Abstract
The human body is home to more than 1 trillion microbes, with the gastrointestinal tract alone harboring a diverse array of commensal microbes that are believed to contribute to host nutrition, developmental regulation of intestinal angiogenesis, protection from pathogens, and development of the immune response. Recent advances in genome sequencing technologies and metagenomic analysis are providing a broader understanding of these resident microbes and highlighting differences between healthy and disease states. The aim of this review is to provide a detailed summary of current pediatric microbiome studies in the literature, in addition to highlighting recent findings and advancements in studies of the adult microbiome. This review also seeks to elucidate the development of, and factors that could lead to changes in, the composition and function of the human microbiome.
KEY WORDS: human microbiome, metagenome, microbiota, pediatric diseases, dysbiosis, probiotic
The growing recognition that our resident microbes may contribute fundamentally to infant and childhood development and immunity is creating an impetus to understand the importance of the human microbiome in pediatrics.1–5 Global health efforts focused on the first 1000 days of life and developmental origins of disease highlight the potential significance of the human microbiome for human health.6 Bacterial cells outnumber human cells in the body by an estimated factor of 10, with ∼10 to 100 trillion microbes living in the gastrointestinal (GI) tract alone.7,8 The collective genomes and gene products of these resident microbes living within and on humans are referred to as the human microbiome.7,9 In 2007, the National Institutes of Health–sponsored Human Microbiome Project was formed to gain insights into the evolution and composition of the human microbiome, factors that may influence or affect its composition, and whether the human microbiome affects health and tendencies toward particular diseases.7 The advent of new molecular technologies has been useful in the detection of uncultured microbes and may enable more microbes to be cultured in the future.10,11 These culture-independent methods include fluorescence in situ hybridization (FISH), DNA pyrosequencing, microarrays (PhyloChip), and quantitative polymerase chain reaction assays12,13 (Table 1). Advances in DNA sequencing technologies and computational methods have been used to analyze bacterial communities by using the conserved 16S rRNA gene for phylogenetic analysis,7 resulting in a deeper understanding of our commensal residents, beneficial microbes, and their contribution to human health.
TABLE 1.
Glossary of Terms
| Microbiome: Collective genomes and gene products of resident microbes living within and on humans |
| Microbiota: Microbial community |
| Metagenome: Collection of genomes within complex microbial communities and human DNA |
| Culture-independent techniques: Techniques that do not require the growth of bacteria on defined media under controlled laboratory conditions; they include techniques like PhyloChip, FISH, and 16S rRNA gene pyrosequencing |
| PhyloChip: DNA microarray that is unique in its ability to identify multiple bacterial and archaeal organisms from complex microbial samples13 |
| FISH: Technique that uses fluorescent probes designed to bind to specific complementary sequences of DNA, thereby allowing for detection of specific DNA sequences by fluorescence microscopy148 |
| 16S ribosomal RNA gene: 16S ribosomal RNA is a component of prokaryotic ribosomes that is highly conserved between different species of bacteria and is used for phylogenetic studies |
| Pyrosequencing: Method of DNA sequencing that detects the release of pyrophosphate upon nucleotide incorporation rather than chain termination by deoxynucleotides with Sanger sequencing149 |
| UniFrac: β-Diversity measure that is phylogeny based; microbial communities are more similar if they are composed of members that are more closely related, phylogenetically, as this implies a shared evolutionary history15,147 |
| Principal coordinates analysis: Standard multivariate statistic used to analyze and visualize individual or group similarities |
Composition of the Human Microbiome
This review discusses the development and composition of the human microbiome at different body sites and illustrates how changes in composition may have important consequences for human pathophysiology and disease susceptibilities. Human-associated bacterial communities likely play a central role in host nutrition, development of immunity, and protection from diverse pathogens.9,14 The human body contains many different sites that are colonized by microbial communities during neonatal and childhood development and throughout the lifetime of individuals in health and disease states. Predominant bacterial phyla (composed of hundreds of bacterial genera and species) in the human body, regardless of body site, include Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria (Table 2).15 Bacterial species populations vary significantly between individuals, and bacterial community composition appears to be driven primarily by body habitat.16 To highlight the heterogeneity of human microbiome composition, the human skin differs dramatically in terms of predominant bacterial phyla (Actinobacteria, Firmicutes, or Proteobacteria) by virtue of the location of the skin site on the human body and its relative humidity.17 The phylum Bacteroidetes is a minor component of the human microbiome on many different skin sites17 while Firmicutes comprise the major phylum in the vagina.18 Studies of the GI tract, by contrast, have consistently demonstrated the predominance of the same phyla, Bacteroidetes and Firmicutes, in children and adults.16,19 In addition to body habitat, different bacteria may serve as “anchor microbes” in particular individuals. For example, a recent study found 3 distinct identifiable enterotypes in the intestinal microbiome among adults from multiple countries, which were characterized by prominent genera including Bacteroides (enterotype 1), Prevotella (enterotype 2), and Ruminococcus (enterotype 3).20 These enterotypes also appear to be driven by species composition and relative functional capacities of gut bacterial communities. Distinct intestinal enterotypes are yet to be described in the pediatric population.
TABLE 2.
Predominant Bacterial Phyla in the Human Body
| Phylum | Class | Characteristics | Examples |
|---|---|---|---|
| Firmicutes | Bacilli; Clostridia | Gram-positive; diverse in their morphology (rod, coccoid, spiral), physiology (anaerobic, aerobic); include commensal and beneficial bacteria | Lactobacillus; Ruminococcus; Clostridium; Staphylococcus; Enterococcus; Faecalibacterium |
| Bacteroidetes | Bacteroidetes | Gram-negative; composed of 3 large classes widely distributed in the environment, including soil, seawater, and guts of animals | Bacteroides; Prevotella |
| Proteobacteria | Gammaproteobacteria; Betaproteobacteria | Gram-negative; include a wide variety of pathogens | Escherichia; Pseudomonas |
| Actinobacteria | Actinobacteria | Gram-positive; diverse morphology; major antibiotic producers in the pharmaceutical industry | Bifidobacterium; Streptomyces; Nocardia |
The intestinal microbiome undergoes dynamic change during development, with the most dramatic changes in composition believed to occur throughout infancy and childhood.21,22 The diversity and flux of microbes observed during this time are believed to be important for the normal functional development of the immune system and its impact on health later in life.21,23 The predominantly colonizing phyla found in the infant GI tract belong to Firmicutes, Bacteroidetes, and Proteobacteria.10,21 However, the composition of gut-associated bacterial communities was found to be highly variable in individual infants in terms of timing of acquisition and colonization by individual bacterial species.21 A recent study observed significant changes in genomic divergence and relative abundance of 2 Citrobacter strains in a premature infant during a 3-week period, suggesting that fluctuations in strains of a single species may contribute to differences in the “fine” or detailed functional capacity of the microbiome.24 Facultative bacteria like Escherichia coli, Enterococcus spp, α-hemolytic streptococci, and Staphylococcus spp colonize the sterile, aerobic newborn GI tract in the first few days of life.21,25–28 After the first weeks of life, anaerobic conditions have been created in the gut due to the consumption of oxygen by these facultative bacteria.29,30 This environment coupled with the presence of human milk oligosaccharides (HMOs) in breast milk leads to a shift in composition to predominantly anaerobic bacteria such as Bacteroides, Bifidobacterium, and Clostridium spp.21,25,31 Development of a core microbiome, which can refer to a set of microbes or a set of metabolic functions, may occur by the end of infancy.21,32,33 However, recent metagenomic studies suggest that the gut microbiota of school-age and adolescent children differ significantly from that of adults,34,35 indicating that the human microbiome may be evolving during childhood and adolescence.
Environmental Factors Affecting the Composition of the Human Microbiome
Mode of Birth Delivery
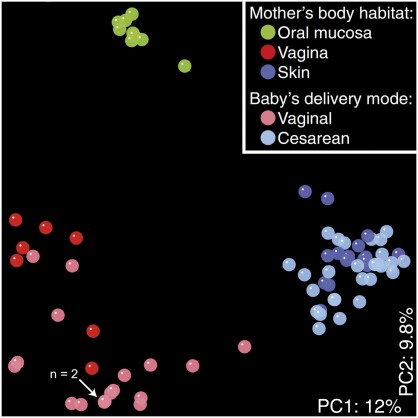
Mode of birth delivery, hospitalization, diet, and nature of feeding are environmental factors that may impact the composition and diversity of the infant microbiota.36 Early microbial colonization of a newborn begins at birth.21,37 The microbiota of vaginally delivered newborns represented the maternal vaginal and intestinal microbiota, while newborns delivered via cesarean delivery exhibited a microbiome representative of the maternal skin microbiota including Staphylococcus spp (Fig 1).38 Vaginally delivered newborns exhibited bacterial communities composed of several prominent genera including Lactobacillus, Prevotella, Escherichia, Bacteroides, Bifidobacterium, and Streptococcus spp.38,39 Different molecular methods confirmed a reduced proportion of Bifidobacterium or Bacteroides spp in the GI tract of infants delivered via cesarean delivery.29,40 Regardless of delivery mode and in contrast to their mothers, bacterial communities among newborns exhibited a uniform distribution across different body sites, including the skin, nasopharynx, intestine, and oral cavity.38 Presumably, it takes weeks or longer for the human microbiome to differentiate into body site–specific microbial communities. A recent study by Capone et al corroborates this argument as site-specific bacterial communities were found on the skin of infants ranging from 1 to 3 months of life.41 As such, the birth process and mode of delivery may have a profound impact on microbial composition early in life, and these factors may help explain 1 aspect of the developmental origins of human microbiomes at different body sites.
FIGURE 1.
16S bacterial rRNA analysis reveals influence of delivery mode on the neonatal microbiome. UniFrac analysis revealed similarities and clustering of bacterial communities based on the mother’s body habitat or the delivery mode of the newborn. Each colored point represents a similar community in specific body sites of the mother and all newborn body habitats.147 The percentage of variation of the principal coordinates analysis is indicated in white text on both axes. (Reproduced with permission from Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107[26]:11973.)
First Foods: Breast Milk and Infant Formula
Beneficial factors in breast milk are widely acknowledged and include immunoglobulins, cytokines, growth factors, lysozyme, lactoferrin, and HMOs.42–44 HMOs are an abundant carbohydrate component in breast milk and function similarly to prebiotics, stimulating the growth of Bifidobacterium sp and thereby selectively altering microbial composition in the infant intestine.45 More than 200 different HMO structures have been characterized in human milk, and HMOs contain a lactose core with diversity generated by covalent modifications such as extensive fucosylation and/or sialylation.46 Particular HMOs also share similar glycan structural motifs, which are believed to protect infants from disease by acting as decoys in preventing pathogens from binding to epithelial cells.47 Anaerobes such as Bifidobacterium appear during the first weeks of life, and species of this genus are well adapted to HMOs.48 While numerous studies reported a higher relative abundance of Bifidobacterium and Lactobacillus in the microbiomes of breastfed infants,21,44,49,50 others have reported no difference in abundance between these 2 genera in breastfed and formula-fed infants.29,51,52 Increased colonization by Clostridium spp and particularly C difficile in formula-fed infants compared with breastfed infants has been reported in several studies.23,29 The greater abundance of C difficile in the intestinal microbiota of formula-fed infants has also been associated with eczema in infants.29,52,53 Breastfed, vaginally delivered term infants exhibited reduced colonization by C difficile and E coli and enhanced colonization by beneficial microbes, like Bifidobacterium spp.29 The proliferation of beneficial microbes supported by breastfeeding may provide protection from disorders such as allergies, neonatal diarrhea,54 necrotizing enterocolitis (NEC),55 obesity,56 and type 2 diabetes.57
Hospitalization and Gestational Age
Hospitalization and gestational age may impact the composition and development of the intestinal microbiota. Preterm infants, exhibiting diverse bacterial communities after birth, acquired similar intestinal bacterial composition during the first weeks of life as a result of cross-transmission during hospitalization.58 Correspondingly, increased hospital stays have been associated with delays in colonization and development of the infant intestinal microbiota, which could result from exposure to different microbes or antibiotic treatment.59,60 For example, increased colonization by C difficile was observed in both preterm infants and infants hospitalized after birth, which could be attributed to a high carriage rate and the persistence of C difficile spores in the environment.29 Furthermore, the intestinal microbiota of preterm infants with a gestational age of <33 weeks exhibited significantly reduced bacterial diversity.61,62 In particular, recurrent C difficile infection and other disease states have been associated with reduced bacterial diversity in the intestine.63
Effects of Diet
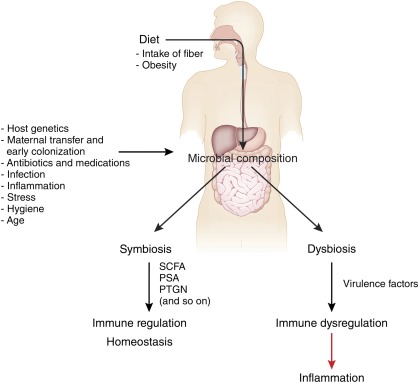
Major shifts of taxonomic groups in the microbiome have been observed with major life events including changes in diet, such as weaning to solid foods.31 In fact, diet may be a primary factor involved in generating compositional change and diversity in the microbiome.64 Studies performed in germ-free mice colonized with human microbial communities revealed that the initial colonizing bacterial communities can be rapidly altered by diet.65 Alterations of fiber and fat/protein content in the diets of a small cohort of children and adults also yielded changes in the composition of the microbiome within a 24-hour period, which then remained stable over the duration of the study.66 Longer-term changes in diet may be necessary to effect more substantial changes. Moreover, the Bacteroides enterotype was associated with consumption of animal protein and saturated fat, whereas the Prevotella enterotype was associated with a carbohydrate-rich diet.66 Comparisons of intestinal microbiota from children in rural Africa and Europe exhibited similar patterns with a greater abundance of Bacteroidetes and lower abundance of Firmicutes in the Africa cohort compared with the European cohort.67 Bacteroides spp produce beneficial molecules like polysaccharide A and short-chain fatty acids.65,68 Polysaccharide A yielded a protective effect in a mouse colitis model,69 while short-chain fatty acids have demonstrated beneficial effects for the host including the maintenance of the colonic epithelium, provision of energy for host metabolism, and regulation of immunity14 (Fig 2). Bacteroides spp may affect the maturation of humoral immunity in early infancy and the balance of the Th1 and Th2 cell immunity.2,70–73 A study also reported an abundance of 2 genera, Prevotella and Xylanibacter, which contain genes involved in the hydrolysis of cellulose and xylan. These findings support the hypothesis that the intestinal microbiota is altered by differences in diet, allowing for enhanced energy extraction from a polysaccharide-rich diet and anti-inflammatory effects.67 Microbial communities in the intestinal microbiota were first shown to influence host energy homeostasis and fat storage by Backhed et al.74 Subsequent studies revealed the ability of intestinal microbiota to suppress the expression of fasting-induced adipose factor, resulting in the increased storage of triglycerides in adipocytes.74 A humanized gnotobiotic mouse model supported this observation with mice developing more adiposity within 2 weeks of being fed a typical Western diet, high in fat and sugar and low in plant polysaccharides, compared with control mice.65 Together, these studies suggest that the human diet may impact the phylogenetic diversity and functional capacity of the human microbiome with downstream effects on disease risk and disease penetrance.
FIGURE 2.
Effects of diet, host, and environmental factors on the microbiome. Antibiotic use, diet, host, and environmental factors can affect the composition of the microbiota. In this model, balanced microbial composition may result in symbiosis among resident microbes, production of immunomodulatory compounds, and subsequent regulation of the immune response. Disruption or alteration of the microbiota by environmental factors such as diet and antibiotic use could result in dysbiosis and dysregulation of the immune response. (Reproduced with permission from Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat Immunol. 2011;12[1]:6.)
Effects of Antibiotics
Antimicrobial agents can drastically alter the composition of the intestinal and oral microbiota contingent on the spectrum and dosage, route of administration, and treatment duration.75–77 Reduction in microbial diversity is often observed within days of ingestion of antibiotics, and complete recovery of initial bacterial community composition is rarely achieved.75,77 Moreover, profound alterations of microbial communities have been shown within days of treatment with the fluoroquinolone ciprofloxacin. The lack of recovery from this perturbation by several organisms emphasizes the potential impact of excessive antimicrobial therapy.75 The impact on the native gut microbiota is pronounced in infants <1 year of age, with significant reductions in Bifidobacterium and Bacteroides as well as overall reductions of bacterial community diversity.29,78 Additional risks associated with antimicrobial treatment include the selection of antibiotic-resistant strains of bacteria and the development of C difficile–associated diarrhea.76,77,79–81 Several studies have shown the persistent increase of erythromycin B (ermB) gene levels in fecal samples after antibiotic treatment with macrolides like clarithromycin.77,82 The occurrence of C difficile infections is increasing in the United States, and this disease pattern may be due to the expansion of preexisting C difficile populations after antibiotic treatment or the acquisition of spores from a hospital environment.83,84 Additionally, reduced diversity of the intestinal microbiome due to widespread use of antibiotics may be placing more children at risk for C difficile infections and other causes of antibiotic-associated diarrhea/colitis.63 These reports demonstrate the importance of judicious application of antibiotics to minimize potentially deleterious effects on the composition and function of the human microbiome.
Altered States of the Human Microbiome and Pediatric Diseases
Skin Microbiome, Dermatologic, and Immune-Mediated Disorders
Actinobacteria, Proteobacteria, Firmicutes, and Bacteroidetes represent the predominant phyla colonizing the adult human skin, and considerable bacterial diversity was observed at the species level.17 Metagenomic sequencing also revealed significant interpersonal variation among individuals and temporal variation dependent on the specific body site.17 The phylum Firmicutes predominates at specific skin sites in the infant microbiome, possibly as a result of differences in the structure and composition of infant skin compared with adult skin.41,85 Changes in the microbiota linked to skin diseases have been found in children, including psoriasis, atopic dermatitis, and acne.17,86–90 A study of psoriatic lesions on adult skin revealed significantly overrepresented Firmicutes, while the Proteobacteria and Actinobacteria phyla were significantly underrepresented compared with healthy skin.86 Furthermore, sequence-based analysis identified the presence of several species not previously associated with atopic dermatitis, such as Stenotrophomonas maltophilia.89
The frequency of atopic diseases such as eczema, asthma, and food allergies is rising in incidence and linked to alterations of the intestinal microbiota.53 The hygiene hypothesis, proposed by Strachan in 1989, suggested that the lack of infections in early infancy led to this observed rise in atopic disease.91 As the interaction of immune cells with microbial antigens is fundamental to the function and development of the adaptive immune response, the lack of immune stimulation during early life in developed countries could account for increased immune dysregulation observed in asthma and atopic diseases.2,42,51 Delays or changes in the core microbiome could also potentially affect the development of the immune response.70 Epidemiologic data provide further evidence that infants delivered via cesarean delivery have higher incidences of atopic diseases such as asthma and type 1 diabetes and food allergies compared with vaginally delivered infants.92–94
Pulmonary Microbiome and Diseases of the Respiratory Tract
Few metagenomic studies in the literature have described the microbiome of the human respiratory tract. While some studies have reported stable oral microbial communities in adults and children,16,95 others have found highly variable and diverse bacterial communities in the nasopharynx of children that were independent of antibiotic use.96 Bacterial communities in the respiratory tract of intubated patients with ventilator-associated pneumonia demonstrated infection by the pathogen Pseudomonas aeruginosa associated with concomitant loss of microbial diversity after antibiotic administration.97 Compared with culture-based studies, pyrosequencing studies identified a more diverse and comprehensive set of microbes in cystic fibrosis.98 Pyrosequencing also revealed greater interpersonal variability of bacterial community compositions in the lungs of patients with cystic fibrosis, which may be influenced by colonization of bacterial communities in the oral cavity.99,100 Sequencing-based studies are expanding our appreciation of diverse and abundant microbial communities in the respiratory tracts of healthy patients and those with cystic fibrosis.100,101
GI Microbiome and Intestinal Disorders
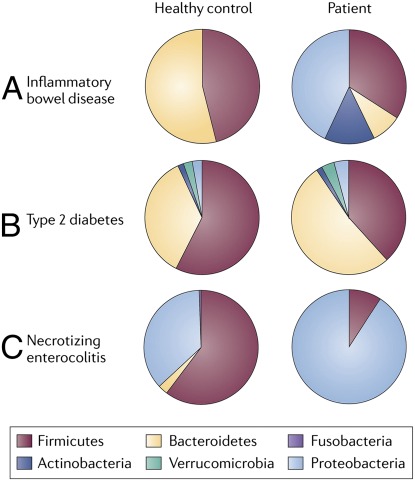
The pathophysiology of NEC appears to be multifactorial, with premature birth being the most pronounced risk factor.102 Other factors in the development of NEC include intestinal immaturity, an excessive intestinal inflammatory response to microbial stimuli, and colonization by disease-predisposing microbial populations in the GI tract.103 Several studies using metagenomic comparisons of fecal microbiota reported a reduction in microbial diversity in preterm infants with NEC compared with healthy preterm infants.15,104 However, other studies reported similar overall microbial profiles between infants with NEC and control infants.105,106 Recent studies of infants with NEC found increased abundances of Proteobacteria (Fig 3) including Citrobacter sp in fecal microbiota.15,105 Furthermore, Neu et al recently described a greater proportion of Gammaproteobacteria in fecal microbiota prior to the diagnosis of NEC in infants.107 Treatment regimens for NEC currently include the prolonged use of parenteral antibiotics, which may reduce intestinal microbial diversity and preclude colonization by a diverse community of microbes.104,108,109 Human-associated microbial communities with reduced microbial diversity may be supplemented by probiotics or expressed breast milk, as several studies have shown a reduced incidence of NEC in preterm infants after breast milk and probiotic consumption.110–113
FIGURE 3.
Disease states reveal phylum-level differences compared with healthy controls. Comparisons of the relative abundances of predominant bacterial phyla in IBD, type 2 diabetes, and NEC compared with healthy controls. Fecal samples from infants with NEC and patients with type 2 diabetes were compared with healthy controls revealing a predominance of Proteobacteria in patients with NEC. Cecal samples from patients with IBD were compared with healthy controls, and relative abundances were assessed. (Reproduced with permission from Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9[4]:281.)
The composition of the intestinal microbiome differs between healthy individuals and individuals with inflammatory bowel disease (IBD) with respect to phylogenetic diversity and relative abundances of microbial taxa.114–116 This imbalance or disruption of the host microbiota, termed dysbiosis, can induce an inflammatory response by the host as evidenced in Crohn disease.117,118 Studies have shown that the gut microbiota of individuals with IBD exhibited reduced proportions of Firmicutes and Bacteroidetes and an increased proportion of Proteobacteria compared with healthy individuals (Fig 3).115,119 Present evidence suggests that IBD may result from abnormal interactions between indigenous microbiota and the host immune system.120–122 A reduced abundance of Faecalibacterium prausnitzii, a member of the Firmicutes phyla, has been associated with Crohn disease activity.115 Anti-inflammatory effects of F prausnitzii were demonstrated by cytokine studies in vitro and a murine TNBS-induced colitis model.115 In addition, metabolomic studies yielded signature microbial metabolites possibly involved in the pathogenesis of Crohn disease.123
Irritable bowel syndrome (IBS) is a functional GI disorder that includes recurrent abdominal pain and changes in defecation patterns ranging from hard to watery stool.124 Although the pathogenesis of IBS is also not well understood, dysbiosis has been associated with diarrhea-predominant IBS and constipation-predominant IBS. The intestinal microbiota of patients with diarrhea-predominant IBS differs from that of healthy subjects with respect to the relative prevalence of genera, including Lactobacillus, Streptococcus, Ruminococcus, and Veillonella.125,126 Specific microbial signatures in school-age, preadolescent children with IBS compared with healthy controls were recently described and included a greater abundance of Gammaproteobacteria and an association of Alistipes with greater pain frequency. Pediatric IBS subtypes were distinguished by using microbial feature selection and compositional differences of the human intestinal microbiome.34
Treatment and Manipulation of the Human Microbiome
Manipulation of the human microbiome may include microbial supplements (probiotics or synbiotics), foods or substrates (diet or prebiotics), and microbial suppression or elimination (antibiotics) strategies. Beneficial microbes such as Lactobacillus, Bifidobacterium, and Streptococcus are commonly used as probiotics, which are defined as “live microorganisms which when administered in adequate amounts confer a health benefit on the host.”127 Probiotics are believed to assist the resident microbiota in preventing pathogen adherence,128,129 downregulating proinflammatory cytokines,130 inducing immunoglobulin A production,131 and enhancing intestinal mucosal barrier function and epithelial integrity.129 Recent studies have demonstrated that probiotic L reuteri targets sensory nerves in the enteric nervous system, thereby affecting pain perception and gut motility.132 Probiotic formulations are generally believed to be safe, and the American Academy of Pediatrics has supported the administration of probiotics for the treatment of acute gastroenteritis and the prevention of antibiotic-associated diarrhea.108,133 Prebiotics, which are nondigestible food ingredients that stimulate the growth and activity of designated species of beneficial bacteria, may enhance the treatment efficacy of other anti-inflammatory medications by stimulating butyrate production in humans and suppressing production of proinflammatory cytokines.134,135 Synbiotics, a combination of both probiotics and prebiotics, have also been used to treat inflammatory diseases.136 Probiotics and prebiotics may be applied in the regulation and homeostasis of intestinal microbial composition and as a therapeutic strategy for various disorders.137,138 Other therapies have been effective in restoring normal bacterial communities including the transplantation of fecal microbiota from a healthy donor to a patient. Fecal transplantation has been increasingly used in the last 2 decades for C difficile infection, with a success rate of >90%.139–141 Analysis of the microbiota could result in the development of naturally derived drugs to treat chronic inflammation, and additional evidence suggests that enteric bacteria produce immunomodulatory molecules that have anti-inflammatory properties.142–144 For example, a recent study demonstrated that a polysaccharide of Bacteroides fragilis had immunomodulatory properties and prevented intestinal inflammation in mice.69 More examples of these naturally derived substances include bacteriocins, which are antimicrobial peptides produced by bacteria that inhibit the growth of other bacteria in the microbial community. Broad- and narrow-spectrum bacteriocins have been effective against C difficile.145,146 In addition to bacteriocins, antibiotics may selectively target classes of organisms in the human microbiome. Combinations of antibiotics, probiotics, and diet may yield potent strategies to manipulate and reshape disease-prone microbiomes.
Conclusions
The importance of the microbiota to many aspects of human health and the realization that its foundation is established in early infancy are becoming increasingly recognized. The rapidly advancing knowledge of the human microbiome, through metagenomic analysis, has yielded information regarding the differences observed between healthy and disease states and factors that influence the composition and diversity of the microbiome. Future studies may lead to improved health benefits for pediatric patients through the manipulation of the intestinal microbiota.
Glossary
- FISH
fluorescence in situ hybridization
- GI
gastrointestinal
- HMO
human milk oligosaccharide
- IBD
inflammatory bowel disease
- IBS
irritable bowel syndrome
- NEC
necrotizing enterocolitis
- TNBS
2,4,6-trinitrobenzene sulfonic acid
Footnotes
FINANCIAL DISCLOSURE: Dr Versalovic receives unrestricted research support from BioGaia AB; Dr Johnson has indicated she has no financial relationships relevant to this article to disclose.
FUNDING: Funded by the National Institutes of Health (NIH).
References
- 1.Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312(5778):1355–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122(1):107–118 [DOI] [PubMed] [Google Scholar]
- 3.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313(5790):1126–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci USA. 2002;99(24):15451–15455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hooper LV, Falk PG, Gordon JI. Analyzing the molecular foundations of commensalism in the mouse intestine. Curr Opin Microbiol. 2000;3(1):79–85 [DOI] [PubMed] [Google Scholar]
- 6.InterAction and the Global Alliance for Improved Nutrition (GAIN) iccwtUSDoS. 1,000 Days. 2011. Available at: www.thousanddays.org/. Accessed August 30, 2011
- 7.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449(7164):804–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu J, Gordon JI. Honor thy symbionts. Proc Natl Acad Sci USA. 2003;100(18):10452–10459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292(5519):1115–1118 [DOI] [PubMed] [Google Scholar]
- 10.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlisle EM, Poroyko V, Caplan MS, Alverdy JA, Liu D. Gram negative bacteria are associated with the early stages of necrotizing enterocolitis. PLoS ONE. 2011;6(3):e18084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morelli L. Postnatal development of intestinal microflora as influenced by infant nutrition. J Nutr. 2008;138(9):1791S–1795S [DOI] [PubMed] [Google Scholar]
- 13.DeSantis TZ, Brodie EL, Moberg JP, Zubieta IX, Piceno YM, Andersen GL. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb Ecol. 2007;53(3):371–383 [DOI] [PubMed] [Google Scholar]
- 14.Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat Immunol. 2011;12(1):5–9 [DOI] [PubMed] [Google Scholar]
- 15.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9(4):279–290 [DOI] [PubMed] [Google Scholar]
- 16.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grice EA, Kong HH, Conlan S, et al. NISC Comparative Sequencing Program . Topographical and temporal diversity of the human skin microbiome. Science. 2009;324(5931):1190–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ravel J, Gajer P, Abdo Z, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA. 2011;108(suppl 1):4680–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124(4):837–848 [DOI] [PubMed] [Google Scholar]
- 20.Arumugam M, Raes J, Pelletier E, et al. MetaHIT Consortium . Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5(7):e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tiihonen K, Ouwehand AC, Rautonen N. Human intestinal microbiota and healthy ageing. Ageing Res Rev. 2010;9(2):107–116 [DOI] [PubMed] [Google Scholar]
- 23.Vael C, Desager K. The importance of the development of the intestinal microbiota in infancy. Curr Opin Pediatr. 2009;21(6):794–800 [DOI] [PubMed] [Google Scholar]
- 24.Morowitz MJ, Denef VJ, Costello EK, et al. Strain-resolved community genomic analysis of gut microbial colonization in a premature infant. Proc Natl Acad Sci USA. 2011;108(3):1128–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stark PL, Lee A. The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year of life. J Med Microbiol. 1982;15(2):189–203 [DOI] [PubMed] [Google Scholar]
- 26.Rotimi VO, Duerden BI. The development of the bacterial flora in normal neonates. J Med Microbiol. 1981;14(1):51–62 [DOI] [PubMed] [Google Scholar]
- 27.Favier CF, Vaughan EE, De Vos WM, Akkermans AD. Molecular monitoring of succession of bacterial communities in human neonates. Appl Environ Microbiol. 2002;68(1):219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benno Y, Sawada K, Mitsuoka T. The intestinal microflora of infants: composition of fecal flora in breast-fed and bottle-fed infants. Microbiol Immunol. 1984;28(9):975–986 [DOI] [PubMed] [Google Scholar]
- 29.Penders J, Thijs C, Vink C, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118(2):511–521 [DOI] [PubMed] [Google Scholar]
- 30.Bezirtzoglou E. The intestinal microflora during the first weeks of life. Anaerobe. 1997;3(2-3):173–177 [DOI] [PubMed] [Google Scholar]
- 31.Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108(suppl 1):4578–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587(pt 17):4153–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saulnier DM, Riehle K, Mistretta TA, et al. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology. 2011;141(5):1782–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agans R, Rigsbee L, Kenche H, Michail S, Khamis HJ, Paliy O. Distal gut microbiota of adolescent children is different from that of adults. FEMS Microbiol Ecol. 2011;77(2):404–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murgas Torrazza R, Neu J. The developing intestinal microbiome and its relationship to health and disease in the neonate. J Perinatol. 2011;31(suppl 1):S29–S34 [DOI] [PubMed] [Google Scholar]
- 37.Vaishampayan PA, Kuehl JV, Froula JL, Morgan JL, Ochman H, Francino MP. Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. Genome Biol Evol. 2010;2:53–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107(26):11971–11975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hyman RW, Fukushima M, Diamond L, Kumm J, Giudice LC, Davis RW. Microbes on the human vaginal epithelium. Proc Natl Acad Sci USA. 2005;102(22):7952–7957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biasucci G, Rubini M, Riboni S, Morelli L, Bessi E, Retetangos C. Mode of delivery affects the bacterial community in the newborn gut. Early Hum Dev. 2010;86(suppl 1):13–15 [DOI] [PubMed] [Google Scholar]
- 41.Capone KA, Dowd SE, Stamatas GN, Nikolovski J. Diversity of the human skin microbiome early in life. J Invest Dermatol. 2011;131(10):2026–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vassallo MF, Walker WA. Neonatal microbial flora and disease outcome. Nestle Nutr Workshop Ser Pediatr Program. 2008;61:211–224 [DOI] [PubMed] [Google Scholar]
- 43.Hanson LA, Korotkova M, Telemo E. Breast-feeding, infant formulas, and the immune system. Ann Allergy Asthma Immunol. 2003;90(6 suppl 3):59–63 [DOI] [PubMed] [Google Scholar]
- 44.Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr. 2000;30(1):61–67 [DOI] [PubMed] [Google Scholar]
- 45.Ward RE, Niñonuevo M, Mills DA, Lebrilla CB, German JB. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl Environ Microbiol. 2006;72(6):4497–4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ninonuevo MR, Park Y, Yin H, et al. A strategy for annotating the human milk glycome. J Agric Food Chem. 2006;54(20):7471–7480 [DOI] [PubMed] [Google Scholar]
- 47.Newburg DS, Ruiz-Palacios GM, Morrow AL. Human milk glycans protect infants against enteric pathogens. Annu Rev Nutr. 2005;25:37–58 [DOI] [PubMed] [Google Scholar]
- 48.Sela DA, Chapman J, Adeuya A, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci USA. 2008;105(48):18964–18969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bullen CL, Tearle PV, Willis AT. Bifidobacteria in the intestinal tract of infants: an in-vivo study. J Med Microbiol. 1976;9(3):325–333 [DOI] [PubMed] [Google Scholar]
- 50.Knol J, Scholtens P, Kafka C, et al. Colon microflora in infants fed formula with galacto- and fructo-oligosaccharides: more like breast-fed infants. J Pediatr Gastroenterol Nutr. 2005;40(1):36–42 [DOI] [PubMed] [Google Scholar]
- 51.Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Acta Paediatr. 2009;98(2):229–238 [DOI] [PubMed] [Google Scholar]
- 52.Penders J, Vink C, Driessen C, London N, Thijs C, Stobberingh EE. Quantification of Bifidobacterium spp., Escherichia coli and Clostridium difficile in faecal samples of breast-fed and formula-fed infants by real-time PCR. FEMS Microbiol Lett. 2005;243(1):141–147 [DOI] [PubMed] [Google Scholar]
- 53.Penders J, Thijs C, van den Brandt PA, et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut. 2007;56(5):661–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ruiz-Palacios GM, Calva JJ, Pickering LK, et al. Protection of breast-fed infants against Campylobacter diarrhea by antibodies in human milk. J Pediatr. 1990;116(5):707–713 [DOI] [PubMed] [Google Scholar]
- 55.McGuire W, Anthony MY. Donor human milk versus formula for preventing necrotising enterocolitis in preterm infants: systematic review. Arch Dis Child Fetal Neonatal Ed. 2003;88(1):F11–F14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Owen CG, Martin RM, Whincup PH, Smith GD, Cook DG. Effect of infant feeding on the risk of obesity across the life course: a quantitative review of published evidence. Pediatrics. 2005;115(5):1367–1377 [DOI] [PubMed] [Google Scholar]
- 57.Owen CG, Martin RM, Whincup PH, Smith GD, Cook DG. Does breastfeeding influence risk of type 2 diabetes in later life? A quantitative analysis of published evidence. Am J Clin Nutr. 2006;84(5):1043–1054 [DOI] [PubMed] [Google Scholar]
- 58.Schwiertz A, Gruhl B, Löbnitz M, Michel P, Radke M, Blaut M. Development of the intestinal bacterial composition in hospitalized preterm infants in comparison with breast-fed, full-term infants. Pediatr Res. 2003;54(3):393–399 [DOI] [PubMed] [Google Scholar]
- 59.Hällström M, Eerola E, Vuento R, Janas M, Tammela O. Effects of mode of delivery and necrotising enterocolitis on the intestinal microflora in preterm infants. Eur J Clin Microbiol Infect Dis. 2004;23(6):463–470 [DOI] [PubMed] [Google Scholar]
- 60.el-Mohandes AE, Keiser JF, Johnson LA, Refat M, Jackson BJ. Aerobes isolated in fecal microflora of infants in the intensive care nursery: relationship to human milk use and systemic sepsis. Am J Infect Control. 1993;21(5):231–234 [DOI] [PubMed] [Google Scholar]
- 61.Rougé C, Goldenberg O, Ferraris L, et al. Investigation of the intestinal microbiota in preterm infants using different methods. Anaerobe. 2010;16(4):362–370 [DOI] [PubMed] [Google Scholar]
- 62.Jacquot A, Neveu D, Aujoulat F, et al. Dynamics and clinical evolution of bacterial gut microflora in extremely premature patients. J Pediatr. 2011;158(3):390–396 [DOI] [PubMed] [Google Scholar]
- 63.Chang JY, Antonopoulos DA, Kalra A, et al. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis. 2008;197(3):435–438 [DOI] [PubMed] [Google Scholar]
- 64.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–1920 [DOI] [PubMed] [Google Scholar]
- 65.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 2009;1(6):6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107(33):14691–14696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 2011;10(4):311–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453(7195):620–625 [DOI] [PubMed] [Google Scholar]
- 70.Grönlund MM, Arvilommi H, Kero P, Lehtonen OP, Isolauri E. Importance of intestinal colonisation in the maturation of humoral immunity in early infancy: a prospective follow up study of healthy infants aged 0–6 months. Arch Dis Child Fetal Neonatal Ed. 2000;83(3):F186–F192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Björkstén B. The gut microbiota: a complex ecosystem. Clin Exp Allergy. 2006;36(10):1215–1217 [DOI] [PubMed] [Google Scholar]
- 72.Adlerberth I, Lindberg E, Aberg N, et al. Reduced enterobacterial and increased staphylococcal colonization of the infantile bowel: an effect of hygienic lifestyle? Pediatr Res. 2006;59(1):96–101 [DOI] [PubMed] [Google Scholar]
- 73.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003;4(3):269–273 [DOI] [PubMed] [Google Scholar]
- 74.Bäckhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101(44):15718–15723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA. 2011;108(suppl 1):4554–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology. 2010;156(pt 11):3216–3223 [DOI] [PubMed] [Google Scholar]
- 77.Jakobsson HE, Jernberg C, Andersson AF, Sjölund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS ONE. 2010;5(3):e9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alm B, Erdes L, Möllborg P, et al. Neonatal antibiotic treatment is a risk factor for early wheezing. Pediatrics. 2008;121(4):697–702 [DOI] [PubMed] [Google Scholar]
- 79.Nyberg SD, Osterblad M, Hakanen AJ, et al. Long-term antimicrobial resistance in Escherichia coli from human intestinal microbiota after administration of clindamycin. Scand J Infect Dis. 2007;39(6-7):514–520 [DOI] [PubMed] [Google Scholar]
- 80.McFarland LV. Epidemiology, risk factors and treatments for antibiotic-associated diarrhea. Dig Dis. 1998;16(5):292–307 [DOI] [PubMed] [Google Scholar]
- 81.De La Cochetière MF, Durand T, Lepage P, Bourreille A, Galmiche JP, Doré J. Resilience of the dominant human fecal microbiota upon short-course antibiotic challenge. J Clin Microbiol. 2005;43(11):5588–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sjölund M, Wreiber K, Andersson DI, Blaser MJ, Engstrand L. Long-term persistence of resistant Enterococcus species after antibiotics to eradicate Helicobacter pylori. Ann Intern Med. 2003;139(6):483–487 [DOI] [PubMed] [Google Scholar]
- 83.Wilson KH. The microecology of Clostridium difficile. Clin Infect Dis. 1993;16(suppl 4):S214–S218 [DOI] [PubMed] [Google Scholar]
- 84.Marciniak C, Chen D, Stein AC, Semik PE. Prevalence of Clostridium difficile colonization at admission to rehabilitation. Arch Phys Med Rehabil. 2006;87(8):1086–1090 [DOI] [PubMed] [Google Scholar]
- 85.Stamatas GN, Nikolovski J, Luedtke MA, Kollias N, Wiegand BC. Infant skin microstructure assessed in vivo differs from adult skin in organization and at the cellular level. Pediatr Dermatol. 2010;27(2):125–131 [DOI] [PubMed] [Google Scholar]
- 86.Gao Z, Tseng CH, Strober BE, Pei Z, Blaser MJ. Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PLoS ONE. 2008;3(7):e2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Balci DD, Duran N, Ozer B, Gunesacar R, Onlen Y, Yenin JZ. High prevalence of Staphylococcus aureus cultivation and superantigen production in patients with psoriasis. Eur J Dermatol. 2009;19(3):238–242 [DOI] [PubMed] [Google Scholar]
- 88.Bek-Thomsen M, Lomholt HB, Kilian M. Acne is not associated with yet-uncultured bacteria. J Clin Microbiol. 2008;46(10):3355–3360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dekio I, Sakamoto M, Hayashi H, Amagai M, Suematsu M, Benno Y. Characterization of skin microbiota in patients with atopic dermatitis and in normal subjects using 16S rRNA gene-based comprehensive analysis. J Med Microbiol. 2007;56(pt 12):1675–1683 [DOI] [PubMed] [Google Scholar]
- 90.Kong HH. Skin microbiome: genomics-based insights into the diversity and role of skin microbes. Trends Mol Med. 2011;17(6):320–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299(6710):1259–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Negele K, Heinrich J, Borte M, et al. LISA Study Group . Mode of delivery and development of atopic disease during the first 2 years of life. Pediatr Allergy Immunol. 2004;15(1):48–54 [DOI] [PubMed] [Google Scholar]
- 93.Debley JS, Smith JM, Redding GJ, Critchlow CW. Childhood asthma hospitalization risk after cesarean delivery in former term and premature infants. Ann Allergy Asthma Immunol. 2005;94(2):228–233 [DOI] [PubMed] [Google Scholar]
- 94.Laubereau B, Filipiak-Pittroff B, von Berg A, et al. GINI Study Group . Caesarean section and gastrointestinal symptoms, atopic dermatitis, and sensitisation during the first year of life. Arch Dis Child. 2004;89(11):993–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Crielaard W, Zaura E, Schuller AA, Huse SM, Montijn RC, Keijser BJ. Exploring the oral microbiota of children at various developmental stages of their dentition in the relation to their oral health. BMC Med Genomics. 2011;4:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bogaert D, Keijser B, Huse S, et al. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS ONE. 2011;6(2):e17035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Flanagan JL, Brodie EL, Weng L, et al. Loss of bacterial diversity during antibiotic treatment of intubated patients colonized with Pseudomonas aeruginosa. J Clin Microbiol. 2007;45(6):1954–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Harris JK, De Groote MA, Sagel SD, et al. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc Natl Acad Sci USA. 2007;104(51):20529–20533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Klepac-Ceraj V, Lemon KP, Martin TR, et al. Relationship between cystic fibrosis respiratory tract bacterial communities and age, genotype, antibiotics and Pseudomonas aeruginosa. Environ Microbiol. 2010;12(5):1293–1303 [DOI] [PubMed] [Google Scholar]
- 100.van der Gast CJ, Walker AW, Stressmann FA, et al. Partitioning core and satellite taxa from within cystic fibrosis lung bacterial communities. ISME J. 2011;5(5):780–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sibley CD, Grinwis ME, Field TR, et al. Culture enriched molecular profiling of the cystic fibrosis airway microbiome. PLoS ONE. 2011;6(7):e22702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fell JM. Neonatal inflammatory intestinal diseases: necrotising enterocolitis and allergic colitis. Early Hum Dev. 2005;81(1):117–122 [DOI] [PubMed] [Google Scholar]
- 103.Martin CR, Walker WA. Intestinal immune defences and the inflammatory response in necrotising enterocolitis. Semin Fetal Neonatal Med. 2006;11(5):369–377 [DOI] [PubMed] [Google Scholar]
- 104.Wang Y, Hoenig JD, Malin KJ, et al. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 2009;3(8):944–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mshvildadze M, Neu J, Shuster J, Theriaque D, Li N, Mai V. Intestinal microbial ecology in premature infants assessed with non-culture-based techniques. J Pediatr. 2010;156(1):20–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mshvildadze M, Neu J, Mai V. Intestinal microbiota development in the premature neonate: establishment of a lasting commensal relationship? Nutr Rev. 2008;66(11):658–663 [DOI] [PubMed] [Google Scholar]
- 107.Mai V, Young CM, Ukhanova M, et al. Fecal microbiota in premature infants prior to necrotizing enterocolitis. PLoS ONE. 2011;6(6):e20647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shane AL. Applications of probiotics for neonatal enteric diseases. J Perinat Neonatal Nurs. 2008;22(3):238–243 [DOI] [PubMed] [Google Scholar]
- 109.Cotten CM, Taylor S, Stoll B, et al. NICHD Neonatal Research Network . Prolonged duration of initial empirical antibiotic treatment is associated with increased rates of necrotizing enterocolitis and death for extremely low birth weight infants. Pediatrics. 2009;123(1):58–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lin HC, Su BH, Chen AC, et al. Oral probiotics reduce the incidence and severity of necrotizing enterocolitis in very low birth weight infants. Pediatrics. 2005;115(1):1–4 [DOI] [PubMed] [Google Scholar]
- 111.Meinzen-Derr J, Poindexter B, Wrage L, Morrow AL, Stoll B, Donovan EF. Role of human milk in extremely low birth weight infants’ risk of necrotizing enterocolitis or death. J Perinatol. 2009;29(1):57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lin HC, Hsu CH, Chen HL, et al. Oral probiotics prevent necrotizing enterocolitis in very low birth weight preterm infants: a multicenter, randomized, controlled trial. Pediatrics. 2008;122(4):693–700 [DOI] [PubMed] [Google Scholar]
- 113.Deshpande G, Rao S, Patole S, Bulsara M. Updated meta-analysis of probiotics for preventing necrotizing enterocolitis in preterm neonates. Pediatrics. 2010;125(5):921–930 [DOI] [PubMed] [Google Scholar]
- 114.Dicksved J, Halfvarson J, Rosenquist M, et al. Molecular analysis of the gut microbiota of identical twins with Crohn’s disease. ISME J. 2008;2(7):716–727 [DOI] [PubMed] [Google Scholar]
- 115.Sokol H, Seksik P, Furet JP, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15(8):1183–1189 [DOI] [PubMed] [Google Scholar]
- 116.Qin J, Li R, Raes J, et al. MetaHIT Consortium . A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Collins SM, Denou E, Verdu EF, Bercik P. The putative role of the intestinal microbiota in the irritable bowel syndrome. Dig Liver Dis. 2009;41(12):850–853 [DOI] [PubMed] [Google Scholar]
- 118.Gori A, Tincati C, Rizzardini G, et al. Early impairment of gut function and gut flora supporting a role for alteration of gastrointestinal mucosa in human immunodeficiency virus pathogenesis. J Clin Microbiol. 2008;46(2):757–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104(34):13780–13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Garrett WS, Gordon JI, Glimcher LH. Homeostasis and inflammation in the intestine. Cell. 2010;140(6):859–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134(2):577–594 [DOI] [PubMed] [Google Scholar]
- 123.Jansson J, Willing B, Lucio M, et al. Metabolomics reveals metabolic biomarkers of Crohn’s disease. PLoS ONE. 2009;4(7):e6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rasquin A, Di Lorenzo C, Forbes D, et al. Childhood functional gastrointestinal disorders: child/adolescent. Gastroenterology. 2006;130(5):1527–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kassinen A, Krogius-Kurikka L, Mäkivuokko H, et al. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology. 2007;133(1):24–33 [DOI] [PubMed] [Google Scholar]
- 126.Malinen E, Rinttilä T, Kajander K, et al. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol. 2005;100(2):373–382 [DOI] [PubMed] [Google Scholar]
- 127.FAO/WHO. Health and Nutritional Properties of Probiotics in Food Including Powder Milk With Live Lactic Acid Bacteria. 2001
- 128.Servin AL, Coconnier MH. Adhesion of probiotic strains to the intestinal mucosa and interaction with pathogens. Best Pract Res Clin Gastroenterol. 2003;17(5):741–754 [DOI] [PubMed] [Google Scholar]
- 129.Resta-Lenert S, Barrett KE. Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC). Gut. 2003;52(7):988–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dotan I, Rachmilewitz D. Probiotics in inflammatory bowel disease: possible mechanisms of action. Curr Opin Gastroenterol. 2005;21(4):426–430 [PubMed] [Google Scholar]
- 131.Kaila M, Isolauri E, Soppi E, Virtanen E, Laine S, Arvilommi H. Enhancement of the circulating antibody secreting cell response in human diarrhea by a human Lactobacillus strain. Pediatr Res. 1992;32(2):141–144 [DOI] [PubMed] [Google Scholar]
- 132.Kunze WA, Mao YK, Wang B, et al. Lactobacillus reuteri enhances excitability of colonic AH neurons by inhibiting calcium-dependent potassium channel opening. J Cell Mol Med. 2009;13(8B):2261–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Thomas DW, Greer FR. Probiotics and prebiotics in pediatrics. Pediatrics. 2010;126(6):1217–1231 [DOI] [PubMed] [Google Scholar]
- 134.Lewis S, Brazier J, Beard D, Nazem N, Proctor D. Effects of metronidazole and oligofructose on faecal concentrations of sulphate-reducing bacteria and their activity in human volunteers. Scand J Gastroenterol. 2005;40(11):1296–1303 [DOI] [PubMed] [Google Scholar]
- 135.Kanauchi O, Oshima T, Andoh A, Shioya M, Mitsuyama K. Germinated barley foodstuff ameliorates inflammation in mice with colitis through modulation of mucosal immune system. Scand J Gastroenterol. 2008;43(11):1346–1352 [DOI] [PubMed] [Google Scholar]
- 136.Furrie E, Macfarlane S, Kennedy A, et al. Synbiotic therapy (Bifidobacterium longum/Synergy 1) initiates resolution of inflammation in patients with active ulcerative colitis: a randomised controlled pilot trial. Gut. 2005;54(2):242–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Andoh A, Fujiyama Y. Therapeutic approaches targeting intestinal microflora in inflammatory bowel disease. World J Gastroenterol. 2006;12(28):4452–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yadav H, Jain S, Sinha PR. Antidiabetic effect of probiotic dahi containing Lactobacillus acidophilus and Lactobacillus casei in high fructose fed rats. Nutrition. 2007;23(1):62–68 [DOI] [PubMed] [Google Scholar]
- 139.van Nood E, Speelman P, Kuijper EJ, Keller JJ. Struggling with recurrent Clostridium difficile infections: is donor faeces the solution? Euro Surveill. 2009;14(34): pii=19316. [DOI] [PubMed] [Google Scholar]
- 140.Yoon SS, Brandt LJ. Treatment of refractory/recurrent C. difficile-associated disease by donated stool transplanted via colonoscopy: a case series of 12 patients. J Clin Gastroenterol. 2010;44(8):562–566 [DOI] [PubMed] [Google Scholar]
- 141.Rohlke F, Surawicz CM, Stollman N. Fecal flora reconstitution for recurrent Clostridium difficile infection: results and methodology. J Clin Gastroenterol. 2010;44(8):567–570 [DOI] [PubMed] [Google Scholar]
- 142.Rachmilewitz D, Katakura K, Karmeli F, et al. Toll-like receptor 9 signaling mediates the anti-inflammatory effects of probiotics in murine experimental colitis. Gastroenterology. 2004;126(2):520–528 [DOI] [PubMed] [Google Scholar]
- 143.Yan F, Cao H, Cover TL, Whitehead R, Washington MK, Polk DB. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology. 2007;132(2):562–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shanahan F. Therapeutic implications of manipulating and mining the microbiota. J Physiol. 2009;587(pt 17):4175–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rea MC, Clayton E, O’Connor PM, et al. Antimicrobial activity of lacticin 3,147 against clinical Clostridium difficile strains. J Med Microbiol. 2007;56(pt 7):940–946 [DOI] [PubMed] [Google Scholar]
- 146.Rea MC, Sit CS, Clayton E, et al. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc Natl Acad Sci USA. 2010;107(20):9352–9357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lengauer C, Eckelt A, Weith A, et al. Painting of defined chromosomal regions by in situ suppression hybridization of libraries from laser-microdissected chromosomes. Cytogenet Cell Genet. 1991;56(1):27–30 [DOI] [PubMed] [Google Scholar]
- 149.Ronaghi M. Pyrosequencing sheds light on DNA sequencing. Genome Res. 2001;11(1):3–11 [DOI] [PubMed] [Google Scholar]