Abstract
Since the 1980s, several small RNA motifs capable of chemical catalysis have been discovered. These small ribozymes, composed of between approximately 40 and 200 nucleotides, have been found to play vital roles in the replication of subviral and viral pathogens, gene regulation in prokaryotes, and have recently been discovered in noncoding eukaryotic RNAs. All of the known natural small ribozymes – the hairpin, hammerhead, hepatitis delta virus, Varkud satellite, and glmS ribozymes – catalyze the same self-cleavage reaction as RNAse A, resulting in two products, one bearing a 2′–3′ cyclic phosphate and the other a 5′-hydroxyl group. Although originally thought to be obligate metalloenzymes like the group I and II self-splicing introns, the small ribozymes are now known to support catalysis in a wide variety of cations that appear to be only indirectly involved in catalysis. Nevertheless, under physiologic conditions, metal ions are essential for the proper folding and function of the small ribozymes, the most effective of these being magnesium. Metal ions contribute to catalysis in the small ribozymes primarily by stabilizing the catalytically active conformation, but in some cases also by activating RNA functional groups for catalysis, directly participating in catalytic acid-base chemistry, and perhaps by neutralizing the developing negative charge of the transition state. Although interactions between the small ribozymes and cations are relatively nonspecific, ribozyme activity is quite sensitive to the types and concentrations of metal ions present in solution, suggesting a close evolutionary relationship between cellular metal ion homeostasis and cation requirements of catalytic RNAs, and perhaps RNA in general.
Keywords: hairpin ribozyme, hammerhead ribozyme, hepatitis delta virus ribozyme, Varkud satellite ribozyme, glmS ribozyme, general acid-base catalysis, electrostatic screening
1. INTRODUCTION
Since the discovery that RNA can catalyze chemical reactions [1], RNA enzymes (ribozymes) have been found to perform many essential functions in nature, including protein biosynthesis [2], RNA processing [1, 3–4], regulation of gene expression [5], and genomic processing in pathogens [6–10]. While some of these functional RNAs operate within the context of large ribonucleoprotein complexes, many ribozymes can support catalysis without protein cofactors [1, 5–10]. The naturally occurring small self-cleaving ribozymes, each comprising fewer than 200 nucleotides, demonstrate the capability of RNA to efficiently and economically catalyze biologically important chemistry. These include the hairpin [7], hammerhead [6, 9], hepatitis delta virus (HDV) [8, 11–12], Varkud satellite (VS) [10], and glmS [5] ribozymes. Although all of these were initially isolated from bacteria [5], viruses [7, 11–12], or subviral pathogens [6, 9–10], structural and functional homologs of the hammerhead and HDV ribozymes have recently been discovered within the genomes of several eukaryotes, including mammals [13–15], revealing the exciting possibility that small catalytic RNAs may help regulate eukaryotic gene expression. The versatility of small ribozymes as catalysts has been demonstrated by the discovery of many non-natural small ribozymes through in vitro selection, including a lead-dependent self-cleaving RNA [16], a ribozyme that catalyzes the synthetically useful Diels-Alder cycloaddition [17], and ribozymes that exploit allosteric binding of particular classes of metal ions [18–19]. The discovery of a tiny 29-nucleotide RNA that catalyzes aminoacyl-RNA synthesis [20] and a recently reported self-replicating RNA enzyme [21] support the notion that RNA could have served as the original catalyst of life [22].
In spite of this versatility, all of the currently known natural small ribozymes catalyze the same internal phosphodiester isomerization reaction as RNase A, resulting in two cleavage products: one bearing a 2′,3′-cyclic phosphate, and the other a 5′-hydroxyl group (see Figure 1). This reaction involves deprotonation of the 2′-OH nucleophile by a general base, attack of the activated 2′-oxyanion on the adjacent phosphate, and protonation of the 5′-oxygen leaving group by a general acid [23–26]. Although the small ribozymes were originally thought to be obligate metalloenzymes like the group I and II self-splicing introns, utilizing site-bound magnesium to directly coordinate and stabilize negatively charged groups in the transition state, this is no longer the prevailing view [27]. However, like all functional RNA, ribozymes require cations to counterbalance the abundant negative charge of their phosphate backbone as they fold into their functional three-dimensional conformations, and may also utilize cations in long-range electrostatic catalysis or general acid-base chemistry. At physiologic ionic strength, multivalent cations are essential for the small ribozymes to adopt their native tertiary structures [27], though a number of other metallic and non-metallic cations support catalysis to varying degrees [19, 28–38].
Figure 1. General mechanism of self-cleavage by the natural small ribozymes.
(A) A Brønsted-Lowry base (β) abstracts a proton to activate the 2′-OH nucleophile, which then attacks the adjacent phosphate, forming a pentacoordinate transition state (B) with approximate collinearity between the 2′-oxygen, phosphorus atom, and 5′-oxygen leaving group – the in-line attack geometry. The negative charge of the transition state may be stabilized by one or several metal cations (Mn+) that interact through inner- or outer-sphere contacts with the non-bridging oxygen atoms or by long-distance coulombic stabilization. A Brønsted-Lowry acid (α) donates a proton to the 5′-oxygen leaving group, resulting in a 5′-product bearing a 2′,3′-cyclic phosphate and a 3′-product bearing a 5′-OH (C).
The composition of free metal ions in the cell is well-tuned to the function of small ribozymes. The predominant metal cations in the cytosol are typically K+, with an activity of ~100 mM [39] and Mg2+, with an activity of ~1 mM [40]. It has recently been revealed that magnesium concentration is regulated by homeostasis in both bacterial [41–44] and eukaryotic [45] cells. In turn, under physiologic conditions magnesium ions are expected to play the most important role in stabilizing RNA tertiary structure and, by extension, facilitating the function of ribozymes. Perhaps not coincidentally, then, the natural small ribozymes have evolved to be functional at intracellularly available Mg2+ concentrations [5, 35, 46–47], and the likely enhancement of metal binding due to molecular crowding in the cytosol suggests an even tighter correlation [48]. In addition, it can be argued that the total cellular Mg2+ concentration of ~20 mM [49] is buffered by the large amounts of nucleic acids and in particular RNAs (typically 1–6% of the cellular mass [50]) that bind the divalent with typical affinities in the low millimolar range. A picture emerges of an intimate relationship between the metal ion composition of the cell and the metal ion dependence of functional RNAs, analogous to the correlation between intracellular availability of metal ion cofactors and the affinity of protein enzymes for these cofactors [51]. The small ribozymes provide illuminating examples of how RNA structure, function, and dynamics have coevolved to take advantage of a carefully maintained entourage of metal cations.
2. INTERACTIONS BETWEEN METAL IONS AND THE SMALL RIBOZYMES
2.1 Modes of Interaction
Near neutral pH, the phosphodiester backbone of RNA carries copious negative charge. In order for functional RNAs to fold into their compact native conformations, this negative charge must be at least partially neutralized. The necessary countercharge is supplied largely by metal ions, whose interactions with RNA span a continuum between two extremes: diffuse interactions, which are transient (with typical residence times thought to be in the millisecond regime [52–53]) and poorly localized, forming a kind of dynamic ionic atmosphere or “cloud” of positive electrostatic potential around the RNA; and specific interactions, which involve relatively tight (and longer-lived) binding to precise sites on the RNA molecule (Figure 2) [54]. Diffuse ion binding to RNA has been described theoretically using Hill-type binding formalisms and continuum treatments such as the Nonlinear Poisson-Boltzmann equation [54–55], and accounts for the majority of the electrostatic stabilization in RNA [56]. Consistent with this observation, monovalent ions, which generally bind only weakly to RNA, can induce proper folding and activity in the small ribozymes [35, 57–58]. Nevertheless, because the three-dimensional structure of RNA can develop concentrated pockets of negative electrostatic potential (−15 to −20 kT/e in the major groove, as low as −100 kT/e at some metal-binding sites) [59], entropy favors stabilization of compact native folds by divalent metal ions at physiologic concentrations [56].
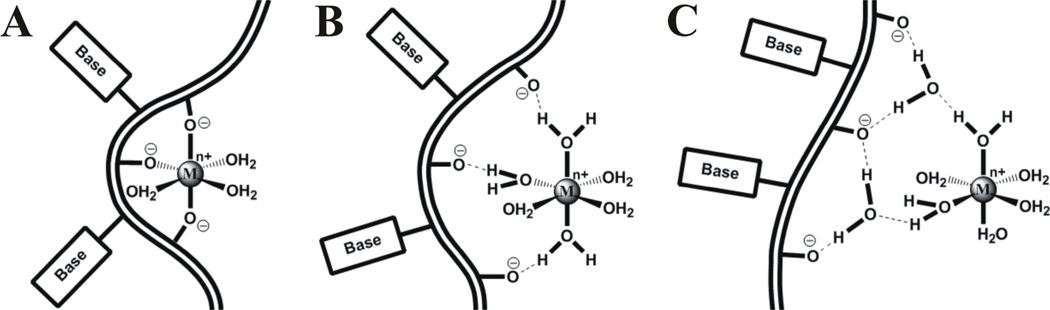
Figure 2. Modes of metal ion binding to RNA.
Metal cations (Mn+) can associate with RNA via long-lived, specific interactions (A, B) requiring at least partial dehydration of the metal ion and RNA, or transient, diffuse interactions between the solvated RNA and metal ion (C). Specific interactions can involve direct chelation of the metal ion by RNA functional groups such as non-bridging phosphate oxygens (A), contacts mediated by inner-sphere water molecules (B), or a combination of the two.
Tightly bound metal ions are observed in crystal structures of all of the small ribozymes (Figure 3), and can associate with RNA through either inner-sphere interactions involving direct coordination to electronegative RNA functional groups (Figure 2A), or outer-sphere interactions mediated by water ligands (Figure 2B). Due to the great enthalpic penalty for completely dehydrating the metal ion and RNA, inner-sphere complexation generally requires a very dense pocket of buried negative charge, such as that provided by close proximity of several negatively charged oxygen atoms at the interior of an RNA molecule [56]. Direct coordination of divalent cations to phosphoryl oxygens or to the N7 of purine bases is frequently observed, but outer-sphere interactions are far more common. In fact, all metal ions observed in crystal structures for the hammerhead, HDV, hairpin, and glmS ribozymes remain at least partly hydrated, even if they make some inner-sphere contacts as judged primarily by their distance to potential ligands on the RNA [60–66]. Compared with other divalent ions such as Ca2+, Mn2+, and Zn2+, a magnesium ion has a greater propensity for outer-sphere interactions, in accordance with its small ionic radius and high charge density that give rise to a large hydration energy and relatively slow rate of water exchange [67–68].
Figure 3. Three-dimensional structures and metal ion binding sites of the natural small ribozymes.
The ribozyme structures are shown in silver, divalent cations or probable binding sites in black, and the cleavage site in each ribozyme indicated by a black arrow. Crystal structures of (A) a hairpin ribozyme in the presence of Ca2+ ions [65], (B) a hammerhead ribozyme with Mn2+ ions [64], (C) the HDV ribozyme with Mg2+ ions [62], and (D) the glmS ribozyme in Mg2+ ions, with the necessary glucosamine-6-phosphate cofactor shown in dark gray [63]. Cocrystallized proteins and protein-binding domains of RNA used for crystallization purposes are not shown in these structures. E, Partial three-dimensional structure of the VS ribozyme derived from two similar low-resolution models [142–143] (courtesy of Richard A. Collins and Ricardo Zamel), with black spheres indicating phosphates having probable direct contacts with divalent metal ions as revealed by phosphorothioate rescue with Mn2+ [72].
Fully hydrated, tightly bound divalent metal ions have been resolved in X-ray crystal structures of the HDV, hammerhead, glmS, and hairpin ribozymes [61–65]. In the latter three of these, the exchange-inert complex cobalt(III) hexammine (Co(NH3)63+), used as a rough proxy for fully hydrated divalent ions [69], supports efficient self-cleavage [28, 32, 36, 58], suggesting that outer-sphere coordination of metal ions is sufficient for activity. Usually, though, site-bound divalent ions make at least one inner-sphere contact with RNA functional groups in crystal structures of the small ribozymes. For instance, all three Mn2+ ions that are bound to conserved regions in a crystal structure of the hammerhead ribozyme form inner sphere contacts, including an ion at the active site (Figure 3B) [64]. The fact that cobalt(III) hexammine inhibits this ribozyme in the presence of Mn2+ [70] suggests that some of these inner-sphere contacts are functionally important. Although the more biologically available magnesium may be expected not to coordinate to the same functional groups as the softer manganese(II) ion, molecular dynamics (MD) simulations suggest that a Mg2+ ion could effectively promote catalysis by occupying nearly the same site as a specific Mn2+ ion observed in the active site [71]. While no crystal structure exists for the VS ribozyme, phosphorothioate interference-rescue experiments point to direct metal ion coordination to four phosphate groups in and around the catalytic core (Figure 3E) [72]. This ribozyme cannot efficiently self-cleave in the sole presence of cobalt(III) hexammine, suggesting that inner-sphere coordination may be important to activity [73]. Interestingly, cobalt(III) hexammine can cooperatively promote VS ribozyme activity in the presence of Mg2+ [73], consistent with the presence of at least some orthogonal outer-sphere and inner-sphere binding sites.
2.2 Selectivity of Metal Interactions
Most of the small ribozymes bind a variety of cations with different affinities, allowing them to fold and perform efficient self-cleavage with varying maximal rates. The glmS, hammerhead, HDV, and hairpin ribozymes can all self-cleave in a variety of divalent cations [5, 31, 34, 74–75]. Consistent with its prevalence in the cell, the magnesium ion is among the most efficient of divalent ions at promoting catalysis in all of the natural small ribozymes, though this preference is only mild in many cases. The hairpin ribozyme cleaves in Mg2+ more than twice as efficiently as in Sr2+ and ten times as efficiently as in Ca2+, and cannot cleave in Mn2+, Co2+, Ni2+, or Cd2+ without facilitation by other cations [34, 76]. The hammerhead ribozyme cleaves more rapidly in Mg2+ than other group IIA ions, but is actually more strongly activated by certain divalent transition metals (Mn2+, Co2+, Zn2+, and Cd2+) at concentrations of 1 mM divalent ion and 100 mM NaCl [74–75]. One inner-sphere site in the hammerhead ribozyme appears to accommodate Mg2+, Mn2+, Co2+, and Cd2+ ions [64, 77–78]. In the HDV ribozyme, a catalytically important site for hydrated Mg2+ appears to bind Ca2+, Ba2+, and Sr2+ with similar affinity, though catalytic activity is lower in barium and strontium ions [79]. This difference in activity may be tied to a structurally important site of inner-sphere metal ion coordination, observed biochemically, and shown to have selectivity for a magnesium ion over calcium, barium, and strontium [79], consistent with Raman crystallographic studies showing ~5 direct Mg2+-phosphate contacts per HDV molecule [80]. Interestingly, while the active site of the genomic HDV ribozyme shows a slight preference for binding Mg2+ over Ca2+, this preference is reversed in the antigenomic ribozyme, and can be switched by mutation of a single nucleotide [81].
Some binding sites not only accommodate divalent cations other than Mg2+, but also some trivalent ions. While the exchange-inert cobalt(III) hexammine complex effectively binds and supports catalysis in several small ribozymes [28, 32, 36, 58], it is not a perfect substitute for Mg2+, generally giving rise to maximal cleavage rates 10- to 100-fold smaller than in magnesium. This may be the result of displacing functionally important magnesium ions. For example, cobalt(III) hexammine has been observed to compete for the binding site of an outer-sphere coordinated magnesium ion at the active site of the HDV ribozyme (Figure 3C) [62], and even displaces some inner-sphere coordinated metal ions in the HDV and hammerhead ribozymes, consistent with its inhibitory effect on the activity of those ribozymes in the presence of divalent ions [70, 82]. In other cases, site-bound ions may inhibit ribozymes by inducing alternate, inactive conformations, as has been suggested in the case of hammerhead, hairpin, and HDV ribozyme inhibition by terbium(III) ions [83–86].
Monovalent salts can at least partly substitute for divalent cations in all of the small ribozymes, as they are almost as active in molar concentrations of NaCl, LiCl, or even the non-metallic NH4OAc as in millimolar MgCl2 [35, 57–58]. Comparison of several modified hammerhead ribozymes suggests that the RNA adopts a similar conformation in monovalent and divalent metal ions, albeit with some subtle differences at the interaction site of a catalytically important divalent ion [87]. Monovalent and divalent cations directly compete for some of the same interactions with the hairpin [36, 55] and HDV ribozymes [88], though they act synergistically in promoting self-cleavage of the VS ribozyme [35]. Thus, none of the natural small ribozymes have a strict requirement for Mg2+ or other divalent cations, and they exhibit varying degrees of overlap between monovalent and divalent cation binding sites.
Intriguingly, small synthetic ribozymes have been engineered with strong functional selectivity for ions of transition metals or heavy metals over Mg2+ [16, 19], yet such selectivity is not common in nature. This may be the combined result of the low intracellular activity and frequent toxicity of such metals, which are closely controlled by cellular homeostasis. It has been proposed that the 5S rRNA contains a natural lead-dependent ribozyme that may partly account for the cytotoxicity of lead [89]. The more acidic hydrated cations of transition metals, heavy metals, and lanthanides compared to Mg2+ also result in more rapid nonspecific degradation of RNA through general base catalysis from their hydroxo complexes [90], making them a poor evolutionary choice for site-specific catalysis in ribozymes. The low free concentration of such ions in the cell and the paucity of natural ribozymes selective for them support the notion of a coevolution between cellular metal ion composition and functional RNAs.
In summary, metal ions stabilize the structure of small ribozymes by binding diffusely or at specific sites, with generally low structural discrimination between divalent metal ions but stricter ion requirements for efficient catalysis. Water molecules mediate some or all of the contacts between a given metal ion and RNA functional groups because of the very unfavorable enthalpy of dehydration, especially for Mg2+. Direct chelation of metal ions by RNA functional groups is occasionally required for optimal catalysis, and both labile and inert complexes of various metal ions compete for many of the same binding sites.
3. ROLES OF METAL IONS IN SMALL RIBOZYMES
3.1 Structural Roles
3.1.1 Stabilization of the Active Global Conformation
As for all functional RNA, the folding of the small ribozymes into their native conformations may be coarsely viewed as a hierarchical two-step process, where the two steps are distinguished by their temporal and spatial regimes. In the first step, an unfolded RNA rapidly acquires local secondary structure by the formation of hydrogen bonds between nucleobases, resulting in a combination of base-paired helices, junctions, loops, and pseudoknots. In the second, slower step, pre-formed helices and loops establish longer-range interactions in three-dimensional space to form the native tertiary structure, sometimes accompanied by small base-pairing rearrangements [91–94]. Metal ions facilitate both of these processes, but formation of tertiary structure requires much higher ionic strength than that of secondary structure [56, 94].
While monovalent and divalent metal cations both facilitate folding by neutralizing the negative charge of the phosphate backbone, they influence the folding pathway somewhat differently. Most obviously, the formation of tertiary structure requires much higher concentrations of monovalent cations. For example, the hairpin ribozyme self-cleaves (and presumably folds) with similar efficiency in 0.5 mM Co(NH3)3+, 10 mM Mg2+, or 1 M monovalent salts [35]. More interestingly, in the presence of Mg2+, Na+ ions actually destabilize secondary structure by preferentially associating with the unfolded random coil, and destabilize tertiary structure by competing with Mg2+ [53]. This raises the interesting possibility that monovalent cations may help a ribozyme to find the correct minimum-energy native structure by destabilizing alternative misfolds, as was recently suggested for the HDV ribozyme [88] as well as for larger group I intron ribozymes [95–96]. Since very dense electronegative pockets can form within the tertiary structure of RNA [59], divalent and trivalent cations, with their high charge density and ability to bridge pairs of negatively charged phosphates, promote the folding of RNA particularly well. Their ability to form stable inner-sphere contacts with electronegative functional groups, while not conferring much additional stability compared to outer-sphere electrostatic screening, has been proposed to make a larger range of backbone conformations available to RNA [56]. While the potassium ion has been observed to make stable direct contacts with RNA functional groups at highly electronegative sites [97], even replacing a site-bound Mg2+ in the active site of a group I intron [98], such tightly bound monovalent ions have not been routinely noted in the small ribozymes, perhaps in part due to the difficulty of distinguishing fractionally occupied monovalent ion sites from water molecules in X-ray crystal structures [99]. However, one recent crystallographic study of the HDV ribozyme found that two thallium (Tl+) ions bind weakly at a location previously seen to be occupied by a hydrated Mg2+, as predicted by MD simulations [66], and a third Tl+ binds tightly at a new site with direct coordination to the 2′-OH nucleophile [100]. The similar charge, ionic radius and coordination geometry of the thallium compared to the potassium ion, and the ability of Tl+ to occupy sites different than Mg2+ near the active site, suggest that monovalent ions may play more important structural (and perhaps even catalytic) roles in the natural small ribozymes than is currently appreciated.
Structural stabilization, mostly of an electrostatic nature, may be the most important role for metal ions in the small ribozymes. Most can achieve near-maximal activity (within ~30 fold) in a variety of monovalent, divalent, and trivalent salts [5, 35, 57]. In addition, the vast majority of well-structured metal ions found in crystal structures of the small ribozymes are located tens of angstroms from the site of cleavage chemistry, including all metal ions observed in the glmS and hairpin ribozymes (Figure 3) [61–65], suggesting that these ions contribute to activity either indirectly through structural stabilization or by long-distance electrostatic interactions with the site of cleavage chemistry (see sections 3.4 and 3.5, below).
3.1.2 Influence on Conformational Changes
Metal ions have been linked to catalytically important conformational changes in the hairpin, VS, and HDV ribozymes [26]. In the hairpin ribozyme, catalysis requires docking of two internal loops of nucleobases located in separate helical stems [101–104]. At equilibrium, the docked and undocked states are both populated, and the rate constants of their interconversion are sensitive to the concentrations of monovalent and divalent cations. The docking reaction is accompanied by an uptake of sodium and/or magnesium ions, which can compete with each other in promoting this transition [55]. The VS ribozyme exhibits analogous docking behavior that includes the metal cation-dependent formation of a loop-loop “kissing” interaction [105–106] that induces a critical change in the base pairing pattern of the substrate stem-loop in the wild-type ribozyme prior to catalysis [91]. In the HDV ribozyme, the self-cleavage reaction is accompanied by the dissociation of a divalent ion from the active site (Figure 3C) and significant conformational changes that reposition important active site residues [62, 85– 86, 107–109]. Such conformational changes are common, but not universal, features of the folding landscapes of the small ribozymes: for instance, the precatalytic pocket in the glmS ribozyme is essentially rigid once it is formed in divalent ion-containing buffer, undergoing little change even upon binding of the glucosamine-6-phosphate cofactor and self-cleavage [63, 110]; the addition of Mg2+ together with cofactor does, however, induce a catalytically rate-limiting conformational change in this ribozyme [111].
3.1.3 Organization of the Active Site
In some cases, metal ions appear to organize residues or solvent molecules within active sites of the small ribozymes. To achieve self-cleavage the small ribozymes must adopt a so-called in-line attack configuration, with an approximately 180-degree angle between the 2′-oxyanion nucleophile, the phosphorus atom of the scissile phosphate, and the 5′-OH leaving group (Figure 1) [23–24, 26]. In the hammerhead ribozyme, diffusely bound Mg2+ ions have been proposed to help properly align the catalytic core from a distance, presumably by twisting its stems I and II that intersect at the core, especially in variants that lack tertiary kissing loop interactions between these stems [53]. In addition, MD simulations suggest that threshold occupancy of a cation-binding pocket near the active site (Figure 3B) is required to sample the correct in-line attack geometry for self-cleavage. The cation facilitates formation of the correct geometry by neutralizing negative charge and possibly by coordinating with particular RNA functional groups [71]. A divalent ion observed crystallographically at this site has also been proposed to organize a network of water molecules that may, in turn, facilitate proton transfer in the cleavage reaction [64]. Such a role for well-ordered water molecules in small ribozymes is an active area of investigation [24]. A cation binding site in the genomic HDV ribozyme could also play a role in organizing the active site, although the geometry around the cleavage site does not appear to depend specifically on the presence of Mg2+ [60, 62, 100].
3.2 Mechanistic Roles
3.2.1 Electrostatic Activation of Catalytic Residues
The small ribozymes are all thought to perform their catalysis by acid/base chemistry in which a general base abstracts a proton from the 2′-OH of the nucleotide 5′ of the cleavage site, activating the nucleophile for attack on the adjacent phosphate, and a general acid donates a proton to the 5′-OH leaving group of the nucleotide 3′ of the cleavage site (Figure 1). For the hammerhead, hairpin, glmS, and VS ribozymes, the general acid and base appear to be functional groups of the RNA itself [23–24, 112]. However, free nucleobases possess pKa values far from neutral pH – for example, 3.5–4.2 for adenosine (N1H)+ and cytidine (N3H)+; 9.2–9.5 for guanosine (N1)H and uridine (N3)H, and ~12.5 for the 2′-OH of ribose – at first glance seeming to preclude them as efficient proton donors or acceptors near physiologic conditions [113–116].
One possible way for metal ions to stimulate catalysis in ribozymes is by electrostatic modulation of ground-state active site functional groups so as to shift their effective pKa values towards neutrality. Electrostatic modulation of catalytic residues is common in protein enzymes: nearby positive charges have been observed to lower the pKa of serine or cysteine residues in serine and cysteine proteases [117–118], and in ribonuclease H, the binding of a Mg2+ cofactor induces a pKa shift of almost two units in an aspartate residue [119]. These effects can be significant over distances as great as 15 Å [67,120]. The long-distance impact of multiple charges on the acid dissociation constant of an amino acid residue can be partially additive, as well [121]. It is therefore plausible that multiple associated metal cations, or even a diffuse ion atmosphere, could have a significant impact on the reactivity of catalytic residues in some or all of the small ribozymes.
Characterization of electrostatic contributions to catalysis is complicated by the relatively weak binding of most metal ions to RNA, as well as the complex dependence of electrostatic effects on the environment in and around a macromolecule, especially in water with its highly dipolar character [122]. However, metal ion-dependent pKa values have been observed in some of the small ribozymes. In the VS ribozyme, pH-rate profiles suggest that most or all of the ion-specific rate enhancement may result from differential modulation of nucleobase pKa by different cations, rather than from effects on the intrinsic bond breaking rate constant [123]. As there is no crystal structure of the VS ribozyme, it is not clear in what manner the metal ions may be modulating the effective pKa, whether through direct or indirect coordination to RNA functional groups, or long-distance interactions. This phenomenon is not universal, however, as the apparent pKa of the general base in the hammerhead ribozyme appears to be independent of metal ion identity [30]. Effective pKa shifts toward neutrality have been observed in a catalytically important adenosine of the hairpin ribozyme [124] and an essential cytosine in the HDV ribozyme [25, 57], but there is no evidence of direct metal ion participation in these perturbations. In both of these latter cases, the shifts towards higher pKa could be mediated by the negative electrostatic environment created by RNA functional groups such as phosphoryl oxygens. Accordingly, in case of the HDV ribozyme Mg2+ appears to compete with this pKa shift [57].
3.2.2 Direct Participation in Catalysis
In principle, direct participation by metal ions in the chemical step of self-cleavage in small ribozymes could include (Figure 1): 1) deprotonation of the upstream 2′-OH by a metal hydroxide, 2) electrostatic stabilization of the developing negative charge in the transition state, and/or 3) protonation of the leaving group 5′-oxygen by a hydrated metal ion [23–24, 26, 125]. In contrast to the group I and II introns [126–129], however, direct participation of metal ions in catalysis by the small ribozymes has not been clearly demonstrated. In all cases, any specific contribution of divalent cations to catalysis is minor, accounting for a modest ~20–30-fold rate enhancement over non-acidic monovalent cations [35, 130].
Active-site divalent metal ions have been proposed to play non-obligatory, even if important catalytic roles in the HDV and hammerhead ribozymes (Figure 3, B and C). Solution kinetics data are consistent with participation of a single hydrated Mg2+ ion as a general base in the HDV ribozyme, and cytosine 75 (C76 in the antigenomic ribozyme) as the general acid [57, 131–132], or vice-versa [62, 133–135]. A Mg2+ ion poised for a role as the general base has not been found in the X-ray crystal structures of the cleavage product or non-cleavable mutant forms of the HDV ribozyme [61–62] but was suggested by Raman spectroscopy of a two-stranded HDV ribozyme bearing an inactivating 2′-O-methyl modification at the cleavage site [60]. In contrast, one of the above X-ray crystal structures shows a hydrated Mg2+ ion poised to act as a general acid, although residue 75 is not positioned to act as a general acid or base [62]. Thus, direct participation of a magnesium ion in the chemistry of cleavage by the HDV ribozyme is possible, but not conclusively demonstrated. A crystal structure of the full-length hammerhead ribozyme in Mn2+ shows no metal cations in position to participate in acid-base chemistry, but suggest that a divalent ion at the active site may stabilize the transition state by solvent-mediated charge withdrawal or direct coordination to nonbridging oxygens of the scissile phosphate (Figure 3B) [64, 67]. While enhancement by charge withdrawal is supported by the crystal structure in Mn2+, MD simulations suggest that Mg2+ could facilitate the in-line attack angle by directly coordinating a nonbridging oxygen of the scissile phosphate [71].
An intriguing possibility is that cations may stabilize the negatively charged transition state in small ribozymes through long-distance electrostatic interactions. For instance, although the crystal structure of the hairpin ribozyme showed no divalent cation at the immediate active site [65], it revealed six calcium ions within 16 Å of the scissile bond (Figure 3A), likely close enough to strongly stabilize the transition state [67]. This mode of activation from a distance could also help to explain why aminoglycoside antibiotics and the polyamine spermine support hairpin ribozyme activity approaching that in magnesium ions [33]. A similar function has been proposed for the divalent ion found in the active site of the full-length hammerhead ribozyme [64, 67, 136], and may also apply to the Mg2+ found in the active site of the HDV ribozyme [60, 62]. Such long-range stabilization is consistent with the generally small specific rate enhancements conferred by divalent cations, and suggests that transiently bound monovalent cations may even help to stabilize the developing charge of the transition state when present at sufficient concentrations to efficiently populate cation binding sites on the RNA.
3.2.3 Influence of Metal Ions on Reaction Pathways
While it is convenient to conceptualize the self-cleavage of small ribozymes as occurring via a unique reaction trajectory, there is evidence that at least some ribozymes may make use of a variety of reaction channels that are differentially populated (and effectively compete with one another) as a function of reaction conditions. An intriguing example is found in the HDV ribozyme, where kinetic studies revealed three possible reaction pathways in the presence of varying concentrations of NaCl and MgCl2 [130]. At very low magnesium ion concentrations (<10−7 M), the rate of self-cleavage is independent of Mg2+ concentration, with a pH-rate profile suggesting that solvent and hydroxide ions operate as general and specific bases in the reaction. At intermediate Mg2+ concentrations (10−7−10−4 M), the observed cleavage rate constant exhibits log-linear dependence on magnesium ion concentration, with pH-rate profiles consistent with the binding of at least one structural divalent cation. Finally, at physiologic Mg2+ concentrations and higher, a second metal ion binding site becomes saturated, yielding an inverted pH-rate profile consistent with a role of a metal hydroxide or solvent hydroxide as the base in catalysis. However, a subsequent study found that the cleavage reaction of the HDV ribozyme in 4 M Li+ exhibits a similar pH-rate profile in the presence and absence of Mg2+, albeit with a smaller observed rate constant, suggesting that Li+ can at least partially substitute for Mg2+ in determining pathway preference [29]. Furthermore, due to the modest specific contribution of Mg2+ to catalysis [130], the absence of a magnesium hydroxide poised for general base catalysis from the published X-ray crystal structures [62], and the existence of other pH-dependent conformational changes in the absence of divalent ions that affect activity [137], the nature of these apparent reaction channels requires further elucidation.
Scenarios involving multiple metal cation-dependent reaction pathways have also been proposed for other ribozymes. Kinetic characterization of a tertiary-stabilized form of the hammerhead ribozyme suggests that magnesium ions and cobalt(III) hexammine may support separate catalytic pathways with incompatible RNA conformations [138–139]. Furthermore, recent work has demonstrated multiple catalytically active conformations of the hairpin ribozyme [140] as well as the Tetrahymena group I intron ribozyme [141] that are all populated near physiological conditions. In the case of the group I intron, interconversion between the different native conformations occurs slowly in the presence Mg2+, but rapidly in its absence. These results raise the interesting possibility that some small ribozymes may operate via multiple reaction pathways in a metal ion-dependent fashion. For example, the rate of catalysis by individual subpopulations of ribozymes could be limited by different chemical steps dependent on subtly different conformations or differentially occupied cation binding sites. If this is the case, it will reveal a striking flexibility in the folding and function of ribozymes.
4. CONCLUDING REMARKS AND FUTURE DIRECTIONS
In summary, metal cations are critical to the intramolecular phosphodiester isomerization reaction catalyzed by the small self-cleaving ribozymes known in nature. Much as water is an obligatory solvent for proper folding of many macromolecules, appropriate combinations of metal ions are required for optimal activity in the small ribozymes. They universally facilitate catalysis through structural stabilization, but in certain cases may also help to organize active site functional groups and water molecules through hydrogen bonding, activate the catalytic acid or base, or participate directly in catalysis through acid-base chemistry or transition state stabilization. Proper folding and efficient self-cleavage occur with generally low selectivity in a variety of monovalent and divalent cations, but under physiological conditions Mg2+ is the most important of these, and is generally preferred over less naturally abundant divalent cations. While small ribozymes could, in principle, use transition metals and heavy metals for catalysis, their toxicity and generally low free concentrations in the cell preclude these metals from playing an important role as cofactors for the natural small ribozymes.
Future work should further elucidate any direct catalytic roles played by metal ions in the HDV and hammerhead ribozymes, long-distance interactions between the active site and metal ions, potentially overlooked roles of monovalent ions (including the physiologically most relevant K+), the nature and metal ion-dependence of alternate reaction pathways, and the impact of metal ions on local conformational changes and solvent organization during catalysis. This will require a combination of increasingly sophisticated methods of chemical modification, spectroscopic techniques, and theoretical models. As small ribozymes increasingly appear to be widespread in nature, understanding their manifold interactions with metal ions will yield a more complete understanding of the roles of RNA in life and its origins.
ABBREVIATIONS AND DEFINITIONS
- Divalent cation
An ion with a net electronic charge of +2.
- Enthalpy of dehydration
The enthalpy change accompanying the removal of all water molecules to an infinite distance from a fully hydrated ion.
- General acid
A functional group or moiety that catalyzes a chemical reaction by donating a proton to a reactive group.
- General base
A functional group or moiety that catalyzes a chemical reaction by accepting a proton from a reactive group.
- glmS ribozyme
A ribozyme found in numerous Gram-positive bacteria that self-cleaves in the presence of a glucosamine-6-phosphate cofactor, regulating translation of the glmS gene in E. coli.
- HDV
Hepatitis delta virus
- Inner-sphere interaction
Direct interaction between a metal ion and an electronegative ligand such as the oxygen atom of water or an oxygen or nitrogen atom of an RNA molecule.
- Monovalent cation
Ion with a net electronic charge of +1.
- Outer-sphere interaction
Interaction between a metal ion and another species mediated by water molecules or other ligands of the metal ion.
- Secondary structure
The ensemble of hydrogen bonding interactions between nucleobases (base pairs, triples, and occasionally quartets) in an RNA molecule, resulting in the formation of base-paired stems, unpaired loops, and junctions between these.
- Tertiary structure
The three-dimensional structure of an RNA molecule, including all base pairs as well as additional interactions between helical stems and loops.
- VS
Varkud satellite
Contributor Information
Alexander E. Johnson-Buck, Email: alebuck@umich.edu.
Sarah E. McDowell, Email: semcdowe@umich.edu.
Nils G. Walter, Email: nwalter@umich.edu.
REFERENCES
- 1.Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 2.Cech TR. Science. 2000;289:878–879. doi: 10.1126/science.289.5481.878. [DOI] [PubMed] [Google Scholar]
- 3.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 4.Toor N, Keating KS, Taylor SD, Pyle AM. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 6.Forster AC, Symons RH. Cell. 1987;49:211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- 7.Hampel A, Tritz R. Biochemistry. 1989;28:4929–4933. doi: 10.1021/bi00438a002. [DOI] [PubMed] [Google Scholar]
- 8.Lai MMC. Annu. Rev. Biochem. 1995;64:259–286. doi: 10.1146/annurev.bi.64.070195.001355. [DOI] [PubMed] [Google Scholar]
- 9.Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G. Science. 1986;231:1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- 10.Saville BJ, Collins RA. Cell. 1990;61:685–696. doi: 10.1016/0092-8674(90)90480-3. [DOI] [PubMed] [Google Scholar]
- 11.Kuo MYP, Sharmeen L, Dintergottlieb G, Taylor J. J. Virol. 1988;62:4439–4444. doi: 10.1128/jvi.62.12.4439-4444.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu HN, Lin YJ, Lin FP, Makino S, Chang MF, Lai MMC. P. Natl. Acad. Sci. USA. 1989;86:1831–1835. doi: 10.1073/pnas.86.6.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martick M, Horan LH, Noller HF, Scott WG. Nature. 2008;454 doi: 10.1038/nature07117. 899-U857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Webb CHT, Riccitelli NJ, Ruminski DJ, Luptak A. Science. 2009;326:953–953. doi: 10.1126/science.1178084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salehi-Ashtiani K, Luptak A, Litovchick A, Szostak JW. Science. 2006;313:1788–1792. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- 16.Pan T, Uhlenbeck OC. Biochemistry. 1992;31:3887–3895. doi: 10.1021/bi00131a001. [DOI] [PubMed] [Google Scholar]
- 17.Agresti JJ, Kelly BT, Jaschke A, Griffiths AD. P. Natl. Acad. Sci. USA. 2005;102:16170–16175. doi: 10.1073/pnas.0503733102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reymond C, Beaudoin JD, Perreault JP. Cell. Mol. Life Sci. 2009;66:3937–3950. doi: 10.1007/s00018-009-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zivarts M, Liu Y, Breaker RR. Nucleic Acids Res. 2005;33:622–631. doi: 10.1093/nar/gki182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Illangasekare M, Yarus M. RNA. 1999;5:1482–1489. doi: 10.1017/s1355838299991264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lincoln TA, Joyce GF. Science. 2009;323:1229–1232. doi: 10.1126/science.1167856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilbert W. Nature. 1986;319:618–618. [Google Scholar]
- 23.Cochrane JC, Strobel SA. Acc. Chem. Res. 2008;41:1027–1035. doi: 10.1021/ar800050c. [DOI] [PubMed] [Google Scholar]
- 24.Walter NG. Mol. Cell. 2007;28:923–929. doi: 10.1016/j.molcel.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oyelere AK, Kardon JR, Strobel SA. Biochemistry. 2002;41:3667–3675. doi: 10.1021/bi011816v. [DOI] [PubMed] [Google Scholar]
- 26.Walter NG, Perumal S. In: Non-Protein Coding RNAs. Walter NG, Woodson SA, Batey RT, editors. Berlin: Springer; 2009. pp. 103–127. [Google Scholar]
- 27.Fedor MJ. Curr. Opin. Struct. Biol. 2002;12:289–295. doi: 10.1016/s0959-440x(02)00324-x. [DOI] [PubMed] [Google Scholar]
- 28.Curtis EA, Bartel DP. RNA. 2001;7:546–552. doi: 10.1017/s1355838201002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perrotta AT, Been MD. Biochemistry. 2006;45:11357–11365. doi: 10.1021/bi061215+. [DOI] [PubMed] [Google Scholar]
- 30.Roychowdhury-Saha M, Burke DH. RNA. 2006;12:1846–1852. doi: 10.1261/rna.128906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suh YA, Kumar PKR, Taira K, Nishikawa S. Nucleic Acids Res. 1993;21:3277–3280. doi: 10.1093/nar/21.14.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young KJ, Gill F, Grasby JA. Nucleic Acids Res. 1997;25:3760–3766. doi: 10.1093/nar/25.19.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Earnshaw DJ, Gait MJ. Nucleic Acids Res. 1998;26:5551–5561. doi: 10.1093/nar/26.24.5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chowrira BM, Berzal-Herranz A, Burke JM. Biochemistry. 1993;32:1088–1095. doi: 10.1021/bi00055a014. [DOI] [PubMed] [Google Scholar]
- 35.Murray JB, Seyhan AA, Walter NG, Burke JM, Scott WG. Chem. Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- 36.Hampel A, Cowan JA. Chem. Biol. 1997;4:513–517. doi: 10.1016/s1074-5521(97)90323-9. [DOI] [PubMed] [Google Scholar]
- 37.Nesbitt S, Hegg LA, Fedor MJ. Chem. Biol. 1997;4:619–630. doi: 10.1016/s1074-5521(97)90247-7. [DOI] [PubMed] [Google Scholar]
- 38.Walter NG, Burke JM. Curr. Opin. Chem. Biol. 1998;2:303–303. doi: 10.1016/s1367-5931(98)80073-2. [DOI] [PubMed] [Google Scholar]
- 39.Cayley S, Lewis BA, Guttman HJ, Record MT. J. Mol. Biol. 1991;222:281–300. doi: 10.1016/0022-2836(91)90212-o. [DOI] [PubMed] [Google Scholar]
- 40.Froschauer EM, Kolisek M, Dieterich F, Schweigel M, Schweyen RJ. FEMS Microbiol. Lett. 2004;237:49–55. doi: 10.1016/j.femsle.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 41.Cromie MJ, Shi YX, Latifi T, Groisman EA. Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 42.Dann CE, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- 43.Ramesh A, Winkler WC. RNA Biol. 2010;7 doi: 10.4161/rna.7.1.10490. [DOI] [PubMed] [Google Scholar]
- 44.Wakeman CA, Ramesh A, Winkler WC. J. Mol. Biol. 2009;392:723–735. doi: 10.1016/j.jmb.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voets T, Nilius B, Hoefs S, van der Kemp AWCM, Droogmans G, Bindels RJM, Hoenderop JGJ. J. Biol. Chem. 2004;279:19–25. doi: 10.1074/jbc.M311201200. [DOI] [PubMed] [Google Scholar]
- 46.Khvorova A, Lescoute A, Westhof E, Jayasena SD. Nat. Struct. Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- 47.De la Peña M, Gago S, Flores R. EMBO J. 2003;22:5561–5570. doi: 10.1093/emboj/cdg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakano S, Karimata HT, Kitagawa Y, Sugimoto N. J. Am. Chem. Soc. 2009;131:16881–16888. doi: 10.1021/ja9066628. [DOI] [PubMed] [Google Scholar]
- 49.Dai LJ, Quamme LJ. J. Clin. Invest. 1991;88:1255–1264. doi: 10.1172/JCI115429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. Molecular Biology of the Cell. New York: Garland Publishing; 1994. [Google Scholar]
- 51.Cowan JA. Chem. Rev. 1998;98:1067–1087. doi: 10.1021/cr960436q. [DOI] [PubMed] [Google Scholar]
- 52.Pecoraro VL, Hermes JD, Cleland WW. Biochemistry. 1984;23:5262–5271. doi: 10.1021/bi00317a026. [DOI] [PubMed] [Google Scholar]
- 53.Rueda D, Wick K, McDowell SE, Walter NG. Biochemistry. 2003;42:9924–9936. doi: 10.1021/bi0347757. [DOI] [PubMed] [Google Scholar]
- 54.Draper DE, Grilley D, Soto AM. Annu. Rev. Biophys. Biom. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- 55.Bokinsky G, Rueda D, Misra VK, Rhodes MM, Gordus A, Babcock HP, Walter NG, Zhuang XW. P. Natl. Acad. Sci. USA. 2003;100:9302–9307. doi: 10.1073/pnas.1133280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Draper DE. Biophys. J. 2008;95:5489–5495. doi: 10.1529/biophysj.108.131813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakano S, Chadalavada DM, Bevilacqua PC. Science. 2000;287:1493–1497. doi: 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- 58.Roth A, Nahvi A, Lee M, Jona I, Breaker RR. RNA. 2006;12:607–619. doi: 10.1261/rna.2266506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chin K, Sharp KA, Honig B, Pyle AM. Nat. Struct. Biol. 1999;6:1055–1061. doi: 10.1038/14940. [DOI] [PubMed] [Google Scholar]
- 60.Chen JH, Gong B, Bevilacqua PC, Carey PR, Golden BL. Biochemistry. 2009;48:1498–1507. doi: 10.1021/bi8020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ferre-D'Amare AR, Zhou KH, Doudna JA. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 62.Ke AL, Zhou KH, Ding F, Cate JHD, Doudna JA. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 63.Klein DJ, Ferre-D'Amare AR. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- 64.Martick M, Lee TS, York DM, Scott WG. Chem. Biol. 2008;15:332–342. doi: 10.1016/j.chembiol.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rupert PB, Ferré-D'Amaré AR. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 66.Krasovska MV, Sefcikova J, Reblova K, Schneider B, Walter NG, Sponer J. Biophys. J. 2006;91:626–638. doi: 10.1529/biophysj.105.079368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sigel RKO, Pyle AM. Chem. Rev. 2007;107:97–113. doi: 10.1021/cr0502605. [DOI] [PubMed] [Google Scholar]
- 68.Lincoln SF. Helv. Chim. Acta. 2005;88:523–545. [Google Scholar]
- 69.Cowan JA. J. Inorg. Biochem. 1993;49:171–175. doi: 10.1016/0162-0134(93)80002-q. [DOI] [PubMed] [Google Scholar]
- 70.Horton TE, DeRose VJ. Biochemistry. 2000;39:11408–11416. doi: 10.1021/bi001141g. [DOI] [PubMed] [Google Scholar]
- 71.Lee TS, Giambasu GM, Sosa CP, Martick M, Scott WG, York DM. J. Mol. Biol. 2009;388:195–206. doi: 10.1016/j.jmb.2009.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sood VD, Beattie TL, Collins RA. J. Mol. Biol. 1998;282:741–750. doi: 10.1006/jmbi.1998.2049. [DOI] [PubMed] [Google Scholar]
- 73.Maguire JL, Collins RA. J. Mol. Biol. 2001;309:45–56. doi: 10.1006/jmbi.2001.4625. [DOI] [PubMed] [Google Scholar]
- 74.Dahm SC, Uhlenbeck OC. Biochemistry. 1991;30:9464–9469. doi: 10.1021/bi00103a011. [DOI] [PubMed] [Google Scholar]
- 75.Boots JL, Canny MD, Azimi E, Pardi A. RNA. 2008;14:2212–2222. doi: 10.1261/rna.1010808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walter F, Murchie AIH, Thomson JB, Lilley DMJ. Biochemistry. 1998;37:14195–14203. doi: 10.1021/bi981513+. [DOI] [PubMed] [Google Scholar]
- 77.Pley HW, Flaherty KM, Mckay DB. Nature. 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- 78.Scott WG, Murray JB, Arnold JRP, Stoddard BL, Klug A. Science. 1996;274:2065–2069. doi: 10.1126/science.274.5295.2065. [DOI] [PubMed] [Google Scholar]
- 79.Nakano S, Cerrone AL, Bevilacqua PC. Biochemistry. 2003;42:2982–2994. doi: 10.1021/bi026815x. [DOI] [PubMed] [Google Scholar]
- 80.Gong B, Chen Y, Christian EL, Chen JH, Chase E, Chadalavada DM, Yajima R, Golden BL, Bevilacqua PC, Carey PR. J. Am. Chem. Soc. 2008;130:9670–9672. doi: 10.1021/ja801861s. [DOI] [PubMed] [Google Scholar]
- 81.Perrotta AT, Been MD. Biochemistry. 2007;46:5124–5130. doi: 10.1021/bi602569x. [DOI] [PubMed] [Google Scholar]
- 82.Gong B, Chen JH, Bevilacqua PC, Golden BL, Carey PR. Biochemistry. 2009;48:11961–11970. doi: 10.1021/bi901091v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Walter NG, Yang N, Burke JM. J. Mol. Biol. 2000;298:539–555. doi: 10.1006/jmbi.2000.3691. [DOI] [PubMed] [Google Scholar]
- 84.Feig AL, Scott WG, Uhlenbeck OC. Science. 1998;279:81–84. doi: 10.1126/science.279.5347.81. [DOI] [PubMed] [Google Scholar]
- 85.Jeong S, Sefcikova J, Tinsley RA, Rueda D, Walter NG. Biochemistry. 2003;42:7727–7740. doi: 10.1021/bi034627g. [DOI] [PubMed] [Google Scholar]
- 86.Harris DA, Tinsley RA, Walter NG. J. Mol. Biol. 2004;341:389–403. doi: 10.1016/j.jmb.2004.05.074. [DOI] [PubMed] [Google Scholar]
- 87.O'Rear JL, Wang SL, Feig AL, Beigelman L, Uhlenbeck OC, Herschlag D. RNA. 2001;7:537–545. doi: 10.1017/s1355838201002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brown TS, Chadalavada DM, Bevilacqua PC. J. Mol. Biol. 2004;341:695–712. doi: 10.1016/j.jmb.2004.05.071. [DOI] [PubMed] [Google Scholar]
- 89.Barciszewska MZ, Wyszko E, Bald R, Erdmann VA, Barciszewski J. J. Biochem. 2003;133:309–315. doi: 10.1093/jb/mvg042. [DOI] [PubMed] [Google Scholar]
- 90.Huff JW, Sastry KS, Wacker WEC, Gordon MP. Biochemistry. 1964;3:501–506. doi: 10.1021/bi00892a006. [DOI] [PubMed] [Google Scholar]
- 91.Andersen AA, Collins RA. Mol. Cell. 2000;5:469–478. doi: 10.1016/s1097-2765(00)80441-4. [DOI] [PubMed] [Google Scholar]
- 92.Brion P, Westhof E. Annu. Rev. Biophys. Biom. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 93.Onoa B, Tinoco I. Curr. Opin. Chem. Biol. 2004;14:374–379. doi: 10.1016/j.sbi.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 94.Tinoco I, Bustamante C. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 95.Jiang YF, Xiao M, Yin P, Zhang Y. RNA. 2006;12:561–566. doi: 10.1261/rna.2188306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Russell R, Das R, Suh H, Traver KJ, Laederach A, Engelhardt MA, Herschlag D. J. Mol. Biol. 2006;363:531–544. doi: 10.1016/j.jmb.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 97.Conn GL, Gittis AG, Lattman EE, Misra VK, Draper DE. J. Mol. Biol. 2002;318:963–973. doi: 10.1016/S0022-2836(02)00147-X. [DOI] [PubMed] [Google Scholar]
- 98.Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Nature. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- 99.Tereshko V, Wilds CJ, Minasov G, Prakash TP, Maier MA, Howard A, Wawrzak Z, Manoharan M, Egli M. Nucleic Acids Res. 2001;29:1208–1215. doi: 10.1093/nar/29.5.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ke AL, Ding F, Batchelor JD, Doudna JA. Structure. 2007;15:281–287. doi: 10.1016/j.str.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 101.Hampel KJ, Walter NG, Burke JM. Biochemistry. 1998;37:14672–14682. doi: 10.1021/bi981083n. [DOI] [PubMed] [Google Scholar]
- 102.Walter NG, Hampel KJ, Brown KM, Burke JM. EMBO J. 1998;17:2378–2391. doi: 10.1093/emboj/17.8.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Walter NG, Harris DA, Pereira MJB, Rueda D. Biopolymers. 2001;61:224–242. doi: 10.1002/bip.10144. [DOI] [PubMed] [Google Scholar]
- 104.Zhuang XW, Kim H, Pereira MJB, Babcock HP, Walter NG, Chu S. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 105.Pereira MJB, Nikolova EN, Hiley SL, Jaikaran D, Collins RA, Walter NG. J. Mol. Biol. 2008;382:496–509. doi: 10.1016/j.jmb.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rastogi T, Beattie TL, Olive JE, Collins RA. EMBO J. 2006;15:2820–2825. [PMC free article] [PubMed] [Google Scholar]
- 107.Tinsley RA, Harris DA, Walter NG. Biochemistry. 2004;43:8935–8945. doi: 10.1021/bi049471e. [DOI] [PubMed] [Google Scholar]
- 108.Harris DA, Rueda D, Walter NG. Biochemistry. 2002;41:12051–12061. doi: 10.1021/bi026101m. [DOI] [PubMed] [Google Scholar]
- 109.Pereira MJB, Harris DA, Rueda D, Walter NG. Biochemistry. 2002;41:730–740. doi: 10.1021/bi011963t. [DOI] [PubMed] [Google Scholar]
- 110.Tinsley RA, Furchak JRW, Walter NG. RNA. 2007;13:468–477. doi: 10.1261/rna.341807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Brooks KM, Hampel KJ. Biochemistry. 2009;48:5669–5678. doi: 10.1021/bi900183r. [DOI] [PubMed] [Google Scholar]
- 112.Ditzler MA, Sponer J, Walter NG. RNA. 2009;15:560–575. doi: 10.1261/rna.1416709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Da Costa CP, Sigel H. Inorg. Chem. 2003;42:3475–3482. doi: 10.1021/ic020672l. [DOI] [PubMed] [Google Scholar]
- 114.Knobloch B, Linert W, Sigel H. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7459–7464. doi: 10.1073/pnas.0501446102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Christen JJ, Rytting JH, Izatt RM. J. Chem. Soc. B. 1970:4907–4913. [Google Scholar]
- 116.Christen JJ, Rytting JH, Izatt RM. J. Chem. Soc. B. 1970:1643–1646. [Google Scholar]
- 117.Lewis SD, Johnson FA, Shafer JA. Biochemistry. 1981;20:48–51. doi: 10.1021/bi00504a009. [DOI] [PubMed] [Google Scholar]
- 118.Storer AC, Menard R. Methods Enzymol. 1994;244:486–500. doi: 10.1016/0076-6879(94)44035-2. [DOI] [PubMed] [Google Scholar]
- 119.Oda Y, Yamazaki T, Nagayama K, Kanaya S, Kuroda Y, Nakamura H. Biochemistry. 1994;33:5275–5284. doi: 10.1021/bi00183a034. [DOI] [PubMed] [Google Scholar]
- 120.Jackson SE, Fersht AR. Biochemistry. 1993;32:13909–13916. doi: 10.1021/bi00213a021. [DOI] [PubMed] [Google Scholar]
- 121.Russell AJ, Fersht AR. Nature. 1987;328:496–500. doi: 10.1038/328496a0. [DOI] [PubMed] [Google Scholar]
- 122.Fersht AR, Sternberg MJE. Protein Eng. 1989;2:527–530. doi: 10.1093/protein/2.7.527. [DOI] [PubMed] [Google Scholar]
- 123.Smith MD, Mehdizadeh R, Olive JE, Collins RA. RNA. 2008;14:1942–1949. doi: 10.1261/rna.1102308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Guo M, Spitale RC, Volpini R, Krucinska J, Cristalli G, Carey PR, Wedekind JE. J. Am. Chem. Soc. 2009;131:12908–12909. doi: 10.1021/ja9060883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Emilsson GM, Nakamura S, Roth A, Breaker RR. RNA. 2003;9:907–918. doi: 10.1261/rna.5680603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gordon PM, Fong R, Piccirilli JA. Chem. Biol. 2007;14:607–612. doi: 10.1016/j.chembiol.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 127.Shan SO, Yoshida A, Sun SG, Piccirilli JA, Herschlag D. P. Natl. Acad. Sci. USA. 1999;96:12299–12304. doi: 10.1073/pnas.96.22.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Stahley MR, Strobel SA. Science. 2005;309:1587–1590. doi: 10.1126/science.1114994. [DOI] [PubMed] [Google Scholar]
- 129.Zhang L, Doudna JA. Science. 2002;295:2084–2088. doi: 10.1126/science.1069268. [DOI] [PubMed] [Google Scholar]
- 130.Nakano S, Proctor DJ, Bevilacqua PC. Biochemistry. 2001;40:12022–12038. doi: 10.1021/bi011253n. [DOI] [PubMed] [Google Scholar]
- 131.Cerrone-Szakal AL, Siegfried NA, Bevilacqua PC. J. Am. Chem. Soc. 2008;130:14504–14520. doi: 10.1021/ja801816k. [DOI] [PubMed] [Google Scholar]
- 132.Das SR, Piccirilli JA. Nat. Chem. Biol. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- 133.Banas P, Rulisek L, Hanosova V, Svozil D, Walter NG, Sponer J, Otyepka M. J. Phys. Chem. B. 2008;112:11177–11187. doi: 10.1021/jp802592z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Krasovska MV, Sefcikova J, Spackova N, Sponer J, Walter NG. J. Mol. Biol. 2005;351:731–748. doi: 10.1016/j.jmb.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 135.Perrotta AT, Shih IH, Been MD. Science. 1999;286:123–126. doi: 10.1126/science.286.5437.123. [DOI] [PubMed] [Google Scholar]
- 136.Martick M, Scott WG. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wadkins TS, Shih IH, Perrotta AT, Been MD. J. Mol. Biol. 2001;305:1045–1055. doi: 10.1006/jmbi.2000.4368. [DOI] [PubMed] [Google Scholar]
- 138.Roychowdhury-Saha M, Burke DH. RNA. 2007;13:841–848. doi: 10.1261/rna.339207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Takagi Y, Inoue A, Taira K. J. Am. Chem. Soc. 2004;126:12856–12864. doi: 10.1021/ja031991u. [DOI] [PubMed] [Google Scholar]
- 140.Ditzler MA, Rueda D, Mo J, Håkansson K, Walter NG. Nucleic Acids Res. 2008;36:7088–7099. doi: 10.1093/nar/gkn871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Solomatin SV, Greenfeld M, Chu S, Herschlag D. Nature. 2010;463:681–684. doi: 10.1038/nature08717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hiley SL, Collins RA. EMBO J. 2001;20:5461–5469. doi: 10.1093/emboj/20.19.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lafontaine DA, Norman DG, Lilley DM. EMBO J. 2002;21:2461–2471. doi: 10.1093/emboj/21.10.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]