Abstract
Emerging evidence indicates that RUNX3 is a tumor suppressor in breast cancer. RUNX3 is frequently inactivated in human breast cancer cell lines and cancer samples by hemizygous deletion of the Runx3 gene, hypermethylation of the Runx3 promoter, or cytoplasmic sequestration of RUNX3 protein. Inactivation of RUNX3 is associated with the initiation and progression of breast cancer. Female Runx3+/− mice spontaneously develop ductal carcinoma, and overexpression of RUNX3 inhibits the proliferation, tumorigenic potential, and invasiveness of breast cancer cells. This review is intended to summarize these findings and discuss the tumor suppressor function of RUNX3 in breast cancer.
Keywords: BREAST CANCER, ESTROGEN RECEPTOR, INACTIVATION, RUNX3, TUMOR SUPPRESSOR
INTRODUCTION
Breast cancer is the most frequently diagnosed cancer and the leading cause of cancer death in females worldwide. It accounted for 23% of the total new cases and 14% of the total cancer deaths in 2008 [Jemal et al., 2011]. In the United States, breast cancer is also a major cause of cancer death, accounting for 28% of the total new cases and 15% of the total cancer death in 2010 [Jemal et al., 2010]. Breast carcinogenesis is a complex, multistep and multifactorial process arising from abnormal proliferation of epithelium via the steps of precursor lesions, atypical ductal hyperplasia, and in situ carcinoma hyperplasia, followed by invasive carcinoma [Beckmann et al., 1997]. Even though the multistep nature of breast cancer carcinogenesis makes it difficult to identify the causing factors, various etiological factors have been found to contribute to the development and progression of breast cancer. These factors include the activation of oncogenes, inactivation of tumor suppressors, exposure to endogenous or exogenous steroid hormones, and variations in individuals’ genetic backgrounds [Osborne et al., 2004].
RUNX3 belongs to the Runt-related transcription factor family that regulates gene expression in several important developmental pathways. Studies from Runx3 knockout mice reveal that RUNX3 is required for T cell development during thymopoiesis and has a primary role in determining the dorsal-root ganglion proprioceptive neuron function [Inoue et al., 2002; Levanon et al., 2002; Taniuchi et al., 2002]. In addition to its ability to regulate the lineage-specific gene expression in developmental processes, RUNX3 has been shown to be involved in the formation of a variety of cancers [Ito, 2004]. RUNX3 was first suggested to be a tumor suppressor in gastric cancer due to the causal relationship between the loss of RUNX3 and the genesis and progression of gastric cancer [Li et al., 2002]. Gastric epithelium of Runx3−/− mice exhibited hyperplasia due to increased proliferation and diminished apoptosis from the insensitivity of gastric epithelia cells to the growth inhibitory effect of TGF-β [Li et al., 2002]. Since the discovery of the potential role of RUNX3 in the initiation and progression of gastric cancer, RUNX3 has been found to be involved in the development of a variety of cancers, including colorectal cancer, liver cancer, lung cancer, and breast cancer [Subramaniam et al., 2009b].
The Runx3 gene is located in 1p36, a region of frequent genomic loss in a wide variety of human carcinomas, including breast cancer [Weith et al., 1996]. A role for RUNX3 as a tumor suppressor in breast cancer emerged when its inactivation was seen in many breast cancer cell lines and breast cancer tissues. Like in other cancers, RUNX3 is inactivated in breast cancer by reduced copy number, promoter hypermethylation, hemizygous deletion, and protein mislocalization [Chen et al., 2007; Hwang et al., 2007; Jiang et al., 2008; Lau et al., 2006; Subramaniam et al., 2009a]. RUNX3 inactivation is considered to be an early event in breast cancer progression, and its expression generally decreases during this process [Subramaniam et al., 2009a]. RUNX3 expression in tumors is associated with a more favorable prognosis with reduced recurrence and better survival rates in breast cancer patients [Finak et al., 2008; Jiang et al., 2008]. In addition to the strong pathophysiological link between RUNX3 inactivation and breast cancer, recent studies from Runx3+/− animals provide further genetic evidence to support the notion that RUNX3 is a tumor suppressor in breast cancer [Huang et al., 2011]. Inactivation of RUNX3 in mammary epithelial cells leads to the development of ductal carcinoma in more than 20% of Runx3+/− female mice [Huang et al., 2011]. Consistent with its tumor suppressor activity, reintroduction of RUNX3 into breast cancer cells suppresses their tumorigenic potentials [Chen et al., 2007; Huang et al., 2011; Lau et al., 2006].
INACTIVATION OF RUNX3 BY PROMOTER HYPERMETHYLATION IN BREAST CANCER
Hypermethylation of promoter CpG islands of tumor suppressors is known to be a frequent and early event in carcinogenesis and is a common mechanism for the heritable maintenance of gene silencing in human cancer [Jones and Baylin, 2002]. In breast cancer, hypermethylation of promoter CpG islands has been described for various genes controlling different aspects of cellular function, including DNA replication, cell proliferation, programmed cell death, and cell migration and tissue invasion [Jovanovic et al., 2010; Widschwendter and Jones, 2002].
While methylation frequency varies in different studies, it has become clear that methylation of the Runx3 promoter is prevalent and tumor specific in breast cancer. Kim et al first reported that methylation of the Runx3 promoter was detected in 25% of human breast cancer samples (case number (n)=25) [Kim et al., 2004]. In an effort to identify the role of gene silencing via aberrant methylation in the TGF-β signaling pathway in human cancers, Suzuki et al also found that methylation of the Runx3 promoter occurred in 22% of breast cancer samples (n=37). A higher methylation frequency is found in another study with 52% of breast cancer samples (n=44) and 50% of breast cancer cell lines (n=19) showing Runx3 promoter hypermethylation [Lau et al., 2006]. In a separate study, Hwang et al showed a similar frequency of hypermethylation of the Runx3 promoter in 53% of breast cancer tissues (n=40) and in 57% of breast cancer cell lines (n=13) [Hwang et al., 2007].
Like the methylation for other tumor suppressors, the frequent hypermethylation of the Runx3 promoter appears to be an early event in breast cancer. No methylation was detected in the normal tissues and the earlier stages in breast cancer progression, including atypical ductal hyperplasia and flat epithelial atypia [Kim et al., 2004; Park et al., 2011; Subramaniam et al., 2009a; Suzuki et al., 2005]. Methylation of the Runx3 promoter starts to appear in ductal carcinoma in situ (DCIS), the precursor lesion of the breast, and remains at a similar frequency in invasive ductal carcinoma (IDC) [Park et al., 2011; Subramaniam et al., 2009a]. Furthermore, Runx3 promoter hypermethylation could also be detected in breast cancer patient sera. Tan et al demonstrated that Runx3 promoter hypermethylation was detected in 47% (9 out of 19) of patient serum samples [Tan et al., 2007]. Runx3 promoter hypermethylation appears to be a common feature in breast cancer.
While it is clear that hypermethylation of the Runx3 promoter plays a major role in the inactivation of RUNX3 in breast cancer, the detailed molecular mechanism by which this aberrant methylation is initiated is not clear. Nevertheless, estrogen, the major trigger for breast cancer, seems to play a role in this process (Fig. 1A). Prolonged exposure to nuclear hormones, especially estrogen, is known to cause aberrant imprinting and increases the risk of developing breast cancer. They disrupt normal growth of breast epithelia and trigger breast cancer development partially through estrogen-mediated gene silencing [Yager and Davidson, 2006]. Runx3 promoter hypermethylation was induced by estrogen in mammosphere-derived cells [Cheng et al., 2008]. This estrogen-dependent epigenetic silencing of Runx3 is likely mediated via the estrogen receptor (ER) signaling, since the expression of ERα in these breast progenitor cells inversely correlates with the expression of RUNX3 [Cheng et al., 2008]. Further supporting this possibility, Suzuki et al found that the frequency of Runx3 promoter methylation was higher in ER-positive samples than in ER-negative samples. Thirty-six percent of ER-positive samples had Runx3 promoter hypermethylation (8 out of 22), while none of the ER-negative samples had detectable hypermethylation of the Runx3 promoter (0 out of 8) [Suzuki et al., 2005]. Consistently, Subramaniam et al also reported that Runx3 promoter hypermethylation correlated significantly with positive ER expression in invasive carcinomas [Subramaniam et al., 2009a]. However, it remains to be determined how estrogen receptor signaling initiates the methylation of the Runx3 promoter. One possibility could be that estrogen receptor signaling regulates the expression or activity of the enzymes involved in the process of epigenetic gene silencing (Fig. 1A).
Fig. 1.
Inactivation of tumor suppressor RUNX3 in breast cancer. (A) Hypermethylation of Runx3 promoter. Estrogen or other cellular stress might induce hypermethylation by facilitating the recruitment or activation of histone- or DNA-modifying enzymes. Deacetylation of histones by HDACs or the concomitant methylation of histones by HMTs may result in the recruitment of the DNMT-containing repression complex, which induces the methylation of DNA and the silencing of Runx3. Ac=acetylation, Me=methylation, ○=CpG, ●=mCpG. (B) Inactivation of RUNX3 by protein mislocalization. Cytoplasmic relocalization of RUNX3 might result from 1) the masking of the NLS by a binding partner; 2) Src- or Pim-1-mediated RUNX3 phosphorylation; or 3) HDACs- or G9a-mediated deacetylation or methylation of RUNX3. Sequestration of RUNX3 in the cytoplasm prevents the expression of RUNX3 target genes.
In breast cancer, DNA hypermethylation together with abnormal histone modifications is frequently associated with epigenetic silencing of tumor suppressor genes and genomic instability [Jones and Baylin, 2007; Jovanovic et al., 2010]. Several enzymes involved in DNA methylation and histone modifications are found to regulate the expression of Runx3 (Fig. 1A). DNA methylation is conferred by DNA methyltransferases (DNMTs), which catalyze the transfer of the methyl group from S-adenosine methionine to the 5-carbon of the cytosine ring within CpG dinucleotides [Jones and Baylin, 2007; Jovanovic et al., 2010]. Therefore, it is not surprising to see that DNMTs are directly involved in the methylation of the Runx3 promoter. Jung et al showed that depletion of DNMT1 but not DNMT3b by siRNA reactivated the expression of RUNX3 by increasing the unmethylated levels of the Runx3 promoter [Jung et al., 2007]. DNA methylation may also be indirectly regulated by histone modifications such as histone methylation [Tamaru and Selker, 2001]. G9a, a lysine methyltransferase for histone H3 lysine 9 (H3K9) methylation, induces Runx3 silencing [Lee et al., 2009]. This G9a-mediated H3 methylation might contribute to Runx3 promoter methylation since G9a has been shown to be required for de novo DNA methylation [Leung et al., 2011]. Enhancer of zeste homologue 2 (EZH2), a highly conserved histone methyltransferase that specifically targets histone H3K27 [van der Vlag and Otte, 1999], also suppresses Runx3 expression [Fujii et al., 2008]. EZH2 bound to the promoter of Runx3 and induced the trimethylation of H3K27 in breast cancer cells [Fujii et al., 2008]. This binding might eventually trigger the methylation of the Runx3 promoter since EZH2-mediated histone methylation serves as a recruitment platform for the DNMTs [Vire et al., 2006]. Inhibiting the activity of EZH2 either by siRNA or microRNA successfully reduced H3K27 trimethylation and increased expression of Runx3 [Fujii et al., 2008; Varambally et al., 2008].
INACTIVATION OF RUNX3 BY PROTEIN MISLOCALIZATION IN BREAST CANCER
In addition to promoter hypermethylation of Runx3, mislocalization of RUNX3 is often observed in cancer and accounts for a significant proportion of RUNX3 inactivation in cancer. Cytoplasmic re-localization of RUNX3 is found in 38% of gastric cancer and in 15% of colorectal cancer [Ito et al., 2008; Ito et al., 2005]. In breast cancer, low-level cytoplasmic mislocalized RUNX3 was observed in more than 80% of DCIS and IDC samples, while normal breast epithelia had only nuclear RUNX3 [Lau et al., 2006; Subramaniam et al., 2009a], indicating that inactivation of RUNX3 by cytoplasmic relocalization is also an early event in the carcinogenic process. Promoter hypermethylation and mislocalization, two likely independent events, seem to account for the loss of RUNX3 function in nearly all breast cancer samples [Lau et al., 2006; Subramaniam et al., 2009a]. While one allele of Runx3 could be inactivated through promoter hypermethylation, the remaining wild-type allele could be inactivated through cytoplasmic relocalization of its product, RUNX3 protein.
Mistargeting of tumor suppressors have significant cellular consequences and potentially lead to the initiation and progression of cancer [Fabbro and Henderson, 2003]. Being a transcription factor, nuclear localization of RUNX3 protein is essential for its proper function, and relocalization of RUNX3 from the nucleus to the cytoplasm is known to be associated with loss of its function and is implicated in tumorigenesis [Chuang and Ito, 2010]. Proper subcellular localization represents an important regulatory mechanism for controlling the functions of RUNX3 protein. However, the underlying mechanisms for RUNX3 inactivation by mislocalization in breast cancer remain obscure.
RUNX3 is a downstream target of TGF-β signaling and cooperates with the SMADs to regulate the expression of its target genes [Ito and Miyazono, 2003]. Activation of TGF-β signaling induces the nuclear translocalization of RUNX3 and the subsequent growth inhibition [Ito et al., 2005]. It is well documented that TGF-β signaling is frequently impaired in cancer tissues. This impaired TGF-β signaling might result in the aberrant cytoplasmic localization of RUNX3.
Recognition of the protein import or export signals by their respective receptors is a prerequisite for nucleocytoplasmic protein shuttling [Nigg, 1997]. The masking of these transport signals, either by changes in protein conformation or by binding of a partner protein, provides a quick and efficient mechanism for preventing signal recognition by the nuclear transport machinery [Nigg, 1997]. Although a clear nuclear localization signal (NLS) for RUNX3 has not yet been identified, it is believed that such signals exist both in the Runt domain and in the C-terminus of RUNX3 [Adya et al., 1998; Kanno et al., 1998]. Binding of protein to the Runt domain or C-terminus might mask the NLS and prevent the nuclear import of RUNX3 (Fig. 1B). Supporting this hypothesis, Chi et al showed that binding of MDM2 to the Runt domain sequestered RUNX3 in the cytoplasm [Chi et al., 2009]. Similarly, binding of Jun-activation domain-binding protein 1 (Jab1/CSN5) to the Runt domain of RUNX3 led to the cytoplasmic localization of RUNX3 [Kim et al., 2009]. Interestingly, RUNX3 nuclear export was often coupled with its degradation [Chi et al., 2009; Kim et al., 2009], indicating that cytoplasmic relocalization might be an essential step for the degradation and inactivation of RUNX3 by the cytoplasmic proteasome machinery.
RUNX3 is subject to a variety of posttranslational modifications, including phosphorylation, acetylation and ubiquitination [Bae and Lee, 2006]. These modifications could also alter the subcellular localization of RUNX3 if such a modification occurs within or proximal to the nuclear import or export signals (Fig. 1B). Phosphorylation has been shown to play an important role in the redistribution of RUNX3. Tyrosine phosphorylation of RUNX3 by oncogenic protein Src kinase, which is overexpressed in many human cancer cells, results in the cytoplasmic localization of RUNX3. In Src-activated breast cancer cell lines, including BT20 and MDA-MB-468, endogenous RUNX3 is phosphorylated and is localized in the cytoplasm due to the enhanced nuclear export of RUNX3 [Goh et al., 2010]. Another oncogenic protein, Pim-1, a serine/threonine kinase, phosphorylates the Runt domain of RUNX3 and induces the nuclear export of RUNX3 [Kim et al., 2008]. Hypoxia has also been shown to induce the cytoplasmic relocalization of RUNX3 through histone deacetylase HDAC1 and histone methyltransferase G9a [Lee et al., 2009]. The hypoxia-induced relocalization likely results from the HDAC1-mediated deacetylation of RUNX3 and G9a-mediated methylation of RUNX3 (Fig. 1B). Both of these modifications have been shown to be involved in the nucleocytoplasmic shuttling of RUNX3 [Lee, 2011].
RUNX3 TARGETS ESTROGEN RECEPTOR α FOR PROTEASOME-MEDIATED DEGRADATION
Estrogen receptor signaling plays a critical role in normal mammary gland development through the regulation of genes involved in the cell cycle and apoptosis [Katzenellenbogen and Katzenellenbogen, 2000]. Abnormal estrogen receptor signaling is associated with initiation and progression of breast cancer [Cheskis et al., 2007]. Our recent studies demonstrate that RUNX3 suppresses estrogen receptor signaling by inhibiting the transcriptional activity of ERα and reducing ERα-dependent cancer cell proliferation and tumorigenic potential [Huang et al., 2011]. Importantly, about 20% of female Runx3+/− mice spontaneously developed ductal carcinoma with an enhanced expression of ERα and proliferation marker Ki-67 [Huang et al., 2011]. Overexpression of RUNX3 in breast cancer cells reduces the cellular levels of ERα, whereas depletion of RUNX3 by siRNA enhances the cellular levels of ERα. Consistently, expression of RUNX3 and ERα is inversely correlated in breast cancer cell lines and human breast cancer samples [Huang et al., 2011]. Reduced ERα expression is mechanistically linked with RUNX3-mediated ubiquitination and degradation of ERα [Huang et al., 2011].
Currently, it is not clear how RUNX3 induces the ubiquitination and degradation of ERα. One possibility could be that binding of RUNX3 to ERα alters its posttranslational modification, thus changing its stability. We found that RUNX3 binds to the hinge region of ERα (Huang and Chen, unpublished data), which is subject to a variety of posttranslational modifications and is critical for the stability of ERα [Berry et al., 2008; Subramanian et al., 2008]. It is also possible that binding of RUNX3 to ERα facilitates the recruitment of an E3 ligase for ERα. E3 ligases such as carboxyl terminus of Hsc70-interacting protein (CHIP) and MDM2 have been shown to mediate the ubiquitination of ERα [Duong et al., 2007; Fan et al., 2005], and could potentially be involved in RUNX3-induced ubiquitination and degradation of ERα. In fact, RUNX3 has been found to be associated with MDM2 and with Smurfs, which are also E3 ligases [Chi et al., 2009; Jin et al., 2004], and may utilize these E3 ligases for the ubiquitination of ERα. The detailed molecular mechanisms need to be further clarified.
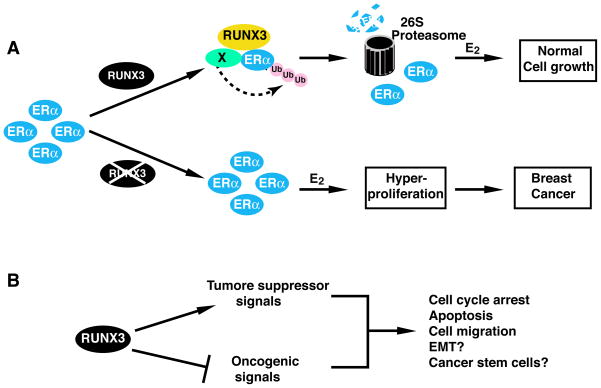
How does RUNX3 act as tumor suppressor by controlling the cellular levels of ERα? Tight control of ERα level by proteasome-mediated degradation of ERα is important in maintaining normal estrogen responsiveness [Duong et al., 2007; Fan et al., 2004; Tateishi et al., 2004]. In normal mammary tissues where RUNX3 is present, the association of RUNX3 with ERα might limit the levels of ERα and estrogen responsiveness by promoting the ubiquitination and degradation of ERα. RUNX3, by modulating ERα turnover, might dictate the cellular response to circulating estrogen levels and prevent the excessive proliferation of ERα-positive cells. However, when RUNX3 is inactivated, cellular ERα levels and subsequent estrogen-stimulated cell proliferation would be enhanced due to the increased stability of ERα. Enhanced ERα expression in normal breast epithelium is associated with increased risk of breast cancer [Khan et al., 1994; Shoker et al., 2000]. Therefore, RUNX3 might function as a “gate-keeper” for breast cancer by controlling the cellular level of ERα (Fig. 2A).
Fig. 2.
RUNX3 integrates with various cellular signaling pathways for its tumor suppressor activity. (A) RUNX3 suppresses ERα signaling in breast cancer. Binding of RUNX3 to ERα induces the ubiquitination and degradation of ERα by an unidentified E3 ligase (X). The low levels of ERα maintain the normal growth of mammary epithelial cells. When RUNX3 is inactivated, the cellular levels of ERα are enhanced, leading to the enhanced response to estradiol (E2), the hyperproliferation of breast cells, and eventually the formation of breast cancer. (B) RUNX3 could either act as a downstream target of a tumor suppressor pathway (e.g, TGF-β signaling) or function as a suppressor in an oncogenic pathway (e.g., Wnt signaling) to regulate cell proliferation, apoptosis, and cell migration. By interacting with various signaling pathways, RUNX3 might also have a role in breast cancer carcinogenesis by regulating the EMT and breast cancer stem cell development.
It has to be noticed that RUNX3 can also suppress the tumorigenic potential of ERα negative breast cancer cells without affecting the proliferation of the cells [Lau et al., 2006]. While the exact mechanism remains unidentified, the anti-tumor activity of RUNX3 in the ERα-negative cells might result from RUNX3-mediated cell apoptosis. RUXN3 has been shown to cooperate with other factors, such as FOXO3a or receptor-regulated SMADs (R-SMAD), to induce cellular apoptosis by activating genes involved in apoptosis and cell cycle arrest [Yamamura et al., 2006; Yano et al., 2006]. Additionally, RUNX3 might also target other cellular signaling pathways to exert its anti-tumor activity in these ERα negative cells.
RUNX3 INTEGRATES WITH OTHER CELLULAR SIGNALING PATHWAYS IN BREAST CANCER
Besides the ERα signaling pathway, RUNX3 has been shown to be associated with some other major signaling pathways essential for cancer development. RUNX3 serves either as a downstream target of tumor suppressor signaling pathways or as an antagonist of oncogenic pathways. The ability of RUX3 to positively or negatively modulate these pathways also contributes to its anti-tumor activity.
TGF-β regulates all phases of postnatal mammary gland development, including branching morphogenesis, lactation, and involution. TGF-β also plays a key role in suppressing mammary tumorigenesis by preventing mammary epithelial cell proliferation or by inducing their apoptosis [Drabsch and ten Dijke, 2011]. When TGF-β is bound, TGF-β receptors phosphorylate and activate R-SMADs. Activated R-SMADs associate with common mediator SMAD4 and enter the nucleus, where they bind to different transcription factors to regulate target gene expression [Ito and Miyazono, 2003]. RUNX3 has been shown to directly bind to SMADs and functions as an integral part of the TGF-β signaling pathway [Ito and Miyazono, 2003]. TGF-β receptors and their downstream signaling components are frequently inactivated in cancers, resulting in the resistance of cancer cells to TGF-β-mediated cell death or cell cycle arrest [Cohen, 2003]. Since the downstream SMAD pathway remains active in a majority of breast cancer cells [Xie et al., 2002], the resistance of these cells to the growth-inhibitory effects of TGF-β might be derived from the lack of functional RUNX3 in these breast cancer cells (Fig. 2B). In support of this possibility, Yano et al showed that reintroduction of RUNX3 into RUNX3-deficient cancer cells re-established cells’ sensitivity to TGF-β by up-regulating the expression of proapoptotic gene Bim [Yano et al., 2006]. As such, TGF-β-mediated cell apoptosis or cell cycle arrest might rely on the ability of a functional RUNX3 (Fig. 2B), likely in cooperation with SMADs, to regulate target gene expression [Ito and Miyazono, 2003].
On the other hand, RUNX3 might elicit its anti-tumor activity by interfering with some oncogenic cellular signaling pathways. Wnt oncogenic singling pathway is critical for the normal development of the mammary gland. Wnt signaling regulates cell proliferation and cell survival by increasing β-catenin levels and alters gene expression via transcription factors such as Lef/TCF [Reya and Clevers, 2005]. RUNX3 attenuates Wnt signaling by directly inhibiting β-catenin/TCF4 in colon cancer and gastric cancer [Ito, 2011]. RUNX3 forms a complex with β-catenin/TCFs and inhibits the transactivation of β-catenin/TCFs by preventing β-catenin/TCFs DNA binding [Ito et al., 2008]. Therefore, RUNX3 is critical for maintaining the normal function of Wnt signaling (Fig. 2B). Dysregulation of Wnt signaling is found to promote breast cancer, and inactivation of RUNX3 might alter the proper function of Wnt signaling and trigger breast carcinogenesis [Boras-Granic and Wysolmerski, 2008].
RUNX3 might also target oncogenic Notch signaling to suppress mammary tumor formation. Notch signaling is important for normal breast development, and abnormal Notch signaling is associated with the development of breast cancer [Guo et al., 2011]. RUNX3 was found to directly associate with the intracellular domain of Notch1 and suppress Notch signaling in hepatocellular carcinoma cells [Gao et al., 2010]. It is possible that RUNX3 utilizes a similar mechanism to antagonize Notch signaling in breast cancer (Fig. 2B). Dysregulation of different signaling pathways might be involved in the formation of different subtypes of breast cancer with distinct gene expression profiles and different clinical outcomes [Sorlie et al., 2001]. It remains to be determined whether inactivation of RUNX3 is associated with the initiation and progression of a particular breast cancer subtype by interacting with a specific signaling pathway or whether RUNX3 targets various pathways concomitantly to achieve its ultimate tumor suppressor activity.
CONCLUSING REMARKS AND FUTURE PROSPECTS
The frequent inactivation of RUNX3 in breast cancer and the development of mammary ductal carcinoma in Runx3+/− female mice strongly support the notion that RUNX3 is a breast cancer tumor suppressor. Hypermethylation of the Runx3 promoter and cytoplasmic relocalization account for the majority of RUNX3 inactivation in breast cancer. It is important to note that these events are reversible and that RUNX3 could be re-activated by inhibitors of DNMTs or nuclear export receptor CRM1 [Goh et al., 2010; Guo et al., 2002]. Reintroduction of RUNX3 into RUNX3-deficient breast cancer cells suppressed cancer cell proliferation and their tumorigenic potential [Chen et al., 2007; Huang et al., 2011; Lau et al., 2006]. Therefore, restoring RUNX3 activation by specific small molecules or inhibitors to block these two events might constitute a novel therapeutic strategy for the treatment of breast cancer.
RUNX3 has been suggested to be a potential immunohistochemical marker for use in diagnostic histopathology, since inactivation of RUNX3 is tumor specific and represents an early event during breast cancer development [Subramaniam et al., 2009b]. A number of studies illustrate the potential for the use of methylation markers in the early detection of a variety of cancers including breast cancer [Brooks et al., 2009]. Interestingly, methylation of the Runx3 promoter could be detected in serum samples of breast cancer patients [Tan et al., 2007]. It is plausible to speculate that RUNX3 could be a promising diagnostic and prognostic marker in breast cancer.
While many of the recent findings have suggested a tumor suppressor role for RUNX3 in breast cancer, the function of RUNX3 and its regulation in normal mammary gland development and in breast cancer carcinogenesis remain largely elusive. Future studies using Runx3 deletion animals might provide new insights into these unknown aspects. It is also worthy of notice that Runx3+/− female mice developed mammary gland tumors at 14 or 15 months of life, an age corresponding to age 40 to 50 in human years, making Runx3+/− mice a potential mouse model of spontaneously occurring mammary tumor. Additionally, RUNX3 integrates with various signaling pathways, which play important roles in breast cancer carcinogenesis, including epithelial-mesenchymal transition (EMT), the development of breast cancer stem cells, and breast cancer metastasis. In addition to its ability to regulate breast cancer cell proliferation, cell apoptosis and cell migration, RUNX3 might be associated with these various aspects of breast cancer carcinogenesis. Furthermore, the development of resistance to hormone therapy is a severe limitation in the treatment of ERα-positive breast tumors. Since RUNX3 directly associates with ERα and regulates its stability, whether restoring RUNX3 expression may sensitize endocrine refractory breast tumors is also an interesting question and remains to be further explored.
Acknowledgments
This work is supported in part by fund provided by UIUC (to L.F.C.) and NIH grant DK-085158 (to L.F.C.).
References
- Adya N, Stacy T, Speck NA, Liu PP. The leukemic protein core binding factor beta (CBFbeta)-smooth-muscle myosin heavy chain sequesters CBFalpha2 into cytoskeletal filaments and aggregates. Mol Cell Biol. 1998;18:7432–43. doi: 10.1128/mcb.18.12.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae SC, Lee YH. Phosphorylation, acetylation and ubiquitination: the molecular basis of RUNX regulation. Gene. 2006;366:58–66. doi: 10.1016/j.gene.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Beckmann MW, Niederacher D, Schnurch HG, Gusterson BA, Bender HG. Multistep carcinogenesis of breast cancer and tumour heterogeneity. J Mol Med (Berl) 1997;75:429–39. doi: 10.1007/s001090050128. [DOI] [PubMed] [Google Scholar]
- Berry NB, Fan M, Nephew KP. Estrogen receptor-alpha hinge-region lysines 302 and 303 regulate receptor degradation by the proteasome. Mol Endocrinol. 2008;22:1535–51. doi: 10.1210/me.2007-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boras-Granic K, Wysolmerski JJ. Wnt signaling in breast organogenesis. Organogenesis. 2008;4:116–22. doi: 10.4161/org.4.2.5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks J, Cairns P, Zeleniuch-Jacquotte A. Promoter methylation and the detection of breast cancer. Cancer Causes and Control. 2009;20:1539–50. doi: 10.1007/s10552-009-9415-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Salto-Tellez M, Palanisamy N, Ganesan K, Hou Q, Tan LK, Sii LH, Ito K, Tan B, Wu J, Tay A, Tan KC, Ang E, Tan BK, Tan PH, Ito Y, Tan P. Targets of genome copy number reduction in primary breast cancers identified by integrative genomics. Genes, Chromosomes and Cancer. 2007;46:288–301. doi: 10.1002/gcc.20411. [DOI] [PubMed] [Google Scholar]
- Cheng AS, Culhane AC, Chan MW, Venkataramu CR, Ehrich M, Nasir A, Rodriguez BA, Liu J, Yan PS, Quackenbush J, Nephew KP, Yeatman TJ, Huang TH. Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome. Cancer Res. 2008;68:1786–96. doi: 10.1158/0008-5472.CAN-07-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheskis BJ, Greger JG, Nagpal S, Freedman LP. Signaling by estrogens. J Cell Physiol. 2007;213:610–7. doi: 10.1002/jcp.21253. [DOI] [PubMed] [Google Scholar]
- Chi XZ, Kim J, Lee YH, Lee JW, Lee KS, Wee H, Kim WJ, Park WY, Oh BC, Stein GS, Ito Y, van Wijnen AJ, Bae SC. Runt-related transcription factor RUNX3 is a target of MDM2-mediated ubiquitination. Cancer Res. 2009;69:8111–9. doi: 10.1158/0008-5472.CAN-09-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang LS, Ito Y. RUNX3 is multifunctional in carcinogenesis of multiple solid tumors. Oncogene. 2010;29:2605–15. doi: 10.1038/onc.2010.88. [DOI] [PubMed] [Google Scholar]
- Cohen MM., Jr TGF beta/Smad signaling system and its pathologic correlates. Am J Med Genet A. 2003;116A:1–10. doi: 10.1002/ajmg.a.10750. [DOI] [PubMed] [Google Scholar]
- Drabsch Y, ten Dijke P. TGF-beta signaling in breast cancer cell invasion and bone metastasis. Journal of Mammary Gland Biology and Neoplasia. 2011;16:97–108. doi: 10.1007/s10911-011-9217-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong V, Boulle N, Daujat S, Chauvet J, Bonnet S, Neel H, Cavailles V. Differential regulation of estrogen receptor alpha turnover and transactivation by Mdm2 and stress-inducing agents. Cancer Res. 2007;67:5513–21. doi: 10.1158/0008-5472.CAN-07-0967. [DOI] [PubMed] [Google Scholar]
- Fabbro M, Henderson BR. Regulation of tumor suppressors by nuclear-cytoplasmic shuttling. Experimental Cell Research. 2003;282:59–69. doi: 10.1016/s0014-4827(02)00019-8. [DOI] [PubMed] [Google Scholar]
- Fan M, Nakshatri H, Nephew KP. Inhibiting proteasomal proteolysis sustains estrogen receptor-alpha activation. Mol Endocrinol. 2004;18:2603–15. doi: 10.1210/me.2004-0164. [DOI] [PubMed] [Google Scholar]
- Fan M, Park A, Nephew KP. CHIP (carboxyl terminus of Hsc70-interacting protein) promotes basal and geldanamycin-induced degradation of estrogen receptor-alpha. Mol Endocrinol. 2005;19:2901–14. doi: 10.1210/me.2005-0111. [DOI] [PubMed] [Google Scholar]
- Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, Hallett M, Park M. Stromal gene expression predicts clinical outcome in breast cancer. Nature Medicine. 2008;14:518–27. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- Fujii S, Ito K, Ito Y, Ochiai A. Enhancer of zeste homologue 2 (EZH2) down-regulates RUNX3 by increasing histone H3 methylation. J Biol Chem. 2008;283:17324–32. doi: 10.1074/jbc.M800224200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Chen Y, Wu KC, Liu J, Zhao YQ, Pan YL, Du R, Zheng GR, Xiong YM, Xu HL, Fan DM. RUNX3 directly interacts with intracellular domain of Notch1 and suppresses Notch signaling in hepatocellular carcinoma cells. Experimental Cell Research. 2010;316:149–57. doi: 10.1016/j.yexcr.2009.09.025. [DOI] [PubMed] [Google Scholar]
- Goh YM, Cinghu S, Hong ET, Lee YS, Kim JH, Jang JW, Li YH, Chi XZ, Lee KS, Wee H, Ito Y, Oh BC, Bae SC. Src kinase phosphorylates RUNX3 at tyrosine residues and localizes the protein in the cytoplasm. J Biol Chem. 2010;285:10122–9. doi: 10.1074/jbc.M109.071381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Liu M, Gonzalez-Perez RR. Role of Notch and its oncogenic signaling crosstalk in breast cancer. Biochimica et Biophysica Acta. 2011;1815:197–213. doi: 10.1016/j.bbcan.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo WH, Weng LQ, Ito K, Chen LF, Nakanishi H, Tatematsu M, Ito Y. Inhibition of growth of mouse gastric cancer cells by Runx3, a novel tumor suppressor. Oncogene. 2002;21:8351–5. doi: 10.1038/sj.onc.1206037. [DOI] [PubMed] [Google Scholar]
- Huang B, Qu Z, Ong CW, Tsang YH, Xiao G, Shapiro D, Salto-Tellez M, Ito K, Ito Y, Chen LF. RUNX3 acts as a tumor suppressor in breast cancer by targeting estrogen receptor alpha. Oncogene. 2011 doi: 10.1038/onc.2011.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang KT, Han W, Bae JY, Hwang SE, Shin HJ, Lee JE, Kim SW, Min HJ, Noh DY. Downregulation of the RUNX3 gene by promoter hypermethylation and hemizygous deletion in breast cancer. J Korean Med Sci. 2007;22(Suppl):S24–31. doi: 10.3346/jkms.2007.22.S.S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Ozaki S, Shiga T, Ito K, Masuda T, Okado N, Iseda T, Kawaguchi S, Ogawa M, Bae SC, Yamashita N, Itohara S, Kudo N, Ito Y. Runx3 controls the axonal projection of proprioceptive dorsal root ganglion neurons. Nat Neurosci. 2002;5:946–54. doi: 10.1038/nn925. [DOI] [PubMed] [Google Scholar]
- Ito K. RUNX3 in oncogenic and anti-oncogenic signaling in gastrointestinal cancers. J Cell Biochem. 2011;112:1243–9. doi: 10.1002/jcb.23047. [DOI] [PubMed] [Google Scholar]
- Ito K, Lim AC, Salto-Tellez M, Motoda L, Osato M, Chuang LS, Lee CW, Voon DC, Koo JK, Wang H, Fukamachi H, Ito Y. RUNX3 attenuates beta-catenin/T cell factors in intestinal tumorigenesis. Cancer Cell. 2008;14:226–37. doi: 10.1016/j.ccr.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Ito K, Liu Q, Salto-Tellez M, Yano T, Tada K, Ida H, Huang C, Shah N, Inoue M, Rajnakova A, Hiong KC, Peh BK, Han HC, Ito T, Teh M, Yeoh KG, Ito Y. RUNX3, a novel tumor suppressor, is frequently inactivated in gastric cancer by protein mislocalization. Cancer Res. 2005;65:7743–50. doi: 10.1158/0008-5472.CAN-05-0743. [DOI] [PubMed] [Google Scholar]
- Ito Y. Oncogenic potential of the RUNX gene family: ‘overview’. Oncogene. 2004;23:4198–208. doi: 10.1038/sj.onc.1207755. [DOI] [PubMed] [Google Scholar]
- Ito Y, Miyazono K. RUNX transcription factors as key targets of TGF-beta superfamily signaling. Current Opinion in Genetics and Development. 2003;13:43–7. doi: 10.1016/s0959-437x(03)00007-8. [DOI] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: A Cancer Journal for Clinicians. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA: A Cancer Journal for Clinicians. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Tong D, Lou G, Zhang Y, Geng J. Expression of RUNX3 gene, methylation status and clinicopathological significance in breast cancer and breast cancer cell lines. Pathobiology. 2008;75:244–51. doi: 10.1159/000132385. [DOI] [PubMed] [Google Scholar]
- Jin YH, Jeon EJ, Li QL, Lee YH, Choi JK, Kim WJ, Lee KY, Bae SC. Transforming growth factor-beta stimulates p300-dependent RUNX3 acetylation, which inhibits ubiquitination-mediated degradation. J Biol Chem. 2004;279:29409–17. doi: 10.1074/jbc.M313120200. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic J, Ronneberg JA, Tost J, Kristensen V. The epigenetics of breast cancer. Mol Oncol. 2010;4:242–54. doi: 10.1016/j.molonc.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y, Park J, Kim TY, Park JH, Jong HS, Im SA, Robertson KD, Bang YJ. Potential advantages of DNA methyltransferase 1 (DNMT1)-targeted inhibition for cancer therapy. J Mol Med. 2007;85:1137–48. doi: 10.1007/s00109-007-0216-z. [DOI] [PubMed] [Google Scholar]
- Kanno Y, Kanno T, Sakakura C, Bae SC, Ito Y. Cytoplasmic sequestration of the polyomavirus enhancer binding protein 2 (PEBP2)/core binding factor alpha (CBFalpha) subunit by the leukemia-related PEBP2/CBFbeta-SMMHC fusion protein inhibits PEBP2/CBF-mediated transactivation. Mol Cell Biol. 1998;18:4252–61. doi: 10.1128/mcb.18.7.4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzenellenbogen BS, Katzenellenbogen JA. Estrogen receptor transcription and transactivation: Estrogen receptor alpha and estrogen receptor beta: regulation by selective estrogen receptor modulators and importance in breast cancer. Breast Cancer Res. 2000;2:335–44. doi: 10.1186/bcr78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SA, Rogers MA, Obando JA, Tamsen A. Estrogen receptor expression of benign breast epithelium and its association with breast cancer. Cancer Res. 1994;54:993–7. [PubMed] [Google Scholar]
- Kim HR, Oh BC, Choi JK, Bae SC. Pim-1 kinase phosphorylates and stabilizes RUNX3 and alters its subcellular localization. J Cell Biochem. 2008;105:1048–58. doi: 10.1002/jcb.21906. [DOI] [PubMed] [Google Scholar]
- Kim JH, Choi JK, Cinghu S, Jang JW, Lee YS, Li YH, Goh YM, Chi XZ, Lee KS, Wee H, Bae SC. Jab1/CSN5 induces the cytoplasmic localization and degradation of RUNX3. J Cell Biochem. 2009;107:557–65. doi: 10.1002/jcb.22157. [DOI] [PubMed] [Google Scholar]
- Kim TY, Lee HJ, Hwang KS, Lee M, Kim JW, Bang YJ, Kang GH. Methylation of RUNX3 in various types of human cancers and premalignant stages of gastric carcinoma. Lab Invest. 2004;84:479–84. doi: 10.1038/labinvest.3700060. [DOI] [PubMed] [Google Scholar]
- Lau QC, Raja E, Salto-Tellez M, Liu Q, Ito K, Inoue M, Putti TC, Loh M, Ko TK, Huang C, Bhalla KN, Zhu T, Ito Y, Sukumar S. RUNX3 is frequently inactivated by dual mechanisms of protein mislocalization and promoter hypermethylation in breast cancer. Cancer Res. 2006;66:6512–20. doi: 10.1158/0008-5472.CAN-06-0369. [DOI] [PubMed] [Google Scholar]
- Lee SH, Kim J, Kim WH, Lee YM. Hypoxic silencing of tumor suppressor RUNX3 by histone modification in gastric cancer cells. Oncogene. 2009;28:184–94. doi: 10.1038/onc.2008.377. [DOI] [PubMed] [Google Scholar]
- Lee YM. Control of RUNX3 by histone methyltransferases. J Cell Biochem. 2011;112:394–400. doi: 10.1002/jcb.22969. [DOI] [PubMed] [Google Scholar]
- Leung DC, Dong KB, Maksakova IA, Goyal P, Appanah R, Lee S, Tachibana M, Shinkai Y, Lehnertz B, Mager DL, Rossi F, Lorincz MC. Lysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencing. Proc Natl Acad Sci U S A. 2011;108:5718–23. doi: 10.1073/pnas.1014660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon D, Bettoun D, Harris-Cerruti C, Woolf E, Negreanu V, Eilam R, Bernstein Y, Goldenberg D, Xiao C, Fliegauf M, Kremer E, Otto F, Brenner O, Lev-Tov A, Groner Y. The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J. 2002;21:3454–63. doi: 10.1093/emboj/cdf370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, Kim HM, Kim WJ, Yamamoto H, Yamashita N, Yano T, Ikeda T, Itohara S, Inazawa J, Abe T, Hagiwara A, Yamagishi H, Ooe A, Kaneda A, Sugimura T, Ushijima T, Bae SC, Ito Y. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113–24. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- May CD, Sphyris N, Evans KW, Werden SJ, Guo W, Mani SA. Epithelial-mesenchymal transition and cancer stem cells: a dangerously dynamic duo in breast cancer progression. Breast Cancer Res. 2011;13:202. doi: 10.1186/bcr2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–87. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Osborne C, Wilson P, Tripathy D. Oncogenes and tumor suppressor genes in breast cancer: potential diagnostic and therapeutic applications. The Oncologist. 2004;9:361–77. doi: 10.1634/theoncologist.9-4-361. [DOI] [PubMed] [Google Scholar]
- Park SY, Kwon HJ, Lee HE, Ryu HS, Kim SW, Kim JH, Kim IA, Jung N, Cho NY, Kang GH. Promoter CpG island hypermethylation during breast cancer progression. Virchows Archiv. 2011;458:73–84. doi: 10.1007/s00428-010-1013-6. [DOI] [PubMed] [Google Scholar]
- Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–50. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- Shoker BS, Jarvis C, Clarke RB, Anderson E, Munro C, Davies MP, Sibson DR, Sloane JP. Abnormal regulation of the oestrogen receptor in benign breast lesions. J Clin Pathol. 2000;53:778–83. doi: 10.1136/jcp.53.10.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam MM, Chan JY, Soong R, Ito K, Ito Y, Yeoh KG, Salto-Tellez M, Putti TC. RUNX3 inactivation by frequent promoter hypermethylation and protein mislocalization constitute an early event in breast cancer progression. Breast Cancer Res Treat. 2009a;113:113–21. doi: 10.1007/s10549-008-9917-4. [DOI] [PubMed] [Google Scholar]
- Subramaniam MM, Chan JY, Yeoh KG, Quek T, Ito K, Salto-Tellez M. Molecular pathology of RUNX3 in human carcinogenesis. Biochim Biophys Acta. 2009b;1796:315–31. doi: 10.1016/j.bbcan.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Subramanian K, Jia D, Kapoor-Vazirani P, Powell DR, Collins RE, Sharma D, Peng J, Cheng X, Vertino PM. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol Cell. 2008;30:336–47. doi: 10.1016/j.molcel.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Shigematsu H, Shames DS, Sunaga N, Takahashi T, Shivapurkar N, Iizasa T, Frenkel EP, Minna JD, Fujisawa T, Gazdar AF. DNA methylation-associated inactivation of TGFbeta-related genes DRM/Gremlin, RUNX3, and HPP1 in human cancers. Br J Cancer. 2005;93:1029–37. doi: 10.1038/sj.bjc.6602837. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–83. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- Tan SH, Ida H, Lau QC, Goh BC, Chieng WS, Loh M, Ito Y. Detection of promoter hypermethylation in serum samples of cancer patients by methylation-specific polymerase chain reaction for tumour suppressor genes including RUNX3. Oncology Reports. 2007;18:1225–30. [PubMed] [Google Scholar]
- Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, Ito Y, Littman DR. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–33. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- Tateishi Y, Kawabe Y, Chiba T, Murata S, Ichikawa K, Murayama A, Tanaka K, Baba T, Kato S, Yanagisawa J. Ligand-dependent switching of ubiquitin-proteasome pathways for estrogen receptor. EMBO J. 2004;23:4813–23. doi: 10.1038/sj.emboj.7600472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet. 1999;23:474–8. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, Brenner JC, Yu J, Kim JH, Han B, Tan P, Kumar-Sinha C, Lonigro RJ, Palanisamy N, Maher CA, Chinnaiyan AM. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–9. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–4. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Weith A, Brodeur GM, Bruns GA, Matise TC, Mischke D, Nizetic D, Seldin MF, van Roy N, Vance J. Report of the second international workshop on human chromosome 1 mapping 1995. Cytogenetics and Cell Genetics. 1996;72:114–44. [PubMed] [Google Scholar]
- Widschwendter M, Jones PA. DNA methylation and breast carcinogenesis. Oncogene. 2002;21:5462–82. doi: 10.1038/sj.onc.1205606. [DOI] [PubMed] [Google Scholar]
- Xie W, Mertens JC, Reiss DJ, Rimm DL, Camp RL, Haffty BG, Reiss M. Alterations of Smad signaling in human breast carcinoma are associated with poor outcome: a tissue microarray study. Cancer Res. 2002;62:497–505. [PubMed] [Google Scholar]
- Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. New England Journal of Medicine. 2006;354:270–82. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- Yamamura Y, Lee WL, Inoue K, Ida H, Ito Y. RUNX3 cooperates with FoxO3a to induce apoptosis in gastric cancer cells. J Biol Chem. 2006;281:5267–76. doi: 10.1074/jbc.M512151200. [DOI] [PubMed] [Google Scholar]
- Yano T, Ito K, Fukamachi H, Chi XZ, Wee HJ, Inoue K, Ida H, Bouillet P, Strasser A, Bae SC, Ito Y. The RUNX3 tumor suppressor upregulates Bim in gastric epithelial cells undergoing transforming growth factor beta-induced apoptosis. Mol Cell Biol. 2006;26:4474–88. doi: 10.1128/MCB.01926-05. [DOI] [PMC free article] [PubMed] [Google Scholar]