Abstract
Human respiratory syncytial virus (HRSV) is the leading viral cause of severe respiratory illness for infants and young children worldwide. Two major antigenic groups (A and B) of HRSV exist, and viruses from both subgroups can cocirculate during epidemics; however, their frequencies might differ between seasons. The subgroup prevalence and genotype distribution patterns of HRSV strains were investigated in a community in Belgium during 10 successive epidemic seasons (1996 to 2006). A regular 3-year cyclic pattern of subgroup dominance was observed, consisting of two predominant HRSV-A seasons, followed by a single HRSV-B-dominant year. HRSV infections with both subgroups were more prevalent among children younger than 6 months and had a peak incidence in December. The most frequently detected genotypes were GA5 and GB13, the latter including strains with the 60-nucleotide duplication in the G gene. Furthermore, GA5 remained the dominant HRSV genotype in two consecutive epidemic seasons twice during the study period. Additional variability was detected among the GB13 isolates, due to the usage of a novel termination codon in the G gene. Dual infections with both HRSV subgroups were detected for 9 patients, and subsequent infections with the heterologous HRSV subgroup were documented for 15 patients. Among five patients with homologous reinfections, only one was caused by HRSV-B. Our results support the hypothesis that the overall prevalence of HRSV-A over HRSV-B could be due to a more-transient subgroup A-specific immune protection.
Human respiratory syncytial virus (HRSV) is the leading viral cause of pneumonia and bronchiolitis for infants and young children worldwide (4, 8, 16). It is also recognized as an important respiratory pathogen for the elderly, individuals with cardiopulmonary disease, and the immunocompromised (9). By the age of 2 years, virtually all infants have experienced at least one HRSV infection. Reinfections are very common throughout life, and in older children and adults they are usually associated with milder disease, indicating that HRSV infections induce only partial immunity (14, 19).
HRSV is an enveloped virus with a nonsegmented, negative-stranded RNA genome and is classified in the family Paramyxoviridae (8). Two major subgroups, A (HRSV-A) and B (HRSV-B), have been initially identified on the basis of distinct reactivity patterns with panels of monoclonal antibodies (2, 27). Subsequent sequencing data revealed that these antigenic groups correspond to genetically distinct viruses. The most extensive antigenic and genetic differences between and within the two subgroups are found in the attachment G glycoprotein (4, 21, 38). The G attachment protein is also one of the main targets for inducing neutralizing and protective antibodies (29), and it has been suggested that antigenic differences within this protein could facilitate repeated HRSV infections (21).
Epidemiological surveys have revealed that strains from both HRSV subgroups can be present in the same community during outbreaks; however, their relative proportions may differ between epidemics (3, 4, 10, 20). HRSV has complex circulation patterns, with multiple genotypes or lineages cocirculating within the same community and replacement of the predominating genotypes with new ones over successive epidemic seasons (1, 5, 6, 13, 18, 33). HRSV genotype distribution patterns can be distinctive for each community, and it has been suggested that they are determined by local factors such as the level of herd immunity to certain strains (1, 5, 32).
A better understanding of the subgroup prevalence shift and genotype distribution patterns of HRSV infections within the same community could be important for the prediction of future outbreak strains and for the development of effective vaccines or antiviral therapy. The aim of the present study was to investigate the subgroup prevalence shift and genotype circulation patterns of HRSV-A and -B infections in Belgium during 10 successive epidemic seasons.
MATERIALS AND METHODS
Patients and clinical specimens.
Respiratory samples including nasal swabs, bronchoalveolar lavage specimens, and nasopharyngeal and tracheal aspirates were collected from patients with severe respiratory tract illness who were admitted or who consulted at the Gasthuisberg University Hospital in Leuven, Belgium. During the period from November 1996 to February 2006, a total of 1,412 specimens, obtained from 1,389 patients, were determined to be positive for HRSV antigen by using the TestPack RSV (Abbott Laboratories, Abbott Park, IL) or NOW RSV (Binax) immunoassay and were included in this study. For 552 (40%) of the samples, HRSV infection was also confirmed by exhibition of a characteristic syncytial cytopathic effect in HeLa or HEp-2 cell cultures. The ages of the HRSV-infected patients ranged from 8 days to 78 years (median, 7 months), and the majority (97%) were infants and children younger than 5 years. The respiratory specimens were stored at −80°C until further use.
RNA extraction and multiplex RT-PCR.
RNA was extracted directly from the respiratory samples or from the supernatants of the cell-cultured specimens by using the QIAmp viral RNA minikit according to the manufacturer's manual (QIAGEN, Benelux, The Netherlands). HRSVs were typed as belonging to group A or group B by using a multiplex reverse transcriptase PCR (RT-PCR) assay. RT-PCR was performed with the OneStep RT-PCR kit (QIAGEN) in a 25-μl final volume containing 0.6 μM (each) HRSV-A (G267-G511)- and HRSV-B (BGF-BGR)-specific forward and reverse primers, 10 U of RNase inhibitor (RNaseOUT; Invitrogen, Merelbeke, Belgium), and 5 μl of the extracted RNA (Table 1). Thermocycling was performed on a GeneAmp PCR system 9600 thermal cycler (Applied Biosystems, Foster City, CA) under the following conditions: 55°C for 30 min; 95°C for 15 min; 40 cycles of 94°C for 30 s, 60°C for 1 min, and 72°C for 1 min; and a final extension step at 72°C for 10 min. HRSV-A and -B amplicons had expected sizes of 283 bp and 800 bp, respectively. For certain samples, the initial results were validated by repeating the RT-PCR in separate singleplex reactions for subgroups A and B, with annealing temperatures of 61°C and 63°C, respectively.
TABLE 1.
HRSV-A and -B oligonucleotide primers and probes used in this study
| Primer or probe | Gene | Positionsa | Sequence (5′→3′) | Polarity | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| RT-PCR primers | ||||||||||
| HRSV-A | ||||||||||
| G267 | G | 248-267 | ATGCAACAAGCCAGATCAAG | + | 39 | |||||
| G511 | G | 511-531 | GGGTACAAAGTTAAACACTTC | − | 15 | |||||
| F164 | F | 164-186 | GTTATCACACTGGTATACC AACC | − | 39 | |||||
| HRSV-B | ||||||||||
| BGF | G | 169-192 | GCAGCCATAATATTCATCATCTCT | + | 48 | |||||
| BGR | G-F | 5637-5658 | TGCCCCAGRTTTAATTTCGTTC | − | 48 | |||||
| qRT-PCR primers | ||||||||||
| HRSV-A | ||||||||||
| AFF | F | 669-695 | CTGTGATAGARTTCCAACAAAAGAACA | + | This study | |||||
| AFR | F | 718-745 | AGTTACACCTGCATTAACACTAAATTCC | − | This study | |||||
| HRSV-B | ||||||||||
| BNF | N | 435-458 | GGCTCCAGAATATAGGCATGATTC | + | This study | |||||
| BNR | N | 480-508 | TGGTTATTACAAGAGCAGCTATACACAGT | − | This study | |||||
| MGB probes | ||||||||||
| HRSV-A, F-TP | F | 697-715 | CAGACTACTAGAGATTACC | + | This study | |||||
| HRSV-B, N-TP | N | 461-477 | TATCATCCCACAGTCTG | − | This study | |||||
| Sequencing primers | ||||||||||
| HRSV-A, G599 | G | 599-619 | GCTTGGTGGTGGTTTTCTTTC | − | 49 | |||||
| HRSV-B | ||||||||||
| G640 | G | 640-662 | AGAGACCCAAAAACACYAGCCAA | + | 48 | |||||
| G495 | G | 495-519 | ACAGGGAACGAAGTTGAACACTTCA | − | 48 | |||||
HRSV-A RT-PCR.
A 900-bp fragment of the G gene of HRSV-A was amplified with the OneStep RT-PCR kit (QIAGEN) and the G267-F164 primer set (Table 1). The reaction was performed in a 50-μl final volume containing 0.6 μM forward and reverse primers, 20 U of RNaseOUT RNase inhibitor (Invitrogen), and 10 μl of the extracted RNA (Table 1). Amplification was carried out on a GeneAmp PCR system 9600 thermal cycler (Applied Biosystems) as follows: 50°C for 30 min; 95°C for 15 min; 40 cycles of 94°C for 30 s, 53°C for 1 min, and 72°C for 1 min; and a final extension step at 72°C for 10 min.
Nucleotide sequencing.
Fifty-six randomly selected HRSV-A-specific amplicons and 21 randomly selected HRSV-B-specific amplicons of 800 bp and 900 bp, respectively, were purified with the Invitek MSB Spin PCRapace purification kit (Westburg, Leusden, The Netherlands). The amplicons were sequenced in both directions by using the ABI PRISM BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems). In addition to the RT-PCR primers, subgroup A (G599)- and subgroup B (G495 and G640)-specific oligonucleotide primers were used to obtain 629 nucleotides (nt) and 724 nt of the subgroup A and B G genes, respectively. The chromatogram sequencing files were compiled in contigs by using SeqMan II (DNAStar, Madison, WI).
Phylogenetic analyses.
The sequenced HRSV-A and -B strains were compared to previously determined sequences of Belgian HRSV isolates (63 subgroup A and 142 subgroup B isolates). Multiple sequence alignments of HRSV-A and -B G-genes were obtained with CLUSTAL X, version 1.83 (41), and were manually edited with the GeneDoc alignment editor, version 2.6.002 (28). Identical sequences were identified with the DAMBE program, version 4.2.13 (47). Phylogenetic analyses were conducted by using the neighbor-joining method under the Tamura-Nei parameter model with the MEGA (version 3.0) software package (22, 40). The statistical support of the dendrograms was evaluated by bootstrapping (1,000 replicates). HRSV strain designations indicate the country of isolation (Belgium [BE]), the laboratory number of the sample, and the epidemic season (e.g., BE/156/03-04).
Multiplex qRT-PCR assay.
Antigen positive, RT-PCR-negative HRSV samples were further tested for the presence of subgroup A and/or subgroup B strains by using a multiplex real-time RT-PCR assay. An HRSV-A-specific primer set and probe containing a 5′ reporter dye, 6-carboxyfluorescein, were developed from the F gene, while an HRSV-B-specific primer set and probe containing the 5′ reporter dye VIC were designed from the N gene (Table 1). Qualitative real-time RT-PCR (qRT-PCR) was performed in a final volume of 25 μl using 5 μl extracted RNA, 12.5 μl of Eurogentec One-Step RT qPCR master mix containing the internal reference dye 6-carboxy-X-rhodamine as a passive reference, 0.125 μl Euroscript with RT and RNase inhibitor (Eurogentec, Belgium), 900 nM HRSV-A and -B-specific forward and reverse primers, and 100 nM HRSV-A and -B MGB probes. Real-time RT-PCR was carried out in a ABI PRISM 7700 sequence detection system (Applied Biosystems) under the following conditions: an initial reverse transcription at 48°C for 30 min, followed by PCR activation at 95°C for 10 min and 45 cycles of amplification (15 s at 95°C and 1 min at 60°C).
Statistical analysis.
Statistical analysis for comparison of differences between two mean values was performed by the t test for independent groups in the SPSS software package (version 11.5). Differences were considered statistically significant if the P value of the two-tailed significance was <0.05.
Nucleotide sequence accession numbers.
The HRSV nucleotide sequences determined in this study have been deposited in the GenBank database under accession numbers DQ985080 to DQ985156.
RESULTS
HRSV typing and subgroup prevalence.
Twenty samples that were reactive in the antigen assay were determined to be negative for HRSV RNA by using the RT-PCR and qRT-PCR assays. These samples were considered to be HRSV antigen false positives and were therefore excluded from further analysis, resulting in a total number of 1,392 HRSV antigen-positive samples. HRSV strains were characterized as subgroup A or subgroup B for 979 (70%) of the samples, of which 608 (62%) were subtyped by multiplex RT-PCR, 166 (17%) were subtyped by multiplex qRT-PCR, and 205 (21%) strains had been previously subtyped by RT-PCR and sequencing analyses as described elsewhere (48, 49). As a result, 537 (55%) specimens had subgroup A strains, 433 (44%) had subgroup B strains, and in 9 (1%) specimens both HRSV-A and HRSV-B were identified. The proportion of typed HRSV antigen-positive samples from each epidemic season ranged from 59% to 83% (Table 2). The majority of the typed HRSV strains were obtained directly from the clinical samples. Due to the lack of sufficient sample material and the absence of the respective cell cultured virus strain, we were not able to analyze all HRSV antigen-positive samples.
TABLE 2.
Subgroup distributions of HRSV-A and -B infections in Belgium during 10 epidemic seasons
| Epidemic season | No. of samplesa | No. (%) of typed samples | No. (%)b of HRSV infections with the following subgroup(s):
|
||
|---|---|---|---|---|---|
| HRSV-A | HRSV-B | HRSV-A and HRSV-B | |||
| 1996-1997 | 104 | 86 (83) | 57 (66) | 28 (33) | 1 (1) |
| 1997-1998 | 129 | 97 (75) | 62 (64) | 33 (34) | 2 (2) |
| 1998-1999 | 124 | 102 (82) | 26 (25) | 76 (75) | 0 |
| 1999-2000 | 146 | 97 (66) | 73 (75) | 23 (24) | 1 (1) |
| 2000-2001 | 136 | 84 (62) | 69 (82) | 15 (18) | 0 |
| 2001-2002 | 129 | 86 (67) | 13 (15) | 73 (85) | 0 |
| 2002-2003 | 173 | 131 (75) | 72 (55) | 59 (45) | 0 |
| 2003-2004 | 178 | 123 (69) | 75 (61) | 46 (37) | 2 (2) |
| 2004-2005 | 132 | 90 (68) | 20 (22) | 68 (76) | 2 (2) |
| 2005-2006 | 141 | 83 (59) | 70 (84) | 12 (15) | 1 (1) |
Total number of HRSV antigen-positive samples.
Percentages apply to the total number of typed HRSV antigen-positive samples for the season.
HRSV-A and -B strains cocirculated during all 10 epidemic seasons. Subgroup A viruses were predominant during seven epidemic seasons, while subgroup B strains prevailed during the epidemic seasons of 1998 to 1999, 2001 to 2002, and 2004 to 2005. A regular pattern of subgroup predominance replacement was observed, consisting of two consecutive seasons of HRSV-A dominance followed by a single intervening season of HRSV-B dominance.
Monthly and age distributions of HRSV infections.
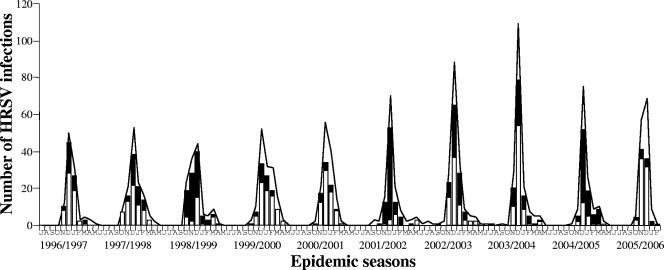
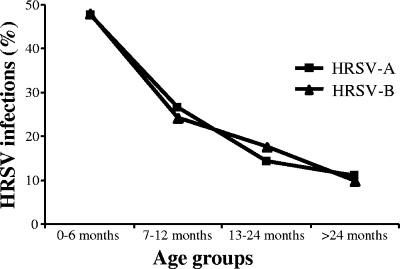
HRSV outbreak activity in Belgium was high during the months of November, December, and January and had a peak incidence in December (Fig. 1). HRSV strains from both subgroups were isolated at higher frequencies during the peak outbreak months. Sixteen sporadic HRSV infections were observed during the period from May to September. The distributions of HRSV-A and -B infections were similar for the four age groups defined in this study. HRSV infections with both subgroups were significantly more frequent (P < 0.05) for children younger than 6 months (Fig. 2).
FIG. 1.
Seasonal frequency of antigen-positive HRSV infections (curves) and monthly distribution of typed HRSV-A (white bars) and HRSV-B (black bars) infections among hospitalized patients in Belgium (1996 to 2006). Initial letters of months are given along the y axis, beginning with July for each season.
FIG. 2.
Age distributions of HRSV-A and -B infections among hospitalized patients in Belgium from 1996 to 2006.
Phylogenetic analyses and genotype distribution patterns.
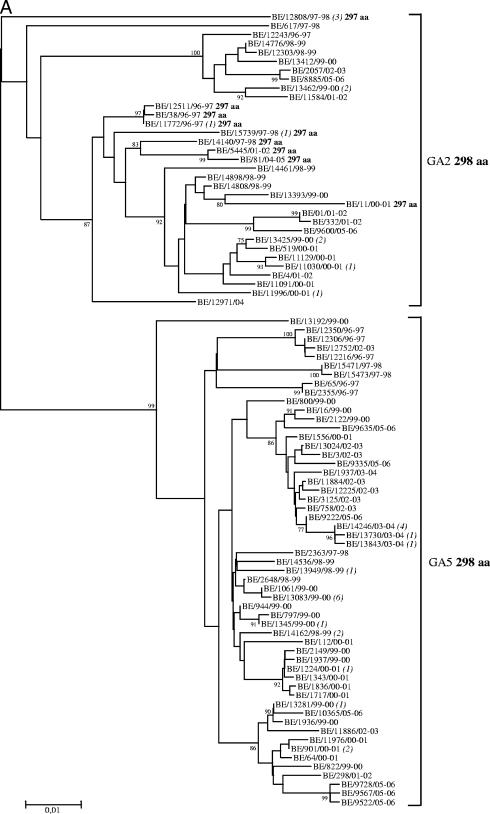
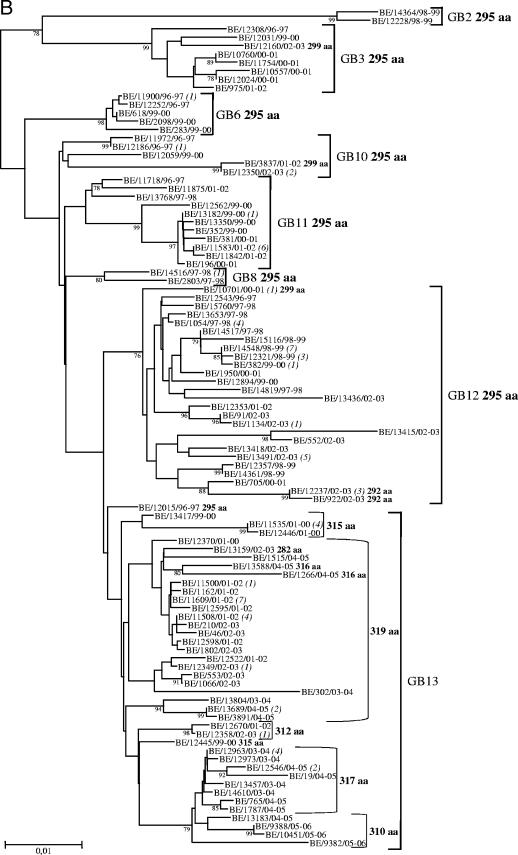
Phylogenetic analyses were conducted on a total of 119 HRSV-A and 163 HRSV-B Belgian G gene sequences, including those of the 56 HRSV-A and 21 HRSV-B strains whose amplicons were processed previously as described above (see Materials and Methods) (48, 49) (Fig. 3A and B). Two major HRSV-A genotypes (GA2 and GA5) and eight HRSV-B genotypes (GB2, GB3, GB6, GB8, and GB10 to GB13) were detected among the Belgian HRSV strains during the 10-year study period. Multiple identical sequences were identified, the majority of which were from strains isolated during the same epidemic season. Exceptions to the latter observation included three HRSV-A strains with sequences identical to those of strains that were circulating 5 to 7 epidemic seasons earlier and eight HRSV-B strains with sequences identical to those of strains obtained 1 to 3 epidemic seasons earlier. The HRSV-A and -B genotype distributions during each epidemic season are shown in Table 3. Two to seven genotypes were identified in a single outbreak, and the dominant HRSV genotype shifted in every 1 to 2 following seasons. HRSV-A genotypes prevailed for 1 to 3 successive epidemic seasons, whereas HRSV-B genotypes predominated for 1 to 4 consecutive seasons. GA5 was the prevailing subgroup A genotype during 7 epidemics and was the only HRSV genotype to dominate in consecutive seasons (1999 to 2000 and 2000 to 2001; 2002 to 2003 and 2003 to 2004). GA2 was the dominant HRSV genotype in 1997 to 1998 and the prevailing subgroup A genotype during the epidemic seasons of 2001 to 2002 and 2004 to 2005. GA5 strains had deduced G proteins of 298 amino acids (aa), while the GA2 genotype also included strains with G proteins of 297 aa. The most frequently isolated HRSV-B genotypes were GB12 and GB13, which were observed in 7 and 6 epidemic seasons, respectively, and were also the prevailing subgroup B genotypes in 5 and 4 outbreaks, respectively. GB13, which included strains with the 60-nt duplication, was the dominant HRSV genotype during the two subgroup B-prevalent outbreaks (2001 to 2002 and 2004 to 2005) and was also the only subgroup B genotype observed during the last three seasons (2003 to 2004, 2004 to 2005, and 2005 to 2006). In addition to the five different predicted protein lengths (282, 312, 315, 317, and 319 aa) that we have previously reported among Belgian GB13 strains (48), we identified for the first time strains with predicted proteins of 310 and 316 aa. The 310-aa HRSV-B strains had a 6-nt deletion and used the termination codon at nucleotide position 877, while the 316-aa strains had a stop codon-inducing nucleotide substitution at position 890 (TCA→TAA) (according to the numbering of strain B1 [GenBank accession number AF013254]). GB13 strains of different deduced G protein lengths were dominant in different seasons. In 1999 to 2000, two 315-aa strains were detected. In 2001 to 2002, the predominant strains were 319 aa long; these were replaced by 315-aa strains in 2002 to 2003. During the subsequent 2 epidemic seasons (2003 to 2004 and 2004 to 2005), the prevailing strains had 317 aa, and in 2005 to 2006, only 310-aa strains were detected (Fig. 3B).
FIG. 3.
Unrooted neighbor-joining phylogenetic trees of the G genes of 119 HRSV-A (A) and 163 HRSV-B (B) strains obtained in Belgium from 1996 to 2006. Only bootstrap values above 75% are shown. The italicized numbers in parentheses at the terminal nodes correspond to the numbers of identical sequences. Brackets on the right delimit the HRSV-A and HRSV-B genotypes. The predicted length of the G protein is given in boldface after the genotype and, for exceptions, after the strain designation or grouping. The nomenclature is based on phylogenetic clustering with sequences previously assigned to specific genotypes. HRSV-A genotypes GA2 and GA5 are designated according to the classification systems of Peret et al. (32, 33) and Venter et al. (45). HRSV-B genotype designation GB2 represents the classification systems of Peret et al. (32) and Venter et al. (44, 45), while GB3, GB6, GB8, and GB10 to GB13 are based on the classification of Zlateva et al. (48).
TABLE 3.
Genotype distributions of HRSV-A and -B strains isolated in Belgium during 10 consecutive epidemic seasons, 1996 to 2006
| Epidemic season | No. (%)a of genotyped HRSV isolates
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GA2 | GA5 | GB2 | GB3 | GB6 | GB8 | GB10 | GB11 | GB12 | GB13 | |
| 1996-1997 | 4 (44) | 5 (56) | 1 (11) | 3 (33) | 3 (33) | 1 (11) | 1 (11) | |||
| 1997-1998 | 7 (70) | 3 (30) | 3 (23) | 1 (8) | 9 (69) | |||||
| 1998-1999 | 5 (42) | 7 (58) | 2 (17) | 10 (83) | ||||||
| 1999-2000 | 7 (26) | 20 (74) | 1 (5) | 3 (16) | 1 (5) | 5 (26) | 7 (37) | 2 (11) | ||
| 2000-2001 | 9 (41) | 13 (59) | 4 (40) | 2 (20) | 4 (40) | |||||
| 2001-2002 | 5 (83) | 1 (17) | 1 (3) | 1 (3) | 9 (24) | 1 (3) | 25 (68) | |||
| 2002-2003 | 2 (20) | 8 (80) | 1 (3) | 3 (9) | 19 (54) | 12 (34) | ||||
| 2003-2004 | 10 (100) | 10 (100) | ||||||||
| 2004-2005 | 3 (75) | 1 (25) | 15 (100) | |||||||
| 2005-2006 | 2 (22) | 7 (78) | 3 (100) | |||||||
Percentages are calculated separately for HRSV-A and -B strains in each epidemic season.
Patients with repeated HRSV infections.
Twenty-two patients tested positive for HRSV antigen more than once during the study period within a time interval ranging from 10 to 70 months. Eight of these patients had underlying conditions such as asthma (two patients), spastic diplegia, simultaneous trisomy 4p and monosomy 5p, pyelonephritis, mitral insufficiency, epilepsy, and quadriplegia. Another five children were born prematurely with a gestational age of <37 weeks. The antigenic groups for both infections were determined for 20 patients, including also a third infection experienced by one patient, resulting in a total of 21 infections. Heterologous-subgroup reinfections were observed for 15 patients, of whom 8 had their first infection with an HRSV-A strain and 7 with an HRSV-B strain. Homologous reinfections were detected for five patients; four of these reinfections were caused by HRSV-A and one by HRSV-B (Table 4). One patient had an initial dual infection with both HRSV subgroups and was reinfected with a subgroup A strain after 11 months. More than two infections were detected for each of the two asthmatic patients. One of them had four HRSV infections over 5 years. The first infection (not typed) was encountered at the age of 4 months; the second infection occurred after 13 months and was caused by HRSV-B, followed by two more infections caused by HRSV-A, encountered, respectively, after 24 and 60 months. The G gene sequences of the two HRSV-A strains (BE/14807/98-99 and BE/14616/03-04) from this patient clustered within genotype GA5 and differed at 16 (2.5%) nucleotide positions, resulting in 6 (3%) differences on the deduced amino acid level. The second asthmatic child experienced three infections during 3 subsequent epidemic seasons; the initial infection was not typed, and the following two infections were caused by HRSV-A. The homologous HRSV-B infections identified for one patient occurred 70 months apart and were caused by a GB12 strain (BE/1054/97-98) and a GB13 strain with the 60-nt duplication and 6-nt deletion (BE/12963/03-04). The numbers of genetic differences identified between the two subgroup B strains were 83 (11%) on the nucleotide level and 30 (12%) on the deduced amino acid level.
TABLE 4.
Subgroup occurrence and timing of HRSV reinfections for hospitalized patients in Belgium
| Reinfection interval (mo) | Total no. of reinfections | No. of HRSV reinfectionsa
|
||||
|---|---|---|---|---|---|---|
| A-A | B-B | A-B | B-A | A/B-A | ||
| 10-12 | 12 | 3 | 5 | 3 | 1 | |
| 13-24 | 3 | 2 | 1 | |||
| 25-36 | 2 | 2 | ||||
| 37-48 | 1 | 1 | ||||
| 49-60 | 2 | 1 | 1 | |||
| 61-72 | 1 | 1 | ||||
The first letter of each designation is the HRSV subgroup responsible for the first infection, and the second letter is the subgroup responsible for the second infection. For example, A-B represents a first infection with HRSV-A and reinfection with HRSV-B. A/B indicates coinfection with both subgroups.
DISCUSSION
HRSVs from both subgroups were cocirculating during each of the 10 epidemic seasons investigated in Belgium; however, HRSV-A strains were isolated more frequently and were predominant during 7 seasons. The monthly distributions of HRSV-A and -B infections was similar, with a high prevalence during the winter months (November to January) and a peak incidence in December. These findings indicate that viruses from the two subgroups have similar temporal occurrences. The occasional isolation of HRSV in May to September suggests that the virus is present in the community continuously throughout the year. HRSV is highly transmissible and is spread either via respiratory secretions by close contact with infected individuals or indirectly, through exposure to contaminated surfaces (17). Thus, the very low incidence of HRSV infections during the summer months is probably due to less indoor crowding, which can limit further spread of the virus. Infections with both subgroups were more prevalent in children younger than 6 months.
More-frequent detection of HRSV-A strains during outbreaks has been observed worldwide (4, 12, 26, 31). In a community, a shift in the HRSV subgroup dominance can occur at different time intervals, and different patterns of subgroup A and B prevalence have been described. In Finland, the HRSV subgroup dominance changed every 2 years over 10 consecutive years (46), while in Rochester, NY, a cyclic pattern, consisting of 1 to 2 consecutive years of high subgroup A dominance followed by a single intervening year of subgroup B dominance or by codominance of both subgroups, was observed over a 15-year period (18). Similar subgroup occurrence patterns have been seen in Kenya, Uruguay, and the United States, where HRSV-B became predominant after 2 consecutive seasons of high subgroup A prevalence (3, 7, 37). During the 10 successive epidemic seasons investigated in Belgium, a regular 3-year cyclic pattern of subgroup occurrence was observed. It consisted of 2 consecutive HRSV-A-predominant seasons, followed by a single HRSV-B-predominant season. Multiple genotypes were circulating simultaneously in the Belgian community during the study period. However, the subgroup A genotype GA5 and the subgroup B genotype GB13, which included strains with the 60-nt duplicated region (42), were isolated more frequently during the study period and were identified as the predominant HRSV genotypes in 5 and 2 epidemic seasons, respectively. Interestingly, GB13 strains with different predicted G protein lengths were prevalent during these 2 seasons. Previous investigations have reported that a replacement of the predominating HRSV genotype occurs each year (5, 11, 33). However, we did not identify such an annual shift of the prevailing genotype among hospitalized patients in Belgium. GA5 remained the dominant HRSV genotype in 2 consecutive epidemic seasons twice during the study period. This genotype was reported to be predominant among HRSV-A strains during 3 consecutive seasons in a community in Japan (35) and in India (30) and has been the most frequently detected HRSV-A genotype in Sweden (34) and New Zealand (24) in recent years. Higher antigenic diversity among the GA5 strains is most likely responsible for the ability of this genotype to persist and prevail more frequently in certain communities (3, 30). This study included only one hospital in Belgium, and the majority of patients were inhabitants of the Flemish part of Belgium. Nevertheless, taking into account the facts that Belgium is a small country with a dense population and that people frequently commute to another city for work, we believe that our findings on HRSV subgroup and genotype distribution patterns are likely to be representative for the whole country.
It has been speculated that the global prevalence of HRSV-A is due to a higher degree of divergence among viruses from this antigenic subgroup (7). However, recent studies have demonstrated that subgroup B viruses have a greater potential for divergence by accumulating substantial modifications in their attachment G protein genes, such as 60-nt duplications, 6-nt and 3-nt deletions and insertions, frameshift mutations, and premature stop codons (24, 42, 43, 48). Additional diversity among the subgroup B viruses is accomplished by the usage of three alternative termination codons (23, 43, 48). This is the major mechanism responsible for the high degree of G protein length polymorphisms observed among the Belgian GB13 strains possessing the 60-nt duplicated region. Moreover, we identified a new stop codon position at the C-terminal end of the G protein gene in two GB13 strains from the epidemic season 2004 to 2005, resulting in a predicted G protein of 316 aa. The increased genetic variability observed among the HRSV-B GB13 strains and their recent prevalence in the Belgian community imply that this genotype is probably evolving under stronger selective pressure than other HRSV-B genotypes. This is consistent with the increased average substitution rate reported for HRSV-B strains possessing the 60-nt duplication (43).
Another possible explanation for the more frequent occurrence of subgroup A is that subgroup B may elicit a more complete and/or long-lasting subgroup-specific immune response than subgroup A and therefore may cause disease less often (18). In support of this speculation, only one of the five homologous HRSV reinfections observed in this study was caused by subgroup B viruses. This is in accordance with the findings of previous studies, in which more subgroup A than subgroup B homologous reinfections have been detected (25, 31, 35, 36). Most of the repeated infections observed in this study occurred in two consecutive epidemic seasons and were caused by strains of different subgroups. In addition, we identified nine patients who were coinfected with HRSV-A and -B strains. Interestingly, one (amino acid position 117) and three (amino acid positions 223, 247, and 258) of the amino acid changes identified between the subgroup A and B strains that caused homologous infections, respectively, were at positions that have been determined to be under positive selection pressure (48, 49).
In summary, a regular 3-year periodic cycle of HRSV subgroup occurrence was observed among hospitalized patients in a community in Belgium during 10 successive epidemic seasons. This pattern consisted of 2 seasons in which HRSV-A was prevalent, followed by a single season of HRSV-B predominance. Multiple genotypes were cocirculating during each epidemic, and successful genotypes predominated for more than one season. Higher genetic and antigenic diversity among GA5 and GB13 strains is most likely responsible for the abilities of these genotypes to prevail for a prolonged period in the Belgian community. We speculate that the observed overall prevalence of HRSV-A over HRSV-B strains could be due to more-transient subgroup A-specific immune protection rather than to higher HRSV-A genetic diversity as has been previously suggested.
Acknowledgments
K. Zlateva and L. Vijgen were supported by the K.U. Leuven Research Fund.
We thank the colleagues of the Laboratory of Clinical and Epidemiological Virology, Rega Institute, University of Leuven, for helpful comments and critical reading of the manuscript.
Footnotes
Published ahead of print on 3 July 2007.
REFERENCES
- 1.Anderson, L. J., R. M. Hendry, L. T. Pierik, C. Tsou, and K. McIntosh. 1991. Multicenter study of strains of respiratory syncytial virus. J. Infect. Dis. 163:687-692. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, L. J., J. C. Hierholzer, C. Tsou, R. M. Hendry, B. F. Fernie, Y. Stone, and K. McIntosh. 1985. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J. Infect. Dis. 151:626-633. [DOI] [PubMed] [Google Scholar]
- 3.Arbiza, J., A. Delfraro, and S. Frabasile. 2005. Molecular epidemiology of human respiratory syncytial virus in Uruguay: 1985-2001—a review. Mem. Inst. Oswaldo Cruz 100:221-230. [DOI] [PubMed] [Google Scholar]
- 4.Cane, P. A. 2001. Molecular epidemiology of respiratory syncytial virus. Rev. Med. Virol. 11:103-116. [DOI] [PubMed] [Google Scholar]
- 5.Cane, P. A., D. A. Matthews, and C. R. Pringle. 1994. Analysis of respiratory syncytial virus strain variation in successive epidemics in one city. J. Clin. Microbiol. 32:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi, E. H., and H. J. Lee. 2000. Genetic diversity and molecular epidemiology of the G protein of subgroups A and B of respiratory syncytial viruses isolated over 9 consecutive epidemics in Korea. J. Infect. Dis. 181:1547-1556. [DOI] [PubMed] [Google Scholar]
- 7.Coggins, W. B., E. J. Lefkowitz, and W. M. Sullender. 1998. Genetic variability among group A and group B respiratory syncytial viruses in a children's hospital. J. Clin. Microbiol. 36:3552-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins, P. L., R. M. Chanock, and B. R. Murphy. 2001. Respiratory syncytial virus, p. 1443-1485. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 9.Falsey, A. R., and E. E. Walsh. 2000. Respiratory syncytial virus infection in adults. Clin. Microbiol. Rev. 13:371-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freymuth, F., J. Petitjean, P. Pothier, J. Brouard, and E. Norrby. 1991. Prevalence of respiratory syncytial virus subgroups A and B in France from 1982 to 1990. J. Clin. Microbiol. 29:653-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galiano, M. C., V. Luchsinger, C. M. Videla, L. De Souza, S. S. Puch, C. Palomo, C. Ricarte, B. Ebekian, L. Avendano, and G. Carballal. 2005. Intragroup antigenic diversity of human respiratory syncytial virus (group A) isolated in Argentina and Chile. J. Med. Virol. 77:311-316. [DOI] [PubMed] [Google Scholar]
- 12.Galiano, M. C., C. Palomo, C. M. Videla, J. Arbiza, J. A. Melero, and G. Carballal. 2005. Genetic and antigenic variability of human respiratory syncytial virus (groups A and B) isolated over seven consecutive seasons in Argentina (1995 to 2001). J. Clin. Microbiol. 43:2266-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García, O., M. Martin, J. Dopazo, J. Arbiza, S. Frabasile, J. Russi, M. Hortal, P. Perez-Brena, I. Martinez, B. Garcia-Barreno, and J. A. Melero. 1994. Evolutionary pattern of human respiratory syncytial virus (subgroup A): cocirculating lineages and correlation of genetic and antigenic changes in the G glycoprotein. J. Virol. 68:5448-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glezen, W. P., L. H. Taber, A. L. Frank, and J. A. Kasel. 1986. Risk of primary infection and reinfection with respiratory syncytial virus. Am. J. Dis. Child. 140:543-546. [DOI] [PubMed] [Google Scholar]
- 15.Gottschalk, J., R. Zbinden, L. Kaempf, and I. Heinzer. 1996. Discrimination of respiratory syncytial virus subgroups A and B by reverse transcription-PCR. J. Clin. Microbiol. 34:41-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall, C. B. 2001. Respiratory syncytial virus and parainfluenza virus. N. Engl. J. Med. 344:1917-1928. [DOI] [PubMed] [Google Scholar]
- 17.Hall, C. B., R. G. Douglas, Jr., and J. M. Geiman. 1980. Possible transmission by fomites of respiratory syncytial virus. J. Infect. Dis. 141:98-102. [DOI] [PubMed] [Google Scholar]
- 18.Hall, C. B., E. E. Walsh, K. C. Schnabel, C. E. Long, K. M. McConnochie, S. W. Hildreth, and L. J. Anderson. 1990. Occurrence of groups A and B of respiratory syncytial virus over 15 years: associated epidemiologic and clinical characteristics in hospitalized and ambulatory children. J. Infect. Dis. 162:1283-1290. [DOI] [PubMed] [Google Scholar]
- 19.Henderson, F. W., A. M. Collier, W. A. Clyde, Jr., and F. W. Denny. 1979. Respiratory-syncytial-virus infections, reinfections and immunity. A prospective, longitudinal study in young children. N. Engl. J. Med. 300:530-534. [DOI] [PubMed] [Google Scholar]
- 20.Hendry, R. M., A. L. Talis, E. Godfrey, L. J. Anderson, B. F. Fernie, and K. McIntosh. 1986. Concurrent circulation of antigenically distinct strains of respiratory syncytial virus during community outbreaks. J. Infect. Dis. 153:291-297. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, P. R., M. K. Spriggs, R. A. Olmsted, and P. L. Collins. 1987. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc. Natl. Acad. Sci. USA 84:5625-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 23.Martínez, I., O. Valdes, A. Delfraro, J. Arbiza, J. Russi, and J. A. Melero. 1999. Evolutionary pattern of the G glycoprotein of human respiratory syncytial viruses from antigenic group B: the use of alternative termination codons and lineage diversification. J. Gen. Virol. 80:125-130. [DOI] [PubMed] [Google Scholar]
- 24.Matheson, J. W., F. J. Rich, C. Cohet, K. Grimwood, Q. S. Huang, D. Penny, M. D. Hendy, and J. R. Kirman. 2006. Distinct patterns of evolution between respiratory syncytial virus subgroups A and B from New Zealand isolates collected over thirty-seven years. J. Med. Virol. 78:1354-1364. [DOI] [PubMed] [Google Scholar]
- 25.Mufson, M. A., R. B. Belshe, C. Orvell, and E. Norrby. 1987. Subgroup characteristics of respiratory syncytial virus strains recovered from children with two consecutive infections. J. Clin. Microbiol. 25:1535-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mufson, M. A., R. B. Belshe, C. Orvell, and E. Norrby. 1988. Respiratory syncytial virus epidemics: variable dominance of subgroups A and B strains among children, 1981-1986. J. Infect. Dis. 157:143-148. [DOI] [PubMed] [Google Scholar]
- 27.Mufson, M. A., C. Orvell, B. Rafnar, and E. Norrby. 1985. Two distinct subtypes of human respiratory syncytial virus. J. Gen. Virol. 66:2111-2124. [DOI] [PubMed] [Google Scholar]
- 28.Nicholas, K. B., H. B. Nicholas, Jr., and D. W. Deerfield II. 1997. GeneDoc: analysis and visualization of genetic variation. EMBnet News 4:1-4. [Google Scholar]
- 29.Norrby, E., M. A. Mufson, H. Alexander, R. A. Houghten, and R. A. Lerner. 1987. Site-directed serology with synthetic peptides representing the large glycoprotein G of respiratory syncytial virus. Proc. Natl. Acad. Sci. USA 84:6572-6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parveen, S., S. Broor, S. K. Kapoor, K. Fowler, and W. M. Sullender. 2006. Genetic diversity among respiratory syncytial viruses that have caused repeated infections in children from rural India. J. Med. Virol. 78:659-665. [DOI] [PubMed] [Google Scholar]
- 31.Parveen, S., W. M. Sullender, K. Fowler, E. J. Lefkowitz, S. K. Kapoor, and S. Broor. 2006. Genetic variability in the G protein gene of group A and B respiratory syncytial viruses from India. J. Clin. Microbiol. 44:3055-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peret, T. C., C. B. Hall, G. W. Hammond, P. A. Piedra, G. A. Storch, W. M. Sullender, C. Tsou, and L. J. Anderson. 2000. Circulation patterns of group A and B human respiratory syncytial virus genotypes in 5 communities in North America. J. Infect. Dis. 181:1891-1896. [DOI] [PubMed] [Google Scholar]
- 33.Peret, T. C., C. B. Hall, K. C. Schnabel, J. A. Golub, and L. J. Anderson. 1998. Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J. Gen. Virol. 79:2221-2229. [DOI] [PubMed] [Google Scholar]
- 34.Rafiefard, F., B. Johansson, T. Tecle, and C. Orvell. 2004. Molecular epidemiology of respiratory syncytial virus (RSV) of group A in Stockholm, Sweden, between 1965 and 2003. Virus Res. 105:137-145. [DOI] [PubMed] [Google Scholar]
- 35.Sato, M., R. Saito, T. Sakai, Y. Sano, M. Nishikawa, A. Sasaki, Y. Shobugawa, F. Gejyo, and H. Suzuki. 2005. Molecular epidemiology of respiratory syncytial virus infections among children with acute respiratory symptoms in a community over three seasons. J. Clin. Microbiol. 43:36-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott, P. D., R. Ochola, M. Ngama, E. A. Okiro, N. D. James, G. F. Medley, and P. A. Cane. 2006. Molecular analysis of respiratory syncytial virus reinfections in infants from coastal Kenya. J. Infect. Dis. 193:59-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scott, P. D., R. Ochola, M. Ngama, E. A. Okiro, D. J. Nokes, G. F. Medley, and P. A. Cane. 2004. Molecular epidemiology of respiratory syncytial virus in Kilifi district, Kenya. J. Med. Virol. 74:344-354. [DOI] [PubMed] [Google Scholar]
- 38.Sullender, W. M. 2000. Respiratory syncytial virus genetic and antigenic diversity. Clin. Microbiol. Rev. 13:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sullender, W. M., and G. W. Wertz. 1991. Synthetic oligonucleotide probes differentiate respiratory syncytial virus subgroups in a nucleic acid hybridization assay. J. Clin. Microbiol. 29:1255-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamura, K., and M. Nei. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512-526. [DOI] [PubMed] [Google Scholar]
- 41.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trento, A., M. Galiano, C. Videla, G. Carballal, B. Garcia-Barreno, J. A. Melero, and C. Palomo. 2003. Major changes in the G protein of human respiratory syncytial virus isolates introduced by a duplication of 60 nucleotides. J. Gen. Virol. 84:3115-3120. [DOI] [PubMed] [Google Scholar]
- 43.Trento, A., M. Viegas, M. Galiano, C. Videla, G. Carballal, A. S. Mistchenko, and J. A. Melero. 2006. Natural history of human respiratory syncytial virus inferred from phylogenetic analysis of the attachment (G) glycoprotein with a 60-nucleotide duplication. J. Virol. 80:975-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venter, M., M. Collinson, and B. D. Schoub. 2002. Molecular epidemiological analysis of community circulating respiratory syncytial virus in rural South Africa: comparison of viruses and genotypes responsible for different disease manifestations. J. Med. Virol. 68:452-461. [DOI] [PubMed] [Google Scholar]
- 45.Venter, M., S. A. Madhi, C. T. Tiemessen, and B. D. Schoub. 2001. Genetic diversity and molecular epidemiology of respiratory syncytial virus over four consecutive seasons in South Africa: identification of new subgroup A and B genotypes. J. Gen. Virol. 82:2117-2124. [DOI] [PubMed] [Google Scholar]
- 46.Waris, M. 1991. Pattern of respiratory syncytial virus epidemics in Finland: two-year cycles with alternating prevalence of groups A and B. J. Infect. Dis. 163:464-469. [DOI] [PubMed] [Google Scholar]
- 47.Xia, X., and Z. Xie. 2001. DAMBE: software package for data analysis in molecular biology and evolution. J. Hered. 92:371-373. [DOI] [PubMed] [Google Scholar]
- 48.Zlateva, K. T., P. Lemey, E. Moes, A. M. Vandamme, and M. Van Ranst. 2005. Genetic variability and molecular evolution of the human respiratory syncytial virus subgroup B attachment G protein. J. Virol. 79:9157-9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zlateva, K. T., P. Lemey, A. M. Vandamme, and M. Van Ranst. 2004. Molecular evolution and circulation patterns of human respiratory syncytial virus subgroup A: positively selected sites in the attachment G glycoprotein. J. Virol. 78:4675-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]