Abstract
Menopause not only affects fertility but also has widespread impact on systemic health. Yet, the molecular mechanisms underlying this process are not fully understood, partly due to the absence of robust, age-relevant preclinical models with comprehensive molecular and phenotypic characterization. To address this, we systematically compared three candidate mouse models of menopause: (1) intact aging, (2) chemical ovarian follicle depletion using 4-vinylcyclohexene diepoxide (VCD) administered at multiple ages, and (3) Foxl2 haploinsufficiency, a genetic model based on a transcription factor linked to human premature ovarian failure. Through histology, serum hormone profiling, single-cell transcriptomics and machine-learning approaches, we uncovered both shared and model-specific features of follicle loss, endocrine disruption, and transcriptional remodeling. The VCD and Foxl2 haploinsufficiency models revealed distinct patterns of hormonal and immune alterations not captured by intact aging alone. This comparative framework enables informed selection of context-appropriate preclinical rodent models to study menopause and the broader physiological consequences of ovarian aging.
Keywords: Menopause, ovarian aging, preclinical models, single-cell RNAseq, ovarian age clock
Introduction
Women are born with a finite ovarian reserve, and its progressive depletion with age leads to the irreversible cessation of reproductive function, known as menopause1–3. Epidemiological studies have shown that later age-at-menopause is associated with increased lifespan and reduced risk of age-related diseases, including cardiovascular disease, osteoporosis, and neurodegeneration4–6. Conversely, the onset of menopause is accompanied by a sharp rise in the incidence of age-associated pathologies, underscoring the widespread physiological consequences of ovarian aging7. Despite its clinical relevance, the molecular mechanisms linking ovarian failure to systemic aging remain poorly understood.
Menopause marks the end of female fertility and is characterized by significant shifts in the hormonal milieu, which exert systemic effects8. As the ovarian follicular pool becomes depleted, declining steroidogenic activity disrupts feedback regulation of the hypothalamic–pituitary–gonadal (HPG) axis, leading to pronounced changes in reproductive hormones8. For example, circulating levels of estrogen and anti-Müllerian hormone (AMH) decline to near-undetectable levels, while follicle-stimulating hormone (FSH) and Inhibin A (INHBA) exhibit marked elevations9–11. This hormonal reprogramming impacts not only reproductive tissues but also multiple extragonadal systems, including the metabolic, skeletal, and central nervous systems12,13. Accurately modeling this complex endocrine transition in preclinical systems is essential for delineating the mechanisms through which ovarian dysfunction impacts systemic aging.
Currently available rodent models for menopause research each possess distinct limitations. Intact aging female mice remain the most widely used model due to their physiological relevance and ease of implementation. Moreover, decades of research across diverse systems have established a robust foundational understanding of age-related biological changes, providing a valuable reference point for comparative analyses. Yet, intact aging mice do not undergo true menopause; instead, they enter estropause, retaining low but detectable estrogen levels and lacking the postmenopausal hormonal milieu observed in humans14,15. Another popular preclinical model is the use of surgical removal of ovaries, known as ovariectomy (OVX). While OVX achieves complete estrogen depletion, the surgery bypasses the gradual endocrine transition associated with menopause, and eliminates post-reproductive ovarian tissue, which retains androgenic activity and contributes to aging phenotypes16. Thus, despite its popularity, OVX is a poor preclinical model of menopause.
An alternative approach involves chemical ovarian follicle depletion using 4-vinylcyclohexene diepoxide (VCD), which selectively targets small pre-antral follicles (primordial and primary follicles), and induces follicle atresia, and leads to progressive ovarian failure14. This strategy allows for temporal control of ovarian depletion and results in estrus acyclicity, estrogen deficiency, and compensatory FSH elevation, features that closely mirror human menopause17,18. However, prior applications of the VCD model have been limited to young mice (~2–3 months old), restricting its relevance for studying menopause in the context of organismal aging19,20. Given that age alters the metabolic, immune, and endocrine landscapes21–23, it is critical to evaluate how the timing of ovarian failure intersects with aging processes. For example, heterochronic parabiosis experiments have demonstrated that the aging systemic milieu can impair tissue function in young animals, with circulating factors from aged animals negatively affecting neurogenesis and cognitive function24. Thus, evaluating VCD exposure in older animals may yield important insights into how age-related changes in the systemic environment shape the physiological consequences of menopause.
The genetics of premature menopause in humans may also help expand the toolkit to study menopause in preclinical models25,26. Specifically, haploinsufficiency of Foxl2, a transcription factor essential for granulosa cell identity and ovarian function25–28, is a great candidate model for menopause in mice. Indeed, heterozygous FOXL2 mutations are causally linked to premature ovarian insufficiency in humans29–31. Although most research efforts have focused on full Foxl2 knockout mice25,26, which show high perinatal lethality and primary ovarian defects, previous studies did report anecdotal subfertility phenotypes of heterozygous carriers. Despite these promising reports, the physiological consequences of Foxl2 haploinsufficiency on ovarian health and function have not been characterized in mice in vivo. Unlike surgical or chemically induced models, Foxl2 haploinsufficiency enables the study of endogenous, genetically encoded ovarian dysfunction and its downstream systemic effects. This model offers a unique opportunity to interrogate gradual follicular depletion and endocrine dysregulation in the absence of exogenous perturbation (surgical or chemical insult).
In this study, we characterized and systematically compared three complementary mouse models that reflect distinct mechanisms of ovarian decline as candidate models of menopause: (1) the intact aging model, (2) a VCD-induced follicle depletion initiated at different ages spanning young adulthood to early middle-age, and (3) a genetic model of Foxl2 haploinsufficiency. Using these models, we sought to identify shared and model-specific features of ovarian functional decline across multiple biological scales and phenotypic layers. We integrated histological analyses, serum hormone profiling, and single-cell RNA-sequencing (scRNA-seq) to assess changes in ovarian structure, endocrine function, and transcriptional landscape across menopause models. Additionally, we developed hormone-based and transcriptome-based ovarian aging clocks to evaluate functional trajectories of reproductive aging. Together, our data provides a comprehensive framework for systematically evaluating and comparing physiologically relevant models of ovarian aging, thereby defining their distinct advantages and applications for studying systemic and molecular effects of menopause.
Results
Selection of mouse models of menopause for the study
To systematically assess and compare physiologically relevant menopause models, we characterized three rodent models that represent distinct mechanisms of ovarian dysfunction: (1) the intact aging model (“Aging model”), (2) a chemically-induced model using repeated intraperitoneal injections of 4-vinylcyclohexene diepoxide (VCD, “VCD model”)14,32, and (3) a genetic model of Foxl2 haploinsufficiency (“Foxl2 +/− model”)27. Our aim was to establish a comparative framework to evaluate these models in terms of histological, hormonal, and integrated ovarian health outcomes to evaluate their suitability for modeling key aspects of ovarian aging (Fig. 1a). The Aging model involved comparison of young adult (4-month-old) and old (20-month-old) female C57BL/6JNia mice. We selected these timepoints to capture the transition from adult reproductive with stable estrus cycling (4 months) to post-estropausal age (20 months), as C57BL/6 females typically remain cyclic until ~12 months and show pronounced ovarian aging phenotypes by 20 months33. The VCD model included C57BL/6J female mice injected with vehicle control (safflower oil, hereafter referred to as “CTL”) or VCD starting at 3, 6, 8, or 10 months of age. This scheme allowed us to examine how the timing of ovarian insult (potentially modeling varying “age-at-menopause”) can impact ovarian health outcomes, particularly when VCD exposure occurs closer to natural estropause transition in mice (~12 months)33. The Foxl2 haploinsufficiency model compared Foxl2+/+ and Foxl2+/− female littermate mice from our transgenic colony at young (3–4 months) and early middle-age (8–10 months), prior to the onset of natural estropause, to test whether partial loss of Foxl2 can recapitulate age-associated ovarian changes at earlier ages than control mice.
Fig 1. Characterization of ovarian health markers of animals from Aging, VCD and Foxl2 haploinsufficiency models.
a, Schematic of the experimental design. b-d, Representative hematoxylineosin staining images of ovarian tissues from Aging, VCD, and Foxl2 haploinsufficiency model mice. For the VCD model, ovarian tissues were analyzed 5 months post-injection. e-g, Combined follicle counts (primordial, primary, secondary and antral follicles, and corpus luteum) from Aging, VCD, and Foxl2 haploinsufficiency model mice. h-j, Serum AMH level quantification from Aging, VCD, and Foxl2 haploinsufficiency model mice. k-m, Serum FSH level quantification from Aging, VCD, and Foxl2 haploinsufficiency model mice. n-p, Serum INHBA level quantification from Aging, VCD, and Foxl2 haploinsufficiency model mice. q-s, Ovarian health index from Aging, VCD, and Foxl2 haploinsufficiency model mice. For panels e-s, statistical significance was assessed using the Wilcoxon test, and p-values are reported. Open data points indicate data used for ovarian health index calculation. Scale bar: 250 μm.
For the Foxl2 haploinsufficiency model, we generated Foxl2 haploinsufficient mice using a regenerated floxed targeting allele, as previously described27, which introduces a null mutation in the Foxl2 gene via deletion of the gene single exon, using services from Cyagen Biomodels LLC (Extended Data Fig. 1a; see Methods). Unlike prior studies that employed this allele for temporally controlled, conditional deletion in adulthood, our study used this strategy to achieve constitutive Foxl2 haploinsufficiency throughout life (see Methods). To confirm that reduced Foxl2 expression was indeed observed in ovarian tissue from Foxl2+/− animals, as required to observe phenotypic consequences of haploinsufficiency, we performed RT-qPCR using ovaries from young and early middle-age matched Foxl2+/+ and Foxl2+/− female mice from our colony (Extended Data Fig. 1b). While the young group showed only a non-significant trend towards decreased expression, the early middle-age Foxl2+/− ovaries showed a significant reduction in expression (p-value ~0.032; Extended Data Fig. 1b). To confirm whether Foxl2 haploinsufficiency had an impact on ovarian function in these mice, we examined fertility outcomes for Foxl2+/+ and Foxl2+/− females from our breeding colony (see Methods; Extended Data Fig. 1c,d; Extended Data Table 1). Interestingly, Foxl2+/− females showed a trend towards a smaller first litter size (p-value ~0.075; Extended Data Fig. 1c), as well as a significantly increased latency to first pregnancy compared to matched Foxl2+/+ females (p-value ~0.035; Extended Data Fig. 1d), suggesting impaired reproductive potential consistent with ovarian dysfunction or premature ovarian aging.
Histological and hormonal characterization of mouse menopause models
A decline in ovarian follicle numbers is a well-established hallmark of ovarian aging and menopause34. Thus, we performed histological analysis of ovarian follicle populations using hematoxylin and eosin (H&E) staining (Fig. 1b–g; Extended Data Fig. 2). As expected, the Aging model showed a significant reduction in follicles at all developmental stages in old females, including primordial, primary, secondary, and antral follicles, as well as corpora lutea (Fig. 1b,e; Extended Data Fig. 2a), consistent with the established progressive follicular depletion that accompanies natural ovarian aging35,36. At five months post-injections, VCD-treated animals also showed significant depletion of follicle numbers, regardless of age-at-injection (Fig. 1c,f; Extended Data Fig. 2b). Due to the terminal nature of histological analysis, ovaries were collected for H&E staining at the single 5 months post-injection time point. This endpoint was selected to align with the conclusion of our longitudinal serum profiling, wherein monthly hormone measurements enabled tracking of endocrine changes over time (see below). In contrast, Foxl2+/− mice did not show any reduction in combined follicle counts; instead, we observed a trend toward increased follicle counts in the middle-aged Foxl2+/− animals (p-value ~0.08; Fig. 1d,g; Extended Data Fig. 2c).
To further assess ovarian function, we measured serum levels of AMH, FSH and INHBA, key markers of ovarian reserve and hypothalamic–pituitary–gonadal (HPG) axis regulation37,38 (Fig. 1h–p; Extended Data Table 2). In the Aging model, old females had decreased AMH and INHBA and increased FSH compared to young females, consistent with diminished ovarian reserve38,39 (Fig. 1h,k,n). A similar pattern was observed in the VCD model, whereby AMH and INHBA were decreased and FSH was increased compared to the vehicle control groups at 5 months post-injection timepoint (Fig. 1i,l,o). To enable a comprehensive analysis of endocrine responses across reproductive life stages, we also performed longitudinal hormone profiling over a five-month period following VCD exposure at various ages (3, 6, 8 and 10 months; Extended Data Fig. 3a–c). Interestingly, VCD injections at older ages resulted in attenuated hormonal shifts compared to younger counterparts, suggesting that ovarian or systemic factors at midlife may modulate sensitivity to VCD-induced insult to ovarian reserve. In the Foxl2+/− mice, we observed distinct hormonal trends: AMH levels were increased, FSH levels were mildly elevated, and INHBA levels were reduced compared to wild-type controls, although none of these changes reached statistical significance (Fig. 1j,m,p). The increased AMH levels are consistent with our histological findings of increased follicle counts in the Foxl2+/− animals (Fig. 1d,g). Importantly, FOXL2 has been shown to interact with AMH to modulate follicle recruitment in humans40. Thus, the increased follicle counts and AMH levels in the Foxl2+/− animals likely reflect altered gene regulatory network driven by Foxl2 deficiency, rather than true ovarian rejuvenation (see Discussion).
Next, we applied our previously described composite ovarian health index, which integrates information from follicle counts and serum hormone levels to yield a holistic measure of ovarian health41 (Fig. 1q–s). As expected, this index was significantly lower in old vs. young animals and VCD-treated animals vs. CTL (regardless of age-at-injection), reflecting impaired ovarian health (Fig. 1q,r). In contrast, Foxl2+/− mice did not differ significantly from wild-type animals at either age group, consistent with their preserved histological and hormonal profiles (Fig. 1s).
Together, these results provide a comprehensive characterization of three distinct candidate mouse models of menopause. Both the Aging and VCD models exhibit robust phenotypic hallmarks of ovarian decline, including depletion of follicular reserves and disrupted endocrine profiles, with the VCD model offering unique advantages for temporally controlled induction of ovarian dysfunction. In contrast, the Foxl2 haploinsufficiency model presents an unexpected phenotype characterized by preserved or even elevated follicle counts and increased AMH levels, opposite to typical features of ovarian aging9–11, despite clear evidence of reproductive dysfunction (Extended Data Fig. 1c,d). These findings raise the possibility that Foxl2 haploinsufficiency alters ovarian physiology through mechanisms distinct from conventional aging, warranting deeper molecular investigation.
A hormone-based clock, “OvAge”, for non-terminal prediction of ovarian aging
Although the ovarian health index is a useful metric of ovarian health, its partial reliance on post-mortem histological measurements limits its applicability in longitudinal studies. Thus, we aimed at developing a complementary non-terminal metric of ovarian aging, using a hormone-based predictive machine-learning model, which we termed “OvAge”, capable of estimating ovarian age from circulating serum hormone levels (Fig. 2a). By incorporating AMH, FSH, and INHBA levels, we sought to develop a model that provides a more comprehensive and integrative assessment of endocrine function than evaluating each hormone individually. To increase the generalizability of the clock, we trained a random forest regression model using serum hormone data (AMH, FSH, and INHBA) from animals with intact ovarian function and no experimental perturbation, including animals from the Aging model, vehicle control (CTL) groups from the VCD model, Foxl2+/+ animals from the Foxl2 haploinsufficiency model, and data from Fshr+/+ animals from our previously published Fshr haploinsufficiency study42 (Fig. 2a). This hormone level data was then randomly partitioned into training (n = 190) and testing (n = 62) sets to enable robust evaluation of model performance (Fig. 2a).
Fig 2. Development and application of OvAge, a hormone-based ovarian aging clock.
a, Schematic representation of the OvAge clock development pipeline. Data from all animals from the Aging model, vehicle control (CTL) from the VCD model, Foxl2+/+ from the Foxl2 haploinsufficiency model, and Fshr+/+ from the Fshr haploinsufficiency model mice were used to train and test OvAge. b, Scatter plot comparing predicted and chronological age, reported in weeks, in the test dataset. c-d, Scatter plots comparing predicted and chronological age for VCD and Foxl2 haploinsufficiency model mice. e-g, Age acceleration analysis for VCD model animals at 30-, 90- and 150-days post-injection. h, Age acceleration analysis for Foxl2 haploinsufficiency animals. Age acceleration was measured as the difference between predicted OvAge and chronological age. Open data points indicate data from out-of-bag predictions. For panels e-h, statistical significance was assessed using the Wilcoxon test, and p-values are reported.
Model performance evaluation showed strong agreement between predicted and chronological age in the withheld test dataset (Spearman Rho ~0.621; p-value ~7.4 × 10−8), confirming the robustness of our hormone-based OvAge clock (Fig. 2b). We then applied OvAge to assess ovarian age of VCD-injected and Foxl2+/− animals (Fig. 2c,d). In the VCD model, predicted ovarian age was consistently higher than chronological age across all age-at-injection groups, suggesting that VCD exposure accelerates ovarian aging (Fig. 2c). Notably, the gap between predicted and chronological age narrowed as the age-at-injection increased, a pattern consistent with both the hormone profiles and ovarian health index (Fig. 1l,o,r). In contrast, no consistent pattern was observed in the Foxl2 haploinsufficiency model; predicted ages of Foxl2+/− animals varied and did not seem to differ substantially from their wild-type counterparts (Fig. 2d). To quantify the degree of divergence between predicted and chronological age, we computed hormonal age acceleration, defined as the difference between OvAge-predicted and true chronological age (Fig. 2e–h). For the VCD model, we analyzed animals at 30-, 90-, and 150-days post-injection (Fig. 2e–g). We observed significant increases in age acceleration in VCD-treated animals compared to controls at both 30 and 90-days post-injection across all age-at-injection groups (Fig. 2e,f). Interestingly, by 150 days post-injection, animals injected at 10 months of age (now aged ~15 months) no longer showed significant age acceleration (Fig. 2g). This may reflect convergence in ovarian aging trajectories in both vehicle control and VCD-treated animals as they approach or enter the estropausal interval (12–15 months in the C57BL/6 strain33). In contrast, animals from the Foxl2 haploinsufficiency model did not show a consistent increase in OvAge or age acceleration (Fig. 2h). Instead, middle-age Foxl2+/− animals exhibited a statistically significant reduction in age acceleration (p-value ~0.011), consistent with other observed phenotypes in this model, including preserved follicle counts and higher AMH levels (Fig. 1d,g,j). These findings may reflect a unique regulatory trajectory in this model, which could offer a complementary perspective on ovarian aging as well as premature ovarian failure.
Together, our results demonstrate that OvAge effectively captures both accelerated and attenuated ovarian aging trajectories in response to different biological perturbations. The clock recapitulates expected aging patterns in intact naturally aged animals, reveals premature ovarian aging following VCD exposure, and highlights a distinct endocrine aging profile in the Foxl2 haploinsufficiency model. These findings underscore the utility of OvAge as a non-terminal framework for assessing ovarian aging and provide insight into model-specific differences that may be leveraged to explore various facets of reproductive senescence.
Although the Foxl2 haploinsufficiency model did not exhibit overt signs of ovarian dysfunction based on the ovarian health index or OvAge (Fig. 1s and 2d,h), we reasoned that its preserved endocrine profile does not preclude underlying molecular perturbations consistent with accelerated ovarian aging. Given the established role of FOXL2 in granulosa cell identity and ovarian maintenance, its association with premature menopause in humans29,30,43, and the fertility phenotypes that we observed (Extended Data Fig. 1c,d), Foxl2 haploinsufficiency may still disrupt transcriptional programs in the ovary before functional decline becomes apparent.
Thus, we next examined the transcriptional landscape of ovaries from each model using single-cell RNA-sequencing (scRNA-seq), enabling deeper investigation of molecular aging dynamics across natural, chemical, and genetic models of menopause.
Single-cell transcriptomic profiling of ovarian cell types across menopause models
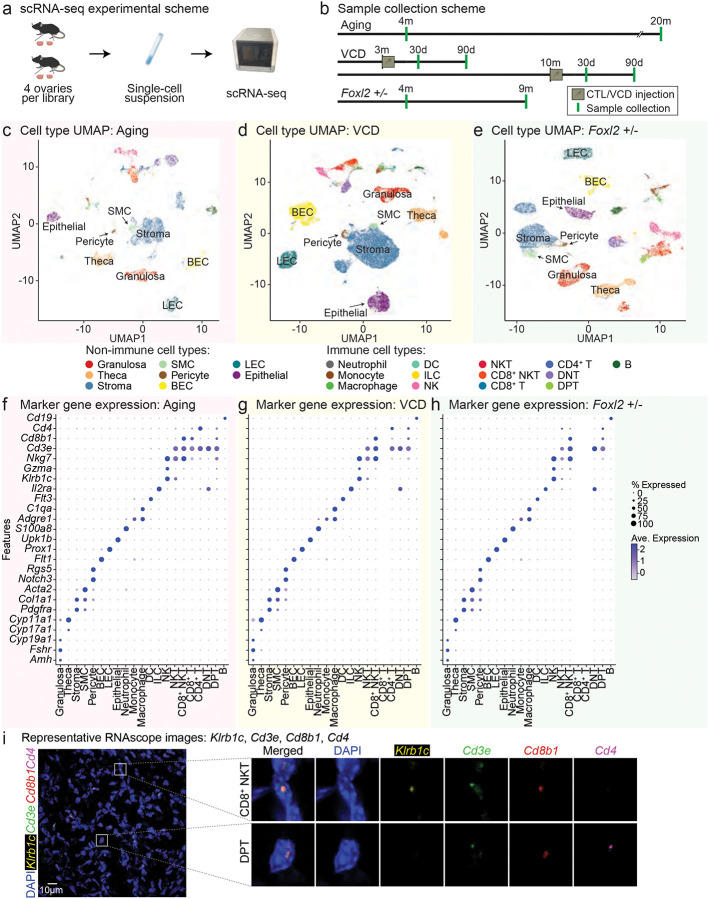
To characterize the ovarian transcriptional landscapes across candidate mouse menopause models, we performed scRNA-seq on ovaries collected from the Aging, VCD, and Foxl2 haploinsufficiency models (Fig. 3a). For the Aging model, samples were collected from young (4-month-old) and old (20-month-old) animals to capture transcriptional changes associated with intact aging (Fig. 3b). For the VCD model, we focused on two age-at-injection groups, 3 and 10 months, to capture the extremes of VCD responsiveness observed in prior analyses, as well as two post-injection time points, 30 and 90 days, based on differential effects on age acceleration at these stages (Fig. 3b). For the Foxl2 haploinsufficiency model, ovaries were collected from Foxl2+/+ and Foxl2+/− animals at young adult (~4 months) and early middle-age (~9 months), enabling the assessment of gene expression changes driven by partial loss of Foxl2 function across adulthood (Fig. 3b). The resulting datasets yielded 9,559, 47,386, and 21,301 high-quality single cells from the Aging, VCD, and Foxl2 haploinsufficiency models, respectively (Fig. 3a–e; Extended Data Table 3). Ovarian cell populations were defined through dimensionality reduction, unsupervised clustering, and annotation using both established marker genes and automated reference mapping to publicly available single-cell datasets (Fig. 3c–h; Extended Data Table 4; see Methods).
Fig 3. scRNA-seq profiling of ovaries from Aging, VCD, and Foxl2 haploinsufficiency model mice.
a, Schematic of the experimental design. b, Schematic of sample collection timepoints. c-e, UMAP plots of scRNA-seq datasets from Aging, VCD and Foxl2 haploinsufficiency models. f-h, Dotplot representation of expression of marker genes of cell types detected in scRNA-seq datasets from Aging, VCD and Foxl2 haploinsufficiency models. i, Representative RNAscope images of DAPI, Klrb1c, Cd3e, Cd8b1 and Cd4 probes. Images were enhanced for visualization. The shown image is from a middle-aged Foxl2+/+ animal.
After annotation of our scRNA-seq datasets, we were able to identify all major ovarian cell types (Fig. 3c–h). Among the abundant non-immune populations, granulosa cells (Amh, Fshr, Cyp19a1), theca cells (Cyp17a1, Cyp11a1), stromal cells (Pdgfra, Col1a1), smooth muscle cells (SMCs; Acta2), pericytes (Notch3, Rgs5), blood endothelial cells (BECs; Flt1), lymphatic endothelial cells (LECs; Prox1), and epithelial cells (Upk1b) were identified. The immune compartment included neutrophils (S100a8), monocytes (Adgre1), macrophages (C1qa), dendritic cells (Flt3), innate lymphoid cells (ILCs; Il2ra), natural killer (NK) cells (Klrb1c, Gzma, Nkg7), NKT cells (Cd3e, Klrb1c, Gzma, Nkg7), CD8+ NKT cells (Cd3e, Klrb1c, Gzma, Nkg7, Cd8b1), CD8+ T cells (Cd3e, Cd8b1), CD4+ T cells (Cd3e, Cd4), double-negative T (DNT) cells (Cd3e), double-positive T (DPT) cells (Cd3e, Cd8b1, Cd4), and B cells (Cd19) (Fig. 3f–h).
Given the known ovotoxic effects of VCD, we performed additional histological analyses on condition-matched ovaries (matched for age-at-injection and time post-injection) to confirm the presence of follicular structures in animals processed for scRNA-seq (Extended Data Fig. 4a). H&E staining of condition-matched samples revealed that despite extensive follicle depletion, residual follicles were present in most VCD-treated animals (Extended Data Fig. 4a). These findings were consistent with the detection of follicle-associated cell types in our scRNA-seq data (Fig. 3d,g). Quantification of total follicles by blinded observers further supported the persistence of follicular structures in the VCD model animals (Extended Data Fig. 4b). We also performed serum hormone quantification (AMH, FSH and INHBA) on age- and condition-matched samples corresponding to those used for scRNA-seq (Extended Data Fig. 5a–c). Consistent with previous findings, AMH and INHBA levels were reduced in the VCD groups (Extended Data Fig. 5a,c). To note, FSH levels were measured using an updated assay kit implemented by the University of Virginia Ligand core facility, distinct from the kit used in prior analyses (see Methods). Despite this change, we observed a trend toward increased FSH levels in the VCD-treated group relative to matched controls, although statistical significance was not reached, likely due to limited sample size (Extended Data Fig. 5b). Then, we evaluated the ovarian health index in these animals (Extended Data Fig. 5e). Because FSH was measured using a different assay than the one used to establish the ovarian health index, we implemented a previously reported correction procedure41 to ensure consistency and comparability of FSH values across cohorts (Extended Data Fig. 5d; see Methods). We observed a trend for lower ovarian health index scores in the VCD group compared to matched controls, although it did not reach statistical significance, again likely due to small sample size (Extended Data Fig. 4e).
To validate cell type assignments, we performed RNAscope, a high sensitivity in situ hybridization assay for both well-characterized and less frequently profiled ovarian cell populations (Fig. 3i; Extended Data Fig. 6). Granulosa cells (Fshr), theca cells (Cyp11a1), stromal cells (Pdgfra) and SMCs (Acta2) were validated using established marker probes, confirming their presence in the dataset (Extended Data Fig. 6a). Epithelial cells (Upk1b), BECs (Flt1) and LECs (Prox1) were similarly detected, supporting the robustness of annotations (Extended Data Fig. 6b). Importantly, we also validated the presence of various immune cell subsets, including NK (Klrb1c), NKT (Klrb1c, Cd3e), CD8+ T (Cd3e, Cd8b1), CD4+ T cells (Cd3e, Cd4), DNT (Cd3e, with no Cd8b1/ Cd4 signal) and B (Cd19) cells (Extended Data Fig. 6c). We also confirmed presence of less commonly reported ovarian cell types, including CD8+ NKT (Klrb1c, Cd3e, Cd8b1) and DPT (Cd3e, Cd8b1, Cd4) cells (Fig. 3i). These results further support the accuracy of the single-cell transcriptomic annotations and highlight the diverse somatic and immune microenvironments present across the aging models.
To assess cell type consistency across models, we compared annotated cell populations across datasets. A total of 19 cell types, 8 non-immune and 11 immune types, were shared across all three models (Extended Data Fig. 6d). Some cell types were not shared across models, potentially due to technical factors (e.g., capture efficiency and sequencing depth), or underlying biological variation inherent to each mouse model. To assist researchers in navigating the data efficiently, we developed an interactive R shiny app that makes the annotated datasets directly accessible and explorable through a graphical interface: https://minhooki.shinyapps.io/shinyappmulti/.
scRNA-seq analysis reveals shifts in ovarian cell composition in menopause models
Next, we examined changes in cell type proportions across our datasets (Fig. 4a; Extended Data Table 5). We first assessed whether the proportion of immune cells shifted with ovarian aging by analyzing the expression of Ptprc, which encodes Cd45, a pan-immune marker (Fig. 4b–d). In the Aging model, we observed a significant increase in Ptprc+ cells, indicating an expansion of the immune cell population with age (Fig. 4b). This is consistent with previous scRNA-seq studies in intactly aged mouse ovaries, which reported an increase in immune cell abundance at even younger ages (3 vs. 9 months; 4.5 vs. 10.5 vs. 15.5 months)44,45. In contrast, single-nucleus RNA-sequencing data from human ovaries did not identify a similar shift in immune cell proportions with age, consistent with potential species-specific differences in immune remodeling for the aging ovary46. However, this discrepancy could also reflect differences in age ranges examined or technical factors such as dissociation and capture methods inherent to single-nucleus versus single-cell approaches.
Fig 4. Characterization of cell type proportions in Aging, VCD, and Foxl2 haploinsufficiency model scRNA-seq datasets.
a, Schematic of the experimental design. b-d, Proportion differences of Ptprc− and Ptprc+ cells between young vs. old in Aging model, CTL (vehicle control) vs. VCD in VCD model, and Foxl2+/+ vs. Foxl2+/− in the Foxl2 haploinsufficiency model datasets. e-g, Flow cytometry analysis of CD45+ cell proportions in ovaries of Aging, VCD, and Foxl2 haploinsufficiency model mice. h-j, Proportion differences of non-immune cell types detected in Aging, VCD, and Foxl2 haploinsufficiency model scRNA-seq datasets. k-m, Representative RNAscope images of Fshr (granulosa) and Cyp11a1 (theca) probes from Aging, VCD, and Foxl2 haploinsufficiency model mice. Scale bar: 100μm. n-o, Cell abundance quantification data from RNAscope image analysis of Fshr+ and Cyp11a1+ cells from Aging, VCD, and Foxl2 haploinsufficiency model mice. For panels e-g and n-p, statistical significance was assessed using the Wilcoxon test, and p-values are reported.
VCD-treated mice exhibited a consistent reduction in Ptprc+ cells across all conditions, suggesting a depletion of immune cells following exposure to VCD (Fig. 4c). This reduction may result from the loss of follicular structures, alterations in immune recruitment, or uncharacterized systemic effects of VCD on immune homeostasis. Additionally, broader tissue remodeling processes, including fibrosis, may contribute to the altered immune cell landscape observed in VCD-treated ovaries. Thus, the VCD model does not seem to recapitulate immune features of intact aging ovaries in mice, regardless of age-at-injection. In the Foxl2 haploinsufficiency model, we detected a significant increase in Ptprc+ cells in the Foxl2+/− animals, regardless of age (Fig. 4d). These findings were further validated by flow cytometric analysis of CD45+ cells, using ovarian single-cell preparations generated with the same dissociation protocol as the scRNA-seq experiments (Fig. 4e–g; Extended Data Fig. 7a). Intriguingly, this suggests that the Foxl2 haploinsufficiency model may better recapitulate immune shifts in the ovary seen with intact aging compared to the VCD model.
Then, we conducted a comprehensive analysis of all ovarian cell populations detected in the datasets to identify shifts in cellular composition in response to intact aging, VCD exposure, and Foxl2 haploinsufficiency (Fig. 4h–j; Extended Data Fig. 7b–d). In the Aging model, nearly all non-immune cell populations showed a decline in proportion with age, except for stromal cells and LECs, which remained unchanged (Fig. 4h). In contrast, several immune cell populations increased with age, including neutrophils, DCs, ILCs, CD8+ NKT, CD4+ T, DNT, and DPT cells (Extended Data Fig. 7b). Notably, DNT cells have been reported to increase in aging mouse ovaries, and a potential role for these cells in regulating ovarian aging has been proposed44,47. Conversely, monocytes, NK, CD8+ T, and B cells exhibited a significant decrease with age (Extended Data Fig. 7b).
In the VCD model, cell proportion shifts were largely consistent within the treatment group, regardless of age-at-injection or time post-injection (Fig. 4i; Extended Data Fig. 7c). The VCD-treated group showed an increasing trend in stromal cells, BECs, LECs, epithelial cells, and CD4+ T cells. In contrast, granulosa cells, theca cells, SMC, and pericytes were significantly reduced in proportion (Fig. 4i). Among immune cells, neutrophils, monocytes, macrophages, DCs, NKT, DPT, and B cells were all decreased following VCD treatment (Extended Data Fig. 7c).
In the Foxl2 haploinsufficiency model, granulosa and epithelial cell proportions were consistently reduced in Foxl2+/− animals, regardless of age (Fig. 4j). Conversely, monocytes, NK, CD8+ NKT, and B cells were increased in Foxl2+/− animals (Extended Data Fig. 7d). Interestingly, we observed opposing trends in stromal cells, macrophages, and DNT cells. Stromal cell proportions increased in Foxl2+/+ young mice but decreased in Foxl2+/+ animals at early middle-age, whereas macrophages and DNT cells showed the opposite trend. These findings highlight model-specific patterns of non-immune and immune cell remodeling, underscoring distinct trajectories shaped by chronological, chemical, and genetic perturbations of ovarian function.
To validate the cell abundance findings from our scRNA-seq data, we leveraged our RNAscope in situ hybridization data for representative non-immune and immune populations (see above; Fig. 4k–p; Extended Data Fig. 8,9). We examined granulosa cells, theca cells, stromal cells, SMCs, BECs, LECs, and epithelial cells, along with NK, NKT, CD8+ NKT, CD8+ T, CD4+ T, DNT, DPT, and B cells. In the Aging model, RNAscope confirmed a decrease in granulosa and theca cell abundances with intact aging, consistent with our scRNA-seq results (Fig. 4k,n). To note, there was non-specific staining for granulosa and theca markers (which are histologically absent in the old), which did not colocalize with DAPI, consistent with known technical limitations of the assay (Fig. 4k, right panel). Stromal cell proportions remained unchanged, again in agreement with the scRNA-seq data (Extended Data Fig. 8a). While the scRNA-seq data suggested a decrease in SMCs with age, we did not detect a significant difference by RNAscope, likely due to low detection sensitivity of the Acta2 probe. While epithelial cells showed a decreasing trend with age in the RNAscope dataset without reaching statistical significance, BECs exhibited a significant decline, consistent with scRNA-seq observations. We also examined LECs and did not observe any significant age-associated changes, which aligns with findings from the scRNA-seq data. For immune cells, RNAscope detected decreased abundances of NK, NKT, and B cells, and increased abundances of CD8+ NKT, CD4+ T, DNT, and DPT cells, closely recapitulating the scRNA-seq trends (Extended Data Fig. 9a). However, we did not observe a significant change in CD8+ T cells by RNAscope, despite a decrease in the scRNA-seq dataset (Extended Data Fig. 7b,9a). This discrepancy may stem from differences in detection sensitivity or spatial distribution of CD8+ T cells that may have limited probe accessibility in situ.
In the VCD model, we focused our RNAscope analysis on samples from 3-month age-at-injection and 30 days post-injection, given the consistent trends observed across conditions (Fig. 4l,o; Extended Data Fig. 8b,9b). Among non-immune cells, we observed a significant decrease in granulosa and increase in stromal with VCD exposure, consistent with scRNA-seq data (Fig. 4l,o; Extended Data Fig. 8b). We also observed some non-specific staining that did not colocalize with DAPI in the VCD-treated ovaries, where granulosa is histologically absent (Fig. 4l, right panel). Theca cells showed a decreasing trend in proportion with VCD exposure, but did not reach statistical significance (Fig. 4l,o). For SMCs, BEC, LECs and epithelial cells, RNAscope revealed no significant change in proportions with age, which contrasts with the scRNA-seq findings (Extended Data Fig. 8b). These inconsistencies may be attributable to a numbers of technical factors: (i) reduced probe efficacy, (ii) limited marker expression or (iii) under sampling of rare cell types in unique tissue slices compared to whole dissociated ovaries, all potentially leading to under-detection of rare cell types by RNAscope. For immune populations, RNAscope detected consistent decreases in NK, NKT, CD8+ NKT, CD8+ T and DNT cells, generally aligning with findings from our scRNA-seq results (Extended Data Fig. 7c,9b). CD8+ T and B cells did not show any differences in proportion with VCD exposure (Extended Data Fig. 9b).
In the Foxl2 haploinsufficiency model, RNAscope analysis revealed a reduction in granulosa cells in Foxl2+/− animals across both age groups, recapitulating the scRNA-seq findings (Fig. 4j,m,p). We also detected a non-significant decrease in epithelial cell proportions in Foxl2+/− animals in both age groups, consistent with the trends observed in scRNA-seq data (Fig. 4j; Extended Data Fig. 8c). A significant increase in SMCs and decrease in LECs in young and middle-age Foxl2+/− animals were observed, respectively, which aligns with the trends observed in our scRNA-seq datasets (Extended Data Fig. 8c). Theca cells, stromal cells and BECs did not show significant changes in abundance, consistent with scRNA-seq observations (Extended Data Fig. 8c). In the immune compartment, RNAscope identified patterns that were generally consistent with our scRNA-seq results (Extended Data Fig. 9c). For example, we observed increased proportions of NK and NKT cells in Foxl2+/− animals. Additionally, we observed increased DNT abundance in middle-age Foxl2+/− animals, which agrees with our scRNA-seq data (Extended Data Fig. 9c). As with other models, some cell types did not reach statistical significance in RNAscope, often due to lower detection rates, but the overall trends were consistent. Together, these findings further support model-specific shifts in immune and non-immune ovarian cell populations, as captured by both scRNA-seq and RNAscope-based validation.
Global transcriptional perturbations of ovarian cell types across menopause models
To systematically evaluate how different models of menopause influence global transcriptional responses in the ovary, we applied Augur48, a computational framework that quantifies cell type–specific separability across experimental conditions based on single-cell level gene expression profiles (Fig. 5a). Augur scores (derived from underlying machine-learning algorithm performance, reported as Area Under the Curve [AUC] values) represent the ability within each cell type to distinguish between biological groups within a given model based on overall transcriptional landscapes48. In the Aging model, granulosa cells exhibited the highest AUC score (AUC ~0.79), reinforcing their role as key transcriptional responders to chronological aging (Fig. 5b,c). Among other non-immune cell types, BECs and LECs also showed relatively high transcriptional divergence, suggesting that vascular and lymphatic compartments may undergo age-associated remodeling. Indeed, vascular decline has been closely linked to ovarian aging in multiple species, including humans, non-human primates, and mice46,49,50. In the immune compartment, CD4+ T, NK, and B cells ranked highest (Fig. 5b,c). Interestingly, DNT cells, despite their expansion in the aging ovary, showed the lowest AUC score (AUC ~0.53), indicating limited transcriptional remodeling with age (Fig. 5b,c, Extended Data Fig. 7b).
Fig 5. Comparative analysis of global gene expression in Aging, VCD and Foxl2 haploinsufficiency models via Augur.
a, Schematic of global gene expression analysis comparisons using Augur. b-c, UMAP visualization of AUC scores and lollipop plot of AUC quantification from the Aging model. d-e, UMAP visualization of AUC scores and scatter plot of AUC quantification from the VCD model, comparing CTL vs. VCD at 3m and 10m age-at-injection. For the scatter plot, Ptprc− and Ptprc+ cells were separately plotted to improve visualization. All data are from the 90d post-injection timepoint. f-g, UMAP visualization of AUC scores and scatter plot of AUC quantification from the Foxl2 haploinsufficiency model comparing Foxl2+/+ vs. Foxl2+/− at young and mid-age. For the scatter plot, Ptprc− and Ptprc+ cells were separately plotted to improve visualization. For e and g, data points with NA AUC scores (due to low cell count or QC filtering) were assigned an AUC of 0.5 and colored gray to improve visualization.
In the VCD model, we observed marked transcriptional perturbations in theca, stromal, BEC, and LEC cells, especially at 90 days post-injection, with these effects largely consistent across both 3-month and 10-month age-at-injection groups (Fig. 5d,e). However, epithelial and granulosa cells displayed more pronounced age-at-injection-dependent differences: epithelial cells were more responsive in younger animals, while granulosa cells were notably absent in the 10-month cohort, likely reflecting more severe follicular depletion (Fig. 5e, left panel). Among immune cell types, NK, DNT, and CD8+ NKT cells showed higher transcriptional sensitivity in younger animals (Fig. 5e, right panel). At 30 days post-injection, transcriptional divergence was generally more divergent in the VCD model compared to its 90-day counterpart (Extended Data Fig. 10a,b). For example, LECs in the 10-month group displayed strong divergence (AUC ~0.81), suggesting age-related vascular remodeling may emerge early after VCD exposure.
In the Foxl2 haploinsufficiency model, transcriptional divergence was more pronounced at middle-age, suggesting that the effects of Foxl2 loss may not fully manifest in the young ovary (Fig. 5f,g). Among non-immune populations, granulosa and theca cells showed the highest AUC scores at mid-age, while epithelial cells were more divergent in the young group (Fig. 5g, left panel). In immune populations, neutrophils, NKT, CD8+ NKT, and B cells exhibited the strongest divergence, with all showing elevated AUC scores at middle-age, indicating potential immune activation or heightened sensitivity of specific immune compartments to Foxl2 insufficiency (Fig. 5g, right panel).
Together, these results reveal both common and model-specific overall transcriptional trajectories across ovarian aging paradigms. Granulosa cells consistently emerge as sensitive indicators of ovarian dysfunction across all three models. Immune cells show particularly pronounced divergence in Foxl2+/− animals (Fig. 5g, right panel), suggesting that immune alterations may precede, accompany, or even drive ovarian changes in this genetic model. These comparisons underscore the importance of contextualizing cell type–specific transcriptional changes within each model's mechanistic framework and highlight how distinct perturbations – such as chronological aging, chemical ablation, or genetic loss – can produce divergent molecular responses across ovarian cell types.
Transcriptional and pathway-level signatures across menopause models
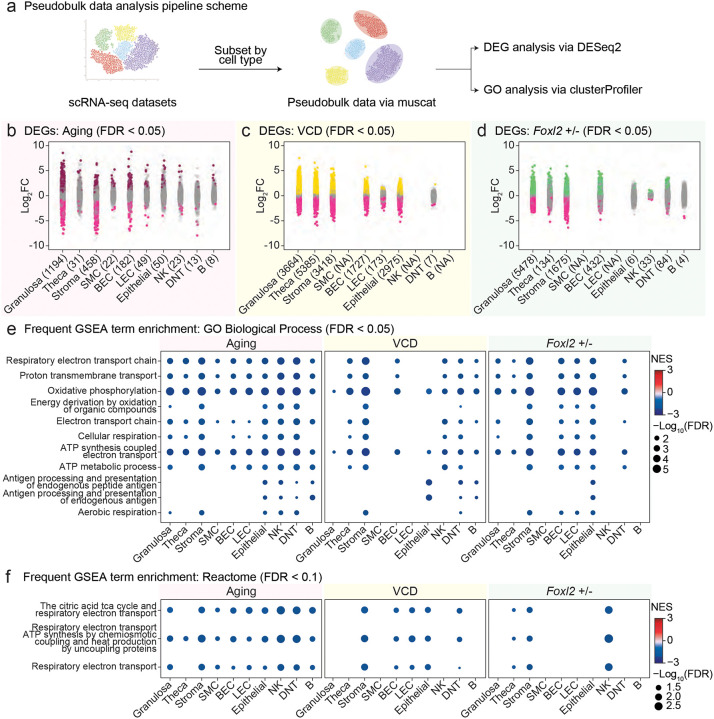
To investigate transcriptional remodeling across menopause models, we performed differential expression and pathway enrichment analyses using data pseudobulked by cell type in each individual scRNA-seq library (Fig. 6a; Extended Data Table 6). Six cell types, granulosa, theca, stromal, BEC, epithelial, and DNT cells, were consistently detected across all three models and thus included in downstream analyses (Extended Data Fig. 11a). DESeq251 was used to identify differentially expressed genes (DEGs) for each model: young vs. old in the Aging model; CTL vs. VCD in the VCD model (using age-at-injection and time post-injection as modeling covariates); and Foxl2+/+ vs. Foxl2+/− in the Foxl2 haploinsufficiency model (using age as a modeling covariate). As expected, we confirmed reduced Foxl2 expression in Foxl2+/− animals at both ages based on our pseudobulked dataset (Extended Data Fig. 11b).
Fig 6. Pseudobulk analysis of differential gene expression and gene ontology analysis across Aging, VCD, and Foxl2 haploinsufficiency models.
a, Schematic of pseudobulk analysis pipeline. b-d, Strip plots of differentially expressed genes (DEGs) from Aging, VCD and Foxl2 haploinsufficiency models. DEGs were identified using DESeq2, comparing young vs. old (Aging model), CTL vs. VCD (VCD model), and Foxl2+/+ vs. Foxl2+/− (Foxl2 haploinsufficiency model). Genes that passed the FDR < 0.05 threshold are colored and the numbers in parentheses following each cell type indicate the total number of DEGs that met the significance threshold. e-f, Frequent Gene Set Enrichment Analysis (GSEA) term enrichment for GO Biological Process (GO BP, FDR < 0.05) and Reactome (FDR < 0.1) pathways across Aging, VCD, and Foxl2 haploinsufficiency models. Terms enriched in at least four and three cell types within each dataset were extracted for GO BP and Reactome, respectively.
Using an FDR cutoff of 0.05, we observed substantial transcriptional changes that were both model- and cell type-specific (Fig. 6b–d). Granulosa cells exhibited the highest number of DEGs in both the Aging (n=1,194) and Foxl2 haploinsufficiency (n=5,478) models, highlighting their sensitivity to physiological and genetic perturbations. In the VCD model, theca cells showed the strongest transcriptional response (n=5,385), followed by granulosa cells (n=3,664), suggesting robust transcriptional remodeling in both steroidogenic cell types following VCD exposure (Fig. 6b–d).
We next performed gene set enrichment analysis (GSEA) to identify gene ontology (GO) biological process terms recurrently enriched across models (Fig. 6e; Extended Data Table 7). Mitochondrial-related gene sets were consistently downregulated across models and cell types, including “Respiratory electron transport chain,” “Oxidative phosphorylation” and “ATP synthesis coupled electron transport” (Fig. 6e; FDR < 0.05). Immune-related processes, such as “Antigen processing and presentation of endogenous peptide antigen,” were also frequently identified as downregulated (Fig. 6e). Using independent gene sets from Reactome52 revealed consistent trends of mitochondria-related gene sets being downregulated (Fig. 6f; FDR < 0.1), suggesting convergent dysregulation of metabolic function during ovarian aging and in menopause models.
Together, these comparative analyses revealed both shared and model-specific transcriptional features of ovarian aging. Granulosa cells consistently exhibited robust transcriptional changes across all three models, underscoring their role as key responders to ovarian aging-related perturbations. Notably, mitochondrial dysfunction and immune dysregulation emerged as common themes across models and cell types, pointing to conserved biological pathways that may underlie the progression of ovarian aging.
Conserved and divergent aging-associated features are observed across menopause models
To identify features that may be broadly shared or uniquely diverge across menopause models, we derived age-associated gene signatures from the Aging model, which reflects natural ovarian physiological decline in mice (Extended Data Fig. 11c). These signatures were then used as a biological reference to explore potential patterns of convergence and divergence across the VCD and Foxl2 haploinsufficiency models (Extended Data Fig. 11c). VCD dataset showed consistent upregulation of aging-associated genes across cell types, whereas the Foxl2 haploinsufficiency model exhibited partially divergent expression patterns (Extended Data Fig. 11c). Notably, granulosa and stromal cells showed decreased expression of genes typically upregulated with age, while BEC cells exhibited opposite trends for both age-associated up- and down-regulated genes. These patterns in a subset of cell types may reflect a distinct aging trajectory, or alternative molecular programs engaged in the setting of partial Foxl2 loss.
We also examined expression of the SenMayo53 gene set, a curated gene set of senescence-associated genes, which can be used as a proxy for senescence burden in transcriptional data (Extended Data Fig. 11d). In the Aging model, all six cell types tested showed significant upregulation of the SenMayo signature. In the VCD model, granulosa, stromal, BEC, and DNT cells mimicked this enrichment, while theca and epithelial cells showed a negative correlation. In the Foxl2 haploinsufficiency model, theca and DNT cells showed consistent upregulation, whereas granulosa, stromal, BEC, and epithelial cells showed an inverse pattern of downregulation (Extended Data Fig. 11d). These contrasting results suggest that the VCD and Foxl2 haploinsufficiency models may each capture distinct cell-specific aspects of an accelerated ovarian aging process.
We next evaluated transcriptional noise by calculating the coefficient of variation (CV) for each cell type (Extended Data Fig. 11e). Increased transcriptional variability is a characteristic of aging and has been observed across multiple tissues, including the ovary45 and hypothalamus54 in mice, reflecting a loss of transcriptional regulation with age. In the Aging model, CV increased across all detected cell types, except in DNT cells. In the VCD model, theca cells consistently exhibited increased CV, while granulosa and LECs showed more variable results. In the Foxl2 haploinsufficiency model, theca cells again showed increased CV regardless of age, although other cell types displayed inconsistent trends in young and middle-aged animals. While increased CV is often linked to age-related loss of regulatory precision54,55, these mixed results in VCD and Foxl2 haploinsufficiency models suggest a more nuanced transcriptional landscape that may reflect compensatory or cell type-specific adaptations.
Lastly, we assessed transposable element (TE) expression using scTE56 and DESeq2 (Extended Data Fig. 11f). TE activity is known to increase with aging across tissues, driven in part by age-associated chromatin remodeling, reduced heterochromatin integrity and impaired epigenetic repression57,58. In the Aging model, granulosa, stromal, SMC, BEC, and LEC cells showed increased TE expression with age, while theca, epithelial, NK, and B cells showed decreased TE expression. In the VCD model, TE expression increased in stromal, BEC, LEC, and epithelial cells, and decreased in theca cells. In the Foxl2 haploinsufficiency model, TE expression increased in most cell types, except stromal cells. Increase in TE expression is recognized as a hallmark of aging, contributing to genomic instability, inflammation, and cellular stress57,59. Rather than reflecting uniform activation of TEs, these cell type–specific shifts suggest that TE regulation may respond differently depending on the mechanism of ovarian perturbation (Extended Data Fig. 11f).
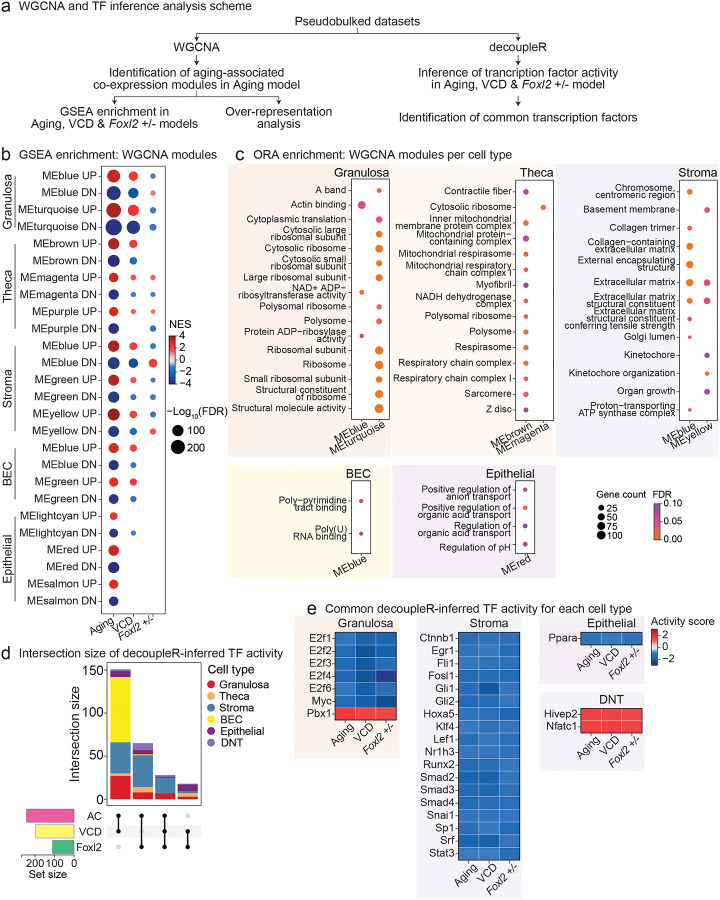
Gene co-expression network and transcription factor activity across menopause models
To identify gene regulatory programs associated with ovarian aging and dysfunction, we performed weighted gene co-expression network analysis (WGCNA)60 and transcription factor (TF) activity inference using decoupleR61 (Fig. 7a). To obtain a comparative reference state, we first applied WGCNA to pseudobulked gene expression data from the Aging model, reflecting naturally occurring physiological changes. Modules with significant trait correlation to age (FDR < 0.1) were retained for downstream analysis (Extended Data Fig. 12,13). GSEA of these gene modules revealed strong enrichment in the Aging dataset (as expected by construction), providing a useful benchmark for evaluating transcriptional convergence and divergence across intact aging and intervention models (Fig. 7b).
Fig 7. Preservation of co-expression modules and transcription factor activity in Aging, VCD, and Foxl2 haploinsufficiency models.
a, Schematic of WGCNA and transcription factor (TF) activity inference analysis. b, GSEA enrichment of WGCNA module genes associated with aging traits across Aging, VCD and Foxl2 haploinsufficiency models. WGCNA modules significantly associated with aging traits (FDR < 0.1) were identified for each cell type from the Aging model. c, Over-representation analysis of GO ALL terms for WGCNA module genes (FDR < 0.1). d, UpSet plot of decoupleR-inferred TF activity, showing the number of TF overlap across datasets and the corresponding cell types. e, Heatmap of TF activity scores of common decoupleR-inferred TFs for each cell type.
In the VCD model, most modules exhibited enrichment patterns consistent with the Aging model, suggesting that VCD-induced ovarian failure recapitulates key transcriptional features observed during normal chronological aging. In contrast, the Foxl2 haploinsufficiency model revealed notable differences in module behavior. For instance, in granulosa cells, genes from both reference modules were downregulated in the Foxl2 haploinsufficiency dataset, despite being upregulated in the Aging model (Fig. 7b). Conversely, the MEblue module, downregulated with age in the Aging model, was upregulated in Foxl2+/− granulosa cells. Theca cells showed broadly consistent enrichment across models, whereas stromal cells exhibited mixed and model-specific patterns (Fig. 7b). These findings illustrate both shared and model-specific regulatory programs and highlight the utility of comparative approaches to dissect diverse aspects of ovarian aging.
To further explore the biological relevance of these co-expression modules, we performed over-representation analysis (ORA) on each module’s genes using Gene Ontology. In granulosa cells, enriched terms included components of the ribosome and translational machinery (e.g., “cytosolic ribosome,” “polysome,” “cytoplasmic translation”), as well as actin binding and structural molecule activity (e.g., “A band,” “actin binding,” and “structural molecule activity”), highlighting coordinated regulation of protein synthesis and cytoskeletal organization. Notably, enrichment for NAD+ ADP-ribosyltransferase activity was also observed. This finding aligns with recent evidence linking the NAD+ pathway to the regulation of ovarian aging62. In theca cells, modules were enriched for mitochondrial and contractile fiber-related terms, including “mitochondrial respiratory chain complex I,” “respirasome,” and “sarcomere,” suggesting that transcriptional remodeling in aging theca cells may involve coordinated changes in energy metabolism and cytoskeletal dynamics. The appearance of the “NADH dehydrogenase complex” term further supports a role for the NAD+ pathway in theca cell aging62. Stromal cell modules were primarily enriched for extracellular matrix (ECM)-related pathways, such as “collagen-containing extracellular matrix,” “external encapsulating structure,” and “basement membrane,” consistent with known stromal remodeling during ovarian aging63,64. In BECs, we observed enrichment of RNA-binding activities, including “poly(U) RNA binding,” while epithelial cell modules were enriched for pathways related to pH regulation and organic acid metabolism, suggesting cell type–specific metabolic adaptations during aging.
To complement co-expression network-based analyses, we also inferred TF activity across cell types using decoupleR (Fig. 7d,e; Extended Data Table 8). Across datasets, we detected shared changed TF activity signatures in multiple cell types (Fig. 7d). In granulosa cells, we consistently observed decreased predicted activity of cell cycle–associated factors (E2f1, E2f2, E2f3, E2f4, E2f6, Myc) and increased predicted activity of Pbx1 (Fig. 7e). Pbx1 has been implicated in anti-aging pathways, as well as tissue and organ homeostasis and regeneration, in hair follicle-derived mesenchymal stem cells and adrenocortical cells, respectively65,66; thus, increased activity in the context of ovarian aging may reflect a compensatory response to maintain cellular function or structural integrity (Fig. 7e). Stromal cells showed consistent decreased predicted activity of several signaling and remodeling-associated TFs, including Runx2, Fosl1, Fli1, Klf4, Smad2/3/4, and Stat3. These factors are broadly involved in ECM remodeling, vascular integrity, immune signaling, and cellular stress responses46,67–69; thus, their coordinated decline may contribute to age-associated stromal dysfunction, fibrosis, and impaired tissue homeostasis. In epithelial cells, Ppara activity was consistently predicted to decrease across menopause models. Ppara has been shown to regulate lipid metabolism and oxidative stress responses70; its reduced activity in aged/treated epithelial cells may contribute to metabolic dysregulation associated with menopause. In DNT cells, Hivep2 and Nfatc1 showed increased activity (Fig. 7e). HIVEP2 has been implicated in regulatory T cell-mediated immunosuppression in humans71, while Nfatc1 is known to control cytotoxic function in CD8+ T cells in mice72,73. Intriguingly, Nfatc1 has been shown to be important for regulation of CD8+ T cell metabolism72. However, the specific roles of these factors in the context of DNT cells warrant further investigation.
These analyses reveal both conserved and model-specific gene regulatory programs underlying ovarian aging. Network-based approaches identified modules and transcription factors linked to protein synthesis, mitochondrial function, and ECM remodeling, with distinct patterns emerging across cell types and models.
Transcriptome-based aging clocks capture evidence of accelerated ovarian aging across models
To gain a complementary and integrative view of ovarian aging, we developed transcriptome-based clocks that predict ovarian age using gene expression data (Fig. 8a; Extended Data Fig. 15a), similar to efforts in other tissues, including blood and brain74,75. Given the central role of granulosa cells in ovarian function and their consistently high responsiveness in our models (see above), we first trained a lasso regression model using single-cell transcriptomes from granulosa cells (Fig. 8a). In parallel, we also constructed a comparable model using theca cell transcriptomes to evaluate performance in another key steroidogenic ovarian cell type (Extended Data Fig. 15a).
Fig 8. Development of a transcriptome-based aging clock for granulosa cells.
a, Schematic of analysis and training workflow for the lasso regression-based clock. b, Scatter plot of predicted age vs. chronological age for the test set, with Spearman’s correlation and p-value displayed. c-d, Age prediction for VCD and Foxl2 haploinsufficiency datasets, comparing predicted age to chronological age in weeks. Offsets were added to improve visualization. e-f, Age acceleration analysis for VCD and Foxl2 haploinsufficiency datasets, calculated as the difference between predicted and chronological age. Statistical significance was assessed using the Wilcoxon test. g, Heatmap of expression lasso features from pseudobulked expression data. h, Over-representation analysis of GO ALL terms of the top 500 lasso features.
Performance evaluation on the test dataset revealed strong predictive accuracy for the granulosa cell-based model, with a Spearman correlation of ~0.76 and p-value of ~3.9 × 10−293 between predicted and true chronological age (Fig. 8b). The theca cell-based model also performed well (Rho ~0.70, p-value ~1.9 × 10−237; Extended Data Fig. 15b), although it showed a dip in prediction accuracy at older ages. Notably, predictive performance in the theca model starts to decline at ~12 months of age, coinciding with the reported onset of estropause in mice of the C57BL/6 strain33 (Extended Data Fig. 15b; age of expected estropause onset highlighted in gray).
Having validated the predictive capacity of the model, we next applied the model to granulosa cell data from the VCD-treated and Foxl2+/− animals (Fig. 8c,d). In the VCD model, transcriptome-predicted ages were consistently higher than chronological age, supporting the notion that VCD exposure accelerates ovarian aging at the molecular level (Fig. 8c). In the Foxl2 haploinsufficiency model, Foxl2+/− animals also showed older predicted transcriptomic ages compared to wild-type controls (Fig. 8d). For granulosa cells, the median predicted age was 27.3 weeks in young Foxl2+/+, 43.0 weeks in young Foxl2+/−, 36.3 weeks in middle-age Foxl2+/+, and 38.5 weeks in middle-age Foxl2+/− (Fig. 8d).
To further quantify these differences, we calculated transcriptome-based age acceleration by subtracting true chronological age from predicted age (Fig. 8e,f). In the VCD dataset, age acceleration decreased as age at VCD exposure increased, a trend also observed in the hormone-based OvAge model, suggesting a reduced magnitude of response to insult in older ovaries (Fig. 2e–g, 8e). Granulosa cells from Foxl2+/− animals were predicted to be older than their chronological age, diverging from the results of OvAge, where condition-matched Foxl2+/− animals were predicted to be younger (Fig. 2h, 8f). This discrepancy suggests that the transcriptome-based clock likely detects early molecular alterations in the ovary that have not yet manifested as systemic hormonal shifts.
We also applied the theca cell-based aging clock to the VCD and Foxl2 haploinsufficiency models to determine its predictions on ovarian transcriptional age (Extended Data Fig. 15c,d). Despite slightly lower testing accuracy, the theca cell clock produced trends highly consistent with the granulosa-based model. Specifically, VCD-treated animals showed elevated transcriptomic age estimates compared to controls, and Foxl2+/− animals similarly exhibited increased predicted ages relative to wild-type counterparts (Extended Data Fig. 15c,d). The median predicted age was 26.7 weeks in young Foxl2+/+, 34.4 weeks in young Foxl2+/−, 34.9 weeks in mid-age Foxl2+/+, and 36.3 weeks in mid-age Foxl2+/−. We also calculated age acceleration for the theca cells and observed similar patterns to those seen in granulosa cells (Extended Data Fig. 15e,f). Notably, the difference between age acceleration estimates was statistically significant for all comparisons, including the middle-age group from the Foxl2 haploinsufficiency model, further supporting its sensitivity in detecting molecular ovarian aging phenotypes across both VCD and Foxl2 haploinsufficiency models. These findings reinforce the capacity of the theca cell-based clock to detect molecular signatures of accelerated aging and suggest that both granulosa and theca cells encode convergent transcriptional readouts of ovarian aging across models.
To explore the transcriptional features contributing to age prediction, features with non-zero β coefficients from the trained lasso model were extracted and their expression trajectories assessed across age in pseudobulked granulosa cell dataset (Fig. 8g). These features exhibited diverse patterns of expression over time, suggesting involvement in distinct age-related regulatory programs (Fig. 8g). To further investigate their biological relevance, the top 500 features, ranked by the absolute value of β coefficients in the final lasso model, were subjected to ORA enrichment (Fig. 8h). Enriched terms were primarily related to immune processes and vascular function, including “leukocyte migration,” “interferon-gamma production,” “positive regulation of antigen processing and presentation,” and “blood circulation” and “vascular remodeling” (Fig. 8h). These findings suggest that early shifts in immune signaling and vascular architecture may play a central role in shaping the transcriptomic landscape of ovarian aging and highlight the sensitivity of the transcriptome-based clock in detecting early molecular changes.
A similar approach was applied to the theca cell-based model, where features with non-zero β coefficients were extracted and their expression trajectories examined across age in the pseudobulked theca cell dataset (Extended Data Fig. 15g). These features also showed heterogeneous age-dependent expression patterns, indicating involvement of diverse regulatory programs during theca cell aging. To evaluate their biological relevance, the top 500 features, ranked by the absolute value of their β coefficients as above, were also subjected to ORA enrichment (Extended Data Fig. 15h). Enriched terms included immune-related processes such as “regulation of myeloid cell apoptotic process,” “regulation of cytokine production involved in immune response,” and “positive regulation of response to external stimulus,” as well as metabolic and structural pathways including “cellular response to lipid,” “collagen-containing extracellular matrix,” and components of the “mitochondrial outer membrane” and “membrane microdomains.” These results suggest that transcriptional aging in theca cells may be shaped by coordinated shifts in immune regulation, lipid metabolism, and mitochondrial and membrane-associated processes.
Together, these data demonstrate the utility of transcriptome-based models for detecting molecular aging trajectories in the mouse ovary. These predictive tools offer a valuable framework for assessing molecular age across experimental models and for uncovering early transcriptomic alterations that precede overt phenotypic decline.
Discussion
Understanding how ovarian decline contributes to systemic aging requires robust experimental models that accurately recapitulate the physiological features of menopause. In this study, we systematically evaluated three mechanistically distinct mouse models of menopause: an intact aging model, a chemical follicle depletion model using VCD exposure, and a genetic model of Foxl2 haploinsufficiency. By integrating histological, hormonal, and transcriptomic profiling across these models, we provide a comparative framework for understanding shared and model-specific trajectories of ovarian aging. Findings from our study provide a critical comparative resource for identifying conserved and divergent features of ovarian aging and evaluating the suitability of each model, with the goal of advancing both mechanistic insights and translational strategies in menopause research.
In the Aging model, we observed hallmark features of physiological ovarian decline, including depletion of all follicle stages and disrupted endocrine profiles. These changes were accompanied by immune cell expansion, increased gene expression variability, elevated TE expression, and significant transcriptional remodeling in somatic and immune cell types. Granulosa cells consistently exhibited the highest transcriptional responsiveness, reinforcing their central role in orchestrating ovarian function and aging. Among immune populations, DNT cells displayed a marked increase in abundance with age. This expansion was also reported in two independent studies using mouse scRNA-seq datasets44,47, reinforcing the emerging role of DNT cells as conserved modulators of ovarian immune remodeling and aging. Mitochondrial dysfunction, immune activation, and vascular remodeling emerged as common transcriptional themes in the aging ovary. Gene regulatory network analyses further implicated downregulation of key transcription factors involved in cell cycle regulation, stress responses, and tissue homeostasis.
In the VCD model, animals displayed features reminiscent of premature ovarian failure, including acute follicular depletion, endocrine disruption, and elevated predicted ovarian age based on both OvAge and transcriptomic clocks. The extent of age acceleration was most pronounced in animals exposed to VCD at younger ages. This is particularly relevant given that early menopause is a strong predictor of systemic aging and reduced lifespan in humans4. By varying the timing of VCD exposure, this model could be leveraged to probe the systemic impact of menopausal timing and explore interactions between reproductive and systemic aging trajectories.
The Foxl2 haploinsufficiency model revealed a more nuanced aging trajectory. Despite normal or elevated follicle counts and serum AMH levels in young and mid-age Foxl2+/− animals, transcriptome-based clocks detected increased molecular age in granulosa and theca cells. Additionally, immune populations, including neutrophil, NKT and B cells showed enhanced transcriptional responsiveness, consistent with an immune-primed ovarian environment35,47. These results suggest that partial loss of Foxl2 function may induce early molecular reprogramming prior to overt histological or endocrine decline. Interestingly, granulosa and stromal cells showed inverse expression of classical aging signatures, and the model exhibited downregulation of typical senescence pathways and aging modules identified in the Aging model. FOXL2 has been shown to interact with AMH to modulate follicle recruitment in humans40. Thus, the observed histological, endocrine, and transcriptional features may reflect enhanced Amh-mediated reserve preservation in the Foxl2+/− animals. Nonetheless, the preserved endocrine output, coupled with increased transcriptional sensitivity in steroidogenic and immune compartments, positions the Foxl2 haploinsufficiency model as a unique tool for interrogating early or subclinical features of ovarian aging. Based on the increased transcriptional responsiveness of immune cells and the nuanced acceleration in transcriptomic aging of granulosa and theca cells, it may be valuable to assess these animals at older age ranges. The extended timeline could reveal whether early transcriptomic shifts translate into functional decline and help uncover regulatory mechanisms that are obscured in more overt aging models.
Cross-species single-cell analyses of ovarian aging further support the translational relevance of our findings. Studies in mice, non-human primates, and humans have consistently identified conserved molecular signatures associated with ovarian aging, including reduced oxidative phosphorylation, diminished antioxidant gene expression, and elevated immune activation across multiple ovarian cell types44–46,76. These transcriptional features were similarly prominent across all three models examined in our study. Additionally, regulatory network analyses from human ovarian datasets have implicated transcription factors such as FOXL1 and RUNX2 as age-associated regulators46. Although these factors were detected in different cell types in our dataset, their recurrent identification across species suggests that they may represent broader regulators of ovarian homeostasis. These cross-species parallels underscore the robustness of the aging signatures captured by our models and highlight the value of this comparative framework for investigating conserved mechanisms of ovarian aging.
Several mouse genetic models have provided additional insights into molecular features underlying ovarian decline. In a model with impaired interferon-gamma regulation (ARE+/− and ARE−/−), elevated systemic interferon-gamma led to CD8+ T cell infiltration in reproductive tissues and resulted in infertility77. In contrast, we observed a decrease in CD8+ T cell abundance in aged ovaries from our Aging model, raising the possibility that infiltration may occur earlier in the aging process, followed by depletion or exhaustion at advanced stages. CD8+ T cells were also sparse in the VCD and Foxl2 models, suggesting that interferon-mediated immune activation may represent a distinct and context-dependent mechanism.
A separate study investigating Cd38 deletion identified early ovarian inflammation and NAD+ depletion as key features of ovarian aging, with loss of Cd38 rescuing both transcriptional profiles and follicular health62. In the study, immune cell abundance decreased following Cd38 deletion62. In contrast, we observed increased immune cell abundance in the Aging and Foxl2 haploinsufficiency models, and decreased abundance in the VCD model. Despite these differences, our results converge with the Cd38 study on a set of transcription factors potentially involved in regulating ovarian aging. Notably, transcription factors such as Myc, Egr1, Fli1, and Klf4, reported to be restored upon Cd38 deletion, were also identified across all three models in our analysis62. These shared regulatory nodes point to conserved, age-sensitive networks that may facilitate physiological decline.
In summary, our comparative analysis reveals that ovarian aging is shaped by a combination of cell type–specific, model-specific, and shared molecular processes. Intact aging reflects gradual, widespread physiological decline; VCD exposure triggers acute and accelerated failure; and Foxl2 haploinsufficiency initiates early molecular perturbations that may precede functional impairment. By integrating histological, hormonal, and transcriptomic datasets, we provide a multidimensional framework for dissecting the mechanistic landscape of ovarian aging. This platform will be valuable for both basic and translational studies, offering new opportunities to define early biomarkers, and design interventions to delay or reverse reproductive senescence.
Methods and materials
Mouse husbandry
All animals were treated and housed in accordance with the Guide for Care and Use of Laboratory Animals. All experimental procedures were approved by the USC’s Institutional Animal Care and Use Committee (IACUC) and are in accordance with institutional and national guidelines. Samples were derived from animals on approved IACUC protocol numbers 21155 and 21454. All animals were acclimated in the specific-pathogen-free animal facility at USC for two weeks prior to any experimental procedures. Mice were provided PicoLab Rodent Diet 20 (LabDiet, 5053) ad libitum. The facility was maintained on a 12-hour light/dark cycle, with housing rooms set to 72°F and 30% humidity.
For the Aging model, female C57BL/6JNia mice (4- and 20-month-old) were obtained from the National Institute on Aging (NIA) colony at Charles River Laboratories.
For the VCD model, female C57BL/6J mice (2.5-, 5.5-, 7.5-, and 9.5-month-old) were purchased from Jackson Laboratory. Animals received daily intraperitoneal injections of either vehicle (safflower oil; Sigma S8281) or VCD (160 mg/kg/day; Sigma 94956) for 15 consecutive days. All injections were administered between 8:00 and 10:00 AM to minimize variability due to circadian influences.
Foxl2 floxed (Foxl2lox/lox) mice were generated by Cyagen Biosciences Inc. using a previously described targeting strategy25. Briefly, a targeting construct was designed to replace a 2.177 kb fragment containing the entire Foxl2 coding region with a 1.157 kb Neo gene cassette (Extended Data Fig. 1a). The construct was linearized and introduced into C57BL/6NTac embryonic stem (ES) cells via electroporation. Candidate clones were assessed via PCR and Southern blot analysis. A targeted ES cell clone was selected and injected into C57BL/6 NTac albino embryos, which were then implanted into pseudo-pregnant CD-1 females. Founder animals were identified based on coat color, and germline transmission was confirmed through breeding with C57BL/6 females followed by genotyping of the offspring. Mice carrying the desired floxed Foxl2 allele were sent to USC for experiments and downstream phenotyping.
Foxl2 haploinsufficiency mice (Foxl2+/−) were generated by crossing homozygous Foxl2lox/lox C57BL/6NTac mice with heterozygous B6.C-Tg(CMV-cre)1Cgn/J (JAX, stock #006054) mice. Offspring were genotyped for both the Foxl2 alleles and the CMV-Cre transgene. To eliminate the CMV-Cre transgene, Foxl2+/− mice were crossed with wild-type C57BL/6J mice. Progeny were selected to carry the deleted Foxl2 allele (Foxl2+/−), but not the CMV-Cre transgene. Afterwards, Foxl2+/− mice were maintained as a stable heterozygous knockout colony using allele transmission through both parent sexes. Routine genotyping was performed using genomic DNA extracted from tail biopsies using specific PCR primers, as listed in Extended Data Table 9a.
RT-qPCR analysis of Foxl2 expression in Foxl2 haploinsufficiency model mouse ovaries
Flash-frozen ovaries collected from mice were used for RNA extraction, cDNA synthesis and quantitative RT-PCR. Ovaries were resuspended in 600 μL of TRIzol reagent (Thermo-Fisher, 15596018) and lysed using the BeadBug Homogenizer (Benchmark Scientific, D1036). Homogenization was performed at 3,500 rpm in 30-second intervals, repeated for a total of nine cycles. Total RNA was then purified using the Direct-Zol RNA Miniprep kit (Zymo Research, R2052), following the manufacturer’s protocol. RNA was eluted in nuclease-free water sand immediately quantified using the NanoDrop spectrophotometer (Thermo Scientific). The purity of the RNA was assessed by measuring the 260/280nm and 260/230nm absorbance ratios. Samples with ratios close to ~2.0 were considered suitable for downstream applications.
Total RNA was reverse transcribed into cDNA, using the Thermo Scientific™ Maxima H Minus First Strand cDNA Synthesis Kit (Thermo Scientific, K1682), following the manufacturer’s protocol. Quantitative PCR was performed using SensiFAST SYBR no-ROX kit (Bioline, BIO-98020) and the MIC Tubes and Caps kit (Bio Molecular Systems, 71e101) on the Magnetic Induction Cycler (MIC) machine (Bio Molecular Systems, MIC-2). micPCR v2.12.6 was used to capture and quantify Ct values. Gene expression levels were calculated using the ΔΔCt method, normalized to the geometric mean of two housekeeping genes: Ubc and Hprt. Primers and sequences used for RT-qPCR reactions can be found in Extended Data Table 9b.
Fertility assessment of Foxl2 haploinsufficiency model mice
Fertility data were obtained from our breeding colony, where Foxl2+/+ and Foxl2+/− females were paired with Foxl2+/− and Foxl2+/+ males, respectively. Data were included in the fertility assessment only if Foxl2+/+ and Foxl2+/− female pairings were initiated on the same day. Pairings conducted on days when only Foxl2+/+ or only Foxl2+/− females were enrolled were excluded from analysis to avoid any batch-specific effects on fertility assessments due to uncontrolled routine handling fluctuations.
Female mice between 2.5 and 3 months of age were paired one-to-one with age-matched males. Fertility was assessed based on the pup count of the first litter and the latency to the first litter (i.e., the time between pairing and birth of the first litter). Differences in litter size between Foxl2+/+ and Foxl2+/− females were assessed using the non-parametric Wilcoxon rank-sum test. One outlier from the Foxl2+/+ group was removed based on a significant Grubbs' test (p-value < 0.05), using the outliers package (v.0.15)79 in R (v. 4.1.2). Latency to the first litter was analyzed using a log-rank test to compare “survival” curves, using the survival package (v. 3.6–4)80 in R. Raw fertility data can be found in Extended Data Table 1a.
Hematoxylin-eosin staining of mouse ovarian tissues
Ovaries were fixed in Bouin’s solution (Sigma, HT10132) for 24 hours at room temperature before being transferred to 70% ethanol for storage. Paraffin embedding, tissue sectioning, and hematoxylin and eosin (H&E) staining were carried out by the USC Norris Comprehensive Cancer Center Translational Pathology Core Facility. H&E-stained slides were imaged using the Keyence BZ-X All-in-One Fluorescence Microscope platform using 20X objective and automated stitching using default parameters.
Ovarian follicle counts
Follicle counts, including primordial, primary, secondary, and antral follicles, and corpus luteum, were performed on three sections per ovary by three blinded observers. Median values from the three observers were used for data analysis. Statistical differences between groups were determined using the non-parametric Wilcoxon rank-sum test.
Quantification of serum AMH, FSH and Inhibin A concentrations
Blood was collected either immediately following euthanasia via cardiac puncture or from live animals via a facial vein blood draw. Blood was allowed to clot at room temperature for one hour. Serum was then separated using the MiniCollect® Serum Tube (Greiner, 450472) and stored at -80°C until further analysis. The University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core quantified serum levels of AMH (Rat and Mouse Anti-Müllerian Hormone (AMH) ELISA kit, Ansh Labs, AL-113), FSH (Millipore Pituitary Panel Multiplex kit, RPT86K or Ultra-Sensitive Mouse & Rat FSH, UVA Ligand Core, in-house81), and Inhibin A (Inhibin A ELISA kit, Ansh Labs, AL-161), providing standardized normalized values. Statistical differences between groups were evaluated using the non-parametric Wilcoxon rank-sum test. Raw serum hormone quantification data can be found in Extended Data Table 2.
Ovarian health index calculation
The ovarian health index was calculated as previously described41. The index was determined by integrating two components: ovarian hormone levels (AMH, FSH, and Inhibin A) and follicle counts, which included the combined counts of primordial, primary, secondary, and antral follicles, as well as corpus luteum. Each parameter was assigned a score based on a three-tier system. Values exceeding the median of the young female group were assigned a score of 3, values positioned between the medians of the young and old groups were assigned a score of 2, and values below the median of the old group were assigned a score of 1. The hormone score was calculated as the mean of the individual scores for AMH, FSH, and Inhibin A. This hormone score was then combined with the follicle score in a 1:1 ratio to generate the overall ovarian health index, which was subsequently scaled to a 0–100 range for standardization. Statistical differences between groups were evaluated using the Wilcoxon rank-sum test.
Two different FSH assay kits were used due to changes in offered FSH quantitation assays at the UVA core during the time of our study. To ensure compatibility between measurements made using the different kits, we applied a correction procedure as previously described41. Briefly, matched serum samples were analyzed using both kits, and a polynomial regression model was trained to capture the relationship between the two outputs. This model was then applied to adjust FSH values obtained using the ultra-sensitive assay, enabling direct computation of ovarian health index across datasets.
The scripts used for ovarian health index calculation are available on the Benayoun lab GitHub repository: https://github.com/BenayounLaboratory/Ovarian_Aging_Microbiome/tree/main/1_Ovarian_health_index_calculation.
OvAge clock model training
The OvAge clock was trained using serum hormone quantification data from multiple model and cohorts, including animals from aging model, vehicle control animals from VCD model, wild-type animals from Foxl2 haploinsufficiency model (Foxl2+/+) and wild-type animals from a previously published Fshr haploinsufficiency model (Fshr+/+)42. To enhance the applicability of the model across different genetic and experimental backgrounds, animals from multiple models were included in the training dataset. Hormone data included serum levels of AMH, FSH, and INHBA, with corresponding age at blood collection recorded in weeks.
The dataset was partitioned into training (75%) and testing (25%) subsets using stratified sampling based on age, using the createDataPartition function from R package caret (v. 6.0–91)82. A random forest (RF) regression model was trained using the training set, with out-of-bag (OOB) error correction applied for internal validation, using caret (v. 6.0–91)82 and randomForest packages (v. 4.7–1)83. The model was optimized by tuning the mtry parameter (best-performing value = 2), yielding an OOB root mean square error (RMSE) of 12.31 weeks and an OOB R2 of 0.67. A total of 500 trees were used in the final model. Model performance was evaluated using Spearman correlation between chronological and predicted ages in the test dataset. The RF model achieved a Spearman correlation coefficient of 0.62 with a p-value of 7.35 × 10−8. Age acceleration was calculated as difference between predicted and chronological age. The final OvAge clock R object containing the trained RF model is available on the Benayoun lab GitHub repository: https://github.com/BenayounLaboratory/Mouse_Menopause_Models. We have also developed an interactive R shiny app to enable users to input hormone values and estimate predicted ovarian age using the OvAge clock: https://minhooki.shinyapps.io/OvAge_Predictor/.
Single-cell RNAseq sample and library preparation
Ovaries from two animals, totaling four ovaries, were used for each single-cell RNA sequencing (scRNA-seq) sample and library preparation to enable optimal cellular viability and yield. Single-cell suspensions were generated using Miltenyi’s Multi Tissue Dissociation Kit (Miltenyi Biotec, 130-110-201) with the gentleMACS Octo Dissociator with Heaters (Miltenyi Biotec, 130-096-427), running the 37_m_LDK1 program. To maximize the recovery of viable single cells, the program was deliberately aborted 10 sec after start of the highspeed phase, approximately 20 sec before program completion. After dissociation, red blood cells were removed using Red Blood Cell Lysis Buffer (Miltenyi Biotec, 130-094-183) according to the manufacturer’s instructions. Dead cells were subsequently eliminated using the EasySep Dead Cell Removal (Annexin V) Kit (STEMCELL Technologies, 17899).
Cell count and viability were assessed via Viobility™ 405/452 Fixable Dye (Miltenyi Biotec, J66584-AB) staining and flow cytometry using a MACSQuant Analyzer 10 (Miltenyi Biotec, 130-130-420). Flow cytometry data were analyzed with FlowLogic v8 (Inivai Technologies). Cells were gated based on forward scatter area (FSC-A) vs. side scatter area (SSC-A), and singlets were identified by evaluating the linear grouping of cells in FSC-A vs. FSC-H plots (Extended Data Fig. 7a).
Single-cell libraries were prepared using the Chromium Next GEM Single Cell 3 GEM, Library & Gel Bead Kit v3.1 (10X Genomics, PN-1000121) according to the manufacturer’s instructions. Based on flow cytometry estimates, cell suspensions were loaded to achieve a targeted recovery of 6,000 cells per sample. Samples were processed on a Chromium Next GEM Chip G (10X Genomics, 2000177) as per the manufacturer’s protocol.
Completed single-cell libraries were assessed for quality using the 4200 TapeStation system (Agilent Technologies, G2991A) with High Sensitivity D1000 DNA ScreenTape (Agilent Technologies, 50675584). Libraries were sequenced on an Illumina NovaSeq 6000, generating 150 bp paired-end reads at Novogene USA. Raw FASTQ reads have been deposited in the Sequence Read Archive under accession PRJNA863443.
Single-cell RNAseq data analysis
Data processing
Raw sequencing reads were processed using Cell Ranger (v. 7.1.0)84 from 10x Genomics (Extended Data Table 3). Reads were aligned to the mm10 reference genome. To remove contamination from ambient RNA, decontamination was performed using DecontX from the celda (v. 1.14.2)85 package. Raw and filtered feature-barcode matrices were used to estimate ambient RNA levels, and corrected expression matrices were generated for each sample. Doublet removal was performed using a two-step approach. First, DoubletFinder (v. 2.0.3)86 was applied to each sample independently, estimating doublet rates based on the expected multiplet rates from 10x Genomics. The optimal pK parameter was determined using a parameter sweep, and predicted doublets were labeled. In parallel, a second doublet detection method, scds (v. 1.10.0)87, was applied using the cxds_bcds_hybrid function. Cells identified as doublets by either method were excluded from further analysis.
The filtered single-cell expression matrices were loaded into Seurat (v. 4.3.0)88, and initial quality control (QC) steps were applied. Cells with fewer than 500 detected features, a total RNA count outside the range of 500–100,000, mitochondrial RNA content exceeding 15%, or a DecontX contamination score above 0.25 were removed. Following QC, datasets were normalized using SCTransform, regressing out the number of detected features, mitochondrial content, DecontX contamination score and batch effects.
Data integration
To correct for batch effects in the VCD and Foxl2 haploinsufficiency datasets, integration was performed using Harmony (v. 1.0)89. First, datasets were processed separately by normalizing gene expression using SCTransform, regressing out number of detected features (nFeature_RNA), mitochondrial percentage (percent.mito), ambient RNA contamination (decontX_contamination), and sequencing library (Library). Highly variable genes were selected using vst (nfeatures=2000 genes). The preprocessed datasets were merged using Seurat (v. 4.3.0)88, ensuring shared variable features were maintained. Principal component analysis (PCA) was performed on the merged dataset, and batch effects were assessed by visualizing PCA embeddings colored by batch identity. Batch correction was applied using Harmony, where batch information was explicitly modeled as a covariate. The corrected embeddings were used for downstream analysis, including UMAP visualization, clustering, and differential expression analysis.
Cell type annotation
Cell type annotation was conducted using a multi-step approach that combined automated reference-based methods with manual marker-based annotation. First, Ptprc+ and Ptprc− cell populations were identified using scGate (v. 1.0.1)90. Cells expressing Ptprc (Cd45) were classified as immune cells, while those lacking Ptprc expression were designated as non-immune cells. The two subsets were then processed separately for further annotation. Each subset underwent independent cell type annotation using three complementary computational methods: SingleR (v. 1.8.1)91, scSorter (v. 0.0.2)92, and scType (v. 1.0)93. The ImmGen dataset94 was used as a reference for Ptprc+ cells, while two publicly available ovarian single-cell RNA-seq datasets (Open Science Framework ID 924fz95 and NCBI GEO accession no. GSE23230944) were used for Ptprc− cells. To supplement these automated methods, manual annotation was conducted based on the expression of canonical marker genes specific to ovarian cell types (Extended Data Table 4). The final annotation was determined using a majority voting approach – cell type labels from SingleR, scSorter, scType, and manual annotation were compared, and the most frequently assigned label was selected as the final classification. All analyses were performed using Seurat (v. 4.3.0)88 within R.
Cell type proportion and Augur analysis
Cell type proportions were quantified using scProportionTest (v. 0.0.0.9000)96 within R. This method performs a permutation-based test to compare cell-type abundances between two groups, estimating the relative change in proportions along with a confidence interval for each cell type. The analysis was conducted at two levels: the broader classification of Ptprc+ vs. Ptprc− populations and the more detailed sub-cell type level within each category.
We used Augur (v. 1.0.3)48 to identify cell types that exhibited the most transcriptional changes associated with aging. Pairwise comparisons were conducted for each experimental condition, assessing differences between (1) young and old females, (2) vehicle control (CTL) and VCD-treated groups, and (3) Foxl2+/+ and Foxl2+/− genotypes.
Pseudobulk analysis for differential gene expression
Pseudobulk differential gene expression analysis was conducted separately for three datasets: the Aging, VCD, and Foxl2 haploinsufficiency models. Single-cell transcriptomic data were aggregated at the cell type level using muscat (v. 1.18.0)97 to generate pseudobulk expression profiles. To ensure robust analysis, cell types were filtered based on dataset-specific criteria. In the Aging model, cell types with at least 25 cells per sample across all samples were retained. For the VCD model, cell types were included if they contained at least 25 cells in at least 12 samples, representing 75% of the dataset. In the Foxl2 haploinsufficiency model, the threshold was set at a minimum of 25 cells in at least 8 samples, or 80% of the dataset.
Differential gene expression analysis was performed using DESeq2 (v. 1.44.0)51, with comparisons made between young and old females in the Aging model, vehicle control (CTL) and VCD-treated samples in the VCD model, and Foxl2+/+ and Foxl2+/− samples in the Foxl2 haploinsufficiency model. Batch effects in the VCD and Foxl2 haploinsufficiency models were assessed using sva (v. 3.52.0)98 to determine the number of surrogate variables (SVs), and batch correction was applied using the removeBatchEffect() function from limma (v. 3.60.6)99. The adjusted counts were subsequently used for DESeq2 analysis, incorporating age-at-injection and time post-injection as covariates for the VCD model and age as a covariate for the Foxl2 haploinsufficiency model. Variance-stabilized transformed counts were computed using getVarianceStabilizedData(), and differentially expressed genes (FDR < 0.05) were visualized using strip plots. The complete pseudobulk DESeq2 output, including log fold changes, adjusted p-values, and base mean expression values for each dataset and cell type, can be found in Extended Data Table 6.
GO analysis for differential gene expression
Gene set enrichment analysis (GSEA) was performed to identify Gene Ontology Biological Process (GO BP) and Reactome terms enriched in differentially expressed genes across cell types in the Aging, VCD, and Foxl2 haploinsufficiency models. The analysis was conducted using the clusterProfiler package (v. 4.2.2)100 and the Molecular Signatures Database (MSigDB) gene sets retrieved via msigdbr (v. 7.4.1)101. Differential expression results from pseudobulk DESeq2 analysis were ranked by t-statistic and used as input for GSEA. Enrichment analysis was performed separately for each cell type in each dataset, with an FDR cutoff of 0.1 applied to identify significant terms. To identify recurrent biological processes across datasets, GO-BP terms enriched in at least four cell types within each dataset were extracted. The list of significantly enriched GO-BP terms (FDR < 0.1) for all datasets can be found in Extended Data Table 7.
GSEA of aging-associated transcriptional signatures across models
GSEA was performed using differentially expressed genes (DEGs) from the Aging model dataset as input gene sets with the clusterProfiler (v4.12.6)100 package. Upregulated and downregulated gene sets (FDR < 0.05) were extracted from the Aging model dataset and tested for enrichment in the Aging, VCD and Foxl2 haploinsufficiency model datasets. Genes were ranked based on the DESeq2-derived t-statistic for analysis. Additionally, GSEA was conducted to assess the enrichment of the SenMayo53 gene set within the dataset. Differential expression results from the Aging, VCD, and Foxl2 datasets were used for GSEA, with genes ranked using the DESeq2-derived t-statistic.
Coefficient of variation analysis
Gene expression variability across cell types was assessed by calculating the coefficient of variation (CV) for each gene in the Aging, VCD, and Foxl2 haploinsufficiency models. CV was computed as the ratio of the standard deviation to the mean expression level per gene within each condition (young vs. old for Aging, CTL vs. VCD for VCD, and Foxl2+/+ vs. Foxl2+/− for Foxl2 haploinsufficiency model) using variance-stabilized transformed counts obtained from DESeq2 (v1.44.0)51 pseudobulk analysis. Statistical differences in CV between groups were evaluated using the Wilcoxon rank-sum test.
Transposable element (TE) analysis
Transposable element (TE) expression was quantified using scTE (v1.0)56 with default parameters. BAM files generated from Cell Ranger (v. 7.1.0)84 outputs were processed using the mm10 genome index provided by scTE. Features detected by scTE but absent in the single-cell gene expression data were extracted and merged, generating a Seurat object containing both gene and TE expression profiles.
Cells were aggregated at the cell type level using muscat (v1.18.0)97, and differential expression analysis was performed using DESeq2 (v1.44.0)51, following the same approach as described above. The VCD and Foxl2 haploinsufficiency datasets underwent batch correction as before.
To assess functional enrichment of TE families, GSEA was conducted on ranked gene lists derived from DESeq2 results. TE annotations were obtained from the UCSC database for mm10, the same reference used by scTE. TE subfamilies were stratified to broader categories, including LINE, SINE, LTR, and DNA transposons. TE family annotations were obtained from the rmsk.txt file downloaded from the UCSC genome annotation database for the mm10 reference genome. For each cell type, genes were ranked by t-statistic from differential expression analysis, and enrichment of TE families was assessed using clusterProfiler (v4.12.6)100. Significant TE families (FDR < 0.1) were reported for visualization.
WGCNA and ORA enrichment analysis
Weighted gene co-expression network analysis (WGCNA)60 was performed on the Aging model dataset to identify gene co-expression modules. Variance-stabilized transformed counts from DESeq2 (v1.44.0)51 were used as input, with analyses conducted separately for each cell type. For each cell type, an optimal soft-thresholding power was determined using pickSoftThreshold, followed by the construction of an adjacency matrix and topological overlap matrix (TOM). Hierarchical clustering was performed to identify gene modules, which were assigned colors using cutreeDynamic, and eigengenes were computed to summarize module expression patterns. Gene sets from each module were classified into upregulated and downregulated groups based on eigengene expression differences between young and old samples. These gene sets were then subjected to GSEA via clusterProfiler (v. 4.2.2)100, using differential expression results from the VCD and Foxl2 haploinsufficiency model datasets to assess whether the identified modules were significantly enriched in these datasets. Modules significantly associated with aging (FDR < 0.1) were further analyzed through over-representation analysis using clusterProfiler (v. 4.2.2)100 and the org.Mm.eg.db annotation package.
Transcription factor activity inference using decoupleR
Transcription factor (TF) activity inference was performed using decoupleR (v2.10.0)61 with the CollecTRI transcriptional regulatory network for mouse. Differential expression results from the Aging, VCD, and Foxl2 haploinsufficiency models were used as input. For each dataset, fgsea scoring was applied to rank TF activity based on DESeq2-derived t-statistics, and significant TFs were identified at a threshold of FDR < 0.05. To compare TF activity across datasets, TFs were filtered to retain those with consistent activity direction (positive or negative) in at least two datasets.
R shiny application generation
An interactive R shiny application of Aging, VCD and Foxl2 haploinsufficiency scRNA-seq datasets was generated using ShinyCell (v. 2.1.0)102, and made available at https://minhooki.shinyapps.io/shinyappmulti/.
RNAscope sample preparation, imaging and data analysis
In situ Hybridization Protocol
Frozen ovaries were mounted in Tissue-Tek O.C.T Compound (Sakura, 4583) and sliced into 8 μm slices from the same ovarian region using PTFE-coated microtome blades (Duraedge, 7223) at −20°C on a Cryostat CM1860 (Leica, 14-0491-46884). 3 ovarian sections from the same region were mounted in succession on VWR Premium Superfrost Plus Microscope slides (VWR, 48311–703). All batches of sample processing were prepared using the fresh-frozen sample preparation protocol along with negative and positive controls (Advanced Cell Diagnostics, UM 323100-USM). 12 probes were tested in 3 sets, with Set 1 containing markers for theca (Cyp11a1; C1), granulosa (Fshr; C2), smooth muscle (Acta2; C3), and stromal (Pdgfra; C4) cells. Set 2 had markers for epithelial cells (Upk1b; C1), lymphatic endothelial cells (Prox1; C2), blood endothelial cells (Flt1; C3), and B cells (Cd19; C4). Set 3 contained probes for T (Cd3e; C1), natural killer (Klr1b1c; C2), CD8+ (Cd8b1; C3), and CD4+ (Cd4; C4) cells. Per the protocol, the RNAscope™ Multiplex Fluorescent Reagent Kit v2 (Advanced Cell Diagnostics, 323100) was used. The fluorescent signals TSA Vivid Fluorophore 520 (Advanced Cell Diagnostics, 323271), TSA Vivid Fluorophore 570 (Advanced Cell Diagnostics, 323272), TSA Vivid Fluorophore 650 (Advanced Cell Diagnostics, 323273), and Opal 690 (Akoya Biosciences, FP1497001KT) were used to stain the probes in each set. The slides were preserved in ProLong Gold Antifade Mountant (Thermo Fisher Scientific, P36930).
Imaging
Imaging was performed using a Leica Stellaris 5 confocal microscope (Leica Microsystems) equipped with a HC PL APO 20x/0.75 IMM CORR CS2 objective (Leica, 11506343) and Leica Microsystems Type F Immersion liquid oil (Leica Microsystems, 11513859). Images were acquired using Leica Application Suite X software (LAS X v4.4.0).
Imaging analysis
To quantify probe expression, the images were analyzed using the QuPath program v0.5.1. Cell boundaries based on DAPI signal were calculated using the following settings modified from the default parameters – Requested pixel size: 0 μm, Nucleus background radius: 10 μm, Maximum area: 400 μm2, Threshold: 0.5, and Cell expansion: 2. Sets 1 and 2 were analyzed using subcellular detection to determine their expression levels relative to the number of cells. All thresholding values were uniform across the date of processing of each ovarian tissue. For Set 3, single measurement classifiers were created for each probe to generate colocalization data. Cells below the detection threshold were not counted for analysis.
Cell-type proportion analysis using flow cytometry
Single-cell suspensions of ovarian cells were prepared following the same protocol used for single-cell RNA sequencing sample preparation, as described above. Cells were stained for APC-CD45 antibody (Miltenyi, 130-123-784) according to the manufacturer’s recommendations. Flow cytometry data were acquired using the MACSQuant10 flow cytometer (Miltenyi Biotec, 130-096-343) and analyzed using FlowLogic v.8 (Inivai Technologies). The raw flow cytometry dataset was deposited to Figshare (DOI: 10.6084/m9.figshare.28733222).
Granulosa and theca transcriptional clock model training and feature assessment
Granulosa and theca cells were independently subsetted from scRNA-seq datasets encompassing multiple experimental models, including natural aging, VCD, and Foxl2 haploinsufficiency. Additionally, an independent middle-aged female dataset (13, 76, and 86 weeks) was included to ensure age coverage for intermediate ages not well represented in the datasets.
To develop the cell transcriptional clocks, cells from aging model, middle-aged females, vehicle controls from the VCD model, and Foxl2+/+ animals from the Foxl2 haploinsufficiency model were extracted from the dataset. These cells were used to train and evaluate the model. Data were partitioned into training (75%) and testing (25%) sets using stratified sampling, using the createDataPartition function from the caret package (v. 6.0–91). Training data were used for model optimization, while the test set served as an independent validation dataset.
Predictive models were trained using L1-regularized lasso regression with the glmnet package (v. 4.1–7) to estimate transcriptional age (in weeks) based on gene expression profiles. Five-fold nested cross-validation was performed on the training dataset, and the optimal regularization parameter (lambda.min) was determined based on cross-validation results. Spearman correlation analysis was performed to assess the correlation between predicted and chronological age.
Transcriptional clocks were applied to experimental test groups, including VCD-treated and Foxl2+/− animals. Model predictions for test groups were compared to those of control samples to assess differences in predicted age patterns. Additionally, age acceleration was calculated as the difference between predicted and chronological age. Statistical significance of group differences in predicted age and age acceleration was assessed using the Wilcoxon rank-sum test.
To examine the age-associated expression dynamics of lasso features selected by the models, we performed pseudobulk analysis using control samples. For each cell type, raw counts were aggregated per sample using muscat (v. 1.18.0)97, and DESeq2 (v. 1.44.0)51 was used to compute variance-stabilized transformed expression values. Batch effects were corrected using removeBatchEffect, and expression heatmaps of lasso-selected features were generated to visualize age-dependent expression trends.
The top 500 lasso features were selected based on the absolute values of their coefficients. Over-representation analysis was performed using clusterProfiler (v. 4.2.2) and org.Mm.eg.db (v. 3.14.0), analyzing “ALL” Gene Ontology terms. A p-value cutoff of 0.05 was applied to identify significantly enriched pathways.
Quantification and statistical analysis
All statistical analysis was performed using the R software, version 4.1.2. For all boxplots, the data is shown with the median, the 25th and 75th percentile of the data, and the whiskers represent 1.5 * the inter-quartile range (IQR). Individual datapoints are overlayed, when possible, for transparency and rigor. Specific statistical tests, number of biological replicates and animals used are indicated in the corresponding figure legends and associated methods.
Supplementary Material
Extended Data Table 1. Raw fertility data from Foxl2 haploinsufficiency model mice.
Extended Data Table 2. Raw serum AMH, FSH and INHBA quantification data. Raw data from a, Aging, b, VCD and c, Foxl2 haploinsufficiency models.
Extended Data Table 3. 10xGenomics CellRanger QC metrix.
Extended Data Table 4. List of canonical marker genes used to annotate cell types in scRNA-seq datasets.
Extended Data Table 5. Relative proportion of detected cell types from scRNA-seq datasets. Proportion data from a, Aging, b, VCD and c, Foxl2 haploinsufficiency models.
Extended Data Table 6. DESeq2 output from pseudobulk analysis. DESeq2 output from a, Aging, b, VCD and c, Foxl2 haploinsufficiency model datasets.
Extended Data Table 7. Significant GSEA enrichment terms (FDR < 10%) for GO Biological Process and Reactome terms. GSEA enrichment terms for Biological Process from a, Aging, b, VCD and c, Foxl2 haploinsufficiency model datasets. GSEA enrichment terms for Reactome from d, Aging, e, VCD and f, Foxl2 haploinsufficiency model datasets.
Extended Data Table 8. List of decoupleR TF activity scores (FDR < 5%) for each cell type.
Extended Data Table 9. Primers used in the study. a, Primers used for genotyping Foxl2 haploinsufficiency model animals. b, Primers used for Foxl2 expression quantification in Foxl2 haploinsufficiency model animals via RT-qPCR.
Extended Data Fig. 1. Characterization of the Foxl2 haploinsufficiency model. a, Schematic representation of constructs for Foxl2 haploinsufficiency mouse line generation. b, Foxl2 expression levels in Foxl2+/− and Foxl2+/− mice ovaries from young and mid-age groups, measured by RT-qPCR. P-values were calculated using the Wilcoxon test. c-d, Fertility assessment of Foxl2 haploinsufficiency females, comparing first litter pup counts and latency to first pregnancy. P-values were calculated using the Wilcoxon and log-rank tests, respectively.
Extended Data Fig. 2. Follicle counts across Aging, VCD and Foxl2 haploinsufficiency models. a-c, Follicle counts for primordial, primary, secondary and antral follicles, and corpus luteum, from Aging, VCD and Foxl2 haploinsufficiency model mice. P-values were calculated using the Wilcoxon test. Illustrations of ovarian follicles were obtained from Generic Diagramming Platform78.
Extended Data Fig. 3. Longitudinal analysis of serum AMH, FSH, and Inhibin A levels in the VCD model. a-c, Serum levels of AMH, FSH, and Inhibin A (INHBA) measured from 0 to 5 months post-injection in VCD model mice.
Extended Data Fig. 4. Histological characterization of the VCD model mice. a, Representative hematoxylin and eosin staining images of ovarian tissues from condition-matched animals used in the VCD model scRNA-seq analysis. b, Follicle counts for the condition-matched animals used in the VCD model scRNA-seq analysis. Satistical significance was assessed using the Wilcoxon test, and p-values are reported.
Extended Data Fig. 4. Serum hormone quantification and ovarian health index calculation of the VCD model mice. a-c, Serum levels of AMH, FSH, and Inhibin A (INHBA) measured from condition-matched animals used in the VCD model scRNA-seq analysis. d, Adjusted FSH values to account for inter-assay differences between measurement assays. e, Ovarian health index from condition-matched animals used in the VCD model scRNA-seq analysis. Statistical significance was assessed using the Wilcoxon test, and p-values are reported.
Extended Data Fig. 6. Validation of detected ovarian cell types by in situ hybridization. a, Representative RNAscope images for Fshr, Cyp11a1, Pdgfra and Acta2 probes. b, Representative RNAscope images for Upk1b, Cd19, Flt1 and Prox1 probes. c, Representative RNAscope images for Klrb1c, Cd3e, Cd8b2 and Cd4. Images were enhanced for visualization. d, Heatmap of detected cell types in the Aging, VCD and Foxl2 haploinsufficiency scRNA-seq datasets. For panels a-c, shown images are from a middle-aged Foxl2+/+ animal.
Extended Data Fig. 7. Cell type proportion analysis of ovarian cells via flow cytometry and scRNA-seq analysis. a, Gating strategy of CD45+ cells in flow cytometry analysis. b-d, Proportion differences of Ptprc+ cell types detected in Aging, VCD and Foxl2 haploinsufficiency model scRNA-seq datasets. Asterisks (*) indicate comparisons in which the observed log2 fold change is infinite due to complete absence of the cell type in one of the groups.
Extended Data Fig. 8. Cell type proportion analysis of non-immune ovarian cells via in situ hybridization assays. a-c, Cell type abundance analysis of stromal, SMC, BEC, LEC and epithelial cells via RNAscope in situ hybridization assays from Aging, VCD and Foxl2 haploinsufficiency model mice.
Extended Data Fig. 9. Cell type proportion analysis of immune ovarian cells via in situ hybridization assays. a-c, Cell type abundance analysis of NK, NKT, CD8+ NKT, CD8+ T, CD4+ T, DNT, DPT and B cells via RNAscope in situ hybridization assays from Aging, VCD and Foxl2 haploinsufficiency model mice.
Extended Data Fig. 10. Comparative analysis of global gene expression in VCD model via Augur. a-b, UMAP visualization of AUC scores and scatter plot of AUC quantification in the VCD model, comparing CTL vs. VCD at 3m and 10m age-at-injection. For the scatter plot, Ptprc− and Ptprc+ cells were separately plotted to improve visualization. All data are from the 30d post-injection timepoint. For b, data points with NA AUC scores (due to low cell count or QC filtering) were assigned an AUC of 0.5 and colored gray to improve visualization.
Extended Data Fig 11. Aging-associated signatures and transposable element enrichment analysis across Aging, VCD and Foxl2 haploinsufficiency models. a, Heatmap of cell types detected in pseudobulked scRNA-seq datasets. b, DESeq2 normalized log2 counts of Foxl2 from young and mid-age Foxl2+/+ and Foxl2+/− mice. c-d, GSEA enrichment analysis of differentially expressed genes (DEGs) identified in the Aging model (FDR < 0.05) and SenMayo gene list. e, Dot plot showing the coefficient of variation analysis results. f, GSEA enrichment analysis of transposable element (TE) families across Aging, VCD and Foxl2 haploinsufficiency models.
Extended Data Fig 12. Module trees of WGCNA analysis. a, WGCNA module trees identified in the Aging model for granulosa, theca, stromal, BEC, epithelial, and DNT cells.
Extended Data Fig 13. Association analysis of WGCNA modules and ovarian aging traits from the Aging model. a, Heatmap of trait significance for WGCNA modules in the Aging model. Modules that passed the FDR < 0.1 threshold are shown in bold.
Extended Data Fig 14. Analysis of recurrent transcription factor activity inferred by decoupleR. a-c, Heatmap of transcription factor (TF) activity for factors detected in at least three cell types within each model with an FDR < 0.05.
Extended Data Fig 15. Development of a transcriptome-based aging clock for theca cells. a, Schematic of analysis and training workflow for the lasso regression-based clock. b, Scatter plot of predicted age vs. chronological age for the test set, with Spearman’s correlation and p-value displayed. c-d, Age prediction for VCD and Foxl2 haploinsufficiency datasets, comparing predicted age to chronological age in weeks. Offsets were added for visualization purposes. e-f, Age acceleration analysis for VCD and Foxl2 haploinsufficiency datasets, calculated as the difference between predicted and chronological age. Statistical significance was assessed using the Wilcoxon test. f-g, ORA enrichment analysis of GO ALL and Reactome terms of the top 500 lasso features.
Acknowledgements
We thank Dr. Gilbert Garcia for his feedback on confocal imaging. We also thank Dr. Lynae M. Brayboy for providing the ovarian single-cell dissociation protocol and for her valuable feedback on optimization. We thank Kristen Mehalko for maintaining the Foxl2 haploinsufficiency colony and contributing to mouse tissue collection. We thank Younggyun Kim, Kelly Koh, and Sanjana Paye for assisting with follicle counting from hematoxylin and eosin-stained ovarian sections. We thank Dr. Victor Ansere, Clayton Baker and Aaron J.J. Lemus for feedback on our manuscript.
This work was supported by the GCRLE-2020 post-doctoral fellowship from the Global Consortium for Reproductive Longevity and Equality at the Buck Institute, made possible by the Bia-Echo Foundation to M.K.; USC Provost's Undergraduate Research Fellowship to J.W.; a Diana Jacobs Kalman/AFAR Scholarships for Research in the Biology of Aging, NIA T32 AG052374, and AthenaDAO student leader award to R.J.L.; GCRLE-0520 Junior Scholar Award from the Global Consortium for Reproductive Longevity and Equality at the Buck Institute, made possible by the Bia-Echo Foundation, Pew Biomedical Scholar award #00034120 from the Pew Charitable Trust, and generous gifts from Kathleen Gilmore and Dr. Eric Hennigan to B.A.B.
Ovarian histological analysis was performed by the Translational Pathology Core at the USC Norris Comprehensive Cancer Center (supported by NCI P30 CA014089). Serum hormone quantification was performed by the University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core (supported by the Eunice Kennedy Shriver NICHD/NIH Grant R24HD102061).
Some panels were generated using the Generic Diagramming Platform78.
Footnotes
Declaration of interests
The authors declare no competing interests.
Data availability
The raw FASTQ files have been deposited in the Sequence Read Archive under accession number PRJNA863443. Raw microscopy images of ovarian hematoxylin and eosin (H&E) staining, compressed RNAscope images (due to file size constraints), and raw flow cytometry datasets are available on Figshare (DOIs: 10.6084/m9.figshare.28745765, 10.6084/m9.figshare.28745792, 10.6084/m9.figshare.28745813, 10.6084/m9.figshare.29260391 and 10.6084/m9.figshare.28733222). The final annotated Seurat objects have been deposited to Figshare (10.6084/m9.figshare.28934126) and can be explored via an interactive R shiny application: https://minhooki.shinyapps.io/shinyappmulti/. All analyses were performed using R version 4.1.2. Any additional information required to reproduce or reanalyze the data presented in this study is available from the corresponding author upon reasonable request.
Code availability
All new scripts used to analyze the datasets are available on the Benayoun lab GitHub at https://github.com/BenayounLaboratory/Mouse_Menopause_Models.
References
- 1.Davis S. R. et al. Menopause. Nat Rev Dis Primers 1, 15004, doi: 10.1038/nrdp.2015.4 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Broekmans F. J., Soules M. R. & Fauser B. C. Ovarian aging: mechanisms and clinical consequences. Endocr Rev 30, 465–493, doi: 10.1210/er.2009-0006 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Faddy M. J. & Gosden R. G. A model conforming the decline in follicle numbers to the age of menopause in women. Hum Reprod 11, 1484–1486, doi: 10.1093/oxfordjournals.humrep.a019422 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Ossewaarde M. E. et al. Age at menopause, cause-specific mortality and total life expectancy. Epidemiology 16, 556–562, doi: 10.1097/01.ede.0000165392.35273.d4 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Hong J. S. et al. Age at menopause and cause-specific mortality in South Korean women: Kangwha Cohort Study. Maturitas 56, 411–419, doi: 10.1016/j.maturitas.2006.11.004 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Shadyab A. H. et al. Ages at menarche and menopause and reproductive lifespan as predictors of exceptional longevity in women: the Women's Health Initiative. Menopause 24, 35–44, doi: 10.1097/GME.0000000000000710 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gubbels Bupp M. R. Sex, the aging immune system, and chronic disease. Cell Immunol 294, 102–110, doi: 10.1016/j.cellimm.2015.02.002 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Santoro N., Roeca C., Peters B. A. & Neal-Perry G. The Menopause Transition: Signs, Symptoms, and Management Options. J Clin Endocrinol Metab 106, 1–15, doi: 10.1210/clinem/dgaa764 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Randolph J. F. Jr. et al. Change in follicle-stimulating hormone and estradiol across the menopausal transition: effect of age at the final menstrual period. J Clin Endocrinol Metab 96, 746–754, doi: 10.1210/jc.2010-1746 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelsey T. W., Wright P., Nelson S. M., Anderson R. A. & Wallace W. H. A validated model of serum anti-mullerian hormone from conception to menopause. PLoS One 6, e22024, doi: 10.1371/journal.pone.0022024 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiss G., Skurnick J. H., Goldsmith L. T., Santoro N. F. & Park S. J. Menopause and hypothalamic-pituitary sensitivity to estrogen. JAMA 292, 2991–2996, doi: 10.1001/jama.292.24.2991 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Xiong J. et al. FSH blockade improves cognition in mice with Alzheimer's disease. Nature 603, 470–476, doi: 10.1038/s41586-022-04463-0 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao L., Wang L., Bennett S., Xu J. & Zou J. Effects of follicle-stimulating hormone on fat metabolism and cognitive impairment in women during menopause. Front Physiol 13, 1043237, doi: 10.3389/fphys.2022.1043237 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Kempen T. A., Milner T. A. & Waters E. M. Accelerated ovarian failure: a novel, chemically induced animal model of menopause. Brain Res 1379, 176–187, doi: 10.1016/j.brainres.2010.12.064 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blenck C. L., Harvey P. A., Reckelhoff J. F. & Leinwand L. A. The Importance of Biological Sex and Estrogen in Rodent Models of Cardiovascular Health and Disease. Circ Res 118, 1294–1312, doi: 10.1161/CIRCRESAHA.116.307509 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laszczynska M., Brodowska A., Starczewski A., Masiuk M. & Brodowski J. Human postmenopausal ovary--hormonally inactive fibrous connective tissue or more? Histol Histopathol 23, 219–226, doi: 10.14670/HH-23.219 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Grub J., Suss H., Willi J. & Ehlert U. Steroid Hormone Secretion Over the Course of the Perimenopause: Findings From the Swiss Perimenopause Study. Front Glob Womens Health 2, 774308, doi: 10.3389/fgwh.2021.774308 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burger H. G., Dudley E. C., Robertson D. M. & Dennerstein L. Hormonal changes in the menopause transition. Recent Prog Horm Res 57, 257–275, doi: 10.1210/rp.57.1.257 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Mangoni A. A. & Jackson S. H. Age-related changes in pharmacokinetics and pharmacodynamics: basic principles and practical applications. Br J Clin Pharmacol 57, 6–14, doi: 10.1046/j.1365-2125.2003.02007.x (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conboy I. M. & Rando T. A. Heterochronic parabiosis for the study of the effects of aging on stem cells and their niches. Cell Cycle 11, 2260–2267, doi: 10.4161/cc.20437 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chahal H. S. & Drake W. M. The endocrine system and ageing. J Pathol 211, 173–180, doi: 10.1002/path.2110 (2007). [DOI] [PubMed] [Google Scholar]
- 22.van den Beld A. W. et al. The physiology of endocrine systems with ageing. Lancet Diabetes Endocrinol 6, 647–658, doi: 10.1016/S2213-8587(18)30026-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rando T. A. & Jones D. L. Regeneration, Rejuvenation, and Replacement: Turning Back the Clock on Tissue Aging. Cold Spring Harb Perspect Biol 13, doi: 10.1101/cshperspect.a040907 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villeda S. A. et al. The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature 477, 90–94, doi: 10.1038/nature10357 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uda M. et al. Foxl2 disruption causes mouse ovarian failure by pervasive blockage of follicle development. Hum Mol Genet 13, 1171–1181, doi: 10.1093/hmg/ddh124 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Schmidt D. et al. The murine winged-helix transcription factor Foxl2 is required for granulosa cell differentiation and ovary maintenance. Development 131, 933–942, doi: 10.1242/dev.00969 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Uhlenhaut N. H. et al. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell 139, 1130–1142, doi: 10.1016/j.cell.2009.11.021 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Benayoun B. A., Dipietromaria A., Bazin C. & Veitia R. A. FOXL2: at the crossroads of female sex determination and ovarian function. Adv Exp Med Biol 665, 207–226, doi: 10.1007/978-1-4419-1599-3_16 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Gersak K., Harris S. E., Smale W. J. & Shelling A. N. A novel 30 bp deletion in the FOXL2 gene in a phenotypically normal woman with primary amenorrhoea: case report. Hum Reprod 19, 2767–2770, doi: 10.1093/humrep/deh496 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Harris S. E. et al. Identification of novel mutations in FOXL2 associated with premature ovarian failure. Mol Hum Reprod 8, 729–733, doi: 10.1093/molehr/8.8.729 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Laissue P. et al. Functional evidence implicating FOXL2 in non-syndromic premature ovarian failure and in the regulation of the transcription factor OSR2. J Med Genet 46, 455–457, doi: 10.1136/jmg.2008.065086 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Borman S. M., Christian P. J., Sipes I. G. & Hoyer P. B. Ovotoxicity in female Fischer rats and B6 mice induced by low-dose exposure to three polycyclic aromatic hydrocarbons: comparison through calculation of an ovotoxic index. Toxicol Appl Pharmacol 167, 191–198, doi: 10.1006/taap.2000.9006 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Finch C. E. The menopause and aging, a comparative perspective. J Steroid Biochem Mol Biol 142, 132–141, doi: 10.1016/j.jsbmb.2013.03.010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gosden R. G. & Faddy M. J. Ovarian aging, follicular depletion, and steroidogenesis. Exp Gerontol 29, 265–274, doi: 10.1016/0531-5565(94)90006-x (1994). [DOI] [PubMed] [Google Scholar]
- 35.Lliberos C. et al. Evaluation of inflammation and follicle depletion during ovarian ageing in mice. Sci Rep 11, 278, doi: 10.1038/s41598-020-79488-4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sokalska A. & Valentin L. Changes in ultrasound morphology of the uterus and ovaries during the menopausal transition and early postmenopause: a 4-year longitudinal study. Ultrasound Obstet Gynecol 31, 210–217, doi: 10.1002/uog.5241 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Jiao X. et al. Ovarian Reserve Markers in Premature Ovarian Insufficiency: Within Different Clinical Stages and Different Etiologies. Front Endocrinol (Lausanne) 12, 601752, doi: 10.3389/fendo.2021.601752 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Danforth D. R. et al. Dimeric inhibin: a direct marker of ovarian aging. Fertil Steril 70, 119–123, doi: 10.1016/s0015-0282(98)00127-7 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Kevenaar M. E. et al. Serum anti-mullerian hormone levels reflect the size of the primordial follicle pool in mice. Endocrinology 147, 3228–3234, doi: 10.1210/en.2005-1588 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Park M., Suh D. S., Lee K. & Bae J. Positive cross talk between FOXL2 and antimullerian hormone regulates ovarian reserve. Fertil Steril 102, 847–855 e841, doi: 10.1016/j.fertnstert.2014.05.031 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Kim M. et al. Estropausal gut microbiota transplant improves measures of ovarian function in adult mice. bioRxiv, doi: 10.1101/2024.05.03.592475 (2025). [DOI] [Google Scholar]
- 42.Mehalko K. et al. Lack of accelerated ovarian aging in a follicle-stimulating hormone receptor haploinsufficiency model. Transl Med Aging 7, 1–8, doi: 10.1016/j.tma.2023.01.001 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crisponi L. et al. The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nat Genet 27, 159–166, doi: 10.1038/84781 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Isola J. V. V. et al. A single-cell atlas of the aging mouse ovary. Nat Aging 4, 145–162, doi: 10.1038/s43587-023-00552-5 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X. et al. Transcriptomic signatures of mouse ovarian aging and estropausal transition at single cell resolution. bioRxiv, 2024.2008.2012.607592, doi: 10.1101/2024.08.12.607592 (2024). [DOI] [Google Scholar]
- 46.Jin C. et al. Molecular and genetic insights into human ovarian aging from single-nuclei multi-omics analyses. Nat Aging 5, 275–290, doi: 10.1038/s43587-024-00762-5 (2025). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ben Yaakov T., Wasserman T., Aknin E. & Savir Y. Single-cell analysis of the aged ovarian immune system reveals a shift towards adaptive immunity and attenuated cell function. Elife 12, doi: 10.7554/eLife.74915 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Squair J. W., Skinnider M. A., Gautier M., Foster L. J. & Courtine G. Prioritization of cell types responsive to biological perturbations in single-cell data with Augur. Nat Protoc 16, 3836–3873, doi: 10.1038/s41596-021-00561-x (2021). [DOI] [PubMed] [Google Scholar]
- 49.Xu X. et al. Imaging and tracing the pattern of adult ovarian angiogenesis implies a strategy against female reproductive aging. Sci Adv 8, eabi8683, doi: 10.1126/sciadv.abi8683 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu H. et al. Aging hallmarks of the primate ovary revealed by spatiotemporal transcriptomics. Protein Cell 15, 364–384, doi: 10.1093/procel/pwad063 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Love M. I., Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gillespie M. et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res 50, D687–D692, doi: 10.1093/nar/gkab1028 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saul D. et al. A new gene set identifies senescent cells and predicts senescence-associated pathways across tissues. Nat Commun 13, 4827, doi: 10.1038/s41467-022-32552-1 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hajdarovic K. H. et al. Single-cell analysis of the aging female mouse hypothalamus. Nat Aging 2, 662–678, doi: 10.1038/s43587-022-00246-4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harris S. E. et al. Age-related gene expression changes, and transcriptome wide association study of physical and cognitive aging traits, in the Lothian Birth Cohort 1936. Aging (Albany NY) 9, 2489–2503, doi: 10.18632/aging.101333 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He J. et al. Identifying transposable element expression dynamics and heterogeneity during development at the single-cell level with a processing pipeline scTE. Nat Commun 12, 1456, doi: 10.1038/s41467-021-21808-x (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bravo J. I., Nozownik S., Danthi P. S. & Benayoun B. A. Transposable elements, circular RNAs and mitochondrial transcription in age-related genomic regulation. Development 147, doi: 10.1242/dev.175786 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Cecco M. et al. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging (Albany NY) 5, 867–883, doi: 10.18632/aging.100621 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herr L. M., Schaffer E. D., Fuchs K. F., Datta A. & Brosh R. M. Jr. Replication stress as a driver of cellular senescence and aging. Commun Biol 7, 616, doi: 10.1038/s42003-024-06263-w (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langfelder P. & Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559, doi: 10.1186/1471-2105-9-559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Badia I. M. P. et al. decoupleR: ensemble of computational methods to infer biological activities from omics data. Bioinform Adv 2, vbac016, doi: 10.1093/bioadv/vbac016 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang Q. et al. NADase CD38 is a key determinant of ovarian aging. Nat Aging 4, 110–128, doi: 10.1038/s43587-023-00532-9 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dipali S. S. et al. Proteomic quantification of native and ECM-enriched mouse ovaries reveals an age-dependent fibro-inflammatory signature. Aging (Albany NY) 15, 10821–10855, doi: 10.18632/aging.205190 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ouni E. et al. A blueprint of the topology and mechanics of the human ovary for next-generation bioengineering and diagnosis. Nat Commun 12, 5603, doi: 10.1038/s41467-021-25934-4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Y. et al. PBX1 Attenuates Hair Follicle-Derived Mesenchymal Stem Cell Senescence and Apoptosis by Alleviating Reactive Oxygen Species-Mediated DNA Damage Instead of Enhancing DNA Damage Repair. Front Cell Dev Biol 9, 739868, doi: 10.3389/fcell.2021.739868 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lichtenauer U. D. et al. Pre-B-cell transcription factor 1 and steroidogenic factor 1 synergistically regulate adrenocortical growth and steroidogenesis. Endocrinology 148, 693–704, doi: 10.1210/en.2006-0681 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Shen L., Liu J., Luo A. & Wang S. The stromal microenvironment and ovarian aging: mechanisms and therapeutic opportunities. J Ovarian Res 16, 237, doi: 10.1186/s13048-023-01300-4 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hao X., Reyes Palomares A., Anastacio A., Liu K. & Rodriguez-Wallberg K. A. Evidence of apoptosis as an early event leading to cyclophosphamide-induced primordial follicle depletion in a prepubertal mouse model. Front Endocrinol (Lausanne) 15, 1322592, doi: 10.3389/fendo.2024.1322592 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ravelojaona M. et al. Oncostatin M and STAT3 Signaling Pathways Support Human Trophoblast Differentiation by Inhibiting Inflammatory Stress in Response to IFNgamma and GM-CSF. Cells 13, doi: 10.3390/cells13030229 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Devchand P. R., Ziouzenkova O. & Plutzky J. Oxidative stress and peroxisome proliferator-activated receptors: reversing the curse? Circ Res 95, 1137–1139, doi: 10.1161/01.RES.0000151331.69399.b2 (2004). [DOI] [PubMed] [Google Scholar]
- 71.Schumann K. et al. Functional CRISPR dissection of gene networks controlling human regulatory T cell identity. Nat Immunol 21, 1456–1466, doi: 10.1038/s41590-020-0784-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Klein-Hessling S. et al. NFATc1 controls the cytotoxicity of CD8(+) T cells. Nat Commun 8, 511, doi: 10.1038/s41467-017-00612-6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kostel Bal S. et al. Biallelic NFATC1 mutations cause an inborn error of immunity with impaired CD8+ T-cell function and perturbed glycolysis. Blood 142, 827–845, doi: 10.1182/blood.2022018303 (2023). [DOI] [PubMed] [Google Scholar]
- 74.Buckley M. T. et al. Cell-type-specific aging clocks to quantify aging and rejuvenation in neurogenic regions of the brain. Nat Aging 3, 121–137, doi: 10.1038/s43587-022-00335-4 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peters M. J. et al. The transcriptional landscape of age in human peripheral blood. Nat Commun 6, 8570, doi: 10.1038/ncomms9570 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang S. et al. Single-Cell Transcriptomic Atlas of Primate Ovarian Aging. Cell 180, 585–600 e519, doi: 10.1016/j.cell.2020.01.009 (2020). [DOI] [PubMed] [Google Scholar]
- 77.Bafor E. E. et al. Aberrant CD8(+)T cells drive reproductive dysfunction in female mice with elevated IFN-gamma levels. Front Immunol 15, 1368572, doi: 10.3389/fimmu.2024.1368572 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang S. et al. Generic Diagramming Platform (GDP): a comprehensive database of high-quality biomedical graphics. Nucleic Acids Res 53, D1670–D1676, doi: 10.1093/nar/gkae973 (2025). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Komsta L. outliers: Tests for Outliers., <https://CRAN.R-project.org/package=outliers> (2022).
- 80.Therneau T. A Package for Survival Analysis in R, <https://CRAN.R-project.org/package=survival> (2022).
- 81.Ongaro L. et al. Development of a Highly Sensitive ELISA for Measurement of FSH in Serum, Plasma, and Whole Blood in Mice. Endocrinology 162, doi: 10.1210/endocr/bqab014 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kuhn M. caret: Classification and Regression Training. (2022).
- 83.Breiman L., Cutler A., Liaw A. & Wiener M. Breiman and Cutlers Random Forests for Classification and Regression, <https://www.stat.berkeley.edu/~breiman/RandomForests/> (
- 84.Zheng G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun 8, 14049, doi: 10.1038/ncomms14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Campbell J., Yang S., Wang Z., Corbett S. & Koga Y. celda: CEllular Latent Dirichlet Allocation, 2023).
- 86.DoubletFinder: DoubletFinder is a suite of tools for identifying doublets in single-cell RNA sequencing data, 2022).
- 87.Bais A. & Kostkam D. scds: Computational Annotation of Doublets in Single Cell RNA Sequending Data. (2019). [DOI] [PMC free article] [PubMed]
- 88.Hao Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 e3529, doi: 10.1016/j.cell.2021.04.048 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Korsunsky I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 16, 1289–1296, doi: 10.1038/s41592-019-0619-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Andreatta M., Berenstein A. J. & Carmona S. J. scGate: marker-based purification of cell types from heterogeneous single-cell RNA-seq datasets. Bioinformatics 38, 2642–2644, doi: 10.1093/bioinformatics/btac141 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aran D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 20, 163–172, doi: 10.1038/s41590-018-0276-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guo H. & Li J. scSorter: assigning cells to known cell types according to marker genes. Genome Biol 22, 69, doi: 10.1186/s13059-021-02281-7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nader K. et al. ScType enables fast and accurate cell type identification from spatial transcriptomics data. Bioinformatics 40, doi: 10.1093/bioinformatics/btae426 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Heng T. S., Painter M. W. & Immunological Genome Project C. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 9, 1091–1094, doi: 10.1038/ni1008-1091 (2008). [DOI] [PubMed] [Google Scholar]
- 95.Morris M. E. et al. A single-cell atlas of the cycling murine ovary. Elife 11, doi: 10.7554/eLife.77239 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Miller S. A. et al. LSD1 and Aberrant DNA Methylation Mediate Persistence of Enteroendocrine Progenitors That Support BRAF-Mutant Colorectal Cancer. Cancer Res 81, 3791–3805, doi: 10.1158/0008-5472.CAN-20-3562 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Crowell H. L. et al. muscat detects subpopulation-specific state transitions from multi-sample multi-condition single-cell transcriptomics data. Nat Commun 11, 6077, doi: 10.1038/s41467-020-19894-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Leek J. et al. sva: Surrogate Variable Analysis.
- 99.Ritchie M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47, doi: 10.1093/nar/gkv007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wu T. et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2, 100141, doi: 10.1016/j.xinn.2021.100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dolgalev I. msigdbr: MSigDB Gene Sets for Multiple Organisms in a Tidy Data Format, <https://CRAN.R-project.org/package=msigdbr> (2021).
- 102.Ouyang J. F., Kamaraj U. S., Cao E. Y. & Rackham O. J. L. ShinyCell: simple and sharable visualization of single-cell gene expression data. Bioinformatics 37, 3374–3376, doi: 10.1093/bioinformatics/btab209 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Extended Data Table 1. Raw fertility data from Foxl2 haploinsufficiency model mice.
Extended Data Table 2. Raw serum AMH, FSH and INHBA quantification data. Raw data from a, Aging, b, VCD and c, Foxl2 haploinsufficiency models.
Extended Data Table 3. 10xGenomics CellRanger QC metrix.
Extended Data Table 4. List of canonical marker genes used to annotate cell types in scRNA-seq datasets.
Extended Data Table 5. Relative proportion of detected cell types from scRNA-seq datasets. Proportion data from a, Aging, b, VCD and c, Foxl2 haploinsufficiency models.
Extended Data Table 6. DESeq2 output from pseudobulk analysis. DESeq2 output from a, Aging, b, VCD and c, Foxl2 haploinsufficiency model datasets.
Extended Data Table 7. Significant GSEA enrichment terms (FDR < 10%) for GO Biological Process and Reactome terms. GSEA enrichment terms for Biological Process from a, Aging, b, VCD and c, Foxl2 haploinsufficiency model datasets. GSEA enrichment terms for Reactome from d, Aging, e, VCD and f, Foxl2 haploinsufficiency model datasets.
Extended Data Table 8. List of decoupleR TF activity scores (FDR < 5%) for each cell type.
Extended Data Table 9. Primers used in the study. a, Primers used for genotyping Foxl2 haploinsufficiency model animals. b, Primers used for Foxl2 expression quantification in Foxl2 haploinsufficiency model animals via RT-qPCR.
Extended Data Fig. 1. Characterization of the Foxl2 haploinsufficiency model. a, Schematic representation of constructs for Foxl2 haploinsufficiency mouse line generation. b, Foxl2 expression levels in Foxl2+/− and Foxl2+/− mice ovaries from young and mid-age groups, measured by RT-qPCR. P-values were calculated using the Wilcoxon test. c-d, Fertility assessment of Foxl2 haploinsufficiency females, comparing first litter pup counts and latency to first pregnancy. P-values were calculated using the Wilcoxon and log-rank tests, respectively.
Extended Data Fig. 2. Follicle counts across Aging, VCD and Foxl2 haploinsufficiency models. a-c, Follicle counts for primordial, primary, secondary and antral follicles, and corpus luteum, from Aging, VCD and Foxl2 haploinsufficiency model mice. P-values were calculated using the Wilcoxon test. Illustrations of ovarian follicles were obtained from Generic Diagramming Platform78.
Extended Data Fig. 3. Longitudinal analysis of serum AMH, FSH, and Inhibin A levels in the VCD model. a-c, Serum levels of AMH, FSH, and Inhibin A (INHBA) measured from 0 to 5 months post-injection in VCD model mice.
Extended Data Fig. 4. Histological characterization of the VCD model mice. a, Representative hematoxylin and eosin staining images of ovarian tissues from condition-matched animals used in the VCD model scRNA-seq analysis. b, Follicle counts for the condition-matched animals used in the VCD model scRNA-seq analysis. Satistical significance was assessed using the Wilcoxon test, and p-values are reported.
Extended Data Fig. 4. Serum hormone quantification and ovarian health index calculation of the VCD model mice. a-c, Serum levels of AMH, FSH, and Inhibin A (INHBA) measured from condition-matched animals used in the VCD model scRNA-seq analysis. d, Adjusted FSH values to account for inter-assay differences between measurement assays. e, Ovarian health index from condition-matched animals used in the VCD model scRNA-seq analysis. Statistical significance was assessed using the Wilcoxon test, and p-values are reported.
Extended Data Fig. 6. Validation of detected ovarian cell types by in situ hybridization. a, Representative RNAscope images for Fshr, Cyp11a1, Pdgfra and Acta2 probes. b, Representative RNAscope images for Upk1b, Cd19, Flt1 and Prox1 probes. c, Representative RNAscope images for Klrb1c, Cd3e, Cd8b2 and Cd4. Images were enhanced for visualization. d, Heatmap of detected cell types in the Aging, VCD and Foxl2 haploinsufficiency scRNA-seq datasets. For panels a-c, shown images are from a middle-aged Foxl2+/+ animal.
Extended Data Fig. 7. Cell type proportion analysis of ovarian cells via flow cytometry and scRNA-seq analysis. a, Gating strategy of CD45+ cells in flow cytometry analysis. b-d, Proportion differences of Ptprc+ cell types detected in Aging, VCD and Foxl2 haploinsufficiency model scRNA-seq datasets. Asterisks (*) indicate comparisons in which the observed log2 fold change is infinite due to complete absence of the cell type in one of the groups.
Extended Data Fig. 8. Cell type proportion analysis of non-immune ovarian cells via in situ hybridization assays. a-c, Cell type abundance analysis of stromal, SMC, BEC, LEC and epithelial cells via RNAscope in situ hybridization assays from Aging, VCD and Foxl2 haploinsufficiency model mice.
Extended Data Fig. 9. Cell type proportion analysis of immune ovarian cells via in situ hybridization assays. a-c, Cell type abundance analysis of NK, NKT, CD8+ NKT, CD8+ T, CD4+ T, DNT, DPT and B cells via RNAscope in situ hybridization assays from Aging, VCD and Foxl2 haploinsufficiency model mice.
Extended Data Fig. 10. Comparative analysis of global gene expression in VCD model via Augur. a-b, UMAP visualization of AUC scores and scatter plot of AUC quantification in the VCD model, comparing CTL vs. VCD at 3m and 10m age-at-injection. For the scatter plot, Ptprc− and Ptprc+ cells were separately plotted to improve visualization. All data are from the 30d post-injection timepoint. For b, data points with NA AUC scores (due to low cell count or QC filtering) were assigned an AUC of 0.5 and colored gray to improve visualization.
Extended Data Fig 11. Aging-associated signatures and transposable element enrichment analysis across Aging, VCD and Foxl2 haploinsufficiency models. a, Heatmap of cell types detected in pseudobulked scRNA-seq datasets. b, DESeq2 normalized log2 counts of Foxl2 from young and mid-age Foxl2+/+ and Foxl2+/− mice. c-d, GSEA enrichment analysis of differentially expressed genes (DEGs) identified in the Aging model (FDR < 0.05) and SenMayo gene list. e, Dot plot showing the coefficient of variation analysis results. f, GSEA enrichment analysis of transposable element (TE) families across Aging, VCD and Foxl2 haploinsufficiency models.
Extended Data Fig 12. Module trees of WGCNA analysis. a, WGCNA module trees identified in the Aging model for granulosa, theca, stromal, BEC, epithelial, and DNT cells.
Extended Data Fig 13. Association analysis of WGCNA modules and ovarian aging traits from the Aging model. a, Heatmap of trait significance for WGCNA modules in the Aging model. Modules that passed the FDR < 0.1 threshold are shown in bold.
Extended Data Fig 14. Analysis of recurrent transcription factor activity inferred by decoupleR. a-c, Heatmap of transcription factor (TF) activity for factors detected in at least three cell types within each model with an FDR < 0.05.
Extended Data Fig 15. Development of a transcriptome-based aging clock for theca cells. a, Schematic of analysis and training workflow for the lasso regression-based clock. b, Scatter plot of predicted age vs. chronological age for the test set, with Spearman’s correlation and p-value displayed. c-d, Age prediction for VCD and Foxl2 haploinsufficiency datasets, comparing predicted age to chronological age in weeks. Offsets were added for visualization purposes. e-f, Age acceleration analysis for VCD and Foxl2 haploinsufficiency datasets, calculated as the difference between predicted and chronological age. Statistical significance was assessed using the Wilcoxon test. f-g, ORA enrichment analysis of GO ALL and Reactome terms of the top 500 lasso features.
Data Availability Statement
The raw FASTQ files have been deposited in the Sequence Read Archive under accession number PRJNA863443. Raw microscopy images of ovarian hematoxylin and eosin (H&E) staining, compressed RNAscope images (due to file size constraints), and raw flow cytometry datasets are available on Figshare (DOIs: 10.6084/m9.figshare.28745765, 10.6084/m9.figshare.28745792, 10.6084/m9.figshare.28745813, 10.6084/m9.figshare.29260391 and 10.6084/m9.figshare.28733222). The final annotated Seurat objects have been deposited to Figshare (10.6084/m9.figshare.28934126) and can be explored via an interactive R shiny application: https://minhooki.shinyapps.io/shinyappmulti/. All analyses were performed using R version 4.1.2. Any additional information required to reproduce or reanalyze the data presented in this study is available from the corresponding author upon reasonable request.
All new scripts used to analyze the datasets are available on the Benayoun lab GitHub at https://github.com/BenayounLaboratory/Mouse_Menopause_Models.