The Arabidopsisgene ABI3 shows developmentally regulated alternative splicing. ABI3-α and ABI3-β splice variants encode full-length and truncated ABI3 proteins, respectively. The conserved splicing factor SUA reduces splicing of a cryptic ABI3 intron, which leads to the accumulation of ABI3-α. Mutations in sua suppress the frameshift mutant abi3-5 by restoring its reading frame.
Abstract
ABSCISIC ACID INSENSITIVE3 (ABI3) is a major regulator of seed maturation in Arabidopsis thaliana. We detected two ABI3 transcripts, ABI3-α and ABI3-β, which encode full-length and truncated proteins, respectively. Alternative splicing of ABI3 is developmentally regulated, and the ABI3-β transcript accumulates at the end of seed maturation. The two ABI3 transcripts differ by the presence of a cryptic intron in ABI3-α, which is spliced out in ABI3-β. The suppressor of abi3-5 (sua) mutant consistently restores wild-type seed features in the frameshift mutant abi3-5 but does not suppress other abi3 mutant alleles. SUA is a conserved splicing factor, homologous to the human protein RBM5, and reduces splicing of the cryptic ABI3 intron, leading to a decrease in ABI3-β transcript. In the abi3-5 mutant, ABI3-β codes for a functional ABI3 protein due to frameshift restoration.
INTRODUCTION
Seeds are essential for the spread and survival of most plant species and constitute a major food source. Seed features like desiccation tolerance, dormancy, and the accumulation of storage proteins are established during seed maturation. In Arabidopsis thaliana, the phytohormone abscisic acid (ABA) controls seed maturation and dormancy by preventing germination and reserve mobilization. ABA signaling at this stage is concomitant with the expression of four major regulatory genes of seed maturation with partially redundant functions: LEAFY COTYLEDON1 (LEC1), LEC2, FUSCA3 (FUS3), and ABSCISIC ACID INSENSITIVE3 (ABI3) (Kroj et al., 2003; To et al., 2006). ABI3 is a main component of the ABA signaling pathway and is highly conserved among plant species. The ABI3 protein contains four functional domains (Giraudat et al., 1992; Suzuki et al., 1997). The A1 domain is an acidic transcriptional activator (McCarty et al., 1991), and B1 can interact with the seed-specific transcription factor ABI5 (Nakamura et al., 2001). B2 and B3 are two basic DNA binding domains responsible for the ABA-dependent activation of seed maturation genes (Suzuki et al., 1997; Ezcurra et al., 2000; Nag et al., 2005).
Several abi3 mutant alleles were isolated in Arabidopsis. One of the most severe is abi3-5, which was originally identified by its stay-green seed phenotype. abi3-5 seeds are insensitive to ABA during germination, are desiccation intolerant, and have reduced longevity, similar to other strong abi3 alleles (Ooms et al., 1993).
ABI3 transcription is promoted by LEC1, LEC2, FUS3, ABI3 (To et al., 2006), and ABA (Lopez-Molina et al., 2002). During germination, ABI3 is repressed by the chromatin remodeling factor PICKLE (Perruc et al., 2007) and the ABI3 protein is targeted to 26S proteasome degradation by the ABI3-INTERACTING PROTEIN2 (Zhang et al., 2005). The identification of several splice variants of ABI3 homologs in monocotyledon and dicotyledon species (McKibbin et al., 2002; Fan et al., 2007; Gagete et al., 2009) implies that alternative splicing also has an important role in controlling ABI3 expression. However, splicing variants of ABI3 were not observed in Arabidopsis.
Although alternative splicing of mRNA is an important component of posttranscriptional regulation in higher eukaryotes, its relevance and mechanisms in plants are poorly understood. In Arabidopsis, ∼42% of all transcripts from intron-containing genes are alternatively spliced (Filichkin et al., 2010). Alternative splicing can produce transcripts that encode for proteins with altered or lost function. Furthermore, it can lead to tissue-specific transcripts or affect mRNA stability and turnover via nonsense-mediated decay (McGlincy and Smith, 2008). Splicing is directed by the spliceosome, a dynamic RNA-protein multicomponent machinery that is conserved among eukaryotes. In Arabidopsis, only a few splicing-related proteins have been characterized (Lopato et al., 1999; Ali et al., 2007; Tanabe et al., 2007; Zhang and Mount, 2009), and the information on their biochemical function and their targets in relevant developmental and environmental contexts is limited. We identified SUPPRESSOR OF ABI3-5 (SUA) as a novel plant splicing factor that influences seed maturation by controlling alternative splicing of ABI3. SUA is an evolutionary conserved protein that suppresses splicing of a cryptic ABI3 intron. Splicing of this intron leads to a transcript that encodes a truncated ABI3 protein in the wild type but a functional protein in the abi3-5 mutant background.
RESULTS
Isolation of the abi3-5 sua-1 Double Mutant
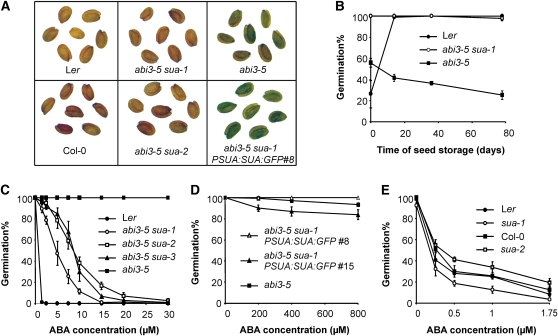
Seeds of the abi3-5 glabra1 (gl1) transparent testa5-1 (tt5-1) triple mutant were mutagenized by γ-irradiation to isolate mutants involved in ABI3 signaling. The gl1 and tt5-1 mutations are located on both sides of the ABI3 locus and were used as phenotypic markers to distinguish suppressor mutants from wild-type contaminants. A strong suppressor mutant of abi3-5 was identified in the M2 generation and named suppressor of abi3-5 (sua-1). The gl1-1 and tt5-1 mutations were removed from this line by backcrossing with its wild-type Landsberg erecta (Ler) genetic background and subsequent selection for abi3-5 and sua-1 in the progeny. Ripe abi3-5 seeds are green due to the presence of chlorophyll, but abi3-5 sua-1 seeds are yellow-brown, similar to the wild type (Figure 1A). In addition, abi3-5 seeds are nondormant and sensitive to desiccation, which causes reduced longevity. Seeds of abi3-5 sua-1 are also nondormant, but their longevity is strongly improved and they still germinate nearly 100% after 10 weeks of storage (Figure 1B). Finally, abi3-5 sua-1 seeds show an increased sensitivity to ABA and cannot germinate on 15 μM ABA, whereas viable abi3-5 mutant seeds show 100% germination on 30 μM ABA (Figure 1C).
Figure 1.
The sua Mutation Suppresses abi3-5 Phenotypes.
(A) Seeds of wild type (Ler and Col-0), abi3-5 (in Ler background), abi3-5 sua-1 (in Ler background), abi3-5 sua-2 (in Ler/Col-0 background) and abi3-5 sua-1 PSUA:SUA:GFP #8 (in Ler background).
(B) Germination of Ler, abi3-5, and abi3-5 sua-1 seeds after different periods of dry storage. Harvested seeds were stored at 20°C and 42% relative humidity. Percentages are means (±se) of three biological replicates.
(C) Germination of Ler, abi3-5, abi3-5 sua-1, abi3-5 sua-2, and abi3-5 sua-3 seeds, imbibed at different ABA concentrations. Seeds were 1 week after-ripened and stratified 4 d. Percentages are means (±se) of four biological replicates.
(D) Germination of 1-week-old abi3-5, abi3-5 sua-1 PSUA:SUA:GFP #8, and abi3-5 sua-1 PSUA:SUA:GFP #15 seeds at different ABA concentrations. Percentages are means (±se) of three biological replicates.
(E) Germination of Ler, sua-1, Col-0, and sua-2 seeds, imbibed at different ABA concentrations. Seeds were 6 months after-ripened and 4 d stratified. Percentages are means (±se) of four biological replicates.
Identification of the SUA Gene
Initial mapping indicated that the sua mutation is located on chromosome 3. Fine-mapping was performed using an F2 mapping population of ∼4000 individuals derived from a cross between the abi3-5 sua-1 double mutant (in Ler background) and Columbia (Col-0). The abi3-5 sua-1 double mutant was identified in this mapping population by its yellow-brown seed color trait in combination with the ability to germinate in the presence of 5 μm ABA. The location of the sua-1 mutation was narrowed down to a region of 64 kb at the bottom of chromosome 3 between two markers located at 20.056 and 20.120 Mb. This region contains 17 genes and did not show recombination in our mapping population. Comparison of sequenced candidate genes with sequences in The Arabidopsis Information Resource (Garcia-Hernandez et al., 2002) revealed a 47-bp deletion in the 15th exon of At3G54230 in the abi3-5 sua-1 double mutant (Figure 2A).
Figure 2.
Genetic Structure, Domain Organization, and Phylogenetic Relationships of SUA.
(A) Schematic structure of the SUA gene. Triangles indicate the T-DNA insertion sites of sua-2 and sua-3, and the dashed region represents the 47-bp deletion of the sua-1 allele. UTRs are shown in white, exons in gray, and introns as thick lines.
(B) Domain structure of the SUA protein. aa, amino acids; RRM, RNA recognition motif; Zn, zinc finger; OCRE, octamer repeat; G-p, Gly patch.
(C) Phylogram of SUA and its closest related proteins. FCA is an RNA binding protein that was added to the tree to emphasize the similarity between SUA and its homologs in evolutionary distant species. Populus trichocarpa (Pt), Vitis vinifera (Vv), Oryza sativa (Os), Physcomitrella patens (Pp), Chlamydomonas reinhardtii (Cr), Xenopus laevis (Xl), Mus musculus (Mm), and Homo sapiens (Hs). Bootstrap values are shown when higher than 50.
The identity of At3G54230 as the SUA gene was confirmed by complementation of the sua-1 mutant in the abi3-5 background. A construct containing the SUA cDNA, expressed from a 2711-bp putative SUA promoter and fused with a C-terminal green fluorescent protein (GFP) tag (PSUA:SUA:GFP), was used to transform abi3-5 sua-1 plants. Two independent T2 transformants, containing a single insertion event, both complemented sua-1 and showed the abi3-5 phenotype. One of these transformants, abi3-5 sua-1 PSUA:SUA:GFP #8, even showed an enhanced abi3-5 phenotype, yielding seeds with a more intense green color and stronger ABA insensitivity (Figures 1A and 1D).
Additional mutant alleles of SUA in the Col-0 background (sua-2 and sua-3) were obtained from the Salk insertion mutant collection and from the GABI-Kat collection. These lines contain T-DNA insertions in the fourth and the ninth intron and were named sua-2 and sua-3, respectively (Figure 2A). Both alleles lack full-length SUA expression and were crossed with abi3-5. The double mutants abi3-5 sua-2 and abi3-5 sua-3 were selected in the resulting F2, and all of them showed suppression of the abi3-5 phenotypes, similar to abi3-5 sua-1 (Figures 1A and 1C).
The sua single mutants did not have any obvious visual phenotype. Detailed analysis revealed that sua-1 seeds are more susceptible to ABA germination inhibition compared with wild-type Ler. By contrast, sua-2 seeds germinated better than wild-type Col-0 in the presence of ABA (Figure 1E).
SUA Encodes an RNA Binding Protein Located in the Nucleus and Expressed in All Plant Tissues
SUA encodes a protein with a conserved domain architecture that suggests a function in RNA metabolism. SUA contains two RNA recognition motifs surrounding a Zinc finger domain, an octamer repeat domain, and a Gly-rich domain close to the carboxy end (Figure 2B). The Arabidopsis genome does not contain a second gene with this combination of domains. SUA homologs, however, can be found throughout the eukaryotic kingdom (Figure 2C). SUA has 45% sequence similarity with the human RNA Binding Motif Protein 5 (RBM5), which was originally identified as a putative tumor suppressor gene that is part of a small gene family (Edamatsu et al., 2000).
Publicly available microarray data (Zimmermann et al., 2004) show ubiquitous SUA expression in Arabidopsis, with a moderate enrichment in seeds. Quantitative real-time RT-PCR analysis confirmed that the relative abundance of SUA transcripts is comparable in most Arabidopsis tissues, but highest in siliques toward the end of seed maturation (Figure 3A). The subcellular localization of the SUA protein was studied using the PSUA:SUA:GFP lines. A GFP signal was detected in the nucleus of vegetative and reproductive tissues (Figure 3B). The SUA_GFP chimeric protein showed diverse patterns. Speckles of different size were observed in some nuclei, but fluorescence was diffuse and rather weak in others (Figure 3B). We did not observe a correlation between the SUA_GFP fluorescence pattern and tissue or developmental stages.
Figure 3.
SUA Is Expressed in All Tissues and Its Protein Is Localized in the Nucleus.
(A) Quantitative real-time RT-PCR analysis of SUA expression in different tissues. SUA mRNA levels are normalized to ACTIN8 mRNA levels. S6D, seedlings 6 d after germination; R, roots; RL, rosette leaves; CL, cauline leaves; FB, flower buds; S10 to S20, siliques 10, 12, 14, 16, 18, and 20 d after pollination. Data are from two independent biological replicates. Error bars represent se.
(B) Confocal analysis of subcellular localization of SUA:GFP in developing embryo tissue from transgenic abi3-5 sua plants containing the PSUA:SUA:GFP construct. Three nuclei with different GFP patterns are shown. Bar = 2 μM.
SUA Interacts with the Prespliceosomal Component U2AF65
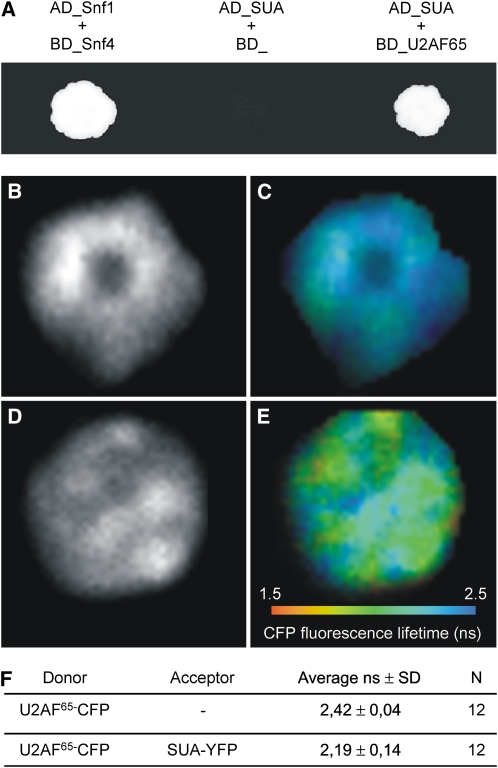
RBM5, the human homolog of SUA, is a member of the prespliceosomal complex (Behzadnia et al., 2007) and interacts with U2AF65 in vivo (Bonnal et al., 2008). U2AF65 is the larger subunit of the conserved pre-mRNA splicing factor U2AF. It guides splice site selection during the formation of the spliceosomal complex (Zamore et al., 1992; Sickmier et al., 2006). In a yeast two-hybrid GAL4 assay, we detected interaction between SUA and Arabidopsis U2AF65 (BAH19725) (Domon et al., 1998; Figure 4A). To confirm the SUA-U2AF65 interaction in planta, we performed a fluorescence resonance energy transfer/fluorescence lifetime imaging (FRET/FLIM) assay. Arabidopsis leaf protoplasts were cotransfected with two vectors for the overexpression of SUA_YFP (yellow fluorescent protein) and U2AF65_CFP chimerical proteins. FRET/FLIM analysis of protoplasts coexpressing SUA_YFP and U2AF65_CFP (cyan fluorescent protein) showed a significant reduction of the mean CFP fluorescence lifetime compared with those expressing the U2AF65-CFP alone (Figures 4B to 4F), confirming interaction of both proteins in planta.
Figure 4.
SUA Interacts with U2AF65.
(A) Interaction between SUA and U2AF65 detected with the yeast two-hybrid assay. Cotransformed yeast strains were grown on SD-L-W-H with 5 mM 3AT. Snf1 and Snf4 are yeast proteins that strongly interact (Jiang and Carlson, 1997).
(B)to (E) Interaction between SUA and U2AF65 based on FRET measured by FLIM. FLIM analysis of protoplasts transiently expressing U2AF65-CFP ([B]and [C]) and coexpressing U2AF65-CFP and SUA-YFP ([D]and [E]). Intensity channel ([B]and [D]) and false color code ([C]and [E]). The absence of interaction results in a long lifetime, visible as a dark-blue color. Interaction leads to a reduction in donor lifetime, visible as a shift toward orange. A representative protoplast nucleus is shown.
(F) Average CFP fluorescence lifetime values for the FRET/FLIM analysis. N, number of nuclei analyzed.
The Suppression of abi3-5 by sua-1 Is Allele Specific
The abi3-5 mutant is one of the strongest abi3 alleles, which all show reduced seed dormancy and decreased sensitivity to ABA during germination (Bies-Etheve et al., 1999). Seeds of the abi3-4 and abi3-6 mutants are nondormant, highly insensitive to ABA, and show reduced longevity and a high chlorophyll content similar to abi3-5. To study the suppression effect of the sua-1 mutant on different abi3 mutant alleles, double mutants were constructed. The ABA-insensitive abi3-4 and abi3-6 alleles, as well as the weak abi3-1 and abi3-7 alleles (Figure 5A), were combined with sua-1, sua-2, and sua-3. Surprisingly, none of these combinations showed any suppression phenotype, indicating that the suppression of abi3-5 by sua mutants is allele specific (Figure 5B).
Figure 5.
sua Is an Allele-Specific Suppressor of the abi3-5 Allele.
(A) Schematic structure of the ABI3 gene. The locations and nature of the abi3-1, abi3-4, abi3-5, abi3-6, and abi3-7 mutations are indicated. UTRs are shown in white, exons in gray, and introns as thick lines. The box with diagonal stripes represents the cryptic intron.
(B) Table showing the suppression of the abi3 phenotype in different combinations of sua and abi3 mutant alleles. A check mark indicates abi3 suppression; the “x” indicates absence of abi3 suppression. ND, not determined.
(C) Sequence of the ABI3 cryptic intron and surrounding region. The asterisk indicates the single base pair deleted in abi3-5, and the subsequent stop codon is underlined. The cryptic intron is shown in lowercase letters.
Detection of Functional ABI3 Protein in the abi3-5 sua-1 Double Mutant
The abi3-5 mutation causes a frameshift leading to a premature stop codon after 34 erroneous codons. The abi3-4 mutant has a single nucleotide mutation that causes a stop codon at approximately the same position (Bies-Etheve et al., 1999; Figure 5A). Therefore, abi3-4 and abi3-5 produce ABI3 transcripts that translate into truncated ABI3 proteins with similar sizes. Nevertheless, the phenotype of the abi3-5 mutant is strongly suppressed by sua, whereas that of abi3-4 is not. To understand this discrepancy, we analyzed the ABI3 protein in dry seeds of Ler, sua-1, abi3-4, abi3-5, and the double mutants abi3-4 sua-1 and abi3-5 sua-1 by immunoblotting. A specific antibody, targeted to the amino end of ABI3, was used for detection. The ABI3 protein (720 amino acids) migrates as a 116-kD polypeptide (Parcy et al., 1997). We detected two bands of approximately this size for the ABI3 protein in Ler and sua-1 seeds. One of these two bands probably represents a modified version of ABI3. A truncated ABI3 protein corresponding to a 428–amino acid polypeptide and migrating as a 70-kD band, was observed in the abi3-5 mutant (Figure 6). A similar sized (416 amino acids) highly abundant ABI3 protein was found in abi3-4 and abi3-4 sua-1. The high abundance of the truncated ABI3 protein in abi3-4 seeds was previously observed by Parcy et al. (1997). In the abi3-5 sua-1 double mutant, two weak bands of comparable size to full-length ABI3 were detected, along with the smaller truncated abi3-5 mutant protein (Figure 6). The presence of full-length ABI3 protein in abi3-5 sua-1 seeds was consistent with all the observed suppression phenotypes and predicts the presence of an ABI3 transcript with a restored reading frame that has lost the abi3-5 premature stop codon.
Figure 6.
Detection of Full-Length ABI3 Protein in the abi3-5 sua-1 Double Mutant.
(A) Immunoblot analysis of ABI3 protein. Total protein was extracted from freshly harvested seeds and separated on a Tris-Gly SDS 4 to 12% polyacrylamide gradient gel. The ABI3 protein is identified as a double band of ∼116 kD in Ler, sua-1, and abi3-5 sua-1. The truncated ABI3 proteins (ΔABI3) produced by abi3-4, abi3-5, and abi3-5 sua-1 and the novel splicing variant of ABI3 are ∼70 kD. Asterisk indicates a nonspecific band that is used as loading control. Sizes of the molecular markers (in kilodaltons) are shown next to the blot.
(B) Predicted ABI3 protein isoforms. Gray boxes represent the conserved functional motifs of ABI3 (from left to right: A1, B1, B2, and B3). Boxes with diagonal stripes represent erroneous amino acids stretches.
Identification of a Novel ABI3 Splice Variant
The abi3-5 transcripts were analyzed in detail by RT-PCR and sequencing. In the abi3-5 sua-1 double mutant, we identified, besides the expected full-length abi3-5 transcript, an alternatively spliced novel abi3-5 transcript that lacks a cryptic intron of 77 nucleotides. This cryptic intron is located shortly downstream of the abi3-5 mutation and includes the premature abi3-5 stop codon (Figure 5C). The combination of the 1-bp abi3-5 deletion and the removal of the 77-nucleotide cryptic intron results in a transcript that restores the reading frame of abi3-5 after 21 erroneous and 26 deleted codons. We named this transcript abi3-5-β and named the transcript with the retained intron abi3-5-α. The translated abi3-5-β polypeptide (abi3-5-β) is predicted to be 694 amino acids. This protein contains all four ABI3 protein domains (Figure 6B), and the phenotype of the abi3-5 sua-1 seeds indicated that abi3-5-β largely retains the ABI3 molecular functions (Figure 1).
The ABI3-β transcript only encodes a functional protein in the abi3-5 mutant background. In the wild type, it causes a frameshift and codes for a truncated protein of 429 amino acids. This predicted truncated polypeptide was immunodetected in the sua-1 single mutant and also, at lower levels, in wild-type Ler seed protein extracts. The wild-type ABI3-β protein migrates with a similar speed in the gel as the proteins encoded by abi3-4 (α and β splicing forms) and abi3-5 (α splicing form) mutants (Figure 6).
In addition to the accumulation of the ABI3-β splice variant, the sua-1 mutant also shows an overall increase in ABI3 expression. The amount of ABI3-α transcript, coding for full-length ABI3, is higher in sua-1 than in the wild type (Figure 7A). This could explain the increased ABA sensitivity of sua-1 seeds. Instead, overall ABI3 expression in sua-2 seeds is similar to that in wild-type Col-0, but the portion of the transcript coding for full-length ABI3 is reduced, resulting in a decrease of ABA sensitivity (Figure 1E). We tested the possibility that sua-1 has a gain-of-function effect on ABI3 expression by transforming the sua-1 mutant allele, expressed from the endogenous SUA promoter, into the sua-2 mutant. Indeed, the obtained transformants showed a higher ABI3 expression than the sua-2 mutant and had increased ABA sensitivity (see Supplemental Figure 1 online). This indicates that sua-1 is a gain-of-function mutant regarding ABI3 expression.
Figure 7.
Quantification of ABI3 Splicing Variants.
Quantitative real-time RT-PCR analysis of ABI3-α (white) and ABI3-β (gray) expression in Ler, sua-1, abi3-5, abi3-5 sua-1, Col-0, and sua-2 (A) and in Ler developing siliques 10 to 20 d after pollination (DAP) (B). For (A), mRNA was extracted from siliques 16 d after pollination. ABI3 mRNA levels are normalized to ACTIN8 mRNA levels. Data are from two independent biological replicates. Error bars represent se.
ABI3 Alternative Splicing Is Developmentally Regulated
The relative abundance of ABI3-α and ABI3-β transcripts was quantified in wild-type seeds by real-time RT-PCR. Developing siliques 16 d after pollination showed a very low abundance of ABI3-β transcript in Ler and Col-0 (1.53 ± 1.36% and 0.95 ± 0.83%, respectively, of the overall ABI3 transcripts; Figure 7A). During progressive development of wild-type siliques, the ratio between both ABI3 transcripts shifted toward ABI3-β. At 20 d after pollination, the amount of ABI3-β exceeded that of ABI3-α (Figure 7B). The observed change in ratio between ABI3-α and ABI3-β transcripts during seed maturation indicates that alternative splicing of ABI3 is developmentally regulated.
DISCUSSION
ABI3 Is Regulated by Alternative Splicing
The transcription factor ABI3 regulates seed maturation and influences seed quality. The abundance of ABI3 is tightly regulated at different levels. In addition to complex genetic interactions with LEC1, LEC2, and FUS3 at the transcriptional level (To et al., 2006), ABI3 expression is controlled posttranscriptionally. Alternative splicing of ABI3 homologs in cereal species (Triticum aestivum and Oryza sativa) and dicots (Pisum sativum) (McKibbin et al., 2002; Fan et al., 2007; Gagete et al., 2009) generates multiple mis-spliced transcripts that often code for truncated polypeptides. This has been linked to reduced grain quality in rice and wheat (McKibbin et al., 2002; Fan et al., 2007). Here, we show that the ABI3 gene of Arabidopsisis also regulated by alternative splicing. A 77-bp cryptic ABI3 intron is alternatively spliced, which leads to the occurrence of two transcripts. The ABI3-α transcript encodes a full-length ABI3 protein, and the ABI3-β transcript encodes a truncated protein that contains two of the four functional domains. Splicing of the cryptic intron of ABI3 is developmentally regulated, and ABI3-β accumulates only at the end of seed maturation. This probably contributes to a fast downregulation of full-length ABI3 in ripe seeds, which is necessary to inhibit the seed maturation program in germinating seeds.
Transcripts with a long 3′ untranslated region (UTR) or with 3′UTR-located introns can be detected and degraded by the nonsense-mediated decay machinery in plants (Kerényi et al., 2008). To distinguish a natural stop codon from a premature stop codon, nonsense-mediated decay requires a second signal that has not been identified yet in plants (van Hoof and Green, 2006). The ABI3-β transcript contains a premature stop codon but probably lacks this second signal because it is not affected by nonsense-mediated decay. The protein encoded by the ABI3-β transcript contains the A1 acidic transcriptional activation domain and the first basic domain and might still mediate ABA signaling during late seed maturation.
The prevalent model of splicing in Arabidopsis is intron definition, in which intronic sequences are recognized by the spliceosomal complex. The features of a canonical plant intron are a consensus 5′ splice site (AG/GU, where GU is the more conserved dinucleotide), a U-rich sequence, and a consensus 3′ splice site (CAG/G where AG is invariant) (Simpson and Filipowicz, 1996; Lorković et al., 2000). Arabidopsis exons contain on average 29% U, while introns have on average 42% U (Reddy, 2007). It was shown that U-rich elements can function as splicing signals (Simpson et al., 2004), and short introns and introns with low AU content are more likely to be retained (Wang and Brendel, 2006). The ABI3 cryptic intron has sequence similarities with canonical plant introns, in particular with the consensus sequences at the two borders (Figure 5C), but it has a U content of only 29%, while the other ABI3 introns have, on average, 46% U. Because of that, the cryptic ABI3 intron may not be easily recognized by the spliceosomal complex.
SUA Controls Alternative Splicing of ABI3
SUA suppresses splicing of the cryptic ABI3 intron and thereby influences the ratio between the ABI3-α and ABI3-β transcripts. Reduced suppression of the cryptic intron in the sua mutant leads to an increased amount of ABI3-β transcript and decreased levels of the ABI3-α transcript. However, a substantial amount of ABI3-α transcript could still be detected in the sua mutant. Other splicing factors probably act redundantly with SUA in the suppression of the cryptic ABI3 intron.
Alternative splicing in plants is regulated by tissue-specific developmental cues and stresses and might provide a means for optimal adaptation to the environment (Ali and Reddy, 2008). Alternative splicing of ABI3 could also be regulated by specific environmental conditions. In this respect, it is interesting to note that publicly available microarray data show an upregulation of SUA expression by senescence (Zimmermann et al., 2004). The water content of seeds strongly decreases during the maturation phase until ∼7% in mature seed (Baud et al., 2002). This process is comparable to senescence and also coincides with increased SUA mRNA levels (Figure 3A). Higher SUA abundance will favor cryptic intron retention and increase the full-length ABI3 protein levels during seed maturation. Consistent with that, our experiments showed a correlation between increased levels of SUA transcript and a reduction in ABI3-β levels in transgenic plants. The abi3-5 sua-1 PSUA:SUA:GFP #8 line, for instance, showed increased levels of SUA transcript and reduced amounts of abi3-5-β, resulting in an enhanced abi3-5 phenotype (see Supplemental Figure 2 online; Figures 1A and 1D). SUA-mediated alternative splicing of ABI3 could represent a system to fine-tune seed maturation. However, in wild-type plants, the ABI3-β transcript accumulates at the end of seed maturation when SUA is still substantially expressed. Possibly, the SUA protein is not active or degraded at the end of seed maturation. Alternatively, other factors could counteract the role of SUA in retention of the cryptic intron at this time.
The sua-1 single mutant showed increased ABA sensitivity during germination. This is probably caused by upregulation of ABI3 expression, which does not occur in sua-2. This difference between sua-1 (in a Ler background) and sua-2 (in a Col-0 background) could be explained by natural genetic variation between Ler and Col-0 that modifies the sua mutant phenotype. However, sua-2 plants transformed with sua-1 also showed an increased ABI3 expression and enhanced ABA sensitivity. Therefore, it is more likely that sua-1 is a gain-of-function allele, which is translated into a truncated protein. This predicted polypeptide includes the RNA recognition motifs and the Zn finger motif but lacks the G patch domain at the C terminus. The nonfunctional mutant sua-1 protein might compete for substrates with other proteins of the mRNA splicing machinery and could therefore function as a dominant-negative allele.
The abi3-5 mutant still contains a small amount of abi3-5-β transcript, which encodes a functional ABI3 protein. Consequently, abi3-5 is not a complete loss-of-function mutant. The sua mutant can suppress abi3-5 because it enhances the amount of abi3-5-β transcript.
SUA Has a Conserved Role in Splicing
The conserved domain architecture of SUA and its role in the suppression of the cryptic ABI3 intron indicate a function in mRNA processing. Moreover, the speckled fluorescence patterns observed in nuclei expressing the chimeric SUA:GFP gene are similar to those obtained with Ser/Arg-rich GFP proteins, which are involved in RNA metabolism in plants (Lorković and Barta, 2004). The SUA protein has two RNA recognition motifs, which are also found in many eukaryotic RNA processing proteins (Burd and Dreyfuss, 1994). Based on its functional motifs, SUA could bind directly to specific RNA targets. However, SUA might also interact with the mRNA targets indirectly and be part of the spliceosome, which is composed of ∼300 proteins in Arabidopsis (Reddy, 2007).
The SUA protein shares a high sequence similarity with the human tumor suppressor RBM5, which regulates alternative splicing of the apoptosis genes Fas and c-FLIP (Bonnal et al., 2008). Previous work on RBM5 showed that this protein can interfere with the progression of spliceosomal assembly after formation of the prespliceosomal complex. Assuming that conservation of domain architecture indicates conservation of function, it is plausible that SUA prevents excision of the cryptic ABI3 intron after formation of the prespliceosomal complex. The confirmed interaction of SUA with the spliceosomal factor U2AF65 by yeast two-hybrid assay and in vivo suggests that SUA, similarly to RBM5, plays a role in early spliceosome formation.
Mutations in SUA have little impact on the visual plant phenotype, similar to other Arabidopsis ABA signaling mutants that affect conserved, single copy genes with a role in RNA processing. The aba-hypersensitive1 (abh1) and supersensitive to aba and drought1 (sad1) mutants, for instance, show mild phenotypes and altered expression of only a few genes (Fedoroff, 2002). These mutants show hypersensitivity to ABA at germination. ABH1 functions as the large subunit of an Arabidopsis mRNA cap binding complex (Hugouvieux et al., 2001), and SAD1 is an ortholog of the yeast subunit of the U6 small nuclear RNP complex, which is involved in nuclear RNA processing (Bouveret et al., 2000). The lack of strong phenotypes in these mutants could be due to redundancy with nonhomologous genes that share the same function. It is also possible that genes like SUA play an essential role under specific, not yet identified, environmental conditions.
With the isolation of SUA, we identified a splicing factor in plants that has a function in seed maturation. This function can be entirely explained by the influence of SUA on alternative splicing of ABI3. However, a conserved gene like SUA is likely to have a broader role, which could be revealed by the identification of additional genes whose splicing is controlled by SUA.
METHODS
Plant Materials and Growth Conditions
The mutants abi3-5 (Ooms et al., 1993; Bies-Etheve et al., 1999), abi3-4 (Giraudat et al., 1992), abi3-1, abi3-7 (Ooms et al., 1993; Bies-Etheve et al., 1999), gl1 (Oppenheimer et al., 1991), and tt5 (Shirley et al., 1995) are all in the Ler accession background, and abi3-6 (Nambara et al., 1994) is in the Col background. All mutants were obtained from the research groups in which they were isolated. The sua-1 mutant is in the Ler background. The sua-2 (SALK T-DNA insertion line, SALK_054379; Alonso et al., 2003) and sua-3 (GABI-KAT T-DNA insertion line, GK-815C12; Rosso et al., 2003) mutants are in the Col background and were obtained from the Nottingham Arabidopsis Stock Centre. All plants were grown on soil containing a mixture of substrate and vermiculite (3:1) in a greenhouse where the temperature was maintained close to 20°C; 16 h of light was provided daily.
Germination Tests
About 100 to 150 seeds from each batch were sown into 6-cm-wide plastic Petri dishes on round filter papers (Macherey and Nagel) soaked with 580 μL demineralized water. The longevity of seeds was determined after a stratification treatment, consisting of 5 d incubation at 4°C in the dark. Seeds were subsequently put into an incubator, in long-day conditions (16 h light at 25°C, followed by 8 h darkness at 20°C). After 6 d of incubation, the germinated seeds were counted. For dormancy experiments, seeds were processed in the same way, without stratification treatment. In the experiments for assessing the ABA hormone sensitivity, seeds were stratified and incubated as described above on filter papers soaked with solutions of different ABA concentrations. ABA (Sigma-Aldrich) was dissolved in methanol and diluted with water.
Mapping
Simple sequence length polymorphisms and cleaved amplified polymorphic sequences were used as polymorphic PCR markers in the mapping experiment (see Supplemental Table 1 online). Most of the primers were designed to flank short polymorphic sequences referring to the Monsanto Landsberg-Columbia polymorphisms database (https://www.Arabidopsis.org/cgi-bin/cereon/).
RNA Extraction and Quantitative Real-Time RT-PCR
mRNA was extracted from developing Arabidopsis thaliana siliques with the Magnetic mRNA Isolation Kit (New England Biolabs). First-strand cDNA was synthesized using the Superscript II Reverse Transcriptase (Invitrogen) and oligo dT(16) primers, starting from 200 ng of mRNA. Quantitative real-time RT-PCR reactions were performed with the SYBR Green Master Mix (Applied Biosystems) with the following primer sets for ABI3-α (5′-CCGGGTTTTGGATACATGC-3′, 5′-CGGATTCATGTTGTATCCATTG-3′), ABI3-β (5′-ATGCCGCCAACCTCGCAGAC-3′, 5′-ACAGGTTTCTCCGATTTGGG-3′), ACT8 (5′-CTCAGGTATTGCAGACCGTATGAG-3′, 5′-CTGGACCTGCTTCATCATACTCTG-3′), SUA (5′-CAGCAATTGCATCAGAGAAGAG-3′, 5′-ACGTTGTATTTGGGTTTCATGG-3′), and sua-1 (5′-TGACAATCCACCTACAGTTTCG-3′, 5′-CACCCAAAGTTTCTCCCACTC-3′) and analyzed on a Mastercycler realplex2 system (Eppendorf). Quantification of ACT8, ABI3-α, ABI3-β, SUA, and sua-1 was determined by the standard curve method. For each of the two biological replicates, the amounts of target transcripts were normalized to the value of ACT8. For the quantification of each target, the values of two biological replicates were averaged.
Plant Transformation
The binary vectors carrying PSUA:SUA_GFP and PSUA:sua-1 were prepared using standard molecular cloning techniques in combination with the Gateway technology (Invitrogen). PSUA consisted of 2711-bp genomic sequence upstream of the SUA start codon. The SUA and sua-1 cDNAs comprised the sequences from the start codon to the first stop codon. The constructs were first transformed in Escherichia coli strain DH5α (Hanahan, 1983) and subsequently in Agrobacterium tumefaciens strain GV3101, carrying the helper plasmid pMP90RK (Koncz and Schell 1986). Transgenic plants were produced through the floral dip method as described by Clough and Bent (1998).
Protein Extraction and Immunoblot
Twenty milligrams of dry seeds were weighed and ground in liquid nitrogen and then extracted with 200 μL of a buffer containing 8 M Urea, 0.2% Triton X-100, 0.2% Sarkosyl, and 100 mM Tris-Cl, pH 7.5. After separation on a Tris-glycine SDS 4 to 12% polyacrylamide gradient gel (Anamed), according to Laemmli (1970), the proteins were blotted on a polyvinylidene fluoride membrane (Millipore) through semidry electrotransfer for 75 min at 2.8 mA/cm2. The immunological reactions of primary and secondary antibodies with the immobilized target proteins were done in 10 to 15 mL of a buffer containing 50 mM Tris-Cl, 150 mM NaCl, 0.25% Tween 20, and 5% skim milk. The primary polyclonal antibody was targeted to the amino end of the ABI3 protein and was kindly provided by Kazumi Nakabayashi and Eiji Nambara (RIKEN Plant Science Center, Yokohama, Japan). The secondary antibody was an alkaline phosphatase–conjugated anti rabbit-IgG. The blots were developed in a substrate buffer with 0.12 mM nitro blue tetrazolium, 0.15 mM 5-bromo-4-chloro-3-indolylphosphate, and 4 mM MgCl2 in 0.1 M Tris-Cl buffer, pH 9.5.
Phylogenetic Analysis
Protein sequences with the highest similarity to SUA were chosen after blasting its full-length protein sequence with the database of the National Center for Biotechnology Information. An unrooted phylogenetic analysis was conducted using MEGA version 4 (Tamura et al., 2007), using the alignment shown in Supplemental Data Set 1 online. Bootstrap test of phylogeny was performed using the neighbor-joining method from 1000 replications for each branch.
Yeast Two-Hybrid Analysis
SUA and U2AF65 (AT4G36690) coding sequences were cloned from Ler into pDONR 207 and pENTR/D-TOPO (Invitrogen), respectively, and then in the pACT2-gateway (GAL4 AD fusion) and pAS2-gateway (GAL4 BD fusion) vectors (modified from Clontech). Yeast two-hybrid assays were performed in yeast strain PJ69-4A (James et al., 1996) that was grown at 30°C. Yeast transformation was performed using a LiAc/SS carrier DNA/PEG method as described by Gietz et al. (1997). Cotransformed colonies were selected on synthetic dropout medium (SD) lacking Leu (L) and Trp (W). Interaction tests were performed on SD lacking L, W, and His (H) and on increasing concentrations of 3-aminotriazole (5, 10, and 20 mM). Interaction between SUA and U2AF65 was detected when SUA was expressed fused to GAL4 AD and U2AF65 to GAL4 BD. SUA fused to GAL4 BD could activate the transcription of the reporter gene alone.
FRET/FLIM Analysis
SUA and U2AF65 in entry clones were recombined into pENSG-YFP-gateway and pENSG-CFP-gateway destination vectors, respectively (Wenkel et al., 2006), producing YFP/CFP N-terminal fusions under control of the 35S promoter. FLIM was performed as described by Kwaaitaal et al. (2010). Arabidopsis protoplasts (from Col) were obtained from rosette leaves of 4-week-old plants grown under short-day conditions (8 h light/16 h darkness) and were transfected using 20 to 30 μg of pENSG_SUA_YFP and pENSG_U2AF65_CFP plasmids. Before imaging, they were incubated for 16 h in the light at 25°C.
Analysis of nuclear fluorescence was performed by confocal laser scanning microscopy using an LSM510 META confocal microscope (Zeiss). The histograms were analyzed with the SPC-Image 2.9.1 software (Becker and Hickl). Samples of cells expressing U2AF65-CFP alone were fitted with a single-exponential model to estimate the donor lifetime. For the analysis of protoplasts coexpressing U2AF65-CFP and SUA-YFP, a double exponential model was used. The second component was fixed to the average lifetime of the donor to the value found for U2AF65-CFP (2.5 ns) (Tonaco et al., 2006). About 15 nuclei per transformation were analyzed for two biological replicates.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers GU735482 and ADE44117 (At_SUA cds and protein), XP_002327561 (Pt_SUA homolog), CBI33920 (Vv_SUA homolog), EEC72564 (Os_SUA homolog), XP_001759968 (Pp_SUA homolog), XP_001691823 (Cr_SUA homolog), NP_001090434 (Xl_RBM5), AAH43058 (Mm_RBM5), NP_005769 (Hs_RBM5), and NP_849543 (At_FCA).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Analysis of sua-2 Plants Transformed with PSUA:sua-1 (#A2, #A12, and #A14).
Supplemental Figure 2. Analysis of abi3-5 sua-1 Plants Transformed with PSUA:SUA:GFP (#8 and #15).
Supplemental Table 1. Polymorphic Markers Used for Fine Mapping the sua-1 Mutation.
Supplemental Data Set 1. Text File of Alignment Used to Generate Figure 2C.
Acknowledgments
We thank Hetty Blankestijn-de Vries and Gerda Ruys for technical assistance during the selection of the suppressor line, Regina Gentges for the propagation of the plant material, Isabella Nougalli Tonaco and Mark Kwaaitaal for help with the confocal microscope, and Martijn van Zanten for comments on the manuscript. We also thank Kazumi Nakabayashi and Eiji Nambara for providing the ABI3 antibody. This work was supported by a Marie Curie Host Fellowship for Early Stage Researchers Training grant to M.S., by an Alexander von Humboldt postdoctoral fellowship to V.B., by Grant WBI.4737 from the Technology Foundation STW to E.J.M.C., and by the Max Planck Society.
References
- Ali G.S., Palusa S.G., Golovkin M., Prasad J., Manley J.L., Reddy A.S. (2007). Regulation of plant developmental processes by a novel splicing factor. PLoS One 2: e471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali G.S., Reddy A.S. (2008). Regulation of alternative splicing of pre-mRNAs by stresses. Nuclear pre-mRNA Processing in Plants, Reddy A.S.N., Golovkin M., (Berlin, Heidelberg, Germany: Springer; ), pp. 257–275 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Baud S., Boutin J.P., Miquel M., Lepiniec L., Rochat C. (2002). An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol. Biochem. 40: 151–160 [Google Scholar]
- Behzadnia N., Golas M.M., Hartmuth K., Sander B., Kastner B., Deckert J., Dube P., Will C.L., Urlaub H., Stark H., Lührmann R. (2007). Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 26: 1737–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bies-Etheve N., da Silva Conceicao A., Giraudat J., Koornneef M., Leon-Kloosterziel K., Valon C., Delseny M. (1999). Importance of the B2 domain of the Arabidopsis ABI3 protein for Em and 2S albumin gene regulation. Plant Mol. Biol. 40: 1045–1054 [DOI] [PubMed] [Google Scholar]
- Bonnal S., Martínez C., Förch P., Bachi A., Wilm M., Valcárcel J. (2008). RBM5/Luca-15/H37 regulates Fas alternative splic site pairing after exon definition. Mol. Cell 32: 81–95 [DOI] [PubMed] [Google Scholar]
- Bouveret E., Rigaut G., Shevchenko A., Wilm M., Séraphin B. (2000). A Sm-like protein complex that participates in mRNA degradation. EMBO J. 19: 1661–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C.G., Dreyfuss G. (1994). Conserved structures and diversity of functions of RNA-binding proteins. Science 265: 615–621 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Domon C., Lorkovic Z.J., Valcarcel J., Filipowicz W. (1998). Multiple forms of the U2 small nuclear ribonucleoprotein auxiliary factor U2AF subunits expressed in higher plants. J. Biol. Chem. 273: 34603–34610 [DOI] [PubMed] [Google Scholar]
- Edamatsu H., Kaziro Y., Itoh H. (2000). LUCA15, a putative tumour suppressor gene encoding an RNA-binding nuclear protein, is down-regulated in ras-transformed Rat-1 cells. Genes Cells 5: 849–858 [DOI] [PubMed] [Google Scholar]
- Ezcurra I., Wycliffe P., Nehlin L., Ellerstrom M., Rask L. (2000). Transactivation of the Brassica napus napin promoter by ABI3 requires interaction of the conserved B2 and B3 domains of ABI3 with different cis-elements: B2 mediates activation through an ABRE, whereas B3 interacts with an RY/G-box. Plant J. 24: 57–66 [DOI] [PubMed] [Google Scholar]
- Fan J., Niu X., Wang Y., Ren G., Zhuo T., Yang Y., Lu B.R., Liu Y. (2007). Short, direct repeats (SDRs)-mediated post-transcriptional processing of a transcription factor gene OsVP1 in rice (Oryza sativa). J. Exp. Bot. 58: 3811–3817 [DOI] [PubMed] [Google Scholar]
- Fedoroff N.V. (2002). RNA-binding proteins in plants: The tip of an iceberg? Curr. Opin. Plant Biol. 5: 452–459 [DOI] [PubMed] [Google Scholar]
- Filichkin S.A., Priest H.D., Givan S.A., Shen R., Bryant D.W., Fox S.E., Wong W.K., Mockler T.C. (2010). Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagete A.P., Riera M., Franco L., Rodrigo M.I. (2009). Functional analysis of the isoforms of an ABI3-like factor of Pisum sativum generated by alternative splicing. J. Exp. Bot. 60: 1703–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Hernandez M., et al. (2002). TAIR: A resource for integrated Arabidopsis data. Funct. Integr. Genomics 2: 239–253 [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Triggs-Raine B., Robbins A., Graham K.C., Woods R.A. (1997). Identification of proteins that interact with a protein of interest: applications of the yeast-two-hybrid system. Mol. Cell. Biochem. 172: 67–79 [PubMed] [Google Scholar]
- Giraudat J., Hauge B.M., Valon C., Smalle J., Parcy F., Goodman H.M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. (1983). Studies on transformation of E. coli with plasmids. J. Mol. Biol. 166: 557–580 [DOI] [PubMed] [Google Scholar]
- Hugouvieux V., Kwak J.M., Schroeder J.I. (2001). An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106: 477–487 [DOI] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R., Carlson M. (1997). The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol. Cell. Biol. 17: 2099–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerényi Z., Mérai Z., Hiripi L., Benkovics A., Gyula P., Lacomme C., Barta E., Nagy F., Silhavy D. (2008). Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J. 27: 1585–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Schell J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204: 383–396 [Google Scholar]
- Kroj T., Savino G., Valon C., Giraudat J., Parcy F. (2003). Regulation of storage protein gene expression in Arabidopsis. Development 130: 6065–6073 [DOI] [PubMed] [Google Scholar]
- Kwaaitaal M., Keinath N.F., Pajonk S., Biskup C., Panstruga R. (2010). Combined bimolecular fluorescence complementation and Förster resonance energy transfer reveals ternary SNARE complex formation in living plant cells. Plant Physiol. 152: 1135–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lopato S., Kalyna M., Dorner S., Kobayashi R., Krainer A.R., Barta A. (1999). atSRp30, one of two SF2/ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev. 13: 987–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., McLachlin D.T., Chait B.T., Chua N.H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Lorković Z.J., Barta A. (2004). Compartmentalization of the splicing machinery in plant cell nuclei. Trends Plant Sci. 9: 565–568 [DOI] [PubMed] [Google Scholar]
- Lorković Z.J., Kirk D.A.W., Lambermon M.H.L., Filipowicz W. (2000). Pre-mRNA splicing in higher plants. Trends Plant Sci. 5: 160–167 [DOI] [PubMed] [Google Scholar]
- McCarty D.R., Hattori T., Carson C.B., Vasil V., Lazar M., Vasil I.K. (1991). The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66: 895–905 [DOI] [PubMed] [Google Scholar]
- McGlincy N.J., Smith C.W. (2008). Alternative splicing resulting in nonsense-mediated mRNA decay: What is the meaning of nonsense? Trends Biochem. Sci. 33: 385–393 [DOI] [PubMed] [Google Scholar]
- McKibbin R.S., Wilkinson M.D., Bailey P.C., Flintham J.E., Andrew L.M., Lazzeri P.A., Gale M.D., Lenton J.R., Holdsworth M.J. (2002). Transcripts of Vp-1 homeologues are misspliced in modern wheat and ancestral species. Proc. Natl. Acad. Sci. USA 99: 10203–10208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag R., Maity M.K., Dasgupta M. (2005). Dual DNA binding property of ABA insensitive 3 like factors targeted to promoters responsive to ABA and auxin. Plant Mol. Biol. 59: 821–838 [DOI] [PubMed] [Google Scholar]
- Nakamura S., Lynch T.J., Finkelstein R.R. (2001). Physical interactions between ABA response loci of Arabidopsis. Plant J. 26: 627–635 [DOI] [PubMed] [Google Scholar]
- Nambara E., Keith K., McCourt P., Naito S. (1994). Isolation of an internal deletion mutant of the Arabidopsis thaliana ABI3 gene. Plant Cell Physiol. 35: 509–513 [PubMed] [Google Scholar]
- Ooms J., Leon-Kloosterziel K.M., Bartels D., Koornneef M., Karssen C.M. (1993). Acquisition of desiccation tolerance and longevity in seeds of Arabidopsis thaliana (a comparative study using abscisic acid-insensitive abi3 mutants). Plant Physiol. 102: 1185–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer D.G., Herman P.L., Esch J., Sivakumaran S., Marks M.D. (1991). A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67: 483–493 [DOI] [PubMed] [Google Scholar]
- Parcy F., Valon C., Kohara A., Miséra S., Giraudat J. (1997). The ABSCISIC ACID-INSENSITIVE 3 (ABI3), FUSCA 3 (FUS3) and LEAFY COTYLEDON 1 (LEC1) loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell 9: 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perruc E., Kinoshita N., Lopez-Molina L. (2007). The role of chromatin-remodeling factor PKL in balancing osmotic stress responses during Arabidopsis seed germination. Plant J. 52: 927–936 [DOI] [PubMed] [Google Scholar]
- Reddy A.S.N. (2007). Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 58: 267–294 [DOI] [PubMed] [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Shirley B.W., Kubasek W.L., Storz G., Bruggemann E., Koornneef M., Ausubel F.M., Goodman H.M. (1995). Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 8: 659–671 [DOI] [PubMed] [Google Scholar]
- Sickmier E.A., Frato K.E., Shen H., Paranawithana S.R., Green M.R., Kielkopf C.L. (2006). Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol. Cell 23: 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson C.G., Jennings S.N., Clark G.P., Thow G., Brown J.W. (2004). Dual functionality of a plant U-rich intronic sequence element. Plant J. 37: 82–91 [DOI] [PubMed] [Google Scholar]
- Simpson G.G., Filipowicz W. (1996). Splicing of precursors to mRNA in higher plants: Mechanism, regulation and sub-nuclear organisation of the spliceosomal machinery. Plant Mol. Biol. 32: 1–41 [DOI] [PubMed] [Google Scholar]
- Suzuki M., Kao C.Y., McCarty D.R. (1997). The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 9: 799–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tanabe N., Yoshimura K., Kimura A., Yabuta Y., Shigeoka S. (2007). Differential expression of alternatively spliced mRNAs of Arabidopsis SR protein homologs, atSR30 and atSR45a, in response to environmental stress. Plant Cell Physiol. 48: 1036–1049 [DOI] [PubMed] [Google Scholar]
- To A., Valon C., Savino G., Guilleminot J., Devic M., Giraudat J., Parcy F. (2006). A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18: 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonaco I.A.N., Borst J.W., De Vries S.C., Angenent G.C., Immink R.G.H. (2006). In vivo imaging of MADS box transcription factor interactions. J. Exp. Bot. 57: 33–42 [DOI] [PubMed] [Google Scholar]
- van Hoof A., Green P.J. (2006). NMD in plants. Nonsense-Mediated mRNA Decay. Maquat L., (New York: Landes Bioscience; ), pp. 167–172 [Google Scholar]
- Wang B.B., Brendel V. (2006). Genomewide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. USA 103: 7175–7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkel S., Turck F., Singer K., Gissot L., Le Gourrierec J., Samach A., Coupland G. (2006). CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18: 2971–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P.D., Patton J.G., Green M.R. (1992). Cloning and domain structure of the mammalian splicing factor U2AF. Nature 355: 609–614 [DOI] [PubMed] [Google Scholar]
- Zhang X., Garreton V., Chua N.H. (2005). The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 19: 1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.N., Mount S.M. (2009). Two alternatively spliced isoforms of the Arabidopsis SR45 protein have distinct roles during normal plant development. Plant Physiol. 150: 1450–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]