Abstract
The variability between respiratory syncytial virus (RSV) strains is one of the features of RSV infections that might contribute to the ability of the virus to infect people repeatedly and cause yearly outbreaks. To study the molecular epidemiology of RSV, more than 1,400 RSV isolates from human nasopharyngeal aspirates or nasal or throat swabs from patients with respiratory illness were identified and differentiated by TaqMan reverse transcription-PCR into groups A and B. RSV group A was dominant in seven out of nine epidemic seasons. Phylogenetic analysis revealed that RSV group A genotypes GA2 and GA5 circulated from 1998 to 2007. Genotype GA7 was present in only two seasons (1999 to 2000 and 2002 to 2003). Comparison of the synonymous mutation/nonsynonymous mutation ratios showed greater evidence for selection pressure for genotype GA2 (1.18) than for GA5 (4.34). Partial protein sequences were predicted to encode G proteins of 298 amino acids in length and in a few cases of G proteins of 297 amino acids in length. Amino acid analysis also revealed genotype-specific amino acid substitutions: two substitutions for genotype GA2, seven for GA5, and three for GA7. Two to four putative, genotype-specific N-linked glycosylation sites were determined. Predicted O-glycosylation sites included 22 to 34 residues. This study provides for the first time data on the circulation pattern of RSV group A genotypes and their molecular characterization in Germany during nine consecutive epidemic seasons.
Worldwide, the respiratory syncytial virus (RSV) is the major pathogen of lower respiratory tract infections in infants and young children in both developing and developed countries (46). By the age of 2 years, virtually all children have been infected at least once with RSV (16). Nevertheless, reinfections are very common throughout life. In older children and adults, they are usually associated with milder disease, indicating that RSV infections induce only partial immunity (19).
RSV strains are separated into two major groups based on antigenic and genetic variability. The main differences between RSV groups A and B were found in the attachment glycoprotein G (1, 6, 30). Variability in this protein is greater than that in the other proteins, both between and within the major antigenic groups of RSV. The G protein is a type II glycoprotein of 289 to 299 amino acids in length, consisting of two hypervariable regions in the extracellular domain. Diversity occurs mainly in the G ectodomain which has only 44% amino acid identity between the two groups compared with 83% for the transmembrane and cytoplasmic domains (21). The G protein is heavily glycosylated with N-linked and O-linked sugars (60). Mainly, the ectodomain of the G protein has a high content of serine and threonine residues which are potential acceptor sites for O-linked sugars. However, the amino acid sequence positions of potential N-linked and O-linked glycosylation sites are poorly conserved, although the general locations of the latter are similar (21). Amino acid variations of the protein exist in both groups, but variations are more pronounced in RSV group A (39, 51). Most molecular analyses include the second variable region of the G protein, coding for the C-terminal end of the protein. Eight RSV group A genotypes, named GA1 to GA7 and South Africa A1 (SAA1) have been described so far (38, 39, 51). Genotype analysis of RSV group A is limited to particular countries worldwide. In Europe, for instance, genotypic RSV group A data are provided only from Belgium and Sweden (40, 62, 63).
Variability between RSV strains is one of the features of RSV infections that might contribute to the ability of the virus to infect people repeatedly and cause yearly outbreaks (38). RSV has a clear seasonality. In temperate climates, outbreaks occur yearly in the late fall, winter, or spring but not in the summer (22, 29, 61). Both RSV groups can be present in the same community, and their relative proportions may differ between epidemics (2, 4, 13, 20), although group A viruses tend to predominate (8).
There is limited information regarding the molecular epidemiology of RSV in Germany. For the first time, we describe both the genetic variability of RSV group A viruses and the circulation pattern of different RSV group A genotypes during previous years in Germany.
MATERIALS AND METHODS
Clinical samples.
Human nasopharyngeal aspirates or nasal or throat swabs from patients with respiratory illness were sent to the Robert Koch-Institut from two hospitals and about 150 medical practices between October 1998 and September 2007. The medical practices were located all over Germany. Specimens collected at the time of admission were forwarded undiluted. Immediately upon arrival, swabs were resuspended in 5 ml of sterile minimal essential medium with HEPES (Gibco BRL, Eggenstein, Germany) and 100 U/ml penicillin-streptomycin (GIBCO), aliquoted, and stored at −70°C. Nasopharyngeal aspirates were adjusted to a volume of 3 ml with the minimal essential medium described above.
During the period of examination, a total of 6,115 samples were collected and tested for RSV. These included samples from 713 (12%) infants less than 3 months old, 497 samples (8%) of those aged 3 to 6 months, 640 samples (10%) of those aged 7 to 12 months, and 1,719 samples (28%) of children 1 to 3 years old; 2,328 (38%) samples were from patients more than 3 years old, and 218 samples (4%) were from individuals of unknown age.
RNA preparation and cDNA synthesis.
RNA extraction was performed from 400 μl of the diluted specimens using RTP DNA/RNA virus minikit (Invitek, Berlin, Germany) according to the manufacturer's instructions. cDNA was synthesized with 25 μl of RNA in a mixture containing a 500 nM concentration of random hexamer primers, a 200 μM concentration of each deoxynucleoside triphosphate (Invitrogen, Karlsruhe, Germany), 2.5 mM dithiothreitol (Invitrogen), 40 U RNasin (Promega, Madison, WI), 200 U Moloney murine leukemia virus reverse transcriptase (Invitrogen), and first-strand buffer (Invitrogen), containing 250 mM Tris-HCl (pH 8.3), 375 mM KCl, and 15 mM MgCl2, in a final volume of 40 μl. The synthesis of cDNA was carried out for 5 min at 42°C, followed by 35 min at 37°C, and finally for 5 min at 94°C.
Real-time PCR for RSV detection.
For detection of RSV, cDNA was first analyzed by a generic RSV TaqMan PCR targeting the RSV N-protein gene. PCR was performed in a 25-μl reaction mixture using 5 μl of cDNA product, 250 nM of each of the primer pairs RSV-N15-F and RSV-N184-R, 200 nM of the RSV-N106-S probe (Table 1), 1 μM 6-carboxy-X-rhodamine, 100 μM deoxynucleoside triphosphates, 5 mM MgCl2 (Invitrogen), 0.5 U of Platinum Taq DNA polymerase (Invitrogen), and PCR buffer (200 mM Tris-HCl [pH 8.4], 500 mM KCl). Amplification was carried out at 95°C for 5 min, followed by 45 cycles of PCR, with 1 cycle consisting of 15 s at 95°C and 30 s at 60°C, and a final extension step of 15 min at 72°C.
TABLE 1.
RSV oligonucleotide primers and probes used in this study
| Method(s) (purpose) and oligonucleotide primer or probe | Oligonucleotide sequence (5′-3′) | Positionsa | Gene | Polarity | Reference |
|---|---|---|---|---|---|
| Real-time PCR | |||||
| Generic PCR (detection of RSV group A and B viruses targeting the N gene) | |||||
| RSV-N15-F | GATGGCTCTTAGCAAAGTCAAGTT | 15-38 | N | + | This study |
| RSV-184-R | CATCTTCWGTGATTAATARCATRCCACATA | 155-184 | N | − | This study |
| RSV-N106-S | F-CTGTCATCYAGCAAATACACYATYCAACGKAGYACAGGAG-BHQ1 | 67-106 | N | + | This study |
| Specific PCR (detection of RSV group A viruses targeting the G gene) | |||||
| RSVA-G409-F | AAGACCAAAAACACAACAACAA | 409-430 | G | + | This study |
| RSVA-G586-R | TTGGTATTCTTTTGCAGATAGTRGCC | 564-586 | G | − | This study |
| RSVA-G556-S | F-TTGGATTGTTGCTGCATATGCTGCTXTPH | 532-556 | G | − | This study |
| Conventional PCR and sequencing | |||||
| External PCR | |||||
| RSVA-G513-F | AGTGTTCAACTTTGTACCCTGC | 513-534 | G | + | This study |
| RSVA-F131-R | CTGCACTGCATGTTGATTGAT | 111-131 | F | − | This study |
| Nested PCR | |||||
| RSVA-G606-F | AACCACCACCAAGCCCACAA | 606-625 | G | + | 39 |
| RSV-F22-R | CAACTCCATTGTTATTTGCC | 3-22 | F | − | 39 |
Nucleotide positions are given according to the gene positions in RSV strain A2 (GenBank accession number U50362). All oligonucleotides were purchased from TibMolbiol GmbH (Berlin, Germany). Abbreviations: F, 6′-carboxyfluorescein (FAM); BHQ1, black hole quencher 1; X, 6-carboxytetramethylrhodamine (TAMRA); PH, phosphate.
Following this, RSV-positive samples were differentiated into RSV group A and B by group-specific real-time PCRs targeting the large glycoprotein gene (G gene). Since the focus is on RSV group A, only these conditions are described here. cDNA product (5 μl) was amplified in a 25-μl reaction mixture and the same amplification conditions as mentioned above. Specific RSV group A amplification was carried out using the primer pairs RSVA-G409-F and RSVA-G586-R as well as the RSVA-G556-S probe (Table 1).
All reactions were carried out using an ABI PRISM 7700 sequence detection system and 7900HT sequence detection system (Applied Biosystems, Darmstadt, Germany).
Analysis of RSV epidemic seasons.
For each season, RSV-positive samples were estimated per calendar week. In order to determine the beginning (onset) and ending (offset) of an epidemic season, sporadic RSV cases were calculated. Since there were no RSV-positive samples detected during the summer months, the time of beginning was determined as that time when the first positive samples were detected in at least three consecutive weeks. Conversely, the time of ending was defined as the time when the last samples in a cascade of positive samples were detected. The duration was defined as the number of weeks between the determined onset and offset weeks, and the epidemic peak was defined as the week after onset with the highest number of positive test results. The cutoff for determination of early and late seasons was set at week 49. Differences between weak and strong seasons were statistically analyzed using the chi-square test in the SPSS version 16.0.2.
PCR for molecular analysis of the G-protein gene.
A representative number of group A RSV-positive samples (at least 20%) were selected for amplification of the second hypervariable region of the G-protein gene by either external or nested PCR. The first amplification was performed with 5 μl cDNA in a 50-μl reaction mixture by using 250 nM of each of the primer pairs RSVA-G513-F and RSVA-F131-R (Table 1), a 100 μM concentration of each deoxynucleoside triphosphate, 4 mM MgCl2, 0.5 U Platinum Taq DNA polymerase (Invitrogen), and PCR buffer (200 mM Tris-HCl [pH 8.4], 500 mM KCl). Amplification was carried out at 94°C for 5 min, followed by 40 cycles of PCR, with 1 cycle consisting of 30 s at 94°C, 30 s at 58°C, and 1 min at 72°C, and a final extension step of 10 min at 72°C. The amplified products of 583 bp were analyzed by electrophoresis on a 1.5% agarose gel. In the case of negative results, 5 μl of the external PCR mixture was used for nested PCR, which was performed in a 50-μl reaction mixture with 250 nM of RSVA-G606-F and RSV-F22-R (Table 1). The cycling protocol was the same as for the external PCR, except for the annealing temperature, which was 53°C. The nested amplicons of 391 bp were visualized by agarose gel electrophoresis as well.
DNA sequencing.
The PCR products were purified either directly with QIAquick PCR purification kit (Qiagen, Hilden, Germany) or from agarose gels with JETquick spin column technique (Genomed, Löhne, Germany) according to the manufacturer's instructions. Purified PCR products were cycle sequenced in the forward direction and the reverse direction with primer pair RSVA-G513-F and RSVA-F131-R or primer pair RSVA-G606-F and RSV-F22-R (Table 1), respectively, in a 3130xl genetic analyzer (Applied Biosystems) by using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems).
Phylogenetic analysis.
A multiple-sequence alignment was compiled from the second variable region of the G gene (270 bp) of 221 sequences using ClustalW in the Bioedit version 7.0.9 (17). For determination of RSV group A genotypes, phylogenetic analyses were performed using the PHYLIP version 3.64 package (11). Sequence distances were calculated from alignments using F84, and clustering was done by the neighbor-joining algorithm of the PHYLIP program package. Unique representative sequences of each of the eight RSV group A genotypes were included in the phylogenetic analysis. GenBank accession numbers and the year and country of isolation of these sequences are given in Table 2. Phylogenetic relationships among German viruses were reviewed, and closely related strains were based on nucleotide similarity values of ≥96.6% abridged to one consensus sequence, which is now represented as a subtype in the tree. Evaluation of the robustness of the tree was performed by bootstrap analysis carried out with 1,000 replicates. The tree was plotted using TREEVIEW (33) and was manually edited in Corel Draw 12 program.
TABLE 2.
RSV group A sequences used in this study
| Strain | GenBank accession no. | Country of isolation | Reference |
|---|---|---|---|
| Ab4026B01 | AY146435 | South Africa | 25 |
| Ab5076Pt01 | AY146437 | South Africa | 25 |
| AL19376-1 | AF233900 | United States | 38 |
| AL19452-2 | AF233901 | United States | 38 |
| AL19471-5 | AF233902 | United States | 38 |
| AL19556-3 | AF233903 | United States | 38 |
| AUS/A2/61 | M11486 | United States | 59 |
| CH09 | AF065254 | United States | 39 |
| CH17 | AF065255 | United States | 39 |
| CH28 | AF065256 | United States | 39 |
| CH34 | AF065257 | United States | 39 |
| CN1973 | AF233904 | Canada | 38 |
| CN2395 | AF233905 | Canada | 38 |
| CN2708 | AF233906 | Canada | 38 |
| CN2851 | AF233907 | Canada | 38 |
| LLC235-267 | AY114149 | Singapore | 24 |
| LLC242-282 | AY114150 | Singapore | 24 |
| LLC62-111 | AY114151 | Singapore | 24 |
| MAD/1/89 | Z33412 | Spain | 15 |
| MAD/3/92 | Z33455 | Spain | 15 |
| MAD/5/92 | Z33417 | Spain | 15 |
| MO01 | AF233909 | United States | 38 |
| MO02 | AF233910 | United States | 38 |
| MO16 | AF233913 | United States | 38 |
| MO48 | AF233914 | United States | 38 |
| MO55 | AF233915 | United States | 38 |
| MON/1/87 | Z33421 | Uruguay | 15 |
| MON/1/90 | Z33494 | Uruguay | 15 |
| MON/1/94 | AF448498 | Uruguay | 12 |
| MON/3/88 | Z33425 | Uruguay | 15 |
| MON/4/90 | Z33426 | Uruguay | 15 |
| MON/5/90 | Z33427 | Uruguay | 15 |
| MON/8/92 | Z33430 | Uruguay | 15 |
| MON/9/91 | Z33431 | Uruguay | 15 |
| MON/9/92 | Z33432 | Uruguay | 15 |
| NG/009/02 | AB175815 | Japan | 44 |
| NY103 | AF233916 | United States | 38 |
| NY108 | AF233917 | United States | 38 |
| NY20 | AF233918 | United States | 38 |
| SA97D1289 | AF348803 | South Africa | 51 |
| SA98V603 | AF348807 | South Africa | 51 |
| SA99V1239 | AF348808 | South Africa | 51 |
| SA99V360 | AF348804 | South Africa | 51 |
| Sal/173/99 | AY472094 | Brazil | 28 |
| Sal/87/99 | AY472086 | Brazil | 28 |
| TX67951 | AF233919 | United States | 38 |
| TX68481 | AF233920 | United States | 38 |
| TX68532 | AF233921 | United States | 38 |
| TX69564 | AF233923 | United States | 38 |
| USA/Long/56 | M17212 | United States | 21 |
| WV/6973/82 | AF065407 | United States | 49 |
Comparisons between nucleotide or amino acid sequences were calculated using the DNADist or Protdist tool, respectively, of the Bioedit software, and the divergence values determined were described in terms of mean and standard deviation. To estimate the numbers of potentially O-glycosylated serine and threonine residues, NetOglyc software (version 3.1) (18) was used. Synonymous and nonsynonymous mutations were analyzed by the method of Nei and Gojobori (31). The program SNAP (Synonymous/Nonsynonymous Analysis Program) provided by the human immunodeficiency virus (HIV) sequence database website (http://www.hiv.lanl.gov/content/sequence/SNAP/SNAP.html) was used for analysis of synonymous mutations versus nonsynonymous mutations.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the nucleotide sequences obtained in the present study are FJ391237 to FJ391295, FJ391297 to FJ391327, and FJ391329 to FJ391459. Details concerning their subtype distribution are shown in Table S1 in the supplemental material.
RESULTS
Frequencies and distribution of RSV throughout consecutive seasons.
Over 6,100 respiratory samples were tested for RSV from October 1998 to September 2007, and 23% (1,426) of the samples were found to be positive. To determine the prevalence of RSV infections in certain age groups, the percentage of RSV-positive samples was calculated out of all specimens tested per age group. As expected, the majority of positive individuals were infants younger than 3 months (51%) and those aged between 3 to 6 months (47%). The prevalence of RSV decreased with increasing age, as observed for infants aged between 7 and 12 months (34%) and between 1 to 3 years (23%). Patients older than 3 years were characterized by a positive rate of 7%. Moreover, 28% positive samples were detected in individuals of unknown age. Since the prevalence was highest in children between 0 and 3 years old, it is assumed that the majority of patients of unknown age were younger than 3 years old.
RSV infections occurred in all seasons from late autumn to spring, and RSV peak activity varied within the seasons (Fig. 1). There were three out of eight seasons (2000 to 2001, 2002 to 2003, and 2006 to 2007) with a very intensive circulation of RSV in Germany (estimated using chi-square test, P < 0.0001). The onset, offset, peak, and seasonal pattern of RSV epidemics are documented in Table 3. A regular 2-year cyclic pattern was observed for two consecutive late and early seasons. An exception was the period between 2003 and 2005 where the pattern was changing from one season to the other. There was no correlation between onset and severity during the RSV epidemic season.
FIG. 1.
Number of total samples investigated and number of RSV-positive samples from 1999 to 2007. Results are shown for each month of eight epidemic seasons (from left to right, October [O], November [N], December [D], January [J], February [F], March [M], April [A], May [M], June [J], July [J], August [A], and September [S]).
TABLE 3.
Characteristics of RSV epidemic seasons from 1999 to 2007
| Epidemic season (yr) | Pattern | Onset of epidemic (wk) | Peak of epidemic (wk) | Offset of epidemic (wk) | Duration of epidemic (wk) |
|---|---|---|---|---|---|
| 1999-2000 | Late | 52 | 4 | 17 | 18 |
| 2000-2001 | Late | 49 | 8 | 17 | 21 |
| 2001-2002 | Early | 47 | 13 | 30 | 36 |
| 2002-2003 | Early | 42 | 51 | 14 | 25 |
| 2003-2004 | Late | 1 | 12 | 18 | 18 |
| 2004-2005 | Early | 47 | 7 | 14 | 21 |
| 2005-2006 | Late | 7 | 15 | 19 | 13 |
| 2006-2007 | Late | 50 | 8 | 17 | 20 |
Distribution of RSV group A in epidemic seasons.
All 1,426 RSV-positive samples were first detected by a generic real-time reverse transcription-PCR, and subsequently, the majority (Table 4) was differentiated by specific real-time reverse transcription-PCRs into RSV group A and B viruses. Table 4 shows the distribution of RSV group A viruses in nine epidemic seasons throughout the study period, and RSV group A viruses predominated in all epidemic seasons except for the season from 1998 to 1999 and that from 2002 to 2003. Indeed, there is no correlation between the distribution of RSV group A and the appearance of an early or late epidemic season. A representative number of more than 20% of group A viruses were randomly selected for sequencing and phylogenetic analyses irrespective of the severity of the epidemic season.
TABLE 4.
Distribution of RSV group A viruses in nine epidemic seasons
| Epidemic season (yr) | No. (%) of samples
|
|||
|---|---|---|---|---|
| RSV positive | With differentiated RSVa | With RSV group A viruses | With sequenced RSV group A viruses | |
| 1998-1999 | 24 | 20 (83) | 4 (20) | 2 (50) |
| 1999-2000 | 92 | 90 (98) | 84 (93) | 19 (23) |
| 2000-2001 | 286 | 262 (92) | 152 (58) | 32 (21) |
| 2001-2002 | 128 | 126 (98) | 84 (67) | 18 (21) |
| 2002-2003 | 271 | 244 (90) | 82 (34) | 23 (28) |
| 2003-2004 | 68 | 65 (96) | 49 (75) | 18 (37) |
| 2004-2005 | 143 | 136 (95) | 109 (80) | 37 (34) |
| 2005-2006 | 104 | 103 (99) | 66 (64) | 19 (29) |
| 2006-2007 | 310 | 224 (72) | 118 (53) | 53 (45) |
Number of samples differentiated by specific real-time reverse transcription-PCRs into RSV group A and B viruses.
Phylogenetic analysis of RSV group A sequences.
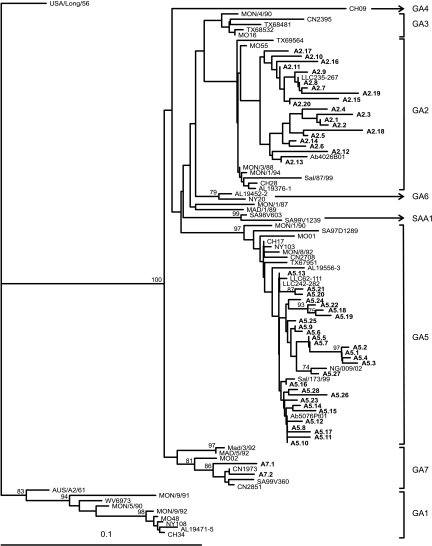
A total of 221 group A RSV samples were sequenced and analyzed in a neighbor-joining tree. Analysis revealed that all RSV group A viruses belonged to three genotypes: 74 (33%) viruses to genotype GA2, 138 (62%) to genotype GA5, and 12 (5%) to GA7. Groups of identical sequences were identified for each genotype. Phylogenetic relationships among strains were, therefore, reviewed, and closely related strains, based on nucleotide similarity values of ≥96.6%, were further divided into groups that we arbitrarily designated subtypes (according to Peret et al. [39]). Genotypes GA2 and GA5 showed a high variability with 20 and 28 subtypes, respectively. GA7 includes only two subtypes (Fig. 2).
FIG. 2.
Phylogenetic tree of the second variable region of RSV group A genotypes and subtypes. The prototype strain USA/Long/56 was used as the outgroup in the analysis. Reference sequences describing the eight different genotypes were retrieved from GenBank and included in the tree. The tree was constructed using the neighbor-joining algorithm with 1,000 replicates through PHYLIP. Subtypes are shown in boldface. The genotypes are indicated by the brackets to the right of the figure. Only bootstrap values greater than 70% are displayed at the branch nodes. The scale bar indicates the number of nucleotide substitutions per site.
Seasonal circulation pattern of RSV group A genotypes and subtypes.
Phylogenetic analyses revealed that three out of eight different genotypes circulated in Germany during the last seasons: GA2, GA5, and GA7. All seasons were characterized by the appearance of genotypes GA2 and GA5. GA7 was present only during the seasons from 1999 to 2000 and 2002 to 2003. Indeed, the extent of each genotype differed from one season to the other, but GA5 was the dominant genotype in the years from 1999 to 2007 (Fig. 3).
FIG. 3.
Distribution of RSV group A genotypes over eight consecutive seasons. The number of samples is shown on the abscissa.
The subtypes identified were analyzed for determination of the temporal circulation pattern of RSV throughout nine consecutive seasons (Table 5). There were several subtypes (A2.2, A2.4 to A2.7, A2.10, A5.2, A5.11, A5.12, A5.14, A5.15, A5.18, A5.19, A5.21, and A7.1) that showed identical sequences only within one season. In contrast, other subtypes contained identical sequences in either two (A2.1, A2.3, A2.8, A2.9, A2.12, A2.14, A2.15, A2.17, and A5.3 to A5.5), three (A5.10 and A5.25), or four (A5.13) consecutive seasons. Sequences of subtypes A5.7 and A5.8 were found in seven or eight consecutive seasons. There were also some subtypes representing single sequences not clustering with other sequences (A2.18 toA2.20, A5.27, and A7.2). Two subtypes are characterized by a biennial (A2.13) or triennial (A5.16) rhythm. In total, there were two genotypes, GA2 and GA5, circulating over nine consecutive seasons. The appearance, disappearance, or rather recurrence of genotype-specific viruses was, however, a random event.
TABLE 5.
Subtype distribution of RSV group A genotypes from 1998 to 2007
| Subtypea | No. of isolates (n)b | % Similarity in subtype | Epidemic season (n)c |
|---|---|---|---|
| A2.1 | 8 (7) | 99.62 | 2005-2006 (1) |
| 2006-2007 (7) | |||
| A2.2 | 8 (7) | 99.62 | 2006-2007 (8) |
| A2.3 | 3 (0) | 97.4 | 2005-2006 (1) |
| 2006-2007 (2) | |||
| A2.4 | 2 (2) | 100 | 2004-2005 (2) |
| A2.5 | 3 (3) | 100 | 2001-2002 (3) |
| A2.6 | 2 (2) | 100 | 1999-2000 (2) |
| A2.7 | 6 (5) | 99.62 | 2001-2002 (6) |
| A2.8 | 4 (2) | 99.25 | 1999-2000 (3) |
| 2000-2001 (1) | |||
| A2.9 | 2 (0) | 98.51 | 2003-2004 (1) |
| 2004-2005 (1) | |||
| A2.10 | 2 (2) | 100 | 2000-2001 (2) |
| A2.11 | 4 (2) | 98.51 | 2000-2001 (4) |
| A2.12 | 10 (3, 2) | 97.4 | 2005-2006 (2) |
| 2006-2007 (8) | |||
| A2.13 | 3 (0) | 97.77 | 1998-1999 (1) |
| 2000-2001 (1) | |||
| 2002-2003 (1) | |||
| A2.14 | 3 (2) | 97.77 | 1999-2000 (2) |
| 2000-2001 (1) | |||
| A2.15 | 4 (3) | 99.25 | 2003-2004 (3) |
| 2004-2005 (1) | |||
| A2.16 | 3 (0) | 98.51 | 2000-2001 (1) |
| 2003-2004 (1) | |||
| 2006-2007 (1) | |||
| A2.17 | 4 (2) | 98.14 | 2005-2006 (1) |
| 2006-2007 (3) | |||
| A2.18 | 1 | 2006-2007 (1) | |
| A2.19 | 1 | 2000-2001 (1) | |
| A2.20 | 1 | 2000-2001 (1) | |
| A5.1 | 10 (9) | 99.25 | 1999-2000 (1) |
| 2004-2005 (7) | |||
| 2006-2007 (2) | |||
| A5.2 | 3 (3) | 100 | 2004-2005 (3) |
| A5.3 | 7 (5) | 98.14 | 2005-2006 (1) |
| 2006-2007 (6) | |||
| A5.4 | 4 (3) | 98.8 | 2005-2006 (1) |
| 2006-2007 (3) | |||
| A5.5 | 4 (2, 2) | 99.25 | 2004-2005 (2) |
| 2005-2006 (2) | |||
| A5.6 | 4 (2) | 98.8 | 2000-2001 (4) |
| A5.7 | 15 (2, 9) | 96.6 | 2000-2001 (1) |
| 2001-2002 (3) | |||
| 2002-2003 (3) | |||
| 2003-2004 (2) | |||
| 2004-2005 (3) | |||
| 2005-2006 (3) | |||
| A5.8 | 20 (17) | 96.6 | 1999-2000 (1) |
| 2000-2001 (5) | |||
| 2001-2002 (2) | |||
| 2002-2003 (3) | |||
| 2003-2004 (3) | |||
| 2004-2005 (4) | |||
| 2005-2006 (1) | |||
| 2006-2007 (1) | |||
| A5.9 | 3 (0) | 97.77 | 1998-1999 (1) |
| 1999-2000 (1) | |||
| 2005-2006 (1) | |||
| A5.10 | 3 (0) | 98.8 | 2000-2001 (1) |
| 2001-2002 (1) | |||
| 2002-2003 (1) | |||
| A5.11 | 2 (2) | 100 | 2004-2005 (2) |
| A5.12 | 2 (2) | 100 | 2003-2004 (2) |
| A5.13 | 11 (7) | 97 | 1999-2000 (6) |
| 2000-2001 (2) | |||
| 2001-2002 (2) | |||
| 2002-2003 (1) | |||
| A5.14 | 3 (3) | 100 | 2004-2005 (3) |
| A5.15 | 2 (2) | 100 | 2006-2007 (2) |
| A5.16 | 4 (2) | 98.51 | 2000-2001 (1) |
| 2003-2004 (2) | |||
| 2006-2007 (1) | |||
| A5.17 | 7 (4) | 97 | 2004-2005 (6) |
| 2006-2007 (1) | |||
| A5.18 | 3 (3) | 100 | 2004-2005 (3) |
| A5.19 | 3 (3) | 100 | 2006-2007 (3) |
| A5.20 | 6 (4) | 99.25 | 2003-2004 (1) |
| 2005-2006 (4) | |||
| 2006-2007 (1) | |||
| A5.21 | 3 (3) | 100 | 2003-2004 (3) |
| A5.22 | 3 (0) | 98.14 | 2000-2001 (3) |
| A5.23 | 2 (0) | 97.77 | 2002-2003 (2) |
| A5.24 | 2 (0) | 98.8 | 2000-2001 (1) |
| 2005-2006 (1) | |||
| A5.25 | 5 (0) | 97.77 | 1999-2000 (1) |
| 2000-2001 (3) | |||
| 2001-2002 (1) | |||
| A5.26 | 2 (0) | 98.8 | 2006-2007 (2) |
| A5.27 | 1 | 1999-2000 (1) | |
| A5.28 | 2 (0) | 97.4 | 2002-2003 (1) |
| 2006-2007 (1) | |||
| A7.1 | 11 (9) | 99.25 | 2002-2003 (11) |
| A7.2 | 1 | 1999-2000 (1) |
Subtypes were defined as ≥96.6% nucleotide similarity and denoted with a three-character code.
n is the number of identical sequences (subtypes A2.12, A5.5 and A5.7 showed two distinct groups of identical sequences).
n is the number of isolates found in that season.
Intragenotype divergence of German RSV group A sequences.
The nucleotide sequence and the deduced amino acid sequence of the second variable region of all German viruses were compared within each genotype and the mean percentage of nucleotide and amino acid divergences are shown in Table 6. Nucleotide divergence ranged between 0.5% and 3.7%, whereas the most similar viruses belonged to genotype GA7. In contrast, the amino acid divergence ranged from 1.5% to 12.5%. All sequences revealed a higher degree of amino acid divergence compared to the degree of nucleotide divergence.
TABLE 6.
Nucleotide and amino acid divergence of the carboxy-terminal variable region of the G protein of German RSV group A viruses
| Genotype | No. of strains | Nucleotide divergence (%) (mean ± SEM) | Amino acid divergence (%) (mean ± SEM) |
|---|---|---|---|
| GA2 | 74 | 3.67 ± 0.0067 | 12.45 ± 0.018 |
| GA5 | 138 | 2.96 ± 0.0043 | 7.45 ± 0.0098 |
| GA7 | 12 | 0.50 ± 0.0014 | 1.52 ± 0.0011 |
Synonymous-to-nonsynonymous mutations.
To study the evolutionary divergence of the RSV sequences, the number of synonymous (silent) and nonsynonymous (amino acid altering) nucleotide substitutions was estimated. On average, the synonymous mutation/nonsynonymous mutation (ds/dn) ratio for the three RSV group A genotypes were 1.18 for genotype GA2, 4.34 for genotype GA5, and 1.92 for genotype GA7. Ratios of ≫one demonstrate a high abundance of synonymous mutations. This implicates a neutral selection pressure on the variable region for genotype GA5 and GA7. A positive selection pressure can be suggested for genotype GA2, for which more nonsynonymous mutations have been observed.
Amino acid analysis.
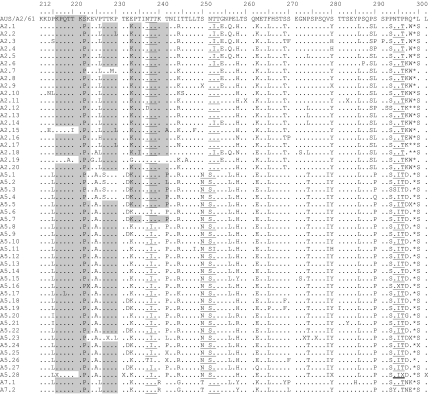
A consensus sequence was generated including all nucleotide sequences of a subtype and was translated into one amino acid sequence. Predicted amino acid sequences of all subtypes were then compared to prototype strain A2 (Fig. 4 and Table 7). Nearly all partial G-protein gene sequences were predicted to encode G proteins of 298 amino acids in length. Only three GA2 subtypes (A2.12, A2.17, and A2.18) and one of GA5 (A5.5) encoded a G protein with 297 amino acids. The difference in the length of the deduced amino acid sequences of subtypes A2.17 and A2.18 as well as of A5.5 resulted from a mutation at the second nucleotide position and a mutation at the first nucleotide position in the first stop codon in the carboxyl terminus of the G-protein gene.
FIG. 4.
Deduced amino acid alignment of the second variable region of the G-protein gene from RSV group A subtypes. Consensus sequences are generated from each subtype, and the alignment is shown relative to the sequence of prototype strain A2 (GenBank accession number M11486). The amino acids shown correspond to strain A2 G-protein positions 212 to 298. Identical residues are indicated by dots, and stop codons by asterisks. The presence of an X at an amino acid position indicates that there was not a consensus for the subtype at that position and both amino acids were presented in equivalent proportion among the studied strains. The corresponding amino acids and positions are further described in Table 7. Potential N-glycosylation sites (NXT, where X is not proline) are underlined. Potential sites for extensive O-glycosylation KPXnTTKXn motifs (where X is any amino acid) are indicated by gray shading. Half of the sequences of subtype A5.28 contain either a potential O-glycosylation site (double underline) and/or an N-glycosylation site (thick underline).
TABLE 7.
Amino acid positions of RSV group A subtypes when two amino acids were presented in equivalent proportion in the deduced amino acid sequence of RSV group A subtypes (Fig. 4)
| Subtype | Amino acids at position
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 216 | 223 | 228 | 231 | 241 | 250 | 260 | 273 | 276 | 280 | 283 | 295 | 296 | 298 | |
| A2.9 | S/T | P/L | ||||||||||||
| A2.11 | S/I | S/T | ||||||||||||
| A5.5 | Q/*a | |||||||||||||
| A5.16 | E/K | I/T | ||||||||||||
| A5.23 | T/A | N/T | P/S | Q/L | ||||||||||
| A5.25 | P/L | |||||||||||||
| A5.26 | P/L | |||||||||||||
| A5.28 | K/Q | T/I | Y/H | I/T | ||||||||||
An amino acid coding for a stop codon is indicated by an asterisk.
Amino acid substitutions that were specific for each of the three analyzed genotypes were identified. Each genotype could be distinguished from the others based on particular amino acid substitutions at different positions along the deduced amino acid sequence. For instance, genotype GA2 could be distinguished by Thr269and Ser289, and genotype GA5 could be distinguished by Ala225, Asn250, Ser251, Thr274, and Ile279. Amino acid substitutions Ile295 and Asp297 were also specific for genotype GA5 with the exception of subtypes A5.14, A5.16, and A5.20. Genotype GA7 could be differentiated by certain amino acid substitutions as well. These were Arg240, Gly244, and Thr250 (Fig. 4).
Glycosylation sites.
The G protein is heavily glycosylated with both N-linked and O-linked sugars (60). We identified four putative N-glycosylation sites (NXT, where X is not Pro) among group A subtypes, whereas the positions of the first and fourth sites were nearly conserved among all subtypes (Fig. 4). Only subtypes A2.12 and A5.26 as well as subtypes A5.3 and A5.28 lacked the first glycosylation site or rather the fourth glycosylation site. The second site was genotype specific and was present only among GA5 subtypes. In contrast, the third site was identified among genotype GA2 and GA7 viruses.
Most of the carbohydrates of the G protein are O glycosylated. Thereby serine and threonine residues are potential attachment sites for O-linked sugars, and the G protein has an unusually high content of both (9). The analysis of 89 amino acids by the program NetOglyc predicted 22 to 34 serine and threonine residues to be potentially O glycosylated with score predicators (G scores) of 0.5 and 0.8. Interestingly, there were two O-glycosylation sites that were either unique to genotype GA2 and GA7 (T241) or GA5 (T274). In addition to serine and threonine residues, one to three repeats of the motif KPXnTTKXn (Fig. 4) were present among the subtypes and may be associated with extensive O glycosylation of the G protein (6).
DISCUSSION
RSV is the most common viral pathogen causing lower respiratory tract infections among infants and young children (48). Infection rates for RSV of 68.8/100 children during the first year of life and 82.6/100 during the second year have been observed (16). The incidence of RSV-associated bronchiolitis and pneumonia was highest in 2- to 3-month-old infants, whereas with increasing age, the incidence decreased (36, 47). In this study, RSV was detected in over 1,400 respiratory samples over a period of nine consecutive seasons. Most infected individuals were babies 0 to 3 months old accounting for 51% of all samples analyzed and babies between 3 and 6 months accounting for 47% of all samples analyzed in these age groups. With increasing age, the RSV prevalence decreased to levels of 23% in children between 1 and 3 years old. Long-term studies from Germany analyzing the impact of RSV in hospitalized children confirm our findings (3, 57). These data revealed that 22.8% to 42.8% were positive for RSV in the age group between 0 and ≤12 months (54, 56). In children older than 1 year, the prevalence of RSV was decreasing (54). The frequency of RSV particularly in small children might be caused by their immune status. Although high levels of maternal antibodies are protective against the disease, the magnitude of such immune response in the early years is low. Thus, the majority of children become infected during the first months of life. Both serum and secretory antibodies are produced in response to infection, even by very young infants; in this group, however, titers achieved are usually low (7, 26, 55). Significant boosting, particularly for the immunoglobulin G and immunoglobulin A responses, will develop soonest when children are reinfected for the second or third time (43).
RSV was isolated from autumn to spring in all nine seasons investigated herein. Thus, RSV is circulating mainly in the winter months as determined by other studies of the temperate zone (23). Monitoring of RSV activity in Germany revealed a 2-year cyclic pattern for two consecutive late and early seasons. An exception was the period between 2003 and 2005 when the seasonal pattern was changing from one season to the other. Other European countries, such as Switzerland, Sweden, and Finland, observed a yearly varying pattern of early and late seasons, whereas early seasons are associated with high incidences and vice versa (10, 41, 53). Previous reports from Germany including regions around Kiel, Stuttgart, and Freiburg also described a biennial rhythm of severe early and weak late seasons (3, 50, 58). An explanation for the absence of the biennial pattern in our study might be due to missed cases during summer months which would influence the onset of a season. Apart from that, we observed no frequency in severe or weak RSV seasons as described previously. So far, this finding cannot be explained since, first, in every season a huge number of cases were both detected and examined and, second, the hospitals and physicians included in the study remained nearly unchanged during the nine seasons.
Consistent shifts in RSV group dominance have been reported worldwide in which RSV group A viruses are more frequently detected. In Germany, RSV group A was dominant in seven out of nine epidemic seasons predominating over a period of three (1999 to 2000 to 2001 to 2002) and four (2003 to 2004 to 2006 to 2007) seasons. During the same periods, mainly RSV group A dominated in several other countries (Table 8). A regular three-year cyclic pattern of group dominance was observed in Belgium and Argentina where RSV group A viruses predominated for two consecutive seasons (52, 63). A study in India reported on RSV group A dominance for three consecutive epidemic seasons (37). RSV group B predominated for a single season in the countries investigated and at the same or similar time as observed in Germany. Thus, it can be concluded that RSV group A predominated within similar seasons, implicating that most RSV infections were caused by RSV group A worldwide at the same time.
TABLE 8.
Prevalence of the predominating RSV group in different geographic areas
| Epidemic season or yr | Predominant RSV group (prevalence [%]) in:
|
||||||
|---|---|---|---|---|---|---|---|
| European countries
|
Other countries
|
||||||
| Germany | Swedena | Belgiumb | Argentinac | Japand | Kenyae | Indiaf | |
| 1998-1999 | B (80) | B (76) | |||||
| 1999 | B (63) | ||||||
| 1999-2000 | A (93) | A (73) | |||||
| 2000 | A (100) | ||||||
| 2000-2001 | A (58) | A (82) | |||||
| 2001 | A (90) | A (100) | |||||
| 2001-2002 | A (67) | B (73) | A (94) | B (100) | |||
| 2002 | B (71) | A (98) | |||||
| 2002-2003 | B (66) | B (65) | A (72) | B (83) | A (100) | ||
| 2003 | A (74) | B (61) | |||||
| 2003-2004 | A (75) | A (61) | A (84) | A (91) | |||
| 2004 | A (82) | ||||||
| 2004-2005 | A (80) | B (76) | A (53) | ||||
| 2005 | |||||||
| 2005-2006 | A (64) | A (84) | |||||
| 2006 | |||||||
| 2006-2007 | A (53) | ||||||
RSV group A is classified into several genotypes (38, 39, 51). Three out of eight genotypes circulated in Germany between 1998 and 2007, and more than one genotype was detected within each season. Most of the German viruses belonged to the genotypes GA2 and GA5 demonstrating a dominance of GA5 in two and then four consecutive epidemic seasons. The finding of GA2 and GA5 as the most common genotypes of RSV group A is in agreement with the results of long-term studies from Europe and other geographic areas. Between 1964 and 1996, genotypes GA1, GA3, and GA4 circulated sporadically in addition to the genotypes GA2 and GA5 in Sweden, Belgium, Uruguay, Argentina, and the United States (12, 14, 39, 40, 62). In these and other countries, mainly the genotypes GA2 and GA5 and in a few cases genotype GA7 have been circulating since 1998 (12, 32, 37, 44). The continuous and predominant circulation of genotypes GA2 and GA5 demonstrate that these genotypes are stable and have become epidemic in many countries.
Viruses isolated in Germany comprise sets of similar sequences within each of the identified genotypes. Some of them reemerged in subsequent seasons, and viruses of subtypes A5.7 and A5.8 were circulating over long periods of six and eight seasons, respectively. It can be surmised that viruses circulating for years caused yearly new infections. Otherwise, there are RSV group A viruses appearing only once during the study period in Germany. One-third of those viruses emerged during the seasons 2000 to 2001, 2002 to 2003, as well as 2006 to 2007 and might be responsible for severe outbreaks. Presumably, strains came from outside Germany and were transmitted more efficiently due to a lack of immunity in the German population. Indeed, viruses isolated in Germany are closely related to viruses from distinct places (e.g., A2.8, A2.9, and LLC235-267 from Singapore; A2.13 and Ab4026B01 from South Africa; A5.27 and NG/009/02 from Japan; A5.16 and Sal/173/99 from Brazil; and A7.2 and CN1973 from Canada), suggesting that RSV group A strains are distributed worldwide as described previously (5, 15).
A reason for the long-lasting circulation of genotypes GA2 and GA5 in Germany might be due to mutations in their amino acid sequences. The nucleotide divergence determined for genotype GA2 was 3.7%, and for GA5, it was 3% (both values representing low levels). Since amino acid divergence of the genotypes GA2 (12.5%) and GA5 (7.5%) was greater than the nucleotide divergence, mutations in nucleotides resulted in amino acid changes. For genotype GA2, more nonsynonymous mutations were observed, indicating a positive selection pressure for those viruses, which may be reasonable for the long-lasting circulation. Contrary to this, selection pressure for GA5 is neutral due to the existence of more synonymous mutations. The significance of this observation is not known so far. In Stockholm, Sweden, the genotype GA5 also has dominated over a couple of epidemic seasons, and it is speculated that a variation in the A-terminal part of the protein may result in the prolonged dominance of this genotype, whereas in the C-terminal part, little divergence was detected (40). This might be also a reason for the circulation pattern of the German GA5 viruses. At present, there is not sufficient information about the A-terminal part of the protein to support the data from Stockholm, Sweden, or rather the hypothesis.
The amino acid sequences of German subtypes were predicted to encode G proteins of 297 or 298 amino acids in length. This is in agreement with data from Uruguay and Argentina where both these changes in protein length were observed and were correlated with genotypes. Viruses with a G-protein length of 297 amino acids grouped into GA1 and GA2. Viruses representing genotypes GA3 and GA5 had a G-protein length of 298 amino acids (12). We found that subtypes A2.12, A2.17, and A2.18 of genotype GA2 and subtype A5.5 of genotype GA5 code for the 297-amino-acid variant. All other subtypes possessing 298-amino-acid G proteins were found to belong to genotypes GA2, GA5, and GA7. German group A RSV viruses differed not only with regard to the length of amino acids but also revealed genotype-specific amino acid substitutions as described previously (14, 39, 40, 51, 52). Interestingly, most substitutions found worldwide were different between geographical locations, suggesting that RSV strains develop independently. Amino acid substitutions at positions 269, 274, and 295 observed in German subtypes were also found in Argentinean and South African viruses and strains from Rochester, New York (14, 39, 51) as a consequence of global circulation of strains.
N and O glycosylation of the G protein is suggested to help the virus evade the host immune response (6). Variation in the number and distribution patterns of glycosylation sites can influence the expression of certain epitopes, and therefore, recognition of RSV by carbohydrate-specific antibodies can be inhibited (27, 34, 35). The predicted O-glycosylation sites in the second hypervariable region of the G-protein gene analyzed in this study included 22 to 34 residues. For the same region, 8 to 10 potential O-linked glycosylation sites were identified in group A RSV strains from India (37). On the basis of RSV strains from Belgium, serine and threonine residues were predicted to be O-glycosylated with a high potential at the positive selected sites at positions 290 and 225 (62). German viruses did not possess such potential glycosylation sites, and a serine or threonine residue was not located at these sites. However, potential N- and O-glycosylation motifs among German viruses were observed, which have been described previously (37, 42). Two to four putative N-glycosylation sites have been estimated for the German viruses of genotype GA2, GA5, and GA7. The number of potential O-linked acceptor sites varies considerably among viruses within a genotype. Consequently, glycosylation of the G-protein gene is highly variable in RSV strains. The modulation of the number and location of the glycosylation sites may cause the long-lasting circulation of certain genotypes, as the viruses circumvent neutralization by preexisting antibodies. In general, further analyses are necessary to understand the functional importance of glycosylation sites. This information might help to predict possible changes in virulence and immune responses to RSV strains in future vaccination programs.
In summary, RSV was mainly detected among small children from autumn to spring during nine epidemic seasons in Germany. Molecular characterization of RSV revealed the predominance of RSV group A viruses for three and then four consecutive seasons. Most German viruses belonged to the genotypes GA2 and GA5, which are the predominant genotypes circulating worldwide. Positive selection pressure or modulation of glycosylation sites among genotypes are most likely responsible for the prolonged circulation of these genotypes in Germany. At present, detailed data on the circulation of RSV group A genotypes over a couple of seasons are available for only some countries. This study thus contributes to a better knowledge on the molecular epidemiology of RSV group A in Europe and provides data for a comparative analysis with group A strains circulating in other regions of the world. Comprehensive information on the circulation profile of RSV is important for the selection of appropriate vaccine strains and thus may contribute to vaccine development.
Supplementary Material
Acknowledgments
We thank all medical practices and hospitals taking part in our study. We are obliged to Susi Hafemann, Ingrid Zadow, Heidi Lehmann, Madelaine Rüschpler, and Ute Hopf-Guevara for their excellent technical assistance.
Footnotes
Published ahead of print on 22 April 2009.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Anderson, L. J., J. C. Hierholzer, C. Tsou, R. M. Hendry, B. F. Fernie, Y. Stone, and K. McIntosh. 1985. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J. Infect. Dis. 151626-633. [DOI] [PubMed] [Google Scholar]
- 2.Arbiza, J., A. Delfraro, and S. Frabasile. 2005. Molecular epidemiology of human respiratory syncytial virus in Uruguay: 1985-2001—a review. Mem. Inst. Oswaldo Cruz 100221-230. [DOI] [PubMed] [Google Scholar]
- 3.Berner, R., F. Schwoerer, R. F. Schumacher, M. Meder, and J. Forster. 2001. Community and nosocomially acquired respiratory syncytial virus infection in a German paediatric hospital from 1988 to 1999. Eur. J. Pediatr. 160541-547. [DOI] [PubMed] [Google Scholar]
- 4.Cane, P. A. 2001. Molecular epidemiology of respiratory syncytial virus. Rev. Med. Virol. 11103-116. [DOI] [PubMed] [Google Scholar]
- 5.Cane, P. A., and C. R. Pringle. 1995. Evolution of subgroup A respiratory syncytial virus: evidence for progressive accumulation of amino acid changes in the attachment protein. J. Virol. 692918-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cane, P. A., and C. R. Pringle. 1991. Respiratory syncytial virus heterogeneity during an epidemic: analysis by limited nucleotide sequencing (SH gene) and restriction mapping (N gene). J. Gen. Virol. 72349-357. [DOI] [PubMed] [Google Scholar]
- 7.Chanock, R. M., H. W. Kim, A. J. Vargosko, A. Deleva, K. M. Johnson, C. Cumming, and R. H. Parrott. 1961. Respiratory syncytial virus. I. Virus recovery and other observations during 1960 outbreak of bronchiolitis, pneumonia, and minor respiratory diseases in children. JAMA 176647-653. [PubMed] [Google Scholar]
- 8.Collins, C. L., and A. J. Pollard. 2002. Respiratory syncytial virus infections in children and adults. J. Infect. 4510-17. [DOI] [PubMed] [Google Scholar]
- 9.Collins, P. L., R. M. Chanock, and B. R. Murphy. 2001. Respiratory syncytial virus, p. 1443-1485. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 10.Duppenthaler, A., M. Gorgievski-Hrisoho, U. Frey, and C. Aebi. 2003. Two-year periodicity of respiratory syncytial virus epidemics in Switzerland. Infection 3175-80. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 2004. PHYLIP (Phylogeny Inference Package) version 3.6.4. Department of Genome Sciences, University of Washington, Seattle.
- 12.Frabasile, S., A. Delfraro, L. Facal, C. Videla, M. Galiano, M. J. de Sierra, D. Ruchansky, N. Vitureira, M. Berois, G. Carballal, J. Russi, and J. Arbiza. 2003. Antigenic and genetic variability of human respiratory syncytial viruses (group A) isolated in Uruguay and Argentina: 1993-2001. J. Med. Virol. 71305-312. [DOI] [PubMed] [Google Scholar]
- 13.Freymuth, F., J. Petitjean, P. Pothier, J. Brouard, and E. Norrby. 1991. Prevalence of respiratory syncytial virus subgroups A and B in France from 1982 to 1990. J. Clin. Microbiol. 29653-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galiano, M. C., C. Palomo, C. M. Videla, J. Arbiza, J. A. Melero, and G. Carballal. 2005. Genetic and antigenic variability of human respiratory syncytial virus (groups A and B) isolated over seven consecutive seasons in Argentina (1995 to 2001). J. Clin. Microbiol. 432266-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García, O., M. Martín, J. Dopazo, J. Arbiza, S. Frabasile, J. Russi, M. Hortal, P. Perez-Breña, I. Martínez, B. Garcia-Barreno, and J. A. Melero. 1994. Evolutionary pattern of human respiratory syncytial virus (subgroup A): cocirculating lineages and correlation of genetic and antigenic changes in the G glycoprotein. J. Virol. 685448-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glezen, W. P., L. H. Taber, A. L. Frank, and J. A. Kasel. 1986. Risk of primary infection and reinfection with respiratory syncytial virus. Am. J. Dis. Child. 140543-546. [DOI] [PubMed] [Google Scholar]
- 17.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Window 95/98/NT. Nucleic Acids Symp. Ser. 4195-98. [Google Scholar]
- 18.Hansen, J. E., O. Lund, J. Engelbrecht, H. Bohr, J. O. Nielsen, and J. E. Hansen. 1995. Prediction of O-glycosylation of mammalian proteins: specificity patterns of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase. Biochem. J. 308801-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson, F. W., A. M. Collier, W. A. Clyde, Jr., and F. W. Denny. 1979. Respiratory syncytial virus infections, reinfections and immunity. A prospective, longitudinal study in young children. N. Engl. J. Med. 300530-534. [DOI] [PubMed] [Google Scholar]
- 20.Hendry, R. M., A. L. Talis, E. Godfrey, L. J. Anderson, B. F. Fernie, and K. McIntosh. 1986. Concurrent circulation of antigenically distinct strains of respiratory syncytial virus during community outbreaks. J. Infect. Dis. 153291-297. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, P. R., M. K. Spriggs, R. A. Olmsted, and P. L. Collins. 1987. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc. Natl. Acad. Sci. USA 845625-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, H. W., J. G. Canchola, C. D. Brandt, G. Pyles, R. M. Chanock, K. Jensen, and R. H. Parrott. 1969. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am. J. Epidemiol. 89422-434. [DOI] [PubMed] [Google Scholar]
- 23.Knipe, D. M., P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.). 2001. Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 24.Lim, C. S., G. Kumarasinghe, and V. T. Chow. 2003. Sequence and phylogenetic analysis of SH, G, and F genes and proteins of human respiratory syncytial virus isolates from Singapore. Acta Virol. 4797-104. [PubMed] [Google Scholar]
- 25.Madhi, S. A., M. Venter, R. Alexandra, H. Lewis, Y. Kara, W. F. Karshagen, M. Greef, and C. Lassen. 2003. Respiratory syncytial virus associated illness in high-risk children and national characterisation of the circulating virus genotype in South Africa. J. Clin. Virol. 27180-189. [DOI] [PubMed] [Google Scholar]
- 26.McIntosh, K., H. B. Masters, I. Orr, R. K. Chao, and R. M. Barkin. 1978. The immunologic response to infection with respiratory syncytial virus in infants. J. Infect. Dis. 13824-32. [DOI] [PubMed] [Google Scholar]
- 27.Melero, J. A., B. Garcia-Barreno, I. Martinez, C. R. Pringle, and P. A. Cane. 1997. Antigenic structure, evolution and immunobiology of human respiratory syncytial virus attachment (G) protein. J. Gen. Virol. 782411-2418. [DOI] [PubMed] [Google Scholar]
- 28.Moura, F. E., A. Blanc, S. Frabasile, A. Delfraro, M. J. de Sierra, L. Tome, E. A. Ramos, M. M. Siqueira, and J. Arbiza. 2004. Genetic diversity of respiratory syncytial virus isolated during an epidemic period from children of northeastern Brazil. J. Med. Virol. 74156-160. [DOI] [PubMed] [Google Scholar]
- 29.Mufson, M. A., H. D. Levine, R. E. Wasil, H. E. Mocega-Gonzalez, and H. E. Krause. 1973. Epidemiology of respiratory syncytial virus infection among infants and children in Chicago. Am. J. Epidemiol. 9888-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mufson, M. A., C. Orvell, B. Rafnar, and E. Norrby. 1985. Two distinct subtypes of human respiratory syncytial virus. J. Gen. Virol. 662111-2124. [DOI] [PubMed] [Google Scholar]
- 31.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3418-426. [DOI] [PubMed] [Google Scholar]
- 32.Ostlund, M. R., A. T. Lindell, S. Stenler, H. M. Riedel, B. Z. Wirgart, and L. Grillner. 2008. Molecular epidemiology and genetic variability of respiratory syncytial virus (RSV) in Stockholm, 2002-2003. J. Med. Virol. 80159-167. [DOI] [PubMed] [Google Scholar]
- 33.Page, R. D. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12357-358. [DOI] [PubMed] [Google Scholar]
- 34.Palomo, C., P. A. Cane, and J. A. Melero. 2000. Evaluation of the antibody specificities of human convalescent-phase sera against the attachment (G) protein of human respiratory syncytial virus: influence of strain variation and carbohydrate side chains. J. Med. Virol. 60468-474. [PubMed] [Google Scholar]
- 35.Palomo, C., B. Garcia-Barreno, C. Penas, and J. A. Melero. 1991. The G protein of human respiratory syncytial virus: significance of carbohydrate side-chains and the C-terminal end to its antigenicity. J. Gen. Virol. 72669-675. [DOI] [PubMed] [Google Scholar]
- 36.Parrott, R. H., H. W. Kim, J. O. Arrobio, D. S. Hodes, B. R. Murphy, C. D. Brandt, E. Camargo, and R. M. Chanock. 1973. Epidemiology of respiratory syncytial virus infection in Washington, D.C. II. Infection and disease with respect to age, immunologic status, race and sex. Am. J. Epidemiol. 98289-300. [DOI] [PubMed] [Google Scholar]
- 37.Parveen, S., W. M. Sullender, K. Fowler, E. J. Lefkowitz, S. K. Kapoor, and S. Broor. 2006. Genetic variability in the G protein gene of group A and B respiratory syncytial viruses from India. J. Clin. Microbiol. 443055-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peret, T. C., C. B. Hall, G. W. Hammond, P. A. Piedra, G. A. Storch, W. M. Sullender, C. Tsou, and L. J. Anderson. 2000. Circulation patterns of group A and B human respiratory syncytial virus genotypes in 5 communities in North America. J. Infect. Dis. 1811891-1896. [DOI] [PubMed] [Google Scholar]
- 39.Peret, T. C., C. B. Hall, K. C. Schnabel, J. A. Golub, and L. J. Anderson. 1998. Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J. Gen. Virol. 792221-2229. [DOI] [PubMed] [Google Scholar]
- 40.Rafiefard, F., B. Johansson, T. Tecle, and C. Orvell. 2004. Molecular epidemiology of respiratory syncytial virus (RSV) of group A in Stockholm, Sweden, between 1965 and 2003. Virus Res. 105137-145. [DOI] [PubMed] [Google Scholar]
- 41.Reyes, M., M. Eriksson, R. Bennet, K. O. Hedlund, and A. Ehrnst. 1997. Regular pattern of respiratory syncytial virus and rotavirus infections and relation to weather in Stockholm, 1984-1993. Clin. Microbiol. Infect. 3640-646. [DOI] [PubMed] [Google Scholar]
- 42.Roca, A., M. P. Loscertales, L. Quinto, P. Perez-Brena, N. Vaz, P. L. Alonso, and J. C. Saiz. 2001. Genetic variability among group A and B respiratory syncytial viruses in Mozambique: identification of a new cluster of group B isolates. J. Gen. Virol. 82103-111. [DOI] [PubMed] [Google Scholar]
- 43.Ruuskanen, O., and P. L. Ogra. 1993. Respiratory syncytial virus. Curr. Probl. Pediatr. 2350-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato, M., R. Saito, T. Sakai, Y. Sano, M. Nishikawa, A. Sasaki, Y. Shobugawa, F. Gejyo, and H. Suzuki. 2005. Molecular epidemiology of respiratory syncytial virus infections among children with acute respiratory symptoms in a community over three seasons. J. Clin. Microbiol. 4336-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scott, P. D., R. Ochola, M. Ngama, E. A. Okiro, D. J. Nokes, G. F. Medley, and P. A. Cane. 2004. Molecular epidemiology of respiratory syncytial virus in Kilifi district, Kenya. J. Med. Virol. 74344-354. [DOI] [PubMed] [Google Scholar]
- 46.Selwyn, B. J., on behalf of the Coordinated Data Group of BOSTID Researchers. 1990. The epidemiology of acute respiratory tract infection in young children: comparison of findings from several developing countries. Rev. Infect. Dis. 12(Suppl. 8)S870-S888. [DOI] [PubMed] [Google Scholar]
- 47.Sims, D. G., M. A. Downham, J. McQuillin, and P. S. Gardner. 1976. Respiratory syncytial virus infection in north-east England. Br. Med. J. 2(6044)1095-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sullender, W. M. 2000. Respiratory syncytial virus genetic and antigenic diversity. Clin. Microbiol. Rev. 131-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sullender, W. M., M. A. Mufson, G. A. Prince, L. J. Anderson, and G. W. Wertz. 1998. Antigenic and genetic diversity among the attachment proteins of group A respiratory syncytial viruses that have caused repeat infections in children. J. Infect. Dis. 178925-932. [DOI] [PubMed] [Google Scholar]
- 50.Terletskaia-Ladwig, E., G. Enders, G. Schalasta, and M. Enders. 2005. Defining the timing of respiratory syncytial virus (RSV) outbreaks: an epidemiological study. BMC Infect. Dis. 520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Venter, M., S. A. Madhi, C. T. Tiemessen, and B. D. Schoub. 2001. Genetic diversity and molecular epidemiology of respiratory syncytial virus over four consecutive seasons in South Africa: identification of new subgroup A and B genotypes. J. Gen. Virol. 822117-2124. [DOI] [PubMed] [Google Scholar]
- 52.Viegas, M., and A. S. Mistchenko. 2005. Molecular epidemiology of human respiratory syncytial virus subgroup A over a six-year period (1999-2004) in Argentina. J. Med. Virol. 77302-310. [DOI] [PubMed] [Google Scholar]
- 53.Waris, M. 1991. Pattern of respiratory syncytial virus epidemics in Finland: two-year cycles with alternating prevalence of groups A and B. J. Infect. Dis. 163464-469. [DOI] [PubMed] [Google Scholar]
- 54.Wasem, S., S. Weichert, S. Walther, J. A. Weigl, W. Puppe, G. Ihorst, H. J. Schmitt, and J. Forster. 2008. Lower respiratory tract disease in children: constant pathogens - constant management?! Klin. Padiatr. 220291-295. [DOI] [PubMed] [Google Scholar]
- 55.Watt, P. J., M. Zardis, and P. R. Lambden. 1986. Age related IgG subclass response to respiratory syncytial virus fusion protein in infected infants. Clin. Exp. Immunol. 64503-509. [PMC free article] [PubMed] [Google Scholar]
- 56.Weigl, J. A., W. Puppe, B. Grondahl, and H. J. Schmitt. 2000. Epidemiological investigation of nine respiratory pathogens in hospitalized children in Germany using multiplex reverse-transcriptase polymerase chain reaction. Eur. J. Clin. Microbiol. Infect. Dis. 19336-343. [DOI] [PubMed] [Google Scholar]
- 57.Weigl, J. A., W. Puppe, C. U. Meyer, R. Berner, J. Forster, H. J. Schmitt, and F. Zepp. 2007. Ten years' experience with year-round active surveillance of up to 19 respiratory pathogens in children. Eur. J. Pediatr. 166957-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weigl, J. A., W. Puppe, and H. J. Schmitt. 2002. Seasonality of respiratory syncytial virus-positive hospitalizations in children in Kiel, Germany, over a 7-year period. Infection 30186-192. [DOI] [PubMed] [Google Scholar]
- 59.Wertz, G. W., P. L. Collins, Y. Huang, C. Gruber, S. Levine, and L. A. Ball. 1985. Nucleotide sequence of the G protein gene of human respiratory syncytial virus reveals an unusual type of viral membrane protein. Proc. Natl. Acad. Sci. USA 824075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wertz, G. W., M. Krieger, and L. A. Ball. 1989. Structure and cell surface maturation of the attachment glycoprotein of human respiratory syncytial virus in a cell line deficient in O glycosylation. J. Virol. 634767-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winter, G. F., and J. M. Inglis. 1987. Respiratory viruses in children admitted to hospital in Edinburgh 1972-1985. J. Infect. 15103-107. [DOI] [PubMed] [Google Scholar]
- 62.Zlateva, K. T., P. Lemey, A. M. Vandamme, and M. Van Ranst. 2004. Molecular evolution and circulation patterns of human respiratory syncytial virus subgroup A: positively selected sites in the attachment G glycoprotein. J. Virol. 784675-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zlateva, K. T., L. Vijgen, N. Dekeersmaeker, C. Naranjo, and M. Van Ranst. 2007. Subgroup prevalence and genotype circulation patterns of human respiratory syncytial virus in Belgium during ten successive epidemic seasons. J. Clin. Microbiol. 453022-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.