analysis. Antibodies specific for CD69 (#IM1943U, Clone TP1.55.3, Beckman Coulter), CD122 (#562887, Clone Mik-B3, BD Biosciences), CD25 (#563159, BD Biosciences), TIGIT (#17-9500-42, Clone MBA43, In Vitrogen), NKG2D (#561815, Clone 1D11, BD Biosciences), Dnam-1 (#338323, Clone 11A8, BioLegend), and Nkp30 (#325217, Clone p30-15, BioLegend) were used.
Statistical analysis
The data were tested for statistical significance using ANOVA and paired student t-test. All numerical data are expressed as mean with SD. Analysis was performed using PRISM software (GraphPad).
Data and code availability
Data that support the findings of this study are available upon reasonable request with the permission of all authors.
Acknowledgments
The authors acknowledge an ANRT-CIFRE (Association Nationale Recherche Technologie-Conventions Industrielles de Formation par la Recherche, France) grant and Sanofi fellowship for CD and IC and Sanofi France. The authors thank Virginie Malard for purifying lipids used for formulations and David Gosset of MO2VING platform facility, in the Center for Molecular Biophysics for assistance with confocal imaging and flow cytometry. This work was also supported by the CNRS and University of Orléans. PLB and CCdS were supported by a doctoral fellowship from Ligue Nationale contre le Cancer and. GG was supported by the Canceropole PACA - Emergence, the Janssen Horizon Fonds de dotation. JN was supported by the fondation Bristol Myers Squibb pour la recherche en Immuno-oncology. CP acknowledges Institut Universitaire de France, Fondation de l'Avenir and PEPR Biotherapies and bioproduction of Innovative therapies France 2030 for their financial support.
Author contributions
Conceptualization and methodology: C.P., C.D., I.C., N.R., A.B., L.E., P.M., and G.G. Investigations: C.D., I.C., P.L.B., C.D.S., J.I., M.H., N.L., and N.R. Writing – original draft: C.P., C.D., I.C., N.R., L.E., and G.G. Writing – review & editing: All authors. Supervision: C.P., L.E., N.R., and G.G.
Declaration of interests
N.R. and L.E. are Sanofi employees and may hold shares and/or stock options in the company. All the experiments involving cells were conducted in the academic laboratory. The company had no role in the design of the study and the decision to publish the results. Lipids used in this work are related to the patent invention # WO2009050372 owned only by the academic researchers.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2024.102263.
Supplemental information
References
- 1.Maskalenko N.A., Zhigarev D., Campbell K.S. Harnessing natural killer cells for cancer immunotherapy: dispatching the first responders. Nat. Rev. Drug Discov. 2022;21:559–577. doi: 10.1038/s41573-022-00413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma P., Kumar P., Sharma R. Natural killer cells - Their role in tumour immunosurveillance. J. Clin. Diagn. Res. 2017;11:BE01–BE05. doi: 10.7860/JCDR/2017/26748.10469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trinchieri G. Biology of Natural Killer Cells. Adv. Immunol. 1989;47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sivori S., Vacca P., Del Zotto G., Munari E., Mingari M.C., Moretta L. Human NK cells: surface receptors, inhibitory checkpoints, and translational applications. Cell. Mol. Immunol. 2019;16:430–441. doi: 10.1038/s41423-019-0206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caligiuri M.A. Human natural killer cells. Blood. 2008;112:461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vivier E., Raulet D.H., Moretta A., Caligiuri M.A., Zitvogel L., Lanier L.L., Yokoyama W.M., Ugolini S. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paolini R., Bernardini G., Molfetta R., Santoni A. NK cells and interferons. Cytokine Growth Factor Rev. 2015;26:113–120. doi: 10.1016/j.cytogfr.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Murugan D., Murugesan V., Panchapakesan B., Rangasamy L. Nanoparticle Enhancement of Natural Killer (NK) Cell-Based Immunotherapy. Cancers. 2022;14:5438. doi: 10.3390/cancers14215438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu H., Zhao X., Li Z., Hu Y., Wang H. From CAR-T Cells to CAR-NK Cells: A Developing Immunotherapy Method for Hematological Malignancies. Front. Oncol. 2021;11:720501. doi: 10.3389/fonc.2021.720501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oberschmidt O., Morgan M., Huppert V., Kessler J., Gardlowski T., Matthies N., Aleksandrova K., Arseniev L., Schambach A., Koehl U., Kloess S. Development of Automated Separation, Expansion, and Quality Control Protocols for Clinical-Scale Manufacturing of Primary Human NK Cells and Alpharetroviral Chimeric Antigen Receptor Engineering. Hum. Gene Ther. Methods. 2019;30:102–120. doi: 10.1089/hgtb.2019.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernard P.L., Delconte R., Pastor S., Laletin V., Costa Da Silva C., Goubard A., Josselin E., Castellano R., Krug A., Vernerey J., et al. Targeting CISH enhances natural cytotoxicity receptor signaling and reduces NK cell exhaustion to improve solid tumor immunity. J. Immunother. Cancer. 2022;10:e004244. doi: 10.1136/jitc-2021-004244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robbins G.M., Wang M., Pomeroy E.J., Moriarity B.S. Nonviral genome engineering of natural killer cells. Stem Cell Res. Ther. 2021;12:350. doi: 10.1186/s13287-021-02406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Mayta R., Zhang Z., Hamilton A.G., Mitchell M.J. Delivery technologies to engineer natural killer cells for cancer immunotherapy. Cancer Gene Ther. 2021;28:947–959. doi: 10.1038/s41417-021-00336-2. [DOI] [PubMed] [Google Scholar]
- 14.Wilk A.J., Weidenbacher N.L.B., Vergara R., Haabeth O.A.W., Levy R., Waymouth R.M., Wender P.A., Blish C.A. Charge-altering releasable transporters enable phenotypic manipulation of natural killer cells for cancer immunotherapy. Blood Adv. 2020;4:4244–4255. doi: 10.1182/BLOODADVANCES.2020002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura T., Nakade T., Sato Y., Harashima H. Delivering mRNA to a human NK cell line, NK-92 cells, by lipid nanoparticles. Int. J. Pharm. 2023;636:122810. doi: 10.1016/j.ijpharm.2023.122810. [DOI] [PubMed] [Google Scholar]
- 16.Douka S., Brandenburg L.E., Casadidio C., Walther J., Garcia B.B.M., Spanholtz J., Raimo M., Hennink W.E., Mastrobattista E., Caiazzo M. Lipid nanoparticle-mediated messenger RNA delivery for ex vivo engineering of natural killer cells. J. Control. Release. 2023;361:455–469. doi: 10.1016/j.jconrel.2023.08.014. [DOI] [PubMed] [Google Scholar]
- 17.Abbas A.K. The Surprising Story of IL-2: From Experimental Models to Clinical Application. Am. J. Pathol. 2020;190:1776–1781. doi: 10.1016/j.ajpath.2020.05.007. [DOI] [PubMed] [Google Scholar]
- 18.Mitra S., Leonard W.J. Biology of IL-2 and its therapeutic modulation: Mechanisms and strategies. J. Leukoc. Biol. 2018;103:643–655. doi: 10.1002/JLB.2RI0717-278R. [DOI] [PubMed] [Google Scholar]
- 19.Gotthardt D., Trifinopoulos J., Sexl V., Putz E.M. JAK/STAT Cytokine Signaling at the Crossroad of NK Cell Development and Maturation. Front. Immunol. 2019;10:2590. doi: 10.3389/fimmu.2019.02590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Widowati W., Jasaputra D.K., Sumitro S.B., Widodo M.A., Mozef T., Rizal R., Kusuma H.S.W., Laksmitawati D.R., Murti H., Bachtiar I., Faried A. Effect of interleukins (Il-2, il-15, il-18) on receptors activation and cytotoxic activity of natural killer cells in breast cancer cell. Afr. Health Sci. 2020;20:822–832. doi: 10.4314/ahs.v20i2.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K.S., Frank D.A., Ritz J. Interleukin-2 enhances the response of natural killer cells to interleukin-12 through up-regulation of the interleukin-12 receptor and STAT4. Blood. 2000;95:3183–3190. doi: 10.1182/blood.V95.10.3183. [DOI] [PubMed] [Google Scholar]
- 22.Hernandez R., Põder J., LaPorte K.M., Malek T.R. Engineering IL-2 for immunotherapy of autoimmunity and cancer. Nat. Rev. Immunol. 2022;22:614–628. doi: 10.1038/s41577-022-00680-w. [DOI] [PubMed] [Google Scholar]
- 23.Dudley M.E., Wunderlich J.R., Yang J.C., Sherry R.M., Topalian S.L., Restifo N.P., Royal R.E., Kammula U., White D.E., Mavroukakis S.A., et al. Adoptive Cell Transfer Therapy Following Non-Myeloablative but Lymphodepleting Chemotherapy for the Treatment of Patients With Refractory Metastatic Melanoma. J. Clin. Oncol. 2005;23:2346–2357. doi: 10.1200/JCO.2005.00.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clark P.R., Stopeck A.T., Parker S.E., Hersh E.M. Cationic lipid gene transfer of an IL-2 transgene leads to activation of natural killer cells in a SCID mouse human tumor xenograft. Cell. Immunol. 2000;204:96–104. doi: 10.1006/cimm.2000.1699. [DOI] [PubMed] [Google Scholar]
- 25.Xiong Q., Zhang H., Ji X., Zhang Y., Shi G., Dai L., Cheng F., Wang H., Luo J., Xu J., et al. A novel membrane-bound interleukin-2 promotes NK-92 cell persistence and anti-tumor activity. OncoImmunology. 2022;11:2127282. doi: 10.1080/2162402X.2022.2127282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brader M.L., Williams S.J., Banks J.M., Hui W.H., Zhou Z.H., Jin L. Encapsulation state of messenger RNA inside lipid nanoparticles. Biophys. J. 2021;120:2766–2770. doi: 10.1016/j.bpj.2021.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki Y., Ishihara H. Difference in the lipid nanoparticle technology employed in three approved siRNA (Patisiran) and mRNA (COVID-19 vaccine) drugs. Drug Metab. Pharmacokinet. 2021;41:100424. doi: 10.1016/j.dmpk.2021.100424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen J.S., Griffin J.H., Schechter A.N. Nuclear magnetic resonance titration curves of histidine ring protons. IV. The effects of phosphate and sulfate on ribonuclease. J. Biol. Chem. 1973;248:4305–4310. [PubMed] [Google Scholar]
- 29.Kumar V.V., Pichon C., Refregiers M., Guerin B., Midoux P., Chaudhuri A. Single histidine residue in head-group region is sufficient to impart remarkable gene transfection properties to cationic lipids: Evidence for histidine-mediated membrane fusion at acidic pH. Gene Ther. 2003;10:1206–1215. doi: 10.1038/sj.gt.3301979. [DOI] [PubMed] [Google Scholar]
- 30.Shalini K., Sharma K., Kumar N. Imidazole and its biological activities: A review. Chem. Sin. 2010;1:36–47. [Google Scholar]
- 31.Miao L., Li L., Huang Y., Delcassian D., Chahal J., Han J., Shi Y., Sadtler K., Gao W., Lin J., et al. Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat. Biotechnol. 2019;37:1174–1185. doi: 10.1038/s41587-019-0247-3. [DOI] [PubMed] [Google Scholar]
- 32.Ripoll M., Bernard M.C., Vaure C., Bazin E., Commandeur S., Perkov V., Lemdani K., Nicolaï M.C., Bonifassi P., Kichler A., et al. An imidazole modified lipid confers enhanced mRNA-LNP stability and strong immunization properties in mice and non-human primates. Biomaterials. 2022;286:121570. doi: 10.1016/j.biomaterials.2022.121570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hassett K.J., Higgins J., Woods A., Levy B., Xia Y., Hsiao C.J., Acosta E., Almarsson Ö., Moore M.J., Brito L.A. Impact of lipid nanoparticle size on mRNA vaccine immunogenicity. J. Control. Release. 2021;335:237–246. doi: 10.1016/j.jconrel.2021.05.021. [DOI] [PubMed] [Google Scholar]
- 34.Cui S., Wang Y., Gong Y., Lin X., Zhao Y., Zhi D., Zhou Q., Zhang S. Correlation of the cytotoxic effects of cationic lipids with their headgroups. Toxicol. Res. 2018;7:473–479. doi: 10.1039/c8tx00005k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akhter S., Berchel M., Jaffrès P.A., Midoux P., Pichon C. mRNA Lipoplexes with Cationic and Ionizable α-Amino-lipophosphonates: Membrane Fusion, Transfection, mRNA Translation and Conformation. Pharmaceutics. 2022;14:581. doi: 10.3390/pharmaceutics14030581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel S., Ashwanikumar N., Robinson E., Xia Y., Mihai C., Griffith J.P., Hou S., Esposito A.A., Ketova T., Welsher K., et al. Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nat. Commun. 2020;11:983. doi: 10.1038/s41467-020-14527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel S., Ashwanikumar N., Robinson E., Duross A., Sun C., Murphy-Benenato K.E., Mihai C., Almarsson Ö., Sahay G. Boosting Intracellular Delivery of Lipid Nanoparticle-Encapsulated mRNA. Nano Lett. 2017;17:5711–5718. doi: 10.1021/acs.nanolett.7b02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akinc A., Querbes W., De S., Qin J., Frank-Kamenetsky M., Jayaprakash K.N., Jayaraman M., Rajeev K.G., Cantley W.L., Dorkin J.R., et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarode A., Fan Y., Byrnes A.E., Hammel M., Hura G.L., Fu Y., Kou P., Hu C., Hinz F.I., Roberts J., et al. Predictive high-throughput screening of PEGylated lipids in oligonucleotide-loaded lipid nanoparticles for neuronal gene silencing. Nanoscale Adv. 2022;4:2107–2123. doi: 10.1039/d1na00712b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eygeris Y., Patel S., Jozic A., Sahay G., Sahay G. Deconvoluting Lipid Nanoparticle Structure for Messenger RNA Delivery. Nano Lett. 2020;20:4543–4549. doi: 10.1021/acs.nanolett.0c01386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delehedde C., Even L., Midoux P., Pichon C., Perche F. Intracellular routing and recognition of lipid-based mRNA nanoparticles. Pharmaceutics. 2021;13:945. doi: 10.3390/pharmaceutics13070945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li B., Luo X., Dong Y. Effects of Chemically Modified Messenger RNA on Protein Expression. Bioconjug. Chem. 2016;27:849–853. doi: 10.1021/acs.bioconjchem.6b00090. [DOI] [PubMed] [Google Scholar]
- 43.Linares-Fernández S., Moreno J., Lambert E., Mercier-Gouy P., Vachez L., Verrier B., Exposito J.Y. Combining an optimized mRNA template with a double purification process allows strong expression of in vitro transcribed mRNA. Mol. Ther. Nucleic Acids. 2021;26:945–956. doi: 10.1016/j.omtn.2021.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roy B. Methods in Molecular Biology. Humana Press Inc.; 2021. Effects of mRNA Modifications on Translation: An Overview; pp. 327–356. [DOI] [PubMed] [Google Scholar]
- 45.Bernard M.C., Bazin E., Petiot N., Lemdani K., Commandeur S., Verdelet C., Margot S., Perkov V., Ripoll M., Garinot M., et al. The impact of nucleoside base modification in mRNA vaccine is influenced by the chemistry of its lipid nanoparticle delivery system. Mol. Ther. Nucleic Acids. 2023;32:794–806. doi: 10.1016/j.omtn.2023.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mackensen A., Haanen J.B.A.G., Koenecke C., Alsdorf W., Wagner-Drouet E., Borchmann P., Heudobler D., Ferstl B., Klobuch S., Bokemeyer C., et al. CLDN6-specific CAR-T cells plus amplifying RNA vaccine in relapsed or refractory solid tumors: the phase 1 BNT211-01 trial. Nat. Med. 2023;29:2844–2853. doi: 10.1038/s41591-023-02612-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reinhard K., Rengstl B., Oehm P., Michel K., Billmeier A., Hayduk N., Klein O., Kuna K., Ouchan Y., Wöll S., et al. An RNA vaccine drives expansion and efficacy of claudin-CAR-T cells against solid tumors. Science. 2020;367:446–453. doi: 10.1126/science.aay5967. [DOI] [PubMed] [Google Scholar]
- 48.Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mulroney T.E., Pöyry T., Yam-Puc J.C., Rust M., Harvey R.F., Kalmar L., Horner E., Booth L., Ferreira A.P., Stoneley M., et al. N 1-methylpseudouridylation of mRNA causes +1 ribosomal frameshifting. Nature. 2024;625:189–194. doi: 10.1038/s41586-023-06800-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang A., Harding F. The challenges and molecular approaches surrounding interleukin-2-based therapeutics in cancer. Cytokine X. 2019;1:100001. doi: 10.1016/j.cytox.2018.100001. [DOI] [Google Scholar]
- 51.Raftery M.J., Franzén A.S., Pecher G. CAR NK Cells: The Future Is Now. Annu. Rev. Cancer Biol. 2023;7:229–246. doi: 10.1146/annurev-cancerbio-061521. [DOI] [Google Scholar]
- 52.Daher M., Melo Garcia L., Li Y., Rezvani K. CAR-NK cells: the next wave of cellular therapy for cancer. Clin. Transl. Immunol. 2021;10:e1274. doi: 10.1002/cti2.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rurik J.G., Tombácz I., Yadegari A., Méndez Fernández P.O., Shewale S.V., Li L., Kimura T., Soliman O.Y., Papp T.E., Tam Y.K., et al. CAR T cells produced in vivo to treat cardiac injury. Science. 2022;375:91–96. doi: 10.1126/science.abm0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Delehedde C., Ciganek I., Rameix N., Laroui N., Gonçalves C., Even L., Midoux P., Pichon C. Impact of net charge, targeting ligand amount and mRNA modification on the uptake, intracellular routing and the transfection efficiency of mRNA lipopolyplexes in dendritic cells. Int. J. Pharm. 2023;647:123531. doi: 10.1016/j.ijpharm.2023.123531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data that support the findings of this study are available upon reasonable request with the permission of all authors.