Introduction
Since the discovery and sequencing of 6-deoxyerythronolide B synthase 20 years ago, this exceptionally large, multifunctional protein remains the paradigm for the understanding of the structure and biochemical function of modular polyketide synthases. The broad-spectrum macrolide antibiotic erythromycin is one of several hundred closely related, branched chain, polyoxygenated polyketides, many of which are widely used in human and veterinary medicine as antibiotic, immunosuppressant, antitumor, antifungal, and antiparasitic agents. The multistep assembly of the parent macrocyclic aglycon, 6-deoxyerythronolide B (6-dEB),2 is controlled by a large (2 MDa) modular protein known as 6-dEB synthase (DEBS) (1–4). The biosynthetic intermediates never leave the polyketide synthase (PKS) but are passed along the DEBS assembly line from one acyl carrier protein (ACP) domain to the next. In fact, DEBS has served as the prototype of modular PKS gene clusters, dozens of which of both known and unknown function have now been sequenced from bacterial sources (5–7).
Each homodimeric DEBS subunit contains two 160–200-kDa protein modules, each responsible for a single round of polyketide chain extension and functional group modification. Within each module are several catalytic domains of 100–400 amino acids each that are analogous in structure, function, and organization to the corresponding fatty acid synthase (FAS) components (8–10). All six DEBS modules contain three core domains consisting of 1) a β-ketoacyl-ACP synthase (the ketosynthase (KS) domain) that catalyzes the key polyketide chain-building reaction, a decarboxylative condensation of a methylmalonyl-ACP building block with the polyketide chain provided by the upstream PKS module (see Figs. 1 and 2); 2) an acyltransferase (AT) domain that specifically loads the methylmalonyl-CoA extender unit onto the flexible 18-Å phosphopantetheine arm of the ACP domain; and 3) the ACP domain itself, which carries the polyketide biosynthetic intermediates from domain to domain and then delivers the resulting product to the KS domain of the downstream module. Additional FAS-like domains are responsible for modification of the oxidation state and stereochemistry of the growing ACP-bound intermediates: a β-ketoacyl-ACP reductase (KR) domain, a dehydratase (DH) domain, and an enoyl reductase (ER) domain. At the N terminus of the most upstream module is a loading didomain that primes the KS domain of module 1 with the propionyl starter unit. Finally, cyclization of the full-length macrocyclic polyketide and release of 6-dEB are controlled by a dedicated thioesterase domain located at the C terminus of the most downstream module.
FIGURE 1.
Modular organization of DEBS. In addition to the KS, AT, and ACP domains, individual extension modules carry varying combinations of KR, DH, and ER domains. The loading didomain primes DEBS module 1 with the propionyl starter unit, and the thioesterase (TE) domain at the C terminus of module 6 catalyzes release and cyclization of the full-length polyketide to give the parent macrolide aglycon, 6-dEB. Two dedicated P450 oxygenases, two glycosyltransferases, and a methyltransferase then generate the mature antibiotic, erythromycin A. (2S)-MeMal-CoA, (2S)-methylmalonyl-CoA.
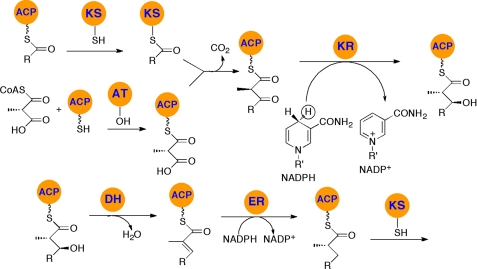
FIGURE 2.
Biochemical function of individual PKS domains, illustrated by polyketide chain extension and functional group modification mediated by the domains of DEBS module 4. The AT domain loads the (2S)-methylmalonyl group onto the pantetheinyl side chain of the ACP domain, whereas the KS domain catalyzes self-acylation of its active-site Cys residue by the tetraketide chain donated by the upstream ACP domain. The KS domain then catalyzes a decarboxylative condensation to generate a d-2-methyl-3-ketoacyl-ACP intermediate, which then undergoes successive KR-catalyzed reduction, DH-catalyzed dehydration, and ER-catalyzed reduction to the reduced pentaketide, which is then transferred to the KS domain of DEBS module 5. Configurational assignments are based on the known or predicted stereochemistry of individual PKS reactions.
Programming of Polyketide Biosynthesis
The chain length, substitution pattern, and oxidation level of the initially generated, full-length heptaketide 6-dEB are the direct consequence of the number of DEBS modules as well as the domain composition of each module (Fig. 1) (8–11). The striking colinearity between the organization of the constituent biosynthetic domains encoded by the DEBS genes and the order of the biochemical reactions that generate 6-dEB had originally encouraged the hope that the sequence of any newly discovered PKS would allow the ready prediction of the chemical structure of the derived polyketide product. Unfortunately, such straightforward correlations have proven to be elusive. Unlike the unidirectionally transcribed DEBS cluster, a variety of modular PKS genes are either divergently or convergently transcribed such that the sequential order of the open reading frames found in the genome does not correspond to the temporal order in which they exercise their biochemical function (12). Recent progress in understanding the structural basis for pairwise interactions of the docking domains that direct the proper intermodular transfer of polyketide intermediates may allow better correlations of deduced sequences and temporal function (13–16). Further obscuring an overly simplistic linear interpretation is the presence in individual PKS modules of inactive DH or ER domains, whereas an additional complication comes from PKS modules that lack integrated AT domains altogether and therefore require the in trans AT-catalyzed priming of ACP domains with the requisite extender units. Broken modules in which the domains mediating a single round of chain elongation are located in adjacent PKS modules, as well as the phenomena of module skipping or iteration (17), further obscure the relationship of PKS gene sequence and biochemical function. The existence of unusual polyketide-building blocks such as methoxymethylmalonyl-CoA and hydroxymethylmalonyl-CoA and the recognition of polyketide chain branching have added additional layers of biochemical complexity (18). Despite promising new bioinformatics approaches (15), knowledge of the specific structure of the eventually formed polyketide has therefore turned out to be critical to the proper assignment of PKS organization and function. Nonetheless, the well studied molecular genetics and biochemistry of DEBS continue to provide the dominant paradigm for the understanding of modular PKS biochemistry (17).
The 14-member ring of 6-dEB displays an impressive aggregate of 10 stereogenic centers. Similar stereochemical patterns are found in a wide range of closely related 12-, 14-, and 16-member ring macrolides (19, 20). Only recently has the mechanistic basis for the remarkable stereochemical complexity of the polyketides generated by DEBS and other modular PKS proteins begun to be unraveled.
PKS Modules and Domains
Heterologous expression of engineered PKS modules has played a central role in the elucidation of the biochemical mechanisms, substrate specificity, protein-protein recognition features, and structures of DEBS modules and domains (21, 22). DEBS modules, including those with inactivated, mutated, or deleted domains, have been expressed in both native and heterologous bacterial hosts, with the structure and stereochemistry of the resulting polyketide products serving as a readout of the role of individual modules or domains (21–23). Recent progress in the development of large deletion mutants of Streptomyces avermitilis as hosts for the heterologous expression of biosynthetic gene clusters (24) should provide a powerful new tool for the assignment of biochemical function to cryptic PKS gene clusters and the discovery of new polyketides.
Individual modules or domains can be expressed using both native and heterologous Streptomyces hosts as well as strains of Escherichia coli harboring a chromosomally integrated copy of the sfp gene from Bacillus subtilis that encodes the surfactin phosphopantetheinyl transferase (25) necessary to convert the ACP domains to their active form (26). Investigations of the specificity and function of PKS domains have frequently utilized chimeric modules harboring engineered combinations of domains derived from a variety of parent modules (22, 27–29). The labor-intensive construction of such hybrid systems has for the most part relied on domain boundaries deduced from multiple sequence alignments. Unfortunately, the resulting hybrids have often suffered from a severe loss in overall catalytic efficiency compared with wild-type modules. Recently, detailed structural studies of PKS modular components have revealed highly conserved functional domain boundaries that can now be exploited for the expression of individual active recombinant PKS domains (30–32). Not only are these stand-alone domains excellent vehicles for protein structural and mechanistic study, but reconstituted mixtures of these PKS domains allow investigation of reaction mechanisms, protein-small molecule specificity, and the role of protein-protein recognition in the programming and control of PKS function.
KS Domains
The irreversible KS-catalyzed chain elongation reaction involves the initial formation of a covalent acyl-enzyme intermediate between the growing polyketide chain, donated by the ACP of the upstream module, and the active-site Cys thiol of the KS, followed by a decarboxylative condensation of this acyl thioester with methylmalonyl-ACP (33, 34) with inversion of configuration at C2 of the (2S)-methylmalonyl moiety to produce the corresponding d-2-methyl-3-ketoacyl-ACP intermediate (Fig. 2) (35). Eventual generation of the l-methyl configuration found at C8 and C12 of 6-dEB requires an epimerization reaction at some stage subsequent to KS-catalyzed chain elongation. Besides the active-site Cys (36), each KS domain harbors a pair of universally conserved His residues (for example, His334 and His374 in DEBS KS5) that facilitate the decarboxylation and condensation reactions (37, 38). (2S,3R)-2-Methyl-3-hydroxypentanoyl-N-acetylcysteamine thioester (NDK-SNAc), the N-acetylcysteamine acyl thioester analog of the natural ACP-bound syn-diketide substrate of DEBS module 2, can conveniently be used as a surrogate substrate for DEBS KS domains (39, 40).
Structures of the DEBS [KS5][AT5] and [KS3][AT3] Didomains
The 2.7-Å x-ray crystal structure of a 194-kDa homodimeric KS and AT didomain of DEBS module 5, [KS5][AT5], provided the first direct insights into the topology and three-dimensional organization of a modular PKS (Fig. 3a) (31). These findings have been reinforced by the 2.6-Å x-ray crystal structure of the homologous 190-kDa homodimeric [KS3][AT3] didomain from DEBS module 3 with the inhibitor cerulenin covalently bound to the active-site Cys of the KS3 domain (32). In both structures, the homodimeric KS domains and the monomeric AT domains at the end of each protein arm closely resemble the structures of previously characterized type II FAS- and PKS-derived KS and AT domains. The [KS5][AT5] homodimer carries the N-terminal docking domain with the homodimeric coiled-coil structure previously deduced from mutant complementation and NMR studies (13, 14). Although the truncated [KS3][AT3] construct lacks the corresponding N-terminal docking domain, two well organized interdomain linker regions are apparent in both structures: 1) a highly ordered KS-to-AT linker domain with a previously uncharacterized protein fold and 2) a 30-amino acid post-AT linker appended to the C terminus of the AT domain that wraps back over both the AT domain and the KS-to-AT linker and terminates on the face of the KS domain distal to the N-terminal docking domain. The relative orientations of the paired AT and KS domains and of the post-AT linker support earlier observations that KS and ACP domains from opposite subunits of a homodimeric module preferentially interact (36, 41). Computational docking has predicted that the ACP should dock to the KS domain within the deep cleft between the KS and AT domains, with interactions that span across both subunits of the KS homodimer (32). The two crystal structures also revealed that the active-site Cys199 of the KS is separated from the active-site Ser642 of the nearest AT domain by ∼80 Å. This distance is obviously too great to be spanned by a statically anchored, fully extended 18-Å phosphopantetheine arm of the ACP domain. Thus, the interaction of the ACP domain with each of the catalytic domains of a modular PKS must involve substantial segmental motion of the entire ACP domain, rather than simple reorientation of the pantetheinyl prosthetic group, contrary to the classical “swinging arm” model of PKS and FAS action. Similar conclusions have been drawn from the 3.2-Å structure of the complete pig FAS protein (42, 43). The similarity of these two structures suggests a model for the entire DEBS module 4 with an analogous X-shaped architecture (Fig. 3e) (2).
FIGURE 3.
Structures of DEBS catalytic domains. a, DEBS [KS5][AT5] didomain, including the homodimeric KS domain (light blue and dark blue), AT domains (green), N-terminal docking domain (orange-brown), KS-AT linkers (yellow), and post-AT linkers (red). b, DEBS KR1, with the structural domain (cyan), catalytic domain (blue), and Leu-Asp-Asp (LDD) loop. c, DEBS DH4, with the double hot dog fold and catalytic His-Asp dyad. d, DEBS apo-ACP2. e, predicted organization and topology of DEBS module 4, analogous to the structure of porcine FAS. Each ACP domain (not shown) must be able to interact with the individual KS, AT, KR, DH, and ER domains. ppant, phosphopantetheine; act, active.
The recombinant DEBS [KS1][AT1], [KS3][AT3], and [KS5][AT5] didomains have been used to study polyketide chain elongation in combination with both homologous and heterologous ACP domains (30, 44). Each [KS][AT] didomain plus an ACP domain can be incubated with NDK-SNAc and methylmalonyl-CoA to give a triketide ketolactone after hydrolytic release (Fig. 4a). The [KS3][AT3] and [KS6][AT6] didomains both exhibit a distinct ACP recognition profile, with strong preference for the ACP domain from their parent modules. Using chemoenzymatically prepared methylmalonyl-ACP and malonyl-ACP as substrates, KS3 has a 4:1 preference for its native methylmalonyl extender unit, whereas KS6 shows a smaller 1.5:1 preference. These findings highlight the key gatekeeping role of the AT domains that must charge the ACP with the correct chain extender unit while demonstrating the intrinsic ability of KS domains to utilize unnatural chain extension units. The reconstituted DEBS domain mixtures allow quantitative kinetic comparisons that are not possible using intact chimeric modules (45). The addition of native or foreign C-terminal docking domains to the ACP domains has negligible effect on intramodular KS-ACP recognition during KS-catalyzed condensation. By contrast, intermodular self-priming of DEBS KS(n) domains by acyl donors carried on upstream ACP(n−1) domains is relatively insensitive to overall ACP structure but strongly dependent on a proper match of the complementary C-terminal ACP(n−1) and N-terminal KS(n) docking domains.
FIGURE 4.
Stereochemistry of DEBS KR-catalyzed reduction and methyl group epimerization using mixtures of individual recombinant domains and didomains and gas chromatography-mass spectrometry analysis of the derived triketide lactones (a) and diketide acids (b).
The [KS3][AT3] didomain can also be dissected into its component KS3 and AT3 domains (46). The recombinant AT3 domain, either with or without the appended post-AT linker, can catalyze both self-acylation by exogenous methylmalonyl-CoA and transfer of this methylmalonyl moiety to the ACP3 domain. Similarly, the dissected KS3 domain undergoes self-catalyzed acylation by NDK-SNAc. By contrast, formation of the triketide ketolactone by KS-catalyzed chain elongation requires that the added AT3 domain carry an appended post-AT linker. The extensive interaction between this post-AT linker and the KS domain that is observed in both the [KS3][AT3] and [KS5][AT5] crystal structures therefore must play a critical functional role in proper alignment of the KS and AT domains and productive interaction with the ACP-bound substrate.
AT Domains
The AT domain of each DEBS module plays a special role as gatekeeper, being strictly specific for (2S)-methylmalonyl-CoA (22, 47, 48). The universally conserved active-site Ser in each DEBS AT domain, found in a signature GHSQGE motif, is part of a canonical catalytic dyad in partnership with the conserved active-site His. The active-site His is believed to act as a general base that activates nucleophilic attack by the Ser hydroxyl on the acyl group of the methylmalonyl thioester and then assists in the transfer of the methylmalonyl-O-Ser intermediate to the phosphopantetheinate side chain of the ACP domain (Fig. 2). The AT domain contains an α/β-hydrolase-like core domain and an appended smaller subdomain with a ferredoxin-like structure (Fig. 3a) (31, 32). The AT domains from each of the six DEBS modules are completely specific for (2S)-methylmalonyl-CoA (47). By contrast, the AT domain in the AT-ACP loading didomain strongly prefers propionyl-CoA but can also use acetyl-CoA to prime the paired ACP domain (49).
KR Domains
The DEBS KR domains, in common with all other PKS and FAS KR proteins, belong to the large family of short chain dehydrogenase/reductases, all of which harbor a conserved active-site Ser-Tyr-Lys triad responsible for binding and activation of the target carbonyl group of the β-ketoacyl-ACP substrate while simultaneously orienting and activating the nicotinamide ring of the NADPH cofactor (50). The stereochemical course of each β-ketoacyl-ACP reduction is an intrinsic property of the responsible KR domain and is independent of either modular context or substrate structure, including chain length and substitution pattern (51). DEBS KR1, KR2, KR5, and KR6 all utilize the H4si hydride of the NADPH cofactor (Fig. 2), in common with FAS KR domains (52, 53). A conserved Leu-Asp-Asp triad found in many PKS KR domains is strongly correlated with the generation of d-hydroxy products (54, 55). Crystal structures of the DEBS KR1 domain and the homologous KR1 domain from module 1 of tylactone synthase have indicated that the loop harboring this Leu-Asp-Asp motif may play a key role in mediating proper access and orientation of the phosphopantetheinyl-bound β-ketoacyl thioester substrate in the common substrate-binding groove (Fig. 3b) (56, 57). In KR domains such as DEBS KR2, KR5, and KR6, which lack this Leu-Asp-Asp motif but harbor instead a conserved Trp at an alternative site, the phosphopantetheinyl-bound β-ketoacyl substrate is thought to be constrained to enter the substrate-binding groove from the opposite end, giving rise to the observed l-hydroxy product. Keatinge-Clay (57) and Starcevic et al. (58) have also proposed that the redox-inactive DEBS KR30 domains may be responsible for the epimerization of the 2-methyl group to give the l-2-methyl-3-ketoacyl-ACP tetraketide intermediate produced by DEBS module 3.
The solution of the 1.8-Å structure of the DEBS KR1 domain also resulted in a major redefinition of the KR domain boundaries that had been previously erroneously inferred solely from multiple sequence alignments (56). Each monomeric KR domain consists of two principal subdomains: an N-terminal structural subdomain and a C-terminal catalytic subdomain, each with a modified Rossmann fold (Fig. 3b). The structural subdomain, which is thought to stabilize the catalytic subdomain, lacks the characteristic GXGXXG NADPH-binding motif but may provide a docking site for the ACP domain (56, 57).
Stereochemistry of KR-catalyzed 3-Ketoacyl-ACP Reductions
The reduction of the 2-methyl-3-ketoacyl-ACP substrate by a KR domain also fixes the configuration of the 2-methyl substituent. Recombinant DEBS KR1 reduces racemic 2-methyl-3-ketopentanoyl-SNAc exclusively to a syn-(2S,3R)-2-methyl-3-hydroxy diketide (NDK-SNAc), thereby demonstrating that this reductase is completely specific not only for reduction on the re-face of the ketone carbonyl but also for the natural l-configuration of the adjacent 2-methyl group, consistent with the overall stereospecificity of DEBS module 1 (59, 60).
Co-incubation of recombinant DEBS KR2 or KR6 plus NADPH with reconstituted DEBS [KS1][AT1] + ACP1, [KS3][AT3] + ACP3, or [KS6][AT6] + ACP6 in the presence of NDK-SNAc and methylmalonyl-CoA gives the corresponding reduced triketide lactone (Fig. 4a) (61, 62). DEBS KR2, KR5, and KR6 are completely diastereoselective, generating a product with the identical (2R,3S)-2-methyl-3-hydroxy stereochemistry as that of the parent DEBS modules from which they had been derived (Figs. 1 and 4a). Reductive quenching with NaBH4 confirmed that DEBS [KS1][AT1], [KS3][AT3], and [KS6][AT6] all generated exclusively the unepimerized d-2-methyl-3-ketoacyl-ACP triketide intermediate (61, 62). Unexpectedly, this ACP-bound 2-methyl-3-ketoacyl triketide thioester was configurationally remarkably stable, undergoing <5–15% epimerization even after 1 h, >15–45-fold slower than the measured rate for buffer-catalyzed deuterium exchange of untethered methyl-2-methyl acetoacetate. This remarkable enhancement in configurational stability is thought to be due to sequestration of the bound polyketide in the ACP cleft between helices 2 and 3 and consequent conformational restriction that significantly reduces the acidity of the H2 proton of the 3-ketoacyl-ACP substrate.
When propionyl-SNAc is used as the primer for DEBS [KS1][AT1], [KS3][AT3], or [KS6][AT6] and their cognate ACP domains, DEBS KR1 generates only the reduced and epimerized (2S,3R)-l-2-methyl-d-3-hydroxypentanoyl-ACP product (62). Alternatively, incubation with DEBS KR6 gives the predicted (2R,3S)-diketide acid. For each reconstituted incubation mixture, the absolute and relative stereochemistry of each chain elongation and reduction product is strictly correlated with the intrinsic hydroxyl and methyl group stereospecificity of the particular DEBS KR domain, independent of whether the KS domain utilized is derived from a module that normally produces a reduced unepimerized product (DEBS KS6), an unreduced epimerized product (DEBS KS3), or a reduced epimerized product (DEBS KS1). These results are all consistent with KR1-catalyzed epimerization following KS-catalyzed chain elongation. The mechanism of this epimerization, including the amino acid residues that are responsible for this process, is currently unknown.
DH Domains
Although the vast majority of polyketides generated by modular PKSs contain one or more double bonds, in DEBS only module 4 harbors a DH domain. The transiently generated enoylacyl-ACP pentaketide produced by DEBS DH4 is ordinarily not observed due to coupled reduction by the paired ER4 enoyl reductase domain. Disruption of the NADPH-binding motif of this ER4 domain in the complete DEBS protein resulted in the accumulation of a derivative of the corresponding (E)-Δ6,7-anhydro-6-dEB (63). The DH4-catalyzed dehydration is expected to take place with syn-stereochemistry by analogy to the known stereospecificity of the DH domain of yeast FAS (Fig. 2) (64). The DEBS DH4 domain carries a conserved HXXXGXXXXP motif that carries part of the established His-Asp catalytic dyad, in which the active-site His acts as a general base to remove the C2 proton, whereas the Asp donates a proton to promote departure of the 3-hydroxyl group (65). Indeed, the H2409F mutant of DEBS module 4 did not produce 6-dEB (66). The 1.85-Å structure of recombinant DEBS DH4 displays the double hot dog fold also observed in the DH domain of pig FAS (Fig. 3c) (67). The active-site His and Asp dyad of DEBS DH4 can be accessed by a tunnel leading from the surface of the protein adjacent to a proposed ACP-binding region.
ER Domains
The ER domain of DEBS module 4 is inserted between the structural and catalytic subdomains of the KR4 domain. The DEBS ER4 domain reduces its unsaturated pentaketide substrate by net conjugate addition of a hydride from NADPH to C3 of the E-enoylacyl-ACP, with addition of a proton to C2. Disruption of the putative NADPH-binding motif of ER4, 2964HAAAGGVGMA, by the double mutant HAAASPVGMA abolishes enoyl reduction (63). An active-site Tyr influences the stereochemistry of the reduction (68).
ACP Domains
The DEBS ACP domains serve as carriers of the growing polyketide chain, which is covalently tethered as the acyl thioester to the phosphopantetheinyl prosthetic group that is in turn attached to the universally conserved serine found within the LGXDS motif of each ACP. It is now evident that the ACP domains of modular PKSs and FASs must undergo considerable segmental motion to access the active sites of the individual domains (31, 32, 42, 43, 69). The NMR solution structure of the 10-kDa recombinant DEBS apo-ACP2 reveals a three-helix bundle connected by two loops, with an additional short helix in the second loop also contributing to core helical packing (Fig. 3d) (70). The overall protein surface of DEBS ACP2 appears to be less charged compared with the type II FAS homologs, which are more highly acidic. Homology models for each of the five remaining DEBS ACP domains have been calculated (70). Although the overall topology is largely conserved, there are much greater differences in the calculated electrostatic potential surfaces, which may account for the observed discrimination in functional interaction of DEBS ACP domains with their cognate KS domains.
Unanswered Questions
Twenty years after the sequencing of the 6-deoxyerythronolide synthase, we now have a broad and deep understanding of the fundamentals of the molecular enzymology and structural biology of DEBS and related modular PKS megaenzymes. Numerous unanswered questions remain. 1) What is the mechanistic basis for the KR-catalyzed methyl group epimerizations that take place during the formation of 6-dEB and other polyketides? 2) What is the balance between substrate specificity and protein-protein recognition in the programming of polyketide biosynthesis? 3) What are the structural and dynamic details of the interaction of KS acceptor domains and upstream ACP donor domains? 4) What is the structural basis for the interaction of ACP domains with the constituent KS, AT, KR, DH, and ER domains of each module, and what are the detailed dynamics by which charged ACP domains find and interact with each β-carbon-processing domain? 5) What is the molecular basis for programming and specificity of type I PKSs that vary from the canonical DEBS paradigm? Unraveling the workings of these complex and fascinating synthetic machines will ultimately require the development of new physical-biochemical techniques and the continued collaboration of chemists, biochemists, microbial geneticists, and structural biologists.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants GM22172 (to D. E. C.) and CA87394 (to C. Khosla). This is the third article in the Thematic Minireview Series on Antibacterial Natural Products. This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- 6-dEB
- 6-deoxyerythronolide B
- DEBS
- 6-dEB synthase
- PKS
- polyketide synthase
- ACP
- acyl carrier protein
- FAS
- fatty acid synthase
- KS
- β-ketoacyl-ACP synthase
- AT
- acyltransferase
- KR
- β-ketoacyl-ACP reductase
- DH
- dehydratase
- ER
- enoyl reductase
- NDK-SNAc
- natural diketide (2S,3R)-2-methyl-3-hydroxypentanoyl-N-acetylcysteamine thioester.
REFERENCES
- 1.Hopwood D. A. (ed) (2009) Methods in Enzymology: Complex Enzymes in Microbial Natural Product Biosynthesis, part B: Polyketides, Aminocoumarins, and Carbohydrates, Vol. 459, Academic Press, New York: [DOI] [PubMed] [Google Scholar]
- 2.Khosla C., Tang Y., Chen A. Y., Schnarr N. A., Cane D. E. (2007) Annu. Rev. Biochem. 76, 195–221 [DOI] [PubMed] [Google Scholar]
- 3.Khosla C., Kapur S., Cane D. E. (2009) Curr. Opin. Chem. Biol. 13, 135–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khosla C. (2009) J. Org. Chem. 74, 6416–6420 [DOI] [PubMed] [Google Scholar]
- 5.Meier J. L., Burkart M. D. (2009) Chem. Soc. Rev. 38, 2012–2045 [DOI] [PubMed] [Google Scholar]
- 6.Van Lanen S. G., Shen B. (2008) Curr. Opin. Drug Discov. Devel. 11, 186–195 [PubMed] [Google Scholar]
- 7.Hertweck C. (2009) Angew. Chem. Int. Ed. Engl. 48, 4688–4716 [DOI] [PubMed] [Google Scholar]
- 8.Donadio S., Katz L. (1992) Gene 111, 51–60 [DOI] [PubMed] [Google Scholar]
- 9.Bevitt D. J., Cortes J., Haydock S. F., Leadlay P. F. (1992) Eur. J. Biochem. 204, 39–49 [DOI] [PubMed] [Google Scholar]
- 10.Caffrey P., Bevitt D. J., Staunton J., Leadlay P. F. (1992) FEBS Lett. 304, 225–228 [DOI] [PubMed] [Google Scholar]
- 11.Cortes J., Haydock S. F., Roberts G. A., Bevitt D. J., Leadlay P. F. (1990) Nature 348, 176–178 [DOI] [PubMed] [Google Scholar]
- 12.Ikeda H., Omura S. (1997) Chem. Rev. 97, 2591–2610 [DOI] [PubMed] [Google Scholar]
- 13.Broadhurst R. W., Nietlispach D., Wheatcroft M. P., Leadlay P. F., Weissman K. J. (2003) Chem. Biol. 10, 723–731 [DOI] [PubMed] [Google Scholar]
- 14.Kumar P., Li Q., Cane D. E., Khosla C. (2003) J. Am. Chem. Soc. 125, 4097–4102 [DOI] [PubMed] [Google Scholar]
- 15.Yadav G., Gokhale R. S., Mohanty D. (2009) PLoS Comput. Biol. 5, e1000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsuji S. Y., Cane D. E., Khosla C. (2001) Biochemistry 40, 2326–2331 [DOI] [PubMed] [Google Scholar]
- 17.Katz L. (2009) Methods Enzymol. 459, 113–142 [DOI] [PubMed] [Google Scholar]
- 18.Chan Y. A., Podevels A. M., Kevany B. M., Thomas M. G. (2009) Nat. Prod. Rep. 26, 90–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Celmer W. D. (1965) J. Am. Chem. Soc. 87, 1801–1802 [DOI] [PubMed] [Google Scholar]
- 20.Celmer W. D. (1971) Pure Appl. Chem. 28, 413–453 [DOI] [PubMed] [Google Scholar]
- 21.Staunton J., Weissman K. J. (2001) Nat. Prod. Rep. 18, 380–416 [DOI] [PubMed] [Google Scholar]
- 22.Khosla C., Gokhale R. S., Jacobsen J. R., Cane D. E. (1999) Ann. Rev. Biochem. 68, 219–253 [DOI] [PubMed] [Google Scholar]
- 23.Gokhale R. S., Tsuji S. Y., Cane D. E., Khosla C. (1999) Science 284, 482–485 [DOI] [PubMed] [Google Scholar]
- 24.Komatsu M., Uchiyama T., Omura S., Cane D. E., Ikeda H. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 2646–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfeifer B. A., Admiraal S. J., Gramajo H., Cane D. E., Khosla C. (2001) Science 291, 1790–1792 [DOI] [PubMed] [Google Scholar]
- 26.Walsh C. T., Gehring A. M., Weinreb P. H., Quadri L. E., Flugel R. S. (1997) Curr. Opin. Chem. Biol. 1, 309–315 [DOI] [PubMed] [Google Scholar]
- 27.Kellenberger L., Galloway I. S., Sauter G., Böhm G., Hanefeld U., Cortés J., Staunton J., Leadlay P. F. (2008) ChemBioChem 9, 2740–2749 [DOI] [PubMed] [Google Scholar]
- 28.Menzella H. G., Reid R., Carney J. R., Chandran S. S., Reisinger S. J., Patel K. G., Hopwood D. A., Santi D. V. (2005) Nat. Biotechnol. 23, 1171–1176 [DOI] [PubMed] [Google Scholar]
- 29.Kim B. S., Sherman D. H., Reynolds K. A. (2004) Prot. Eng. Des. Sel. 17, 277–284 [DOI] [PubMed] [Google Scholar]
- 30.Kim C. Y., Alekseyev V. Y., Chen A. Y., Tang Y., Cane D. E., Khosla C. (2004) Biochemistry 43, 13892–13898 [DOI] [PubMed] [Google Scholar]
- 31.Tang Y., Kim C. Y., Mathews I. I., Cane D. E., Khosla C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11124–11129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang Y., Chen A. Y., Kim C. Y., Cane D. E., Khosla C. (2007) Chem. Biol. 14, 931–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sedgwick B., Cornforth J. W. (1977) Eur. J. Biochem. 75, 465–479 [DOI] [PubMed] [Google Scholar]
- 34.Sedgwick B., Cornforth J. W., French S. J., Gray R. T., Kelstrup E., Willadsen P. (1977) Eur. J. Biochem. 75, 481–495 [DOI] [PubMed] [Google Scholar]
- 35.Weissman K. J., Timoney M., Bycroft M., Grice P., Hanefeld U., Staunton J., Leadlay P. F. (1997) Biochemistry 36, 13849–13855 [DOI] [PubMed] [Google Scholar]
- 36.Kao C. M., Pieper R., Cane D. E., Khosla C. (1996) Biochemistry 35, 12363–12368 [DOI] [PubMed] [Google Scholar]
- 37.Witkowski A., Joshi A. K., Smith S. (2002) Biochemistry 41, 10877–10887 [DOI] [PubMed] [Google Scholar]
- 38.von Wettstein-Knowles P., Olsen J. G., McGuire K. A., Henriksen A. (2006) FEBS J. 273, 695–710 [DOI] [PubMed] [Google Scholar]
- 39.Wu N., Kudo F., Cane D. E., Khosla C. (2000) J. Am. Chem. Soc. 122, 4847–4852 [Google Scholar]
- 40.Wu J., Kinoshita K., Khosla C., Cane D. E. (2004) Biochemistry 43, 16301–16310 [DOI] [PubMed] [Google Scholar]
- 41.Joshi A. K., Witkowski A., Smith S. (1997) Biochemistry 36, 2316–2322 [DOI] [PubMed] [Google Scholar]
- 42.Leibundgut M., Maier T., Jenni S., Ban N. (2008) Curr. Opin. Struct. Biol. 18, 714–725 [DOI] [PubMed] [Google Scholar]
- 43.Maier T., Leibundgut M., Ban N. (2008) Science 321, 1315–1322 [DOI] [PubMed] [Google Scholar]
- 44.Chen A. Y., Schnarr N. A., Kim C. Y., Cane D. E., Khosla C. (2006) J. Am. Chem. Soc. 128, 3067–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hans M., Hornung A., Dziarnowski A., Cane D. E., Khosla C. (2003) J. Am. Chem. Soc. 125, 5366–5374 [DOI] [PubMed] [Google Scholar]
- 46.Chen A. Y., Cane D. E., Khosla C. (2007) Chem. Biol. 14, 784–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marsden A. F., Caffrey P., Aparicio J. F., Loughran M. S., Staunton J., Leadlay P. F. (1994) Science 263, 378–380 [DOI] [PubMed] [Google Scholar]
- 48.Lau J., Fu H., Cane D. E., Khosla C. (1999) Biochemistry 38, 1643–1651 [DOI] [PubMed] [Google Scholar]
- 49.Liou G. F., Lau J., Cane D. E., Khosla C. (2003) Biochemistry 42, 200–207 [DOI] [PubMed] [Google Scholar]
- 50.Kallberg Y., Oppermann U., Jörnvall H., Persson B. (2002) Eur. J. Biochem. 269, 4409–4417 [DOI] [PubMed] [Google Scholar]
- 51.Kao C. M., McPherson M., McDaniel R. N., Fu H., Cane D. E., Khosla C. (1998) J. Am. Chem. Soc. 120, 2478–2479 [Google Scholar]
- 52.McPherson M., Khosla C., Cane D. E. (1998) J. Am. Chem. Soc. 120, 3267–3268 [Google Scholar]
- 53.Yin Y., Gokhale R., Khosla C., Cane D. E. (2001) Bioorg. Med. Chem. Lett. 11, 1477–1479 [DOI] [PubMed] [Google Scholar]
- 54.Reid R., Piagentini M., Rodriguez E., Ashley G., Viswanathan N., Carney J., Santi D. V., Hutchinson C. R., McDaniel R. (2003) Biochemistry 42, 72–79 [DOI] [PubMed] [Google Scholar]
- 55.Caffrey P. (2003) ChemBioChem. 4, 654–657 [DOI] [PubMed] [Google Scholar]
- 56.Keatinge-Clay A. T., Stroud R. M. (2006) Structure 14, 737–748 [DOI] [PubMed] [Google Scholar]
- 57.Keatinge-Clay A. T. (2007) Chem. Biol. 14, 898–908 [DOI] [PubMed] [Google Scholar]
- 58.Starcevic A., Jaspars M., Cullum J., Hranueli D., Long P. F. (2007) ChemBioChem 8, 28–31 [DOI] [PubMed] [Google Scholar]
- 59.Siskos A. P., Baerga-Ortiz A., Bali S., Stein V., Mamdani H., Spiteller D., Popovic B., Spencer J. B., Staunton J., Weissman K. J., Leadlay P. F. (2005) Chem. Biol. 12, 1145–1153 [DOI] [PubMed] [Google Scholar]
- 60.Østergaard L. H., Kellenberger L., Cortés J., Roddis M. P., Deacon M., Staunton J., Leadlay P. F. (2002) Biochemistry 41, 2719–2726 [DOI] [PubMed] [Google Scholar]
- 61.Castonguay R., He W., Chen A. Y., Khosla C., Cane D. E. (2007) J. Am. Chem. Soc. 129, 13758–13769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Valenzano C. R., Lawson R. J., Chen A. Y., Khosla C., Cane D. E. (2009) J. Am. Chem. Soc. 131, 18501–18511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Donadio S., McAlpine J. B., Sheldon P. J., Jackson M., Katz L. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 7119–7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sedgwick B., Morris C., French S. J. (1978) J. Chem. Soc. Chem. Commun. 193–194 [Google Scholar]
- 65.Kimber M. S., Martin F., Lu Y., Houston S., Vedadi M., Dharamsi A., Fiebig K. M., Schmid M., Rock C. O. (2004) J. Biol. Chem. 279, 52593–52602 [DOI] [PubMed] [Google Scholar]
- 66.Bevitt D. J., Staunton J., Leadlay P. F. (1993) Biochem. Soc. Trans. 21, 30S. [DOI] [PubMed] [Google Scholar]
- 67.Keatinge-Clay A. (2008) J. Mol. Biol. 384, 941–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kwan D. H., Sun Y., Schulz F., Hong H., Popovic B., Sim-Stark J. C., Haydock S. F., Leadlay P. F. (2008) Chem. Biol. 15, 1231–1240 [DOI] [PubMed] [Google Scholar]
- 69.Leibundgut M., Jenni S., Frick C., Ban N. (2007) Science 316, 288–290 [DOI] [PubMed] [Google Scholar]
- 70.Alekseyev V. Y., Liu C. W., Cane D. E., Puglisi J. D., Khosla C. (2007) Protein Sci. 16, 2093–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.