Abstract
Here, we report the characterization of the Arabidopsis thaliana ocp11 (for overexpressor of cationic peroxidase11) mutant, in which a β-glucuronidase reporter gene under the control of the H2O2-responsive Ep5C promoter is constitutively expressed. ocp11 plants show enhanced disease susceptibility to the virulent bacterium Pseudomonas syringae pv tomato DC3000 (P.s.t. DC3000) and also to the avirulent P.s.t. DC3000 carrying the effector avrRpm1 gene. In addition, ocp11 plants are also compromised in resistance to the nonhost pathogen P. syringae pv tabaci. Genetic and molecular analyses reveal that ocp11 plants are not affected in salicylic acid perception. We cloned OCP11 and show that it encodes ARGONAUTE4 (AGO4), a component of the pathway that mediates the transcriptional gene silencing associated with small interfering RNAs that direct DNA methylation at specific loci, a phenomenon known as RNA-directed DNA methylation (RdDM). Thus, we renamed our ocp11 mutant ago4-2, as it represents a different allele to the previously characterized recessive ago4-1. Both mutants decrease the extent of DNA cytosine methylation at CpNpG and CpHpH (asymmetric) positions present at different DNA loci and show commonalities in all of the molecular and phenotypic aspects that we have considered. Interestingly, we show that AGO4 works independently of other components of the RdDM pathway in mediating resistance to P.s.t. DC3000, and loss of function in other components of the pathway operating upstream of AGO4, such as RDR2 and DCL3, or operating downstream, such as DRD1, CMT3, DRM1, and DRM2, does not compromise resistance to this pathogen.
INTRODUCTION
Plants are constantly challenged by infectious pathogens; however, successful infections are relatively rare, with only a few pathogens being able to damage the plant. This is explained in part by the observation that plants are equipped with a complex network of synergistic defense strategies that are induced either locally or systemically. Fundamental for this inducible defense system to operate is the ability of the plant to recognize potential parasites, a process mediated by the recognition of pathogen-associated molecular patterns by specific pattern recognition receptors in the plant. This recognition triggers an array of defense reactions that limits the pathogen growth. The sum of these plant defense responses may be classified as basal defense or nonhost resistance, the most prevalent form of disease resistance in plants, conferring resistance at the plant species level (Thordal-Christensen, 2003; Jones and Takemoto, 2004; Chisholm et al., 2006). Nonhost resistance is thought to be multigenic, and the inactivation of any one component may not be sufficient to render a plant susceptible. Despite the importance of nonhost resistance, it still remains very poorly understood, with only a few genes involved in the process being described to date.
The Arabidopsis thaliana NHO1 (for NONHOST RESISTANCE1) gene, which encodes a glycerol kinase, is the only gene found to be required for bacterial nonhost resistance, and nho1 mutant plants are more susceptible to nonhost strains of Pseudomonas (Kang et al., 2003). Similarly, Collins et al. (2003) have reported that the Arabidopsis pen1 (for penetration1), pen2, and pen3 mutants are disabled in nonhost penetration resistance against barley (Hordeum vulgare) powdery mildew (Blumeria graminis f. sp hordei). These results suggest that PEN1 syntaxin-mediated secretion and the concentration of PEN2 glycosyl hydrolases at fungal entry sites are required for preinvasion nonhost resistance (Collins et al., 2003; Lipka et al., 2005). In addition to this genetic evidence, several cellular processes have been found to be required for the activation of basal defense, including activation of a mitogen-activated protein kinase signaling pathway, transcriptional induction of pathogen-responsive genes, production of reactive oxygen species, and callose deposition to reinforce the cell wall at sites of infection to restrict microbial growth (Gomez-Gomez et al., 1999; Asai et al., 2002; Navarro et al., 2004; Zipfel et al., 2004). Despite these findings, the genetic basis for a role of plant defense in limiting nonhost pathogen growth still remains largely unknown.
There is a cause-effect relationship between basal defense and plant resistance; thus, basal defense should be a target to be weakened or delayed by the pathogen to succeed during infection. Pathogenic microbes (virulent pathogens) have evolved strategies to suppress this resistance by interfering with pathogen-associated molecular pattern recognition at the plasma membrane or by secreting effector proteins (virulence factors) into the plant cell to alter resistance signaling or manifestations of resistance responses that in turn cause plant disease (Abramovitch and Martin, 2004; Espinosa and Alfano, 2004; Nomura et al., 2005; Chisholm et al., 2006).
Some plant cultivars have developed a more specialized mechanism to detect microbes and to generate resistance, referred to as gene-for-gene resistance or effector-triggered immunity (ETI). ETI involves a disease resistance (R) protein that monitors the integrity of host cellular targets of pathogen effectors (Dangl and Jones, 2001). Activation of R protein–mediated resistance can also suppress microbial growth, but not before the invader has had an opportunity for limited proliferation. As part of the ETI process, the host cell mRNA expression profile changes rapidly and a hypersensitive cell death response occurs in cells immediately adjacent to the pathogen (Tao et al., 2003; Abramovitch and Martin, 2004; Greenberg and Yao, 2004).
The involvement of endogenous small interfering RNAs (siRNA) and microRNAs (miRNAs) in mediating the triggering of defense against pathogens is an area of considerable interest; however, defense regulation mediated by endogenous small RNAs has been reported only in a few cases. In Arabidopsis, >100 miRNAs have been reported (Lu et al., 2005) and shown to be important for plant development and abiotic stress tolerance (Mallory and Vaucheret, 2006; Sunkar et al., 2007), and one miRNA (miR393) was recently shown to contribute to basal defense against bacteria by targeting auxin signaling components (Navarro et al., 2006). In contrast with the relatively limited number of miRNAs, thousands of endogenous siRNAs have been sequenced (Xie et al., 2004; Lu et al., 2005). However, their biological role is largely unknown, except for the functions of transacting siRNAs in plant development and hormone signaling (Fahlgren et al., 2006) and the roles of some chromatin-associated siRNAs in DNA methylation and transcriptional gene silencing (Chan et al., 2004; Xie et al., 2004). Recently, a member of a subclass of endogenous siRNAs derived from the overlapping region of a pair of natural antisense transcripts (nat-siRNAs), designated nat-siRNAATGB2, was identified to be specifically induced by Pseudomonas syringae carrying the effector avrRpt2 (Katiyar-Agarwal et al., 2006). Induction requires the cognate host disease resistance R gene RPS2 (for RESISTANCE TO PSEUDOMONAS SYRINGAE) and the NON-RACE-SPECIFIC DISEASE RESISTANCE1 gene. This nat-siRNA was proposed to repress a negative regulator of the RPS2 resistance pathway, and the authors pointed out that endogenous siRNA-mediated gene silencing may serve as one important mechanism for gene expression reprogramming in plant defense responses.
Here, we present genetic and molecular evidence for the requirement of the ARGONAUTE4 (AGO4) gene for generating an effective resistance in Arabidopsis against P. syringae. AGO4 is one of the critical components in the transcriptional gene-silencing pathway associated with siRNA that directs DNA methylation at specific loci, a phenomenon known as RNA-directed DNA methylation (RdDM). Our data implicate DNA methylation, as regulated by AGO4, as part of the mechanism mediating plant immunity in Arabidopsis.
RESULTS
Identification of the ocp11 Mutant
The Arabidopsis ocp (for overexpressor of cationic peroxidase) mutants were identified in a screen for constitutive expression of the GUS (for β-glucuronidase) gene driven by the promoter of the defense-related Ep5C gene (PEp5C:GUS) (Coego et al., 2005a). The details of this screen have been described previously (Coego et al., 2005b). The Ep5C gene, which encodes an extracellular peroxidase, originally was isolated from tomato (Solanum lycopersicum) plants and shown to be transiently induced by the H2O2 generated during the interaction of both tomato and Arabidopsis plants with a pathogenic strain of P. syringae (Coego et al., 2005a). Rather than being a PR-type response gene, Ep5C is required for susceptibility to P. syringae, as demonstrated in transgenic tomato plants expressing an Ep5C-antisense gene construct (Coego et al., 2005a). Thus, the identified ocp mutants are anticipated to be different from those mutants isolated on the basis of constitutive expression of PR genes.
Here, we report the characterization of the ocp11 mutant, which is in the Columbia-0 (Col-0) background. Macroscopically, ocp11 plants were observed to be similar to wild-type plants in terms of both plant architecture and growth habit (Figure 1A). Figure 1B shows the constitutive expression of the GUS reporter gene in the Arabidopsis ocp11 mutant compared with the parental nonmutagenized transgenic PEp5C:GUS line. Leaves from the parental line showed no GUS activity, whereas the ocp11 mutant showed intense GUS staining throughout the rosette leaves (Figure 1B). F1 hybrid rosette leaves from a backcross between parental plants and the ocp11 plants still showed GUS activity (Figure 1B), indicating that the ocp11 mutation is dominant. Moreover, the GUS (staining pattern) activity segregated in the F2 progeny as a single semidominant Mendelian locus (OCP11:OCP11/ocp11:ocp11, 18:30:14 [P < 0.05, χ2 test]).
Figure 1.
Characterization of ocp11 Plants.
(A) Comparison of the macroscopic appearance of a 5-week-old wild-type plant (left) and an ocp11 plant (right).
(B) Comparative histochemical analysis of GUS activity in fully expanded rosette leaves from a parental wild-type plant carrying the PEp5C:GUS transgene (left), an ocp11 mutant plant (center), and an F1 ocp11/OCP11 plant (right).
(C) Histograms of P.s.t. DC3000 growth rate in wild-type (Col-0), ocp11, npr1, and dth9 plants. The growth of the bacteria was determined at 0 (white bars), 3 (black bars), and 5 (gray bars) DPI. Analysis of variance indicates differences with a significance level of 0.05. FW, fresh weight.
ocp11 Plants Show Enhanced Disease Susceptibility to P.s.t. DC3000
Since Ep5C gene expression was originally found to be upregulated following infection by P. syringae (Coego et al., 2005a), we reasoned that the constitutive expression of PEp5C:GUS observed in ocp11 plants might be accompanied by an altered disease resistance response in the same mutant plants. To test this hypothesis, we inoculated ocp11 plants with the virulent bacterial pathogen P. syringae pv tomato DC3000 (P.s.t. DC3000) and monitored the growth of the bacteria in the inoculated plants. Wild-type plants as well as the more susceptible npr1 (for nonexpressor of pathogenesis-related genes1) (Cao et al., 1997) and dth9 (for detachment9) (Mayda et al., 2000) mutants were similarly inoculated and used as controls. The resulting quantification of bacterial growth in the different genotypes is shown in Figure 1C. The growth of P.s.t. DC3000 in ocp11 plants was higher than that observed in wild-type plants and significantly higher than that observed for npr1 plants, but lower than that recorded for dth9 plants, at 3 to 5 d post inoculation (DPI). This indicates that the identified mutation in the OCP11 locus renders the plant more susceptible to P.s.t. DC3000.
The plant defense hormone salicylic acid (SA) is vital for mounting an appropriate disease resistance response to P.s.t. DC3000. Thus, mutants impaired in SA accumulation are more susceptibility to bacterial pathogens and fail to induce the expression of PR genes (Metraux, 2002).
To investigate the possible role of SA in the observed enhanced susceptibility of ocp11 plants to P.s.t. DC3000, we attempted to complement pharmacologically the ocp11 phenotype with exogenous SA administration. After spraying ocp11 plants with a 200 μM solution of SA, the extent of bacterial growth was reduced ∼10-fold. This induced resistance was of a similar magnitude to that attained in similarly treated wild-type plants (Figure 2A), but it was not enough to bring down to wild-type levels the observed enhanced susceptibility of ocp11 plants. Thus, the results indicate that the ocp11 mutant does not seem to be compromised in the perception of SA. This idea was further reinforced by the observation that the induced expression of SA-responsive genes, PR1 and GST-6 (for GLUTATHIONE S-TRANSFERASE6), following infection with the avirulent P.s.t. DC3000 (avrRpm1) was similar in ocp11 and wild-type plants (Figure 2B), suggesting that correct perception of SA is not altered in ocp11 plants.
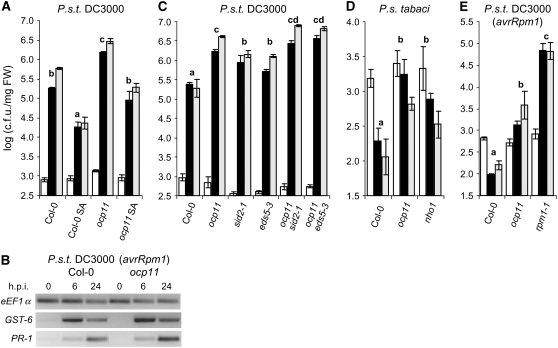
Figure 2.
Effect of SA on P.s.t. DC3000 Growth Rate in ocp11 Plants.
(A) Effect of SA on the growth rate of P.s.t. DC3000 in wild-type (Col-0) and ocp11 plants. Two days prior to the inoculation with P.s.t. DC3000, plants were sprayed once with either water or a 200 μM solution of SA until runoff.
(B) Induced expression of GST-6 and PR-1 following inoculation of wild-type and ocp11 plants with P.s.t. DC3000 (avrRpm1). h.p.i., hours after inoculation.
(C) Histograms of P.s.t. DC3000 growth rate in wild-type (Col-0), ocp11, sid2-1, eds5-3, ocp11 sid2-1, and ocp11 eds5-3 plants.
(D) Histograms of P.s. tabaci growth rate in wild-type (Col-0), ocp11, and nho1 plants.
(E) Histograms of P.s.t. DC3000 (avrRpm1) growth rate in wild-type (Col-0), ocp11, and rpm1-1 plants.
Bacterial growth was determined at 0 (white bars), 3 (black bars), and 5 (gray bars) DPI. Analysis of variance indicates differences with a significance level of 0.05.
To genetically assess the role of SA in the phenotype of ocp11 plants in relation to the observed enhanced susceptibility to P.s.t. DC3000, we crossed ocp11 plants with sid2-1 (SA induction–deficient2) plants and with eds5-3 (enhanced disease susceptibility5) plants and generated the double mutants ocp11 sid2-1 and ocp11 eds5-3. Both sid2-1 (Wildermuth et al., 2001) and eds5-3 (Nawrath et al., 2002) plants are impaired in SA biosynthesis as derived from the isochorismate pathway and thus are more susceptible to P.s.t. DC3000. The rate of bacterial growth observed in ocp11 sid2-1 and ocp11 eds5-3 plants was significantly higher than that measured in the parental sid2-1 or eds5-3 single mutants but only slightly higher than that of the ocp11 mutant (Figure 2C). Although we would have expected a stronger additive effect in the susceptibility of the double mutants to P.s.t. DC3000 if the ocp11 mutant was SA-independent, the fact that the independent mutants are per se more susceptible to the bacteria may impede an unambiguous interpretation of the results. In any case, we always detected an increase in the susceptibility of the double mutants compared with that observed in the single mutant plants. Therefore, it is likely that in the ocp11 mutant, the isochorismate pathway remains intact. This also agrees with the previous observation that the increased susceptibility of ocp11 plants still remains visible and even higher than that of wild-type plants after external SA administration (Figure 2A).
ocp11 Plants Are Compromised in Disease Resistance
It is known that SA produced by the isochorismate pathway is not necessary for resistance against a nonhost pathogen such as P. syringae pv phaseolicola (van Wees and Glazebrook, 2003). Thus, we reasoned that if at least part of the observed enhanced susceptibility in ocp11 plants is controlled in a SA-independent manner, the ocp11 mutant may be similarly compromised in resistance against a nonhost bacterium.
To test whether this may be the case, we challenged ocp11 and wild-type plants with the nonhost bacterium P. syringae pv tabaci (P.s. tabaci) and measured the extent of bacterial growth. As shown in Figure 2D, in ocp11 plants, the rate of growth of P.s. tabaci was ∼10-fold higher than that observed in wild-type plants. The nonhost mutant nho1, previously identified to be compromised in basal defense (Kang et al., 2003), was used as a control and also assayed against P.s. tabaci. As occurred with ocp11, nho1 plants allowed a higher growth rate of the nonhost bacterium (Figure 2D). These results indicate that ocp11 plants are also afflicted in nonhost resistance.
To further extend our analysis of ocp11 plants, we tested the response of this mutant toward P.s.t. DC3000 (avrRpm1), which triggers a gene-for-gene response in Col-0. As expected, inoculation of wild-type plants with P.s.t. DC3000 (avrRpm1) resulted in a disease resistance response that halted bacterial growth (Figure 2E). Conversely, in the inoculated ocp11 plants, the Rpm1-mediated resistance was partially compromised, allowing an ∼50 fold higher bacterial growth than in wild-type plants. This susceptibility of ocp11 plants, albeit significant compared with that of wild-type plants, was less prominent than that of rpm1-1 plants, which allowed a >∼100 fold higher titer of bacterial growth (Figure 2E).
In sum, these results suggest that the observed enhanced susceptibility of ocp11 plants is due to a defect that affects resistance to both host and nonhost bacteria and indicate that OCP11 is an important cellular component mediating resistance to P. syringae.
OCP11 Is At2g27040 and Encodes AGO4
To map the position of ocp11, we outcrossed ocp11 plants to Landsberg erecta (Ler) plants, and F2 plants were scored for cosegregation of high constitutive GUS activity with simple sequence length polymorphisms (SSLPs) (Bell and Ecker, 1994). Analysis of 38 ocp11 plants selected from a Ler × ocp11 F2 population allowed mapping of ocp11 to chromosome 2, at 2.37 Mb between markers g6842 and nga361. Analysis of 423 plants with new polymorphic markers allowed narrowing the position of ocp11 to a 137-kb interval (between BAC clones F12C20 and T20P8) containing 29 annotated genes (Figure 3A). DNA sequencing of this interval allowed us to find one nucleotide substitution corresponding to a G-for-A transition, as expected for a mutation caused by ethyl methanesulfonate mutagenesis. This leads to a Glu-for-Lys substitution at position 641 of the protein encoded by the gene At2g27040 (Figure 3A). Therefore, OCP11 should correspond to At2g27040.
Figure 3.
Positional Cloning of OCP11.
(A) Location of OCP11 on the BAC clone T20P8. OCP11 was positioned in a 137-kb region between BACs F12C20 and T20P8 and corresponds to the gene At2g27040. Lowercase letters mark nucleotide sequences at the beginning of the corresponding PIWI domain of the protein encoded by the At2g27040 gene. The mutant allele is indicated below the wild-type sequence. The G-to-A transition is indicated in boldface uppercase letters. The deduced amino acid sequences are indicated in uppercase single-letter code below each nucleotide triplet, and the boldface letters mark the amino acid changes (E to K) in the protein sequences.
(B) Histochemical staining for GUS activity in fully expanded rosette leaves obtained from wild-type transgenic plants carrying PEp5C:GUS upon transformation with P35S empty vector (left), construct P35S:ocp11 (center), or construct P35S:OCP11 (right).
To test whether the mutation identified in the At2g27040 gene could confer constitutive GUS activity in wild-type plants carrying the PEp5C:GUS transgene, a 3.0-kb At2g27040 cDNA fragment was obtained from ocp11 and wild-type plants. The cDNAs were inserted downstream of a 35S cauliflower mosaic virus promoter to generate P35S:ocp11 (corresponding to At2g27040 cDNA obtained from ocp11 plants) and P35S:OCP11 (corresponding to At2g27040 cDNA obtained from wild-type plants) constructs that were used to transform PEp5C:GUS plants by Agrobacterium tumefaciens–mediated plant transformation. Fifteen of 20 P35S:ocp11 transgenic lines showed constitutive GUS activity (Figure 3B), while none of the 12 P35S:OCP11 lines or the 10 P35S (empty vector) lines generated and analyzed showed constitutive GUS activity (Figure 3B). These results thus support the consideration that At2g27040 is OCP11 and also that the identified mutation in this locus exerts a dominant effect.
Previously, the At2g27040 gene was named AGO4, as it is the gene in which the mutation ago4-1 was identified as a suppressor of the clk-st mutant, an epigenetic allele of SUP (for SUPERMAN) caused by extensive DNA methylation (Zilberman et al., 2003). Hence, from now on, the ocp11 mutation will be named ago4-2.
AGO4 belongs to the ARGONAUTE family of proteins, which are characterized by the presence of two domains: the PAZ domain, which is located at the N-terminal region of the protein (Figure 3A), and the PIWI domain, which is similar to ribonuclease H and localizes at the C-terminal region of the protein (Song et al., 2004).
ago4-1 was identified as a recessive loss-of-function mutation (Zilberman et al., 2003). By contrast, ago4-2 is a dominant mutation that causes the substitution of a Glu at position 641 (located inside the PIWI domain) for a Lys. This Glu-641 is close to the DDH motif (Asp-660–Asp-742–His-874). The DDH motif was shown previously to be necessary for Arabidopsis AGO1 and AGO4, as well as for the human AGO2 protein, to exert their slicer activity (Baumberger and Baulcombe, 2005; Rivas et al., 2005; Qi et al., 2006). It is thus possible that the exchange of the carboxylic group of position 641 (Glu) for an amino group (Lys) in ago4-2 causes an alteration in the distribution of the charge at the protein surface at positions close to the DDH motif, which, in turn, may affect the activity of the protein.
AGO4 is one of the critical components in the transcriptional gene-silencing pathway associated with siRNA that directs DNA methylation at specific loci (Qi and Hannon, 2005), or RdDM. RdDM requires DCL3 (for Dicer-Like3), RDR2 (for RNA-dependant RNA polymerase2), Pol IV (for RNA polymerase IV), and AGO4. Mutations affecting the function of any of these proteins lead to a decreased accumulation of siRNA and, in turn, to a decrease in DNA methylation at many endogenous loci (Zilberman et al., 2003; Xie et al., 2004; Herr et al., 2005; Onodera et al., 2005). Twenty-four-nucleotide siRNA species are probably the main signal for RdDM and are also believed to confer sequence specificity on the process (Xie et al., 2004; Qi et al., 2006). In this scenario, DCL3 functions as the ribonuclease that processes the double-stranded RNA precursors generating the 24-nucleotide siRNAs (Xie et al., 2004; Henderson et al., 2006) and RDR2 presumably functions as a polymerase that generates the double-stranded RNA molecules (Xie et al., 2004) from templates probably arising from the transcription of DNA-associated RNA as mediated by Pol IVa (Pontes et al., 2006). Finally, AGO4 is proposed to be the critical component of the effector complex that directs DNA methylation as guided by siRNA (Qi et al., 2006).
ago4-2 Is Impaired in DNA Methylation and Functions as a Dominant-Negative Mutant
It has been shown that the recessive ago4-1 mutant decreases the extent of DNA cytosine methylation at CpNpG and CpHpH (asymmetric) positions present at different DNA loci corresponding to retroelements, transposons and repetitive DNA sequences (Zilberman et al., 2003).
To ascertain whether ago4-2 may show similar DNA methylation defects, we analyzed the function of the ago4-2 mutation in maintaining CpNpG and asymmetric methylation at an endogenous locus using bisulfite genomic sequencing. Figure 4A shows the percentage of cytosine methylation at the At SN1 retroelement locus in the ago4-1 and ago4-2 mutant backgrounds. ago4-2 plants showed a dramatic and reproducible decrease in CpNpG and asymmetric methylation compared with wild-type plants. This methylation decrease was similar to that reported for ago4-1 plants. The fraction of 5 methyl cytosine (5mC) dropped 25 to 58% at positions CpNpG and 25 to 54% at positions CpHpH in the ago4-2 mutant, while the fraction of 5mC remained almost invariant at CpG positions (0 to 16%). In ago4-1 plants, this decrease appears to be slightly more pronounced compared with that of its parental line (Ler) and is of a magnitude similar to that reported previously (Zilberman et al., 2003). The results thus indicate that ago4-2 phenocopies ago4-1 in terms of methylation defects and suggest that the ago4-1 allele seems to be stronger than ago4-2. Since ago4-1 is a recessive loss-of-function mutation and the ago4-2 allele behaves dominantly, we conclude that ago4-2 is a dominant-negative mutation.
Figure 4.
Comparison of ago4-1 and ago4-2 Phenotypes.
(A) Quantification of 5mC at positions CpG (left), CpHpH (center), and CpNpG (right) present at the At SN1 locus, as detected by bisulfite sequencing of DNA samples from ago4-2, ago4-1, and the corresponding wild-type controls, Col-0 and Ler, respectively.
(B) Histochemical staining of GUS activity in fully expanded rosette leaves obtained from wild-type transgenic PEp5C:GUS plants (left), homozygous ago4-1 PEp5C:GUS plants (center), and heterozygous F1 ago4-1/AGO4-1 plants (right).
(C) Histograms of P.s.t. DC3000 growth rate in ago4-2, ago4-1, and the corresponding wild-type plants, Col-0 and Ler, respectively.
(D) Histograms of P.s.t. DC3000 (avrRpm1) growth rate in wild-type (Ler), ago4-1, and nho1 plants.
(E) Histograms of P.s. tabaci growth rate in wild-type (Ler), ago4-1, and nho1 plants.
Bacterial growth was determined at 0 (white bars), 3 (black bars), and 5 (gray bars) DPI. Analysis of variance indicates differences with a significance level of 0.05.
To extend the parallelism between ago4-1 and ago4-2 beyond the methylation pattern, we tested whether GUS activity as driven by the Ep5C gene promoter would be similarly affected when assayed in an ago4-1 background. For this, we outcrossed ago4-1 to PEp5C:GUS transgenic Arabidopsis and F2 plants were analyzed for GUS activity. Fourteen of 77 F2 plants showed constitutive GUS activity (Figure 4B), as expected for a recessive mutation like ago4-1. Here, F1 plants did not show any constitutive GUS activity (Figure 4B). Thus, from a comparison with the previous results derived from ago4-2 plants (Figure 1), our conclusion was that ago4-2 is a dominant-negative mutation.
We extended this parallelism also to a pathogenic context by studying the disease resistance of ago4-1 plants to bacterial pathogens. As shown in Figure 4C, the P.s.t. DC3000 growth rate in ago4-1 plants was significantly higher than that observed in wild-type plants but of a magnitude similar to that recorded in ago4-2 plants. Furthermore, the enhanced disease susceptibility of ago4-1 plants was also observed upon inoculation with the avirulent P.s.t. DC3000 (avrRpm1) (Figure 4D) and also with the nonhost P.s. tabaci (Figure 4E). In all of the experiments, ago4-1 plants allowed a higher growth rate of the pathogens compared with the rate observed in the parental Ler irrespective of the pathogen used.
These results further demonstrate the requirement of AGO4 for a proper resistance to P. syringae and reinforce the consideration of a dominant-negative behavior for ago4-2, as the enhanced disease susceptibilities attained in ago4-1 and ago4-2 plants are comparable.
AGO4 Works Independently of Other Components of the RdDM Pathway in Mediating Disease Susceptibility to P.s.t. DC3000
Since AGO4 seems to be necessary for mounting an effective disease resistance response, we sought to find a link between RdDM and plant defense. As RDR2 cooperates with DCL3 to form chromatin-associated 24-nucleotide siRNAs that guide AGO4 to direct DNA methylation, we tested whether genetic defects in either of these two genes could reproduce ago4-associated phenotypes.
For this, we crossed rdr2-1 or dcl3-1 plants to PEp5C:GUS transgenic Arabidopsis and analyzed F2 plants for constitutive GUS expression. From the analysis of 84 F2 plants from the rdr2-1 × PEp5C:GUS cross, 16 plants showed constitutive GUS activity (see Supplemental Figure 1 online). Likewise, 12 of 82 F2 plants from the dcl3-1 × PEp5C:GUS cross showed constitutive GUS activity (see Supplemental Figure 1 online). In all cases, the pattern of GUS activity extended along the leaf blade and resembled that observed in ago4-1 and ago4-2 plants. Therefore, it appears that the RdDM pathway negatively regulates PEp5C:GUS transcription, as loss of function in any of the indicated genes confers upregulation of GUS expression.
However, and in marked contrast to what we found for ago4 mutants, rdr2-1 and dcl3-1 plants did not exhibit any defect when assayed for disease susceptibility to P.s.t. DC3000. As shown in Figure 5A, the growth rate of P.s.t. DC3000 in rdr2-1 and dcl3-1 plants was the same as that observed in wild-type plants and dissimilar to the enhanced susceptibility observed in the control ago4-2 plants. These results indicate that while RDR2, DCL3, and AGO4 are important for regulating transcription of the reporter PEp5C:GUS transgene, only AGO4 is essential for the correct plant defense response.
Figure 5.
P.s.t. DC3000 Growth Rate in Different Mutants of the RdDM Pathway.
(A) Histograms of P.s.t. DC3000 growth rate in wild-type (Col-0), dcl3-1, rdr2-1, and ago4-2 plants.
(B) Histograms of P.s.t. DC3000 growth rate in wild-type (Ler), ago4-1, and cmt3-7 plants.
(C) Histograms of P.s.t. DC3000 growth rate in wild-type (Col-0), ago4-2, dmr1-2 dmr2-2, and drd1-6 plants.
Growth of the bacteria was determined at 0 (white bars), 3 (black bars), and 5 (gray bars) DPI. Analysis of variance indicates differences with a significance level of 0.05.
Downstream of AGO4 in the RdDM pathway, different methyltransferases have been described that participate in DNA methylation. In Arabidopsis, cytosine methylation in a CpNpG or CpHpH sequence context is maintained by CMT3 (for Chromomethylase3) and DRM (for Domains Rearranged Methyltransferase1 and -2) (Bartee et al., 2001; Cao and Jacobsen, 2002). The chromatin-remodeling protein DRD1 (for Defective in RNA-directed DNA methylation) is also required for the maintenance of CpNpG and asymmetric methylation at some endogenous loci (Kanno et al., 2004), acting through both DRM2 and CMT3 (Chan et al., 2006). The observation that symmetric and asymmetric methylation at the At SN1 locus is altered in ago4 mutants and that these mutants show enhanced susceptibility to P.s.t. DC3000 prompted us to investigate whether DRD1, CMT3, DRM1, and DRM2 could possibly also play important roles in plant defense. To address this question, we studied the susceptibility of drd1-6 plants, cmt3-7 plants, and drm1-2 drm2-2 double mutant plants to P.s.t. DC3000 compared with their respective parental plants and also compared with ago4-1 or ago4-2 plants. As shown in Figures 5B and 5C, the growth rate of P.s.t. DC3000 measured in drd1-6, drm1-2 drm2-2, and cmt3-7 plants was similar to that observed in their corresponding wild-type parental plants Col-0 and Ler. Again, the enhancement in susceptibility to P.s.t. DC3000 characteristic of ago4-2 and ago4-1 plants could not be reproduced by any of the chromatin-remodeling mutants analyzed. As with RDR2 and DCL3, these results indicate that DRD1, CMT3, DRM1, and DRM2 are not essential for mounting a correct plant defense. Thus, we are left with AGO4 as the component of the RdDM pathway that is critical for keeping resistance intact in Arabidopsis.
Ep5C Gene Expression Is Regulated by Methylation
Since PEp5C:GUS is constitutively expressed in different mutant backgrounds corresponding to critical genes of the RdDM pathway (Figures 1 and 4; see Supplemental Figure 1 online), we reasoned that the observed constitutive activation of the Ep5C gene promoter might be due to a change in the methylation pattern in the promoter sequence. To investigate this, we extracted DNA from wild-type and ago4-1 plants carrying the PEp5C:GUS construct and measured DNA methylation across a 188-nucleotide fragment of the Ep5C promoter using bisulfite genomic sequencing. This region in the promoter of the Ep5C gene was selected due to its absolute requirement for transcriptional activation following pathogen challenge, as deduced from a functional analysis of the Ep5C promoter in Arabidopsis (A. Agorio, A. Coego, and P. Vera, unpublished data). DNA bisulfite sequencing of this region in wild-type plants revealed an extensive methylation at CpG (79%), CpHpH (27%), and CpNpG (31%) sites (Figure 6A). By marked contrast, bisulfite sequencing of the same region in ago4-1 plants, which show constitutive GUS activity, revealed that it was severely hypomethylated (Figure 6A; see Supplemental Figure 2 online). In this mutant, the fraction of 5mC dropped 87% at position CpG, 99% at position CpHpH, and 82% at position CpNpG. Therefore, the methylation status at a 188-nucleotide fragment located at the 5′ promoter region of the Ep5C gene inversely correlates with the transcriptional activation of the downstream encoded gene. These results support the notion that to keep the Ep5C gene repressed under resting conditions, and thus in a highly methylated state at its 5′ promoter region, the role of AGO4 is pivotal. Interestingly, the alteration in the methylation pattern observed for Ep5C in ago4-1 plants differs from that observed for the AtSN1 locus. For the latter, methylation at CpG positions was almost invariable in ago4-1 plants compared with wild-type plants (Figure 4A).
Figure 6.
Methylation Status of the Ep5C 5′ Promoter Region in Arabidopsis and Tomato Plants.
(A) Quantification of 5mC at positions CpG (left), CpHpH (center), and CpNpG (right) in a 188-nucleotide 5′ promoter region of PEp5C:GUS in Arabidopsis transgenic plants, as detected by bisulfite sequencing of DNA samples from ago4-1 PEp5C:GUS and the corresponding wild-type control PEp5C:GUS.
(B) Quantification of 5mC at positions CpG (left), CpHpH (center), and CpNpG (right) in a 188-nucleotide 5′ promoter region of the Ep5C locus in tomato. DNA samples were obtained from leaf tissue at 4 h after inoculation with P.s.t. DC3000. Control plants were inoculated with 10 mM MgSO4 (mock).
Since Ep5C was cloned originally from tomato plants, we also tested the methylation status of the same 188-nucleotide region at the 5′ promoter region of the endogenous Ep5C locus before and after pathogen challenge. Interesting to note here is the difference between the different levels of methylation observed between the Arabidopsis and tomato samples under resting conditions. This difference is likely because the Arabidopsis transgene has not had any maintenance of methylation, in contrast with tomato, in which there is presumably de novo methylation. In DNA samples obtained from tomato plants under resting conditions in which the Ep5C gene is not expressed (Coego et al., 2005a), bisulfite sequencing revealed extensive methylation of 5mC at CpG (82%), CpHpH (47%), and CpNpG (83%) positions (Figure 6B). However, when the same experiments were performed with DNA samples taken at 4 h after inoculation from tomato plants inoculated with P.s.t. DC3000, a time point at which Ep5C mRNA starts to accumulate in the inoculated tissue (Coego et al., 2005a), there was a shift from methylation to demethylation at the same promoter region. While the methylation status at CpG positions remained invariant, those at CpHpH and CpNpG positions showed 31 and 20% reductions, respectively, at 4 h after inoculation with P.s.t. DC3000 (Figure 6B; see Supplemental Figure 2 online). Since these cells cannot have lost methylation through the loss of maintenance, one would have to invoke a mechanism of active demethylation following pathogen attack that, in turn, allows transcriptional activation of Ep5C. Additionally, since it does not seen likely that AGO4 would play a role in actively demethylating DNA, then AGO4 would seem to play a role in maintaining certain genes required for susceptibility in an inactive state, as may be the case of Ep5C. This may offer an explanation of why loss of function of AGO4 leads to plants with enhanced disease susceptibility to Pseudomonas.
In sum, these results suggest that reversible methylation of the Ep5C promoter, irrespective of whether it occurs in the transgene or in the endogenous gene, forms the basis of a mechanism controlling gene expression in which high methylation correlates with gene repression and vice versa. This, in turn, suggests a link with RdDM-mediated methylation under conditions of gene shutdown. Furthermore, this methylation program is modulated and can be partially inhibited by signals generated during the course of a plant–pathogen interaction.
DISCUSSION
The data presented in this work provide evidence for a role of AGO4 in regulating certain aspects of disease resistance in Arabidopsis.
Searching for mutants showing constitutive expression of the pathogen-inducible Ep5C gene and an associated altered disease response, we identified a dominant mutant that was named ocp11 (Coego et al., 2005b). Map-based cloning of this mutant revealed that OCP11 is AGO4. A mutation in this locus (ago4-1) was previously found to mediate suppression of the clk-st mutant phenotype (Zilberman et al., 2003). In view of this, we renamed the ocp11 mutant ago4-2.
We demonstrate that ago4 mutants show enhanced disease susceptibility to the virulent bacterium P.s.t. DC3000 as well as compromised resistance to the avirulent P.s.t. DC3000 (avrRpm1) and to the nonhost P.s. tabaci. Surprisingly, the fact that mutations in the AGO4 locus compromise resistance against the nonhost pathogen P.s. tabaci implies that the ago4 mutants are compromised in nonhost resistance, a poorly understood phenomenon by means of which a particular plant species is resistant to potential pathogens that infect other plant species. To our knowledge, only the NHO1 gene (Kang et al., 2003), encoding a glycerol kinase, has been described as required for basal defense against Pseudomonas. However, at variance with what is observed in ago4 mutants, the nho1 mutant does not show enhanced susceptibility to P.s.t. DC3000, while the resistance to P.s. tabaci and to P.s.t DC3000 (avrRpm1) is compromised. This difference can be interpreted as suggesting that AGO4 may act independently of NHO1 for basal defense in Arabidopsis or, alternatively, may act upstream in the signal pathway.
The resistance of Arabidopsis to P.s.t. DC3000 depends on SA, and mutants compromised in the synthesis (e.g., sid2-1 and eds5-3) or perception (e.g., npr1) of this hormone become more susceptible to this pathogen. In this work, we demonstrate that the ago4-2 mutant is even more susceptible to P.s.t. DC3000 than is the sid2-1, the eds5-3, or even the npr1 mutant. This enhanced susceptibility of ago4-2 plants does not seem to be exerted by a compromised perception of SA. However, despite the observation that ago4-2 plants correctly perceive SA when applied exogenously, exogenous SA does not completely complement the demonstrated enhanced susceptibility to P.s.t. DC3000. These latter observations may indicate that there is a component of the defense response against P.s.t. DC3000 that is not entirely dependent on SA. Interestingly, the previous observation that the defense against nonhost Pseudomonas is independent of SA (at least of that produced by the isochorismatic acid pathway) (van Wees and Glazebrook, 2003) may reinforce our consideration that SA is not essential for the enhanced susceptibility observed in ago4 mutants against P. syringae.
AGO4 is one of the critical components of RdDM (Qi and Hannon, 2005). Recent studies have demonstrated that many AGO4-associated small RNAs are derived from the sense or the antisense strands of genes, pseudogenes, and intergenic regions, indicating that AGO4 might have a previously unrecognized general role in regulating gene expression (Qi et al., 2006). Here, we have found that the methylation status of a 5′ promoter region of Ep5C can influence gene expression in an AGO4-dependent manner. In wild-type plants, the promoter region of Ep5C is highly methylated when gene expression is off. In ago4 plants, the gene is found to be constitutively on and the promoter is hypomethylated. This methylation in Arabidopsis plants seems not to be due to Ep5C being read as a transgene, since a similar methylation pattern is obtained when assayed in tomato plants for the endogenous Ep5C gene. Previous studies have demonstrated that Ep5C gene expression is rapidly activated following pathogen inoculation in both Arabidopsis and tomato (Coego et al., 2005a). Here, we show that this rapid activation of Ep5C following P.s.t. DC3000 infection correlates with demethylation of the promoter region, thus implying that Ep5C activation and demethylation are linked. Hence, for Ep5C, there should be a plastic genetic circuitry that serves to reprogram the methylation pattern under biotic stress and that finally dictates the execution of an effective defense response. For this to take place, AGO4 is required.
Likewise, it has been reported that AGO4 largely affects non-CG methylation at endogenous loci and suggested that AGO4 may mostly affect transcriptional silencing of loci that have a few CG sites, little CG methylation, or both, such as in the SUP gene (Zilberman et al., 2003). Nevertheless, a highly reduced methylation state at CG sites for a few genes that show a higher transcriptional rate in ago4-1 plants has been reported (Zilberman et al., 2004). This could explain why the observed transcriptional activation of PEp5C:GUS coincides with a drastic reduction of methylation at CG sites when assayed in ago4-1 plants.
A change in the methylation pattern of the genome under pathogenic conditions has been described previously, although at a global scale (Pavet et al., 2006). This may indicate that there might be other genes that, like Ep5C, may be regulated by demethylation under pathogenic conditions. These genes may account for the phenotype of enhanced susceptibility observed in ago4 plants. Likewise, some other genes may become repressed by demethylation in their coding regions, as observed to be the case in several mutants altered in the methylation pattern of DNA (Zhang et al., 2006). Thus, in ago4 mutant plants, it is likely that an imbalance between the induction and repression of several or many genes results in a defective defense response. It could also be argued that the sustained demethylation in ago4 plants provokes an alteration in the transcription of genes related to defense that is somehow perceived as inadequate by the plant, which, in turn, develops a compensatory mechanism. As a consequence, the plant is left in a refractory state that impedes the correct perception of initial stimuli derived from the intruder, resulting in a compromised resistance.
We have found that not all of the genes of the RdDM pathway are necessary for the resistance against P.s.t. DC3000. By far, the most abundant and broadly utilized small RNA pathway in Arabidopsis depends on RDR2 and DCL3, resulting in the accumulation of a highly diverse population of primarily 24-nucleotide siRNAs. However, from our observations of the behavior of rdr2-1 and dcl3-1 plants after bacterial inoculation, we suggest that neither of these two genes is essential for mounting a defense response. This does not disregard the possibility that redundant functions of other DCLs and RDRs may compensate for the lack of function associated with rdr2-1 and dcl3-1 mutations. This pathway has a major affiliation with repeated sequences, including transposon elements and retroelements. However, the extent to which the RDR2-DCL3 pathway regulates gene expression is not completely understood (Xie et al., 2004; Henderson et al., 2006; Kasschau et al., 2007).
Only 20% of the annotated Arabidopsis protein-coding genes have identity with at least one small RNA sequence, with RDR2-DCL3–dependent 24-nucleotide siRNAs being the most abundant. In this case, and at variance with what is observed for pseudogenes, loss of RDR2 or DCL3 did not negatively affect all small RNA size classes. The protein-coding gene–derived small RNAs are generated by several distinct pathways, including the RDR2-DCL3 pathway (Kasschau et al., 2007). In summary, and for this study, the existence of alternative pathways for the generation of small RNAs for protein-coding genes that are important for defense may explain why RDR2 and DCL3 are not essential for resistance to P.s.t. DC3000.
In this study, we also investigated the potential participation of DNA methyltransferases in the establishment of disease resistance to P.s.t. DC3000. In particular, we studied the implication of methylases operating downstream of AGO4, such as DRM1, DRM2, and CMT3, as well as the chromatin-remodeling protein DRD1. In no single case did we observe an alteration in the disease resistance response when experiments were performed in drm1-2 drm2-2, cmt3-7, and drd1-6 plants. Again, the possible explanation of functional redundancy may explain this lack of effect. For example, at loci such as At SN1 and at silent alleles of the SUP gene, DRM2 and CMT3 act redundantly to maintain non-CG DNA methylation (Cao and Jacobsen, 2002). Another example of this redundancy is offered by the observation that neither drm1 drm2 nor cmt3 shows any morphological defects, but the drm1 drm2 cmt3 triple mutant shows a pleiotropic suite of developmental abnormalities (Chan et al., 2006). Thus, some degree of redundancy may explain the lack of effect observed for the above-mentioned DNA methylases in the defense response to P.s.t. DC3000.
According to this scenario of redundancies in components operating upstream and downstream of AGO4 in RdDM, it is possible that AGO4 integrates the information coming from different routes mediating chromatin methylation by small RNAs. Although the generation of the majority of 24-nucleotide siRNAs is dependent upon Pol IV, and in turn on RDR2, Zhang et al. (2007) have described a population of 24-nucleotide siRNAs that is Pol IV–independent. Although AGO4 shows preferential association with 24-nucleotide siRNAs dependent on Pol IV, it also associates with 24-nucleotide siRNAs independent of Pol IV (Zhang et al., 2006). Moreover, recent studies have shown that a small fraction of AGO4-associated RNAs are miRNAs (Qi et al., 2006). From these miRNAs, a subset preferentially associates with AGO4 rather than with AGO1. This implies selectivity in how individual miRNAs are processed and passed onto specific AGO-containing RNA-induced silencing complexes (Qi et al., 2006). All of these observations favor the interpretation that AGO4 can funnel not only information coming from the RDR2-DCL3 pathway but also information originating from 24-nucleotide siRNAs and miRNAs generated by different pathways (Figure 7).
Figure 7.
Schematic Representation of the Arabidopsis RdDM Pathway and Proposed Mode of Action of AGO4 in Plant Defense.
RdDM (solid lines) requires RNA polymerase IV (Pol IVa) and RNA-dependant RNA polymerase 2 (RDR2) to form double-stranded RNA (dsRNA) and Dicer-Like3 (DCL3) to form 24-nucleotide siRNAs. The 24-nucleotide siRNAs guide AGO4 to its DNA target, and Chromomethylase3 (CMT3) and Domains Rearranged Methyltransferase2 (DRM2), dependent on Defective in RNA-directed DNA methylation (DRD1), mediate DNA methylation. For the AGO4 mode of action in plant defense, it is proposed (dashed lines) that a source of 24-nucleotide siRNAs different from that generated by DCL3 or a source of miRNAs would be required. This would require a DCL (DCL?) protein different from DCL3. It is also possible that other methyltransferases (¿?), different from DRM2 and CMT3, would be necessary for AGO4 action in plant defense. nt, nucleotide.
Unraveling the mechanism controlling the participation of AGO4 in disease susceptibility and resistance, and identification of other components of the complex network that underlie the biology of small RNAs in plants, are interesting challenges for the future. The understanding of how this information is orchestrated and translated into a reprogramming of the genome by means of DNA methylation should have great potential for providing crop plants with durable resistance to potential pathogens.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana mutants ago4-1 (N6364) and cmt3-7 (N6365) were obtained from the ABRC at Ohio State University. npr1 was provided by X. Dong (Duke University, Durham, NC). rpm1-1 was provided by J. Dangl (University of North Carolina, Chapel Hill). nho1 was provided by J.M. Zhou (National Institute of Biological Sciences, Beijing). sid2-1 and eds5-3 were provided by J.P. Metraux (Université de Fribourg, Switzerland). rdr2-1 and dcl3-1 were provided by J. Carrington (Oregon State University, Corvallis). drd1-6 was provided by M. Matzke (Gregor Mendel Institute of Molecular Plant Biology, Vienna). drm1-2 drm2-2 and the ago4-1 mutant used in the At SN1 bisulfite sequencing assays were provided by S. Jacobsen (University of California, Los Angeles). All of the mutants were in the Col-0 or Ler genetic background, as indicated in the text and figure legends. Arabidopsis and tomato (Solanum lycopersicum) Rio Grande (Pto−) plants were grown in a growth chamber (19 to 23°C, 85% RH, 100 μE·m−2·s−1 fluorescent illumination) in a 10-h light/14-h dark photoperiod.
GUS Staining
For GUS staining, plant leaves were incubated overnight at 37°C in GUS staining buffer as described previously (Coego et al., 2005a).
ago4-2 Mapping and Phenotyping
The ago4-2 mutant was backcrossed twice to the line PEp5C:GUS to confirm its dominant inheritance. To map the ago4-2 mutation, ago4-2 plants were crossed to Ler, and F1 plants were allowed to self. F2 families were screened for segregation of the ago4-2 constitutive GUS activity phenotype. DNA was isolated from 423 F2 plants showing the ago4-2 phenotype and analyzed by PCR using SSLP markers described by Bell and Ecker (1994) or SSLP markers derived from the database of polymorphisms between the Ler and Col-0 ecotypes (http://www.Arabidopsis.org).
The ago4-2 and AGO4 cDNAs were amplified by RT-PCR using RNA from ocp11 plants and from a cDNA template obtained from RIKEN (clone pda01811), respectively, using forward primer AGO4-FL-Fwd (5′-ATCCTCTCTTGTTTCGGCTAGGGT-3′) and reverse primer AGO4-FL-Rev (5′-GCAAACAAGGCTGGCGATAATAGT-3′). The amplified products were cloned into the pCAMBIA1300 vector. These plasmids were introduced into Agrobacterium tumefaciens strain GV3101/pMP90, and PEp5C:GUS transgenic Arabidopsis plants were transformed using the floral dip method (Weigel and Glazebrook, 2002). The ago4-2–conferred phenotype was scored as the appearance of constitutive GUS activity in T1 plants.
ago4 Genetic Markers
A molecular marker used to genotype the ago4-2 mutation was obtained using AGO4-Rev2280 (5′-ATGCCATCGTCTTCAGTTCCA-3′) and AGO4-2-dCAP (5′-GCCTGAACTCAATGTTAAGTCTA-3′) primers and the restriction enzyme XbaI. The molecular marker used to genotype ago4-1 plants was described previously (Zilberman et al., 2003).
DNA Bisulfite Sequencing
DNA bisulfite sequencing was performed as described previously (Clark et al., 1994) with slight modifications using DNA obtained from Arabidopsis or tomato 5-week-old plant leaves. Briefly, after 20 s of sonication, genomic DNA was purified with the Wizard DNA Cleanup system (Promega). One to 2 μg (20 μl) of this DNA was denatured at 94°C for 5 min followed by the addition of 1 μL of fresh 6.3 n NaOH and incubation at 39°C for 30 min. After adding 208 μL of fresh solution A (44% [w/v] sodium bisulfite, 0.0084% [w/v] hydroquinone, and 0.24 n NaOH) to 21 μL of denatured DNA, the reaction was incubated at 55°C for 16 h. The DNA was then purified with the Wizard DNA Cleanup system and dissolved in 50 μL of water. The collected DNA was incubated at 37°C for 15 min after 2.5 μL of 6.3 n NaOH was added, and this was followed by precipitation and dissolving in 30 μL of water.
Converted DNA samples were amplified by PCR, and products were cloned into the pTZ57R vector. Eighteen independent clones were sequenced for each sample.
The region of At SN1 analyzed was amplified using primers AtSN1-BS1 (5′-GTTGTATAAGTTTAGTTTTAATTTTAYGGATYAGTATTAATTT-3′) and AtSN1-BS2 (5′-CAATATACRATCCAAAAAACARTTATTAAAATAATATCTTAA-3′) as described previously (Xie et al., 2004). The region of the Ep5C promoter analyzed corresponds to positions 294 to 482 of EP5C gene top strand (Coego et al., 2005a) and was amplified using primers EP5C-BS1 (5′-ATAGAGTTGAATAAGTAGATATATGAGT-3′) and EP5C-BS2 (5′-CCACAAATCATTTTATTAAAATAATCATTT-3′).
Expression Analysis
To analyze the level of gene expression by RT-PCR, we performed first-strand cDNA synthesis using 5 μg of purified total RNA prepared from leaf tissues and a reverse transcriptase kit (Revert Aid H Minus first-strand cDNA synthesis kit; Fermentas) with oligo(dT)18. The primers used to amplify PR1 (At2g14610) were PR1-Fwd (5′-ATGAATTTTACTGGCTATTC-3′) and PR1-Rev (5′-AACCCACATGTTCACGGCGGA-3′). The primers used to amplify GST-6 (At1g02930) were AtGST6-Fwd (5′-ATGGCAGGAATCAAAGTTTCGGTC-3′) and AtGST6-Rev (5′-GAGATTCACTTAAAGAACCTTCTG-3′). Those used to amplify eEF1α (At5g60390) were eEF1α-Fwd (5′-GCACAGTCATTGATGCCCCA-3′) and eEF1α-Rev (5′-CCTCAAGAAGAGTTGGTCCCT-3′). PCR amplification was programmed for 17 to 18 cycles, with each cycle consisting of 95°C for 0.5 min, 55°C for 0.5 min, and 72°C for 0.5 min.
Bacterial Strains and Bacterial Growth Assays
Bacterial strains used in this study include Pseudomonas syringae pv tomato DC3000, P. syringae pv tomato DC3000 (avrRpm1), and P. syringae pv tabaci. Bacteria were grown overnight at room temperature in King's B solid medium with appropriate antibiotics and diluted to the desired concentration with 10 mM MgSO4 for plant inoculation. These bacteria were used to infect 5-week-old Arabidopsis leaves by infiltration (Mayda et al., 2000) or to infect 2-week-old Arabidopsis plants by dipping using 0.02% Silwet L-77 (Tornero and Dangl, 2001). When infiltrated, P.s.t. DC3000 was used at a suspension of 105 colony-forming units (cfu)/mL, while P.s.t DC3000 (avrRpm1) and P.s. tabaci were used at a suspension of 5 × 105 cfu/mL. For infection by dipping, P.s.t. DC3000 was used at a suspension of 2.5 × 107 cfu/mL, while the others were used at a suspension of 5 × 107 cfu/mL. The bacterial growth was determined at 0, 3, and 5 DPI. For each data point, four samples were used, and the data are represented as means ± se of log (cfu/cm2), when bacteria were infiltrated, or log (mg fresh weight/cm2), when infection was done by dipping. All of the experiments were repeated at least twice with similar results.
Tomato plants were infected by infiltration as described previously (Coego et al., 2005a) with a suspension of 2.5 × 105 cfu/mL P.s.t. T1 (avrPto).
Accession Number
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under accession number At2g27040 (AGO4/OCP11).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Differential Expression Pattern of PEp5C:GUS in Wild-Type, rdr2-1, and dcl3-1 Plants.
Supplemental Figure 2. Histograms of 5mC Number at a 188-Nucleotide Region of the Ep5C 5′ Promoter.
Supplementary Material
Acknowledgments
We thank B. Wulff for comments on the manuscript and for helpful discussion. P. Tornero is acknowledged for helpful comments and A. Coego for his early assistance in mapping. We thank X. Dong for providing seeds of the npr1 mutant, J. Dangl for seeds of rpm1-1, J.M. Zhou for seeds of nho1, J.P. Metraux for seeds of sid2-1 and eds5-3, J. Carrington for seeds of rdr2-1 and dcl3-1, M. Matzke for seeds of drd1-6, and S. Jacobsen for seeds of drm1-2 drm2-2 and the ago4-1 mutants. We acknowledge the support of the Spanish Ministry of Science and Technology (Grant BFU2006-00803 to P.V.) for financial support.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Pablo Vera (vera@ibmcp.upv.es).
Online version contains Web-only data.
References
- Abramovitch, R.B., and Martin, G.B. (2004). Strategies used by bacterial pathogens to suppress plant defenses. Curr. Opin. Plant Biol. 7 356–364. [DOI] [PubMed] [Google Scholar]
- Asai, T., Tena, G., Plotnikova, J., Willmann, M.R., Chiu, W.L., Gomez-Gomez, L., Boller, T., Ausubel, F.M., and Sheen, J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 977–983. [DOI] [PubMed] [Google Scholar]
- Bartee, L., Malagnac, F., and Bender, J. (2001). Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 15 1753–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger, N., and Baulcombe, D.C. (2005). Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 102 11928–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144. [DOI] [PubMed] [Google Scholar]
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88 57–63. [DOI] [PubMed] [Google Scholar]
- Cao, X., and Jacobsen, S.E. (2002). Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci. USA 99 (suppl. 4): 16491–16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S.J., Harrison, J., Paul, C.L., and Frommer, M. (1994). High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22 2990–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coego, A., Ramirez, V., Ellul, P., Mayda, E., and Vera, P. (2005. a). The H2O2-regulated Ep5C gene encodes a peroxidase required for bacterial speck susceptibility in tomato. Plant J. 42 283–293. [DOI] [PubMed] [Google Scholar]
- Coego, A., Ramirez, V., Gil, M.J., Flors, V., Mauch-Mani, B., and Vera, P. (2005. b). An Arabidopsis homeodomain transcription factor, OVEREXPRESSOR OF CATIONIC PEROXIDASE 3, mediates resistance to infection by necrotrophic pathogens. Plant Cell 17 2123–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, N.C., Thordal-Christensen, H., Lipka, V., Bau, S., Kombrink, E., Qiu, J.L., Huckelhoven, R., Stein, M., Freialdenhoven, A., Somerville, S.C., and Schulze-Lefert, P. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425 973–977. [DOI] [PubMed] [Google Scholar]
- Chan, S.W., Henderson, I.R., Zhang, X., Shah, G., Chien, J.S., and Jacobsen, S.E. (2006). RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in Arabidopsis. PLoS Genet 2 e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, S.W., Zilberman, D., Xie, Z., Johansen, L.K., Carrington, J.C., and Jacobsen, S.E. (2004). RNA silencing genes control de novo DNA methylation. Science 303 1336. [DOI] [PubMed] [Google Scholar]
- Chisholm, S.T., Coaker, G., Day, B., and Staskawicz, B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124 803–814. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411 826–833. [DOI] [PubMed] [Google Scholar]
- Espinosa, A., and Alfano, J.R. (2004). Disabling surveillance: Bacterial type III secretion system effectors that suppress innate immunity. Cell. Microbiol. 6 1027–1040. [DOI] [PubMed] [Google Scholar]
- Fahlgren, N., Montgomery, T.A., Howell, M.D., Allen, E., Dvorak, S.K., Alexander, A.L., and Carrington, J.C. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16 939–944. [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez, L., Felix, G., and Boller, T. (1999). A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 18 277–284. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T., and Yao, N. (2004). The role and regulation of programmed cell death in plant-pathogen interactions. Cell. Microbiol. 6 201–211. [DOI] [PubMed] [Google Scholar]
- Henderson, I.R., Zhang, X., Lu, C., Johnson, L., Meyers, B.C., Green, P.J., and Jacobsen, S.E. (2006). Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 38 721–725. [DOI] [PubMed] [Google Scholar]
- Herr, A.J., Jensen, M.B., Dalmay, T., and Baulcombe, D.C. (2005). RNA polymerase IV directs silencing of endogenous DNA. Science 308 118–120. [DOI] [PubMed] [Google Scholar]
- Jones, D.A., and Takemoto, D. (2004). Plant innate immunity—Direct and indirect recognition of general and specific pathogen-associated molecules. Curr. Opin. Immunol. 16 48–62. [DOI] [PubMed] [Google Scholar]
- Kang, L., Li, J., Zhao, T., Xiao, F., Tang, X., Thilmony, R., He, S.Y., and Zhou, J.M. (2003). Interplay of the Arabidopsis nonhost resistance gene NHO1 with bacterial virulence. Proc. Natl. Acad. Sci. USA 100 3519–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno, T., Mette, M.F., Kreil, D.P., Aufsatz, W., Matzke, M., and Matzke, A.J. (2004). Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr. Biol. 14 801–805. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., Fahlgren, N., Chapman, E.J., Sullivan, C.M., Cumbie, J.S., Givan, S.A., and Carrington, J.C. (2007). Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 5 e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal, S., Morgan, R., Dahlbeck, D., Borsani, O., Villegas, A., Jr., Zhu, J.K., Staskawicz, B.J., and Jin, H. (2006). A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. USA 103 18002–18007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka, V., et al. (2005). Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310 1180–1183. [DOI] [PubMed] [Google Scholar]
- Lu, C., Tej, S.S., Luo, S., Haudenschild, C.D., Meyers, B.C., and Green, P.J. (2005). Elucidation of the small RNA component of the transcriptome. Science 309 1567–1569. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C., and Vaucheret, H. (2006). Functions of microRNAs and related small RNAs in plants. Nat. Genet. 38 (suppl.): S31–S36. [DOI] [PubMed] [Google Scholar]
- Mayda, E., Mauch-Mani, B., and Vera, P. (2000). Arabidopsis dth9 mutation identifies a gene involved in regulating disease susceptibility without affecting salicylic acid-dependent responses. Plant Cell 12 2119–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metraux, J.P. (2002). Recent breakthroughs in the study of salicylic acid biosynthesis. Trends Plant Sci. 7 332–334. [DOI] [PubMed] [Google Scholar]
- Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., Voinnet, O., and Jones, J.D. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312 436–439. [DOI] [PubMed] [Google Scholar]
- Navarro, L., Zipfel, C., Rowland, O., Keller, I., Robatzek, S., Boller, T., and Jones, J.D. (2004). The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 135 1113–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath, C., Heck, S., Parinthawong, N., and Metraux, J.P. (2002). EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, K., Melotto, M., and He, S.Y. (2005). Suppression of host defense in compatible plant-Pseudomonas syringae interactions. Curr. Opin. Plant Biol. 8 361–368. [DOI] [PubMed] [Google Scholar]
- Onodera, Y., Haag, J.R., Ream, T., Nunes, P.C., Pontes, O., and Pikaard, C.S. (2005). Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120 613–622. [DOI] [PubMed] [Google Scholar]
- Pavet, V., Quintero, C., Cecchini, N.M., Rosa, A.L., and Alvarez, M.E. (2006). Arabidopsis displays centromeric DNA hypomethylation and cytological alterations of heterochromatin upon attack by Pseudomonas syringae. Mol. Plant Microbe Interact. 19 577–587. [DOI] [PubMed] [Google Scholar]
- Pontes, O., Li, C.F., Nunes, P.C., Haag, J., Ream, T., Vitins, A., Jacobsen, S.E., and Pikaard, C.S. (2006). The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell 126 79–92. [DOI] [PubMed] [Google Scholar]
- Qi, Y., and Hannon, G.J. (2005). Uncovering RNAi mechanisms in plants: Biochemistry enters the foray. FEBS Lett. 579 5899–5903. [DOI] [PubMed] [Google Scholar]
- Qi, Y., He, X., Wang, X.J., Kohany, O., Jurka, J., and Hannon, G.J. (2006). Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443 1008–1012. [DOI] [PubMed] [Google Scholar]
- Rivas, F.V., Tolia, N.H., Song, J.J., Aragon, J.P., Liu, J., Hannon, G.J., and Joshua-Tor, L. (2005). Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 12 340–349. [DOI] [PubMed] [Google Scholar]
- Song, J.J., Smith, S.K., Hannon, G.J., and Joshua-Tor, L. (2004). Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305 1434–1437. [DOI] [PubMed] [Google Scholar]
- Sunkar, R., Chinnusamy, V., Zhu, J., and Zhu, J.K. (2007). Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 12 301–309. [DOI] [PubMed] [Google Scholar]
- Tao, Y., Xie, Z., Chen, W., Glazebrook, J., Chang, H.S., Han, B., Zhu, T., Zou, G., and Katagiri, F. (2003). Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen, H. (2003). Fresh insights into processes of nonhost resistance. Curr. Opin. Plant Biol. 6 351–357. [DOI] [PubMed] [Google Scholar]
- Tornero, P., and Dangl, J.L. (2001). A high-throughput method for quantifying growth of phytopathogenic bacteria in Arabidopsis thaliana. Plant J. 28 475–481. [DOI] [PubMed] [Google Scholar]
- van Wees, S.C., and Glazebrook, J. (2003). Loss of non-host resistance of Arabidopsis NahG to Pseudomonas syringae pv. phaseolicola is due to degradation products of salicylic acid. Plant J. 33 733–742. [DOI] [PubMed] [Google Scholar]
- Weigel, D., and Glazebrook, J. (2002). Arabidopsis. A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Wildermuth, M.C., Dewdney, J., Wu, G., and Ausubel, F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414 562–565. [DOI] [PubMed] [Google Scholar]
- Xie, Z., Johansen, L.K., Gustafson, A.M., Kasschau, K.D., Lellis, A.D., Zilberman, D., Jacobsen, S.E., and Carrington, J.C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2 E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Henderson, I.R., Lu, C., Green, P.J., and Jacobsen, S.E. (2007). Role of RNA polymerase IV in plant small RNA metabolism. Proc. Natl. Acad. Sci. USA 104 4536–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Yazaki, J., Sundaresan, A., Cokus, S., Chan, S.W., Chen, H., Henderson, I.R., Shinn, P., Pellegrini, M., Jacobsen, S.E., and Ecker, J.R. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126 1189–1201. [DOI] [PubMed] [Google Scholar]
- Zilberman, D., Cao, X., and Jacobsen, S.E. (2003). ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299 716–719. [DOI] [PubMed] [Google Scholar]
- Zilberman, D., Cao, X., Johansen, L.K., Xie, Z., Carrington, J.C., and Jacobsen, S.E. (2004). Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr. Biol. 14 1214–1220. [DOI] [PubMed] [Google Scholar]
- Zipfel, C., Robatzek, S., Navarro, L., Oakeley, E.J., Jones, J.D., Felix, G., and Boller, T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.