Abstract
Large Language Models (LLMs) are rapidly being adopted in healthcare, necessitating standardized reporting guidelines. We present TRIPOD-LLM, an extension of the TRIPOD+AI statement, addressing the unique challenges of LLMs in biomedical applications. TRIPOD-LLM provides a comprehensive checklist of 19 main items and 50 subitems, covering key aspects from title to discussion. The guidelines introduce a modular format accommodating various LLM research designs and tasks, with 14 main items and 32 subitems applicable across all categories. Developed through an expedited Delphi process and expert consensus, TRIPOD-LLM emphasizes transparency, human oversight, and task-specific performance reporting. We also introduce an interactive website (https://tripod-llm.vercel.app/) facilitating easy guideline completion and PDF generation for submission. As a living document, TRIPOD-LLM will evolve with the field, aiming to enhance the quality, reproducibility, and clinical applicability of LLM research in healthcare through comprehensive reporting.
Healthcare’s embrace of Large Language Models (LLMs) shows no signs of slowing down, with current and future deployment being considered in several domains across administrative and healthcare delivery use-cases, including generating drafts for patient communication, medical document summarization, question answering, information retrieval, medical diagnosis, treatment recommendations, patient education, and medical education.1–5 The rapid advancements made in LLMs have stretched existing regulatory and governance structures to their limits, exposing a patchwork of solutions that do not fully encompass the nuances of these all-purpose models.6–8 More broadly, the speed of development of LLMs poses a challenge to journal and peer-review publication timelines and to regulatory agencies, who seek to provide timely guidance. To keep pace, researchers publish pre-prints quickly and take an ad-hoc approach to reporting.
Reporting guidelines provide a scalable method for standardizing research, transparent reporting, and the peer review process. The TRIPOD (Transparent Reporting of a Multivariable Model for Individual Prognosis Or Diagnosis) initiative is a critical example that was first introduced in 2015 to establish minimum reporting standards for diagnostic and prognostic prediction model studies (www.tripod-statement.org).9 TRIPOD is one of the core guidelines on the EQUATOR Network, which is an international effort that promotes transparent, accurate reporting of health research literature.10 TRIPOD is widely endorsed and recommended by journals, and is often included in journal instructions to authors. TRIPOD has subsequently been updated to incorporate best practices for AI due to the substantially evolved machine learning landscape, resulting in TRIPOD+AI.11 This is in addition to other guidelines that offer complementary guidance on AI development throughout the model lifecycle.12–14
LLMs represent a distinct frontier within AI, introducing unique challenges and considerations not fully addressed by original TRIPOD guidelines or its newer extensions as we shift from classifier AI models to generative AI. Here, we report the TRIPOD-LLM statement, an extension of TRIPOD+AI11, developed to address these unmet needs and designed to be a living checklist in order to nimbly adapt to the rapidly evolving field. This expansion broaden the TRIPOD scope beyond its original focus on prediction models, and reflects the pervasive impact of LLMs across diverse areas of medical research and practice, from diagnosis to document summarization.
Rationale for TRIPOD-LLM
LLMs as generative AI models are autoregressive, meaning – in the simplest possible terms – that they are trained to predict the next word in a sequence given the words that preceded it. Yet, this foundational training has been shown to equip them with capabilities to perform a wide range of healthcare-related natural language processing (NLP) tasks from a single model. This adaptability is commonly achieved through supervised fine-tuning or few-shot learning methods, which allow LLMs to handle new tasks with minimal examples.15,16 Chatbot solutions (e.g., ChatGPT) use LLMs as their foundation, upon which two more components are added: question-answering (referred to as instruction tuning or supervised fine-tuning) and preference ranking (referred to as alignment). The unique methodological processes involved in LLMs and chatbots are not captured by current reporting guidelines, such as the choice of hyperparameters used for supervised fine-tuning, the intricacies of prompting, variability in model predictions, methods in evaluating natural language outputs, and preference-based learning strategies — which require specific guidance and have a substantial impact on model reliability. In addition, the generalist and generative nature of LLMs require more detailed guidance than covered in prior guidelines. Because LLMs can be applied to a broad range of use cases for which they were not specifically trained for and were not necessarily represented in training data (e.g. disease prevalence typically captured in a task-specific model’s training data for a given use case), they require unique task-specific guidance for robust reporting and downstream reliability and safety.
The selection of appropriate automated and human metrics by which to evaluate generative output remains an open question, and currently a wide range of methodologies are applied to capture various facets of performance. For tasks where the output is truly unstructured text and cannot be resolved to a structured label, such as letter generation or summarization tasks, evaluation is particularly complex. In these cases, most automated metrics prioritize overlap and similarity between input and output text, producing scores that may not capture factual accuracy or relevance of the text produced, and may not detect hallucinations or omissions.17–19 These scores reflect the degree of structural and lexicographical resemblance to reference texts which, though important, capture only a fraction of what constitutes a comprehensive evaluation of performance and safety. Human evaluation of text is a subjective process, complicated by the ambiguity of language and uncertainties inherent to many clinical tasks. These challenges are heightened in medicine, where there is often no single correct answer and both aleatoric and epistemic uncertainty are common. Therefore, specific details are needed to guide reporting of how performance is evaluated. In this paper, we use the term LLM to refer to both LLMs and chatbots. Table 1 highlights key categories of tasks applicable to the healthcare domain and provides notable definitions and examples of existing relevant work.
Table 1.
Research design and LLM task categories for the modular TRIPOD-LLM guideline.
| Task | Definition | Example |
|---|---|---|
| Research design | ||
| De novo LLM development | Building a new language model from scratch or significantly fine-tuning existing base models to develop new functionalities or to adapt to new tasks. | A study pre-training a new LLM on a hospital’s clinical data, e.g., Peng et al. (2023) 56 |
| LLM methods | Quantitative or theoretical investigations that focus on new architectures of model design, new computational methods to understand LLMs, new methods to evaluate LLMs, and/or new methods to optimize LLM prompting. | A study of a new retrieval-augmented generation LLM framework for medicine, e.g., Zakka et al. (2024) 57 |
| LLM evaluation | Assessing or testing an existing LLM to determine its efficacy, accuracy, or suitability for a specific task within healthcare; may also include evaluating the risks and biases arising from using it. | A study investigating biased diagnostic reasoning in an existing LLM, e.g., Zack et al. (2023).21 |
| LLM evaluation in healthcare settings | Evaluating an LLM when used as part of a clinical workflow, focusing on its integration and impact on clinical, administrative, or workforce outcomes. | A study reporting the performance of an LLM deployed in real-time to predict outcomes in hospitalized patients, e.g., Jiang et al. (2023) 6 |
| LLM task | ||
| Text processing | Manipulating and lower-order processing of text data, which includes tasks including but not limited to tokenization, parsing, and entity recognition. | A study investigating a new LLM approach to named entity recognition, e.g., Keloth et al. 2024 58 |
| Classification | Assigning predefined labels to text data. | A study fine-tuning an LLM to determine whether or not a sentence in a clinic note mentions one or several social determinants of health, e.g., Guevara et al. (2024)59 |
| Long-form question-answering | Providing detailed answers to complex queries, which can involve reasoning over multiple documents or pieces of evidence. Note that multi-choice question-answering is subsumed under classification. | A study investigating the ability of an existing LLM to respond to patient portal messages, e.g., Chen et al., (2024) 24 |
| Information retrieval | The process of fetching relevant information from large datasets based on specific queries, which is relevant for tasks like literature review or patient history retrieval. | A study that trained a Transformer model to retrieve biomedical publications relevant to a user’s query, e.g., Jin et al. (2023).60 |
| Conversational agent (chatbot) | Responding to and engaging in conversation with users — often used for patient interaction, health advisories, or as virtual assistants for healthcare providers. | A study investigating whether access to an interactive LLM-based chatbot impacts clinicians’ diagnostic reasoning, e.g., Goh et al. (2024)61 |
| Documentation generation | Automatically creating medical documentation from clinical data, dictations, or recordings. | A study evaluating the quality of clinical notes automatically generated from ambient clinic recordings, e.g., Tierney et al. (2024)5 |
| Summarization and Simplification | Condensing large text documents into shorter versions or simplifying the content for easier comprehension is useful in patient education or in creating executive summaries of medical records. | A study evaluating the ability of LLMs to convert discharge summaries into patient-friendly plain language, e.g., Zaretsky et al. (2024)62 |
| Machine translation | Converting text from one language to another. | A study comparing the ability of smaller language models fine-tuned for translation versus generalist LLMs to translate biomedical text between Spanish and English, e.g., Han et al. (2022)63 |
| Outcome forecasting | Predicting future medical outcomes based on historical data, which can be used in prognosis estimation or treatment effectiveness studies. | A study investigating the ability of LLMs to predict out-of-hospital mortality in patients admitted to intensive care units, e.g., Yoon et al. (2024)64 |
The novel complexities introduced by LLMs include concerns regarding hallucinations, omissions, reliability, explainability, reproducibility, privacy, and biases being propagated downstream, which can adversely affect clinical decision-making and patient care.20–26 Furthermore, growing partnerships between electronic health record (EHR) vendors, technology companies, and healthcare providers have led to deployment horizons that far outpace current regulatory timelines.8,27 To safeguard LLM use and increase transparency, standardization in the development and reporting of LLMs is essential — to ensure consistency, reliability, and verifiability, akin to established clinical grade evaluation in other scientific domains.28–30
The TRIPOD-LLM Statement
The TRIPOD-LLM comprises a checklist of items considered essential for good reporting of studies that are developing, tuning, prompt engineering, or evaluating an LLM (Table 2). Box 1 summarizes noteworthy additions and changes to TRIPOD-2015 and TRIPOD+AI. The TRIPOD-LLM checklist comprises 19 main items about the title (1 item), abstract (1 item), introduction (2 items), methods (8 items), open science practices (1 item), patient and public involvement (1 item), results (3 items), and discussion (2 items). These main items are further divided into 50 subitems. Of these, 14 main items and 32 subitems are applicable to all research designs and LLM tasks. The remaining 5 main items and 18 subitems are specific to particular research designs or LLM task categories. As discussed in the methods section below, the TRIPOD-LLM statement introduces a modular format given the varied nature of LLM studies (Table 1), where some items are only relevant for specific research designs and LLM task categories. These designs and task categories are broad but not mutually exclusive, depending on the context of a specific study, and may need to evolve as application of LLMs evolve.
Table 2:
TRIPOD-LLM Checklist
| Section | Item | Description | Research Design | LLM Task |
|---|---|---|---|---|
| Title | 1 | Identify the study as developing, fine-tuning, and/or evaluating the performance of an LLM, specifying the task, the target population, and the outcome to be predicted. | All | All |
| Abstract | 2 | See TRIPOD-LLM for Abstracts | All | All |
| Introduction | ||||
| Background | 3a | Explain the healthcare context / use case (e.g., administrative, diagnostic, therapeutic, clinical workflow) and rationale for developing or evaluating the LLM, including references to existing approaches and models. | All | All |
| 3b | Describe the target population and the intended use of the LLM in the context of the care pathway, including its intended users in current gold standard practices (e.g., healthcare professionals, patients, public, or administrators). | E H |
All | |
| Objectives | 4 | Specify the study objectives, including whether the study describes the initial development, fine-tuning, or validation of an LLM (or multiple stages). | All | All |
| Methods | ||||
| Data | 5a | Describe the sources of data separately for the training, tuning, and/or evaluation datasets and the rationale for using these data (e.g., web corpora, clinical research/trial data, EHR data, or unknown). | All | All |
| 5b | Describe the relevant data points and provide a quantitative and qualitative description of their distribution and other relevant descriptors of the dataset (e.g., source, languages, countries of origin) | All | All | |
| 5c | Specifically state the date of the oldest and newest item of text used in the development process (training, fine-tuning, reward modeling) and in the evaluation datasets. | All | All | |
| 5d | Describe any data pre-processing and quality checking, including whether this was similar across text corpora, institutions, and relevant socio-demographic groups. | All | All | |
| 5e | Describe how missing and imbalanced data were handled and provide reasons for omitting any data. | All | All | |
| Analytical Methods | 6a | Report the LLM name, version, and last date of training. | All | All |
| 6b | Report details of LLM development process, such as LLM architecture, training, fine-tuning procedures, and alignment strategy (e.g., reinforcement learning, direct preference optimization, etc.) and alignment goals (e.g., helpfulness, honesty, harmlessness, etc.). | M D |
All | |
| 6c | Report details of how text was generated using the LLM, including any prompt engineering (including consistency of outputs), and inference settings (e.g., seed, temperature, max token length, penalties), as relevant. | M D E |
All | |
| 6d | Specify the initial and post-processed output of the LLM (e.g., probabilities, classification, unstructured text). | All | All | |
| 6e | Provide details and rationale for any classification and, if applicable, how the probabilities were determined and thresholds identified. | All | C OF |
|
| LLM Output | 7a | Include metrics that capture the quality of generative outputs, such as consistency, relevance, accuracy, and , and presence/type of errors compared to gold standards. | All | QA IR DG SS MT |
| 7b | Report the outcome metrics’ relevance to downstream task at deployment time and, where applicable, correlation of metric to human evaluation of the text for the intended use. | E H |
All | |
| 7c | Clearly define the outcome, how the LLM predictions were calculated (e.g., formula, code, object, API), the date of inference for closed-source LLMs, and evaluation metrics. | E H |
All | |
| 7d | If outcome assessment requires subjective interpretation, describe the qualifications of the assessors, any instructions provided, relevant information on demographics of the assessors, and inter-assessor agreement. | All | All | |
| 7e | Specify how performance was compared to other LLMs, humans, and other benchmarks or standards. | All | All | |
| Annotation | 8a | If annotation was done, report how text was labeled, including providing specific annotation guidelines with examples. | All | All |
| 8b | If annotation was done, report how many annotators labeled the dataset(s), including the proportion of data in each dataset that were annotated by more than 1 annotator, and the inter-annotator agreement. | All | All | |
| 8c | If annotation was done, provide information on the background and experience of the annotators or characteristics of any models involved in labelling. | All | All | |
| Prompting | 9a | If research involved prompting LLMs, provide details on the processes used during prompt design, curation, and selection. | All | All |
| 9b | If research involved prompting LLMs, report what data were used to develop the prompts. | All | All | |
| Summarization | 10 | Describe any preprocessing of the data before summarization. | All | SS |
| Instruction tuning/Alignment | 11 | If instruction tuning/alignment strategies were used, what were the instructions, data, and interface used for evaluation, and what were the characteristics of the populations doing evaluation? | M D |
All |
| Compute | 12 | Report compute, or proxies thereof (e.g., time on what and how many machines, cost on what and how many machines, inference time, floating-point operations per second (FLOPs)), required to carry out methods. | M D E |
All |
| Ethical Approval | 13 | Name the institutional research board or ethics committee that approved the study and describe the participant-informed consent or the ethics committee waiver of informed consent. | All | All |
| Open Science | 14a | Give the source of funding and the role of the funders for the present study. | All | All |
| 14b | Declare any conflicts of interest and financial disclosures for all authors. | All | All | |
| 14c | Indicate where the study protocol can be accessed or state that a protocol was not prepared. | H | All | |
| 14d | Provide registration information for the study, including register name and registration number, or state that the study was not registered. | H | All | |
| 14e | Provide details of the availability of the study data. | All | All | |
| 14f | Provide details of the availability of the code to reproduce the study results. | All | All | |
| Public Involvement | 15 | Provide details of any patient and public involvement during the design, conduct, reporting, interpretation, or dissemination of the study or state no involvement. | H | All |
| Results | ||||
| Participants | 16a | When using patient/EHR data, describe the flow of text/EHR/patient data through the study, including the number of documents/questions/participants with and without the outcome/label and follow-up time as applicable. | E H |
All |
| 16b | When using patient/EHR data, report the characteristics overall and, for each data source or setting, and for development/evaluation splits, including the key dates, key characteristics, and sample size. | E H |
All | |
| 16c | For LLM evaluation that include clinical outcomes, show a comparison of the distribution of important clinical variables that may be associated with the outcome between development and evaluation data, if available. | E H |
All | |
| 16d | When using patient/EHR data, specify the number of participants and outcome events in each analysis (e.g., for LLM development, hyperparameter tuning, LLM evaluation). | E H |
All | |
| Performance | 17 | Report LLM performance according to pre-specified metrics (see item 7a) and/or human evaluation (see item 7d). | All | All |
| LLM Updating | 18 | If applicable, report the results from any LLM updating, including the updated LLM and subsequent performance. | All | All |
| Discussion | ||||
| Interpretation | 19a | Give an overall interpretation of the main results, including issues of fairness in the context of the objectives and previous studies. | All | All |
| Limitations | 19b | Discuss any limitations of the study and their effects on any biases, statistical uncertainty, and generalizability. | All | All |
| Usability of the LLM in context | 19c | Describe any known challenges in using data for the specified task and domain context with reference to representation, missingness, harmonization, and bias. | E H |
All |
| 19d | Define the intended use for the implementation under evaluation, including the intended input, end-user, level of autonomy/human oversight. | E H |
All | |
| 19e | If applicable, describe how poor quality or unavailable input data should be assessed and handled when implementing the LLM, i.e., what is the usability of the LLM in the context of current clinical care. | E H |
All | |
| 19f | If applicable, specify whether users will be required to interact in the handling of the input data or use of the LLM, and what level of expertise is required of users. | E H |
All | |
| 19g | Discuss any next steps for future research, with a specific view to applicability and generalizability of the LLM. | All | All | |
LLM = large language model; M = LLM methods; D = de novo LLM development; E = LLM evaluation; H = LLM evaluation in healthcare settings; C = classification; OF = outcome forecasting; QA = long-form question-answering; IR = information retrieval; DG = document generation; SS = summarization and simplification; MT = machine translation; EHR = electronic health record. Note: For studies using existing LLMs, users should include reference(s) to reportable information if provided by the original developers or state that this information is not available.
Box 1: TRIPOD-LLM noteworthy changes and additions to TRIPOD-2015 and TRIPOD+AI.
New Checklist for Reporting on LLMs:
A dedicated checklist has been developed to address the unique aspects of reporting large language models (LLMs), reflecting their distinct characteristics and the specific methodologies they employ compared to other AI and prediction models.
Living Guideline:
The checklist is designed as a living document, which will be updated on a regular basis based on review of the literature and input from the community. This approach was taken due to the rapid developments in the field, enabling agile versioning, refinement from user testing, and timely updates as the field advances.
Task-Specific Guidance:
The checklist includes a new section that provides task-specific guidance designed to address the particular challenges and needs associated with different LLM applications in healthcare. This addition ensures that reporting is tailored and relevant to the specific functions and objectives of the LLM under study.
Enhanced Emphasis on Transparency and Fairness:
The new guidelines emphasize ‘transparency’ and ‘fairness,’ highlighting the importance of recognizing and addressing societal biases that may be encoded in clinical models. The checklist integrates these concepts throughout, ensuring that bias and fairness are considered at every stage of the model’s life cycle.
Modular Framework:
The new guidelines are modular, with different requirements based on the Research Design(s) and LLM Task(s) that are reported in a given study. This change was motivated by the wide variety of applications and approaches in biomedical LLM research, from model development through evaluation, necessitating more specialized reporting items.
A separate checklist for journal or conference abstracts of LLM-based studies is included, and the TRIPOD+AI for Abstracts statement18 is revised (TRIPOD-LLM for Abstracts), reflecting new content and maintaining consistency with TRIPOD-LLM (Table 3).
Table 3:
TRIPOD-LLM for Abstracts
| Section | Item | Checklist Item | Research Design | LLM Task |
|---|---|---|---|---|
| Title | 2a | Identify the study as developing, fine-tuning, and/or evaluating the performance of an LLM, specifying the task, the target population, and the outcome to be predicted. | All | All |
| Background | 2b | Provide a brief explanation of the healthcare context, use case and rationale for developing or evaluating the performance of an LLM. | E H |
All |
| Objectives | 2c | Specify the study objectives, including whether the study describes LLMs development, tuning, and/or evaluation | All | All |
| Methods | 2d | Describe the key elements of the study setting. | All | All |
| 2e | Detail all data used in the study, specify data splits and any selective use of data. | M D E |
All | |
| 2f | Specify the name and version of LLM(s) used. | All | All | |
| 2g | Briefly summarize the LLM-building steps, including any fine-tuning, reward modeling, reinforcement learning with human feedback (RLHF), etc. | M D |
All | |
| 2h | Describe the specific tasks performed by the LLMs (e.g., medical QA, summarization, extraction), highlighting key inputs and outputs used in the final LLM. | All | All | |
| 2i | Specify the evaluation datasets/populations used, including the endpoint evaluated, and detail whether this information was held out during training/tuning where relevant, and what measure(s) were used to evaluate LLM performance. | All | All | |
| Results | 2j | Give an overall report and interpretation of the main results. | All | All |
| Discussion | 2k | Explicitly state any broader implications or concerns that have arisen in light of these results. | All | All |
| Other | 2l | Give the registration number and name of the registry or repository (if relevant). | H | All |
LLM = large language model; M = LLM methods; D = de novo LLM development; E = LLM evaluation; H = LLM evaluation in healthcare settings
The recommendations contained within TRIPOD-LLM are for completely and transparently reporting on how LLM-based research was conducted; TRIPOD-LLM does not prescribe how to develop or evaluate LLMs specifically. The checklist is not a quality appraisal tool. Similarly, CANGARU 31 and CHART 32 are complementary guidelines that relate to Generative AI more broadly and Chatbots specifcially.
In addition to the TRIPOD website (www.tripod-statement.org) an accompanying interactive website was developed (https://tripod-llm.vercel.app/) to present the required questions based on research design(s) and task(s) for ease of completion. This site can be used to render a final PDF suitable for submission. Fillable templates for the TRIPOD-LLM checklist can also be downloaded from www.tripod-statement.org. News, announcements, and information relating to TRIPOD-LLM and the release of subsequent statements can be found on the TRIPOD-LLM website and TRIPOD website (www.tripod-statement.org).
An example of a completed TRIPOD-LLM checklist for a previously published study reporting the pre-training, fine-tuning, retrospective evaluation, and clinical deployment of an LLM for clinical and operational hospital tasks is presented in Supplementary Table 1.6 A fillable checklist is provided in Supplementary Table 2 and a new explanation and elaboration document is available in Supplementary Table 3 to aid understanding of new items.
TRIPOD-LLM Statement as a Living Document
Given the rapid pace of the field and the timeline for interaction with healthcare workers and patients, the decision was made to create an accelerated TRIPOD-LLM statement to provide timely guidance for LLM use in (bio)medical and other healthcare applications. This guidance has been designed as part of a living document hosted on an interactive website to facilitate agile versioning, refinement from user testing, updates as the field evolves, and regular meetings to intake and evaluate new standards. Thus, as the reporting recommendations are anticipated to evolve, users are directed to the most current version of the guidelines at https://tripod-llm.vercel.app/.
Our approach to the living TRIPOD-LLM statement is informed by processes established in developing living systematic reviews 33,33,34 and clinical practice guidelines,35,36 which have been adopted to address a similar need to provide updated, timely recommendations based on evolving evidence. Public comments on the statement will be collected from the community via multiple avenues to enhance accessibility: a project-specific GitHub repository, the TRIPOD-LLM website, and the main TRIPOD website (https://www.tripod-statement.org/). We encourage input both on usability, such as language that may be ambiguous or redundant, and on the content of the guidelines themselves. As a few examples, users may suggest a change to an item to make it more feasible in practice, recommend a new item be added, recommend adding or removing items assigned to a given research design of LLM task module, or recommend changes to the research design or LLM task module categories.
An expert panel will convene every three months to discuss updates. Before the meeting, members will review the intercurrent literature to inform any updates. The units for update will be checklist items, research design categories, and LLM task categories delineated in the statement. At the meeting, the panel will discuss the current statement and suggest revisions considering public comments, literature review, and subject matter expertise. The steering committee will revise the statement based on this discussion, and this will be circulated to the expert panel for final review and approval. Review can result in the following action for each component of the TRIPOD-LLM statement (adapted from Mikati et al., 201933) - items, research designs, and LLM tasks:
No modification
Modification of substantive content (small, editorial revisions such as re-wording for clarity and and correcting types will not be considered a modification)
Merging of one or more components together (merging will only take place within a component type)
Splitting one component into two or more components (splitting will only take place within a component type)
Retiring the component from the statement
Release of a new version of the statement will be disseminated to the community through postings on the TRIPOD-LLM website, the main TRIPOD website (https://www.tripod-statement.org/), the EQUATOR Network website (https://www.equator-network.org/reporting-guidelines/), and postings on social media accounts such as X (formerly known as Twitter) @TRIPODStatement. Emails will be sent to journal editors to inform them of the update and ensure that author instructions refer to the most current versioning. Users are requested to cite the version of the statement used in their research.
At each review meeting, the membership of the expert panel will be reviewed for expertise, diversity, and representation, and new members will be solicited if and when gaps are identified. Expert panel members will also have the authority to trigger an ad hoc review of the guidelines to accommodate major, unexpected changes in the field that warrant more urgent discussion.
Discussion
TRIPOD-LLM has been developed to guide researchers, journals, healthcare professionals, LLM developers (commercially and non-commercially), and healthcare institutions in the rapidly evolving field of biomedical and healthcare LLMs. It represents minimum reporting recommendations for studies describing LLMs’ development, tuning, or evaluation. Reporting TRIPOD-LLM items will help users understand and appraise the quality of LLM study methods, increase transparency around study findings, reduce overinterpretation of study findings, facilitate replication and reproducibility, and aid in implementing the LLM.
Transparency throughout the model lifecycle has been strongly emphasized in the guidelines. Detailed documentation is emphasized at each stage of an LLM’s life cycle;37 for example, during the development and fine-tuning phases, there is an emphasis on disclosing the origins and processing of training data. Moreover, the LLM version and specifics of any fine-tuning or alignment modeling processes on top of existing foundation models must be transparently reported to enable fair comparisons of LLMs. This includes specifying the cut-off dates for when training data were collected to clarify the temporal relevance of training datasets and potential for data leakage or contamination during evaluation. In addition, studies should document the model version date and whether the model was frozen or remained dynamic during the data collection phase . Transparency regarding input data is essential because LLMs are typically trained on multiple public large-scale datasets — and thus inherently risk incorporating existing societal biases and inequities in the form of stigmatizing language as well as statistical risk allocation in disparate groups, necessitating a comprehensive and transparent approach to curating data sources and understanding potential biases.21,22,38–41
Human insight and oversight are critical components of the TRIPOD-LLM statement, reflecting an emphasis on components eventually critical for the responsible deployment of LLMs (though deployment reliability and observability are outside the scope of this paper).42–44 The guidelines include requirements for increased reporting of the expected deployment context and specifying the levels of autonomy assigned to the LLM, if applicable. Furthermore, there is a focus on the quality control processes employed in dataset development and evaluation, such as qualifications of human assessors, requirement for dual annotation, and specific details on instructions provided to assessors to ensure that nuances of text evaluation are captured, thus facilitating reliable assessments of safety and performance.
Prompting and task-specific performance are key additions necessitated by the unique characteristics of LLMs. The variability in prompt engineering approaches can have a considerable influence on LLM performance, potentially skewing benchmark comparisons and real-world applicability.45,46 Where relevant, reports must include comprehensive descriptions of data sources used for developing prompts, LLM model name and versions, any preprocessing steps undertaken, and methods employed in prompt engineering. This ensures that prompts are effectively designed to elicit stable and reproducible performance from LLMs. Additionally, the guidelines call for clear reporting on evaluation settings, including instructions and interfaces used and characteristics of populations involved in evaluations. This is intended to emphasize whether LLM performance is assessed under conditions that closely mimic real-world applications, providing a reliable measure of its practical utility.
We anticipate that key users and beneficiaries of TRIPOD-LLM will be (1) academic and industry researchers authoring papers, (2) journal editors and peer reviewers evaluating research papers, and (3) other stakeholders (e.g., the research community in general, academic institutions, policy-makers, funders, regulators, patients, study participants, industry, and the broader public) who will benefit from increased transparency and quality of LLM research. We encourage editors, publishers, and the industry more broadly to support adherence to TRIPOD-LLM by referring to a link within the journal’s instructions to authors, enforcing its use during the submission and peer review process, and making adherence to the recommendations an expectation. We also encourage funders to require applications for LLM studies to include a plan to report their model according to the TRIPOD-LLM recommendations, thereby minimizing research waste and ensuring value for money.
Of note, this guideline was developed with text-only LLMs in mind; however, advances in multi-modal models incorporating LLMs, such as vision-language models,47 are now rapidly emerging - illustrating the need for rapid, nimble approaches for reporting guidelines. Many of the reporting considerations will be shared between text-only LLMs and these multi-modal models. For example, for vision-language models, both text and image preprocessing should be reported. However, unique considerations may arise that merit discussion in future versioning of TRIPOD-LLM or related guidelines. For example, studies reporting the development of LLMs that use imaging data should report details of image acquisition. In the interim, we suggest that studies reporting the development and/or evaluation of a method that includes an LLM as a primary component use the TRIPOD-LLM statement, although we acknowledge this may be subject to interpretation. We advise that users keep in mind the goals of reproducibility, understandability, and transparency to take a common-sense approach to deciding on the appropriate reporting guideline, and to interpreting the relevant components of TRIPOD-LLM statement items to report multimodal LLMs. Users may also refer to methodological guides from multiple AI fields, such as radiomics,48,49 to inform their reporting.
Assurance labs such as the Coalition for Health AI (CHAI)50 and Epic AI Labs,51,52 or internal validation standards are expected to be of importance in the generation, verification, certification, and maintenance of model cards for clinical AI. It is our opinion that the TRIPOD-LLM standard can and should inform approaches by these labs to assure LLMs in ways that meet the required regulatory bar for AI (e.g., the Biden-Harris Administration “Executive Order on Safe, Secure, and Trustworthy Artificial Intelligence”, the United States AI Safety Institute,53 the United States Office of the National Coordinator for Health Information Technology HTI-1 Final Rule,54 and the European Union AI Act55) and build confidence in patients, clinician, and other stakeholders about the utility and trustworthiness of clinical AI.
It must also be emphasized that LLM evaluation and validation requires specialized expertise and resources. To ensure equitable and safe deployment, investments into LLM development should be balanced by investments into infrastructure that enables robust validation beyond large academic settings. Moreover, this checklist should be seen as part of a continuous process for evaluating LLMs due to the temporal and geographic specific contexts these models inherit, which can impact the generalisability of performance and fairness across sites or at the same site over time. These shifts can be even more unpredictable than traditional ML models due to their user-dependent nature, and thus, considerable effort must be placed on understanding trends and heterogeneity of effects instead of single-point estimates that proclaim universal validation.
Limitations of the current TRIPOD-LLM checklist arise from the nearly unprecedented speed of development and publishing in this space, necessitating rapid guideline development to provide guidance to the research community. We carried out an expedited Delphi process to arrive at the initial version of the checklist included here, but acknowledge that this may introduce limitations in consensus and breadth of input. We have therefore implemented the living statement approach to enable more nimble incorporation of feedback and adaptation to rapidly evolving methods. The living statement necessarily means that the checklist included in this manuscript will likely become outdated; users are directed to https://tripod-llm.vercel.app/, which will be kept up-to-date with the most current versioning.
Conclusion
TRIPOD-LLM aims to assist authors in the complete reporting of their study and help LLM developers, researchers, peer reviewers, editors, policymakers, end-users (e.g., healthcare professionals), and patients understand data, methods, findings, and conclusions of LLM-driven research. Adhering to the TRIPOD-LLM reporting recommendations may promote the best and most efficient use of research time, effort, and money, enhancing the value of LLM research to maximize positive impact.
Methods
The TRIPOD-LLM statement was formulated to guide the reporting of studies that develop, tune, or evaluate LLMs for any healthcare application or context and was crafted following pathways utilized in creating the other TRIPOD statements. An expedited Delphi process was implemented given the need for timely reporting guidelines in this field, and was combined with a living statement approach. An accompanying glossary (Box 2) defines essential terms relevant to the TRIPOD-LLM statement.
Box 2: Glossary of terms (in alphabetical order).
- Attention Mechanism
A component in neural networks that allows the model to focus on different parts of the input when producing each part of the output, crucial for handling long-range dependencies in sequence data.
- Chain-of-Thought Prompting
A prompting technique that encourages the model to break down complex reasoning tasks into step-by-step thought processes, often improving performance on logical and mathematical tasks.
- Confabulation
Alternative term to hallucination, highlighting the production of false information that is not intended to deceive.
- Data Leakage
The use of test data during model training and/or fine-tuning, resulting in a model that will perform at a greater level than if it had been fully tested on unseen data.
- Decoder
A component of a model that converts vectorized input data and converts it back into a text sequence.
- Autoregressive model
A type of Transformer-based model that models predict the next component in a sequence, for example the next word in a sentence, based on the preceding sequence. Current state-of-the-art LLMs, including generative pre-trained transformers, are autoregressive models.
- Embedding
A representation of text in a high-dimensional vector space where similar words have similar representation, capturing semantic meaning. See also Vector.
- Encoder
A component of a model that processes the input data, transforming it into a vectorized format or representation that the model can understand.
- Encoder-Decoder
A model architecture framework combining an encoder and decoder to transform input data into an output.
- Few-Shot Learning
A method where the model learns to perform a task proficiently with a very small number of exemplars. In some cases, the number of exemplars are specified in place of “few”, e.g. (one-shot learning).
- Fine-tuning
A process where a pre-trained model is further trained (or fine-tuned) on a smaller, domain-specific dataset to specialize its knowledge for specific tasks.
- Generative Pre-trained Transformer (GPT)
A family of autoregressive Transformer-based models for natural language understanding and generation. These models are pre-trained to predict the next word in a sentence.
- Hallucination
A phenomenon where a language model generates text that is unrelated or loosely related to the input data, often manifesting as fabrications or inaccuracies.
- In Context Learning
The ability of a model to learn a new task from examples provided within this prompt at inference time.
- Instruction Tuning
A fine-tuning approach where models are trained on datasets consisting of natural language instructions and their corresponding desired outputs, improving the model’s ability to follow diverse instructions.
- Prompt
The query or instruction that is input into an LLM to elicit a response.
- Reinforcement learning with human feedback
A machine learning technique commonly used in LLM development that trains a model to optimize its output according to humans’ preferences by providing rewards in response to actions.
- Prompt engineering
A process that guides the models to generate desired outputs. Examples of prompt engineering include prompt iterations, prompting with examples, and chain-of-thought prompting.
- Retrieval-Augmented Generation (RAG)
A technique that combines information retrieval from an external knowledge base with text generation, allowing LLMs to access and incorporate up-to-date or domain-specific information.
- Temperature
A parameter that controls the randomness of predictions by scaling the logits before applying softmax, affecting the diversity of generated text.
- Tokenization
The process of converting text into smaller units, such as words or phrases, to facilitate their processing in NLP tasks.
- Transformer
A commonly used neural network architecture that has significantly advanced the field of NLP. Unlike its predecessors, the transformer relies on self-attention mechanisms to process sequences of data in parallel, improving efficiency and the ability to capture complex dependencies within the text.
- Zero-Shot Learning
The ability of a model to correctly perform tasks it has never explicitly been trained to do, based on its understanding and generalization capabilities.
Note: The definitions and descriptions given relate to the specific context of TRIPOD-LLM and the use of the terms in the guideline. They are not necessarily applicable to other areas of research.
A steering group, including DSB, JG, LAC, GSC, and KGMM, was established to direct the guideline development process. They were joined by an expert panel, including SC, CF, DR, GS, TM, DDF, RU, LHH, YA, JWG, LGM, NM, and RD, and were chosen based on their diverse expertise and experience in natural language processing, artificial intelligence, and medical informatics. This guideline was registered on May 2, 2024, with the EQUATOR Network as a reporting guideline under development (www.equator-network.org).
Ethics
This study received an exemption from the MIT COUHES IRB review board on March 26, 2024 (Exempt ID: E-5705). Delphi survey participants provided electronic informed consent before completing the survey.
Candidate item list generation
The TRIPOD-2015 and TRIPOD+AI guidelines (www.tripod-statement.org) and associated literature on reporting guidelines for LLMs were used to inform the initial candidate item list drafted by DSB and JG.9,11,28,32 The steering group and expert panel expanded this list through additional literature review, ultimately standardizing it to 64 unique items across the following sections: title, abstract, introduction, methods, results, discussion, and others.
Panelist recruitment
Delphi participants were identified by the steering committee from authors of relevant publications and through personal recommendations, including experts recommended by other Delphi participants. The steering group identified participants representing geographical and disciplinary diversity, including key stakeholder groups, e.g., researchers (statisticians, data scientists, epidemiologists, machine learners, clinicians, and ethicists), healthcare professionals, journal editors, funders, policymakers, healthcare regulators, and patient advocate groups. No minimum sample size was placed on the number of participants. A steering group member checked the expertise or experience of each identified person. Participants were then invited to complete a survey via e-mail. Delphi participants did not receive any financial incentive or gift to participate.
Delphi process
The survey was designed to allow individual responses, in English and delivered electronically using Google Forms. All responses were anonymous; no emails or identifying information was collected from participants. Participants were asked to rate each item as ‘can be omitted,’ ‘possibly include,’ ‘desirable for inclusion,’ or ‘essential for inclusion’ as has been conducted in previous TRIPOD guidelines.9 Participants were also invited to comment on any item and suggest new items. DSB and JG collated and analyzed the free text responses then used the themes generated to inform item rephrasing, merging, or suggesting new items. All steering group members were invited to participate in the Delphi surveys.
Round 1 participants
The first round was conducted between 01 March 2024 and 23 April 2024, where the participation link was sent to 56 people. Of the 56 invited, 26 completed the survey. Survey participants came from 9 countries, of which 14 were from North America, five from Europe, two from Asia, one from South America, and one from Australasia. Three participants did not provide this information. Participants reported their primary fields of work and could select more than one field. 20/26 (77 %) had a primary field in AI, Machine Learning, Clinical Informatics, or NLP, and 14/26 selected healthcare.
Consensus Meeting
An online consensus meeting was held on the 22nd and 24th of April, chaired by DSB and JG via Zoom. All steering committee and expert panel members were invited to attend. Recordings and notes were sent immediately after the meetings to enable asynchronous contribution for those who did not attend. The responses to each question were examined in turn, as well as all free-text comments. Items with support from <50% of participants for being ‘Essential to Include’ were highlighted and deliberated for the importance of inclusion. Agreement by consensus was reached in all cases, without needing a third party. To arrive at a consensus, the item was discussed until no panel member had additional comments or disagreements with the final disposition of the item. Transcripts of the discussion, which have been de-identified and any sensitive information removed, are available in supplementary materials for full transparency.
Due to the vast number of applications being developed using LLMs, a modular approach was used to group included items under additional subcategories under the ‘Research Design’ and ‘LLM Task’ headers. This approach was agreed upon during the meeting, and the steering committee approved the final groupings.
To adequately address the variety of studies and uses regarding LLMs, ranging across stages of development, tuning, evaluation, and implementation, as well as across healthcare tasks, items are categorized according to (1) research design and (2) LLM task (Figure 1). The research design categories are: de novo LLM development, LLM methods such as fine-tuning, prompt-engineering techniques and architecture modifications, intrinsic LLM evaluation, and LLM evaluation in dedicated healthcare settings and tasks. The LLM task categories are lower-level text processing (e.g., part-of-speech tagging, relation extraction, named entity recognition), classification (e.g., diagnosis), long-form question-answering, conversational agent, documentation generation, summarization/simplification, machine translation, and outcome forecasting (e.g., prognosis). Items may apply to several design and task categories, and studies may include more than one design and task. Items applicable to every type and task covered in the study should be reported. Definitions and examples of each design and task category are provided in Table 1. We acknowledge that these categorizations are imperfect and overlap may exist across designs and tasks.
Figure 1. TRIPOD-LLM workflow.
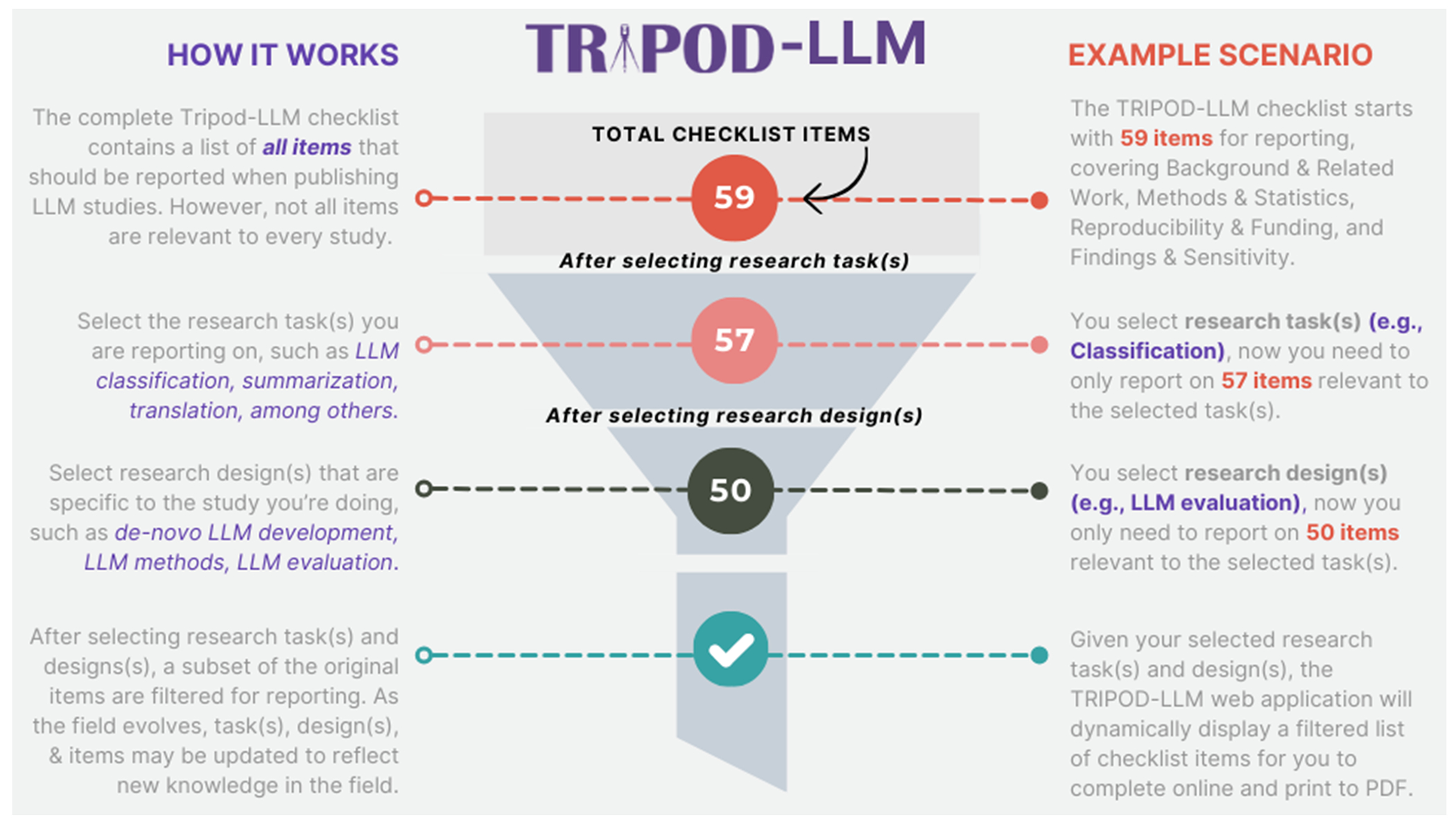
The TRIPOD-LLM checklist workflow starts with 59 reporting items and the number of required items is reduced based on the selection of research tasks (e.g., classification, summarization) and research design (e.g., LLM evaluation). After selecting both, a filtered list is generated for reporting.
Supplementary Material
Acknowledgments
JG is funded by the NIH-USA U54 TW012043-01 and NIH-USA OT2OD032701.
SC was supported by NIH-USA U54CA274516-01A1.
GSC was supported by Cancer Research UK (programme grant: C49297/A27294), and by EPSRC grant for ‘Artificial intelligence innovation to accelerate health research’ (number: EP/Y018516/1, and is a National Institute for Health and Care Research (NIHR) Senior Investigator. The views expressed in this article are those of the author and not necessarily those of the NIHR, or the Department of Health and Social Care.
Yin Aphinyanaphongs was partially supported by NIH 3UL1TR001445-05 and National Science Foundation award #1928614 & #2129076.
LAC is funded by NIH-USA U54 TW012043-01, NIH-USA OT2OD032701, and NIH-USA R01EB017205.
DSB was supported by NIH-USA U54CA274516-01A1 and NIH-USA R01CA294033-01.
Dr. Gichoya is a 2022 Robert Wood Johnson Foundation Harold Amos Medical Faculty Development Program and declares support from RSNA Health Disparities grant (#EIHD2204), Lacuna Fund (#67), Gordon and Betty Moore Foundation, NIH (NIBIB) MIDRC grant under contracts 75N92020C00008 and 75N92020C00021, and NHLBI Award Number R01HL167811.
COI
DSB: Editorial, unrelated to this work: Associate Editor of Radiation Oncology, HemOnc.org (no financial compensation); Research funding, unrelated to this work: American Association for Cancer Research; Advisory and consulting, unrelated to this work: MercurialAI. DDF: Editorial, unrelated to this work: Associate Editor of JAMIA, Editorial Board of Scientific Data, Nature; Funding, unrelated to this work: the intramural research program at the U.S. National Library of Medicine, National Institutes of Health. JWG: Editorial, unrelated to this work: Editorial Board of Radiology: Artificial Intelligence, British Journal of Radiology AI journal and NEJM AI. All other authors declare no conflicts of interest.
References
- 1.Chen Z et al. MEDITRON-70B: Scaling Medical Pretraining for Large Language Models. Preprint at 10.48550/arXiv.2311.16079 (2023). [DOI] [Google Scholar]
- 2.OpenAI. GPT-4 Technical Report. Preprint at 10.48550/arXiv.2303.08774 (2023). [DOI] [Google Scholar]
- 3.Singhal K et al. Large language models encode clinical knowledge. Nature 620, 172–180 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tai-Seale M et al. AI-Generated Draft Replies Integrated Into Health Records and Physicians’ Electronic Communication. JAMA Netw. Open 7, e246565 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tierney AA et al. Ambient Artificial Intelligence Scribes to Alleviate the Burden of Clinical Documentation. NEJM Catal. 5, CAT.23.0404 (2024). [Google Scholar]
- 6.Jiang LY et al. Health system-scale language models are all-purpose prediction engines. Nature 619, 357–362 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Regulating advanced artificial agents | Science. https://www.science.org/doi/10.1126/science.adl0625. [DOI] [PubMed] [Google Scholar]
- 8.Meskó B & Topol EJ The imperative for regulatory oversight of large language models (or generative AI) in healthcare. Npj Digit. Med 6, 1–6 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins GS, Reitsma JB, Altman DG & Moons KGM Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ 350, g7594 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Reporting guidelines | EQUATOR Network. https://www.equator-network.org/reporting-guidelines/. [Google Scholar]
- 11.Collins GS et al. TRIPOD+AI statement: updated guidance for reporting clinical prediction models that use regression or machine learning methods. The BMJ 385, e078378 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X, Cruz Rivera S, Moher D, Calvert MJ & Denniston AK Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. Nat. Med 26, 1364–1374 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vasey B et al. Reporting guideline for the early-stage clinical evaluation of decision support systems driven by artificial intelligence: DECIDE-AI. Nat. Med 28, 924–933 (2022). [DOI] [PubMed] [Google Scholar]
- 14.Norgeot B et al. Minimum information about clinical artificial intelligence modeling: the MI-CLAIM checklist. Nat. Med 26, 1320–1324 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agrawal M, Hegselmann S, Lang H, Kim Y & Sontag D Large Language Models are Few-Shot Clinical Information Extractors. Proceedings of the 2022 Conference on Empirical Methods in Natural Language Processing, pages 1998–2022, Abu Dhabi, United Arab Emirates. Association for Computational Linguistics; (2022). [Google Scholar]
- 16.Liu X et al. Large Language Models are Few-Shot Health Learners. Preprint at 10.48550/arXiv.2305.15525 (2023). [DOI] [Google Scholar]
- 17.Salazar J, Liang D, Nguyen TQ & Kirchhoff K Masked Language Model Scoring. in Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics 2699–2712 (2020). doi: 10.18653/v1/2020.acl-main.240. [DOI] [Google Scholar]
- 18.Wang A et al. GLUE: A Multi-Task Benchmark and Analysis Platform for Natural Language Understanding. Proceedings of the 2018 EMNLP Workshop BlackboxNLP: Analyzing and Interpreting Neural Networks for NLP, pages 353–355, Brussels, Belgium. Association for Computational Linguistics; (2019). [Google Scholar]
- 19.Papineni K, Roukos S, Ward T & Zhu W-J Bleu: a Method for Automatic Evaluation of Machine Translation. in Proceedings of the 40th Annual Meeting of the Association for Computational Linguistics (eds. Isabelle P, Charniak E & Lin D) 311–318 (Association for Computational Linguistics, Philadelphia, Pennsylvania, USA, 2002). doi: 10.3115/1073083.1073135. [DOI] [Google Scholar]
- 20.Goodman KE, Yi PH & Morgan DJ AI-Generated Clinical Summaries Require More Than Accuracy. JAMA (2024) doi: 10.1001/jama.2024.0555. [DOI] [PubMed] [Google Scholar]
- 21.Zack T et al. Assessing the potential of GPT-4 to perpetuate racial and gender biases in health care: a model evaluation study. Lancet Digit. Health 6, e12–e22 (2024). [DOI] [PubMed] [Google Scholar]
- 22.Gallifant J et al. Peer review of GPT-4 technical report and systems card. PLOS Digit. Health 3, 1–15 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wornow M et al. The shaky foundations of large language models and foundation models for electronic health records. Npj Digit. Med. 6, 1–10 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S et al. The effect of using a large language model to respond to patient messages. Lancet Digit. Health 0, (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang CT et al. Red Teaming Large Language Models in Medicine: Real-World Insights on Model Behavior. 2024.04.05.24305411 Preprint at 10.1101/2024.04.05.24305411 (2024). [DOI] [Google Scholar]
- 26.Gallifant J et al. Language Models are Surprisingly Fragile to Drug Names in Biomedical Benchmarks. Findings of the Association for Computational Linguistics: EMNLP 2024, pages 12448–12465, Miami, Florida, USA. Association for Computational Linguistics; (2024). [Google Scholar]
- 27.Boyd E Microsoft and Epic expand AI collaboration to accelerate generative AI’s impact in healthcare, addressing the industry’s most pressing needs. The Official Microsoft Blog https://blogs.microsoft.com/blog/2023/08/22/microsoft-and-epic-expand-ai-collaboration-to-accelerate-generative-ais-impact-in-healthcare-addressing-the-industrys-most-pressing-needs/ (2023). [Google Scholar]
- 28.Moreno AC & Bitterman DS Toward Clinical-Grade Evaluation of Large Language Models. Int. J. Radiat. Oncol. Biol. Phys 118, 916–920 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spann M Welch Medical Library Guides: Evidence Based Medicine: Evidence Grading & Reporting. https://browse.welch.jhmi.edu/EBM/EBM_EvidenceGrading. [Google Scholar]
- 30.Guyatt GH et al. What is “quality of evidence” and why is it important to clinicians? BMJ 336, 995–998 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cacciamani GE, Collins GS & Gill IS ChatGPT: standard reporting guidelines for responsible use. Nature 618, 238–238 (2023). [DOI] [PubMed] [Google Scholar]
- 32.Huo B et al. Reporting standards for the use of large language model-linked chatbots for health advice. Nat. Med 29, 2988–2988 (2023). [DOI] [PubMed] [Google Scholar]
- 33.El Mikati IK et al. A Framework for the Development of Living Practice Guidelines in Health Care. Ann. Intern. Med 175, 1154–1160 (2022). [DOI] [PubMed] [Google Scholar]
- 34.Living systematic reviews | Cochrane Community. https://community.cochrane.org/review-development/resources/living-systematic-reviews. [Google Scholar]
- 35.Akl EA et al. Living systematic reviews: 4. Living guideline recommendations. J. Clin. Epidemiol 91, 47–53 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Fraile Navarro D et al. Methods for living guidelines: early guidance based on practical experience. Paper 5: decisions on methods for evidence synthesis and recommendation development for living guidelines. J. Clin. Epidemiol 155, 118–128 (2023). [DOI] [PubMed] [Google Scholar]
- 37.Bedoya AD et al. A framework for the oversight and local deployment of safe and high-quality prediction models. J. Am. Med. Inform. Assoc. JAMIA 29, 1631–1636 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Omiye JA, Gui H, Rezaei SJ, Zou J & Daneshjou R Large Language Models in Medicine: The Potentials and Pitfalls. Ann. Intern. Med (2024) doi: 10.7326/M23-2772. [DOI] [PubMed] [Google Scholar]
- 39.Chen S et al. Cross-Care: Assessing the Healthcare Implications of Pre-training Data on Language Model Bias. Preprint at 10.48550/arXiv.2405.05506 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansen LH et al. Seeds of Stereotypes: A Large-Scale Textual Analysis of Race and Gender Associations with Diseases in Online Sources. Preprint at 10.48550/arXiv.2405.05049 (2024). [DOI] [Google Scholar]
- 41.Biderman S et al. Pythia: A Suite for Analyzing Large Language Models Across Training and Scaling. Proceedings of the 40th International Conference on Machine Learning, 102, 34, Honolulu, Hawaii, USA, ICML (2023). [Google Scholar]
- 42.Bowman SR et al. Measuring Progress on Scalable Oversight for Large Language Models. (2022) doi: 10.48550/ARXIV.2211.03540. [DOI] [Google Scholar]
- 43.McAleese N et al. LLM Critics Help Catch LLM Bugs. Preprint at 10.48550/arXiv.2407.00215 (2024). [DOI] [Google Scholar]
- 44.Burns C et al. Weak-to-Strong Generalization: Eliciting Strong Capabilities With Weak Supervision. Preprint at 10.48550/arXiv.2312.09390 (2023). [DOI] [Google Scholar]
- 45.Chen S et al. Evaluating the ChatGPT family of models for biomedical reasoning and classification. J. Am. Med. Inform. Assoc ocad256(2024) doi: 10.1093/jamia/ocad256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen S et al. Use of Artificial Intelligence Chatbots for Cancer Treatment Information. JAMA Oncol. 9, 1459–1462 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu MY et al. A visual-language foundation model for computational pathology. Nat. Med 30, 863–874 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kocak B et al. METhodological RadiomICs Score (METRICS): a quality scoring tool for radiomics research endorsed by EuSoMII. Insights Imaging 15, 8 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lambin P et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol 14, 749–762 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Shah NH et al. A Nationwide Network of Health AI Assurance Laboratories. JAMA 331, 245–249 (2024). [DOI] [PubMed] [Google Scholar]
- 51.Diaz Naomi. Epic releases AI validation suite. Becker’s Hospital Review, May 23, 2024. Available at: https://www.beckershospitalreview.com/ehrs/epic-releases-ai-validation-suite.html. [Google Scholar]
- 52.Epic Systems Corporation. Seismometer: AI model evaluation with a focus on healthcare. Epic Open Source, Available at: https://github.com/epic-open-source/seismometer (2024). [Google Scholar]
- 53.U.S. Artificial Intelligence Safety Institute. National Institute of Standards and Technology (NIST), Available at: https://www.nist.gov/aisi (2023).. [Google Scholar]
- 54.Health Data, Technology, and Interoperability: Certification Program Updates, Algorithm Transparency, and Information Sharing. Federal Register https://www.federalregister.gov/documents/2024/01/09/2023-28857/health-data-technology-and-interoperability-certification-program-updates-algorithm-transparency-and (2024). [Google Scholar]
- 55.EU Artificial Intelligence Act. Up-to-date developments and analyses of the EU AI Act. Artificial Intelligence Act Explorer. Future of Life Institute, Available at: https://artificialintelligenceact.eu/ (2024). [Google Scholar]
- 56.Peng C et al. A study of generative large language model for medical research and healthcare. Npj Digit. Med 6, 1–10 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zakka C et al. Almanac — Retrieval-Augmented Language Models for Clinical Medicine. NEJM AI 1, AIoa2300068 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Keloth VK et al. Advancing entity recognition in biomedicine via instruction tuning of large language models. Bioinformatics 40, btae163 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guevara M et al. Large language models to identify social determinants of health in electronic health records. Npj Digit. Med 7, 1–14 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jin Q, Yang Y, Chen Q, & Lu Z (2024). GeneGPT: augmenting large language models with domain tools for improved access to biomedical information. Bioinformatics, 40(2), btae075. 10.1093/bioinformatics/btae075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goh E et al. Large Language Model Influence on Diagnostic Reasoning: A Randomized Clinical Trial. JAMA Netw. Open 7, e2440969 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zaretsky J et al. Generative Artificial Intelligence to Transform Inpatient Discharge Summaries to Patient-Friendly Language and Format. JAMA Netw. Open 7, e240357 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Han L, Erofeev G, Sorokina I, Gladkoff S & Nenadic G Examining Large Pre-Trained Language Models for Machine Translation: What You Don’t Know About It. Preprint at 10.48550/arXiv.2209.07417 (2022). [DOI] [Google Scholar]
- 64.Yoon W et al. LCD Benchmark: Long Clinical Document Benchmark on Mortality Prediction. 2024.03.26.24304920 Preprint at 10.1101/2024.03.26.24304920 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


