Abstract
Shwachman-Diamond syndrome (SDS) is an autosomal recessive disorder characterized by exocrine pancreatic insufficiency and hematologic and skeletal abnormalities. A genomewide scan of families with SDS was terminated at ∼50% completion, with the identification of chromosome 7 markers that showed linkage with the disease. Finer mapping revealed significant linkage across a broad interval that included the centromere. The maximum two-point LOD score was 8.7, with D7S473, at a recombination fraction of 0. The maximum multipoint LOD score was 10, in the interval between D7S499 and D7S482 (5.4 cM on the female map and 0 cM on the male map), a region delimited by recombinant events detected in affected children. Evidence from all 15 of the multiplex families analyzed provided support for the linkage, consistent with a single locus for SDS. However, the presence of several different mutations is suggested by the heterogeneity of disease-associated haplotypes in the candidate region.
Shwachman-Diamond syndrome (SDS [MIM 260400]), also known as “Shwachman syndrome,” “Shwachman-Bodian syndrome,” or “congenital lipomatosis of the pancreas,” is a rare disorder first reported in 1964 (Bodian et al. 1964; Shwachman et al. 1964). Multiple organs are affected, and there is a broad range in severity of presentation even among affected siblings (Burke et al. 1967; Shmerling et al. 1969; Aggett et al. 1980; Savilahti and Rapola 1984), although impaired exocrine pancreatic function and hematologic abnormalities with depressed myeloid cell lineages are the most consistent features (Ginzberg et al. 1999). Exocrine pancreatic insufficiency typically manifests in infancy, although some improvement may be seen with age (Hill et al. 1982; Mack et al. 1996). In addition to persistent or intermittent neutropenia and an increased susceptibility to infections, hematologic problems also can include anemia, thrombocytopenia, and pancytopenia (Smith et al. 1996). SDS is considered to be a genetic bone marrow–failure syndrome; patients with SDS are at increased risk of development of myelodysplasia and hematologic malignancy—in particular, acute myelogenous leukemia (Huijgens et al. 1977; Woods et al. 1981; Smith et al. 1996). Skeletal abnormalities include delayed maturation, metaphyseal chondrodysplasia of the long bones, and thoracic-cage abnormalities with costochondral thickening (Aggett et al. 1980; Dhar and Anderton 1994; Mack et al. 1996). Short stature occurs in 50% of patients (Ginzberg et al. 1999). Other common anomalies include hepatomegaly and abnormal liver-biochemical tests, both found in younger patients (Aggett et al. 1980; Ginzberg et al. 1999). Behavioral and learning difficulties have also been reported in some patients (Aggett et al. 1980; Kent et al. 1990).
An unusual combination of organs are affected in SDS, and, to date, the basic pathophysiological defect is unknown. Segregation analysis of an international collection of families supports an autosomal recessive mode of inheritance (Ginzberg et al. 2000). Molecular analysis (Ikegawa et al. 1999) and family studies (Goobie et al. 1999) have excluded those regions of chromosomes 6 and 12 that were pinpointed in a study of the one patient known to have a constitutional chromosomal abnormality in the form of the balanced de novo translocation t(6;12)(q16.2:q21.2) (Masuno et al. 1995). In the present study, linkage analysis was used to establish the chromosomal location for SDS.
Most of the families with SDS included in the present study have been described elsewhere, and additional families have been obtained through ongoing recruitment (Ginzberg et al. 1999; Goobie et al. 1999). Diagnosis was rigorously determined for all patients; the criterion for inclusion was the presence of both exocrine pancreatic dysfunction and hematologic abnormalities, including neutropenia and other bone marrow failure–associated problems. Consent was obtained from all participating families, and procedural approval was obtained from the human subjects review board of The Hospital for Sick Children, Toronto (HSC). Genomic DNA was extracted either from Epstein-Barr virus–transformed lymphoblastoid cell lines or directly from peripheral white-blood-cell pellets, as described by Miller et al. (1988).
A panel of simple sequence length polymorphic markers from the Cooperative Human Linkage Center Human Screening Set, version 6.0 (Dubovsky et al. 1995), supplemented with selected Généthon markers (Dib et al. 1996), was used to initiate a genomewide scan of 13 multiplex and 8 simplex families. The markers had an average heterozygosity of .75 and an average intermarker distance of 12 cM. In brief, PCRs were set up, with DNA from each family member, with fluorescently labeled oligonucleotides (Research Genetics). Amplified products of an average of eight markers were pooled into groups based on expected allele sizes and labels. Products were analyzed, after separation by electrophoresis on an ABI 377 sequencer (PE Biosystems), as described by Rioux et al. (1998).
Linkage was analyzed under the assumption that SDS is a completely penetrant autosomal recessive trait. We estimated the disease-allele frequency to be .0036, based on an incidence of 1/76,563. The incidence of SDS was obtained by multiplying the cystic fibrosis (CF) incidence (1/2,500) by the ratio of the number of patients with SDS (N=24) to the number of patients with CF (N=735), cared for at HSC during a recent 21-year period. We assumed that levels of ascertainment were the same for both diseases. Marker-allele frequencies were estimated from the genotypes of the founders in the pedigrees, by PEDMANAGER, version 0.9 (M. J. Daly, personal communication). Linkage analysis was performed by the FASTLINK, version 4.1P, of the LINKAGE programs (Lathrop et al. 1984; Cottingham et al. 1993; Schäffer et al. 1994). Two-point LOD scores were computed by MLINK, under the assumption that there is no sex difference in recombination rates. After 228 markers were genotyped, the genomewide scan was terminated because one locus, D7S1830, had a two-point LOD score of 4.2 at a recombination fraction (θ) of .05. Two proximal loci, D7S817 and D7S1808, also gave positive LOD scores, of 0.9 and 1.8, respectively, at θ=.1.
To map the putative SDS locus more precisely, 27 additional chromosome 7 markers, spanning 40 cM (sex-averaged map), were genotyped in families with either two or three affected children (fig. 1 and table 1). Markers were selected from public resources, with a preference for those included in recently assembled genetic maps (Broman et al. 1998). PCRs were set up to use either α[35S]-dATP (Goobie et al. 1999) or γ[33P]-dATP (Amersham Pharmacia Biotech) end-labeled oligonucleotides (Hudson et al. 1997), after optimization for MgCl2 levels (1–1.5 mM) and annealing temperatures. Alleles were assigned after electrophoreses of PCR products on 6% polyacrylamide gels and autoradiography (Goobie et al. 1999). Maximum-likelihood estimates of sex-specific θ values were computed by ILINK. A maximum LOD score of 8.7 was obtained with D7S473, at θ=0 (table 1). LOD scores >3 were obtained over a large region (35 cM on the female map and 8.6 cM on the male map) that includes the centromere. We have no evidence of SDS locus heterogeneity; in each of the 15 families, the expected LOD scores (Leal and Ott 1990) at θ=0 were obtained for at least one marker in the candidate chromosome 7 centromeric region.
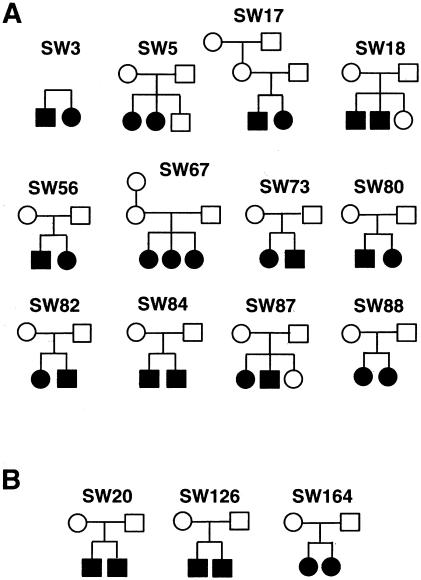
Figure 1.
Pedigrees of families with SDS, indicating individuals from whom DNA was available. The families were from Canada, the United States, England, and The Netherlands; all families were white, and none were known to have consanguineous matings. Affected individuals are denoted by black symbols. A, Multiplex families used to map the SDS locus to chromosome region 7p12-q11. B, Additional families used in the refined mapping (see table 1).
Table 1.
Two-Point Linkage between the SDS Locus and Chromosome 7 Loci
|
Distance on Genetic Mapa(cM) |
|||||
| Locus | Average | Female | Male |
Maximum LOD Score ( / / )b )b
|
Radiation-Hybrid–Map Positionc(cR10000) |
| D7S1808 | 41.69 | 43.59 | 39.84 | 1.49 (.200/.061) | |
| D7S817 | 50.29 | 54.42 | 46.27 | 2.20 (.104/.055) | |
| D7S2206 | … | … | … | 1.64 (.238/.060) | |
| D7S691 | 63.67 | 70.54 | 56.97 | 2.58 (.224/0) | |
| D7S679 | 69.03 | 81.29 | 56.97 | 3.46 (.193/0) | |
| D7S665 | 69.56 | 81.29 | 58.04 | 3.34 (.240/0) | |
| D7S2506 | 69.56 | 81.29 | 58.04 | 5.00 (.068/0) | |
| D7S1818 | 69.56 | 81.29 | 58.04 | 3.77 (.071/0) | |
| D7S2422 | 72.78 | 87.74 | 58.04 | 3.95 (.100/0) | |
| D7S1830d | 72.78 | 87.74 | 58.04 | 4.61 (.170/0) | |
| D7S506 | 73.84 | 89.88 | 58.04 | 7.16 (0/0) | |
| D7S2552 | 74.38 | 90.94 | 58.04 | 3.95 (.052/0) | 2442 |
| EGFRd | … | … | … | 5.32 (.053/0) | |
| D7S2550d | 75.44 | 93.08 | 58.04 | 5.91 (.041/0) | 2458 |
| D7S793d | … | … | … | 4.20 (.068/0) | |
| D7S499d | 75.98 | 94.15 | 58.04 | 6.26 (.085/0) | 2495 |
| D7S659d | 75.98 | 94.15 | 58.04 | 7.19 (0/0) | |
| D7S473d | 76.71 | 95.62 | 58.04 | 8.74 (0/0) | |
| D7S494d | 76.71 | 95.62 | 58.04 | 8.57 (0/0) | |
| D7S2429d | 76.71 | 95.62 | 58.04 | 7.11 (0/0) | 2741 |
| D7S2512d | 77.91 | 98.03 | 58.04 | 7.16 (0/0) | |
| D7S2549d | 77.91 | 98.03 | 58.04 | 5.92 (0/0) | 2836 |
| D7S663d | 78.65 | 99.50 | 58.04 | 7.71 (0/0) | |
| D7S502d | 78.65 | 99.50 | 58.04 | 7.13 (0/0) | 2907 |
| D7S482d | 78.65 | 99.50 | 58.04 | 3.56 (.103/0) | |
| D7S1839d | 78.65 | 99.50 | 58.04 | 7.08 (.072/0) | |
| D7S2503 | 78.65 | 99.50 | 58.04 | 4.61 (.058/0) | |
| D7S2483 | 78.65 | 99.50 | 58.04 | 5.01 (.068/0) | 2960 |
| D7S1816 | 83.99 | 105.92 | 62.34 | 4.13 (.165/0) | |
| D7S2204 | 90.95 | 116.64 | 65.53 | 3.64 (.366/0) | |
From the Center for Medical Genetics Web site, in Kosambi centimorgans (Broman et al. 1998); D7S473 and D7S494 were ordered by recombination in one family, and D7S2512 and D7S2549 were ordered by YAC/BAC mapping. The occurrence of satellite-repeat sequences (D7Z2) localizes the centromere near D7S2429, as indicated in The Genetic Location Database Web site (Collins et al. 1996). D7S793-D7S499 and D7S482-D7S1839-D7S2503, which flank the block of eight nonrecombinant loci, were ordered according to the composite location in The Genetic Location Database. No recombination was observed between the SDS locus and D7S506; D7S506 was not informative in one of the two potentially informative families and was not typed in the other. The sex-averaged genetic map was used for the multipoint analysis by GENEHUNTER 2.0; increments of 0.1 cM were added for subsequent loci assigned to the same position. A locus not included on the Center for Medical Genetics map was ordered according to The Genetic Location Database and was positioned, by linear interpolation, between the closest flanking loci on the Center for Medical Genetics map.
 = maximum-likelihood estimate of the female recombination fraction;
= maximum-likelihood estimate of the female recombination fraction;  = maximum-likelihood estimate of the male recombination fraction.
= maximum-likelihood estimate of the male recombination fraction.
From the G3 radiation-hybrid map (version 2.0) of the Stanford Human Genome Center Web site, selected for LINKMAP multipoint analysis. The markers were assigned to different bins in the same block, in a physical order consistent with genetic mapping. D7S2483 was placed 0.1 cM distal to D7S502 on the female map. Although the average map distance per centiray (cR) along chromosome 7 is 20 kb, the accurate sizes of the bins at the centromere are unknown (Stewart et al. 1997). Both long-range mapping (Wevrick and Willard 1991) and our effort to establish a physical map support a large physical size of the centromeric region of chromosome 7.
Fifteen families were included in the linkage analysis (12 families were included for the remaining loci; see fig. 1).
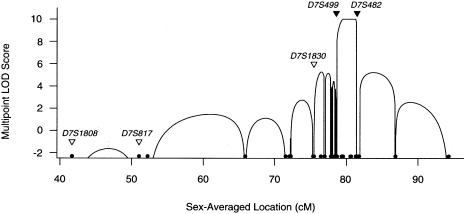
Multipoint linkage analysis, by GENEHUNTER, version 2.0 (Kruglyak et al. 1996; Kruglyak and Lander 1998), using the marker order and sex-averaged map in table 1, indicated a maximum LOD score of 10 in the most likely interval, between D7S499 and D7S482 (fig. 2). The next-most-likely interval, which includes D7S506, had a multipoint LOD score of 5.3. It is noteworthy that male recombination is markedly suppressed (table 1) in the centromeric region, over a female genetic distance of ∼20 cM, with a peak ratio of distance in females to distance in males (female:male distance ratio) of ∼8 at the centromere (Broman et al. 1998). Thus, we selected seven markers (table 1) encompassing the candidate region, for multipoint analysis by the LINKMAP program in the FASTLINK package, which allows for sex differences in recombination rates. The maximum multipoint LOD score with a sliding window of five markers was 10, and there were only small (<0.7) differences between the scores that resulted when a female:male distance ratio of 8 or 1 was used.
Figure 2.
Multipoint LOD scores for the location of the SDS locus. Black circles (●) indicate the positions of the markers. The map of 30 markers appears to be longer than is indicated in table 1 because the Haldane mapping function was used. Black triangles (▾) indicate the positions of the two markers flanking the candidate region, and white triangles (▿) indicate the positions of the markers used in the genomewide scan. LOD scores <−2.5 are not shown.
Haplotypes, from GENEHUNTER 2.0, for the eight markers in the region of no recombination were examined. None of the affected children in the 15 families were homozygous for the eight-marker haplotype. The SDS-associated haplotypes are diverse; of the 30 haplotypes, only 2 were identical. This shared haplotype was found in two families from the United States but was not found among the normal chromosomes; that is, it was not found among parental chromosomes not transmitted to affected children.
The discovery of a locus for SDS indicates that positional cloning will be a feasible approach for identification of the basic defect. A single SDS gene that resides within the critical interval is anticipated, although the diversity of SDS-associated haplotypes is compatible with the existence of a number of different mutations. The generation of a detailed physical map of the SDS critical interval, which would provide a working framework, is complicated by the presence of the centromere and associated satellite-repeat sequences. Characterization of pericentromeric regions of chromosomes has revealed other types of repetitive sequences, as well as recent intra-/interchromosomal sequence duplications, that result in active or inactive remnants of genes, such as pseudogenes or partial genes (Eichler et al. 1996; Ritchie et al. 1998; Eichler 1999; Jackson et al. 1999). Also notable is the limited availability of annotated genomic sequences for these regions, which further hampers the rapid characterization of candidate genes. Nevertheless, >50 gene/expressed-sequence–tag clusters or transcription units that map across the interval delimited by D7S499 and D7S482 (5.4 cM in the female map and 0 cM on the male map) have been identified. Of the known genes—including GBAS, GUSB, CCT6A, and FKBP9, among others—that have been confirmed, by complementary methods, to map to the SDS gene region, none have biological function or defined expression patterns that suggest that they are obvious candidates for the SDS gene. Many of the remaining cDNA clusters are presently insufficiently characterized for direct assessment, but some have recognizable transcription-factor motifs and warrant prioritization in more-detailed investigations.
On chromosome 7, there are local variations in female and male genetic distances, with the most pronounced difference being at the centromere (Broman et al. 1998). In the families included in the present study, we did not observe male recombination with any of the 21 markers assigned to the same position on the male map (table 1), consistent with an absence of male recombination in the families used to build the genetic map (Broman et al. 1998).
From the perspective of bone marrow failure and hematopoietic-cell chromosomal abnormalities, the mapping of the SDS locus to the centromeric region of chromosome 7 is provocative. Both myelodysplastic syndrome (MDS) and acute myelogenous leukemia (AML) are serious complications of SDS, with indications of risk as high as 30%–35% (Smith et al. 1996). Monosomy 7 and deletion of 7q are acquired changes frequently seen in the bone marrow cells of patients with either MDS or AML (Luna-Fineman et al. 1995; Mitelman et al. 1997); isochromosome 7 formation has also been reported (Mertins et al. 1994). Although chromosomal changes generally predict a poor prognosis, both their role in the development of cancer, and patients' susceptibility to their formation, are unknown. Formation of isochromosome 7q has been reported in five patients with SDS; one of these patients has neither MDS nor AML (Dror et al. 1998; Maserati et al. 2000). It is tempting to speculate that mutations in the SDS gene may increase susceptibility to somatic chromosomal changes. However, the way in which this may occur is not readily predictable. Acquired chromosome 7–associated changes of the bone marrow cells are not unique to SDS, nor are there any direct indications that SDS carriers are at increased risk for hematologic problems. Furthermore, some observed alterations to chromosome 7, such as non–whole arm translocations, do not appear to involve the centromere directly. A finding that does hint at events associated with the centromere is the report of chromosome 7 centromeric heteromorphism in bone marrow cells from a patient with SDS who showed no evidence of either MDS or AML (Sokolic et al. 1999). The heteromorphism occurred in an otherwise intact chromosome and was sustained for >6 mo. Although there is one other report of chromosome 7 heteromorphism, involving the bone marrow cells of a 25-year-old woman (without SDS) in remission from pre-B lymphoblastic leukemia (Duval et al. 2000), the true occurrence of centromeric heteromorphism in bone marrow cells is unknown, since whole-chromosome and centromeric analyses are not necessarily performed simultaneously. Heteromorphism may be due to deletions of centromeric repeat sequences, as a result of misalignment and unequal crossing over, but it has not been found for chromosome 7, either in cells of peripheral blood or in established lymphoblastoid lines (Lo et al. 1999). More information is needed to understand whether the SDS locus plays any role in the development of abnormal chromosomes or changes in chromosome numbers.
The identification of a locus for SDS provides the starting point for the identification, by positional cloning, of a gene with SDS-causing mutations and for the application of genetic diagnosis in families at risk. Studies directed toward an understanding of the function of the gene should help explain the variability of disease symptoms and should provide insight into the development of bone marrow failure and of the preleukemic and leukemic states.
Acknowledgments
We thank M. Corey, Ph.D., for discussions and for registry information on patients with CF. We thank S. Clark and C. Darmond-Zwaig, for technical assistance, and J C. Loredo-Osti, for helpful discussions on linkage analysis. We are grateful for the cooperation of the patients with SDS, their families, and their physicians. We also acknowledge support from Shwachman-Diamond Canada, Shwachman Support of Great Britain, The Harrison Wright Appeal, Shwachman-Diamond Support of Australia, Shwachman-Diamond Support International, and the Canadian Institutes of Health Research (CIHR). T.J.H., K.M., and J.M.R. are members of the Networks of Centres of Excellence–Canadian Genetic Diseases Network (CGDN). T.J.H. holds a Clinician Scientist Award from the CIHR. T.M.F. is supported by a gift to McGill University from Alcan Aluminum, Limitée. S.G. was a recipient of a graduate studentship from the Natural Sciences and Engineering Research Council of Canada. M.P. is a recipient of studentships from the Ontario Graduate Program and from the Joint Hospital for Sick Children–CGDN Program. H.G. received Duncan Gordon and Canadian Association of Gastroenterology/Janssen fellowships.
Electronic-Database Information
The accession number and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/ (for genetic maps)
- Genetic Location Database, The, http://cedar.genetics.soton.ac.uk/public_html/ldb.html (for map locations on chromosome 7)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SDS [MIM 260400]) [PubMed]
- Stanford Human Genome Center (SHGC) Web, http://www-shgc.stanford.edu/ (for G3 radiation hybrid map, version 2.0)
References
- Aggett PJ, Cavanagh NP, Matthew DJ, Pincott JR, Sutcliffe J, Harries JT (1980) Shwachman's syndrome: a review of 21 cases. Arch Dis Child 55:331–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodian M, Sheldon W, Lightwood R (1964) Congenital hypoplasia of the exocrine pancreas. Acta Paediatr 53:282–293 [DOI] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke V, Colebatch JH, Anderson CM, Simons MJ (1967) Association of pancreatic insufficiency and chronic neutropenia in childhood. Arch Dis Child 42:147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A, Frezal J, Teague J, Morton NE (1996) A metric map of humans: 23,500 loci in 850 bands. Proc Natl Acad Sci USA 93:14771–14775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham RW Jr, Idury RM, Schäffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Dhar S, Anderton JM (1994) Orthopaedic features of Shwachman syndrome: A report of two cases. J Bone Joint Surg Am 76:278–282 [DOI] [PubMed] [Google Scholar]
- Dib C, Fauré S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morrissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Dror Y, Squire J, Durie P, Freedman MH (1998) Malignant myeloid transformation with isochromosome 7q in Shwachman-Diamond syndrome. Leukemia 12:1591–1595 [DOI] [PubMed] [Google Scholar]
- Dubovsky J, Sheffield VC, Duyk GM, Weber JL (1995) Sets of short tandem repeat polymorphisms for efficient linkage screening of the human genome. Hum Mol Genet 4:449–452 [DOI] [PubMed] [Google Scholar]
- Duval A, Feneux D, Sutton L, Tchernia G, Leonard C (2000) Spurious monosomy 7 in leukemia due to centromeric heteromorphism. Cancer Genet Cytogenet 119:67–69 [DOI] [PubMed] [Google Scholar]
- Eichler EE (1999) Repetitive conundrums of centromere structure and function. Hum Mol Genet 8:151–155 [DOI] [PubMed] [Google Scholar]
- Eichler EE, Lu F, Shen Y, Antonacci R, Jurecic V, Doggett NA, Moyzis RK, Baldini A, Gibbs RA, Nelson DL (1996) Duplication of a gene-rich cluster between 16p11.1 and Xq28: a novel pericentromeric-directed mechanism for paralogous genome evolution. Hum Mol Genet 5:899–912 [DOI] [PubMed] [Google Scholar]
- Ginzberg H, Shin J, Ellis L, Goobie S, Morrison J, Corey M, Durie PR, Rommens JM (2000) Segregation analysis in Shwachman-Diamond syndrome: evidence for recessive inheritance. Am J Hum Genet 66:1413–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginzberg H, Shin J, Ellis L, Morrison J, Ip W, Dror Y, Freedman M, Heitlinger LA, Belt MA, Corey M, Rommens JM, Durie PR (1999) Shwachman syndrome: phenotypic manifestations of sibling sets and isolated cases in a large patient cohort are similar. J Pediatr 135:81–88 [DOI] [PubMed] [Google Scholar]
- Goobie S, Morrison J, Ginzberg H, Ellis L, Corey M, Masuno M, Imaizumi K, Kuroki Y, Fujiwara TM, Morgan K, Durie PR, Rommens JM (1999) Exclusion of linkage of Shwachman-Diamond syndrome to chromosome regions 6q and 12q implicated by a de novo translocation. Am J Med Genet 85:171–174 [DOI] [PubMed] [Google Scholar]
- Hill RE, Durie PR, Gaskin KJ, Davidson GP, Forstner GG (1982) Steatorrhea and pancreatic insufficiency in Shwachman syndrome. Gastroenterology 83:22–27 [PubMed] [Google Scholar]
- Hudson TJ, Clark CD, Gschwend M, Justice-Higgins E (1997) PCR methods of genotyping. In: Dracopoli NC, Haines JL, Korf BR, Moir DT, Morton CC, Seidman CE, Seidman JG, Smith DR, Boyle AL (eds) Current protocols in human genetics. John Wiley & Sons, New York, pp 2.5.1–2.5.23 [Google Scholar]
- Huijgens PC, van der Veen EA, Meijer S, Muntinghe OG (1977) Syndrome of Shwachman and leukaemia. Scand J Haematol 18:20–24 [DOI] [PubMed] [Google Scholar]
- Ikegawa S, Masuno M, Kumano Y, Okawa A, Isomura M, Koyama K, Okui K, Makita Y, Sasaki M, Kohdera U, Okuda M, Koyama H, Ohashi H, Tajiri H, Imaizumi K, Nakamura Y (1999) Cloning of translocation breakpoints associated with Shwachman syndrome and identification of a candidate gene. Clin Genet 55:466–472 [DOI] [PubMed] [Google Scholar]
- Jackson MS, Rocchi M, Thompson G, Hearn T, Crosier M, Guy J, Kirk D, Mulligan L, Ricco A, Piccininni S, Marzella R, Viggiano L, Archidiacono N (1999) Sequences flanking the centromere of human chromosome 10 are a complex patchwork of arm-specific sequences, stable duplications and unstable sequences with homologies to telomeric and other centromeric locations. Hum Mol Genet 8:205–215 [DOI] [PubMed] [Google Scholar]
- Kent A, Murphy GH, Milla P (1990) Psychological characteristics of children with Shwachman syndrome. Arch Dis Child 65:1349–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Lander ES (1998) Faster multipoint linkage analysis using Fourier transforms. J Comput Biol 5:1–7 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1984) Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 81:3443–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal SM, Ott J (1990) Expected lod scores in linkage analysis of autosomal recessive traits for affected and unaffected offspring. Am J Hum Genet Suppl 47:A188 [Google Scholar]
- Lo AW, Liao GC, Rocchi M, Choo KH (1999) Extreme reduction of chromosome-specific alpha-satellite array is unusually common in human chromosome 21. Genome Res 9:895–908 [DOI] [PubMed] [Google Scholar]
- Luna-Fineman S, Shannon KM, Lange BJ (1995) Childhood monosomy 7: epidemiology, biology and mechanistic implications. Blood 85:1985–1999 [PubMed] [Google Scholar]
- Mack DR, Forstner GG, Wilschanski M, Freedman MH, Durie PR (1996) Shwachman syndrome: exocrine pancreatic dysfunction and variable phenotypic expression. Gastroenterology 111:1593–1602 [DOI] [PubMed] [Google Scholar]
- Maserati E, Minelli A, Olivieri C, Bonvini L, Marchi A, Bozzola M, Danesino C, Scappaticci S, Pasquali F (2000) Isochromosome (7)(q10) in Shwachman Syndrome without MDS/AML and role of chromosome 7 anomalies in myeloproliferative disorders. Cancer Genet Cytogenet 121:167–171 [DOI] [PubMed] [Google Scholar]
- Masuno M, Imaizumi K, Nishimura G, Nakamura M, Saito I, Akagi K, Kuroki Y (1995) Shwachman syndrome associated with de novo reciprocal translocation t(6;12)(q16.2;q21.2). J Med Genet 32:894–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins F, Johansson B, Mitelman F (1994) Isochromosomes in neoplasia. Genes Chromosom Cancer 10:221–230 [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitelman F, Mertens F, Johansson B (1997) A breakpoint map of recurrent chromosomal rearrangements in human neoplasia. Nat Genet 15:417–474 [DOI] [PubMed] [Google Scholar]
- Rioux JD, Stone VA, Daly MJ, Cargill M, Green T, Nguyen H, Nutman T, Zimmerman PA, Tucker MA, Hudson T, Goldstein AM, Lander E, Lin AY (1998) Familial eosinophilia maps to the cytokine gene cluster on human chromosomal region 5q31-q33. Am J Hum Genet 63:1086–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie RJ, Mattei MG, Lalande M (1998) A large polymorphic repeat in the pericentromeric region of human chromosome 15q contains three partial gene duplications. Hum Mol Genet 7:1253–260 [DOI] [PubMed] [Google Scholar]
- Savilahti E, Rapola J (1984) Frequent myocardial lesions in Shwachman's syndrome: eight fatal cases among 16 Finnish patients. Acta Paediatr Scand 73:642–651 [DOI] [PubMed] [Google Scholar]
- Schäffer AA, Gupta SK, Shriram K, Cottingham RW Jr (1994) Avoiding recomputation in linkage analysis. Hum Hered 44:225–237 [DOI] [PubMed] [Google Scholar]
- Shmerling DH, Prader A, Hitzig WH, Giedion A, Hadorn ZB, Kühni M (1969) The syndrome of exocrine pancreatic insufficiency, neutropenia, metaphyseal dysostosis and dwarfism. Helvetica Paediatr Acta 24:547–575 [PubMed] [Google Scholar]
- Shwachman H, Diamond LK, Oski FA, Khaw K-T (1964) The syndrome of pancreatic insufficiency and bone marrow dysfunction. J Pediatr 65:645–663 [DOI] [PubMed] [Google Scholar]
- Smith OP, Hann IM, Chessells JM, Reeves BR, Milla P (1996) Haematological abnormalities in Shwachman-Diamond syndrome. Br J Haematol 94:279–284 [DOI] [PubMed] [Google Scholar]
- Sokolic RA, Ferguson W, Mark HF (1999) Discordant detection of monosomy 7 by GTG-banding and FISH in a patient with Shwachman-Diamond syndrome without evidence of myelodysplastic syndrome or acute myelogenous leukemia. Cancer Genet Cytogenet 115:106–113 [DOI] [PubMed] [Google Scholar]
- Stewart EA, McKusick KB, Aggarwal A, Bajorek E, Brady S, Chu A, Fang N, et al (1997) An STS-based radiation hybrid map of the human genome. Genome Res 7:422–433 [DOI] [PubMed] [Google Scholar]
- Wevrick R, Willard HF (1991) Physical map of the centromeric region of human chromosome 7: relationship between two distinct alpha satellite arrays. Nucleic Acids Res 19:2295–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods WG, Roloff JS, Lukens JN, Krivit W (1981) The occurrence of leukemia in patients with the Shwachman syndrome. J Pediatr 99:425–428 [DOI] [PubMed] [Google Scholar]