Abstract
Relatively little is known about the exact mechanisms used by Bacillus subtilis in its behavior as a biocontrol agent on plants. Here, we report the development of a sensitive plant infection model demonstrating that the bacterial pathogen Pseudomonas syringae pv tomato DC3000 is capable of infecting Arabidopsis roots both in vitro and in soil. Using this infection model, we demonstrated the biocontrol ability of a wild-type B. subtilis strain 6051 against P. syringae. Arabidopsis root surfaces treated with B. subtilis were analyzed with confocal scanning laser microscopy to reveal a three-dimensional B. subtilis biofilm. It is known that formation of biofilms by B. subtilis is a complex process that includes secretion of surfactin, a lipopeptide antimicrobial agent. To determine the role of surfactin in biocontrol by B. subtilis, we tested a mutant strain, M1, with a deletion in a surfactin synthase gene and, thus, deficient in surfactin production. B. subtilis M1 was ineffective as a biocontrol agent against P. syringae infectivity in Arabidopsis and also failed to form robust biofilms on either roots or inert surfaces. The antibacterial activity of surfactin against P. syringae was determined in both broth and agar cultures and also by live-dead staining methods. Although the minimum inhibitory concentrations determined were relatively high (25 μg mL-1), the levels of the lipopeptide in roots colonized by B. subtilis are likely to be sufficient to kill P. syringae. Our results collectively indicate that upon root colonization, B. subtilis 6051 forms a stable, extensive biofilm and secretes surfactin, which act together to protect plants against attack by pathogenic bacteria.
Beneficial plant rhizobacteria (PR) are associated with the surfaces of plant roots and may increase plant yield by mechanisms that impart improved mineral nutrient uptake, disease suppression, or phytohormone production (Kloepper et al., 1991; Lutenberg et al., 1991; Costacurta and Vanderleyden, 1995; Defago and Keel, 1995). An important trait of PR is their ability to effectively colonize the rhizosphere and maintain a stable relationship with the surface of plant roots (Lutenberg and Dekkers, 1999). PR may also interact with a variety of soil microorganisms that are normally present in the rhizosphere, in some cases acting as a biocontrol agent against pathogenic bacteria (Pinton et al., 2001). Interestingly, poor root colonization by PR may result in decreased biocontrol activity (Schippers et al., 1987). One beneficial rhizobacterium is Bacillus subtilis, which is ubiquitous in soil, can promote plant growth, protect against fungal pathogen attack (Utkhede and Smith, 1992; Asaka and Shoda, 1996; Emmert and Handelsman, 1999), and play a role in the degradation of organic polymers in the soil (Emmert and Handelsman, 1999). Among the first successful biocontrol agents used against insects and pathogens were members of the genus Bacillus (Powell and Jutsum, 1993). Commercial strains of B. subtilis have been marketed as biocontrol agents for fungal diseases of crops (Emmert and Handelsman, 1999; Warrior et al., 2002). The commercial biofungicide, Serenade, which contains a B. subtilis strain, is reported to be effective against a variety of pathogenic bacteria, including Erwina, Pseudomonas, and Xanthomonas strains (http://www.agraquest.com. The mechanism of this antibacterial effect is uncertain, although it is known that B. subtilis can produce a variety of antibacterial agents, including a broad spectrum of lipopeptides, such as surfactin, that are potent biosurfactants (Zuber et al., 1993; Peypoux et al., 1999).
It is now widely recognized that most bacteria found in natural, clinical, and industrial settings persist in association with surfaces by forming biofilms (Davey and O'Toole, 2000). Biofilms are structured communities of cells adherent to a surface and encased in an extracellular polymeric matrix (Watnick and Kolter, 1999). Furthermore, these microbial communities are often composed of multiple species that interact with each other and their environment (Costerton et al., 1995). The site of one such ecologically beneficial bacterial community is the rhizosphere, where a rich microflora develops around the readily available nutrients released by roots (Weller and Thomashow, 1994). It is hypothesized that in this environment, the microbial populations attached to the roots and the surrounding soil particles may form biofilm communities. Bacteria attach to the root surface using a variety of cell surface components, such as outer membrane proteins, wall polysaccharides (capsules), lipopolysaccharide, cell surface agglutinin, and exopolysaccharide (Michiels et al., 1991; Amellal et al., 1998). Characterization of mutants defective in biofilm formation and development in several genera of both Gram-positive and -negative bacteria have begun to reveal some of the gene products that are involved in biofilm formation; these gene products include motility, cell surface structures, and exopolysaccharide (Pratt and Kolter, 1999; Davey and O'Toole, 2000).
B. subtilis has been a model organism for the study of Gram-positive bacterial physiology. Recently, it has been reported that B. subtilis forms adhering biofilms on inert surfaces under the control of a variety of transcription factors (Hamon and Lazazzera, 2001; Stanley et al., 2003). Of interest for the work presented here, both Branda et al. (2001) and Kinsinger et al. (2003) have noted that biofilm formation is much more robust in wild-type B. subtilis isolates than in highly subcultured laboratory strains and that biofilm-like structures (pellicles on liquid media or on semisolid media) are dependent on the secretion of surfactin, the lipopeptide mentioned above. There is a growing recognition that biosurfactant production not only affects biofilm architecture but can influence the attachment of bacteria to surfaces (Davey et al., 2003). However, there is very little published evidence that B. subtilis uses biofilm formation on plant roots to produce the biocontrol effects noted above. Shoda (2000) summarizes the accepted view that B. subtilis is a common microbe in soils but is not widespread in the rhizosphere except when introduced massively into soil. However, there is emerging evidence that B. subtilis is a common inhabitant of certain types of plant roots, probably because of biofilm formation (Fall et al., 2003).
In this study, we used Arabidopsis as a plant host because it has been shown to be susceptible to Pseudomonas syringae infections (Jakob et al., 2002). P. syringae, a Gram-negative bacteria, is found in the rhizosphere of Arabidopsis, and Arabidopsis plants with a P. syringae-populated rhizosphere showed severe disease symptoms (Jakob et al., 2002). Although P. syringae infections in Arabidopsis are not of economic importance, the interaction of P. syringae with Arabidopsis is well described and considered to be a model system for studying plant-microbe interactions (Tornero and Dangl, 2001). P. syringae is largely an epiphytic foliar bacterium (Bashan and Bashan, 2002). Nevertheless, it is capable of colonizing seeds and roots (Tornero and Dangl, 2001), although P. syringae's root pathogenicity has not been described. In this study, we reasoned that P. syringae-Arabidopsis root interactions would provide an excellent working model system to assess the biocontrol ability of B. subtilis against infection by P. syringae. It was of particular interest to evaluate the interactions of Gram-positive (B. subtilis) and -negative (P. syringae) bacteria on root surfaces because these are likely to occur in the natural rhizosphere. Hence, for this study, our goals were the following: (a) to determine if a wild-type B. subtilis would colonize and form biofilms on Arabidopsis roots, (b) to assess whether such an association would provide biocontrol against a pathogenic P. syringae, and (c) to determine if surfactin formation is essential for B. subtilis biofilm formation and biocontrol in vivo, by using the B. subtilis mutant strain M1 with a deletion in a surfactin synthase gene and, thus, deficient in surfactin production. In addition, this work aimed to develop an experimental system to study P. syringae pathogenicity by using Arabidopsis roots as the host.
RESULTS
Root Pathogenicity of P. syringae pv tomato DC3000
The root pathogenicity of P. syringae pv tomato DC3000 (P. syringae) was tested in vitro and in soil as determined by infection of Arabidopsis roots. Root pathogenicity because of P. syringae infection was assessed by quantifying the mortality rate of infected Arabidopsis plants. When P. syringae was applied to the liquid media in which Arabidopsis plants were grown, the bacterium caused characteristic disease-like symptoms such as black necrotic regions and rotting on the roots submerged in the media (data not shown). Arabidopsis roots infected with P. syringae also displayed other symptoms, including water-soaked translucent spots that later became necrotic, leading to plant mortality 7 d postinoculation (data not shown; Fig. 1A). This result was expected because previous studies have revealed similar disease symptoms and mortality in Arabidopsis leaves exposed to P. syringae (Jakob et al., 2002). In addition to in vitro studies, we tested the ability of P. syringae to infect soil-grown Arabidopsis plants. P. syringae caused plant mortality approximately 7 d postinoculation when infiltrated into the soil immediately surrounding the root system, and plants showed mortality similar to that described above (Fig. 1B). P. syringae also caused plant mortality when infiltrated into Arabidopsis leaves (data not shown).
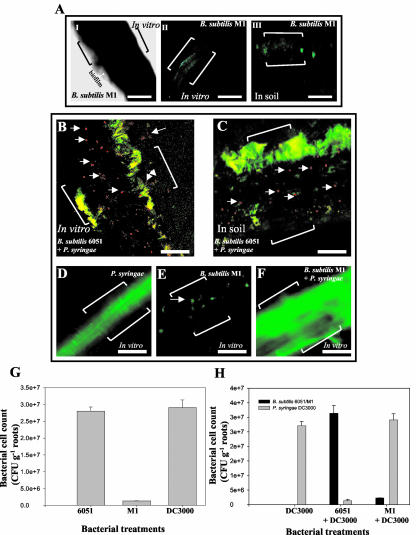
Figure 1.
A, Pathogenicity of the P. syringae strain DC3000 against Arabidopsis in vitro. P. syringae was infiltrated into the liquid Murashige and Skoog medium of in vitro-grown plants, and plant mortality was recorded 7 d postinoculation. B, Pathogenicity of P. syringae strain DC3000 against Arabidopsis in soil. Bacteria were added to sterile soil of plants and plant mortality was again recorded 7 d postinoculation. C, In vitro- and soil-grown Arabidopsis plants cocultivated with B. subtilis 6051. Arabidopsis treated with B. subtilis 6051 alone did not exhibit any plant mortality under in vitro and soil conditions. D, The biocontrol ability of B. subtilis 6051 was checked by inoculating a known concentration of bacterial inoculum into the liquid medium of in vitro- and soil-grown Arabidopsis plants. Arabidopsis plants cocultivated with B. subtilis 6051 and subsequently infected with P. syringae DC3000 under in vitro and soil conditions. P. syringae DC3000 was added on the 4 d post-cocultivation of B. subtilis with Arabidopsis roots. Pretreatment with B. subtilis 6051 reduced plant mortality by approximately 70%.
The P. syringae-Arabidopsis roots pathogenicity system was then used to test the effectiveness of B. subtilis as a biocontrol against P. syringae. The B. subtilis Marburg strain (ATCC 6051) was used as it is arguably the wild-type parent or is closely related to the parent of the B. subtilis 168-derived strains widely used for genetic and genomic studies (Hemphill and Whitely, 1975; Kunst et al., 1997). Arabidopsis treated with B. subtilis 6051 alone did not exhibit any plant mortality (Fig. 1C) under in vitro and soil conditions. Plant mortality induced by P. syringae was reduced in Arabidopsis plants previously cocultivated with B. subtilis 6051 (see “Materials and Methods”); this was true both for plants cultured in vitro and those in sterile soil (Fig. 1, C and D). Further quantified results of this type are presented below.
B. subtilis 6051 Forms a Biofilm on Root surfaces of Arabidopsis
As part of establishing the mechanism of biocontrol by B. subtilis 6051, we determined that this strain adheres to root surfaces of Arabidopsis. Four days post-cocultivation of Arabidopsis roots and B. subtilis 6051 in Murashige and Skoog medium, roots were viewed by phase-contrast and confocal scanning laser microscopy (CSLM). We observed that B. subtilis 6051 cells had colonized virtually the entire root surface (Fig. 2A). Phase contrast and CSLM revealed that roots of Arabidopsis were surrounded by phase-bright material suggestive of an extracellular matrix (Fig. 2A, II and III). Arabidopsis grown in sterile soil with B. subtilis 6051 showed a similar root colonization (Fig. 2A, IV), suggesting that B. subtilis 6051 forms a stable, nonpathogenic biofilm on these roots. For CSLM, roots were stained with a bacterial viable cell procedure (Bianciotto et al., 2001), where green fluorescence revealed an extensive B. subtilis biofilm of live cells under in vitro and soil conditions (Fig. 2A, III and IV).
Figure 2.
A, Phase contrast microscopy and CSLM of Arabidopsis roots cocultivated with B. subtilis 6051 under in vitro and soil conditions for 4 d showed mature biofilm formation by B. subtilis 6051. I, Phase contrast image of untreated roots (the control). II, Roots treated with the B. subtilis 6051 strain under in vitro conditions; note the marked area indicating a phase-bright material suggestive of biofilm surrounding the roots. III, CSLM image of roots treated with B. subtilis 6051 strain under in vitro conditions. IV, CSLM image of roots visualized after cocultivation with B. subtilis 6051 strain under soil conditions. Double brackets in I to IV indicate the roots. Bars = 50 μm. A live-dead BacLight bacterial viability kit was used to detect the live bacteria. I to IV depict a section of the root (from the root tip to the central elongation zone). B, Interaction of Arabidopsis-B. subtilis 6051 and Arabidopsis-B. subtilis M1 cocultures with P. syringae was checked by inoculating a known concentration of P. syringae inoculum under in vitro conditions. B, Lack of biocontrol ability in B. subtilis M1. C, P. syringae was added on the 4th d post-cocultivation of the two B. subtilis strains with Arabidopsis in soil. D, Plant mortality was quantified for each treatment by scoring the percentage of dead plants over the percentage of live ones to measure biocontrol under both in vitro and soil conditions. Bars = one se. Two-way ANOVA for plant mortality: Ftreatment, 12.12; degrees of freedom, 1.26; and P < 0.001.
M1, a Surfactin-Deficient Mutant Strain of B. subtilis, Is Ineffective at Controlling P. syringae Infection
Although plant mortality induced by P. syringae was reduced from about 85% to 10% in plants cocultivated with B. subtilis 6051, both in vitro and in sterile soil (Fig. 2, B-D), a surfactin-deficient mutant known as M1 provided virtually no protection against the pathogen. As reviewed above, biofilm formation in B. subtilis is much more robust in wild-type B. subtilis isolates than in highly subcultured laboratory strains and is dependent on the secretion of the extracellular, antimicrobial lipopeptide surfactin. Recently, one of us has constructed a surfactin-deficient mutant in the B. subtilis 6051 genetic background and determined that this mutant (strain M1) is deficient in both surface motility and biofilm formation (Kinsinger et al., 2003; see “Materials and Methods”). The availability of the M1 mutant allowed us to test whether surfactin secretion is essential for the colonization and/or biocontrol of B. subtilis on Arabidopsis roots. In contrast to the results obtained for the wild-type B. subtilis 6051, Arabidopsis plants cocultivated with B. subtilis M1 and subsequently infected with P. syringae under in vitro conditions showed high plant mortality rates similar to those observed with P. syringae alone (Fig. 2, B and D). A similar lack of protection by strain M1 was seen under soil conditions (Fig. 2, C and D).
B. subtilis M1 Forms Less Biofilm on Root Surfaces of Arabidopsis
To visualize the differences in colonization and biofilm formation in planta between B. subtilis 6051 and the B. subtilis M1 mutant, cocultivated roots were viewed by phase-contrast and CSLM. Unlike the wild-type strain, the B. subtilis M1 mutant failed to colonize the entire root surface (Fig. 3A). Phase contrast microscopy revealed that roots of Arabidopsis colonized with the B. subtilis M1 mutant were surrounded by much less phase-bright material, suggestive of a reduced biofilm formation, compared with B. subtilis 6051 (compare Fig. 2A, II with Fig. 3A, I). Similarly, visualization of viable B. subtilis M1 cells by CSLM using the viable cell procedure (see “Materials and Methods”) revealed that only few and small regions in the roots were colonized by the M1 mutant, and significantly reduced biofilm formation was observed compared with B. subtilis 6051 (Fig. 3A, II and III). Soil-grown Arabidopsis roots cocultivated with the M1 mutant showed similarly reduced biofilm formation (Fig. 3A, III).
Figure 3.
A, Phase-contrast and CSLM of Arabidopsis roots cocultured with the B. subtilis M1 mutant under in vitro and soil conditions showing poor biofilm formation by the B. subtilis M1 mutant. I, Phase contrast image of roots treated with B. subtilis M1; note the marked area indicating a small region of phase-bright material suggestive of poor biofilm surrounding the roots. II, CSLM image of roots treated with B. subtilis M1 under in vitro conditions. III, CSLM image of Arabidopsis roots cocultivated with B. subtilis M1 under soil conditions. Double brackets in I to III indicate the roots. Bars = 50 μm. I to III depict a section of the root (from the root tip to the central elongation zone). B, CSLM of Arabidopsis roots grown in vitro, cocultivated with B. subtilis 6051, and subsequently infected with P. syringae. C, CSLM of Arabidopsis roots grown under soil conditions, cocultivated with B. subtilis 6051, and subsequently infected with P. syringae. A live-dead BacLight bacterial viability kit was used to detect most of the live bacteria. Bacteria on the root surface were stained with propidium iodide and SYTO9 (See “Materials and Methods”) to visualize the polysaccharides. Both the above treatments show intact biofilm formation by wild-type B. subtilis 6051 as represented by green communities. (Note multiple arrows indicating dead bacteria, most likely P. syringae, in red; brackets in the panel indicate the roots). Bars (B and C) = 20 μm. D, CSLM of P. syringae alone infecting Arabidopsis roots. E, CSLM of Arabidopsis roots grown in vitro and cocultured with the B. subtilis M1 strain. Note the arrow indicating poor colonization as represented by green communities in patches. Bars (D and E) = 50 μm. F, CSLM of Arabidopsis roots grown in vitro, cocultured with B. subtilis M1, and subsequently infected with P. syringae. Brackets in the panels indicate the roots. G and H, Quantification of bacterial cell counts on root surfaces of Arabidopsis on the 4th d postinoculation with B. subtilis 6051, M1, and P. syringae under lone and mixed treatments. Specific antibiotic selection with B. subtilis M1 (5 μg mL-1 chloramphenicol) and P. syringae DC3000 (100 μg mL-1 rifampicin) was used for selective plating of the two bacteria for colony-forming units (cfu) counts. Values show the quantitative amount of roots with average (mean ± sd; n = 5) bacterial counts after inoculation of approximately 2.5 × 108 cfu mL-1; five plants of each species were used per treatment.
Interaction of B. subtilis Strains 6051 and M1 with P. syringae on Root Surfaces of Arabidopsis
To further visualize the differences in biocontrol efficiencies of B. subtilis 6051 and M1, P. syringae was inoculated in the liquid growth media in which Arabidopsis plants were grown, 4 d post-treatment with either B. subtilis strain. Viable cell microscopy of roots precultured with the 6051 strain revealed extensive regions of green fluorescence, indicative of viable cells, with numerous clusters of red fluorescence produced by dead bacteria (Fig. 3, B and C). In comparison with roots similarly precultured with strain 6051 and stained this way (Fig. 2A, III and IV) and based on the protective effect of strain 6051 against P. syringae pathogencity, we inferred that the green-stained, viable cells are B. subtilis 6051 (Fig. 3H), and the red-stained dead cells are P. syringae. A control experiment with roots treated with only P. syringae (Fig. 3D) shows that in the absence of B. subtilis 6051, the pseudomonad forms an extensive viable cell biofilm.
In contrast to the cocultivation with B. subtilis 6051, Arabidopsis roots precultured with the B. subtilis M1 mutant and then infected with P. syringae showed no signs of bacterial (P. syringae) mortality, instead revealing an intact pathogenic biofilm under in vitro conditions (Fig. 3F). Again, the M1 mutant showed only small patches of colonization and biofilm formation (Fig. 3E), whereas the subsequent addition of P. syringae resulted in an extensive viable biofilm (Fig. 3F). Similar results were obtained under soil conditions (data not shown).
To evaluate the correlation between plant mortality and the size and quality of bacterial biofilms, we next examined bacterial colonization in plant roots cultivated with lone treatments of B. subtilis 6051, M1, and P. syringae in contrast to roots cultivated with a mixed treatment of B. subtilis 6051 + P. syringae and B. subtilis M1 + P. syringae under in vitro conditions. In accordance with our CSLM and plant mortality results, the B. subtilis M1 demonstrated poor root colonization 4 d post-treatment compared with the colonization ability of B. subtilis 6051 or P. syringae (Fig. 3G). To confirm the likelihood that B. subtilis 6051 out-competed P. syringae multiplication on Arabidopsis roots, we also examined the roots grown under mixed treatments of B. subtilis 6051 with P. syringae. Four days after inoculation, the root-localized bacterial number increased drastically for the nonpathogenic B. subtilis 6051 compared with the P. syringae counts (Fig. 3H). In contrast, Arabidopsis roots precultured with the B. subtilis M1 mutant and then infected with P. syringae showed reduced bacterial counts for the M1 mutant (Fig. 3H), whereas the subsequent addition of P. syringae resulted in an extensive multiplication of P. syringae on the Arabidopsis root surface (Fig. 3H). Specific antibiotic selection for B. subtilis M1 (5 μg mL-1 chloramphenicol) and P. syringae DC3000 (100 μg mL-1 rifampicin) resulted in selective plating of the two bacteria for cfu counts. These results strongly support our hypothesis that B. subtilis 6051 acts as a potent biocontrol against P. syringae infection in Arabidopsis.
Adherence of B. subtilis to Abiotic Surfaces
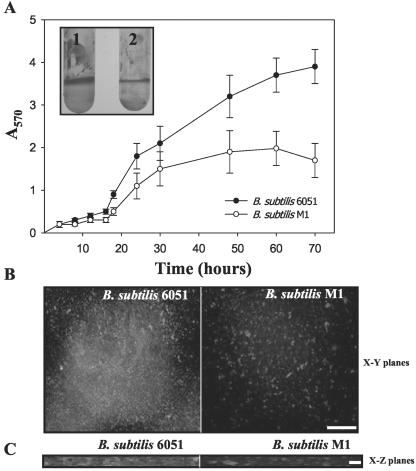
To further test whether the B. subtilis 6051 and M1 strains differ in their ability to form adhering biofilms, we used the recently described methods of Hamon and Lazazzera (2001) to measure adherence of the bacterium to the wells of a microtiter plate. Each strain was grown in the wells of a polyvinylchloride (PVC) microtiter plate in a complex biofilm growth medium, and the amount of adhering cells was quantified by crystal violet (CV) staining (described in “Materials and Methods”). As shown in Figure 4A, measurable levels of adhered cells progressively increased approximately 7-fold from 16 to 30 h of incubation and then plateaued 60 to 70 h after inoculation (Fig. 4A). This increase in CV staining appeared to result from an increase in the number of adhered cells because it corresponded to an increase in the number of cfu of adhered cells (data not shown). The B. subtilis M1 mutant produced approximately one-half the biofilm on microtiter plates compared with the B. subtilis 6051 (Fig. 4A).
Figure 4.
A, OD readings from microtiter plate assays of biofilm formation by B. subtilis 6051 wild-type and M1 mutant strains. OD570 of solubilized CV from microtiter assays over time for the two tested B. subtilis strains; inset shows the solubilized CV in polypropylene tubes depicting adherent biofilms: 1, B. subtilis 6051; and 2, B. subtilis M1 (values are mean ± sd, n = 5). B, CSLM visualization of wild-type 6051 and M1 mutant strains of B. subtilis grown on glass coverslips. C, An artist's rendering of the three-dimensional structure of biofilm formed by wild-type (6051) and M1 mutant strains of B. subtilis on x-z planes as visualized by CSLM. Bars = 5 μm.
To determine if the cells that were adhering to inert surfaces were actually forming a three-dimensional biofilm, cells were analyzed by CSLM. Adhering B. subtilis 6051 cells (grown on glass cover slips) appeared to form a three-dimensional, multicellular structure typical of a biofilm (Fig. 4, B and C). Viewing the cells in an x-y plane, it appears that the adhered cells form a mat of bacteria. From the x-z plane, it can be observed that the adhered cells form a structure with significant depth. As observed with CV staining, CSLM also revealed that the M1 mutant formed less biofilm compared with the B. subtilis strain 6051 (Fig. 4, B and C).
Competition between B. subtilis Strains and P. syringae on Agar Surfaces
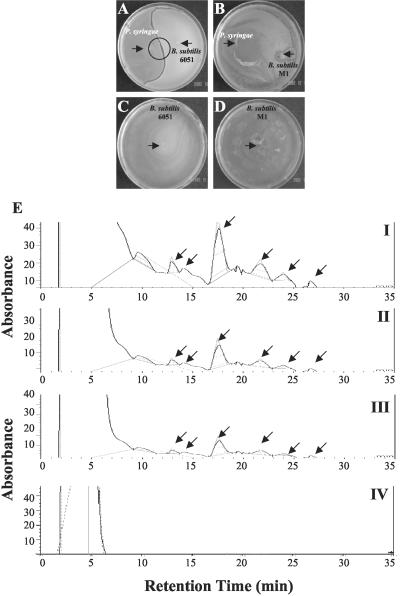
Because of the observed P. syringae mortality on Arabidopsis root surfaces previously cocultivated with B. subtilis 6051, but not those cocultivated with M1, we investigated in situ P. syringae-B. subtilis competition on plates. Nutrient broth (NB) agar medium was co-inoculated with pairs of all three tested bacteria (P. syringae-B. subtilis 6051 and P. syringae-B. subtilis M1). Sixteen hours after inoculation, B. subtilis 6051 and P. syringae each formed a bacterial film over a large surface of their respective plates. Interestingly, when B. subtilis 6051 was cocultured with P. syringae, a distinct inhibition zone between the two bacteria was observed after 16 h of bacterial challenge (Fig. 5A). The inhibition zone remained intact even after 48 h (data not shown). In contrast, the B. subtilis M1 mutant failed to show any inhibition zone against P. syringae and was overgrown by the latter (Fig. 5B). B. subtilis 6051 alone swarmed vigorously on NB agar plates, and the B. subtilis M1 mutant alone swarmed efficiently on NB agar plates as well, suggesting that the surfactin mutation did not impair its growth on this medium (Fig. 5, C and D). This phenomenon of selective inhibition of P. syringae growth by strain 6051 but not M1 was also seen on Luria-Bertani (LB) agar media (data not shown). The essential difference between B. subtilis strains 6051 and M1 was the formation of surfactin, which, based on these results, appears to have an antimicrobial effect on P. syringae (Wei and Chu, 1998). Thus, our results show that the inhibition was clearly produced by B. subtilis 6051 against P. syringae.
Figure 5.
A and B, In situ challenge of B. subtilis 6051 and P. syringae bacterial cultures. Bacterial cultures were inoculated with a sterile toothpick on each one-half of the petri plate. Inhibition zones were visualized and photographed 48 h postinoculation. Plates indicate formation of surface film in NB agar medium (see “Materials and Methods”). A, B. subtilis 6051 shows a clear and distinct inhibition zone repelling P. syringae, independent of the medium used (note the arrow indicating the bacterial inocula). B, B. subtilis M1 was out-competed by P. syringae, a result independent of medium used (note the arrows indicating the bacterial inocula). C and D, Lone B. subtilis 6051 and M1 growth plates on NB agar medium (note the arrows indicating bacterial inocula). E, HPLC spectrograms for surfactin production. Six peaks were used to quantify surfactin content in B. subtilis. (Note the arrows indicating the six different isoforms of surfactin). I, Standard surfactin. II, Surfactin content in B. subtilis 6051 broth cultures. III, Surfactin content isolated from the interface of the inhibition zone observed during competition experiment between B. subtilis 6051 and P. syringae. IV, No surfactin was found from the B. subtilis M1 mutant.
Quantification of Surfactin from B. subtilis Strains 6051 and M1
Using HPLC conditions developed in the present study (see “Materials and Methods”), surfactin production in liquid cultures of B. subtilis 6051 and M1 strains was analyzed (Fig. 5E). As the typical chromatogram (Fig. 5E, I) shows, the commercially purchased standard surfactin produces six different peaks depicting different isomers of surfactin; a similar result is shown by Wei and Chu (1998). We chose these six major peaks through calibration to quantify surfactin production. B. subtilis 6051 produced a substance with a profile very similar to that of standard surfactin (Fig. 5E, II). Interestingly, methanolic extracts from the inhibition zone between B. subtilis 6051 and P. syringae bacterial colonies from the in situ challenge experiments showed a chromatographic profile similar to that observed for B. subtilis 6051 and standard surfactin (Fig. 5E, III), suggesting that the inhibition zone was because of the abundant presence of surfactin. In contrast, the B. subtilis M1 mutant did not appear to produce any of the surfactin isomers seen in the wild-type B. subtilis 6051 (Fig. 5E, IV). This analysis confirms that the ΔsrfA-A mutation in strain M1 blocks formation of all six of the surfactin lipopeptides and that these polipeptides may account for the antibacterial effect of B. subtilis on P. syringae.
Antibacterial Activity of Surfactin against P. syringae
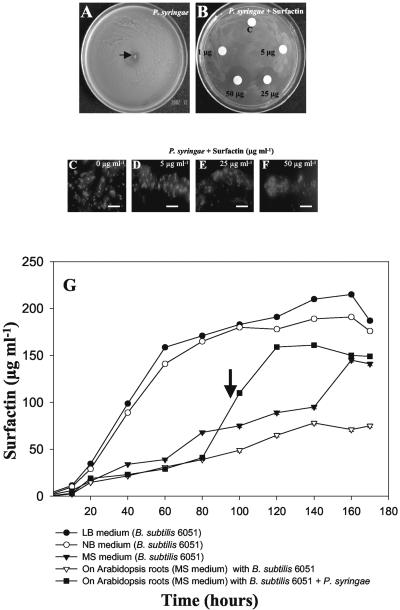
The antibacterial activity of surfactin was tested against P. syringae planktonic cells by the broth microdilution method in 96-well microtiter plates as described in “Materials and Methods.” The minimum inhibitory concentration (MIC) of surfactin for P. syringae was determined to be 25 μg mL-1. The MICs in Arabidopsis Murashige and Skoog basal media were comparable with MICs in cation-adjusted Mueller-Hinton broth (data not shown). Replating of the media from the 96-well microtiter plates (25-100 μg mL-1 surfactin) demonstrated that this lipopeptide is bactericidal (data not shown). Surfactin also showed antibacterial activity against P. syringae on NB agar, evident by increasing zones of inhibition (Fig. 6, A and B). Fluorescence microscopic visualization by live-dead staining exhibited the bactericidal activity of surfactin against P. syringae in the titer plate assay (Fig. 6, C-F). Fluorescence microscopy of the P. syringae treated with MIC levels of surfactin (25 μg mL-1) revealed mortality as shown by red florescence compared with the untreated control (Fig. 6, C-F). These results show that surfactin has bactericidal activity against P. syringae.
Figure 6.
A and B, Antibacterial activity of 1 to 25 μg mL-1 surfactin on the growth of P. syringae. Standard surfactin was applied to the filter discs. A bacterial inoculum (approximately 2 × 108 cells mL-1) of P. syringae was plated and spread on the petri dish, and the radial inhibition was observed on an hourly basis. A filter disc treated with only solvent (2.5% [v/v] dimethyl sulfoxide) was used as a negative control. A, Radial and efficient growth of P. syringae on NB agar plates in the absence of surfactin. B, Radial growth inhibition of P. syringae in the presence of different concentrations of surfactin. C to F, Fluorescence microscopic visualization of live-dead staining to show bactericidal activity of surfactin against P. syringae in the titer plate assay. Bacterial suspensions treated with sub-MIC (0-5 μg mL-1), MIC (25 μg mL-1), and double MIC levels (50 μg mL-1) of surfactin were stained with propidium iodide and SYTO9 (see “Materials and Methods”) to visualize the polysaccharides and nuclei, respectively. The green fluorescence in C and D depict live material surrounding/inside the bacterial colony. Red fluorescence shows dead cells (E and F). Scale bars = 20 μm. G, Surfactin profiles under different media conditions (LB, NB, and Murashige and Skoog) and on the root surface of Arabidopsis grown alone, cocultured with B. subtilis strains, and infected with P. syringae. (Note the arrow indicating 96 h, the time of addition of P. syringae to the Arabidopsis-B. subtilis 6051 coculture). Arabidopsis roots were weighed (50 mg fresh weight) and extracted for surfactin analysis. (Values are mean ± sd, n = 5).
Surfactin Formation by B. subtilis on Root Surfaces
To determine if surfactin was secreted from B. subtilis 6051 when grown on plant roots, we compared the levels of surfactin (a sum of the six isomers) in in vitro cultures and during coculture of B. subtilis with Arabidopsis roots in a defined Murashige and Skoog medium. For comparison, the secretion of surfactin was also measured in typical bacterial growth media, LB and NB, and in Arabidopsis Murashige and Skoog media, which also supported the growth of B. subtilis 6051. Samples (either rinsed roots or culture media supernatants) were collected consecutively for 7 d and analyzed for surfactin by HPLC analysis (see “Materials and Methods”). As shown in Figure 6G, surfactin production by B. subtilis 6051 growing on root surfaces of Arabidopsis was evident, with a final concentration in root extracts of 151.6 μg mL-1 per 50 mg of roots fresh weight (Fig. 6G). Interestingly, surfactin production was elicited approximately 2.0-fold after administration of P. syringae into the Arabidopsis-B. subtilis 6051 system (Fig. 6G; the arrow depicts the sudden increase in surfactin production). Surfactin was not detected in root extracts of Arabidopsis plants cocultivated with B. subtilis M1 and infected with P. syringae (data not shown). Growth of B. subtilis 6051 in common culture media, such as LB and NB, showed an expected linear increase of surfactin concentration during the time course until 160 h, followed by a gradual decrease (Fig. 6G). Substantial surfactin was also secreted by B. subtilis 6051 cultured in the Arabidopsis Murashige and Skoog medium.
DISCUSSION
In this communication, we have described a new root pathogenicity system (Arabidopsis roots-P. syringae) and provide evidence for a unique biocontrol strategy using the ubiquitous soil bacterium B. subtilis—the formation of protective and antibacterial biofilms. First, we developed this experimental system by using in vitro and soil cultures of Arabidopsis to test the root pathogenicity of P. syringae pv tomato DC3000, a strain that has been identified as a potent leaf pathogen in Arabidopsis (Davis et al., 1991). As shown here, the root pathogenicity of P. syringae, as based on mortality rates, was similar both in vitro and in soil. Our experimental system provides a means to disrupt the pathogenic interaction between P. syringae and Arabidopsis roots. We attempted to do this by using the wild-type B. subtilis strain 6051 as a potential biocontrol agent. B. subtilis 6051 was chosen because this species is gaining recognition for biocontrol in a variety of plants, albeit mainly for use as a seed protectant and antifungal agent (Chanway, 2002; Warrior et al., 2002) and because of new evidence that some plant roots contain tightly bound B. subtilis strains that form abundant biofilms in vitro (Fall et al., 2003; Kinsinger et al., 2003).
When we tested the biocontrol efficiency of B. subtilis 6051 against root infection by P. syringae, reduced mortality of Arabidopsis was observed, both in culture and in soil. The reason for this biocontrol efficiency was traced to the formation of an antimicrobial-producing biofilm, allowing for colonization of the root surface of Arabidopsis, and to the secretion of a lipopeptide antibiotic, surfactin. In our studies, we documented that the ability of B. subtilis 6051 to control P. syringae infectivity of Arabidopsis was directly proportional to its ability to colonize and form biofilms on plant root surfaces.
Biofilm formation is a major bacterial adaptive strategy to environmental conditions in aquatic and other settings (Emmert and Handelsman, 1999; Davey and O'Toole, 2000). However, the ability of rhizosphere microorganisms to form protective biofilms is less well understood. Earlier studies suggested that biocontrol mechanisms could be related to biofilm formation on roots, which could protect against pathogenic infection (van Veen et al., 1997; O'Toole and Kolter, 1998). Pseudomonas fluorescens, a Gram-negative soil bacterium and biocontrol agent, has been thought to form biofilms on plant roots, but the relationship between biofilm formation and biocontrol has not been confirmed (O'Toole and Kolter, 1998; Bianciotto et al., 2001). In the case of strains of B. subtilis and its close relatives that have been used as biocontrol agents, numerous mechanisms for biocontrol have been outlined (for review, see Chanway, 2002), but no specific role for biofilm formation by these bacteria on plant roots has been characterized yet. However, in support of our results, it has been found recently that B. subtilis can form biofilms on abiotic surfaces (Branda et al., 2001; Hamon and Lazazzera, 2001; Stanley et al., 2003).
The biocontrol ability of B. subtilis against the fungal pathogen Rhizoctonia solani has been shown to be achieved by virtue of the production of surfactin and iturin A, which are lipopeptides that contain a hydroxy fatty acid connected by an ester peptide linkage to a cyclic heptapeptide (Peypoux et al., 1999). Interestingly, Asaka and Shoda (1996) showed that the persistence of surfactin in soil is better than that of iturin A, suggesting a prolonged stable role for surfactin in the rhizosphere. Here, we demonstrate that the biocontrol of P. syringae by B. subtilis 6051 is related to surfactin formation. We found that surfactin has an MIC of approximately 25 μg mL-1 against P. syringae, which is relatively high for an antimicrobial agent but may be reasonable for the exigencies rhizosphere settings (Vivanco et al., 1999; Park et al., 2002). In the experiments with Arabidopsis roots that were precultured with B. subtilis 6051 (Fig. 6G), levels of surfactin in rinsed roots were substantial (on the order of 151.6 μg mL-1 per 50 mg root fresh weight). Thus, it is possible that at the root surface, the levels of dissolved surfactin are substantially higher than the MIC against P. syringae determined in vitro, suggesting that the biocontrol exhibited against P. syringae is linked to the formation of this antibiotic at the root surface by a B. subtilis biofilm. The exact mechanism by which surfactin acts as an antibacterial agent is not yet known, but is likely to be related to its ability to disrupt membranes (Peypoux et al., 1999) or to alter the physical and chemical properties of the biofilm growth of competing organisms (Neu, 1996). For example, it was shown previously that surfactin inhibits biofilm formation of Salmonella enterica at levels as low as 50 μg mL-1 and Escherichia coli and Proteus mirabilis at higher levels in vitro (Mireles et al., 2001). Taking these observations together, it is possible that the presence of B. subtilis 6051 surfactin may prevent the planktonic cells of other microbes from colonizing biological surfaces, including plant roots. This conclusion could explain the biocontrol of P. syringae seen here in the Arabidopsis root system.
To further verify the role of surfactin in the biocontrol of P. syringae, we utilized a surfactin-minus mutant of B. subtilis, strain M1, constructed using the B. subtilis 6051 background. The B. subtilis M1 mutant showed normal growth in typical laboratory media, but when precultured with Arabidopsis, it was not effective in controlling P. syringae pathogenicity and also exhibited poor biofilm formation on roots, as shown by CSLM. It also formed less robust biofilms than the parent strain on inert surfaces. Although we cannot rule out pleiotropic effects resulting from the deletion in the srfA-A gene of the M1 mutant, these findings strongly suggest that the production of surfactin is essential for biofilm formation and colonization of Arabidopsis roots (and perhaps other plant roots), and surfactin formation may be an essential trait for effective B. subtilis biocontrol strains.
As mentioned above, some plant roots contain tightly bound B. subtilis strains, and we have shown that many of these strains also produce surfactin (Fall et al., 2003; Kinsinger et al., 2003). It is possible that biofilm and surfactin formation may allow B. subtilis and its close relatives to efficiently colonize plant roots and also provide protection to their host. Whether this apparent “symbiotic” relationship is common in the roots of plants is unknown, but it is of interest to note that the roots of some plants contain high populations of B. subtilis and other Bacillus spp. (Lilley et al., 1996; Pandey and Palni, 1997; Germida et al., 1998).
The use of microorganisms to control plant diseases offers an attractive alternative to the use of synthetic chemicals (Emmert and Handelsman, 1999; Shoda, 2000; Warrior et al., 2002). The abundance of a beneficial strain of microorganism in the vicinity of plant roots may suppress plant pathogens without producing lasting effects on the rest of the soil microbial and plant communities (Howarth, 1991; Osburn et al., 1995; van Veen et al., 1997). In accordance, the diversity of microbial communities provides a rich source of potential biocontrol agents. With the root pathogenicity system and methods described here, it should be possible to identify new Bacillus and other bacterial isolates that are effective in formation of protective and antibacterial biofilms.
MATERIALS AND METHODS
Plant Material and Growth Conditions of Arabidopsis in Vitro and in Soil
Seeds of wild-type Arabidopsis ecotype Columbia were obtained from Lehle Seeds (Round Rock, TX). Seeds were surface sterilized using 0.3% (v/v) sodium hypochlorite for 10 to 12 min and then washed four times in sterile double distilled water. For root cultures, seeds were placed on static Murashige and Skoog (1962) basal media in petri dishes for germination and incubated in a growth chamber. Twenty-five-day-old seedlings were individually transferred to 6-mL 12-well culture plates (Fisher Scientific, Loughborough, Leicestershire, UK), each containing 2 mL of liquid Murashige and Skoog basal media. Plant cultures were maintained on an orbital platform shaker (Lab-Line Instruments, Melrose Park, IL) set at 90 rpm with a photoperiod of 16 h of light and 8 h of dark at 25°C ± 2°C.
For in-soil experiments, 25-d-old seedlings were transplanted from static Murashige and Skoog media to 10-cm black plastic pots containing 50 g (dry weight) of PM-O5 Arabidopsis growing medium (Lehle Seeds). Plants were incubated in a growth chamber at 30°C with 12 h of light and watered daily for 2 weeks before inoculation with bacteria.
Bacterial Strains and Culture Conditions
The following Bacillus subtilis strains were used in this study. Wild-type B. subtilis 6051, the Marburg strain, was obtained from the American Type Culture Collection (Manassas, VA). As described in detail elsewhere (Kinsinger et al., 2003), a surfactin-deficient mutant was constructed in the 6051 strain by disruption of the srfA-A gene using the pJM103 integration vector (Perego, 1993). In brief, a 1,040-bp DNA fragment of the srfA-A gene was amplified by PCR from B. subtilis 6051 chromosomal DNA, cloned between BamH1 and EcoR1 sites in pJM103, and the construct was used to transform B. subtilis 6051 using the method described by Anangnostopolous and Spizizen (1961). Although the transformation frequency with the 6051 strain was very low, mutants resistant to chloramphenicol were obtained, indicative of single insertions into and disruption of the srfA-A gene. One mutant, designated M1 (srfA-A::cam), grew normally in LB medium but secreted no surfactin (as confirmed here), consistent with disruption of surfactin synthesis. The surface motility of the M1 mutant (Kinsinger et al., 2003) and its ability to form adhering biofilms on plastic surfaces (R. Kinsinger and R. Fall, unpublished data) were disrupted unless authentic surfactin was added to the growth media; the surfactin-minus phenotype of mutant M1 was quite stable even after repeated subculture without added chloramphenicol. Freshly plated cells from frozen stock cultures were used for all experiments. Each strain was typically grown on LB agar plates and incubated at 37°C; chloramphenicol (5 μg mL-1) was added to maintain the M1 mutant. Plated cells were suspended in 5 mL of LB broth for overnight growth at 37°C and shaken at 250 rpm. Pseudomonas syringae pv tomato DC3000, a wild-type isolate that is pathogenic toward Arabidopsis leaves (Davis et al., 1991), was obtained from the laboratory of Dr. Christopher B. Lawrence (Department of Bioagricultural Sciences and Pest Management, Colorado State University, Fort Collins); it was maintained and grown on LB medium with specified antibiotic selection (Rifampicin 100 μg mL-1) at 28°C. In some experiments, bacterial cells were grown in NB (DIFCO Laboratories, Detroit) or Murashige and Skoog medium as described above.
In Vitro Root Pathogenicity Assay
P. syringae strains were grown to OD600 = 0.2 to 0.4 and added separately to the 2 mL of Murashige and Skoog media supporting each plant to reach an initial OD600 = 0.02 (approximately 2.5 × 107 cfu mL-2). Murashige and Skoog basal media (2 mL) without plant material was inoculated with the same volume of each bacterial strain tested. By inoculation, we refer to the addition of bacterial solution into the Murashige and Skoog medium where the roots were floating. A noninfected plant control was maintained under the same conditions. All the treatments and controls were incubated at 30°C in a controlled environment incubator shaker (New Brunswick Scientific, Edison, NJ) set at 30 rpm with a photoperiod of 16 h of light and 8 h of dark. Ten plants per treatment were used for analysis of mortality rates. Experiments were repeated twice in triplicate to standardize the observations.
Leaf Pathogenicity Assay
For leaf assays, P. syringae strains were grown in LB at 37°C to OD600 = 0.2 to 0.3 and diluted 1:100 (w/v). Diluted suspensions were individually injected with the blunt end of a hypodermic needle into intact leaves of Arabidopsis at a dose of approximately 1 × 103 cfu cm-2 as previously described (Jakob et al., 2002). Infiltrated plants were incubated in a growth chamber at 30°C and 80% relative humidity with 16 h of light and 8 h of dark. Five leaves per plant were used for scoring of disease symptoms.
In-Soil Pathogenicity Assay
For soil infiltration, the 10-cm pots (with 50 g of soil) containing Arabidopsis were each flooded with 10 mL of P. syringae bacterial suspension to give an inoculum concentration of approximately 1 to 5 × 108 cfu g-1 of soil. Plants were incubated under identical conditions as those used for leaf infiltration assays. Ten plants per treatment were used for analysis of mortality rates.
Analyzing the Biocontrol Efficiency of B. subtilis Strains 6051 and M1 against P. syringae
The wild-type B. subtilis 6051 and the M1 mutant were tested for their biocontrol ability on Arabidopsis roots both in vitro and under soil conditions. Bacterial strains were grown to OD600 = 0.3 to 0.4 and added separately to the 2 mL of Murashige and Skoog media of each in vitro plant to reach an initial OD600 = 0.02 (approximately 2.5 × 108 cfu mL-1). For soil infiltration, the 10-cm pots (with 50 g of soil) containing Arabidopsis were each flooded with 10 mL of B. subtilis bacterial suspension to give an inoculum concentration of approximately 5 × 105 cfu g-1 of soil. A noninfected plant control was maintained under the same conditions. All the treatments and controls were incubated at 30°C in a controlled environment incubator shaker (New Brunswick Scientific) set at 30 rpm with a photoperiod of 16 h of light and 8 h of dark. To analyze the biocontrol efficiency of B. subtilis strains, P. syringae was inoculated under in vitro and soil conditions (as previously described) 4 d post-treatment with B. subtilis strains using the inoculum sizes mentioned above. Ten plants per treatment were used for analysis of mortality rates. For bacterial counts on root surfaces, in vitro-grown Arabidopsis root tissues (500 mg fresh weight) with mixed treatments of B. subtilis 6051, M1, and P. syringae were washed with distilled water and homogenized in 1 mL of saline (0.2% [w/v] sodium chloride) with a tissue grinder (size C, Kontes, Rochester, NY), and the suspension was filtered, diluted in saline, and plated on LB agar plates with specified antibiotic selection to determine bacterial cell counts. Specific antibiotic selection with B. subtilis M1 (5 μg mL-1chloramphenicol) and P. syringae DC3000 (100 μg mL-1rifampicin) was used for selective plating of the two bacteria for cfu counts. Each data point represents five replicates. All bacterial growth assays were repeated, and only results that were observed consistently are shown.
Microscopy
CSLM for biofilm formation was performed using the Live-dead BacLight Bacterial Viability Kit (Molecular Probes, Eugene, OR) by incubating B. subtilis-P. syringae colonized Arabidopsis roots at room temperature in the dark for 15 min, according to the manufacturer's manual. The samples were mounted with Citifluor antifading (Sigma, St. Louis) and observed for fluorescence with a confocal laser microscope (Fluroview LGPS-2, Olympus, Minneopolis). For observation of B. subtilis biofilms on glass coverslides by CSLM, B. subtilis biofilms were grown using the method described by Watnick and Kolter (1999). Biofilms of B. subtilis 6051 and M1 strains were grown on glass coverslides (Fisher Scientific) in 6 mL of biofilm growth medium in 50-mL polypropylene conical tubes. To determine whether B. subtilis biofilms were encased in a polysaccharide matrix, we stained the biofilm with Calcofluor, a polysaccharide-binding dye. After rinsing, the slides were stained for 20 min with 10 mL of 75 μg mL-1 Calcofluor (Sigma; fluostain) in wash buffer. The stained biofilm was then analyzed by CSLM as described previously (Hamon and Lazazzera, 2001; Hogan and Kolter, 2002). CSLM scans in both x-y and the x-z planes can be used to view a three-dimensional structure. To view adhered B. subtilis cells by CSLM, both B. subtilis strains were stained with Calcofluor dye adhered to glass slides. Phase contrast images of B. subtilis-colonized root tissues were captured with a 10× objective on an Olympus BX60 microscope equipped with CoolSnap imaging software (San Diego) as described previously (Bianciotto et al., 2001). Phase contrast and CSLM were performed 4 d postinoculation. Samples were analyzed for fluorescence with a confocal laser microscope (Fluroview LGPS-2, Olympus). Samples were viewed using 488 nm as the excitation wavelength.
Microtiter Plate Assay of B. subtilis Biofilm Formation
B. subtilis biofilm formation was monitored separately using a microtiter plate assay based on the methods of O'Toole et al. (1999). B. subtilis cells were grown in 96-well PVC microtiter plates (Fischer Scientific) at 37°C in biofilm growth medium. Biofilm growth medium based on Hamon and Lazazzera (2001) was LB medium plus 0.15 m ammonium sulfate, 100 mm potassium phosphate (pH 7), 34 mm sodium citrate, 1 mm MgSO4, and 0.1% (w/v) Glc. The inocula for the microtiter plates were obtained by growing the cells in biofilm growth medium and shaking to midexponential growth and then diluting the cells to OD600 of 0.01 in fresh biofilm growth medium. Samples of 100 μL of the diluted cells were aliquoted to each well of 96-well PVC microtiter plates. The microtiter plates were incubated at stationary conditions. Cells that had adhered to the wells were stained with 0.1% (w/v) CV in wash buffer (0.15 m ammonium sulfate, 100 mm potassium phosphate [pH 7], 34 mm sodium citrate, and 1 mm MgSO4) at room temperature for 20 min. Excess CV was then removed, and the wells were rinsed with water. The CV that had stained the cells was then solubilized in 200 μL of 80% (v/v) ethanol and 20% (v/v) acetone. Biofilm formation was quantified by measuring the OD570 for each well using an Opsys MR-Dynex plate reader (Chantilly, VA).
In Situ Challenge between B. subtilis 6051 or M1 and P. syringae
LB and NB medium supplemented with 2.5 g L-1 tryptone, Glc (5 g L-1) with 0.4% (w/v) agar was incubated at 37°C. Swarm plates were typically allowed to dry at room temperature overnight before being used. Swarm plates were inoculated with bacteria using a sterile toothpick on both sides of the petri plates to visualize competitive interactions. The plates were then wrapped with plastic wrap to prevent dehydration and incubated at 37°C for 12 to 14 h.
Antibacterial Assays with Surfactin
MICs of surfactin against planktonic cells of P. syringae were determined by the broth microdilution method using an inoculum of approximately 1 × 105 cfu mL-1. Microtiter plates (96 well, Nalge Nunc International, Rochester, NY) were prepared with serial 2-fold dilutions of surfactin (Sigma) in cation-adjusted Mueller-Hinton broth (DIFCO Laboratories). Surfactin was added from a 1 mg mL-1 stock solution in 2.5% (v/v) dimethyl sulfoxide. The MIC was visually defined as the lowest concentration of an antibiotic that completely inhibited cell growth after incubation for 22 h at 37°C. All susceptibility trials were conducted in triplicate. To check the bactericidal activity of surfactin against P. syringae, sub-MIC (0-5 μg mL-1), MIC (25 μg mL-1), and double the MIC levels (50 μg mL-1) of surfactin-treated bacterial cells in microtiter plates were stained with Molecular Probes BacLight Bacterial Viability Kit by incubating bacterial suspension at room temperature in the dark for 20 min, according to the manufacturer's manual. The samples were mounted with Citifluor antifading (Sigma) and observed for fluorescence with a fluorescence microscope (Fluroview LGPS-2, Olympus).
Quantificational Analysis of Surfactin
Surfactin concentration was analyzed by an HPLC procedure. B. subtilis cultures grown at different time points were withdrawn aseptically and centrifuged at 8,000g for 20 min to pellet the cells. The supernatant was extracted in methanol, concentrated and was further analyzed using an HPLC system consisting of P580 pumps (Dionex Co., Sunnyvale, CA) connected to an ASI-100 Automated Sample Injector (Dionex Co.), and a PDA-100 photodiode array variable UV/VIS detector (Dionex Co.). A C18 reverse-phase column (25.8 × 15 × 7 mm) was used for the separation of the extracts. Mobile phase solution A consisted of 3.8 mm trifluoroacetic acid in water and acetonitrile (solution B; Fisher Scientific). Standard surfactin was purchased from Sigma. An isocratic program with 20% (v/v) solution A and 80% (v/v) solution B for 35 min was used for all separations with an initial injection volume of 15 μL and a flow rate of 1 mL min-1. Chromeleon software (Dionex Co.) was used to identify and quantify peaks. In a method similar to the in situ challenge and root colonization experiments, extractions for surfactin were performed by using the interface between the two bacterial colonies by carefully cutting the agar piece (500 mg) and then extracting the agar piece and the intact B. subtilis colonized roots (mainly root tips and elongation zone region; approximately 5 cm long; 50 mg fresh weight) in methanol; post-centrifugation, the supernatant was analyzed by reverse-phase HPLC as described above. The data presented here are from five independent experiments, and quantification of surfactin was performed by combining these experiments to calculate the average mean and to standardize conditions for a representative spectrogram.
This work was supported by the Colorado State University Agricultural Experiment Station (grant to J.M.V.), by the National Science Foundation-CAREER (grant no. MCB 0093014 to J.M.V.), by the State of Colorado (Invasive Weeds Initiative to J.M.V.),by the Lindbergh Foundation (to J.M.V.), and by the U.S. Department of Energy (grant no. DE-FG03-97ER20274 to R.F.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.028712.
References
- Amellal N, Burtin G, Bartoli F, Heulin T (1998) Colonization of wheat roots by an exopolysaccharide-producing pantoea agglomerans strain and its effect on rhizosphere soil aggregation. Appl Environ Microbiol 64: 3740-3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anagnostopolous C, Spizizen J (1961) Requirements for transformation in Bacillus subtilis. J Bacteriol 81: 741-746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaka O, Shoda M (1996) Biocontrol of Rhizoctonia solani damping off of Tomato with Bacillus subtilis RB14. Appl Environ Microbiol 62: 4081-4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashan Y, Bashan LE (2002) Protection of tomato seedlings against infection by Pseudomonas syringae pv. tomato by using the plant growth-promoting bacterium Azospirillum brasilense. Appl Environ Microbiol 68: 2637-2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianciotto V, Andreotti S, Balestrini R, Bonfante R, Perotto S (2001) Mucoid mutants of the biocontrol strain Pseudomonas fluorescens CHA0 show increased ability in biofilm formation on mycorrhizal and nonmycorrhizal carrot roots. Mol Plant-Microbe Interact 14: 255-260 [DOI] [PubMed] [Google Scholar]
- Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R (2001) Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci USA 98: 11621-11626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanway CP (2002) Plant growth promotion by Bacillus and relatives. In R Berkeley, M Heyndrickx, N Logan, P De Vos, eds, B. subtilis for biocontrol in variety of plants. Blackwell Publishing, Malden, MA, pp 219-235
- Costacurta A, Vanderleyden J (1995) Synthesis of phytohormones by plant-associated bacteria. Crit Rev Microbiol 21: 1-18 [DOI] [PubMed] [Google Scholar]
- Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM (1995) Microbial biofilms. Annu Rev Microbiol 49: 711-745 [DOI] [PubMed] [Google Scholar]
- Davey ME, Caiazza NC, O'Toole GA (2003) Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J Bacteriol 185: 1027-1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KR, Schott E, Ausubel FM (1991) Virulence of selected phytopathic pseudomonads in Arabidopsis thaliana. Mol Plant-Microbe Interact 4: 477-488 [Google Scholar]
- Davey ME, O'Toole GA (2000) Microbial biofilm: from ecology to molecular genetics. Microbiol Mol Biol Rev 64: 847-867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defago G, Keel C (1995) Pseudomonads as biocontrol agents of diseases caused by soil borne pathogens. In HMT Hokkanen, JM Lynch, eds, Benefits and Risks of Introducing Biocontrol Agents. University Press, Cambridge, UK, pp 137-148
- Emmert EAB, Handelsman J (1999) Biocontrol of plant disease: a Gram-positive perspective. FEMS Microbiol Lett 171: 1-9 [DOI] [PubMed] [Google Scholar]
- Fall R, Kinsinger RF, Wheeler KA (2003) A simple method to isolate biofilm forming Bacillus subtilis and related species from plant roots. Appl Environ Microbiol (in press) [DOI] [PubMed]
- Germida JJ, Siciliano SD, Frietas JRD, Seib AM (1998) Diversity of root-associated bacteria associated with field-grown canola (Brassica napus L.) and wheat (Triticum aestivum L.). FEMS Microbiol Ecol 26: 43-49 [Google Scholar]
- Hamon MA, Lazazzera BA (2001) The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol Microbiol 42: 1119-1209 [DOI] [PubMed] [Google Scholar]
- Hemphill HE, Whitely HR (1975) Bacteriophages of Bacillus subtilis. Bacteriol Rev 39: 257-315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan A, Kolter R (2002) Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 296: 2229-2232 [DOI] [PubMed] [Google Scholar]
- Howarth FG (1991) Environmental impacts of classical biological control. Annu Rev Entomol 336: 485-509 [Google Scholar]
- Jakob K, Goss EM, Araki H, Van T, Krietman M, Bergelson J (2002) Pseudomonas viridiflava and Pseudomonas syringae-natural pathogens of Arabidopsis thaliana. Mol Plant-Microbe Interact 15: 1195-1203 [DOI] [PubMed] [Google Scholar]
- Kinsinger RF, Shirk MC, Fall R (2003) Rapid surface motility and biofilm formation in Bacillus subtilis is dependent on extracellular surfactin and potassium ion. J Bacteriol 185: 5627-5631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloepper JW, Zablotowick RM, Tipping EM, Lifshitz R (1991) Plant growth promotion mediated by bacterial rhizosphere colonizers. In DL Kliester, PG Cregan, eds, The Rhizosphere and Plant Growth. Kluwer Academic Press, Dordrecht, The Netherlands, pp 315-326
- Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Berkro MG, Bessieres P, Bolotin A, Borchert S (1997) The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390: 249-256 [DOI] [PubMed] [Google Scholar]
- Lilley AK, Fry JC, Bailey MJ, Day MJ (1996) Comparison of aerobic heterotrophic taxa isolated from four root domains of mature sugar beet (Beta vulgaris). FEMS Microbiol 21: 231-242 [Google Scholar]
- Lutenberg BJJ, De Weger LA, Bennett JW (1991) Microbial stimulation of plant growth and protection from disease. Curr Opin Micrbiol 2: 457-464 [Google Scholar]
- Lutenberg BJJ, Dekkers LC (1999) What makes Pseudomonas bacteria rhizosphere competent? Environ Microbiol 1: 9-13 [DOI] [PubMed] [Google Scholar]
- Michiels KW, Croes CL, Vanderleyden L (1991) Two different modes of attachment of Azospirillium brasilense Sp7 to wheat roots. J Gen Microbiol 137: 2241-2246 [Google Scholar]
- Mireles JR-II, Toguchi A, Harshey RM (2001) Salmonella enterica serovar Typhimurium swarming mutants with altered biofilm-forming abilities: surfactin inhibits biofilm formation. J Bacteriol 183: 5848-5854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassay with tissue culture. Physiol Plant 15: 473-497 [Google Scholar]
- Neu TR (1996) Significance of bacterial surface-active compounds in interaction of bacteria with interfaces. Micobiol Rev 60: 151-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osburn RM, Milner JL, Oplinger ES, Smith RS Handelsman J (1995) Effect of Bacillus cereus UW85 on the yield of soybean at two field sites in Wisconsin. Plant Dis 79: 551-556 [Google Scholar]
- O'Toole GA, Kolter R (1998) Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol Microbiol 30: 295-304 [DOI] [PubMed] [Google Scholar]
- O'Toole GA, Pratt LA, Watnick PI, Newman DK, Weaver VB, Kolter R (1999) Genetic approaches to the study of biofilms. Methods Enzymol 310: 91-109 [DOI] [PubMed] [Google Scholar]
- Pandey A, Palni LMS (1997) Bacillus species: the dominant bacteria of the rhizosphere of established tea bushes. Microbiol Res 152: 359-365 [DOI] [PubMed] [Google Scholar]
- Park S-W, Lawrence CB, Linden JC, Vivanco JM (2002) Isolation and characterization of a novel ribosome-inactivating protein from root cultures of pokeweed and its mechanism of secretion from roots. Plant Physiol 130: 164-178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M (1993) Integrational vectors for genetic manipulation in Bacillus subtilis. In AL Sonenshein, JA Hoch, R Losick, eds, Bacillus subtilis and Other Gram-Positive Bacteria: Biochemistry, Physiology, and Molecular Genetics. American Society for Microbiology, Washington, DC, pp 615-624
- Peypoux F, Bonmatin JM, Wallach J (1999) Recent trends in the biochemistry of surfactin. Appl Microbiol Biotechnol 51: 553-563 [DOI] [PubMed] [Google Scholar]
- Pinton R, Varanini Z, Nannipieri P (2001) The rhizosphere as a site of biochemical interactions among soil components, plants and microorganisms. In R Pinton, Z Varanini, P Nannipieri, eds The Rhizosphere: Biochemistry and Organic Substances in the Soil-Plant Interface. Marcel Dekker, New York, pp 1-17
- Powell KA, Jutsum AR (1993) Technical and commercial aspects of biological control products. Pestic Sci 37: 315-321 [Google Scholar]
- Pratt LA, Kolter R (1999) Genetic analyses of bacterial biofilm formation. Curr Opin Microbiol 2: 598-603 [DOI] [PubMed] [Google Scholar]
- Schippers B, Baker AW, Bakker PAHM (1987) Interactions of deleterious and beneficial rhizosphere microorganisms and the effect of cropping practices. Annu Rev Phytopathol 25: 339-358 [Google Scholar]
- Shoda M (2000) Bacterial control of plant diseases. J Biosci Bioeng 89: 515-521 [DOI] [PubMed] [Google Scholar]
- Stanley NR, Britton RA, Grossman AD, Lazazzera BA (2003) Identification of catabolite repression as a physiological regulator of biofilm formation by Bacillus subtilis by use of DNA microarrays. J Bacteriol 185: 1951-1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero P, Dangl JL (2001) A high-throughput method for quantifying growth of phytopathogenic bacteria in Arabidopsis thaliana. Plant J 28: 475-481 [DOI] [PubMed] [Google Scholar]
- Utkhede RS, Smith EM (1992) Promotion of apple tree growth and fruit production by the EBW-4 strain of Bacillus subtilis in apple replant disease soil. Can J Microbiol 38: 1270-1273 [DOI] [PubMed] [Google Scholar]
- van Veen JA, van Overbeek LS, van Elsas JD (1997) Fate and activity of microorganisms introduced into soil. Microbiol Mol Biol Rev 61: 121-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivanco JM, Savary BJ, Flores HE (1999) Characterization of two novel type I ribosome-inactivating proteins from the storage roots of the Andean crop Mirabilis expansa. Plant Physiol 119: 1447-1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrior P, Konduru K, Vasudevan P (2002) Formulation of biological control agents for pest and disease management. In SS Gnanamanickam, ed, Biological Control of Crop Diseases. Marcel Dekker, New York, pp 421-442
- Watnick PI, Kolter R (1999) Steps in the development of a Vibrio cholerae El Tor biofilm. Mol Microbiol 34: 586-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei YH, Chu IM (1998) Enhancement of surfactin production in iron-enriched media by Bacillus subtilis ATCC 21332. Enzy Microbe Technol 22: 724-728 [Google Scholar]
- Weller DM, Thomashow LS (1994) Current challenges in introducing beneficial microorganisms into the rhizosphere. In F O'Gara, DN Dowling, B Boesten, eds, Molecular Ecology of Rhizosphere Microorganisms. VCH, New York, pp 1-18
- Zuber P, Nakano MM, Marahiel MA (1993) Peptide antibiotics. In AL Sonenshein, JA Hoch, R Losick, eds, Bacillus subtilis and Other Gram-Positive Bacteria: Biochemistry, Physiology, and Molecular Genetics. American Society for Microbiology, Washington, DC, pp 897-916