Abstract
Few tools exist to assess replication of chronic pathogens during infection. This has been a considerable barrier to understanding latent tuberculosis, and efforts to develop new therapies generally assume that the bacteria are very slowly replicating or nonreplicating during latency1–3. To monitor Mycobacterium tuberculosis replication within hosts, we exploit an unstable plasmid that is lost at a steady, quantifiable rate from dividing cells in the absence of antibiotic selection. By applying a mathematical model, we calculate bacterial growth and death rates during infection of mice. We show that during chronic infection the cumulative bacterial burden—enumerating total live, dead and removed organisms encountered by the mouse lung—is substantially higher than estimates from colony forming units. Our data show that M. tuberculosis replicates throughout the course of chronic infection of mice and is restrained by the host immune system. This approach may also shed light on the replication dynamics of other chronic pathogens.
Many deadly diseases, such as AIDS, malaria and tuberculosis, result from infections that persist for long periods. Disease processes that unfold over months or years are especially difficult to model in the laboratory. New approaches are sorely needed to generate insight into the pathogenesis and treatment of such agents.
Tuberculosis remains a global scourge, killing two million people per year despite available chemotherapy4. The World Health Organization estimates that 1.8 billion people are infected with M. tuberculosis (Mtb)4, and the vast majority of cases are clinically latent. Despite decades of research, the physiology of Mtb during persistent infection remains poorly understood. One model of tuberculosis pathogenesis holds that bacilli enter a state of very slow replication or nonreplicating persistence insensitive to host and antibiotic-mediated killing5,6. In Mtb-infected mice, bacterial colony-forming units (CFUs) in the lungs increase and then remain stable over time7,8, suggesting that Mtb does not replicate during the chronic phase. In 1961, this idea was tested by comparing Mtb CFUs with the number of acid-fast bacilli in mice over a long period. The researchers determined that CFUs and total bacteria counts remained similar and stable over time and concluded that “bacilli remain viable but do not divide,” a state they termed ‘static equilibrium’9. Another group used quantitative real-time PCR to revisit this question, also concluding that Mtb is in static equilibrium10. However, some studies suggest there is substantial continued replication in vivo. The immune responses of both Mycobacterium marinum–infected zebrafish and Mtb-infected mice are dynamic throughout chronic infection8,11–13. Additionally, isoniazid, the first-line agent used to treat latent tuberculosis, has virtually no efficacy against nonreplicating bacilli in vitro14–16 or in vivo17. Similarly, the rapid reactivation of latent Mtb after anti–tumor necrosis factor-α therapy18 seems inconsistent with infection by bacteria in a nonreplicating state.
To reassess mycobacterial replication, we developed a method based on pBP10, a circular plasmid carrying a kanamycin resistance marker19. Plasmids replicate coordinately with bacterial DNA, sometimes requiring antibiotic selection for their maintenance. Without selection, plasmids are lost from a proportion of daughter cells during cell division, and thus decreasing plasmid numbers can act as a marker of division. We introduced pBP10 into Mycobacterium smegmatis and Mtb to evaluate the feasibility of using this plasmid as a replication clock.
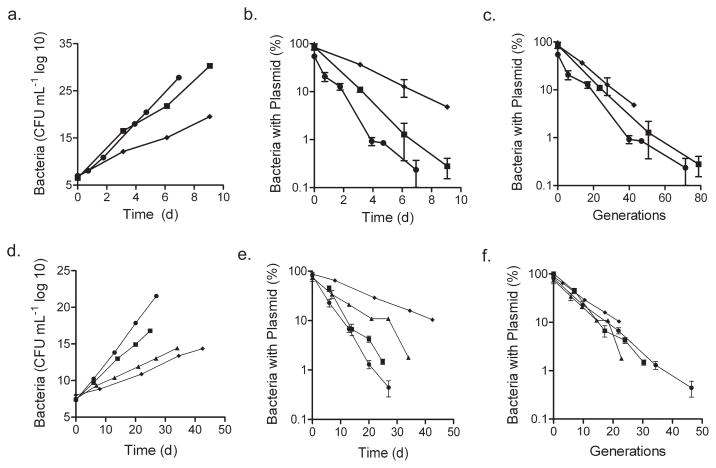
M. smegmatis-pBP10 was maintained in log-phase culture under conditions where doubling times ranged from 2 h to 5 h. Plasmid loss was determined by plating on agar with and without kanamycin. The plasmid was steadily lost as cells divided, with a rate proportional to the growth rate under each condition (Fig. 1a,b). The plasmid loss per generation was constant, regardless of the replication rate (Fig. 1c). We applied a mathematical model (Supplementary Methods online) to determine the segregation constant s — the frequency of daughter cells losing plasmid per generation. This value (0.11 ± 0.0074 (s.d.)) was reproducible across all tested growth conditions.
Figure 1.
In vitro stability of pBP10 in M. smegmatis and Mtb in the absence of antibiotic selection. (a) Total number of M. smegmatis -pBP10, accounting for serial dilutions to show ongoing culture expansion. (b) Frequency of M. smegmatis -pBP10 plasmid-containing bacteria. (c) Frequency of plasmid carriage in M. smegmatis -pBP10 for log-phase cultures versus generations (calculated as (log(OD600t / OD600(t − 1)) / log(2)). (d) Total number of M. tuberculosis -pBP10. (e) Frequency of M. tuberculosis -pBP10 plasmid-containing bacteria. (f) Frequency of plasmid carriage in M. tuberculosis -pBP10 for log-phase cultures versus generations. Data (means ± s.d.) are shown for log-phase cultures in 7H9, 1:1 7H9 to water, 1:3 7H9 to water, 7H9 without shaking and 7H9 in 2% O2. Representative data are shown for two to five experiments performed in triplicate, except for the 2% O2 experiment, which was performed once.
Mtb-pBP10 was maintained in log phase for ~30 generations by subculturing every 2–3 d without antibiotics under a range of conditions. The Mtb doubling time varied between 18 and 54 h (data not shown). Again, the plasmid loss progressed at a rate proportional to the growth rate (Fig. 1d,e), and the plasmid loss per generation remained constant despite altered growth rates (Fig. 1f). The segregation constant s for Mtb was 0.18 ± 0.023.
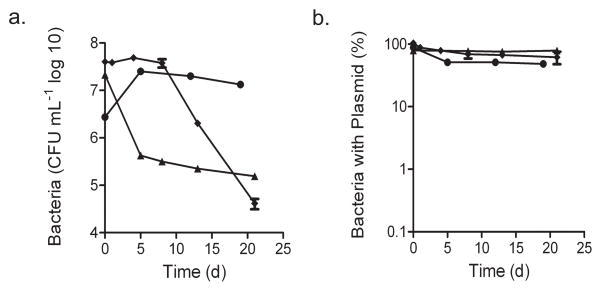
Mtb-pBP10 cultures were maintained in stationary phase for ~20 d without antibiotics, and plasmid loss was reassessed by both CFUs (Fig. 2) and quantitative real-time PCR (Supplementary Fig. 1 online). The plasmid loss ended after bacterial CFUs stabilized (Supplementary Fig. 1), indicating that Mtb replication, unlike Escherichia coli20 and M. smegmatis21 replication, halts in stationary phase. In addition, we measured plasmid loss during anoxia and carbon starvation, conditions putatively linked with nonreplicating persistent Mtb infection in vivo. Under both conditions, CFUs dropped (Fig. 2a), but no plasmid loss was detected among the surviving cells (Fig. 2b). Hence, loss of pBP10 depends on bacterial replication, validating pBP10 as a marker for Mtb division.
Figure 2.
Stability of pBP10 in Mtb in the absence of bacterial replication. (a) Total number of bacteria, as determined by plating for CFUs. (b) Frequency of plasmid-containing bacteria. Data (means ± s.d.) are shown for cultures under starvation, hypoxia or stationary phase. Each independent experiment was performed in triplicate.
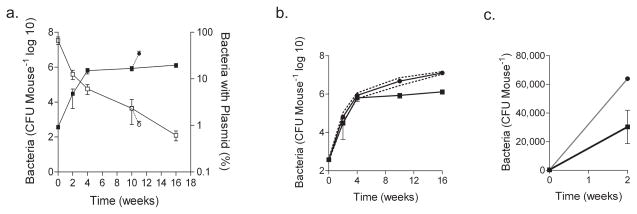
To evaluate bacterial replication in vivo, we aerosol-infected C57BL/6 mice with Mtb-pBP10. At various time points, mice were killed, and the percentage of bacteria carrying plasmid in the lungs was measured (Fig. 3a). Over the initial 4 weeks post-infection, bacterial CFUs increased, and plasmid loss was observed (Fig. 3a). From 4 to 16 weeks, bacterial CFUs remained stable but plasmid loss continued (Fig. 3a). Because CFUs remained constant, replication must have been balanced by bacterial death, which we quantified with our mathematical model (Supplementary Methods and Table 1).
Figure 3. Plasmid loss, bacterial replication and cumulative bacterial burden in mouse lungs.
(a) Bacterial growth by CFU and the percentage of bacteria carrying plasmid in lungs after low-dose aerosol infection of C57BL/6 mice with H37Rv -pBP10. A subset of mice was treated with dexamethasone (dotted line) starting at week 10 for 8 d; lung CFU and the percentage of bacteria carrying plasmid are shown. Data at each time point are the means ± s.d. of five mice. Representative data from one of three experiments are shown, except for the immunosuppression, which reflects representative data from one of two experiments. (b) Mathematical modeling of cumulative bacterial burden (CBB). Using in vivo CFU, measured by nonselective plating, and the ratio of plasmid-bearing and plasmid-free cells, we calculated the CBB in the mouse lung. 95% confidence intervals (dotted lines) are shown for the bootstrap model of 1,000 synthetic datasets. (c) Depicts the same data as in b, but with a decimal y axis to highlight the cumulative bacterial killing during the first 3 weeks of infection. CFUs are the means ± s.d. of five mice. CBB is shown with the 95% confidence interval (gray).
Table 1.
Estimates from bootstrap analysis of in vivo mathematical model.
| Day | Standard plating CFU ml−1a |
Plasmid frequency % |
Per day (95% confidence interval) |
Generation timec h |
Cumulative bacteria CFU ml−1 (95% confidence interval)b |
|
|---|---|---|---|---|---|---|
| r | δd | |||||
| 1 | 3.78 × 102 | 66.68 | 3.78 × 102 (3.03 × 102 – 4.20 × 102) | |||
| 13 | 3.03 × 104 | 12.46 | 0.78 (0.68 – 0.88) | 0.41 (0.33 – 0.51) | 21.42 | 6.39 × 104 (3.18 × 104 – 1.04 × 105) |
| 26 | 6.31 ×105 | 6.06 | 0.31 (0.19 – 0.41) | 0.07 ( 0.05 – 0.18) | 54.04 | 8.56 × 105 (5.79 × 105 – 1.12 × 106) |
| 69 | 8.55 × 105 | 2.33 | 0.12 (0.07 – 0.18) | 0.12 (0.06 – 0.18) | 134.51 | 4.78 × 106 (2.79 × 106 – 7.01 × 106) |
| 111 | 1.31 × 106 | 0.61 | 0.18 (0.12 – 0.24) | 0.17 (0.11 – 0.22) | 94.36 | 1.27 × 107 (1.08 × 107 – 1.45 × 107) |
| 77 | 5.95 ×× 106 | 1.05 | 0.55 (0.27 – 0.83) | 0.31 (0.04 – 0.56) | 30.10 | 1.64 × 107 (1.16 × 107 – 2.15 × 107) |
Day 77 data represent mice treated with dexamethasone for 8 d.
Mean of five mice per time point.
Calculations made with s = 0.18.
Generation time = ln(2) / r; hence, if r = 1, doubling time = 16.6 h; if r = 0.1, doubling time = 166 h.
Variation in the death rate is reflected in the cumulative bacterial burden. For the extreme case that δ= 1, the cumulative burden at day 111 would be 7.76 × 106 CFU ml−1; if δ= 0, the cumulative burden would be the same as that determined by standard plating.
We used the Mtb segregation constant determined in vitro and calculated the growth rate r and death rate δ for each time interval (Table 1). With these parameters, we quantified the total pool of replicating organisms as well as the number removed by death, immune clearance or both to define the cumulative bacterial burden (CBB) — the total number of bacteria live, dead or removed from the mouse lung for the duration of infection (Table 1). We then used statistical ‘bootstrapping’22 to construct 95% confidence intervals for each time point (Table 1 and Fig. 3b). During the first 2 weeks of infection, Mtb growth was rapid (generation time, 21.4 h), similar to that in vitro. The death rate was also substantial, yielding a marked difference between CFUs and CBB (Fig. 3c). Sixteen weeks after infection, the bacterial replication remained considerable, with a calculated generation time of 4 d (Table 1). This growth was balanced by the death rate to yield a plateau in CFUs. After extended infection, at 16 weeks, the CBB in these mice was 1.3 × 107 bacteria, ten times higher than that measured by plating for CFUs (Table 1).
We also treated a subset of infected mice with the immunosuppressant dexamethasone for eight d (80 μg d−1). This resulted in a rapid resurgence of Mtb growth (Table 1 and Fig. 3) and the sudden onset of acute TB disease symptoms (lethargy, trembling and weight loss). Simultaneously, plasmid frequency dropped and the CBB jumped from 4.8 × 106 to 1.6 × 107 bacteria (Table 1 and Fig. 3).
Our mathematical model makes certain assumptions about Mtb growth in vitro (Supplementary Materials online, Mathematical Model of in vitro and in vivo Plasmid Loss). To test how these assumptions affected our results in vivo, we modeled alternative assumptions. If, contrary to our assumptions, bacteria die during log phase in vitro, the result would be lower estimates for s and therefore higher estimates for bacterial turnover in vivo (Supplementary Equation 1 online). In addition, if plasmid maintenance were to exert a fitness cost, the model would predict only a very modest reduction in estimated bacterial turnover in vivo (Supplementary Equation 2 online).
This study describes a versatile tool to analyze pathogen dynamics within hosts. With only standard data collection techniques, we measured plasmid loss as a surrogate for Mtb replication. In contrast to current models of chronic TB that invoke non-replicating persistence, we observed a decline in plasmid frequency during chronic infection that indicates substantial ongoing bacterial replication. We cannot yet distinguish between the possibility that there is slowed but continuous replication of all bacteria and the possibility that some Mtb divide rapidly amid a dormant subset.
Earlier studies using microscopy9 or PCR10 concluded that Mtb is nonreplicating or very slowly replicating during chronic mouse infection. However, both experiments assumed that nonviable bacteria, their chromosomes or both are not removed or degraded over time and therefore can be quantified. But if killed organisms are cleared by the host, both techniques would have underestimated the cumulative bacterial burden. Furthermore, data from both are consistent with continued bacterial replication at a reduced rate.
Our results suggest that some aspects of current tuberculosis persistence models should be revisited. Past studies have indicated deteriorating histopathology during chronic mouse tuberculosis infection despite stable CFUs7,23; such progressive inflammation and tissue damage would be unexpected if the bacilli are dormant but unsurprising if the bacteria still replicate. The current Mtb paradigm8,24 argues that robust killing by host defenses begins only with activation of adaptive immunity at around 3 weeks of infection. In contrast, we found substantial Mtb killing within the first 3 weeks of infection (Fig. 3c), suggesting an early and major role for innate immunity. This is supported by recent work with M. marinum13 and Mtb25 infections. Furthermore, the rapid surge in bacterial CFUs in vivo after immunosuppression (Fig. 3a) is consistent with diminished stress on an already dividing population, rather than a reawakening of dormant organisms.
In conclusion, our results challenge the widespread notion that chronic tuberculosis is in static equilibrium, with replication either very slow or nonexistent. Although no animal model perfectly recapitulates human disease, mice provide the only widely used animal model of chronic tuberculosis and have a predictive role in the development of new therapies26. Recent studies indicate that granulomas in Mtb-infected mice lack hypoxia27–29, whereas lesions in Mtb-infected guinea pigs, rabbits and primates are hypoxic29. The oxygen tension in human tuberculosis granulomas, although unreported, is assumed to be low. We plan to monitor Mtb replication in guinea pigs, rabbits and especially nonhuman primates, as these models offer more relevance to some aspects of human disease. We anticipate that application of this unstable plasmid replication clock may shed light on the in vivo replication dynamics of other persistent agents responsible for widespread human suffering, such as Plasmodium, trypanosomatids and pathogenic fungi.
METHODS
Transformation with plasmid
We transformed M. smegmatis mc2155 (American Type Culture Collection #700084) and Mtb H37Rv (American Type Culture Collection #25618) as previously described30,31 with the plasmid pBP10 (K.G. Papavinasasundaram, University of Massachusetts Medical School). We selected colonies by growth on Middlebrook 7H10 medium (Becton Dickinson) containing 30 μg ml−1 kanamycin.
Bacterial strains and growth conditions
Except where indicated, we grew all strains (M. smegmatis, M. smegmatis-pBP10, Mtb H37Rv and Mtb H37Rv-pBP10 at 37 °C in Middlebrook 7H9 medium (Becton Dickinson) with 0.05% Tween-80 and albumin, dextrose, catalase (Middlebrook ADC Enrichment, BBL Microbiology) with or without 30 μg ml−1 kanamycin or on Middlebrook 7H10 (Becton Dickinson) medium with oleic acid plus albumin, dextrose, catalase (Middlebrook ADC Enrichment, BBL Microbiology) with or without 30 μg ml−1 kanamycin. We grew strains to an optical densityof ~1 at A600 (OD600) and stored them in 15% glycerol at −80 °C. For in vitro log-phase experiments, we maintained replicate rolling cultures in log phase by subculturing every 1–3 d for ~40 generations in the absence of antibiotics. We recorded each dilution to calculate total bacterial numbers. We determined the percentage of mycobacteria carrying plasmid as the number of CFUs on 7H10 agar with kanamycin over the number of CFUs on 7H10 agar without kanamycin. We compared different media conditions to achieve a variety of growth rates, including 7H9 medium diluted with sterile water, with 0.05% Tween to minimize clumping. For the starvation experiments, we grew Mtb in minimal medium consisting of 3.33 mM L-asparagine, 5.74 mM KH2PO4, 10.6 mM Na2HPO4, 40.6 μM MgSO4·7H20, 4.50 μM CaCl2, 0.619 μM ZnSO4 and 50 mg/L ferric ammonium citrate. We measured log-phase growth rates at the initiation of growth (high plasmid frequency) and again at the end (low plasmid frequency) to assess the fitness cost of plasmid carriage. We grew hypoxic cultures in 7H9 medium in spinner flasks with 2% oxygen flow-through as previously described32. Before plating, we brought cultures to their original volumes to account for evaporation from the constant air flow. For in vitro stationary phase and starvation experiments with Mtb, we grew rolling cultures for ~20 d without subculturing. For the hypoxic experiments, we maintained Mtb in 7H9 medium in 96-well plates placed inside airtight bags with a Gaspak EZ Anaerobe Container System Sachet (Becton Dickinson) and a Gaspak Dry Anaerobic Indicator Strip (Becton Dickinson).
Infection of mice
We maintained 6–8-week-old C57BL/6 mice (Jackson Laboratories) in a biosafety level 3 animal facility in accordance with University of Washington Institutional Animal Care and Use Committee protocols. We performed infections in an aerosol infection chamber (Glas-Col) as previously described7. At selected time points, we killed groups of five mice by cervical dislocation. We removed the left lung and homogenized it in 2.5 ml PBS with 0.05% Tween-80, and plated serial dilutions on 7H10 ± 30 μg ml−1 kanamycin. We performed the experiment three times with very similar results. We treated five mice at 10 weeks after aerosol infection with dexamethasone sodium phosphate (Spectrum Chemicals and Lab Products) for 8 d (10 mg dexamethasone per 500-ml water bottle, a calculated daily dose of 80 μg d−1).
Quantitative real-time PCR
We performed quantitative real-time PCR on a Bio-Rad iCycler and quantified products with iQ SYBR Green Supermix (Bio-Rad) following the manufacturer’s instructions. The primers used to quantify pBP10 were 5′-TCAACGGGAAACGTCTTGCTCG-3′ and 5′-ACCTGGAATGCTGTTTTCCCGG-3′. The primers used to quantify Mtb genomes were 5′-CTAGTCTGCCCGTATCGCCC-3′ and 5′-GAAGGTCCGGGTTCTCTCGG-3′, hybridizing to the 16s gene. We generated a standard curve with serial dilutions of Mtb genomic DNA of a known concentration. Quantitative real-time PCR conditions were as follows: 40 cycles of 95 °C (30 s), 60 °C (30 s; data collection) and 72 °C (30 s).
Supplementary Material
Acknowledgments
We would like to thank L. Ramakrishnan, J. Chang, K. Guinn, T. Walker and members of the Sherman lab for review of and contributions to this work, C. DeMille for expert help with manuscript preparation and K.G. Papavinasasundaram (University of Massachusetts Medical School) for providing pBP10. This research was supported by grants from the US National Institutes of Health and the Paul G. Allen Family Foundation.
Footnotes
Statistical analyses. s.]
Note: Supplementary information is available on the Nature Medicine website.
AUTHOR CONTRIBUTIONS
W.P.G., N.S.H., M.R.W., R.P.L. and D.R.S. designed all experiments. W.P.G., N.S.H., R.P.L. and M.R.W. performed all experiments. W.P.G., N.S.H., M.R.W., J..M. and D.R.S. analyzed the data. J.E.M. derived the equations in consultation with W.P.G. and D.R.S. W.P.G. and D.R.S. wrote the paper, which was edited by all authors.
References
- 1.Warner DF, Mizrahi V. Tuberculosis chemotherapy: the influence of bacillary stress and damage response pathways on drug efficacy. Clin Microbiol Rev. 2006;19:558–570. doi: 10.1128/CMR.00060-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho SH, et al. Low-oxygen-recovery assay for high-throughput screening of compounds against nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2007;51:1380–1385. doi: 10.1128/AAC.00055-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lenaerts AJ, et al. Preclinical testing of the nitroimidazopyran PA-824 for activity against Mycobacterium tuberculosis in a series of in vitro and in vivo models. Antimicrob Agents Chemother. 2005;49:2294–2301. doi: 10.1128/AAC.49.6.2294-2301.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frieden TR, Sterling TR, Munsiff SS, Watt CJ, Dye C. Tuberculosis. Lancet. 2003;362:887–899. doi: 10.1016/S0140-6736(03)14333-4. [DOI] [PubMed] [Google Scholar]
- 5.Wayne LG, Sohaskey CD. Nonreplicating persistence of Mycobacterium tuberculosis. Annu Rev Microbiol. 2001;55:139–163. doi: 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- 6.Harries AD, Dye C. Tuberculosis. Ann Trop Med Parasitol. 2006;100:415–431. doi: 10.1179/136485906X91477. [DOI] [PubMed] [Google Scholar]
- 7.Lewis KN, et al. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guerin attenuation. J Infect Dis. 2003;187:117–123. doi: 10.1086/345862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lazarevic V, Nolt D, Flynn JL. Long-term control of Mycobacterium tuberculosis infection is mediated by dynamic immune responses. J Immunol. 2005;175:1107–1117. doi: 10.4049/jimmunol.175.2.1107. [DOI] [PubMed] [Google Scholar]
- 9.Rees RJM, Hart PD. Analysis of the host-parasite equilibrium in chronic murine tuberculosis by total and viable bacillary counts. Br J Exp Pathol. 1961;42:83–88. [PMC free article] [PubMed] [Google Scholar]
- 10.Munoz-Elias EJ, et al. Replication dynamics of Mycobacterium tuberculosis in chronically infected mice. Infect Immun. 2005;73:546–551. doi: 10.1128/IAI.73.1.546-551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cosma CL, Humbert O, Ramakrishnan L. Superinfecting mycobacteria home to established tuberculous granulomas. Nat Immunol. 2004;5:828–835. doi: 10.1038/ni1091. [DOI] [PubMed] [Google Scholar]
- 12.Meijer AH, et al. Identification and real-time imaging of a myc-expressing neutrophil population involved in inflammation and mycobacterial granuloma formation in zebrafish. Dev Comp Immunol. 2008;32:36–49. doi: 10.1016/j.dci.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 13.Swaim LE, et al. Mycobacterium marinum infection of adult zebrafish causes caseating granulomatous tuberculosis and is moderated by adaptive immunity. Infect Immun. 2006;74:6108–6117. doi: 10.1128/IAI.00887-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Comstock GW, Baum C, Snider DE. Isoniazid prophylaxis among alaskan eskimos: a final report of the Bethel isoniazid studies. Am Rev Respir Dis. 1979;119:827–830. doi: 10.1164/arrd.1979.119.5.827. [DOI] [PubMed] [Google Scholar]
- 15.American Thoracic Society & Centers for Disease Control and Prevention. Supplement—American Thoracic Society Centers for Disease Control and Prevention. Targeted tuberculin testing and treatment of latent tuberculosis infection. Am J Respir Crit Care Med. 2000;161:S221–S247. doi: 10.1164/ajrccm.161.supplement_3.ats600. [DOI] [PubMed] [Google Scholar]
- 16.Stover CK, et al. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature. 2000;405:962–966. doi: 10.1038/35016103. [DOI] [PubMed] [Google Scholar]
- 17.Kanai K. Experimental studies on host-parasite equilibrium in tuberculous infection, in relation to vaccination and chemotherapy. Jpn J Med Sci Biol. 1966;19:181–199. doi: 10.7883/yoken1952.19.181. [DOI] [PubMed] [Google Scholar]
- 18.Keane J. TNF-blocking agents and tuberculosis: new drugs illuminate an old topic. Rheumatology (Oxford) 2005;44:714–720. doi: 10.1093/rheumatology/keh567. [DOI] [PubMed] [Google Scholar]
- 19.Bachrach G, et al. A new single-copy mycobacterial plasmid, pMF1, from Mycobacterium fortuitum which is compatible with the pAL5000 replicon. Microbiology. 2000;146:297–303. doi: 10.1099/00221287-146-2-297. [DOI] [PubMed] [Google Scholar]
- 20.Zambrano MM, Siegele DA, Almiron M, Tormo A, Kolter R. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]
- 21.Smeulders MJ, Keer J, Speight RA, Williams HD. Adaptation of Mycobacterium smegmatis to stationary phase. J Bacteriol. 1999;181:270–283. doi: 10.1128/jb.181.1.270-283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davison AC, Hinkley DV. Bootstrap Methods and Their Application. Chapters 2 and 4. Cambridge University Press; Cambridge: 1997. [Google Scholar]
- 23.Rhoades ER, Frank AA, Orme IM. Progression of chronic pulmonary tuberculosis in mice aerogenically infected with virulent Mycobacterium tuberculosis. Tuber Lung Dis. 1997;78:57–66. doi: 10.1016/s0962-8479(97)90016-2. [DOI] [PubMed] [Google Scholar]
- 24.Mogues T, Goodrich ME, Ryan L, LaCourse R, North RJ. The relative importance of T cell subsets in immunity and immunopathology of airborne Mycobacterium tuberculosis infection in mice. J Exp Med. 2001;193:271–280. doi: 10.1084/jem.193.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivas-Santiago B, et al. β-defensin gene expression during the course of experimental tuberculosis infection. J Infect Dis. 2006;194:697–701. doi: 10.1086/506454. [DOI] [PubMed] [Google Scholar]
- 26.Nuermberger E, et al. Rifapentine, moxifloxacin or DNA vaccine improves treatment of latent tuberculosis in a mouse model. Am J Respir Crit Care Med. 2005;172:1452–1456. doi: 10.1164/rccm.200507-1047OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aly S, et al. Oxygen status of lung granulomas in Mycobacterium tuberculosis infected mice. J Pathol. 2006;210:298–305. doi: 10.1002/path.2055. [DOI] [PubMed] [Google Scholar]
- 28.Tsai MC, et al. Characterization of the tuberculous granuloma in murine and human lungs: cellular composition and relative tissue oxygen tension. Cell Microbiol. 2006;8:218–232. doi: 10.1111/j.1462-5822.2005.00612.x. [DOI] [PubMed] [Google Scholar]
- 29.Via LE, et al. Tuberculous granulomas are hypoxic in guinea pigs, rabbits and nonhuman primates. Infect Immun. 2008;76:2333–2340. doi: 10.1128/IAI.01515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wards BJ, Collins DM. Electroporation at elevated temperatures substantially improves transformation efficiency of slow-growing mycobacteria. FEMS Microbiol Lett. 1996;145:101–105. doi: 10.1111/j.1574-6968.1996.tb08563.x. [DOI] [PubMed] [Google Scholar]
- 31.Park HD, et al. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol. 2003;48:833–843. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rustad TR, Harrell MI, Liao R, Sherman DR. The enduring hypoxic response of Mycobacterium tuberculosis. PLoS ONE. 2008;3:e1502. doi: 10.1371/journal.pone.0001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.