Abstract
Phospholipases A2 (PLA2s) catalyze hydrolysis of fatty acids from the sn-2 position of phospholipids. Here we report the identification and characterization of a membrane-associated intracellular calcium-dependent, adipose-specific PLA2 that we named AdPLA (adipose-specific phospholipase A2). We found that AdPLA was highly expressed specifically in white adipose tissue and was induced during preadipocyte differentiation into adipocytes. Clearance of AdPLA by immunoprecipitation significantly decreased PLA activity in white adipose tissue lysates but had no effect on liver lysates, where expression was hardly detectable. In characterizing AdPLA, we employed radiochemical assays with TLC analysis of the enzyme activity of lysates from COS-7 cells overexpressing AdPLA. For kinetic studies, we produced purified recombinant AdPLA for use in a lipoxidase-coupled spectrophotometric assay. AdPLA generated free fatty acid and lysophospholipid from phosphatidylcholine with a preference for hydrolysis at the sn-2 position. Although we found low but detectable lysophospholipase activity, AdPLA showed no significant activity against a variety of other lipid substrates. Calcium was found to activate AdPLA but was not essential for activity. Studies with known phospholipase inhibitors, including bromoenolactone, methyl arachidonyl fluorophosphate, AACOCF3, 7,7-dimethyl-5,8-eicosadienoic acid, and thioetheramide, supported that AdPLA is a phospholipase. Mutational studies showed that His-23 and Cys-113 are critical for activity of AdPLA and suggested that AdPLA is likely a His/Cys PLA2. Overall, although AdPLA is similar to other histidine phospholipases in pH and calcium dependence, AdPLA showed different characteristics in many regards, including predicted catalytic mechanism. AdPLA may therefore represent the first member of a new group of PLA2s, group XVI.
Phospholipases A2 (PLA2)4 catalyze hydrolysis of the sn-2 ester bond of phospholipids (1). They have diverse functions ranging from digestion of dietary phospholipids to membrane remodeling by acylation/deacylation cycles, to cell signaling through the liberation of lysophospholipid and arachidonic acid that is utilized in eicosanoid biosynthesis (2). PLA2s therefore play important biological roles in a variety of tissues. In adipose tissue, arachidonic acid and eicosanoids have been implicated in diverse processes, including regulation of differentiation (3–7), lipolysis (8–10), and glucose transport (11), highlighting the importance of PLA2 in this tissue.
PLA2s are currently classified into 15 major groups that are subclassified into 5 distinct types of enzymes as follows: secretory PLA2s (sPLA2s), cytosolic PLA2s (cPLA2s), and calcium-independent PLA2s (iPLA2s), as well as platelet-activating factor acetylhydrolases and lysosomal PLA2s (1). The sPLA2s are characterized by relatively low molecular weights (typically <20 kDa), millimolar Ca2+ dependence for optimum catalytic activity, and extensive disulfide-stabilized tertiary structure (12). Most sPLA2s contain a catalytic histidine residue within a conserved CCXHDXC motif (12). The cytosolic PLA2s (cPLA2) are much larger, ranging in size to over 100 kDa but have few disulfide bonds and are localized intracellularly (13). They require Ca2+ for translocation from the cytosol to their site of action on membrane phospholipids, but they do not require calcium for catalytic activity per se (14). All contain a conserved Ser/Asp catalytic dyad and Arg that are required for catalytic activity. c-PLA2α is the only PLA2 that shows specificity for arachidonic acid in the sn-2 position (13, 14). iPLA2s are characterized by a GXSXG consensus lipase motif with a catalytic serine, and these are also localized intracellularly. iPLA2sdonot require Ca2+ for membrane association or for catalytic activity (1).
In our efforts to identify novel proteins of significance in adipocyte metabolism, we found a transcript encoding an 18-kDa protein that is highly and differentially expressed only in adipose tissue, and we named it AdPLA for adipose-specific PLA2. Sequence analysis indicated that this protein has been identified previously as HRASLS3 (Ha-RAS like suppressor 3 or H-Rev-107-1), a class II tumor suppressor with no known molecular function (15). This protein has been reported to be ubiquitously expressed at low levels in a variety of tissues, although prior to this study, expression in adipose tissue had not been examined. Thus far, no enzymatic activity has been attributed to this protein. Sequence alignment indicated shared homology with lecithin:retinol acyltransferase (LRAT) (16). However, we found no acyltransferase activity of AdPLA, but rather phospholipase activity. We found that this protein catalyzes the efficient release of free fatty acids (FFA) and lysophospholipid from phosphatidylcholine (PC), indicating that it is a phospholipase A (PLA). Furthermore, we found a preference for hydrolysis at the sn-2 position of phospholipids that classifies AdPLA to the PLA2 superfamily of enzymes. Further characterization of this enzyme has led us to propose that AdPLA may be the first member of an entirely new group of calcium-dependent intracellular PLA2s, group XVI.
EXPERIMENTAL PROCEDURES
Materials—[U-14C]Palmitic acid (specific activity, 850 mCi/mmol) was from PerkinElmer Life Sciences; 1,2-di[1-14C]palmitoyl-sn-glycerol-3-phosphocholine (specific activity, 110 mCi/mmol), 1-palmitoyl-2[1-14C]palmitoyl-sn-glycerol-3-phosphocholine (specific activity, 60 mCi/mmol), 1-[1-14C]palmitoyl-2-hydroxy-sn-glycerol-3-lysophosphocholine (specific activity, 53 mCi/mmol), and [9,10-3H]-triolein (specific activity, 53 Ci/mmol) were from GE Healthcare. 1,2-Dilineoyl-PC, 1-palmitoyl-2-linoleoyl-PC, 1-palmitoyl-2-arachidonyl-PC, 1-palmitoyl-2-linoleoyl-phosphatidylethanolamine, 1-palmitoyl-2-linoleoyl-phosphatidylserine, 1-palmitoyl-2-linoleoyl-phosphatidic acid, 1-palmitoyl-2-linoleoyl-phosphatidylinositol, and egg PC were from Avanti Polar Lipids (Alabaster, AL). Sodium deoxycholate, lipoxidase preparation from soybeans (type V, 701,000 units/mg protein), and pancreatic phospholipase A2 from bovine pancreas were from Sigma. Racemic BEL ((E)-6-(bromomethylene)tetrahydro-3-(1-naphthalenyl)-2H-pyran-2-one), 7,7-dimethyl-5,8-eicosadienoic acid, AACOCF3, methyl arachidonyl fluorophosphate (MAFP), and thioetheramide were from Cayman Chemicals (Ann Arbor, MI). Lipofectamine 2000 was from Invitrogen, and other chemicals and high pressure liquid chromatography grade solvents were from Fisher.
GeneFilter® Microarray Analysis—Identification of genes expressed exclusively in adipose tissue was achieved by comparing the gene expression patterns of different mouse tissues using rat GeneFilter® membranes (Research Genetics) as described previously (17). Briefly, filters were hybridized with 33P-labeled cDNAs synthesized by reverse transcription using 5 μg of total RNA from WAT, brain, muscle, and liver. Only those spots found exclusively in filters hybridized with WAT cDNAs were further analyzed. Candidate EST clones were sequenced, and adipose tissue-specific expression was verified by Northern blot analysis using RNA from liver, brain, muscle, and WAT (data not shown). One of these clones showed exclusive expression in WAT. Using the sequence of the selected EST clone as a query, a BLASTn search of the mouse genome data base of NCBI identified a match with HRASLS3/H-Rev107-1. The full-length cDNA was prepared and sequenced (GenBank™ accession number NM_139269) (18).
Generation of AdPLA Mammalian and Bacterial Expression Vectors and Preparation of Purified Recombinant NUS-AdPLA—We amplified the coding region of AdPLA, including a C-terminal hemagglutinin (HA) tag using mouse cDNA reverse-transcribed from RNA prepared from WAT, and we subcloned this into the BamHI/XhoI sites of pCR3.1. Two AdPLA-GFP constructs bearing either the complete coding region of AdPLA or a truncated form missing the last 36 codons was prepared by subcloning enhanced green fluorescent protein (GFP) in-frame with the C terminus of AdPLA in pCR3.1. Lipofectamine 2000 was used for all transfections. For protein expression in Escherichia coli, HA-tagged AdPLA was subcloned into the NotI-XhoI site of the pET-43.1a prokaryotic expression vector (Novagen) containing the N-terminal His tag for purification and a hydrophilic NUS A tag. AdPLA mutants were prepared using a QuikChange site-directed mutagenesis kit (Stratagene Inc.) according to the vendor's instructions (for primers see supplemental Table S1). For AdPLA and AdPLA mutants, the plasmids were transformed into E. coli BL21STAR-(DE3), expression was induced by 0.5 mm isopropyl 1-thio-β-d-galactopyranoside, and cultures were grown at 27 °C for an additional 3.5 h. His-tagged AdPLA and mutants were affinity-purified using HisPur cobalt resin from Pierce. Protein was visualized by Coomassie staining relative to BSA standards for quantification.
Cell Culture, Differentiation, and Production of Primary Preadipocytes—3T3-L1 and COS-7 cells (American Type Culture Collection) were cultured in DMEM containing 10% fetal bovine serum. To induce differentiation of 3T3-L1 cells into adipocytes, 2-day post-confluent preadipocytes (day 0) were treated with 1 μm dexamethasone, 0.5 mm methylisobutylxanthine (MIX), and 1 μg/ml insulin for 48 h. After the induction period, cells were switched to maintenance medium (DMEM supplemented with 10% fetal bovine serum) and maintained for 5–7 days, at which point 90% of the cells exhibited the typical adipocyte morphology.
Subcutaneous inguinal fat depots from female Zucker rats were dissected, and the lymph nodes were removed. The stromal vascular cells were obtained by collagenase digestion (1 mg/ml) at 37 °C for 45 min in HEPES buffer (10 mm HEPES, pH 7.4, 135 mm NaCl, 2.2 mm CaCl2, 1.25 mm MgSO4, 0.45 mm KH2PO4, 2.17 mm Na2HPO4, 5 mm d-glucose, and 2% w/v BSA). The cell suspension was filtered through a 100-μm nylon filter and centrifuged at 400 × g for 10 min. The pellets were washed, filtered through a 25-μm nylon filter, and plated at a density of 2.5 × 104 cells/cm2 in DMEM with 10% FBS. At confluence, differentiation was initiated by the addition of 0.1 μm dexamethasone, 0.25 mm MIX, and 17 nm insulin. After 2 days, medium was replaced by DMEM with 10% FBS and insulin only. Cells were harvested for RNA isolation at the time points indicated.
Subcellular Localization of AdPLA—COS-7 or 3T3-L1 cells were grown on coverslips. COS-7 cells transfected with AdPLA-GFP expression vector were fixed with 4% paraformaldehyde, incubated with polyclonal antibodies against COX-1 for 1 h (Santa Cruz Biotechnology), washed with phosphate-buffered saline, and incubated with Alexa Fluor 546-conjugated secondary antibody at 5 μg/ml for 3 h (Molecular Probes). For endoplasmic reticulum staining, 3T3-L1 cells transfected with AdPLA-GFP were differentiated into adipocytes and were stained with concanavalin A Alexa Fluor 633 conjugate (Molecular Probes). The samples were mounted on glass microscope slides using Antifade and Prolong mounting media (Molecular Probes), and images were captured using a Zeiss Axiophot LSM 510Meta confocal microscope.
Polyclonal Anti-AdPLA Antiserum and Western Blot Analysis—To generate polyclonal antisera, rabbits were immunized with purified recombinant AdPLA-His expressed in E. coli. For Western blotting, proteins were subjected to 12% SDS-PAGE and transferred to nitrocellulose membrane for immunodetection using primary antibodies against HA (Covance, 1:1000), AdPLA (1:2000), or glyceraldehyde-3-phosphate dehydrogenase (1:1000) (Santa Cruz Biotechnology).
RNA Extraction, Northern Blotting, and Real Time Reverse Transcription-PCR—Total RNA was isolated from tissues using TRIzol Reagent (Invitrogen). For Northern blot analysis, 5–15 μg of total RNA was subjected to electrophoresis on formaldehyde-containing 1.2% agarose gels and transferred onto Hybond N+ nylon membranes (Amersham Biosciences). Hybridization was carried out in ExpressHyb solution (Clontech) using 32P-labeled cDNA-specific probes for AdPLA, adipocyte fatty acid-binding protein, stearoyl-CoA desaturase 1, or peroxisome proliferator-activated receptor-γ. For reverse transcription-quantitative PCR, cDNA was synthesized from 2 μg of total RNA by oligo(dT) priming and Superscript II reverse transcriptase (Invitrogen). Tissue expression of AdPLA, iPLA2β, or cPLA2α was determined with an ABI PRISM 7700 sequence fast detection system (Applied Biosystems) using specific primers from Applied Biosystems. Gene expression level was quantified by measuring the threshold cycle normalized to glyceraldehyde-3-phosphate dehydrogenase and is expressed relative to levels in liver.
In Vitro Phospholipase Activity Assay—The continuous spectrophotometric assay was carried out as described (19) with minor modifications. Briefly, aliquots of stock lipid substrate solutions in chloroform were dried under a stream of N2 and then sonicated into suspension up to a maximum concentration of 100 μm in 50 mm Tris buffer, pH 8, containing 2 mm deoxycholate. The resulting micellar solution was allowed to equilibrate for 10 min at 25 °C. Phospholipase A activity was determined by means of a coupled enzyme assay. Released polyunsaturated fatty acids (i.e. linoleic acid or arachidonic acid) were subsequently oxidized by lipoxygenase (0.36 mg/ml), giving rise to a hydroperoxide derivative that could be measured by spectrophotometric assessment of the increase in absorbance at 234 nm (ε234 = 25,000 m-1 cm-1). The reaction was started by adding purified NUS-AdPLA into the substrate/lipoxidase mixture, and the A234 was monitored continuously for 3 min. Controls without either AdPLA or lipoxygenase were carried out routinely. For studies with inhibitors, the compounds were incorporated at the indicated concentrations into micelles containing 100 μm 1-palmitoyl-2-linoleoyl-PC with 2 mm deoxycholate and 2 mm CaCl2.
For assay of activity from cell lysates, samples were homogenized in Buffer A (0.1 m sucrose, 50 mm KCl, 40 mm KH2PO4, and 30 mm EDTA, pH 7.2) and centrifuged at 100,000 × g for 60 min to separate the supernatant (cytosol) from the membrane fraction. For assay from WAT or liver, homogenates were centrifuged at 20,000 × g and then cleared by incubation with preimmune serum or serum from rabbits immunized against AdPLA (1:100 dilution) followed by immunoprecipitation of antibody-protein complexes on protein A-agarose beads. PLA activity with radioactive substrate was determined essentially as described (20) with minor modifications. Micelles were created by sonification of radiolabeled lipid substrates in assay buffer (50 mm Tris, pH 8, 2 mm deoxycholate, 5 mm EDTA), and hydrolytic activity was monitored by the generation of [14C]palmitate. Reactions were started by the addition of purified enzyme or cell lysates and terminated by the addition of (2:1) methanol:chloroform. Lipids were extracted by the method of Bligh and Dyer (21) and resolved by TLC on a hexane:diethyl ether:acetic acid solvent front (80:20:2). Bands corresponding to [14C]palmitate were identified by comparison with lipid standards and scraped and quantified by liquid scintillation counting. For resolution of phospholipid from lysophospholipid and FFA, lipid extracts were resolved by TLC on a more aqueous chloroform:methanol:acetic acid:water (60:30: 8.4:3.6) solvent front.
For in vitro TAG hydrolase activity, lysates were prepared from COS-7 cells overexpressing AdPLA-HA or desnutrin-HA (positive control) by lysis in Buffer A followed by centrifugation at 16,000 × g. Supernatants (100 μg of protein in 0.1 ml) were incubated with 0.1 ml of substrate containing 100 μm [3H]triolein in mixed micelles with 25 μm egg yolk lecithin, 100 μm taurocholate, 2% BSA (w/v), and 50 mm Tris-HCl, pH 8.0. Reactions were terminated by the addition of 3.25 ml of methanol: chloroform:heptane (10:9:7), and fatty acids were extracted with1 ml of 0.1 m potassium carbonate, 0.1 m boric acid, pH 10.5. Radioactivity in 0.7 ml of upper phase obtained after centrifugation for 20 min at 800 × g was quantified by liquid scintillation counting.
Statistical Analysis—The results are expressed as means ± S.E. Statistically significant differences between two groups were assessed by Student's t test. Differences between multiple groups were assessed by one-way analysis of variance with Bonferroni's post hoc test.
RESULTS AND DISCUSSION
PLA2 has been reported to function in adipocyte cell signaling (22, 23), differentiation (3–7), and in the regulation of important adipocyte metabolic processes such as lipolysis and glucose transport (8–11). Because dysregulated adipocyte differentiation and metabolism are linked to obesity and associated pathologies, understanding phospholipid metabolism in adipose tissue is critical.
Using EST cDNA microarray analysis we identified AdPLA as a differentially expressed gene that was found to be present at high levels specifically in adipocytes. We have used this approach to identify other important adipocyte-specific genes, including adipose-specific secretory factor (ADSF/resistin) (17) and desnutrin (24). Although we have previously found AdPLA to be expressed at low levels in a number of tissues and cultured cell lines (18, 25, 26), prior to this work, expression of AdPLA in adipocytes or in adipose tissue had not been examined. Although we observed that two other phospholipases, iPLA2β(Fig. 1A, right panel) and cPLA2α (Fig. 1A, middle panel), were present at varying levels in a host of tissues, AdPLA expression was clearly predominant in white adipose tissue (WAT) (40 to >150-fold more abundant than in liver) and, to a lesser extent, in brown adipose tissue (Fig. 1A, left panel). Separation of WAT into adipocytes and stromovascular fraction indicated expression of AdPLA exclusively in adipocytes (data not shown). In agreement, AdPLA was not detected in 3T3-L1 or primary rat preadipocytes but was induced upon differentiation (Fig. 1, B and C). Induction of AdPLA tended to occur late in differentiation as compared with the appearance of other marker genes of early to late stage adipocyte differentiation (i.e. peroxisome proliferator-activated receptor-γ, adipocyte fatty acid-binding protein, or stearoyl-CoA desaturase 1). Furthermore, induction of AdPLA most likely resulted from effects occurring during differentiation rather than from transcriptional modulation by either dexamethasone or MIX, because neither agent alone affected AdPLA expression in 3T3-L1 cells (Fig. 1D).
FIGURE 1.
AdPLA is highly expressed in adipose tissue and adipocytes. A, AdPLA mRNA expression (left panel) was 40- to over 150-fold higher in subcutaneous (subcut), renal, and gonadal WAT depots than in liver, and 18-fold higher in brown adipose tissue (BAT)(n = 3). In contrast, cPLA2α (middle panel) and iPLA2β (right panel) were found to be expressed in a wide variety of tissues at varying levels, showing no obvious tissue-specific expression pattern. sk musc, skeletal muscle. B, AdPLA mRNA is induced during late stage differentiation of 3T3-L1 and primary preadipocytes. C, representative immunoblot showing that AdPLA protein is barely detectable in 3T3-L1 preadipocytes but is highly induced upon differentiation. D, AdPLA is induced in 3T3-L1 cells by a differentiation mixture of dexamethasone (DEX) + MIX but not by either agent alone. E, subcellular fractionation of COS-7 cells overexpressing HA-tagged AdPLA indicated localization of AdPLA both to the 100,000 × g membrane fraction as well as in the cytosolic fraction. F, confocal microscopy images showing a punctate localization of GFP-tagged full-length AdPLA predominantly in the perinuclear region of COS-7 cells. Truncated AdPLA lacking the membrane-spanning C terminus domain assumed a more diffuse localization. G, full-length AdPLA-GFP co-localized partially with COX-1 in COS-7 cells. H, full-length AdPLA-GFP showed a perinuclear localization in differentiated 3T3-L1 cells and partially co-localized with the endoplasmic reticulum (ER).
We examined the localization of HA-tagged AdPLA in COS-7 cells and found it both in the 100,000 × g membrane fraction and also in the cytoplasmic fraction (Fig. 1E). Confocal imaging of full-length AdPLA-GFP showed a predominantly perinuclear localization as well as evidence of the presence within the cytoplasm of COS-7 cells (Fig. 1, F and G, left panel) and 3T3-L1 adipocytes (Fig. 1H, left panel). Notably, truncation of the C-terminal hydrophobic domain of AdPLA resulted in a more diffuse localization of the GFP fusion construct in cells (Fig. 1F, right panel). Because some PLA2s act on intracellular membrane phospholipids to provide arachidonic acid to cyclooxygenase 1 (COX-1) for eicosanoid biosynthesis, we examined the localization of AdPLA relative to both the endoplasmic reticulum and COX-1. We found AdPLA-GFP localized, at least in part, in proximity to COX-1 in COS-7 cells (Fig. 1G). Similarly, AdPLA-GFP partly co-localized with the endoplasmic reticulum in differentiated 3T3-L1 adipocytes (Fig. 1H). Taken together, these findings suggest that AdPLA may function in eicosanoid biosynthesis, although further studies will be required to determine the effects of AdPLA on adipose tissue metabolism.
To study the activity of AdPLA, we produced cell lysates from COS-7 cells overexpressing HA-tagged AdPLA (Fig. 2A), as well as bacterially derived recombinant AdPLA for use in a continuous spectrophotometric based assay (Fig. 2B). Recombinant AdPLA tagged with HA and polyhistidine was also tagged with NUS A protein to enhance solubility and expression (27), and was affinity-purified on Co3+-agarose beads to over 90% homogeneity. Because sequence analysis indicated homology with LRAT, we first investigated potential acyltransferase activity of AdPLA. We utilized 1,2-di[1-14C]palmitoyl-PC as the acyl donor, because LRAT utilizes acyl groups from phospholipids, and we utilized unlabeled DAG as the acyl acceptor to focus on a potential role for AdPLA in the synthesis of TAG, the predominant lipid species in adipocytes. However, when we incubated lysates from COS-7 cells overexpressing AdPLA with these compounds, we found no increase in TAG formation but rather the prominent appearance of a band corresponding to radiolabeled FFA (Fig. 2C). This band increased in intensity with increasing reaction time and amount of protein, suggesting phospholipase rather than acyltransferase activity. Additional studies supported that AdPLA may be a phospholipase. Incubation of lysates overexpressing AdPLA with a complex mixture of radiolabeled cellular lipids containing, predominantly, labeled phospholipids, TAG, DAG, and FFA, yielded a notable increase in the production of FFA but no change in any other lipid fraction compared with control cell lysates (Fig. 2D). Furthermore, incubation of membrane or cytosolic fractions from COS-7 cells overexpressing AdPLA with [14C]palmitoyl-CoA yielded significant incorporation but no differences in any lipid groups when compared with control, indicating an absence of any significant acyl-CoA-dependent acyltransferase activity (Fig. 2E). Taken together, these findings indicate that AdPLA is a phospholipase rather than an acyltransferase. When we used a more hydrophilic solvent system to resolve the products of AdPLA hydrolysis of 1,2-di[1-14C]palmitoyl-sn-glycerol-3-PC, we found an increase in the generation not only of FFA but also lysophosphatidylcholine (Fig. 2F). This finding indicates hydrolysis predominantly of a single radiolabeled fatty acyl group and therefore indicates that AdPLA is specifically a PLA.
FIGURE 2.
AdPLA has phospholipase activity but lacks acyltransferase activity. A, representative immunoblot of COS-7 cell lysates transiently transfected with HA-tagged AdPLA. B, NUS-AdPLA tagged with HA and His was affinity-purified on Co3+-agarose beads. Following SDS-PAGE, NUS-AdPLA was detectable as a single band of ∼81 kDa by Coomassie staining (lane 1, molecular weight standards; lane 2, NUS-AdPLA) and by immunoblotting on polyvinylidene difluoride with anti-HA (lane 3). C, COS-7 cell lysates overexpressing AdPLA or control vector were incubated with 1,2-di[1-14C]palmitoyl-sn-glycerol-3-phosphocholine (Sn1*2*PC) and DAG. AdPLA overexpression increased the appearance of [14C]palmitate, indicating phospholipase activity, and this effect was enhanced by increasing the incubation time or amount of protein. There was no evidence of acyltransferase activity (i.e. no 14C-TAG formed). MAG, monoacylglycerol; PL, phospholipid. D, COS-7 cell lysates overexpressing AdPLA caused a greater release of 14C-FFA from a complex mixture of radiolabeled cellular lipids, but otherwise showed no changes in any other lipid groups. E, cytosolic and membrane fractions derived from COS-7 cells transfected with AdPLA or control vector incorporated [14C]palmitoyl-CoA in a similar manner, indicating no apparent acyl-CoA-dependent acyltransferase activity. F, incubation of lysates overexpressing AdPLA with 1,2-di[1-14C]palmitoyl-sn-glycerol-3-phosphocholine (Sn1*2*PC) yielded an increase in the production of radiolabeled FFA and lysophosphatidylcholine (LPC), indicating that AdPLA is a PLA (n = 4). G, COS-7 cells overexpressing AdPLA or control vector incorporated similar amounts of [U-14C]palmitate during an initial 4-h pulse-labeling period. H, overexpression of AdPLA, however, resulted in a significantly greater release of FFA to the medium during the ensuing cold chase period, as determined by liquid scintillation counting of medium, and by extraction of lipids from medium and resolution by TLC (inset)(n = 12). I, clearance by immunoprecipitation (IP) of lysates from WAT with anti-AdPLA antiserum (1:100) decreased levels of AdPLA relative to lysates cleared with preimmune serum (1:100), as shown in a representative immunoblot. AdPLA was essentially undetectable in lysates from liver, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was unchanged by immunoprecipitation. This decrease in immunoreactive AdPLA level in WAT lysates was associated with a significant decrease in PLA activity. No change in PLA activity was evident in lysates from liver (n = 7).
Next, we investigated the phospholipase activity of AdPLA in cultured cells and in tissues. Overexpression of AdPLA in COS-7 cells did not change incorporation of [U-14C]palmitate into cellular lipids during an initial pulse period (Fig. 2G). However, release of labeled FFA from cells overexpressing AdPLA was significantly greater during the cold chase period (Fig. 2H), indicating that hydrolysis of cellular lipids was enhanced, and suggesting that AdPLA may function as a phospholipase also in cells. We have found that AdPLA is most highly expressed in WAT. Partial clearing of AdPLA from WAT homogenates by immunoprecipitation using anti-AdPLA antiserum significantly decreased phospholipase activity under the assay conditions used by ∼20% compared with lysates cleared with preimmune serum (Fig. 2I). Conversely, AdPLA activity was similar in homogenates from liver, where we did not detect significant AdPLA expression, when cleared with either preimmune serum or antiserum, indicating a significant role for AdPLA specifically in adipose tissue phospholipid metabolism. These data further support that AdPLA is a phospholipase.
PLA2s reportedly often exhibit other minor catalytic activities, at least in vitro, although their major function is hydrolysis of phospholipids (12). We investigated the specificity of AdPLA acyl hydrolase activity. Using a lipoxidase-coupled spectrophotometric assay that measures the liberation of polyunsaturated fatty acids, we assessed the hydrolysis of 1,2-dilinoleyl-PC, cholesteryl-linoleate, or 1,3-dilinoleoyl-sn-2-glycerol (DAG) by AdPLA (19). As a positive control for PLA2 activity in these and other experiments, we utilized a crude bovine pancreatic sPLA2 preparation that was obtained from a commercial source. Therefore, it should be noted that values obtained for sPLA2 activity under the assay conditions utilized were thus considerably lower than values reported previously for the purified enzyme (28). Activity of AdPLA against phospholipid substrate was significantly higher than control (NUS A) (p < 0.001) (Fig. 3A). Activity of either phospholipase against DAG or cholesteryl-linoleate, however, was not different from NUS A control levels. Activity of AdPLA in cell lysates against TAG was below levels in control transfected cells (Fig. 3B). Because micellar substrates contained both radiolabeled TAG and cold lecithin, this was likely the result of AdPLA displacement of endogenous lipases on the micellar surface. TAG hydrolase activity in both AdPLA and control transfected cells was well below activity in cells transfected with desnutrin (positive control). Similar to other PLA2s (29), recombinant AdPLA exhibited significant lysophospholipase activity, although PLA activity was approximately 4-fold higher (Fig. 3C).
FIGURE 3.
AdPLA is primarily a phospholipase. A, recombinant AdPLA and sPLA2 (positive control) efficiently hydrolyzed 1,2-dilinoleoyl-PC (PC). Activity of AdPLA and sPLA2 toward cholesteryl-linoleate (CE) or 1,3-dilinoleoyl-sn-2-glycerol (DAG) was not different from control levels (NUS alone) (***, p < 0.001 versus NUS). B, lysates from COS-7 cells transiently transfected with AdPLA or desnutrin were assayed for activity against radiolabeled triolein. **, p < 0.01, ***, p < 0.001, versus NUS. C, similar to other phospholipases, recombinant AdPLA displayed minor hydrolase activity toward 1-[1-14C]palmitoyl-2-hydroxy-sn-glycerol-3-lysophosphocholine (Lyso-PC), but significantly greater activity toward 1-palmitoyl-2-[1-14C]palmitoyl-sn-glycerol-3-phosphocholine (PC) under the same conditions (***, p < 0.001). All data shown are means ± S.E. (n = 3–4).
Various classes of PLA2s differ with regard to optimal conditions for activity, including requirements for pH and calcium ion and with regard to substrate specificity and regioselectivity (12, 14). We utilized the spectrophotometric assay to investigate some of the major characteristics of purified AdPLA. In general, the pH requirement for differing PLA2s varies considerably (12, 14). We found the optimal pH for AdPLA to be ∼8.0 (Fig. 4A). We next investigated dependence of AdPLA on calcium for catalytic activity in vitro and found that whereas calcium was not absolutely required, increasing concentrations up to 1.0 mm resulted in an approximate 2-fold increase in enzyme activity (Fig. 4B). At pH 8.0 and in the presence of 2 mm calcium, a plot of the reaction velocity of AdPLA against increasing concentrations of 1-palmitoyl-2-linoleoyl-PC demonstrated that AdPLA follows first order Michaelis-Menten kinetics (Fig. 4C). Fig. 4C, inset, showed that the Vmax for AdPLA with this substrate is 7.8 μmol/min/mg protein and the Km is 16 μm. We investigated regioselectivity by comparing kinetic curves for AdPLA activity with 1-palmitoyl-2-linoleoyl-PC or 1,2-dilinoleoyl-PC as substrate, where only the hydrolyzed linoleic acid is oxidized and therefore quantified (Fig. 4D). Activity of AdPLA against 1,2-dilinoleoyl-PC was somewhat higher than activity against 1-palmitoyl-2-linoleoyl-PC, but it did not reach a 2-fold increase as would be expected if AdPLA had no positional preference. This finding suggests a preference for hydrolysis of fatty acids at the sn-2 position, indicating that AdPLA is a PLA2, and also demonstrates that this preference is more pronounced at lower concentrations when substrate is limiting. We tested the head group preference of AdPLA and found that choline is preferred over serine, ethanolamine, or inositol (Table 1). The catalytic efficiency of AdPLA (Vmax/Km) was slightly higher for PC with arachidonic acid in the sn-2 position versus linoleic acid because of a lower Km. However, the Vmax was also lower for 1-palmitoyl-2-arachidonyl-PC, indicating that any difference in utilization of these two substrates by AdPLA is not likely significantly different. In this regard, only one PLA2, the 85-kDa cPLA2α, has shown a clear preference for phospholipid containing arachidonic acid (30). The presence of a head group was essential for fatty acid hydrolysis because AdPLA lacked activity against phosphatidic acid.
FIGURE 4.
Characteristics of AdPLA phospholipase activity. Recombinant AdPLA was assayed in 50 mm Tris-HCl with 2 mm deoxycholate (n = 3–4). A, recombinant partially purified AdPLA was found to have a narrow pH range for activity with an optimum pH of 8.0. B, AdPLA activity against 20 μm 1-palmitoyl-2-linoleoyl-PC increased in the presence of increasing concentrations of CaCl2, and this effect was reversed by addition of a calcium chelator (3 mm EGTA). C, concentration-dependent activity of AdPLA against 1-palmitoyl-2-linoleoyl-PC at pH 8.0 with 2 mm CaCl2. Inset, double-reciprocal plot. D, regioselectivity of AdPLA. Concentration-dependent activity of NUS-AdPLA was compared with 1-palmitoyl-2-linoleoyl-PC (1-palm-2-lin-PC) versus 1,2-dilinoleoyl-PC (1,2-dilin-PC), where only hydrolyzed unsaturated fatty acids are quantitated in spectrophotometric readings. A preference for hydrolysis of fatty acids at the sn-2 position was evident, indicating that AdPLA is a PLA2.
TABLE 1.
Kinetic parameters for AdPLA with different substrates
| Compounda | Vmax (μmol/min/mg pro) | Km (μm) | Vmax/Km |
|---|---|---|---|
| 1-Palmitoyl-2 linoleoyl-PC | 3.23 | 18.6 | 0.17 |
| Dilinoleoyl-PC | 5.43 | 17.5 | 0.31 |
| 1-Palmitoyl-2-linoleoyl-PS | 0.56 | 7.25 | 0.08 |
| 1-Palmitoyl-2-linoleoyl-PE | 0.84 | 5.01 | 0.17 |
| 1-Palmitoyl-2-linoleoyl-PA | 0 | ||
| Phosphatidylinositol | 2.90 | 11.64 | 0.25 |
| 1-Palmitoyl-2-arachidonyl-PC | 2.15 | 8.24 | 0.26 |
Recombinant AdPLA was incubated with micelles containing the indicated substrates in 50 mm Tris, pH 8.0 with 2 mm deoxycholate, in the absence of added calcium (n = 3–5). Reactions were monitored by spectrophotometric assay of the change in A234. PS is phosphatidylserine; PA is phosphatidic acid; PE is phosphatidylethanolamine
Sequence analysis of AdPLA indicated the absence of a characteristic serine lipase motif (i.e. GXSXG or GXSGS) (31) but the presence of a histidine at position 23 that is conserved and catalytically active in LRAT family members (supplemental Fig. S1) (16), suggesting that AdPLA may be a histidine PLA2. Histidine PLA2s such as the sPLA2s, however, utilize a His/Asp dyad, whereas catalytic activity of LRAT requires His/Cys residues, as well as other residues that participate in hydrogen bonding (12, 16, 32). Notably, AdPLA contains conserved Cys-113 and His-23 residues that align with the catalytically active amino acids in LRAT (16).
In an effort to better understand and characterize AdPLA catalytic activity, we have performed a series of studies with known PLA2 inhibitors. Inhibition of AdPLA and sPLA2 by the arachidonic acid analogues 7,7-dimethyl-5,8-eicosadienoic acid and AACOCF3 was highly similar (Fig. 5, A and B). 7,7-Dimethyl-5,8-eicosadienoic acid inhibited activity of both AdPLA and sPLA2 proportionately to its content in micelles, suggesting that surface dilution of substrate at the lipid interface may have been a factor, rather than true competitive inhibition. Hydrated ketones such as AACOCF3 form a structural mimetic of the intermediate formed during the hydrolysis of substrates, and are believed to inhibit by direct binding to the enzyme (33). This finding indicates that, like other phospholipases, including (as shown here) sPLA2 and (as shown previously) cPLA2 (33), AdPLA is sensitive to inhibition by AACOCF3. On the other hand, when we treated AdPLA and sPLA2 with BEL, we found no significant effect on activity of either enzyme (Fig. 5C). BEL is an irreversible suicide-substrate inhibitor of iPLA2s that causes a complete loss of catalytic activity at nanomolar to low micromolar concentrations (34). Because BEL is known to inhibit lipases that contain the GXSXG consensus motif through binding at or near the active site serine (34, 35), the lack of effect of BEL was not unexpected. Interestingly, we found an inhibitory effect of MAFP on AdPLA but not sPLA2 activity (Fig. 5D). This compound is believed to inhibit serine and cysteine hydrolases by covalently binding to the enzyme and has been shown to inhibit both iPLA2 and cPLA2 serine esterases, but not sPLA2 that utilizes a His/Asp catalytic dyad (36). In agreement with our predictions from sequence alignment, this finding suggests that catalysis by AdPLA requires an active cysteine residue (i.e. Cys-113) that is inhibited by treatment with MAFP. This finding also supports that AdPLA differs fundamentally from other histidine PLA2s. Results from experiments with thioetheramide-PC, a phospholipid analog containing a thioether at the sn-1 position and an amide at the sn-2 position (37), further support that AdPLA may be functionally distinct from other known histidine PLA2s. We found inhibition of sPLA2 but not AdPLA by thioetheramide treatment (Fig. 5E). It is believed that the amide side chain of thioetheramide forms a hydrogen bond with the water molecule in the sPLA2 His active site, stabilizing binding of the inhibitor and thereby competitively displacing natural substrate that binds with lower affinity (37). An absence of inhibition by thioetheramide highlights differences in the active site motifs of AdPLA and sPLA2s, and further indicates that AdPLA does not fit easily into any pre-existing category of phospholipases.
FIGURE 5.
Treatment of AdPLA with known phospholipase inhibitors. A and B, AdPLA and sPLA2 were inhibited in a similar manner by treatment with 7,7-dimethyl-5,8-eicosadienoic acid or AACOCF3, respectively. C, neither AdPLA nor sPLA2 was inhibited by BEL, a suicide-substrate inhibitor of GXSXG-containing lipases. D, MAFP, an inhibitor of serine and cysteine phospholipases, inhibited AdPLA but not sPLA2. E, thioetheramide inhibited sPLA2 but not AdPLA. Results are means ± S.E. from duplicate or triplicate measurements.
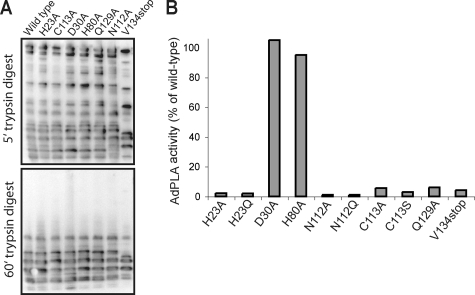
We next performed mutational and deletional studies to investigate the specific requirement for certain residues in AdPLA catalysis. Trypsin digest of mutated and truncated proteins showed a similar pattern of peptide generation, indicating that protein folding was likely unchanged in mutants (Fig. 6A). AdPLA contains His-23 and Cys-113 residues that align with catalytically active amino acids in LRAT (supplemental Fig. S1), and we found that mutation of these caused a complete loss of activity (Fig. 6B). Furthermore, deletion of the C-terminal region containing the membrane-spanning domain also resulted in a total loss of activity. Mutational analysis revealed the essentiality of other highly conserved amino acids, including Gln-129 and Asn-112, for catalysis but also the redundancy of the conserved Asp-30 and His-80 residues. A specific role is therefore indicated for amino acids at positions 23, 112, 113, and 129 in catalytic activity.
FIGURE 6.
Mutational and deletional analysis of AdPLA activity. Sequence alignment indicated that the His-23/Cys-113 motif required by LRAT for catalysis is conserved in AdPLA. A, trypsin digest of recombinant mutants indicates a similar pattern of peptide generation. B, mutation of His-23 or Cys-113 caused a complete loss of activity in vitro, as did truncation of the C-terminal membrane localization domain. Mutation of other highly conserved residues, including Asn-112 and Glu-129, also abolished activity. Mutation of other conserved residues, including Asp-30 and His-80, had no effect, indicating a specific role for residues at positions 23, 112, 113, and 129 in catalytic activity.
Taken together with findings from sequence analysis and inhibitor studies, our results suggest that AdPLA is most likely a His-Cys PLA2. AdPLA was inhibited by MAFP, an inhibitor of cysteine-dependent phospholipases. Furthermore, AdPLA shares characteristics with PLA2s that utilize a catalytic histidine. For example, histidine PLA2s typically have a narrow pH dependence ranging from 7 to 9, because fatty acid hydrolysis requires the activation and orientation of a water molecule by hydrogen bonding to the active site histidine (12). Similarly, AdPLA displayed a narrow optimum pH range with maximum activity at pH 8. AdPLA was also activated in vitro by Ca2+, much like other histidine PLA2s that characteristically require Ca2+ for formation of a positively charged oxyanion hole to stabilize the negatively charged transition state of the PLA2 reaction (12). However, AdPLA differs from currently known histidine PLA2s in a number of ways. First, all known histidine PLA2s are sPLA2s, platelet-activating factor acetylhydrolases, or lysosomal PLA2s (1). Unlike these, AdPLA appears to contain active His/Cys catalytic residues rather than a His/Asp catalytic dyad or Ser/His/Asp catalytic triad. Furthermore, unlike sPLA2s in particular, AdPLA lacks the characteristically conserved CCXXHDXC motif (14). Also, AdPLA localized intracellularly with the perinuclear membrane, and AdPLA contains too few cysteines to support the extensive disulfide bonding characteristic of sPLA2s. Indeed, although AdPLA is most similar to the sPLA2s in terms of size and participation of an active site histidine, AdPLA does not fit readily into any family or group within the currently known and characterized phospholipases.
In summary, we have found that AdPLA that was previously known as H-Rev-107/HRASLS3 is an adipose-specific, calcium-dependent histidine PLA2. Although AdPLA exhibited some but much lower additional lysophospholipase activity, no other acyl hydrolase activity was evident. AdPLA shares some characteristics with other PLA2s, including susceptibility to inhibition by known specific inhibitors, but does not fit neatly into any current PLA2 group. We propose that AdPLA may be the first member of a new group of calcium-dependent intracellular PLA2s, group XVI.
Supplementary Material
Acknowledgments
We thank Chris Lange for assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant DK075682 (to H. S. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
Footnotes
The abbreviations used are: PLA2, phospholipase A2; AdPLA, adipose-specific phospholipase A2; cPLA2, cytosolic phospholipase A2; DAG, diacylglycerol; FFA, free fatty acid; GFP, green fluorescent protein; iPLA2, calcium-independent phospholipase A2; LRAT, lecithin:retinol acyltransferase; PC, phosphatidylcholine; sPLA2, secretory phospholipase A2; TAG, triacylglycerol; WAT, white adipose tissue; HA, hemagglutinin; BEL, bromoenolactone; AACOCF3, arachidonyl trifluoromethyl ketone; DMEM, Dulbecco's modified Eagle's medium; BSA, bovine serum albumin; MAFP, methyl arachidonyl fluorophosphate; MIX, methylisobutylxanthine; EST, expressed sequence tag.
References
- 1.Schaloske, R. H., and Dennis, E. A. (2006) Biochim. Biophys. Acta 1761 1246-1259 [DOI] [PubMed] [Google Scholar]
- 2.Yuan, C., Rieke, C. J., Rimon, G., Wingerd, B. A., and Smith, W. L. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 6142-6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aubert, J., Saint-Marc, P., Belmonte, N., Dani, C., Negrel, R., and Ailhaud, G. (2000) Mol. Cell. Endocrinol. 160 149-156 [DOI] [PubMed] [Google Scholar]
- 4.Fajas, L., Miard, S., Briggs, M. R., and Auwerx, J. (2003) J. Lipid Res. 44 1652-1659 [DOI] [PubMed] [Google Scholar]
- 5.Forman, B. M., Tontonoz, P., Chen, J., Brun, R. P., Spiegelman, B. M., and Evans, R. M. (1995) Cell 83 803-812 [DOI] [PubMed] [Google Scholar]
- 6.Reginato, M. J., Krakow, S. L., Bailey, S. T., and Lazar, M. A. (1998) J. Biol. Chem. 273 1855-1858 [DOI] [PubMed] [Google Scholar]
- 7.Yan, H., Kermouni, A., Abdel-Hafez, M., and Lau, D. C. (2003) J. Lipid Res. 44 424-429 [DOI] [PubMed] [Google Scholar]
- 8.Cohen-Luria, R., and Rimon, G. (1992) Cell. Signal. 4 331-335 [DOI] [PubMed] [Google Scholar]
- 9.Fain, J. N., Leffler, C. W., and Bahouth, S. W. (2000) J. Lipid Res. 41 1689-1694 [PubMed] [Google Scholar]
- 10.Kather, H., and Simon, B. (1979) J. Clin. Investig. 64 609-612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nugent, C., Prins, J. B., Whitehead, J. P., Wentworth, J. M., Chatterjee, V. K., and O'Rahilly, S. (2001) J. Biol. Chem. 276 9149-9157 [DOI] [PubMed] [Google Scholar]
- 12.Six, D. A., and Dennis, E. A. (2000) Biochim. Biophys. Acta 1488 1-19 [DOI] [PubMed] [Google Scholar]
- 13.Ghosh, M., Tucker, D. E., Burchett, S. A., and Leslie, C. C. (2006) Prog. Lipid Res. 45 487-510 [DOI] [PubMed] [Google Scholar]
- 14.Diaz, B. L., and Arm, J. P. (2003) Prostaglandins Leukot. Essent. Fatty Acids 69 87-97 [DOI] [PubMed] [Google Scholar]
- 15.Sers, C., Emmenegger, U., Husmann, K., Bucher, K., Andres, A. C., and Schafer, R. (1997) J. Cell Biol. 136 935-944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jahng, W. J., Xue, L., and Rando, R. R. (2003) Biochemistry 42 12805-12812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, K. H., Lee, K., Moon, Y. S., and Sul, H. S. (2001) J. Biol. Chem. 276 11252-11256 [DOI] [PubMed] [Google Scholar]
- 18.Roder, K., Latasa, M. J., and Sul, H. S. (2002) Biochem. Biophys. Res. Commun. 293 793-799 [DOI] [PubMed] [Google Scholar]
- 19.Jimenez, M., Cabanes, J., Gandia-Herrero, F., Escribano, J., Garcia-Carmona, F., and Perez-Gilabert, M. (2003) Anal. Biochem. 319 131-137 [DOI] [PubMed] [Google Scholar]
- 20.Lucas, K. K., and Dennis, E. A. (2005) Prostaglandins Other Lipid Mediat. 77 235-248 [DOI] [PubMed] [Google Scholar]
- 21.Bligh, E. G., and Dyer, W. J. (1959) Can. J. Biochem. Physiol. 37 911-917 [DOI] [PubMed] [Google Scholar]
- 22.Nazarenko, I., Kristiansen, G., Fonfara, S., Guenther, R., Gieseler, C., Kemmner, W., Schafer, R., Petersen, I., and Sers, C. (2006) Am. J. Pathol. 169 1427-1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nazarenko, I., Schafer, R., and Sers, C. (2007) J. Cell Sci. 120 1393-1404 [DOI] [PubMed] [Google Scholar]
- 24.Villena, J. A., Roy, S., Sarkadi-Nagy, E., Kim, K. H., and Sul, H. S. (2004) J. Biol. Chem. 279 47066-47075 [DOI] [PubMed] [Google Scholar]
- 25.Roder, K., Kim, K. H., and Sul, H. S. (2002) Biochem. Biophys. Res. Commun. 294 63-70 [DOI] [PubMed] [Google Scholar]
- 26.Roder, K., Latasa, M. J., and Sul, H. S. (2002) J. Biol. Chem. 277 30543-30550 [DOI] [PubMed] [Google Scholar]
- 27.Marblestone, J. G., Edavettal, S. C., Lim, Y., Lim, P., Zuo, X., and Butt, T. R. (2006) Protein Sci. 15 182-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambeau, G., and Gelb, M. H. (2008) Annu. Rev. Biochem. 77 21-26 [DOI] [PubMed] [Google Scholar]
- 29.de Carvalho, M. G., Garritano, J., and Leslie, C. C. (1995) J. Biol. Chem. 270 20439-20446 [DOI] [PubMed] [Google Scholar]
- 30.Diez, E., Louis-Flamberg, P., Hall, R. H., and Mayer, R. J. (1992) J. Biol. Chem. 267 18342-18348 [PubMed] [Google Scholar]
- 31.Hirabayashi, T., Murayama, T., and Shimizu, T. (2004) Biol. Pharm. Bull. 27 1168-1173 [DOI] [PubMed] [Google Scholar]
- 32.Xue, L., and Rando, R. R. (2004) Biochemistry 43 6120-6126 [DOI] [PubMed] [Google Scholar]
- 33.Street, I. P., Lin, H. K., Laliberte, F., Ghomashchi, F., Wang, Z., Perrier, H., Tremblay, N. M., Huang, Z., Weech, P. K., and Gelb, M. H. (1993) Biochemistry 32 5935-5940 [DOI] [PubMed] [Google Scholar]
- 34.Hazen, S. L., Zupan, L. A., Weiss, R. H., Getman, D. P., and Gross, R. W. (1991) J. Biol. Chem. 266 7227-7232 [PubMed] [Google Scholar]
- 35.Jenkins, C. M., Mancuso, D. J., Yan, W., Sims, H. F., Gibson, B., and Gross, R. W. (2004) J. Biol. Chem. 279 48968-48975 [DOI] [PubMed] [Google Scholar]
- 36.De Petrocellis, L., Melck, D., Ueda, N., Maurelli, S., Kurahashi, Y., Yamamoto, S., Marino, G., and Di Marzo, V. (1997) Biochem. Biophys. Res. Commun. 231 82-88 [DOI] [PubMed] [Google Scholar]
- 37.Yu, L., Deems, R. A., Hajdu, J., and Dennis, E. A. (1990) J. Biol. Chem. 265 2657-2664 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.