Abstract
PDBsum (http://www.ebi.ac.uk/pdbsum) provides summary information about each experimentally determined structural model in the Protein Data Bank (PDB). Here we describe some of its most recent features, including figures from the structure's key reference, citation data, Pfam domain diagrams, topology diagrams and protein–protein interactions. Furthermore, it now accepts users’ own PDB format files and generates a private set of analyses for each uploaded structure.
INTRODUCTION
Since its inception in 1971 (1) the Protein Data Bank (PDB) has released over 55 000 experimentally determined structural models of proteins and nucleic acids. The archiving, management and quality control of these models is nowadays performed by the worldwide PDB (wwPDB), a consortium whose partners comprise: the Research Collaboratory for Structural Bioinformatics (RCSB), the Macromolecular Structure Database (MSD) at the European Bioinformatics Institute (EBI), the Protein Data Bank Japan (PDBj) at Osaka University and, more recently, the BioMagResBank at the University of Wisconsin-Madison (2). The wwPDB makes the structural models available via ftp together with, in many cases, the original experimental data.
As the formats in which the structural models are supplied can be rather hard to interpret on their own, a number of web sites have been set up to provide ‘atlas’ pages that describe each model in a more readily digestible form. These are usually aimed at a wide variety of users with an interest in protein structure, including structural biologists and bioinformaticians. The pages contain general information, obtained from the relevant PDB file, supplemented by links to other databases and various analyses and schematic diagrams that are often unique to the specific atlas. The best known atlases are: the RCSB PDB (3), the MSD (4), OCA, JenaLib (5), PDBj, MMDB (6) and PDBsum (7). A recent review compared these atlases, or ‘comprehensive information resources’ as it called them, and identified their similarities and differences (8).
Here we provide an update of the last-mentioned atlas, PDBsum (http://www.ebi.ac.uk/pdbsum). We focus on changes made to it since it was last described (9), and specifically concentrate on features that are unique to it.
FIGURES FROM THE LITERATURE
The first major change described here involves the inclusion of figures, plus corresponding figure legends, from the structures’ principal literature references. The aim here is to encapsulate some of the rich information that the given structural model provided when it was solved; and where better to get that information than from the authors’ original publication. A well-chosen figure can often provide much information, and may be especially useful for users who do not have free access to the journal in question. Furthermore, many of the figures are quite beautiful, so their inclusion is likely to draw the user to the original reference.
The figures come either from Open Access publications or from journals whose editors and/or publishers have granted us permission for use of their copyrighted material. In fact, most journals and publishers were very generous and cooperative in this respect, although there were one or two disappointing exceptions. In general, we were given permission to use up to two figures from each relevant paper. The statistics showing the numbers of key references from which figures have been taken are given in PDBsum's ‘Figure Stats’ page.
The actual capture of the figures and their captions from the online copies of the articles has proved something of a challenge. For one thing, the journals keep changing their formats. Furthermore, the very old papers are only available as PDF versions scanned in from the original hard copies. In these cases, each journal page is, in effect, just an image. To process these images requires use of optical character recognition (OCR) utilities to extract the text (a somewhat error-prone procedure) and then each separate block of text has to be categorized as: part of the main body of the paper, a figure legend, other text such as headings or tables, or text within the figures themselves (i.e. labels or parts of the picture misinterpreted as text by the OCR utility). From the placement of the text blocks the likely coordinates of each figure on the page are calculated and the figure spliced out. This procedure has been described in greater detail elsewhere (10). For the more recent papers, the figures can generally be downloaded as image files directly from the online versions of the papers, although this, too, is not 100% reliable.
Selection of which figure, or pair of figures, to use from each paper is made by an algorithm called a support vector machine trained to distinguish interesting figures from dull ones on the basis of the words and word-pairs that appear in the figure legend (10). However, rather than wholly rely on this automated method, the choice of figures is, where possible, e-mailed to the lead author of the paper with a request to review, and possibly change, which figures are to be used and an invitation to add the author's own comment(s). About one in six authors respond to these e-mails and over 200 in all have taken the trouble to annotate their entries with additional information. A useful spin-off from this system has been an increase in the level of feedback from the authors about PDBsum and has led to several improvements, most notably the Pfam domain diagrams described below.
CITATIONS
As well as a PDB entry's key reference, it is also useful to know of more recent literature relating to the structure. To this end, we have started adding citation data to PDBsum; that is, references that cite each structure's key paper. Currently, the data comes from CiteXplore (http://www.ebi.ac.uk/citexplore), and is supplemented by additional citations automatically harvested from the web. Many references are still not captured by either method, but our coverage of the literature should increase with time. As of September 2008, there were over 44 000 citing references for the 25 000 key references in PDBsum.
Pfam DOMAIN DIAGRAMS
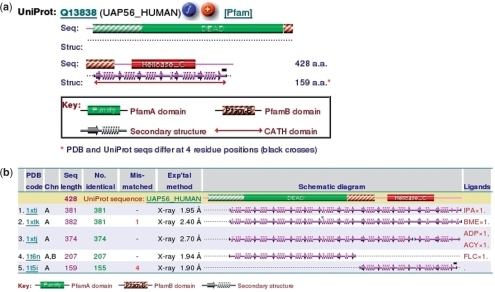
One crucial aspect of PDB structural models that many non-expert users fail to appreciate is that very often the model corresponds to only ‘part’ of the full protein sequence; sometimes it is only a single domain, and occasionally merely a fragment. To show just how the given structure relates to its parent sequence, PDBsum has a little schematic diagram near the top of the structure's page to provide the information at a glance. The diagram represents the full length of the sequence, including any constituent Pfam (11) domains, where known. Below the sequence is shown the full length of the structure in terms of its secondary structure elements. From the diagram one can easily see the correspondence between the structure and the full-length sequence. Figure 1a shows an example, in which the structure is of only the last domain. Also marked on this diagram, where known, are any CATH structural domains and any residue positions where the sequence in the PDB file disagrees with the sequence in UniProt (12). Clicking on a blank part of the image brings up an alignment between the two versions of the sequence, which can be used to identify these mismatches.
Figure 1.
Diagrams illustrating the correspondence between a given structural model and the full-length sequence of the corresponding protein. (a) A schematic Pfam diagram taken from the PDBsum atlas page for PDB entry 1t5i: human pre-mRNA-processing protein (15). The extent of the 3D structural model is shown beneath the Pfam domains and shows that the structural model corresponds to only the C-terminal helicase C domain. Clicking on the orange ‘+’ icon returns all other PDB entries for the given UniProt sequence (UAP56_HUMAN). (b) The top five PDB entries for this sequence. From this one can see that, in addition to the 1t5i partial structural model of this protein shown in (a) (listed here at position 5), there are three models of the complete protein and one model of just the N-terminal domain.
Given that a structural model may be incomplete in this way, PDBsum can illustrate ‘all’ structural models in the PDB for a given UniProt sequence. Figure 1b gives an example. Here one can see that there are in fact three models of the full-length sequence, any of which may be more informative than the single domain model given in Figure 1a.
IMPROVED WIRING DIAGRAMS
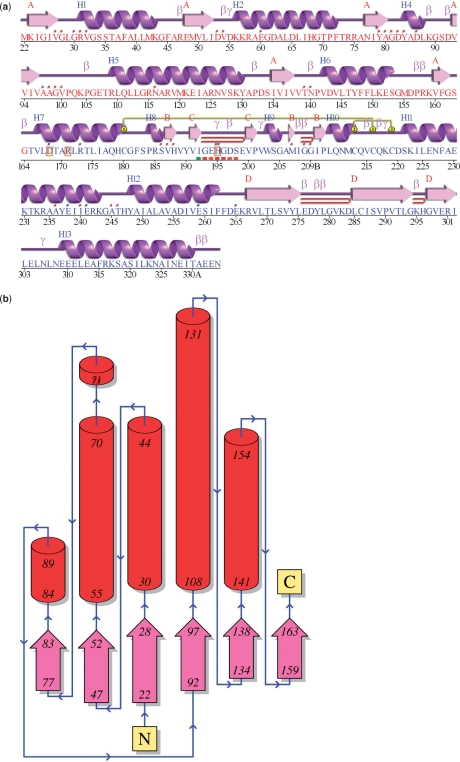
For each unique protein chain in a given structural model, PDBsum provides a ‘protein page’ that includes a schematic diagram of the protein's secondary structure. These ‘wiring diagrams’ have been improved to provide a prettier picture than previously. And now the diagrams can be enlarged to five times their size for publication purposes (Figure 2a).
Figure 2.
Schematic diagrams from the PDBsum ‘Protein page’ for entry 1a5z: lactate dehydrogenase from Thermatoga maritima (16). (a) The ‘wiring diagram’ shows the protein's secondary structure elements (α-helices and β-sheets) together with various structural motifs such as β- and γ-turns, and β-hairpins. The yellow linking bars labelled 1 and 2 represent disulphide bonds. The single-letter amino acid codes showing the protein's sequence are coloured red or blue depending on whether they belong to CATH structural domain 1 or 2, respectively. Catalytic residues are indicated by a box surrounding the amino acid code. Red dots above the single-letter codes signify residues that interact with any bound ligand(s) while coloured lines underneath represent residues belonging to a PROSITE pattern, the redder the colour the more highly conserved the residue in the pattern. (b) Topology diagram of the first (i.e. red) structural domain in 1a5z. The diagram illustrates how the β-strands, represented by the large arrows, join up, side-by-side, to form the domain's central β-sheet. The diagram also shows the relative locations of the α-helices, here represented by the red cylinders. The small arrows indicate the directionality of the protein chain, from the N- to the C-terminus. The numbers within the secondary structural elements correspond to the residue numbering given in the PDB file.
TOPOLOGY DIAGRAMS
Each protein page also includes a topology diagram showing the arrangement and connectivity of the protein's helices and strands (Figure 2b). Where the protein chain consists of more than one domain, as defined by CATH (13), a separate diagram is generated for each and is colour-coded according to the domain colouring on the wiring diagram. The topology diagrams are generated from the hydrogen bonding plots of Gail Hutchinson's HERA program (14).
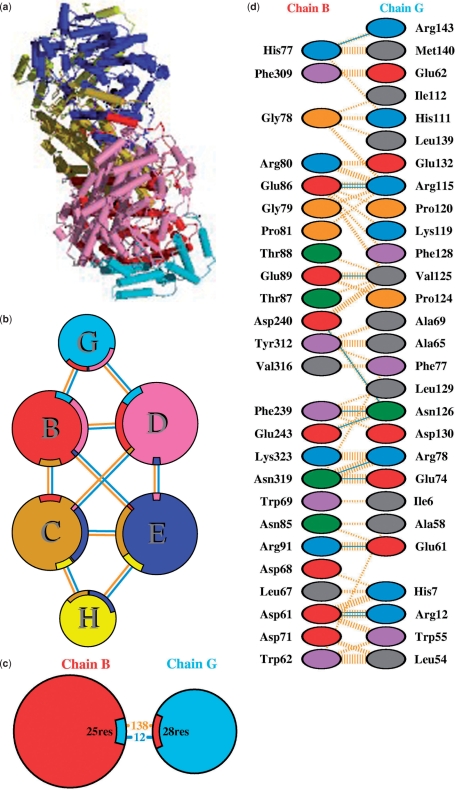
PROTEIN–PROTEIN INTERFACES
Another new feature is the addition of schematic diagrams illustrating the interactions across protein–protein interfaces. Where a structural model contains more than one protein chain (e.g. Figure 3a), the interfaces between the chains are depicted by three types of plot: the first shows an overview of which chains interact with which (Figure 3b), the second summarizes the interactions across any selected interface (Figure 3c), and the third shows in detail which residues actually interact across that interface (Figure 3d).
Figure 3.
Extracts from the protein–protein interaction diagrams in PDBsum for PDB entry 1mmo, a non-haem iron hydroxylase from Methylococcus capsulatus (17). (a) Thumbnail image of the 3D structural model which contains six protein chains: two of MEMB_METCA (chains B and C), two of MEMB_METCA (chains D and E) and two of MEMG_METCA (chains G and H). (b) Schematic diagram showing the interactions between the chains. The area of each circle is proportional to the surface area of the corresponding protein chain. The extent of the interface region on each chain is represented by a coloured wedge whose colour corresponds to the colour of the other chain and whose size signifies the interface surface area. The joining lines are coloured light blue for hydrogen bonds and orange for non-bonded contacts. (c) A schematic diagram showing the numbers of interactions across one of the interfaces, namely the B–G interface, and the numbers of residues involved. (d) Detail of the individual residue–residue interactions across this interface. The colour of the interactions is as above.
PDBSUM PAGES FOR USER-SUBMITTED STRUCTURES
Finally, an upload option has been added to allow users to submit their own PDB-format files to PDBsum and have a set of PDBsum analyses and pages generated for it. The generated pages are password-protected for privacy, and are deleted after about 6 months. Currently the server is receiving an average of around 40 uploads per week.
FUNDING
Funding for open access charge: European Molecular Biology Laboratory (EMBL).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Gail Hutchinson for use of her HERA program to help generate the topology diagrams. Also thanks to the many authors who contributed their time to reviewing the figures included in PDBsum and, in particular, to those who added comments to their pages or provided valuable feedback.
REFERENCES
- 1.Bernstein FC, Koetzle TF, Williams GJB, Meyer E.F., Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: a computer-based archival file of macromolecular structures. J. Mol. Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 2.Berman HM, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 3.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tagari M, Tate J, Swaminathan GJ, Newman R, Naim A, Vranken W, Kapopoulou A, Hussain A, Fillon J, Henrick K, et al. E-MSD: improving data deposition and structure quality. Nucleic Acids Res. 2006;34:D287–D290. doi: 10.1093/nar/gkj163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reichert J, Sühnel J. The IMB Jena Image Library of Biological Macromolecules: 2002 update. Nucleic Acids Res. 2002;30:253–254. doi: 10.1093/nar/30.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J, Anderson JB, DeWeese-Scott C, Fedorova ND, Geer L, He S, Hurwitz DI, Jackson JD, Jacobs AR, Lanczycki CJ, et al. MMDB: Entrez's 3D-structure database. Nucleic Acids Res. 2003;31:474–477. doi: 10.1093/nar/gkg086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laskowski RA. PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res. 2001;29:221–222. doi: 10.1093/nar/29.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hühne R, Koch FT, Sühnel J. A comparative view at comprehensive information resources on three-dimensional structures of biological macro-molecules. Brief. Funct. Genomic. Proteomic. 2007;6:220–239. doi: 10.1093/bfgp/elm020. [DOI] [PubMed] [Google Scholar]
- 9.Laskowski RA, Chistyakov VV, Thornton JM. PDBsum more: new summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Res. 2005;33:D266–D268. doi: 10.1093/nar/gki001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laskowski RA. Enhancing the functional annotation of PDB structures in PDBsum using key figures extracted from the literature. Bioinformatics. 2007;23:1824–1827. doi: 10.1093/bioinformatics/btm085. [DOI] [PubMed] [Google Scholar]
- 11.Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The UniProt Consortium. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greene LH, Lewis TE, Addou S, Cuff A, Dallman T, Dibley M, Redfern O, Pearl F, Nambudiry R, Reid A, et al. The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res. 2007;35:D291–D297. doi: 10.1093/nar/gkl959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutchinson EG, Thornton JM. HERA: a program to draw schematic diagrams of protein secondary structures. Proteins. 1990;8:203–212. doi: 10.1002/prot.340080303. [DOI] [PubMed] [Google Scholar]
- 15.Zhao R, Shen J, Green MR, MacMorris M, Blumenthal T. Crystal structure of UAP56, a DExD/H-box protein involved in pre-mRNA splicing and mRNA export. Structure. 2004;12:1373–1381. doi: 10.1016/j.str.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Auerbach G, Ostendorp R, Prade L, Korndörfer I, Dams T, Huber R, Jaenicke R. Lactate dehydrogenase from the hyperthermophilic bacterium thermotoga maritima: the crystal structure at 2.1 Å resolution reveals strategies for intrinsic protein stabilization. Structure. 1998;6:769–781. doi: 10.1016/s0969-2126(98)00078-1. [DOI] [PubMed] [Google Scholar]
- 17.Rosenzweig AC, Frederick CA, Lippard SJ, Nordlund P. Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane. Nature. 1993;366:537–543. doi: 10.1038/366537a0. [DOI] [PubMed] [Google Scholar]