Abstract
Background and Objective
Voriconazole (VRC), a broad-spectrum antifungal drug, exhibits nonlinear pharmacokinetics (PK) due to saturable metabolic processes, autoinhibition and metabolite-mediated inhibition on their own formation. VRC PK is also characterised by high inter- and intraindividual variability, primarily associated with cytochrome P450 (CYP) 2C19 genetic polymorphism. Additionally, recent in vitro findings indicate that VRC main metabolites, voriconazole N-oxide (NO) and hydroxyvoriconazole (OHVRC), inhibit CYP enzymes responsible for VRC metabolism, adding to its PK variability. This variability poses a significant risk of therapeutic failure or adverse events, which are major challenges in VRC therapy. Understanding the underlying processes and sources of these variabilities is essential for safe and effective therapy. This work aimed to develop a whole-body physiologically-based pharmacokinetic (PBPK) modelling framework that elucidates the complex metabolism of VRC and the impact of its metabolites, NO and OHVRC, on the PK of the parent, leveraging both in vitro and in vivo clinical data in a middle-out approach.
Methods
A coupled parent-metabolite PBPK model for VRC, NO and OHVRC was developed in a stepwise manner using PK-Sim® and MoBi®. Based on available in vitro data, NO formation was assumed to be mediated by CYP2C19, CYP3A4, and CYP2C9, while OHVRC formation was attributed solely to CYP3A4. Both metabolites were assumed to be excreted via renal clearance, with hepatic elimination also considered for NO. Inhibition functions were implemented to describe the complex interaction network of VRC autoinhibition and metabolite-mediated inhibition on each CYP enzyme.
Results
Using a combined bottom-up and middle-out approach, incorporating data from multiple clinical studies and existing literature, the model accurately predicted plasma concentration-time profiles across various intravenous dosing regimens in healthy adults, of different CYP2C19 genotype-predicted phenotypes. All (100%) of the predicted area under the concentration–time curve (AUC) and 94% of maximum concentration (Cmax) values of VRC met the 1.25-fold acceptance criterion, with overall absolute average fold errors of 1.12 and 1.14, respectively. Furthermore, all predicted AUC and Cmax values of NO and OHVRC met the twofold acceptance criterion.
Conclusion
This comprehensive parent-metabolite PBPK model of VRC quantitatively elucidated the complex metabolism of the drug and emphasised the substantial impact of the primary metabolites on VRC PK. The comprehensive approach combining bottom-up and middle-out modelling, thereby accounting for VRC autoinhibition, metabolite-mediated inhibition, and the impact of CYP2C19 genetic polymorphisms, enhances our understanding of VRC PK. Moreover, the model can be pivotal in designing further in vitro experiments, ultimately allowing for extrapolation to paediatric populations, enhance treatment individualisation and improve clinical outcomes.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40262-024-01434-8.
Key Summary Points
| Due to the narrow therapeutic range and complex pharmacokinetics (PK) of the antifungal voriconazole (VRC), therapeutic failure and adverse events that compromise quality of life are frequently observed after standard dosing. This study presents the most comprehensive physiologically-based pharmacokinetic (PBPK) model to date for VRC and its metabolites, voriconazole N-oxide (NO) and hydroxyvoriconazole (OHVRC), leveraging both in vitro and clinical data. The model accurately predicts the complex PK of VRC across a wide range of dosing regimens and CYP2C19 genetic polymorphisms, addressing the knowledge gap in current understanding. |
| By incorporating mechanisms of autoinhibition and metabolite-mediated cytochrome P450 (CYP) inhibition, the model offers new insights into the nonlinear PK of VRC. It underscores the significant impact of metabolites on the PK of the parent, enhancing our understanding of the metabolism of VRC and its variability among individuals. |
| This research exemplifies the effective use of a combined bottom-up and middle-out strategy in PBPK modelling. By iteratively refining the model through a learn-predict-refine paradigm, it showcases a robust methodology for understanding complex PK of VRC, and paving the way for optimised and personalised therapeutic dosing strategies. |
Introduction
Invasive fungal infections (IFIs) are of significant clinical concern due to increasing incidence and high morbidity and mortality rates, particularly in immunocompromised individuals and critically ill children [1]. Early diagnosis and adequate antifungal therapy are crucial for successful treatment outcomes [2]. Voriconazole (VRC), a second-generation triazole agent with broad-spectrum antifungal activity, inhibits cytochrome P450 (CYP)-dependent 14-α-sterol demethylase, disrupting the fungal membrane and preventing fungal growth [3]. The antifungal is listed by the World Health Organization as an essential medicine for adults and children aged 2 years and above [4]. It is recommended for first-line treatment of IFIs such as aspergillosis or candidaemia [5] and as a primary or secondary prophylaxis in immunocompromised patients [6, 7]. In adults, VRC is administered as a body-weight-adapted intravenous infusion over 1–2 h with an initial loading dose (LD) of 6 mg/kg twice daily on the first day, followed by a maintenance dose (MD) of 4 mg/kg twice daily. Yet, switching from intravenous to oral flat dosing is considered feasible due to high bioavailability (96%) reported at therapeutic doses [4, 8, 9].
Although VRC has been on the market in the European Union since 2002 and in the United States since 2003, there are still knowledge gaps concerning its pharmacokinetics (PK). VRC is extensively metabolised, with an apparent total clearance (CL) of approximately 13–36 L/h and a steady-state volume of distribution ranging from 2 to 4.6 L/kg [10, 11]. Additionally, the large intra- and interindividual PK variability observed in clinical practice, often leads to therapeutic failures or adverse drug events (AEs), such as hepatotoxicity or visual and neurological disturbances [9, 11–16].
To ensure safe and effective therapy, it is crucial to enhance the understanding of the underlying processes and sources of PK variabilities. Several factors are reportedly associated with the large exposure variability seen after conventional dosing of VRC, the majority of which are attributed to the extensive and complex metabolism of VRC. Notably, 98% of the VRC dose undergoes metabolic transformation, primarily through the highly polymorphic CYP2C19, as well as CYP3A4 and CYP2C9. Only 2% of the drug is excreted unchanged in urine [10, 17, 18].
Voriconazole N-oxide (NO), the main circulating metabolite in human plasma, is suspected to contribute to AEs linked to VRC treatment, despite lacking significant antifungal activity [19–21]. Other metabolites, such as hydroxyvoriconazole (OHVRC) and dihydroxyvoriconazole, are primarily detected in urine along with their conjugates [11, 14, 18]. Nevertheless, their formation pathway remains elusive, highlighting a significant knowledge gap [12, 14, 17].
Furthermore, VRC, as well as its metabolites, has been shown to be a potent CYP inhibitor of CYP2C19, CYP2C9, and CYP3A4 [22–25], thereby causing (auto)-inhibition of its metabolism and a variety of drug–drug interactions (DDIs) [11, 26]. To unravel these interactions, systematic and quantitative in vitro investigations were performed [17, 27–29]. The investigations characterised the contribution of CYP2C19, 3A4 and 2C9 to the metabolism of VRC, as well as the inhibitory potential of the parent and its metabolites, including the underlying mechanisms of these inhibitory processes. Furthermore, these in vitro analyses have shown that VRC, NO and OHVRC reversibly and time-independently (competitive and noncompetitive) inhibit all three relevant CYP enzymes responsible for VRC metabolism [17]. Notably, these in vitro analyses indicated that OHVRC has an almost equivalent inhibitory effect on CYP3A4 as VRC (a 1.1-fold higher Ki for CYP3A4), suggesting a need for further investigation into its clinical relevance, especially considering its potential to impact local hepatic concentrations. Accordingly, the nonlinear PK of VRC have been attributed to saturable metabolic processes, autoinhibition and metabolite-mediated inhibition of CYP enzymes [17, 30, 31].
In addition, the genetic polymorphisms of CYP2C19 could explain a substantial part of the large interindividual variability observed in VRC PK [32]. The wild-type (CYP2C19*1/*1) is the most common genotype in the Caucasian population, accounting for approximately 40% [33]. Loss-of-function alleles, such as CYP2C19*2 and *3, result in reduced activity, while the presence of the gain-of-function allele CYP2C19*17 increases activity. As a result, based on their CYP2C19 genotype, individuals can be categorised into varying metaboliser phenotypes, from poor (PM) to ultrarapid metabolisers (UM), which may necessitate dose adjustments due to exposure differences and potential AE risks [33].
Although multiple VRC PBPK models have been published to date [34–36], the contribution of each metabolic pathway to VRC CL, and the extent of metabolite-mediated inhibition, remain inadequately defined. Moreover, the pivotal phenomenon of VRC (auto)-inhibition has not been comprehensively integrated into the existing models. Addressing these gaps, our study aims to elucidate the complex PK of VRC and quantify its metabolite-mediated CYP inhibition through the development of a coupled parent-metabolite PBPK model. This model, constructed using a middle-out approach, leverages both in vitro and in vivo clinical data to provide a more accurate and mechanistic understanding of VRC metabolism in healthy adults. The more granular metabolism of VRC described by the model will facilitate more reliable scaling to special populations, such as paediatrics, where research is limited [36], and aids in the identification of DDI mitigation strategies in clinical practice. This enhanced insight fosters a move towards more individualised treatment approaches and improved clinical outcomes, representing a substantial stride towards personalised medicine.
Materials and Methods
In Vitro and Clinical Data
Quantitative in vitro characterisation of the metabolism and inhibition properties of VRC has been previously performed by Schulz et al. [17]. The Michaelis–Menten kinetics of NO formation by CYP2C19, CYP2C9 and CYP3A4 were determined in incubations of human recombinant CYP enzymes and in liver and intestine microsomes. In addition, the inhibitory potential, and the mechanism of inhibition of VRC, NO and OHVRC, were evaluated by their effects on CYP marker reactions. Overall, these quantitative in vitro assays provided a basis for the a priori PBPK modelling of VRC and its metabolites.
Individual clinical data, comprising plasma concentration-time profiles of VRC and its two metabolites (NO and OHVRC), were obtained from three clinical studies [13, 14, 30] conducted in healthy adults at the University Hospital Heidelberg. The studies primarily investigated intravenous subtherapeutic and therapeutic dosing regimens, covering a broad dose range (50–400 mg) of single doses (SDs), and allowed stratification based on CYP2C19 genotype. Specifically, doses of 50 mg infused over 2 h and 100 mg infused over 4 h were categorised as subtherapeutic, while 400 mg infused over 2 h was defined as a therapeutic dose. Moreover, urine samples were collected over a 24-h period in two of the three studies [13, 14], and the concentrations of VRC and both metabolites were measured. These concentrations, along with the recorded urine volumes, were used to calculate the amount of each analyte excreted unchanged in urine. In addition to the SD studies, the model development and evaluation also incorporated digitised mean VRC plasma concentrations from two additional clinical studies [37] that investigated intravenous multiple dosing (MD) regimens. One study (study A) reported individual maximum plasma concentration (Cmax) values for VRC on days 1, 5, 8 and 12, while the other (study B) provided individual minimum plasma concentration (Cmin) from the first to the tenth day of MD, and these data were digitised to further refine and validate the model. A detailed list of all clinical studies used is provided in Table 1.
Table 1.
Clinical studies used for model development and evaluation
| CYP2C19 gXM (genotype) | Dose | Route | N | Male (%) | Age, years | Weight (kg) | Metabolites | Study, year |
|---|---|---|---|---|---|---|---|---|
| PM (*2/*2) | 400 mg (SD) | IV (2 h) | 4 | 50 | 32 (20–37) | 70 (58–79) | NO, OHVRC | Scholz et al. (2009) [14] |
| IM (*1/*2, 2/*17) | 50 mg (SD) | IV (2 h) | 4 | 75 | 29 (24–33) | 74 (56–79) | NO | Hohmann et al. (2016) [13] |
| 100 mg (SD) | IV (4 h) | 2 | 50 | 29 (24–34) | 71 (71–72) | NO | Hohmann et al. (2017) [30] | |
| 400 mg (SD) | IV (2 h) | 2 | 50 | 29 (24–34) | 71 (71–72) | NO | Hohmann et al. (2017) [30] | |
| 400 mg (SD) | IV (2 h) | 4 | 75 | 29 (24–33) | 74 (56–78) | NO | Hohmann et al. (2016 [13] | |
| 400 mg (SD) | IV (2 h) | 8 | 62.5 | 25 (24–32) | 72 (65–103) | NO, OHVRC | Scholz et al. (2009) [14] | |
| NM (*1/*1) | 50 mg (SD) | IV (2 h) | 3 | 100 | 30 (22–38) | 80 (65–86) | NO | Hohmann et al. (2016) [13] |
| 100 mg (SD) | IV (4 h) | 5 | 100 | 31 (23–37) | 89 (64–96) | NO | Hohmann et al. (2017) [30] | |
| 400 mg (SD) | IV (2 h) | 5 | 100 | 31 (23–37) | 89 (64–96) | NO | Hohmann et al. (2017) [30] | |
| 400 mg (SD) | IV (2 h) | 3 | 100 | 30 (22–38) | 80 (65–86) | NO | Hohmann et al. (2016) [13] | |
| 400 mg (SD) | IV (2 h) | 2 | 50 | 31 (24–38) | 76 (69–83) | NO, OHVRC | Scholz et al. (2009) [14] | |
| RM (*1/*17, *17/*17) | 50 mg (SD) | IV (2 h) | 8 | 62.5 | 25 (23–52) | 67 (55–96) | NO | Hohmann et al. (2016) [13] |
| 100 mg (SD) | IV (4 h) | 5 | 100 | 30 (24–52) | 82 (70–95) | NO | Hohmann et al. (2017) [30] | |
| 400 mg (SD) | IV (2 h) | 5 | 100 | 30 (24–52) | 82 (70–95) | NO | Hohmann et al. (2017) [30] | |
| 400 mg (SD) | IV (2 h) | 7 | 71.4 | 26 (23–52) | 68 (58–96) | NO | Hohmann et al. (2016) [13] | |
| 400 mg (SD) | IV (2 h) | 6 | 66.7 | 25 (23–28) | 72 (61–93) | NO, OHVRC | Scholz et al. (2009) [14] | |
| Not reported |
3 mg/kg qd D1; 3 mg/kg bid D3–11 |
IV (1 h) | 9 | 100 | 24a (20–31) | 72a (60–87) | – | Purkins et al. (2003) [59] |
|
6 mg/kg bid D1; 3 mg/kg bid D2–9 |
IV (1 h) | 9 | 100 | 28a (19–41) | 73a (66–80) | – | Purkins et al. (2003) [59] |
bid twice daily, D day of treatment, gXM genotype-predicted phenotype, IM intermediate metaboliser, IV intravenous, N number of individuals, NM normal metaboliser, NO voriconazole N-oxide, OHVRC hydroxyvoriconazole, PM poor metaboliser, qd once daily, RM rapid metaboliser, SD single dose
Demographic aggregate values are reported as median (range) unless otherwise specified
aArithmetic mean (range)
Physiologically-Based Pharmacokinetic (PBPK) Model Development
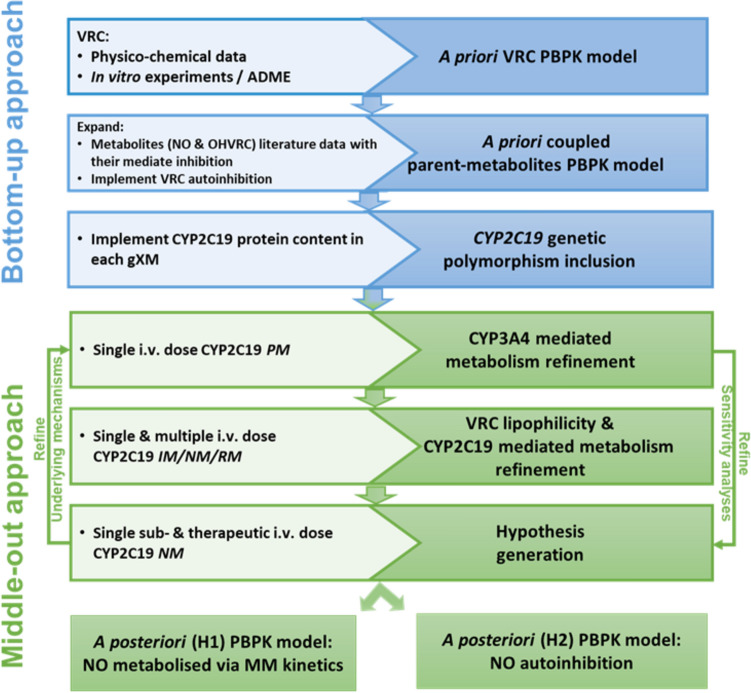
An intravenous adult PBPK model was developed using in vitro data, in silico calculated and in vivo clinical data moving from a pure bottom-up approach to a middle-out approach. These approaches are discussed in the following paragraphs and the input parameters used are summarised in Tables 2 and 3. Figure 1 depicts the entire model development workflow along with the implemented metabolic pathways in Fig. 2 (right panel).
Table 2.
Drug-dependent parameters of the voriconazole a priori PBPK model
| Parameter | Description | Abbreviation | Unit | Value | Source | References |
|---|---|---|---|---|---|---|
| Physicochemical properties | Molar mass | MW | g/mol | 349.3 | Drug label | [4] |
| Effective molar mass | MW* | g/mol | 298.3 | Calculated | – | |
| Acidic dissociation constant | pKa(base) | – | 1.76 | Drug label | [4] | |
| Fraction unbound in plasma | fu | % | 50 | Literature | [60] | |
| Lipophilicity | LogP | – | 1.80 | Drug label | [4] | |
| Distribution | ||||||
| Partition coefficient | Organ-plasma partition coefficients | – | – | Diverse | Berez. | [38] |
| Cellular permeabilities | Permeation across cell membrane | – | cm/min | 0.0026 | PK-Sim | – |
| Metabolising enzymes | ||||||
| CYP2C19 Vmax | Maximum metabolic rate of CYP2C19 | Vmax | pmol/min/pmol | 1.64 | In vitro | [17] |
| CYP2C19 Km | Michaelis–Menten constant of CYP2C19 | Km | µmol/L | 1.31 | In vitro | [17] |
| CYP2C19 Ki | Competitive inhibition constant of CYP2C19 | Ki | µmol/L | 1.90 | In vitro | [17] |
| CYP3A4 Vmax | Maximum metabolic rate of CYP3A4 | Vmax | pmol/min/pmol | 0.00893 | In vitro | [17] |
| CYP3A4 Km | Michaelis–Menten constant of CYP3A4 | Km | µmol/L | 1.20 | In vitro | [17] |
| CYP3A4 Ki | Noncompetitive inhibition constant CYP3A4 | Ki | µmol/L | 2.75 | In vitro | [17] |
| CYP3A4 Vmaxa | Maximum metabolic rate of CYP3A4a | Vmaxa | pmol/min/pmol | 0.10 | Literature | [42] |
| CYP3A4 Kma | Michaelis–Menten constant of CYP3A4a | Kma | µmol/L | 11.0 | Literature | [42] |
| CYP2C9 Vmax | Maximum metabolic rate of CYP2C9 | Vmax | pmol/min/pmol | 0.00705 | In vitro | [17] |
| CYP2C9 Km | Michaelis–Menten constant of CYP2C9 | Km | µmol/L | 4.06 | In vitro | [17] |
| CYP2C9 Ki | Competitive inhibition constant of CYP2C9 | Ki | µmol/L | 2.57 | In vitro | [17] |
| Renal clearance | Plasma clearance | Renal CL | mL/min/kg | 0.022 | Literature | [12, 61] |
Berez. Berezhkovskiy calculation method, CL clearance, CYP cytochrome P450, fu fraction of drug unbound in plasma, Ki inhibition constant (inhibitor concentration causing 50% of maximum inhibition), Km Michaelis-Menten constant (substrate concentration at half-maximal velocity), LogP partition coefficient, MW molecular weight, MW* effective molar mass (VRC contains three fluorine atoms, leading to a reduction of the molar mass), PBPK physiologically-based pharmacokinetic, PKa acid dissociation constant, PK-Sim PK-Sim® standard calculation method, Vmax maximum reaction velocity, VRC voriconazole
aKinetic parameters used to parameterise the formation hydroxyvoriconazole (OHVRC) via recombinant CYP3A4
Table 3.
Drug-dependent parameters of the metabolites (voriconazole N-oxide and hydroxyvoriconazole) a priori PBPK models
| Parameter | Description | Abbreviation | Unit | Value | Source | Reference | Value | Source | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Voriconazole N-oxide | Hydroxyvoriconazole | ||||||||
| Physicochemical properties | Molar mass | MW | g/mol | 365.3 | DrugBank | [4] | 365.3 | DrugBank | [4] |
| Effective molar mass | MW* | g/mol | 314.3 | Calculated | – | 314.3 | Calculated | – | |
| Acidic dissociation constant | pKa(base) | – | 2.00 | DrugBank | [4] | 2.01 | DrugBank | [4] | |
| Fraction unbound in plasma | fu | % | 50 | Assumeda | – | 50 | Assumeda | – | |
| Lipophilicity | LogP | – | 0.51 | ChemAxonb | – | 0.54 | ChemAxonb | – | |
| Distribution | |||||||||
| Partition coefficient | Organ-plasma partition coefficients | – | – | Diverse | Berez. | [38] | Diverse | Berez. | [38] |
| Cellular permeabilities | Permeation across cell membrane | – | cm/min | 2.99E-07 | PK-Sima | – | 3.20E-07 | PK-Sima | – |
| Elimination | |||||||||
| Total hepatic clearance3 | Plasma clearance | CL | L/h/kg | 0.0727 | Literature | [43] | – | – | – |
| Renal clearance | Plasma clearance | Renal CL | mL/min/kg | 0.022 | Literature | [12] | 1.21 | Literature | [12] |
| Inhibition kinetics | |||||||||
| CYP2C19 Ki | Competitive inhibition constant CYP2C19 | Ki | µmol/L | 58.6 | In vitro | [17] | 11.60 | In vitro | [17] |
| CYP3A4 Ki | Noncompetitive inhibition constant CYP3A4 | Ki | µmol/L | 5.24 | In vitro | [17] | 2.53 | In vitro | [17] |
| CYP2C9 Ki | Competitive inhibition constant of CYP2C9 | Ki | µmol/L | 5.47 | In vitro | [17] | 2.80 | In vitro | [17] |
Berez. Berezhkovskiy calculation method, CL clearance, CYP cytochrome P450, fu fraction of drug unbound in plasma, Ki inhibition constant (inhibitor concentration cauing 50% of maximum inhibition), LogP parition coefficient, MW* effective molar mass (NO contains three fluorine atoms, leading to a reduction of the molar mass), NO voriconazole N-oxide, PBPK physiologically based pharmacokinetic, PKa acid dissocation constant, PK-Sim PK-Sim® standard calculation method, VRC voriconazole
aAssumed: Fraction unbound in plasma was assumed as the parent (VRC)
bChemAxon: Calculated by Chemicalize (ChemAxon) [LogD at pH = 7.4]
cTotal hepatic clearance is described as a first-order process in the liver, which eliminates the compound from plasma. Specific clearance is calculated from plasma clearance as the input value
Fig. 1.
Overview of the PBPK model development workflow for voriconazole and its metabolites, illustrating the input/data sources (inside boxes, left side) and the achieved steps (inside boxes, right side). ADME absorption, distribution, metabolism, excretion, CYP cytochrome P450, gXM genotype-predicted phenotype, H hypothesis, IM intermediate metaboliser, i.v. intravenous, MM Michaelis–Menten, NM normal metaboliser, NO voriconazole N-oxide, OHVRC hydroxyvoriconazole, PBPK physiologically-based pharmacokinetic, PM poor metaboliser, RM rapid metaboliser, VRC voriconazole
Fig. 2.
Whole-body intravenous physiologically-based pharmacokinetic model for voriconazole and its metabolites (left) and schematic representation of voriconazole elimination pathways (right). Voriconazole is metabolised by CYP2C19, CYP3A4 and CYP2C9 into voriconazole N-oxide, with subsequent elimination via a nonspecific hepatic route and excreted via renal clearance. The remainder of voriconazole absorbed is transformed via CYP3A4 into hydroxyvoriconazole, which is further metabolised and excreted via renal clearance. The parent compound and both metabolites concomitantly inhibit CYP2C19, CYP3A4 and CYP2C9. CYP cytochrome P450
Bottom-Up Approach
Initially, a pure bottom-up approach was adopted to develop an intravenous a priori PBPK model of the parent compound (VRC), based on literature-derived physicochemical properties and information regarding its distribution, metabolism, and excretion processes (Fig. 1, first blue box). Partitioning into tissues was modelled by the Berezhkovskiy tissue distribution model as implemented in PK-Sim® [38]. The elimination processes involved hepatic metabolism and renal excretion: VRC was assumed to be metabolised via CYP2C19, CYP3A4 and CYP2C9, with a renal CL process implemented to account for the minor excretion of unchanged VRC via the kidneys. Initial values for renal CL were based on literature data using the ratio of the amount excreted in urine to the area under the concentration–time curve (AUC) over 24 h [12, 39]. The relative tissue-specific expression of enzymes was taken from the PK-Sim® expression database based on high-sensitive real-time reverse transcription-polymerase chain reaction (RT-PCR) profiles [40], together with a reference concentration of 0.76 μmol of CYP2C19, 4.32 μmol of CYP3A4 and 3.84 μmol of CYP2C9 per litre of liver tissue [41] that were incorporated in the model.
Subsequently, the a priori parent-model was expanded to include the two metabolites, NO and OHVRC, guided by an exhaustive literature search for their physicochemical properties and distribution, metabolism, and excretion processes (Fig. 1, second blue box). NO was assumed to be formed by the three included CYP enzymes [17], while OHVRC was assumed to be formed via CYP3A4 only [42]. Due to the lack of definitive information regarding the elimination processes of both metabolites, we initially assumed that NO was eliminated through hepatic and renal CL processes. The total hepatic CL of NO was described as a first-order process in the liver derived from plasma CL, back-calculated from a published nonlinear mixed-effects (NLME) model [43], as input value. For OHVRC, we assumed only renal elimination, based on findings from literature suggesting that hydroxy metabolites are more commonly excreted through the kidneys [14] (see Fig. 2, right panel). The renal CL processes for both metabolites were implemented in a similar manner to the parent, with initial values based on literature data, calculated as the amount excreted in urine divided by the corresponding AUC values [12].
Furthermore, to capture the complex interactions, specifically the (auto)-inhibition of VRC and the inhibition mechanisms of NO and OHVRC on each CYP enzyme, equations from multiple inhibition analyses [44] were integrated into the coupled a priori parent-metabolite PBPK model using MoBi®. The reversible competitive inhibition of CYP2C19 and CYP2C9 by VRC and its metabolites was modelled using Eqs. 1 and 2, while the reversible noncompetitive inhibition of CYP3A4 was modelled using Eqs. 1 and 3, based on the respective in vitro data [17].
| 1 |
| 2 |
| 3 |
In Eqs. 1–3, is the velocity of the reaction (rate of metabolism via CYP enzyme), is the unbound concentration of substrate (VRC), is the apparent Michaelis–Menten constant representing half the maximum reaction velocity in the presence of an inhibitor, is the Michaelis–Menten constant representing half the maximum reaction velocity (measured in vitro), is the apparent maximum reaction velocity in presence of an inhibitor, is the maximum reaction velocity (measured in vitro), and is the reversible inhibition constant.
Virtual individuals and populations [45] were created to represent each clinical study included in our modelling workflow based on their respective median data for age, weight, height, and body mass index. The relative expression of enzymes of interest in various organs was determined using the open system pharmacology (OSP) human gene expression database [40]. Different CYP2C19 genotype-predicted phenotypes were incorporated into the model using varying CYP2C19 concentrations (Fig. 1, third blue box). Initially, a virtual population of 1000 individuals was generated based on the demographic characteristics of the clinical studies. For characterising the distribution of hepatic concentrations of CYP2C19 within the simulated population, a log-normal distribution was assumed around the reference CYP2C19 normal metaboliser (NM) concentration value of 0.76 µmol/L with a geometric standard deviation (GeoSD) of 1.79 derived from the OSP human gene expression database [46]. Based on the match between the frequencies of CYP2C19 genotype-predicted phenotype (gXM) in our clinical database and those reported by the Clinical Pharmacogenetics Implementation Consortium (CPIC) for the Caucasian population [33] (Fig. S1 in the electronic supplementary martial [ESM]), four distinct subpopulations, each of 1000 individuals, were constructed. The variability in the CYP2C19 expression of these subpopulations was incorporated into the model using a uniform distribution, constrained by minimum and maximum values determined by calculated percentiles of the reported frequencies of CYP2C19 gXM in the Caucasian population [33]. As a result, the CYP2C19 enzyme concentrations for PM (*2/*2, *2/*3, *3/*2), intermediate metaboliser (IM; *1/*2, *1/*3, *2/*17), NM (*1/*1), and rapid metaboliser (RM)/UM (*1/*17, *17/*17) phenotypes ranged from 0.001 to 0.24, 0.25 to 0.55, 0.56 to 0.95, and 0.96 to 3.55 μmol/L, respectively. Further details on the respective enzyme expression can be found in Table 4.
Table 4.
System-dependent parameters and expression of relevant enzymes for DME
| Enzymes | Reference concentration | Half-life (h) | ||
|---|---|---|---|---|
| Mean [µmol/L]a | GSD | Relative expression | Liver | |
| CYP2C19 | 0.76 [62] | 1.79 [63] | RT-PCRc [64] | 26 [65] |
| CYP2C9 | 3.84 [62] | 2.01 [63] | RT-PCRc [64] | 104 [65] |
| CYP3A4 | 4.32 [62] | 1.18 [63] | RT-PCRc [64] | 36 [65] |
| Dummy hepatic enzyme | 1.00d | 1.40b | ||
DME distribution, metabolism and excretion, CYP cytochrome P450, GSD geometric standard deviation, RT-PCR reverse transcription polymerase chain reaction
aMicromole protein/L in tissue of the highest expression
bGeometric standard deviation with a coefficient of variation of 35% assumed
cPK-Sim® expression database profile
dIf no information is available, the value was set to 1.00 μmol/L and Kcat was estimated according to Meyer et al. [40]
Middle-Out Approach
During model development, a middle-out approach was employed, utilising clinical data to infer (1) model parameters that could not be substantiated by literature or in vitro analyses, as well as (2) influential parameters identified through local sensitivity analyses. Given the model’s complexity, a stepwise approach was adopted within a learn-confirm-predict paradigm to develop the a posteriori coupled parent-metabolite PBPK model.
First, CYP3A4-mediated metabolism was estimated based on data from CYP2C19 PM individuals, assuming that only CYP3A4 contributes to the formation of OHVRC and NO in this population (Fig. 1, first green box). Both the CYP2C19-mediated metabolism and renal CL processes were then estimated using plasma concentration-time profiles and the amount excreted in urine of single and multiple intravenous doses in CYP2C19 IM, NM and RM individuals (Fig. 1, second green box).
In subsequent estimation steps, it became clear that a consistent misprediction of the observed NO PK profiles persisted across different dosing regimens, even upon estimating influential parameters. Therefore, two hypotheses were generated in order to characterise the nonlinear elimination of NO (Fig. 1, third green box).
PBPK Model Evaluation
The parent-metabolite model was evaluated both visually and quantitatively. Predicted concentration-time profiles of VRC, NO and OHVRC were compared with observed data from the corresponding clinical studies. Additionally, predicted plasma concentration values for all studies were compared with their respective individual observed plasma concentrations using goodness-of-prediction (GOP) and goodness-of-fit (GOF) plots. Model performance was further assessed by comparing predicted values with observed values for the AUC from the time of drug administration to the time of the last concentration measurement (AUClast) and for Cmax. In both models, the predicted and observed AUClast were computed using a linear-up log-down trapezoidal method.
Quantitative metrics used to evaluate model performance included the root mean squared error (RMSE) for all predicted and observed plasma concentrations, as well as the average fold error (AFE) and absolute average fold error (AAFE) for all predicted and observed plasma concentrations and PK parameters. These were calculated using Eqs. 4–6.
| 4 |
| 5 |
| 6 |
In Eqs. 4–6, is the ith observed plasma concentration or AUClast or Cmax, is the corresponding predicted plasma concentration or AUClast or Cmax, and n is the number of observed values (studies for PK parameters).
Sensitivity Analysis
Local sensitivity analyses were conducted on both the a priori and a posteriori coupled parent-metabolite PBPK models to assess the impact of single parameter changes. These analyses allowed identifying which of the input parameters significantly influenced the model predictions of VRC, NO and OHVRC exposure. The parameters analysed included those derived from literature, associated with estimated parameters, or assumed to influence the predictions due to calculation methods in the model. The sensitivity of the model to an input parameter was quantified by the ratio of the relative change in the predicted PK metric (AUClast and Cmax) to the relative variation of the input parameter, as outlined in Eq. 7.
| 7 |
In Eq. 7, is the sensitivity index of the PK metric to the evaluated model input parameter, is the change of the predicted PK metric, is the predicted PK metric with original model parameter value, is the change of the assessed model parameter value, and is the original model parameter value (initial input parameter value).
Software
Development of the PBPK model, parameter estimation, simulations, and local sensitivity analyses were conducted using the open-source modelling software PK-Sim® and MoBi® (Open Systems Pharmacology Suite 11.2) [47, 48]. The parameter estimation routine employed the Monte Carlo algorithm, allowing a maximum of 10,000 iterations, and incorporated multiple identification options with randomised start values to ensure the model robustness. Individual PK profiles and amounts excreted in urine for each analyte were used in estimating the input model parameters identified from sensitivity analyses, with the goal of minimising the residual sum of squares (total error) between the simulated and observed data. Additionally, 95% confidence intervals (CIs) for the parameter estimates were derived using the Fisher information matrix. These intervals quantified the uncertainty associated with each parameter estimate, indicating the sensitivity of the total error to changes in parameter values. Published clinical study data were digitised using WebPlotDigitizer (version 4.2 [49]), according to best practices [50]. Model performance metrics and post-processing of simulations were executed in R 4.2.1 (The R Foundation for Statistical Computing, Vienna, Austria) with RStudio 2023.03.0+386 (RStudio Inc., Boston, MA, USA).
Results
Clinical Data
The development and evaluation of the a priori and a posteriori coupled parent-metabolite PBPK models incorporated data from 47 healthy young adults across three clinical studies who were administered intravenous SDs ranging from 50 to 400 mg of VRC. Additionally, data from 18 healthy volunteers across two clinical studies involving intravenous MDs of VRC were considered. In the SD clinical studies, individual VRC and NO plasma concentration-time profiles were reported for all participants, with OHVRC PK data only available for 20 individuals. Urine data were also collected from 35 participants to quantify VRC and both metabolites for the subtherapeutic dose of 50 mg/2 h and the therapeutic dose of 400 mg/2 h, across different CYP2C19 genotypes, while for the MD clinical studies, mean VRC PK data were reported. However, one study provided individual Cmax of VRC from nine individuals, while another reported individual Cmin data from a separate group of nine participants. Notably, no metabolite data were available for the MD studies, with only VRC plasma concentrations reported. Table 1 presents a comprehensive list of all the clinical studies referenced.
PBPK Model Development
The a priori PBPK model structure, depicted in Fig. 2 (left panel), represents a whole-body PBPK model for intravenous administration of VRC, assuming permeation-based tissue distribution kinetics. The kinetic parameters characterising the metabolism of VRC via CYP2C19, CYP3A4 and CYP2C9 were informed by in vitro assays based on recombinant CYP enzymes [17] . VRC and both metabolites were assumed to be excreted through renal CL processes, with NO additionally presumed to be eliminated through total hepatic route. The physicochemical properties, distribution, and elimination processes of VRC and its metabolites, used for the parameterisation of the a priori model, are comprehensively detailed in Tables 2 and 3.
The predictive performance of the coupled parent-metabolite PBPK model following a pure bottom-up approach was evaluated against clinical data from single and multiple intravenous dosing. The a priori model accurately predicted VRC exposure after the subtherapeutic SD (AAFEVRC,50mg/2h = 1.45) but not after the therapeutic dosing regimens. Although a priori predictions closely matched the VRC PK profiles following therapeutic doses for CYP2C19 PM (AAFEVRC,PM = 1.29) and IM (AAFEVRC,IM=1.37), an underprediction was evident for NM (AAFEVRC,NM = 1.46) and RM (AAFE VRC,RM = 1.83). The PK profiles of NO were underpredicted (range: AAFENO = 3.55–1.57), while predictions of OHVRC PK profiles were adequate for CYP2C19 NM and RM (range: AAFEOHVRC = 1.63–1.61) and overpredicted for PM and IM (range: AAFEOHVRC = 3.51–3.05). The model accurately predicted the first VRC Cmax of MD regimens but underpredicted PK profiles during maintenance dosing (range: AAFEMD = 2.33–1.74).
Following a stepwise approach, the a posteriori model was refined using clinical data to improve its predictive performance, as illustrated in Fig. 1. Initially, based on the in vitro data for the formation of NO via CYP3A4 (with a Km value of 1.20 µM), the CYP3A4-based catalytic rate constant (Kcat) was estimated to be 0.24 1/min (95% CI 0.22–0.26 1/min), using the clinical data of intravenous SDs in CYP2C19 PM individuals (Fig. 1, first green box). Subsequently, using clinical data comprising both plasma and urine of intravenous SDs and MDs in CYP2C19 IM, NM and RM individuals (Fig. 1, second green box), the Kcat of CYP2C19 was estimated to be 0.75 1/min (95% CI 0.70–0.80 1/min) and VRC renal CL was estimated to be 0.010 1/min (95% CI 0.009–0.011 1/min). Given the wide range of reported lipophilicity values for VRC (1.65–2.56) in the literature [34, 35, 51, 52], we estimated this parameter to be 2.00 (95% CI 1.99–2.01), and better matches the observed intravenous data. The estimated parameters improved the prediction of VRC PK profiles of SD (range: AFE = 0.87–0.87, AAFE = 1.28–1.35) and MD (range: AFE = 0.90–1.22, AAFE = 1.21–1.37).
During the process of refining the a posteriori model, consistent discrepancies in the predicted NO PK profiles were observed. An exploratory graphical analysis of the observed PK profiles of NO across various intravenous dosing regimens of VRC within our clinical datasets revealed that with increasing doses of VRC, the AUClast of NO increased disproportionately. This pattern indicated nonlinearity in the PK of NO. Based on these observations, we generated two independent hypotheses to characterise the nonlinear elimination of NO (Fig. 1, third green box). The first hypothesis (H1) assumed that NO was metabolised via Michaelis–Menten kinetics, which was tested by implementing a dummy enzyme expressed only in liver, fixing its Km value to 1 µM and estimating its Vmax value according to Eq. 8.
| 8 |
The rate of NO metabolism via dummy enzyme in µmol/L is expressed in Eq. 8, where is the unbound concentration of substrate (NO), is the Michaelis–Menten constant representing half the maximum metabolic rate and is the maximum metabolic rate of the dummy enzyme.
The second hypothesis (H2) assumed autoinhibition of the CL of NO, which was tested by implementing an empirical saturable inhibition equation on the total hepatic CL driven by hepatic intracellular NO concentrations, according to Eq. 9.
| 9 |
In Eq. 9, accounts for (auto-inhibited) hepatic CL of NO, accounts for the maximum (uninhibited) rate of NO metabolism, is the unbound hepatic concentration of NO, is the maximum inhibition, and is the half-maximal inhibitory concentration of NO.
Estimation of the maximum velocity rate of the dummy enzyme (Vmax = 0.25 µM/min) for H1, and the half-maximal inhibitory concentration (IC50 = 1.84 µM) along with the total hepatic CL of NO for H2, independently, provided accurate predictions for plasma NO. Additionally, the NO renal CL pathway was refined for both hypotheses based on the amount of NO excreted in urine and estimated as 0.135 mL/min/kg (95% CI 0.131–0.139 mL/min/kg). The estimated parameters resulted in an overall good prediction of VRC, NO and OHVRC across all dose levels for all CYP2C19 gXM. All parameters were precisely estimated for both a posterior models (Fig. 1, third green box) and are presented in Table 5.
Table 5.
Estimated parameters of the a posteriori PBPK models
| Parameter | Unit | Initial value | Estimates (95% CI) |
|---|---|---|---|
| CYP3A4 Kcat,NO | 1/min | 0.00893 | 0.24 (0.22–0.26) |
| VRC lipophilicity | – | 1.80 | 2.00 (1.99–2.01) |
| CYP2C19 Kcat,NO | 1/min | 1.64 | 0.75 (0.70–0.80) |
| VRC renal CL | mL/min/kg | 0.022 | 0.03 (0.029–0.031) |
| NO renal CL | mL/min/kg | 0.022 | 0.135 (0.131–0.139) |
| H1: Dummy Vmax | µM/min | 1 | 0.25 (0.24–0.26) |
| H2: NO IC50 | µM | 1 | 1.84 (0.73–2.95) |
| H2: NO hepatic CL | L/h/kg | 0.0727 | 0.12 (0.05–0.19) |
CI confidence interval, CL clearance, CYP450 cytochrome P450, CYP3A4 Kcat,NO catalytic rate constant of CYP3A4 forming NO, CYP2C19 Kcat,NO catalytic rate constant of CYP2C19 forming NO, Dummy Vmax maximum metabolic rate of assumed dummy enzyme metabolising NO, H1 hypothesis 1, H2 hypothesis 2, NO IC50 voriconazole N-oxide half-maximal inhibitory concentration in autoinhibition of NO clearance, NO voriconazole N-oxide, VRC voriconazole
PBPK Model Evaluation
Figures 3, 4, 5, 6 and 7 (and ESM Figs. S2–S4), present simulations of VRC administration for both intravenous SD and MD overlaid with the observed data. The initial a priori PBPK model (Fig. 1, third blue box; Figs. 3, 4, 5, 6 and 7, left), which relied solely on in vitro data, demonstrated moderate accuracy in predicting the SDs. However, it significantly underpredicted the MD, particularly during the terminal phase. After estimating selected model parameters using the middle-out framework, both a posteriori PBPK models (Fig. 1, last two green boxes; Figs. 3, 4, 5, 6 and 7, middle and right) accurately described and predicted plasma concentration-time profiles of VRC and its metabolites after SD and MD across different CYP2C19 gXM. While the predicted NO PK profile for PM showed some discrepancies, the a posteriori model of H1 revealed notable improvements in the predictions for this metabolite, increasing the percentage of predicted plasma concentrations within a twofold deviation from observed values to 58%, a significant enhancement from the initial 14%. Incorporating urine data showed that the amount of VRC excreted in urine was predicted within an acceptable range for the subtherapeutic (50 mg/2 h) SD by both the a priori and a posteriori models, with better performance of the a posteriori models at the therapeutic (400 mg/2 h) SD (ESM Fig. S2). The a posteriori models also more accurately predicted NO excretion for both subtherapeutic and therapeutic SD (ESM Fig. S4), while the excretion of OHVRC was equally well predicted by both the a priori and a posteriori models for all doses (ESM Fig. S7). Moreover, for the MD studies, both a posteriori models captured individual variability well, especially for individual Cmax values reported by Study A (Fig. 6b) [37]. Both a posteriori models also better predicted individual Cmin values, particularly during the later phases of the MD regimen (Fig. 7), outperforming the a priori model.
Fig. 3.
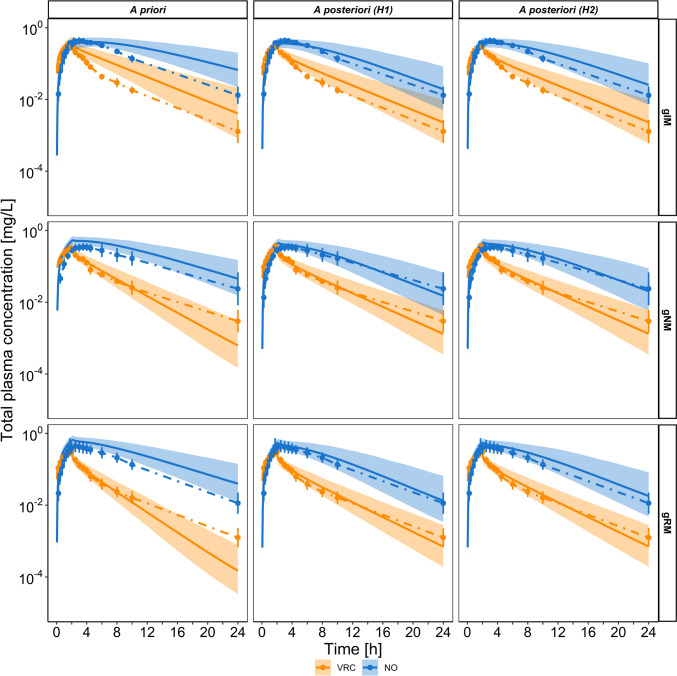
Predicted total plasma concentration-time profiles of VRC (orange) and NO (blue) comparing a priori (left) versus a posteriori models of hypothesis 1 (middle) and hypothesis 2 (right) following administration of a single intravenous dose of 50 mg infused over 2 h. Geometric means of the observed data are shown as dots, with error bars indicating geometric standard deviation for CYP2C19 IM (top), CYP2C19 NM (middle), and CYP2C19 RM (bottom) individuals in the study by Hohmann et al. (N = 15) [13]. Solid lines represent the geometric mean of the respective population predictions (N = 1000) and the shaded area represents the 90% population prediction intervals. CYP cytochrome P450, gXM genotype-predicted phenotype, H hypothesis, IM intermediate metaboliser, N number of individuals, NM normal metaboliser, NO voriconazole N-oxide, RM rapid metaboliser, VRC voriconazole
Fig. 4.
Predicted total plasma concentration-time profiles of VRC (orange) and NO (blue) comparing a priori (left) versus a posteriori models of hypothesis 1 (middle) and hypothesis 2 (right) following administration of a single intravenous dose of 100 mg infused over 4 h. Geometric means of the observed data are shown as dots, with error bars indicating geometric standard deviation for CYP2C19 IM (top), CYP2C19 NM (middle), and CYP2C19 RM (bottom) individuals in the study by Hohmann et al. (N = 12) [30]. Solid lines represent the geometric mean of the respective population predictions (N = 1000) and the shaded area represents the 90% population prediction intervals. CYP cytochrome P450, gXM genotype-predicted phenotype, H hypothesis, IM intermediate metaboliser, N number of individuals, NM normal metaboliser, NO voriconazole N-oxide, RM rapid metaboliser, VRC voriconazole
Fig. 5.
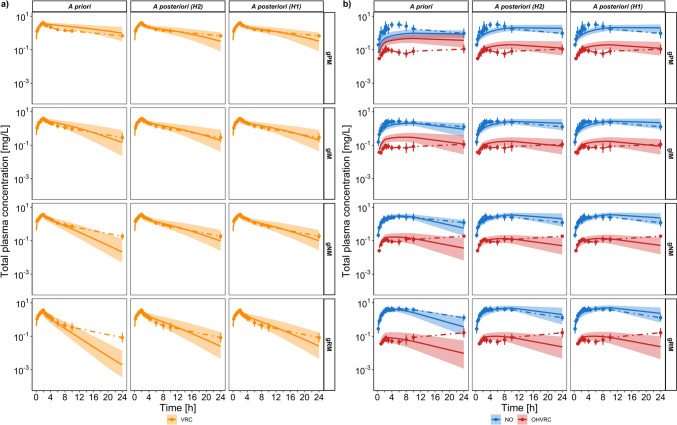
Predicted total plasma concentration-time profiles following a single intravenous dose of 400 mg voriconazole infused over 2 h. a Concentration-time profiles for VRC (orange) comparing a priori versus a posteriori models of hypothesis 1 and hypothesis 2. b Concentration-time profiles for NO (blue) and OHVRC (red) comparing a priori versus a posteriori models of hypothesis 1 and hypothesis 2. Geometric means of the observed data are shown as dots, with error bars indicating the geometric standard deviation for CYP2C19 PM (first row), CYP2C19 IM (second row), CYP2C19 NM (third row), and CYP2C19 RM (bottom row) individuals in the studies by Scholz et al. [14], Hohmann et al. [13] and Hohmann et al. [30] (N = 47). Solid lines represent the geometric mean of the respective population predictions (N = 1000) and the shaded area represents the 90% population prediction intervals. CYP cytochrome P450, gXM genotype-predicted phenotype, H hypothesis, IM intermediate metaboliser, N number of individuals, NM normal metaboliser, NO voriconazole N-oxide, OHVRC hydroxyvoriconazole, PM poor metaboliser, RM rapid metaboliser, VRC voriconazole
Fig. 6.
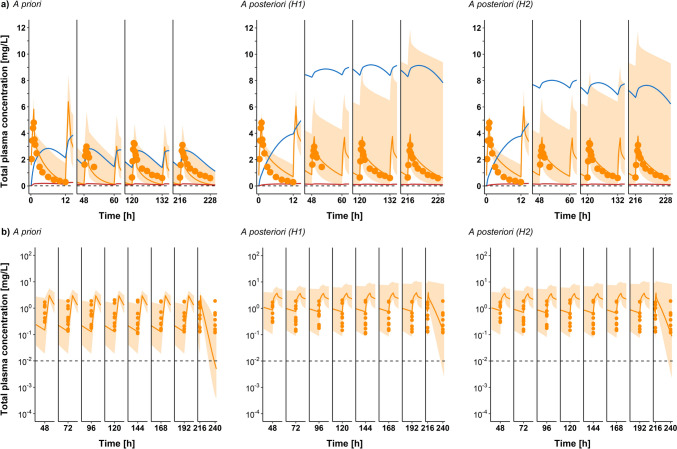
Predicted total plasma concentration-time profiles of VRC (orange) comparing a priori (left) versus a posteriori models of hypothesis 1 (middle) and hypothesis 2 (right) after multiple intravenous administrations of 3 mg/kg qd on the first day, then a MD of 3 mg/kg bid starting day 3 until day 11 of Study A by Purkins et al. [37]. a Arithmetic mean (N = 9) of the observed concentration-time profiles for days 1, 5, 8 and 12. b Individual observed plasma Cmax values for VRC on days 1, 5, 8 and 12. Solid orange lines represent the geometric mean of population predictions (N = 1000); dots represent the observed data for arithmetic means in panel (a) and individual Cmax values in panel (b); shaded area represents the 90% population prediction intervals; solid blue and red lines represent the geometric mean of population predictions (N = 1000) for NO and OHVRC, respectively; and the dashed black line represents the lower limit of quantification for VRC. bid twice daily, Cmax maximum plasma concentration, IV intravenous, MD maintenance dose, N number of individuals, NO voriconazole N-oxide, OHVRC hydroxyvoriconazole, qd once daily, VRC voriconazole
Fig. 7.
Predicted total plasma concentration-time profiles of VRC (orange) comparing a priori (left) versus a posteriori models of hypothesis 1 (middle) and hypothesis 2 (right), after multiple IV administration of an LD of 6 mg/kg bid on the first day, then an MD of 3 mg/kg bid starting on day 2 until day 9 of the Study B by Purkins et al. [37]. a Arithmetic mean (N = 9) of the observed concentration-time profiles for days 1, 3, 6 and 10. b Individual observed plasma Cmin values for VRC for the 1st to 10th days of the MD. Solid orange lines represent the geometric mean of population predictions (N = 1000); the dots represent the observed data for arithmetic means in panel (a) and individual Cmin values in panel (b); shaded area represents the 90% population prediction intervals; solid blue and red lines represent the geometric mean of the population predictions (N = 1000) for NO and OHVRC, respectively; and the dashed black line represents the lower limit of quantification for VRC. bid twice daily, Cmin minimum plasma concentration, H hypothesis, IV intravenous, LD loading dose, MD maintenance dose, N number of individuals, NO voriconazole N-oxide, OHVRC hydroxyvoriconazole, VRC voriconazole
Model performance is demonstrated in Fig. 8 (plasma GOF) as comparisons of the individual observed to predicted plasma concentrations. Both a posteriori models performed equally well and improved the a priori model predictions, with 94%, 84% and 76% of predicted plasma concentrations for VRC, NO, and OHVRC, respectively, falling within a twofold deviation from the observed data. The GOF plots for NO in the H1 and H2 models (Fig. 8b) demonstrate a slightly flatter trend compared with the line of identity, indicating a slight underprediction at higher concentrations. Initially, the a priori model showed, overall, a flatter distribution of data points across the concentration range. However, it significantly underpredicted NO concentrations in the CYP2C19 PM group, especially at therapeutic doses, with only 14% of the predicted data falling within the twofold range of the observed values. In contrast, the a posteriori models (H1 and H2) showed substantial improvements, increasing the accuracy to 58% within the twofold range for the PM group. Despite this improvement, the models still exhibited slight underprediction, suggesting further refinement is needed. The GOF plots for AUClast and Cmax of the parent and its metabolites (Fig. 9) demonstrated satisfactory model performance. All predicted AUClast values, and 94% of predicted Cmax values, for the parent were within a stricter criterion of a 1.25-fold deviation range, while all predicted AUClast and Cmax values for both metabolites were within a twofold deviation from their respective observed values. Moreover, the AAFE of 1.12 for predicted AUClast values and 1.14 for predicted Cmax values from a posteriori predictions of H1 and H2, respectively, for VRC, further confirmed an adequate model performance. The AAFE of AUClast and Cmax for the parent compound and metabolites predicted from H1 and H2 are detailed in ESM Tables S1 and S2, respectively. Additionally, the AFE, AAFE and RMSE for individual plasma concentrations are provided in ESM Tables S3.
Fig. 8.
Goodness-of-fit plots for plasma concentration predictions. A comparison of a priori (left) versus final a posteriori prediction for hypothesis 1 (middle) and hypothesis 2 (right) across CYP2C19 gXM. (a) Individual observed versus predicted VRC total plasma concentrations from all studies (IVSD and IVMD, n = 1765). (b) Individual observed versus predicted NO total plasma concentrations from all IVSD studies (n = 1591). (c) Individual observed versus predicted OHVRC total plasma concentrations from all IVSD studies (n = 372). The solid line represents the identity line, with dotted and dashed lines indicating 1.25-fold and 2-fold deviations, respectively. CYP cytochrome P450, gXM genotype-predicted phenotype, H hypothesis, IM intermediate metaboliser, IVSD intravenous single dose, IVMD intravenous maintenance dose, n number of samples, NO voriconazole N-oxide, NM normal metaboliser, OHVRC hydroxyvoriconazole, PM poor metaboliser, RM rapid metaboliser, VRC voriconazole
Fig. 9.
Pharmacokinetic parameter goodness-of-fit plots comparing a priori (left) versus a posteriori of hypothesis 1 (middle) and hypothesis 2 (right) models. a Predicted AUClast and b Cmax of VRC, NO and OHVRC compared with observed values for all single IV dose studies. The solid line represents the line of identity; dotted lines indicate 1.25-fold, and dashed lines indicate 2-fold deviation. AUClast area under the concentration-time curve from the time of administration to the last measurable concentration, Cmax maximum plasma concentration, IM intermediate metaboliser, IV intravenous, NO voriconazole N-oxide, NM normal metaboliser, OHVRC hydroxyvoriconazole, PM poor metaboliser, RM rapid metaboliser, VRC voriconazole
Sensitivity Analysis
Sensitivity analyses were then performed based on the simulation of the therapeutic intravenous SD regimen (400 mg/2 h), to assess the impact of the parameters on the a priori and a posteriori coupled parent-metabolite models. The VRC model was most sensitive to lipophilicity of VRC, fraction unbound in plasma of VRC, and CYP3A4 and CYP2C19 catalytic rate constant, with sensitivity values ranging from 2.191 to 0.011 (ESM Fig. S5). The NO model was most sensitive to Imax (H2), fraction unbound in plasma of NO, total hepatic CL of NO and lipophilicity of VRC, with sensitivity values ranging from 2.72 to 0.106 (ESM Fig. S6). Lastly, the OHVRC model was most sensitive to lipophilicity of VRC, CYP3A4 catalytic rate constant, fraction unbound in plasma of OHVRC, and renal CL of OHVRC, with sensitivity values ranging from 1.282 to 0.065 (ESM Fig. S7).
Discussion
In the presented work, a whole-body PBPK model of VRC and its metabolites, NO and OHVRC, was successfully developed. The model accurately predicts plasma and urine concentrations across a wide range (50–400 mg) of intravenous SD and MD regimens for healthy adult volunteers, with a focus on the variability introduced by different CYP2C19 genotype-predicted phenotypes, which significantly influence VRC metabolism.
Given the complex nature of the PK of VRC, an iterative workflow was adopted, initiated by a comprehensive collection of in vitro metabolic and clinical data. The first step enabled the precise characterisation of the metabolic and inhibitory properties of VRC and its metabolites, as well as their renal elimination. Leveraging these insights, a bottom-up approach was employed to construct the initial a priori PBPK model. However, the a priori PBPK model encountered limitations in accurately predicting the clinical PK of VRC at therapeutic doses, primarily due to overprediction of CYP2C19-mediated CL. While the model accurately predicted VRC PK in CYP2C19 gPM individuals, it underpredicted VRC PK in gNM, gIM and gRM individuals. Additionally, the initial model performance was suboptimal in predicting the renal CL of VRC and NO. To address these discrepancies and enhance the predictive performance of the model, iterative refinements were performed, particularly focusing on the metabolic pathways mediated by CYP3A4 and CYP2C19, and renal CL. The inclusion of data from multiple clinical studies, featuring a range of SD and MD dosing regimens and intensive sampling of PK data from healthy volunteers, facilitated the development of a robust model, mitigating the uncertainties associated with using data derived from existing literature. Taking into account the urine data, the model was able to capture the amount of parent and metabolites being renally excreted. Furthermore, MD is indicative of drug accumulation within the body; thus, accurately predicting it is paramount, as it implies a comprehensive understanding of complex processes that become evident during MD administration. These processes often pose challenges in clinical settings when striving to determine safe and effective dosing regimens. The resulting coupled parent-metabolite PBPK model showed adequate predictive performance for both intravenous SD and MD across all CYP2C19 genetic variants.
(Auto)-inhibition is a major mechanism for VRC nonlinear PK [53], since VRC is both a substrate and inhibitor of CYP2C19, CYP3A4 and CYP2C9. Based on our previous in vitro analyses [17], time-independent (reversible) inhibition by VRC, NO and OHVRC was observed for CYP2C19, CYP3A4 and CYP2C9, which was also supported by previous investigations [23, 36]. In contrast to published models [36], which faced challenges in directly implementing reversible autoinhibition, it was implemented in a mechanistic way in the current model. Unlike prior studies [34, 35, 54] that opted to simplify their models by excluding the inhibitory effects of VRC metabolites, our model incorporated both metabolites-mediated CYP inhibition, supporting the hypothesis that the primary metabolites of VRC play a crucial role in the overall inhibitory effect of VRC on its own metabolism. This is the first time these mechanisms are implemented in a joint parent-metabolites PBPK model of VRC, expanding on previously developed PBPK models [34–36, 54]. Additionally, our model is the first to incorporate urinary excretion data for VRC and its metabolites, providing a more accurate representation of the renal CL process. Although renal excretion accounts for < 2% of VRC total elimination, incorporating urine data allowed for a more comprehensive validation of the model’s assumptions. Previous models did not include such data, which may leave some uncertainty regarding the accuracy of their renal CL processes.
The incorporation of NO and OHVRC, as well as their inhibition impact, into the PBPK model was crucial in describing the nonlinear PK of VRC. Murayama et al. [42] proposed that OHVRC formation via CYP3A4 might be an imperative metabolic pathway for VRC in individuals with poor CYP2C19 catalytic function. This perspective was further supported by Geist et al. [12]. While OHVRC has not been previously investigated, the in vitro analyses revealed that it has a greater inhibitory effect on CYP3A4 than NO (Ki,OHVRC = 2.53 µM compared with Ki,NO = 5.24 µM) [17]. When we incorporated these data into our PBPK model, we found that the a priori predictions for VRC in the CYP2C19 PM individuals were satisfactory (AFE = 1.21, AAFE = 1.29), with 95% of predicted plasma concentrations falling within a twofold deviation from the observed plasma concentrations, even prior to further model refinements to better characterise the metabolites (OHVRC and NO).
The main enzyme involved in the metabolism of VRC is CYP2C19, particularly in the formation of its primary circulating metabolite, NO [11]. As a highly polymorphic enzyme, CYP2C19 exhibits significant variability in activity depending on genetic variants, which are known to substantially impact VRC metabolism and exposure. In the initial a priori PBPK model, the Kcat value for CYP2C19-mediated NO formation was set at 1.64 1/min, based on our previous in vitro assays using recombinant CYP enzymes [17], which largely represent the wild-type activity of CYP2C19. However, when refining the model using clinical data from individuals across different CYP2C19 genotypes, the estimated Kcat value was reduced by approximately 50%, in line with observed genotype-dependent variability in clinical settings. As initial simulations (a priori predictions) based on the in vitro Kcat value underestimated the accumulation effect observed in study A (one of the MD studies) [37], estimating that parameter based on clinical data enabled the model to accurately predict both Cmax and Cmin during MD regimens. Although genotype information was not reported in these studies, the assumption of only the CYP2C19 NM population captured the observed mean PK profiles. Further exploration of a full CYP2C19 genotype-predicted phenotype distribution in the virtual population allowed for an even better prediction of the variability and accumulation effects observed in study A, particularly for the reported individual Cmax values. This underscores the value of integrating clinical data with in vitro findings, as it improves the predictive performance of the PBPK model to account for population variabilities.
Although the formation of NO has been extensively investigated in various enzymatic systems, including human liver microsomes (HLMs), human intestine microsomes (HIMs) and recombinant human CYP enzymes (rhCYP), its elimination pathway remained elusive in these in vitro analyses [16]. Therefore, a joint NLME model to characterise the PK of VRC and NO, as reported by Li et al. [43], was used to infer the elimination process of NO in our PBPK model. Notably, clinical observations revealed a dose-dependent nonlinearity in NO elimination, challenging our initial model assumptions and leading to the generation of two hypotheses. The first hypothesis assumed that NO undergoes metabolism via Michaelis–Menten kinetics by an unspecified liver enzyme, while the second hypothesis suggested an autoinhibition mechanism for NO, similar to that observed with VRC. These hypotheses aimed to align the observed clinical data with the known metabolic pathways, underscoring the complexity of predicting the NO PK profile.
Upon evaluating the a posteriori predictions, it was observed that both hypotheses successfully captured the NO PK profiles across various dosing regimens, despite their differing mechanistic underpinning. As the first hypothesis offered a simplified semi-mechanistic explanation, the second hypothesis assuming autoinhibition by NO provided a more mechanistic description of the observed dose-dependent nonlinearity of NO elimination. However, a prominent challenge arose in characterising the PK of NO for individuals of CYP2C19 gPM. The difficulty can be attributed to the scarcity of available clinical data for this phenotype, with one study including data from only four such individuals [14]. While the two models presented performed equally and well in characterising single and multiple doses across different CYP2C19 genotype-predicted phenotype groups, such as gRMs, gNMs, and gIMs, they consistently underpredicted NO levels in the CYP2C19 gPM subpopulation, underlining the necessity for more comprehensive data on this particular subpopulation.
The underlying mechanism of the concentration-dependent NO elimination can be further clarified by conducting in vitro analyses, based on the two generated hypotheses. For testing the first hypothesis, model-informed Michaelis–Menten assays using in vitro liver systems, such as primary human hepatocytes, HLMs or rhCYP, can be performed. To investigate the second hypothesis of NO autoinhibition, inhibition assays with respective marker reactions using before-mentioned in vitro liver systems could be conducted [55].
Although the presented PBPK model successfully described the complex metabolism of VRC, we suggest to further expand the model by using the above-mentioned in vitro investigations to elucidate the exact elimination pathway of NO. Conducting further clinical studies in CYP2C19 gPMs with low intravenous SD and MD, including quantification of VRC metabolites [56], would be useful to further improve the model. Key areas for furthur expansion include integrating oral administration data to account for bioavailability and extrahepatic metabolism, and extending the applicability of the developed model to paediatric populations through ontogeny and maturation data. Ultimately, adult and paediatric simulations based on the developed PBPK model can be performed and used to recommend optimal dosing regimens for patients with IFIs or for prophylaxis. This could translate the amalgamated in vitro and clinical research results back into clinics, supporting future therapeutic decisions.
Conclusion
Until now, this is the most comprehensive parent-metabolite PBPK model of VRC, including whole-body PBPK models of VRC, NO and OHVRC, that has been successfully developed. The reversible autoinhibition of VRC and the incorporation of metabolite-mediated CYP inhibition successfully described the clinical data, supporting the hypothesis that primary VRC metabolites play a crucial role in the overall inhibitory effect of VRC on its own metabolism. Furthermore, the ability of the model to generate and refine hypotheses iteratively positions it as a valuable tool for future research, paving the way for more targeted investigations into the PK of VRC and its interaction with other drugs. This model can be used to design experiments to test these hypotheses, allowing further model qualification and ultimately model-based treatment individualisation to reduce therapeutic failure and AEs [57, 58].
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to express their gratitude to Dr. David Busse for his valuable discussions and insights. They also acknowledge the Clinical Trial Center of University Hospital Heidelberg for their support in data generation and sample analysis.
Declarations
Funding
Open Access funding was enabled and organised by Projekt DEAL.
Conflicts of Interest
Charlotte Kloft and Wilhelm Huisinga report grants from an industry consortium (AbbVie Deutschland GmbH & Co. K.G., AstraZeneca, Boehringer Ingelheim Pharma GmbH & Co. K.G., Grünenthal GmbH, F. Hoffmann-La Roche Ltd, Merck KGaA, Novo Nordisk A/S and Sanofi) for the graduate research training programme PharMetrX. In addition, Charlotte Kloft reports research grants from the Innovative Medicines Initiative-Joint Undertaking (‘DDMoRe’), from H2020-EU.3.1.3 (‘FAIR’), Diurnal Ltd and the Federal Ministry of Education and Research within the Joint Programming Initiative on Antimicrobial Resistance Initiative (‘JPIAMR’), all outside the submitted work. Ayatallah Saleh, Josefine Schulz, Jan-Frederik Schlender, Linda B.S. Aulin, Amrei-Pauline Konrad, Franziska Kluwe, Gerd Mikus, and Robin Michelet declare no competing interests that may be relevant to the contents of this work.
Ethics Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. All trial protocols were approved by the responsible Ethics Committees and the respective competent authorities.
Informed Consent
Written informed consent was obtained from all individual study participants before inclusion. Written informed consent was obtained from all individual study participants before inclusion.
Availability of Data and Material
The datasets generated and/or analysed during the current study are not publicly available as patients did not provide consent for sharing their data in a public database. The datasets are available from the corresponding author upon reasonable request.
Availability of Code
The PBPK model will be available on the AK-Kloft GitHub website and can be freely downloaded.
Author Contributions
Conceptualisation: AS, RM, CK. Clinical data collection: GM, CK. In vitro data generation: JS, CK. Planning of analysis: AS, FK, RM, CK. Formal analysis and investigation: AS, RM, JFS, WH, GM, CK. Writing – original draft preparation: AS, RM. Writing – review and editing: All authors contributed to discussion of the results as well as reviewing and editing the manuscript.
Footnotes
Charlotte Kloft and Robin Michelet shared senior authorship on this work.
Contributor Information
Charlotte Kloft, Email: charlotte.kloft@fu-berlin.de.
Robin Michelet, Email: robin.michelet@fu-berlin.de.
References
- 1.von Lilienfeld-Toal M, Wagener J, Einsele H, et al. Invasive fungal infection. Dtsch Arztebl Int. 2019;116:271–8. 10.3238/arztebl.2019.0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perlin DS, Rautemaa-Richardson R, Alastruey-Izquierdo A. The global problem of antifungal resistance: prevalence, mechanisms, and management. Lancet Infect Dis. 2017;17:e383–92. [DOI] [PubMed] [Google Scholar]
- 3.Ghannoum MA, Rice LB. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin Microbiol Rev. 1999;12:501–17. 10.1128/CMR.12.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfizer. VFEND summary of product characteristics (SmPC). Pfizer; 2020. [Google Scholar]
- 5.Pearson MM, Rogers PD, Cleary JD, Chapman SW. Voriconazole: a new triazole antifungal agent. Ann Pharmacother. 2003;37:420–32. 10.1345/aph.1C261. [DOI] [PubMed] [Google Scholar]
- 6.Cordonnier C, Rovira M, Maertens J, et al. Voriconazole for secondary prophylaxis of invasive fungal infections in allogeneic stem cell transplant recipients: results of the VOSIFI study. Haematologica. 2010;95:1762–8. 10.3324/haematol.2009.020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marks DI, Pagliuca A, Kibbler CC, et al. Voriconazole versus itraconazole for antifungal prophylaxis following allogeneic haematopoietic stem-cell transplantation. Br J Haematol. 2011;155:318–27. 10.1111/j.1365-2141.2011.08838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purkins L, Wood N, Ghahramani P, et al. Pharmacokinetics and safety of voriconazole following intravenous- to oral-dose escalation regimens. Antimicrob Agents Chemother. 2002;46:2546–53. 10.1128/AAC.46.8.2546-2553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirbs C, Kluwe F, Drescher F, et al. High voriconazole target-site exposure after approved sequence dosing due to nonlinear pharmacokinetics assessed by long-term microdialysis. Eur J Pharm Sci. 2019;131:218–29. 10.1016/J.EJPS.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Theuretzbacher U, Ihle F, Derendorf H. Pharmacokinetic/pharmacodynamic profile of voriconazole. Clin Pharmacokinet. 2006;45:649–63. 10.2165/00003088-200645070-00002. [DOI] [PubMed] [Google Scholar]
- 11.Schulz J, Kluwe F, Mikus G, et al. Novel insights into the complex pharmacokinetics of voriconazole: a review of its metabolism. Drug Metab Rev. 2019;51:247–65. 10.1080/03602532.2019.1632888. [DOI] [PubMed] [Google Scholar]
- 12.Geist MJP, Egerer G, Burhenne J, et al. Steady-state pharmacokinetics and metabolism of voriconazole in patients. J Antimicrob Chemother. 2013;68:2592–9. 10.1093/jac/dkt229. [DOI] [PubMed] [Google Scholar]
- 13.Hohmann N, Kocheise F, Carls A, et al. Dose-dependent bioavailability and CYP3A inhibition contribute to non-linear pharmacokinetics of voriconazole. Clin Pharmacokinet. 2016;55:1535–45. 10.1007/s40262-016-0416-1. [DOI] [PubMed] [Google Scholar]
- 14.Scholz I, Oberwittler H, Riedel K-D, et al. Pharmacokinetics, metabolism and bioavailability of the triazole antifungal agent voriconazole in relation to CYP2C19 genotype. Br J Clin Pharmacol. 2009;68:906–15. 10.1111/j.1365-2125.2009.03534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gautier-Veyret E, Thiebaut-Bertrand A, Roustit M, et al. Optimization of voriconazole therapy for treatment of invasive aspergillosis: Pharmacogenomics and inflammatory status need to be evaluated. Br J Clin Pharmacol. 2021;87:2534–41. 10.1111/bcp.14661. [DOI] [PubMed] [Google Scholar]
- 16.Gautier-Veyret E, Fonrose X, Tonini J, et al. Variability of voriconazole plasma concentrations after allogeneic hematopoietic stem cell transplantation: impact of cytochrome p450 polymorphisms and comedications on initial and subsequent trough levels. Antimicrob Agents Chemother. 2015;59:2305–14. 10.1128/AAC.04838-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulz J, Thomas A, Saleh A, et al. Towards the elucidation of the pharmacokinetics of voriconazole: a quantitative characterization of its metabolism. Pharmaceutics. 2022;14:477. 10.3390/pharmaceutics14030477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roffey SJ, Cole S, Comby P, et al. The disposition of voriconazole in mouse, rat, rabbit, guinea pig, dog, and human. Drug Metab Dispos. 2003;31:731–41. 10.1124/dmd.31.6.731. [DOI] [PubMed] [Google Scholar]
- 19.Epaulard O, Leccia M-T, Blanche S, et al. Phototoxicity and photocarcinogenesis associated with voriconazole. Med Mal Infect. 2011;41:639–45. 10.1016/j.medmal.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 20.Epaulard O, Villier C, Ravaud P, et al. A multistep voriconazole-related phototoxic pathway may lead to skin carcinoma: results from a French nationwide study. Clin Infect Dis. 2013;57:e182–8. 10.1093/cid/cit600. [DOI] [PubMed] [Google Scholar]
- 21.Zonios D, Yamazaki H, Murayama N, et al. Voriconazole metabolism, toxicity, and the effect of cytochrome P450 2C19 genotype. J Infect Dis. 2014;209:1941–8. 10.1093/infdis/jiu017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeong S, Nguyen PD, Desta Z. Comprehensive in vitro analysis of voriconazole inhibition of eight cytochrome P450 (CYP) enzymes: major effect on CYPs 2B6, 2C9, 2C19, and 3A. Antimicrob Agents Chemother. 2009;53:541–51. 10.1128/AAC.01123-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niwa T, Shiraga T, Takagi A. Effect of antifungal drugs on cytochrome P450 (CYP) 2C9, CYP2C19, and CYP3A4 activities in human liver microsomes. Biol Pharm Bull. 2005;28:1805–8. 10.1248/bpb.28.1805. [DOI] [PubMed] [Google Scholar]
- 24.Niwa T, Inoue-Yamamoto S, Shiraga T, Takagi A. Effect of antifungal drugs on cytochrome P450 (CYP) 1A2, CYP2D6, and CYP2E1 activities in human liver microsomes. Biol Pharm Bull. 2005;28:1813–6. 10.1248/bpb.28.1813. [DOI] [PubMed] [Google Scholar]
- 25.Nassar YM, Hohmann N, Michelet R, et al. Quantification of the time course of CYP3A inhibition, activation, and induction using a population pharmacokinetic model of microdosed midazolam continuous infusion. Clin Pharmacokinet. 2022;61:1595. 10.1007/S40262-022-01175-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pasqualotto AC, Xavier MO, Andreolla HF, Linden R. Voriconazole therapeutic drug monitoring: focus on safety. Expert Opin Drug Saf. 2010;9:125–37. 10.1517/14740330903485637. [DOI] [PubMed] [Google Scholar]
- 27.Schulz J, Michelet R, Zeitlinger M, et al. Microdialysis of voriconazole and its N-oxide metabolite: amalgamating knowledge of distribution and metabolism processes in humans. Pharm Res. 2022;39:3279–91. 10.1007/s11095-022-03407-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simmel F, Kloft C. Microdialysis feasibility investigations with the non-hydrophilic antifungal voriconazole for potential applications in nonclinical and clinical settings. Int J Clin Pharmacol Ther. 2010;48:695–704. 10.5414/CPP48695. [DOI] [PubMed] [Google Scholar]
- 29.Simmel F, Soukup J, Zoerner A, et al. Development and validation of an efficient HPLC method for quantification of voriconazole in plasma and microdialysate reflecting an important target site. Anal Bioanal Chem. 2008;392:479–88. 10.1007/s00216-008-2286-3. [DOI] [PubMed] [Google Scholar]
- 30.Hohmann N, Kreuter R, Blank A, et al. Autoinhibitory properties of the parent but not of the N-oxide metabolite contribute to infusion rate-dependent voriconazole pharmacokinetics. Br J Clin Pharmacol. 2017;83:1954–65. 10.1111/bcp.13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamada T, Mino Y, Yagi T, et al. Saturated metabolism of voriconazole N-oxidation resulting in nonlinearity of pharmacokinetics of voriconazole at clinical doses. Biol Pharm Bull. 2015;38:1496–503. 10.1248/bpb.b15-00241. [DOI] [PubMed] [Google Scholar]
- 32.Weiss J, Ten Hoevel MM, Burhenne J, et al. CYP2C19 genotype is a major factor contributing to the highly variable pharmacokinetics of voriconazole. J Clin Pharmacol. 2009;49:196–204. 10.1177/0091270008327537. [DOI] [PubMed] [Google Scholar]
- 33.Moriyama B, Obeng AO, Barbarino J, et al. Clinical pharmacogenetics implementation consortium (CPIC) guidelines for CYP2C19 and voriconazole therapy. Clin Pharmacol Ther. 2017;102:45–51. 10.1002/cpt.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Frechen S, Moj D, et al. A physiologically based pharmacokinetic model of voriconazole integrating time-dependent inhibition of CYP3A4, genetic polymorphisms of CYP2C19 and predictions of drug-drug interactions. Clin Pharmacokinet. 2020;59:781–808. 10.1007/s40262-019-00856-z. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Zhao S, Wang C, et al. Application of a physiologically based pharmacokinetic model to characterize time-dependent metabolism of voriconazole in children and support dose optimization. Front Pharmacol. 2021;12: 636097. 10.3389/fphar.2021.636097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong J, Liu S-B, Rasheduzzaman JM, et al. Development of physiology based pharmacokinetic model to predict the drug interactions of voriconazole and venetoclax. Pharm Res. 2022;39:1921–33. 10.1007/s11095-022-03289-9. [DOI] [PubMed] [Google Scholar]
- 37.Purkins L, Wood N, Greenhalgh K, et al. The pharmacokinetics and safety of intravenous voriconazole—a novel wide-spectrum antifungal agent. Br J Clin Pharmacol. 2003;56:2. 10.1046/J.1365-2125.2003.01992.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berezhkovskiy LM. Volume of distribution at steady state for a linear pharmacokinetic system with peripheral elimination. J Pharm Sci. 2004;93:1628–40. 10.1002/jps.20073. [DOI] [PubMed] [Google Scholar]
- 39.Rengelshausen J, Banfield M, Riedel K-D, et al. Opposite effects of short-term and long-term St John’s wort intake on voriconazole pharmacokinetics. Clin Pharmacol Ther. 2005;78:25–33. 10.1016/j.clpt.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 40.Meyer M, Schneckener S, Ludewig B, et al. Using expression data for quantification of active processes in physiologically based pharmacokinetic modeling. Drug Metab Dispos. 2012;40:892–901. 10.1124/dmd.111.043174. [DOI] [PubMed] [Google Scholar]
- 41.Achour B, Russell MR, Barber J, Rostami-Hodjegan A. Simultaneous quantification of the abundance of several cytochrome P450 and uridine 5′-diphospho-glucuronosyltransferase enzymes in human liver microsomes using multiplexed targeted proteomics. Drug Metab Dispos. 2014;42:500–10. 10.1124/dmd.113.055632. [DOI] [PubMed] [Google Scholar]
- 42.Murayama N, Imai N, Nakane T, et al. Roles of CYP3A4 and CYP2C19 in methyl hydroxylated and N-oxidized metabolite formation from voriconazole, a new anti-fungal agent, in human liver microsomes. Biochem Pharmacol. 2007;73:2020–6. 10.1016/j.bcp.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 43.Li S, Wu S, Gong W, et al. Application of population pharmacokinetic analysis to characterize CYP2C19 mediated metabolic mechanism of voriconazole and support dose optimization. Front Pharmacol. 2021;12: 730826. 10.3389/fphar.2021.730826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Segel IH. Enzyme kinetics: behavior and analysis of rapid equilibrium and steady-state enzyme systems. Wiley-Blackwell; 1975. [Google Scholar]
- 45.Willmann S, Höhn K, Edginton A, et al. Development of a physiology-based whole-body population model for assessing the influence of individual variability on the pharmacokinetics of drugs. J Pharmacokinet Pharmacodyn. 2007;34:401–31. 10.1007/s10928-007-9053-5. [DOI] [PubMed] [Google Scholar]
- 46.Open Systems Pharmacology Suite Community. PK-Sim® Ontogeny Database Documentation, Version 7.3. 2018. Available at: https://github.com/Open-Systems-Pharmacology/OSPSuite.Documentation/blob/master/PK-SimOntogenyDatabaseVersion7.3.pdf.
- 47.Lippert J, Burghaus R, Edginton A, et al. Open systems pharmacology community-an open access, open source, open science approach to modeling and simulation in pharmaceutical sciences. CPT pharmacometrics Syst Pharmacol. 2019;8:878–82. 10.1002/psp4.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eissing T, Kuepfer L, Becker C, et al. A computational systems biology software platform for multiscale modeling and simulation: integrating whole-body physiology, disease biology, and molecular reaction networks. Front Physiol. 2011;2:4. 10.3389/fphys.2011.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ankit Rohatgi. WebPlotDigitizer. 2022. Available at: https://automeris.io/WebPlotDigitizer/. Accessed 4 Dec 2022
- 50.Wojtyniak J-G, Britz H, Selzer D, et al. Data digitizing: accurate and precise data extraction for quantitative systems pharmacology and physiologically-based pharmacokinetic modeling. CPT Pharmacomet Syst Pharmacol. 2020;9:322–31. 10.1002/psp4.12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zane NR, Thakker DR. A physiologically based pharmacokinetic model for voriconazole disposition predicts intestinal first-pass metabolism in children. Clin Pharmacokinet. 2014;53:1171–82. 10.1007/s40262-014-0181-y. [DOI] [PubMed] [Google Scholar]
- 52.Simon F, Gautier-Veyret E, Truffot A, et al. Modeling approach to predict the impact of inflammation on the pharmacokinetics of CYP2C19 and CYP3A4 substrates. Pharm Res. 2021;38:415–28. 10.1007/s11095-021-03019-7. [DOI] [PubMed] [Google Scholar]
- 53.Muhareb A, Blank A, Meid AD, et al. CYP3A and CYP2C19 activity determined by microdosed probe drugs accurately predict voriconazole clearance in healthy adults. Clin Pharmacokinet. 2023;62:1305–14. 10.1007/S40262-023-01287-7/FIGURES/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang P, Liu S, Yang J. Physiologically based pharmacokinetic modeling to investigate the disease-drug-drug interactions between voriconazole and nirmatrelvir/ritonavir in COVID-19 patients with CYP2C19 phenotypes. Clin Pharmacol Ther. 2024;116(2):363–71. 10.1002/cpt.3222. [DOI] [PubMed] [Google Scholar]
- 55.Nagar S, Argikar UA, Tweedie DJ. Enzyme kinetics in drug metabolism: fundamentals and applications. Methods Mol Biol. 2014;1113:1–6. 10.1007/978-1-62703-758-7_1. [DOI] [PubMed] [Google Scholar]
- 56.Schulz J, Michelet R, Zeitlinger M, et al. Microdialysis of drug and drug metabolite: a comprehensive in vitro analysis for voriconazole and voriconazole N-oxide. Pharm Res. 2022;39:2991–3003. 10.1007/s11095-022-03292-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kluwe F, Michelet R, Huisinga W, et al. Towards model-informed precision dosing of voriconazole: challenging published voriconazole nonlinear mixed-effects models with real-world clinical data. Clin Pharmacokinet. 2023;62(10):1461–77. 10.1007/s40262-023-01274-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kluwe F, Michelet R, Mueller-Schoell A, et al. Perspectives on model-informed precision dosing in the digital health era: challenges, opportunities, and recommendations. Clin Pharmacol Ther. 2021;109:29–36. 10.1002/cpt.2049. [DOI] [PubMed] [Google Scholar]
- 59.Purkins L, Wood N, Greenhalgh K, et al. The pharmacokinetics and safety of intravenous voriconazole—a novel wide-spectrum antifungal agent. Br J Clin Pharmacol. 2003;56(Suppl 1):2–9. 10.1046/j.1365-2125.2003.01992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vanstraelen K, Wauters J, De Loor H, et al. Protein-binding characteristics of voriconazole determined by high-throughput equilibrium dialysis. J Pharm Sci. 2014;103:2565–70. 10.1002/jps.24064. [DOI] [PubMed] [Google Scholar]
- 61.Zubiaur P, Kneller LA, Ochoa D, et al. Evaluation of voriconazole CYP2C19 phenotype-guided dose adjustments by physiologically based pharmacokinetic modeling. Clin Pharmacokinet. 2021;60:261–70. 10.1007/s40262-020-00941-8. [DOI] [PubMed] [Google Scholar]
- 62.Hanke N, Frechen S, Moj D, et al. PBPK models for CYP3A4 and P-gp DDI Prediction: A modeling network of rifampicin, itraconazole, clarithromycin, midazolam, alfentanil, and digoxin. CPT Pharmacometrics Syst Pharmacol. 2018;7:647–59. 10.1002/psp4.12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.PK-Sim Ontogeny Database, Version 7.3. 2018. Available at: https://github.com/Open-Systems-Pharmacology/OSPSuite.Documentation/blob/master/PK-SimOntogeny Database Version 7.3.pdf. Accessed 5 Dec 2020
- 64.Nishimura M, Yaguti H, Yoshitsugu H, et al. Tissue distribution of mRNA expression of human cytochrome P450 isoforms assessed by high-sensitivity real-time reverse transcription PCR. Yakugaku Zasshi. 2003;123:369–75. 10.1248/yakushi.123.369. [DOI] [PubMed] [Google Scholar]
- 65.Ito K, Ogihara K, Kanamitsu S-I, Itoh T. Prediction of the in vivo interaction between midazolam and macrolides based on in vitro studies using human liver microsomes. Drug Metab Dispos. 2003;31:945–54. 10.1124/dmd.31.7.945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.