Abstract
The parasympathetic limb of the autonomic nervous system regulates the activity of multiple organ systems. Muscarinic receptors are G protein coupled receptors (GPCRs) that mediate the response to acetylcholine released from parasympathetic nerves.1–5 Their role in the unconscious regulation of organ and central nervous system function makes them potential therapeutic targets for a broad spectrum of diseases. The M2 muscarinic acetylcholine receptor (M2 receptor) is essential for the physiologic control of cardiovascular function through activation of G protein-coupled inwardly-rectifying potassium channels, and is of particular interest because of its extensive pharmacological characterization with both orthosteric and allosteric ligands. Here we report the structure of antagonist-bound M2 receptor, the first human acetylcholine receptor to be characterized structurally. The antagonist QNB binds in the middle of a long aqueous channel extending approximately two-thirds through the membrane. The orthosteric binding pocket is formed by amino acids that are identical in all 5 muscarinic receptor subtypes, and shares structural homology with other functionally unrelated acetylcholine binding proteins from different species. A layer of tyrosine residues forms an aromatic cap restricting dissociation of the bound ligand. A binding site for allosteric ligands has been mapped to residues at the entrance to the binding pocket near this aromatic cap. The M2 receptor structure provides insights into the challenges of developing subtype-selective ligands for muscarinic receptors and their propensity for allosteric regulation.
The muscarinic receptors constitute a family with five subtypes M1-M51. M1, M3, and M5 subtypes couple with the Gq family of G proteins, and M2 and M4 subtypes with the Gi/Go family of G proteins. The muscarinic acetylcholine receptors were originally defined as a functional concept on the basis of the work by Dale2 and others showing that the muscarinic action by a series of choline esters and other substances in various tissues could be differentiated from their nicotinic action. The muscarinic receptors are now known to be G protein-coupled receptors (GPCRs)3 and the nicotinic receptor a ligand-gated ion channel. Muscarinic receptors were initially defined biochemically as proteins that specifically bound 3-quinuclidinyl-benzilate (QNB) and N-methylscopolamine (NMS). They were among the first GPCRs to be purified from cerebral membranes4, and to be functionally reconstituted with purified G protein in lipid vesicles3. The M1 receptor5 together with the β2 adrenergic receptor6 were the first neurotransmitter-activated GPCRs to be cloned, revealing the seven transmembrane segment (TM) topology initially observed for rhodopsin7, and subsequently found to be common to all members of the GPCR family.
As a consequence of their roles in both the central and parasympathetic nervous systems, muscarinic receptors are targets for treatment of a spectrum of disorders including Alzheimer’s disease, schizophrenia and Parkinson’s disease, and chronic obstructive pulmonary disease8. However, developing highly subtype selective orthosteric drugs for muscarinic receptors has been challenging and thus far largely unsuccessful. Recent drug discovery efforts have therefore shifted to the development of small molecule allosteric modulators. Muscarinic receptors have long been a model system for studying allosteric regulation of GPCR signaling because of their exceptional propensity to bind allosteric ligands9. To better understand the structural basis for challenges in developing orthosteric drugs and the susceptibility for allosteric regulation, we obtained a crystal structure of the M2 receptor.
In our initial efforts to obtain the structure of the M2 receptor we expressed and purified M2 receptor lacking most of the third intracellular loop (IL3) and the native glycosylation sites. The central part of IL3 of the M2 receptor can be removed without impairing its ability to bind to agonists or activate G proteins10, and IL3 was shown to have a flexible structure11. Using this modified M2 receptor bound to the high affinity inverse agonist R-(−)-3-QNB, we performed crystallization by hanging drop vapor diffusion and obtained crystals that diffracted to around 9 Å, but were not able to improve the quality of these crystals. We subsequently replaced IL3 of the M2 receptor with T4-Lysozyme (T4L) as initially described for the β2 adrenergic receptor12 (Supplementary Fig. 1a). This method has been used to obtain crystal structures of four other GPCRs: the adenosine A2A receptor13, the CXCR4 receptor14, the dopamine receptor D315, and most recently the histamine H1 receptor16. The binding properties of M2-T4L with muscarinic ligands were essentially the same as for the wild type M2 receptor (Supplementary Fig. 1b,c), indicating that the overall TM architecture of M2-T4L was minimally affected by introduction of T4L. The M2-T4L was subsequently crystallized in lipidic cubic phase. A 3.0 Å structure was solved by molecular replacement from a data set obtained by merging diffraction data from 23 crystals.
As is typical for proteins crystallized by the lipidic cubic phase method, the lattice for the M2 receptor shows alternating aqueous and lipidic layers with M2 receptor molecules embedded in the latter while T4L is confined to aqueous regions (Supplementary Fig. 2). Within the membrane plane, receptor molecules are packed closely against one another, alternating orientations within the bilayer. There are abundant hydrophobic contacts between receptor molecules within the membrane, while polar interactions primarily involve contacts between T4L molecules as well as receptor-T4L interactions.
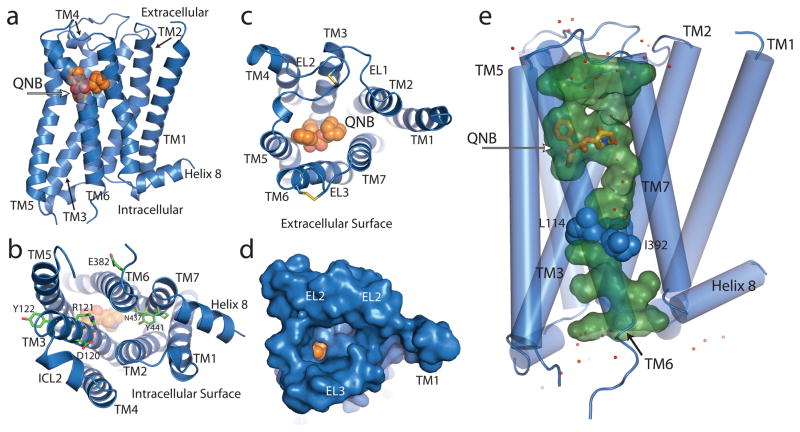
The overall structure of the M2 receptor (Fig. 1a) is similar to that of rhodopsin and other recently crystallized inactive GPCR structures (compared in Supplementary Figure 3). The cytoplasmic surface of the M2 receptor is in an inactive conformation, but as with most other GPCR structures, there is no interaction involving Arg1213.50 in the conserved E/DRY sequence in TM3 and Glu3826.30 in TM6 (Fig. 1b). Instead, the Arg1213.50 side chain forms a salt bridge only with Asp1203.49. In rhodopsin, the homologous residues form part of a charge-charge interaction that stabilizes the cytoplasmic ends of TM3 and TM6 in an inactive state17. The second intracellular loop shows a helical conformation similar to that first seen for the turkey β1 adrenergic receptor18.
Figure 1.
The M2 receptor (blue ribbon) with bound QNB (orange spheres). a, M2 receptor in profile. b, Cytoplasmic surface showing conserved DRY residues in TM3. c, Extracellular view into QNB binding pocket. d, Extracellular view with solvent-accessible surface rendering shows a funnel-shaped vestibule and a nearly buried QNB binding pocket. e, Aqueous channel (green) extending from the extracellular surface into the transmembrane core is interrupted by a layer of three hydrophobic residues (blue spheres). Well-ordered water molecules are shown as red dots.
GPCR crystal structures show the greatest differences in the extracellular surface (Supplementary Fig. 3). The M2 receptor has a relatively simple and open extracellular surface (Fig 1c, d) with the longer extracellular loop (ECL) 2 stabilized by a conserved disulfide with Cys963.25 at the N-terminus of TM3 and Cys176 in the middle of ECL2. In addition, the second disulfide bond was detected between C413 and C416 in the ECL3. The extracellular surface of the M2 receptor most resembles that of the dopamine D3 receptor (Supplementary Fig. 3).
Crystal structures of GPCRs reveal a network of hydrogen bonding interactions that extend from the binding pocket to the cytoplasmic surface. However, a distinctive feature of the M2 receptor is that this network is part of a long, continuous aqueous channel extending from the extracellular surface to a depth of approximately 33 Å when measured from ECL2 (Fig 1e). This channel contains the ligand binding pocket, but extends beyond the ligand and is separated from the cytoplasmic surface by a hydrophobic layer formed by three amino acids: Leu652.46 in TM2, Leu1144.43 in TM4 and Ile3926.40 in TM6. Each of these is absolutely conserved among all five muscarinic subtypes. The dimensions of the channel below the QNB binding site are large enough to accommodate a long, extended orthosteric ligand. Supplementary Figure 4 compares the aqueous channels of other GPCRs.
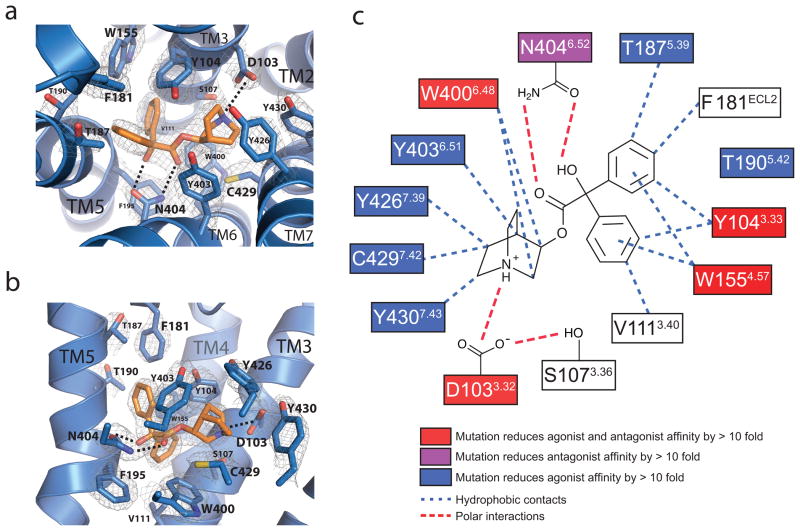
The ligand QNB binds within a deeply buried pocket defined by side chains of TM3, 4, 5, 6 and 7 (Fig. 2a–c, Supplementary Fig. 5, Supplementary table 3). An aromatic cage encloses the amine and forms a lid over the ligand, separating the orthosteric site from the extracellular vestibule. Asp1033.32 and Asn4046.52 serve to orient the ligand in the largely hydrophobic binding cavity, with Asn4046.52 forming paired hydrogen bonds with the hydroxyl and carbonyl groups in QNB while Asp1033.32 engages in a charge-charge interaction with the amine moiety of the ligand (Fig. 2). The transmembrane segment amino acids that form the QNB binding pocket are identical in all five muscarinic receptor subtypes (Supplementary Table 1), consistent with results of QNB binding experiments on M1–M4 receptors, and with site-directed mutagenesis experiments on M119, M220, and M321 receptors. Only Phe181, which extends downward from ECL2 and interacts with one of the two phenyl rings on QNB (Fig. 2), differs from all other muscarinic receptor subtypes which have leucine in the homologous position. The importance of Asp3.32 for both agonist and antagonist binding has been demonstrated in mutagenesis and covalent-labeling experiments and modeling studies19–22. In contrast, mutation of Asn4046.52 to Ala on M123 and M324 receptors was shown to greatly affect binding of QNB but have little effect on binding of or activation by acetylcholine. It is possible that Asn4046.52 is hydrogen-bonded with ester group of QNB but not of acetylcholine.
Figure 2.
Binding interactions between the M2 receptor and QNB.
a, b, Two views of the QNB binding pocket. Amino acids within 4 Å of the ligand are shown as light blue sticks, with QNB in orange. Nitrogen and oxygen atoms are colored dark blue and red, respectively. Polar interactions are indicated by dashed lines. A 2Fo–Fc map is shown in wire at 1.5 σ contour. c, A schematic representation of QNB binding interactions is shown. Mutations of amino acids in red boxes have been shown to reduce both antagonist and agonist binding by more than 10 fold. Mutations of the amino acid in the purple boxes reduce antagonist binding affinity by more that 10 fold. Mutations of amino acids in blue boxes reduce agonist binding by more than 10 fold. Blue dotted lines indicate potential hydrophobic interactions and red lines indicate potential polar interactions.
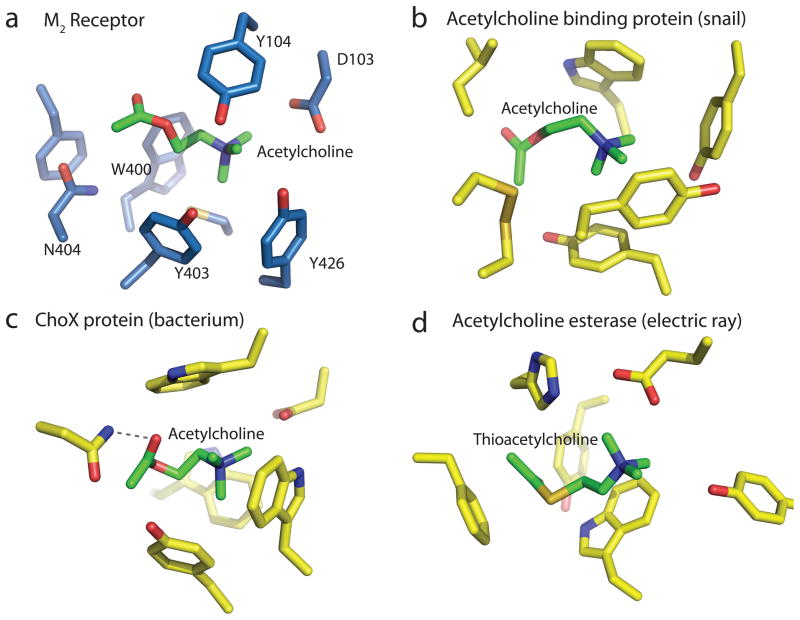
The M2 and other muscarinic receptors represent one of four families of acetylcholine binding proteins to be structurally characterized thus far. Figure 3a shows the orthosteric binding site of the M2 receptor with acetylcholine docked with the gauche form of the O-C2-C1-N dihedral angle, which places the choline group in the aromatic cage interacting with Asp1033.32, while the carbonyl oxygen is tentatively bound to Asn4046.52 (Fig 3a). The natural agonist acetylcholine is much smaller than the bulky antagonist QNB. As described in agonist-bound structure of the β2 adrenergic receptor, the contraction of ligand binding pocket is expected as a result of an inward shift of TM525. This result is consistent with the previous mutation studies showing that Thr1875.39 and Thr1905.42 in TM5 (Fig. 2) alter binding of most agonists but not of antagonists20. Bulky compounds capable of blocking activation-related contraction of the pocket would be very efficient in locking M2 receptor in an inactive conformation as is exemplified here by the antagonist QNB. It has been proposed that the conformational change of M2 receptor upon activation might be accompanied by conformational change of acetylcholine from the gauche to trans form of the O-C2-C1-N dihedral angle26. It remains to be determined in which pose acetylcholine binds to the M2 receptor or to the M2 receptor-G protein complex, and if acetylcholine hydrogen bonds with Asn4046.52 or other residues.
Figure 3.
Convergent evolution of acetylcholine binding sites. a, Acetylcholine is modeled into the crystal structure of the M2 receptor. b, Acetylcholine binding pocket in the crystal structure of the acetylcholine binding protein from the snail Aplysia californica (PDB ID: 2XZ5). c, Acetylcholine binding pocket in the acetylcholine binding protein ChoX from the gram negative bacterium Sinorhizobium meliloti (PDB ID: 2RIN). d, Binding site for thio-acetylcholine in the enzyme acetylcholine esterase from the electric ray Torpedo californica (PDB ID: 2C4H).
In a striking example of convergent evolution, the orthosteric site of M2 receptor exhibits many features noted previously as common structural elements in unrelated acetylcholine binding proteins27. Like the M2 receptor, a nicotinic acetylcholine receptor homologue bound to acetylcholine (Fig. 3b) shows an aromatic cage comprised of three tyrosines and a tryptophan, although it notably lacks a counterion to the choline group28, whereas in the M2 receptor this role is filled by Asp1033.32. A bacterial acetylcholine binding protein ChoX from Sinorhizobium meliloti (Fig. 3c) also possesses an aromatic cage, and like M2 receptor has an aspartate in close proximity to the amine engaging in a charge-charge interaction29. Also like the M2 receptor, ChoX has an asparagine hydrogen bonding to the ligand carbonyl. Like these proteins, the enzyme acetylcholine esterase (Fig. 3d) employs an aromatic cage and a carboxylate to bind the choline group, while the (thio)acetyl group interacts with a phenylalanine, likely through π- π interactions30. Taken together, these structures suggest that an aromatic cage and buried carboxylate are likely to be critical elements for acetylcholine recognition and binding in general.
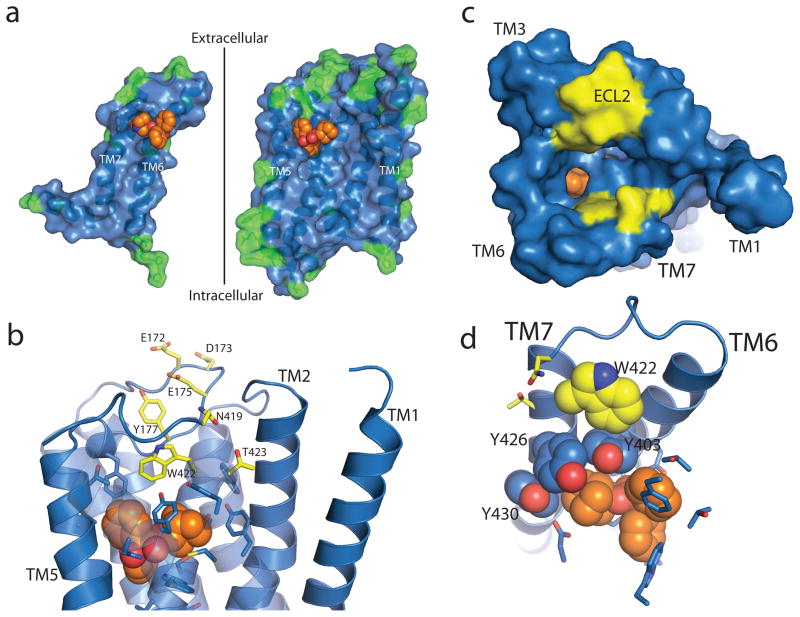
There is a growing interest in the development of allosteric ligands for GPCR targets. This is motivated by the ability to develop more subtype-selective drugs targeted at less conserved regions of the receptor. Moreover, allosteric ligands modulate the effects of natural hormones and neurotransmitters, and may therefore regulate receptor activity in a more physiologic manner. As noted above, the orthosteric binding pocket is highly conserved among all muscarinic receptor subtypes. Allosteric regulation of GPCRs was first observed for the M2 receptor and this receptor has been one of the most extensively characterized allosteric model systems9. Figure 4a shows the inner surface of the M2 receptor highlighting residues that are not conserved with its closest relative, the M4 receptor. It can be seen that the orthosteric binding pocket and transmembrane core are highly conserved. The greatest diversity is observed in the extracellular loops and the extracellular end of TM segments that form the entrance to the orthosteric binding pocket. These amino acids represent structural diversity that could be exploited for the development of more subtype-selective ligands9. Of interest, site-directed mutagenesis and chimeric receptor studies have implicated several of these amino acids in the binding of several well-characterized allosteric modulators9. As shown in Fig 4b-d, these residues are located in ECL2 and N-terminus of TM7 at the entrance to the binding pocket. Trp4227.35, a residue implicated in the binding of several allosteric modulators, appears to form an edge-to-face π –π interaction with Tyr4036.51, part of the aromatic cage surrounding the charged amine of the orthosteric ligand (Fig. 4d). Binding of allosteric ligands to this site would be expected to influence the association and disassociation rates of orthosteric ligands.
Figure 4.
Allosteric binding in the M2 receptor. a, Differences between the M2 and M4 receptors are shown as green residues mapped onto the inner surface of the M2 receptor (blue), with QNB in orange spheres. The sequence conservation within the orthosteric site is apparent, while residues outside show more variability. b–d, Mutations that alter allosteric binding are shown with yellow carbons, and amino acids involved in QNB binding are shown with blue carbons as sticks or spheres. b, c, Different views of possible allosteric binding sites in the M2 receptor. The surface view in c shows the positions of possible allosteric binding sites (yellow) lining the path to the QNB binding pocket. d, Trp422 (yellow spheres), implicated in binding of allosteric ligands, forms an edge-to-face aromatic interaction with Tyr403, part of the aromatic cage (blue spheres) of the orthosteric site.
The structure of the M2 receptor provides insights into both orthosteric and allosteric regulation of muscarinic receptors. The development of more selective drugs for muscarinic receptors will likely require exploitation of the more diverse allosteric surface, either as exclusively allosteric ligands or as ligands that occupy both orthosteric and allosteric sites.
Methods summary
Untagged human M2 muscarinic acetylcholine receptor was expressed in Sf9 cells with the third intracellular loop replaced with T4 lysozyme, then extracted with digitonin and sodium cholate and purified by ligand affinity chromatography, then exchanged into decyl maltoside buffer. Purified receptor was crystallized by the lipidic cubic phase technique following addition of a stabilizing neopentyl glycol detergent. Data collection was performed at Advanced Photon Source beamlines 23ID-B and 23ID-D, and the structure solved by molecular replacement. Refinement statistics are given in Supplementary Table 2.
Methods
Construction of M2-T4L expression vectors for Sf9 cells
The coding sequence of the human M2-T4L was designed to have N-linked glycosylation sites (Asn2, Asn3, Asn6 and Asn9) mutated to aspartic acid and cysteineless T4 lysozyme (C54T, C97A) residues 2 – 161 inserted into the third intracellular loop, replacing M2 residues 218 – 376. This construct was synthesized (TAKARA bio Inc.), and cloned into the pFastbac1 Sf9 expression vector (Invitrogen) as illustrated in Supplementary Figure 1a. A TAA stop codon was placed after the R466 codon, terminating translation. The synthesized M2-T4L described above was confirmed by sequencing.
Expression and membrane preparation
Recombinant baculovirus was made from pFastbac1-M2-T4L using the Bac-to-Bac system (Invitrogen)31. The M2-T4L protein was expressed in baculovirus infected Sf9 insect cell as described previously32. Sf9 insect cells were prepared at a density of 1.0 × 106 cells/ml and suspended in 5 L of the IPL-41/SF900 II complex media or ESF921 insect media. Media containing Sf9 insect cells were transferred into the CELLBAG 22 L/O (GE Healthcare) and cultured for 4 days with the following culture conditions: 20 rpm, 8.5° of rocking angle, 30% O2, 0.25 L/min of air flow rate, and 27°C. After 4 days, 200~300 ml of the M2-T4L baculovirus stock (approximate multiplicity of infection (M.O.I) = 2) and 700~800 ml of IPL-41/SF900 II complex media were transferred into the CELLBAG (final culture volume = 6 L) and infected for 2 days under the following infection conditions: 22 rpm, 8.5° of rocking angle, 50% O2, air flow rate, 0.25 L/min, and 27°C. Two days later, a fraction of the cells was harvested for the binding assay and the remaining cells were centrifuged at 6,000 × g for 10 min and harvested. The cell pellet was washed with 250 ml of Phosphate Buffered Saline without calcium chloride and magnesium chloride (PBS(−)) and resuspended with 100 ml of PBS(−) containing a protease inhibitor cocktail tablet (Roche). Final concentration of protease inhibitors was 2.5 μg/ml pepstatin, 2 μg/ml PMSF, 20 μg/ml leupeptin, and 0.5 mM benzamidine. Cells were quick frozen in liquid nitrogen and stored at −80°C.
The membrane was prepared from the M2-T4L expressing Sf9 insect cells as described previously31. For the preparation of membranes from insect cells, Sf9 insect cells were centrifuged at 1,500 g for 10 min at 4°C. The pellet was washed with PBS(−), then resuspended in 100 ml of hypotonic buffer containing 10 mM HEPES at pH 7.5, 20 mM KCl, 10 mM MgCl2, and protease inhibitor cocktail, followed by Dounce homogenization to resuspend the membranes. Insect cell membranes were centrifuged at 100,000 g for 30 min and the pellets were resuspended in 10 mM HEPES at pH 7.5, 10 mM MgCl2, 20 mM KCl, 40% glycerol, and snap-frozen in liquid nitrogen and then stored at −80 °C until use. Membrane proteins were quantified using the bicinchoninic acid (BCA) method (Pierce) using a BSA standard.
Purification of M2-T4L-QNB
M2-T4L was expressed in Sf9 cells, solubilized with digitonin/Na-cholate solution, and purified by using an affinity column with aminobenztropine (ABT) as a ligand33, as described below. The whole procedure was carried out at 4° C. Sf9 membrane preparations with 2.1 kg of wet weight and approximately 1.5 μmol of [3H]QNB binding sites were solubilized with 1% digitonin/0.35% Na-cholate/10 mM K-phosphate buffer (pH 7.0) (KPB)/50 mM NaCl/1 mM EDTA/a cocktail of protease inhibitors (4 L). The supernatant was applied to two ABT-columns run in parallel (500 ml each), followed by washing with 0.1% digitonin/0.1% Na-cholate/20 mM KPB/150 mM NaCl (2 L x 2) at a rate of approximately 90 ml/hr. M2-T4L was eluted from the ABT columns with 0.5 mM atropine/0.1% digitonin/0.1% Na-cholate/20 mM KPB/150mM NaCl in 2 L elution volume for each column, and was bound to a column of hydroxyapatite (30 ml), which was washed at a rate of 30–50 ml/hr with a series of solutions as follows (1) 0.1% digitonin/0.1% Na-cholate/20 mM KPB (100 ml), (2) 5 μM QNB/0.1% digitonin/0.1% Na-cholate/20 mM KPB (600 ml), (3) 0.35% Na-cholate/20 mM KPB (600 ml), (4) 0.2% decylmaltoside/20 mM KPB (500 ml), (5) 0.2% decylmaltoside/150 mM KPB (100 ml), (6) 0.2% decylmaltoside/500 mM KPB (60 ml). M2-T4L-QNB was finally eluted with 0.2% decylmaltoside/1 M KPB (50 ml). The eluate was concentrated to approximately 1 ml (ca 30 mg protein/ml) with Amicon Ultra (MILLIPORE), followed by dialysis against 0.2% decylmaltoside/20 mM Tris-HCl buffer (pH 7.5) and storage in − 80°C. The yield was estimated to be approximately 50% on the assumption that the recovered protein is pure M2-T4L. Protein concentration was determined using BCA Protein Assay (PIERCE). Since we purified M2-T4L as a complex with QNB we could not estimate the [3H]QNB binding activity because the dissociation rate of QNB is too slow. However, in preliminary experiments using [3H]QNB or dissociable atropine as eluants, we confirmed that the receptor is purified to near homogeneity. The purity of M2-T4L was confirmed by SDS-PAGE and gel permeation chromatography (Supplementary Fig. 6). All QNB used in purification and crystallization was the high affinity enantiomer, R-(–)-3-QNB.
Measurement of ligand binding activity
Ligand binding activity of wild type M2 and M2-T4L receptors was determined as described previously34. Briefly, the receptors solubilized from Sf9 membranes were incubated with 0.1 – 4 nM [3H]QNB with or without 1 μM atropine, or with 2 nM [3H]QNB with various concentrations of carbamylcholine or atropine in 0.1% digitonin/20 mM KPB for 60 min at 30°C (total volume 0.2 ml). The amount of [3H]QNB bound to receptors was assayed by using a small column of Sephadex G50 fine (2 ml). The density of [3H]QNB binding sites in particulate fraction of M2-T4L was 17 pmol/mg of protein in average and ranged from 5.3 to 35 pmol/mg of total protein.
Crystallization
QNB-bound M2-T4L was concentrated to 20 mg/ml in decyl maltoside buffer in a volume of approximately 100 μl. A 10% stock solution of lauryl maltose neopentyl glycol detergent (MNG, Anatrace) with 100 mM NaCl and 20 mM HEPES pH 7.5 was then added to the protein to a final concentration of 1% (w/v) of MNG detergent. The sample was incubated 1 hour on ice, then diluted to 1 ml in 0.1% MNG buffer and reconcentrated to 50 mg/ml prior to reconstitution. The final volume of protein sample at this concentration was typically 20 – 30 μl. Protein was reconstituted in cubic phase by mixing with a 1.5-fold weight excess of a 10:1 monoolein:cholesterol mix by the twin-syringe method35. Briefly, the protein and lipid were mixed by passage through coupled syringes 100 times either by hand or using a Gryphon LCP robot (Art Robbins Instruments). The reconstituted protein was dispensed using a modified ratchet device (Hamilton) or using the Gryphon LCP robot in 40 nl drops to either 24-well or 96-well glass sandwich plates and overlaid with 0.8 μl precipitant solution. A single crystallization lead was initially identified using an in-house screen and then optimized. Crystals for data collection were grown in 25 to 35% PEG 300, 100 mM ammonium phosphate, 2% 2-Methyl-2,4-pentanediol, 100 mM HEPES pH 7.0–7.8. Crystals reached full size and were harvested after 3–4 days at 20°C. Typical crystals are shown in supplementary figure 7.
Data collection and processing
Diffraction data were measured at the Advanced Photon Source beamlines 23 ID-B and 23 ID-D. Several hundred crystals were screened, and a final data set was compiled using diffraction wedges of typically 5 degrees from the 23 most strongly diffracting crystals. Data reduction was performed using HKL200036. Diffraction quality was very heterogeneous, with some crystals diffracting to 2.3 Å while others failed to diffract past 3.5 Å. Among the best crystals, most diffracted to 3.0 – 2.5 Å. Severe radiation damage and anisotropic diffraction resulted in low completeness in higher resolution shells. We report this structure to an overall resolution of 3.0 Å. Despite the low completeness in high resolution bins, inclusion of these reflections significantly improved map quality. Highest shell <I>/<σI> is relatively low, in large part due to anisotropy of the diffraction. The final resolution cutoff was chosen on the basis of completeness and <I>/<σI> in the spherical highest shell, but analysis of average F/σF values along reciprocal space axes suggests resolution limits (based on F/σF > 3) of 3.5, 2.9, and 2.7 Å along a*, b*, and c*, respectively. The real space c axis is normal to the plane of the lipid membrane in the crystal.
Structure solution and refinement
The structure was solved by molecular replacement using Phaser37,38 with the structure of the inactive β2 adrenergic receptor and T4 lysozyme used as search models (PDB ID: 2RH1). The initial molecular replacement model was further fitted by rigid body refinement followed by simulated annealing and restrained refinement in Phenix39. Iterative manual rebuilding and refinement steps were performed with Coot and phenix.refine, respectively. Figures were prepared with PyMOL, and Ramachandran statistics were calculated with MolProbity.
Supplementary Material
Acknowledgments
We thank Prof. So Iwata at Kyoto University for supporting the production of M2 receptor, and we acknowledge support from the Japan Society for the Promotion of Science (Research for Future Program)(T.H.), from the Japan Science and Technology Corporation (CREST) (T.H.), from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Grants-in-Aid for Scientific Research on Priority Area 15083201 (T.H.), from the Japan Science and Technology Corporation (ERATO) (T.K.), from Toray Science Foundation (T.K.), from Takeda Science Foundation (T.K.), from Ichiro Kanehara Foundation (T.K.), from The Sumitomo Foundation (T.K.), from the National Institutes of Health Grants NS028471 and GM083118 (B.K.K), from the Mathers Foundation (B.K.K. and W.I.W.), and from the National Science Foundation (A.C.K.). We thank Tong Sun Kobilka for organizing the GPCR Workshop 2010 that brought together the research groups, and for facilitating this collaboration.
Footnotes
Author Contributions K.H. purified M2 and M2-T4L receptors, characterized their ligand binding activity, and performed attempts to crystallize them with hanging drop and other methods for more than ten years. A.C.K. crystallized the M2-T4L in lipidic cubic phase, collected and processed diffraction data, solved and refined the structure, and assisted with manuscript preparation. H.A. set up the expression system and expressed M2-T4L for in large amounts using the insect cell/baculovirus expression system. T.Y-K. has expressed using a yeast expression system, and has purified and crystallized M2 and M2-T4L receptors for five years. M.S. constructed several mutants of M2-T4L and evaluated their stabilities. C.Z. assisted with data collection and processing. W.I.W. oversaw data processing and refinement. T.O. gave advice to K.H. and T.H. on crystallization of the M2 receptor and interpretation of its structure. B.K. oversaw lipidic cubic phase crystallization, assisted with data collection, and wrote the manuscript together with T.H. and T.K. T.H., together with K.H., has engaged in biochemical studies of muscarinic receptors for more than thirty years, prepared M2 and M2- T4L receptors, and wrote a part of manuscript. T.K. has been collaborating with T.H. for five years, designed the receptor production strategy with T.H., and wrote a part of manuscript.
Author Information Coordinates and structure factors for M2-T4L are deposited in the Protein Data Bank (accession code 3UON). Reprints and permission information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Literature Cited
- 1.Hulme EC, Birdsall NJ, Buckley NJ. Muscarinic receptor subtypes. Annu Rev Pharmacol and Toxicol. 1990;30:633–673. doi: 10.1146/annurev.pa.30.040190.003221. [DOI] [PubMed] [Google Scholar]
- 2.Dale HH. The action of certain esters and ethers of choline, and their relation to muscarine. J Pharmacol Exp Ther. 1914;6:147–190. [Google Scholar]
- 3.Haga K, et al. Functional reconstitution of purified muscarinic receptors and inhibitory guanine nucleotide regulatory protein. Nature. 1985;316:731–733. doi: 10.1038/316731a0. [DOI] [PubMed] [Google Scholar]
- 4.Haga K, Haga T. Purification of the muscarinic acetylcholine receptor from porcine brain. J Biol Chem. 1985;260:7927–7935. [PubMed] [Google Scholar]
- 5.Kubo T, et al. Cloning, sequencing and expression of complementary DNA encoding the muscarinic acetylcholine receptor. Nature. 1986;323:411–416. doi: 10.1038/323411a0. [DOI] [PubMed] [Google Scholar]
- 6.Dixon RA, et al. Cloning of the gene and cDNA for mammalian beta-adrenergic receptor and homology with rhodopsin. Nature. 1986;321:75–79. doi: 10.1038/321075a0. [DOI] [PubMed] [Google Scholar]
- 7.Ovchinnikov Yu A. Rhodopsin and bacteriorhodopsin: structure-function relationships. FEBS letters. 1982;148:179–191. doi: 10.1016/0014-5793(82)80805-3. [DOI] [PubMed] [Google Scholar]
- 8.Wess J, Eglen RM, Gautam D. Muscarinic acetylcholine receptors: mutant mice provide new insights for drug development. Nat Rev Drug Discovery. 2007;6:721–733. doi: 10.1038/nrd2379. [DOI] [PubMed] [Google Scholar]
- 9.Gregory KJ, Sexton PM, Christopoulos A. Allosteric modulation of muscarinic acetylcholine receptors. Curr Neuropharmacol. 2007;5:157–167. doi: 10.2174/157015907781695946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kameyama K, Haga K, Haga T, Moro O, Sadée W. Activation of a GTP-binding protein and a GTP-binding-protein-coupled receptor kinase (beta-adrenergic-receptor kinase-1) by a muscarinic receptor m2 mutant lacking phosphorylation sites. Eur J Biochem/FEBS. 1994;226:267–276. doi: 10.1111/j.1432-1033.1994.tb20050.x. [DOI] [PubMed] [Google Scholar]
- 11.Ichiyama S, et al. The structure of the third intracellular loop of the muscarinic acetylcholine receptor M2 subtype. FEBS Lett. 2006;580:23–26. doi: 10.1016/j.febslet.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 12.Rosenbaum DM, et al. GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 13.Jaakola VP, et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu B, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chien EY, et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimamura T, et al. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011;475:65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 18.Warne T, et al. Structure of a β1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hulme EC, Lu ZL, Bee MS. Scanning mutagenesis studies of the M1 muscarinic acetylcholine receptor. Recept Channels. 2003;9:215–228. [PubMed] [Google Scholar]
- 20.Heitz F, et al. Site-directed mutagenesis of the putative human muscarinic M2 receptor binding site. Eur J Pharmacol. 1999;380:183–195. doi: 10.1016/s0014-2999(99)00439-2. [DOI] [PubMed] [Google Scholar]
- 21.Wess J. Mutational analysis of muscarinic acetylcholine receptors: structural basis of ligand/receptor/G protein interactions. Life Sci. 1993;53:1447–1463. doi: 10.1016/0024-3205(93)90618-d. [DOI] [PubMed] [Google Scholar]
- 22.Goodwin JA, Hulme EC, Langmead CJ, Tehan BG. Roof and floor of the muscarinic binding pocket: variations in the binding modes of orthosteric ligands. Mol Pharmacol. 2007;72:1484–1496. doi: 10.1124/mol.107.038265. [DOI] [PubMed] [Google Scholar]
- 23.Ward SD, Curtis CA, Hulme EC. Alanine-scanning mutagenesis of transmembrane domain 6 of the M1 muscarinic acetylcholine receptor suggests that Tyr381 plays key roles in receptor function. Mol Pharmacol. 1999;56 :1031–1041. doi: 10.1124/mol.56.5.1031. [DOI] [PubMed] [Google Scholar]
- 24.Bluml K, Mutschler E, Wess J. Functional role in ligand binding and receptor activation of an asparagine residue present in the sixth transmembrane domain of all muscarinic acetylcholine receptors. J Biol Chem. 1994;269:18870–18876. [PubMed] [Google Scholar]
- 25.Rasmussen SG, et al. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furukawa H, et al. Conformation of ligands bound to the muscarinic acetylcholine receptor. Mol Pharmacol. 2002;62:778–787. doi: 10.1124/mol.62.4.778. [DOI] [PubMed] [Google Scholar]
- 27.Zacharias N, Dougherty DA. Cation-π interactions in ligand recognition and catalysis. Trends Pharmacol Sci. 2002;23:281–287. doi: 10.1016/s0165-6147(02)02027-8. [DOI] [PubMed] [Google Scholar]
- 28.Brams M, et al. Crystal structures of a cysteine-modified mutant in loop D of acetylcholine-binding protein. J Biol Chem. 2011;286:4420–4428. doi: 10.1074/jbc.M110.188730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oswald C, et al. Crystal structures of the choline/acetylcholine substrate-binding protein ChoX from Sinorhizobium meliloti in the liganded and unliganded-closed states. J Biol Chem. 2008;283:32848–32859. doi: 10.1074/jbc.M806021200. [DOI] [PubMed] [Google Scholar]
- 30.Colletier JP, et al. Structural insights into substrate traffic and inhibition in acetylcholinesterase. EMBO J. 2006;25:2746–2756. doi: 10.1038/sj.emboj.7601175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asada H, et al. Evaluation of the Pichia pastoris expression system for the production of GPCRs for structural analysis. Microb Cell Fact. 2011;10(24) doi: 10.1186/1475-2859-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weber W, Weber E, Geisse S, Memmert K. Optimisation of protein expression and establishment of the Wave Bioreactor for Baculovirus/insect cell culture. Cytotechnology. 2002;38:77–85. doi: 10.1023/A:1021102015070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haga K, Haga T. Affinity chromatography of the muscarinic acetylcholine receptor. J Biol Chem. 1983;258:13575–13579. [PubMed] [Google Scholar]
- 34.Haga T, Haga K, Hulme EC. Solubilization, purification and molecular characterization of muscarinic acetylcholine receptors. In: Hulme EC, editor. Receptor Biochemistry : A Practical Approach. Oxford University Press; 1990. pp. 51–78. [Google Scholar]
- 35.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat Protoc. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 37.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40 :658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Afonine PV, Grosse-Kunstleve RW, Adams PD. A robust bulk-solvent correction and anisotropic scaling procedure. Acta Crystallogr D Biol Crystallogr. 2005;61:850–855. doi: 10.1107/S0907444905007894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.