Abstract
Rab GTPases, key regulators of vesicular transport, hydrolyze GTP very slowly unless assisted by Rab GTPase-activating proteins (RabGAPs). Dysfunction of RabGAPs is involved in many diseases. By combining X-ray structure analysis and time-resolved FTIR spectroscopy we reveal here the detailed molecular reaction mechanism of a complex between human Rab and RabGAP at the highest possible spatiotemporal resolution and in atomic detail. A glutamine residue of Rab proteins (cis-glutamine) that is essential for intrinsic activity is less important in the GAP-activated reaction. During generation of the RabGAP·Rab:GTP complex, there is a rapid conformational change in which the cis-glutamine is replaced by a glutamine from RabGAP (trans-glutamine); this differs from the RasGAP mechanism, where the cis-glutamine is also important for GAP catalysis. However, as in the case of Ras, a trans-arginine is also recruited to complete the active center during this conformational change. In contrast to the RasGAP mechanism, an accumulation of a state in which phosphate is bound is not observed, and bond breakage is the rate-limiting step. The movement of trans-glutamine and trans-arginine into the catalytic site and bond breakage during hydrolysis are monitored in real time. The combination of X-ray structure analysis and time-resolved FTIR spectroscopy provides detailed insight in the catalysis of human Rab GTPases.
Keywords: GTP hydrolysis, dual-finger, isotopic labeling, enzyme catalysis
Rab GTPases are regulators of vesicular trafficking processes in eukaryotic cells and constitute the largest family of small GTPases (1). Interaction with effectors takes place in the GTP-bound “on” state, and the inactivation occurs via GTP hydrolysis to GDP; this is catalyzed by GTPase activating proteins (GAPs), thereby downregulating the GTPase. Most RabGAPs contain Tre2-Bub2-Cdc16 (TBC) domains, and accelerate the intrinsic GTP hydrolysis rate up to 105-fold (2, 3). As regulators of Rab-dependent pathways, RabGAPs are involved in a number of diseases, contributing to viral and bacterial infection, cancer development, predisposition to obesity, and others (1, 2). Therefore, detailed insight into the RabGAP-mediated catalysis with highest possible spatiotemporal resolution is of great interest, and is an important prerequisite for the design of small molecules for molecular therapy.
The stabilization of the nucleophilic-attacking water molecule by the carboxamide moiety of a glutamine or asparagine side chain is crucial for GTP hydrolysis in all small GTPases (4). In Ras (5), Ran (6), and Rho (7), the carboxamide moiety is provided by the conserved glutamine of the so-called “switch II.” In Rap, which has a threonine at the homologous position (amino acid 61), the amide is either not supplied by Rap itself, but by the asparagine thumb of RapGAP (8, 9), or supplied by Gln63 of Rap in the case of catalysis by dual-specificity GAPs (10). In addition, the active site of many small GTPases is completed by a catalytic “arginine finger” that is used, e.g., by Ras-, Rho-, and RabGAPs, but is not needed for catalysis in Ran and Rap. Conventional TBC domains possess conserved arginine and glutamine residues (2, 11). In the case of activation of Rab33 (mouse) by the yeast GAP Gyp1, it was shown that a glutamine residue of Gyp1 (termed trans-glutamine) positions the water molecule rather than the conserved glutamine in Rab33 (termed cis-glutamine) (11). Whether this mechanism applies to bona fide Rab·RabGAP pairs from mammalia has not yet been established.
Ras is the most intensively studied small GTPase. It has been investigated by X-ray crystallography (5, 12–14); biomolecular simulations (15–17); electron paramagnetic resonance and NMR (18); and FTIR (19–22). Time-resolved (tr) FTIR spectroscopy allows monitoring of protein reactions at atomic detail in real time and provides information about charge shifts, protonation states, and protein side-chain movements (19, 21). Based on trFTIR experiments and biomolecular simulations, it has been proposed for RasGAP that the arginine finger movement into the binding pocket displaces water molecules and thereby changes the dielectricum in the active site. This process induces a charge shift from the γ- toward the β-phosphate, drives GTP toward the transition state, and contributes to a reduction of the free activation energy (15, 21). Furthermore, Ras and RasGAP drive GTP into a strained conformation and further reduce the free activation energy (15). In Ras, bond breakage is rapid once the arginine finger enters the active site. The released γ-phosphate and the attacking water form a protein bound H2PO4− in a reversible reaction, and the following Pi-release is the rate-limiting step (19, 21).
Here, we investigated the RabGAP mechanism of a human RabGAP⋅Rab pair, TBC1D20 and Rab1b. TBC1D20 is a Rab1- and Rab2-specific GAP that is involved in hepatitis C virus (HCV) replication and is an efficient GAP in vitro (23, 24). Though the kinetic parameters of most known mammalian RabGAPs either have not been determined or demonstrate low catalytic efficiency (2), we found more than 105-fold acceleration. Rab1 mediates endoplasmatic reticulum–Golgi trafficking and plays a role in Legionella pneumophila pathogenesis (1, 25–28). Here, we describe the structure of the Rab1b·GDP·BeF3−·TBC1D20 and define the catalytic mechanism using complementary FTIR difference spectroscopy with the help of isotopic labeling of Rab, GAP, and the nucleotide. This approach provides a detailed understanding of the catalytic mechanism at the highest spatiotemporal resolution.
Results
Structure of the Isolated TBC1D201–305.
The crystal structure of the isolated TBC1D201–305 was determined at 2.2 Å resolution (see Table S1 for data collection and refinement statistics), and was used as a search model in the structure determination of the Rab1b⋅TBC1D20 complex by molecular replacement. Fig. S1 depicts a comparison between the isolated and the Rab1b-bound state. The arginine-finger residue R105 of the isolated structure has an alternative conformation to that found in the complexed form. The rest of the structure does not undergo other significant structural rearrangement upon binding to Rab1.
Structure of the Rab1·TBC1D20 Complex.
To provide a basis for understanding the mechanism of GTP hydrolysis of Rab1 stimulated by TBC1D20, we determined the crystal structure of Rab1b3–174 in complex with TBC1D201–305, GDP, and BeF3− at 3.3 Å (Table S1). There are five complex molecules in the asymmetric unit that are highly similar to each other, with rmsd values of 0.394–0.716 Å for 2,255 Cα residues. Similar to other TBC domain-containing GAPs, the structure of TBC1D20 can be subdivided into two subdomains. The amino-terminal subdomain comprises helices α1T–α8T, and the carboxyl-terminal subdomain contains helices α9T–α15T (Fig. 1A). In accordance with the previously reported Rab33⋅Gyp1 complex crystal structure, the IxxDxxR and YxQ signature motifs are critical for GAP activity, and they directly contact the nucleotide, whereas the RxxxW motif appears to be important for the structural integrity of the TBC domain (11). In comparison with other TBC domain-containing GAPs, the isoleucine in the IxxDxxR motif is replaced by valine. Helices α5T/α7T of the N-terminal subdomain and α8T/α9T and α11T/α14T of the C-terminal subdomain of TBC1D20 form a groove that is large enough to bind the switch I, switch II, and the P-loop of Rab1.
Fig. 1.
Structure of Rab1b in complex with TBC1D20. (A) Ribbon representation of Rab1b·GDP·BeF3−·TBC1D20 complex with TBC1D20 shown in gray and Rab1b in green, with switch regions colored as indicated by labeling. (B) Interactions in the active site of the complex with coloring as above and functional groups as indicated by labeling. (C) Overlay of the crystal structures of TBC1D20-Rab1b (white) and Gyp1-Rab33 (cyan, PDB ID 2G77). The electron density map contoured at 1.0 σ of the R105T side chain is highlighted in black. The indices G and T are used for Gyp1 and TBC1D20 amino acid legends, respectively.
Additional electron density was found in the interface between Rab1b and TBC1D20 in which GDP, Mg2+, and BeF3− with a trigonal pyramidal conformation could be reliably modeled. In contrast to several other GTPase·GAP structures, no water molecule was found in the active site that was positioned to attack the γ-phosphate group of GTP. However, the structures showing such a water molecule were all with aluminum fluoride (or tetrafluoroaluminate), a transition-state analog, with one exception, the Rap·RapGAP complex, which was also with beryllium fluoride, a ground-state analog (9). In the latter case, there was also no observable attacking water molecule, but the resolution of the solved structure was relatively low (3.3 Å), as in the structure presented here. Therefore, it is possible that a water molecule in this position is not well-defined enough to be seen at this resolution. In analogy to the results seen in aluminum fluoride complexes with other GTPase·GAP pairs (5, 7, 11), the GDP/BeF3− complex is stabilized via polar contacts with the arginine and glutamine fingers, with the glutamine finger coming not from Rab but from the GAP molecule, as seen in the Rab33·Gyp1 structure (11). Residue R105T is located in the α4T helix and mediates bidentate interactions with the α- and β-phosphates of GDP, whereas Q144T is located in loop α5T/α6T and forms interactions via its carboxamide side chain with one of the fluoride atoms of BeF3− and with the backbone amide of Q67R of Rab1b (Fig. 1B). Residue Q67R or its equivalent in the switch II region is involved in the intrinsic hydrolysis of GTP by Rab GTPases, but in the structure presented here, Q67R has moved away from the nucleotide and displays hydrogen bond interactions with the backbone carbonyl of Y142T and the amide of Q144T in the YxQ motif of TBC1D20.
The structure of the complex between Rab1b, TBC1D20, GDP, and beryllium fluoride agrees with the results obtained earlier on a hybrid Rab (mouse)/GAP (yeast) system (11). The superimposition of the protein complex structures of Rab1b⋅TBC1D20 and Rab33⋅Gyp1 reveals analogies in the GTP hydrolytic mechanism (Fig. 1C). In both complexes the significance of the arginine and glutamine fingers for catalysis is a striking feature. R105T in TBC1D20 is the equivalent of R343G in Gyp1 and Q144T of Q378G. The glutamine residues of Rab1b and Rab33 (Q67R1 and Q92R33, respectively) point away from the GDP⋅BeF3− and GDP⋅AlF3 moieties. These residues are oriented toward the α5/α6 and α6/α7 ordered loops, respectively, and are involved in polar interactions with the backbone Y142T/Y376G carbonyl and Q144T/Q378G amino group. Though the GDP moieties and Mg2+ possess nearly identical coordinates, the positions of BeF3− and AlF3 are different. AlF3 shares an axial water with Q378G in the Rab33⋅Gyp1 structure, whereas there is no observable electron density for an axial water of BeF3− in the Rab1⋅TBC1D20 crystal structure. As a consequence, Q144T of TBC1D20 makes polar contacts just to one fluoride atom of BeF3− but not to a water molecule. Residue Q378G of Gyp1 instead contributes two interactions to AlF3, one to the equatorial fluoride and a second indirectly through the axial water molecule.
The conformations of the arginine and glutamine fingers in the Rab1b⋅TBC1D20 complex are essentially identical to those in the Rab33⋅Gyp1 complex structure. The side chain of R343G of Gyp1 forms polar contacts to the α- and β-phosphate, as does R105T of TBC1D20. As shown in Fig. 1C, the arginine conformer of TBC1D20 is supported by the crystallographic data.
Kinetics of GTP Hydrolysis.
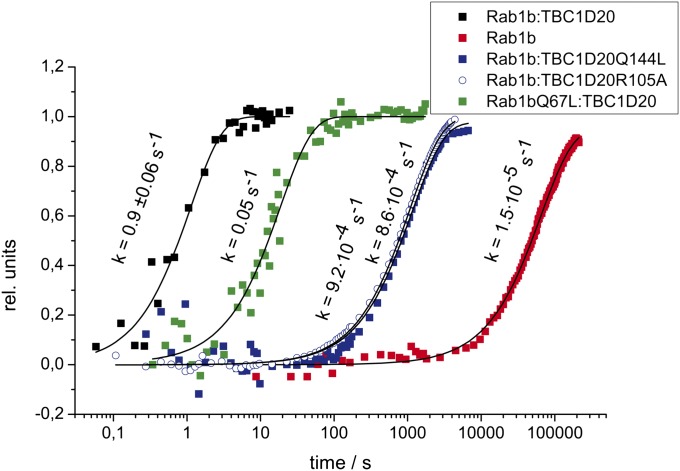
The rate constants of GTP hydrolysis were determined by time-resolved FTIR in single-turnover experiments; this was done for the intrinsic reaction of Rab1b and for the GAP-catalyzed reaction of wild-type and two mutant proteins, TBC1D20Q144L and Rab1bQ67L (Fig. 2). The intrinsic Rab1b GTPase activity is slow (1.5 × 10−5 s−1 at 393 K), but GAP catalyses the reaction by more than five orders of magnitude (0.9 s−1 at 368 K). Mutation of the catalytic fingers (Q144T or R105T of TBC1D20) results in a 1,000-fold decrease in GAP activity, whereas mutation of the cis-glutamine (Q67R of Rab1b) only reduces the hydrolysis rate by one order of magnitude. This result is in agreement with multiple turnover assays conducted at low GAP concentrations using hybrid Rab⋅RabGAP pairs from yeast/mouse (11, 29), and shows that the effect is not due to reduced affinity toward Rab1b, but rather due to a distortion of the active site. The result also confirms the idea that the cis-glutamine is not directly involved in GAP-catalyzed GTP hydrolysis. The intrinsic GTPase activity of Rab1bQ67L is at least 100-fold reduced, which underscores the importance of Q67R for the intrinsic reaction in contrast to GAP-catalyzed hydrolysis.
Fig. 2.
Mutational analysis of the active site of the Rab·RabGAP complex. Kinetics of GTP hydrolysis by Rab1b for the intrinsic (393 K), GAP-catalyzed (368 K), and mutant cases (386 K). The data points represent the normalized difference between adjacent GDP (1,100 cm−1) and GTP (1,129 cm−1) bands, which eliminates baseline drifts for the long measurements. Fast kinetics result from 15 independent measurements, slow kinetics of mutant proteins and the intrinsic reaction from two independent measurements. Errors are given as SDs, which were not calculated for the slow measurements due to the low number of experiments needed to achieve good signal-to-noise ratio. Lines represent monoexponential fits.
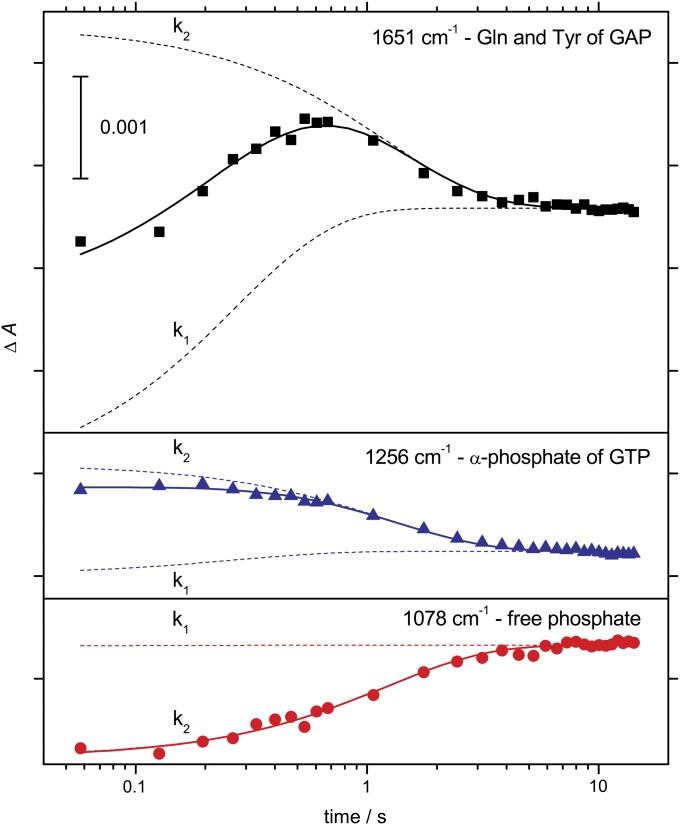
Detailed absorbance changes of the GAP-catalyzed reaction obtained by global fit analysis according to Eq. S1 are shown in Fig. 3. At 1,651 cm−1, an intermediate appears with an apparent rate constant k1 = 7 (± 0.5) s−1 and disappears with k2 = 0.9 (±0.06) s−1 at 368 K (SDs from 15 independent measurements). In the GAP-catalyzed Ras reaction, protein-bound H2PO4− was identified as an intermediate (19). To examine the bond-breaking step, measurements are performed in H218O. The incorporation of the H218O-labeled attacking water molecule into the phosphate leads to band shifts of the phosphate. The protein-bound phosphate in the GAP-catalyzed Ras reaction was identified in this manner (19). The amplitude spectrum of the process occurring with rate constant k1 shows no frequency shifts, and only the free Pi bands at 1,078 cm−1 and 996 cm−1 of the product state are shifted in the spectrum of k2 (Fig. S2A). Therefore, we can conclude that in the Rab1b·TBC1D20 reaction, bond breakage and Pi release occur essentially simultaneously with the rate constant k2, whereas the first rate (k1) corresponds to a conformational change. The release of the Pi and simultaneous disappearance of GTP can be monitored at 1,078 cm−1 and 1,256 cm−1, respectively (Fig. 3).
Fig. 3.
Kinetics of the TBC1D20-catalyzed GTP hydrolysis in Rab1b. Time-dependent absorbance changes of marker bands of the free Pi (1,078 cm−1); overlapping vibrations of the GAP glutamine finger and a tyrosine’s backbone (1,651 cm−1); and the α-phosphate (1,256 cm−1) in 2H2O at 368 K as assigned later in the text. Fifteen independent measurements were averaged. The continuous lines correspond to a global fit with two exponential functions.
The corresponding amplitude spectra reflect the molecular changes within the protein. In intrinsic hydrolysis, bond breakage and Pi release occur as they do for the RabGAP complex in k2, but the amplitude spectrum of k2 differs from that of intrinsic hydrolysis. However, the sum of the amplitude spectra of k1 and k2 of the GAP catalyzed reaction resembles the amplitude spectrum of intrinsic hydrolysis (Fig. S2B), indicating that there are transient TBC1D20 interactions present in the intermediate. In the Ras·RasGAP reaction (19, 21), three reaction rates are resolved, a conformational change from RasoffGTP to RasonGTP, formation of a protein-bound phosphate intermediate, and Pi release as the rate-limiting step. In the RapGAP-catalyzed reaction of Rap, there is also a detectable protein-bound phosphate intermediate (30, 31). In case of Ran (32), there are only two rates, and the observed intermediate still contains GTP and is characterized by a conformational change of both Ran and RanGAP. Therefore, the Rab1b·TBC1D20 reaction is similar to Ran and the accumulating intermediate is the complex of Rab1b·TBC1D20 forming with rate constant k1 and dissociating together with Pi release (k2).
Mechanism of GTP Hydrolysis.
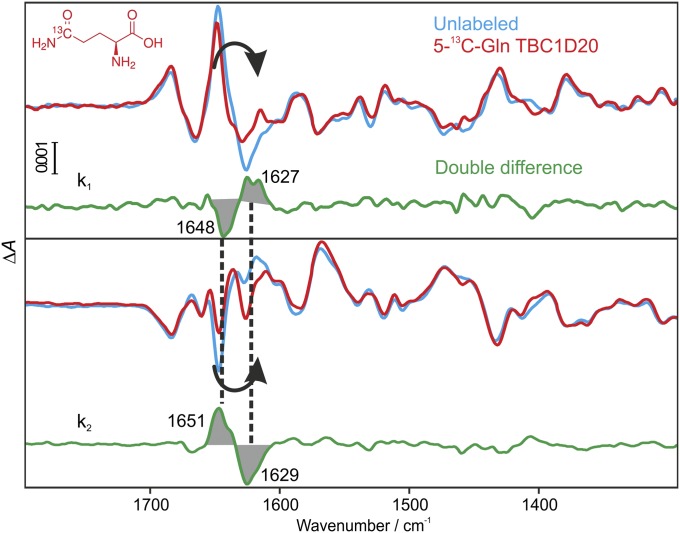
The α- and β-phosphate bands were assigned for the intrinsic and GAP-catalyzed GTPase reaction (Figs. S3 and S4). To further characterize the accumulating reaction intermediate and assign the reaction mechanism of the protein, TBC1D20 was labeled with [5-13C]glutamine. Q144T provides the carboxamide involved in catalysis via a glutamine finger, as seen in the X-ray structure (Fig. 1B). Part of the band at 1,648/1,651 cm−1 is shifted due to labeling, as clearly observed in the double-difference spectra (Fig. 4); it is assigned to the ν(CO) vibration of a glutamine side chain, most likely of Q144, because only this group is crucial for catalysis and no other glutamine band is observed in the amplitude spectra. A clear-cut band assignment can be made in k2, where the reaction is reversed. The extinction coefficient of the glutamine side-chain increases in a hydrophobic environment (33). Therefore, the band indicates the movement of the glutamine finger of TBC1D20 into a more hydrophobic environment toward the active center at rate constant k1. The glutamine finger is relocated back with k2 into the hydrophilic environment, simultaneously with Pi release.
Fig. 4.
Band assignment of the glutamine finger of TBC1D20. Amplitude spectra of the GTPase reaction using unlabeled (cyan) and [5-13C]glutamine-labeled (red) TBC1D20 in 2H2O. In k1, negative bands belong to the GTP state, positive bands to the intermediate state. In k2, negative bands belong to the intermediate state, positive bands to the GDP + Pi state. The double difference (labeled − unlabeled) is shown in green. The band at 1,648/1,651 cm−1 can be assigned to a glutamine side-chain and most likely represents the catalytic glutamine finger moving into the active site with k1 and in the reverse direction with k2.
In the next step, we labeled Rab1b with [5-13C]glutamine to identify the movement of the cis-glutamine (Fig. S5A). This band is also assigned but shows vibrational coupling to the backbone of Y142T and Q144T of TBC1D20, as expected for interacting amide groups (34) (Fig. S5B). Therefore, a pattern with several band-shifts instead of a single marker band is observed. Because Q67R is the only glutamine in the switch regions of Rab1b, the band-shifts indicate the movement of the cis-glutamine that forms the interaction with the YxQ motif in k1 and is released in k2.
In further experiments, TBC1D20 was labeled with [13C915N]tyrosine (Fig. S6A). There are a total of nine tyrosines in TBC1D20 with three tyrosines in the vicinity of the glutamine finger (Y142T, 143T as seen in Fig. 1B, and 146T). As expected, they also cause multiple band-shifts of amide and ring vibrations. The tyrosine bands that appear in k1 are inversed in k2. They probably indicate conformational changes of Tyr142T, 143T, and 146T in the glutamine finger loop. The large band at 1,650 cm−1 also belongs to a tyrosine backbone vibration and overlaps with the band of the glutamine finger. Thus, the band at 1,651 cm−1 presents a convenient marker band for formation and disappearance of the intermediate (Fig. 3).
To elucidate the conformational change between the ground state and the intermediate in more detail, we used complex formation with beryllium fluoride. For Ras, BeF3− induces the same “on” conformation in Ras·GDP as does GTP, as shown by FTIR (35). BeF3− represents an analog of the γ-phosphate (36). The amplitude spectrum of complex formation of Rab1b·GDP with beryllium fluoride and TBC1D20 is very similar to the amplitude spectrum of k1. Therefore, the BeF3− complex clearly mimics the accumulating reaction intermediate. Also, [5-13C]glutamine labeling of Rab1b leads to the same marker pattern as in k1 (Fig. S6B). Therefore, the cis-glutamine still interacts with the YxQ motif in the BeF3− state. The Q144A mutant of TBC1D20 exhibited the same marker pattern on BeF3− complex formation due to isotopic labeling of Rab1b as wild-type TBC1D20 (Fig. S6B), which suggests that the cis-glutamine still interacts with the YxQ motif even if the space in the active site is not occupied by the glutamine finger. Thus, the glutamine finger does not sterically push out the cis-glutamine. The TBC domain of RabGAPs is tailored to pull the cis-glutamine out of the active site so that its own glutamine finger can be inserted.
The glutamine of the DxxGQ motif is crucial for positioning of a water molecule for nucleophilic attack in the intrinsic reaction of most small GTPases. The intrinsic hydrolysis amplitude spectrum of [5-13C]glutamine-labeled Rab1b shows no band-shifts (Fig. S6C), indicating that the cis-glutamine does not change at all. Therefore, we propose that Q67R is rather flexible in the GTP and GDP states of Rab1b and is not able to stabilize the position of the attacking water in the GTP state. Though there is no available crystal structure of Rab1b with a GTP analog, the structure of the slowest Rab GTPase, Rab6a, in the GTP-state revealed higher B-factors for the switch II region than for faster Rab GTPases (37), indicating high flexibility of the switch II and, consequently, of the intrinsic glutamine. High flexibility of Q67R could therefore result in low intrinsic GTPase activity of Rab1b. In contrast, the interaction with GAP leads to conformational changes resulting in a well-defined active site, the glutamine finger taking the position of the cis-glutamine and stabilizing the attacking water molecule. Strongly hydrogen-bonded water molecules lead to continuum absorbances in infrared spectra (38). Indeed, such a continuum band arises in the amplitude spectrum of k1 and disappears in k2, which is a possible marker band for the attacking water molecule stabilized by the glutamine finger (Fig. S7).
In addition to carboxamide side chains, “arginine fingers” complete the catalytic centers of many small GTPases. To determine the contribution of R105T, TBC1D20 was labeled with η-[15N2]arginine. The arginine band was identified as described for RasGAP (21). The arginine band shifts from an aqueous environment to a hydrophobic environment with k1 (Fig. S8A). The band also overlaps with tyrosine bands, as discussed above. The same bands can also be assigned in the amplitude spectrum of AlFx complex formation (Fig. S8A), a transition-state analog. Therefore, we can conclude that the arginine finger enters the active site with k1. In contrast to RasGAP, bond-breaking does not occur with the same rate constant as arginine moves into the catalytic center, but in the next reaction step described by k2. Notably, the spectra of AlFx and BeF3− complex formation differ between 1,580 and 1,540 cm1, indicating a slight conformational difference, whereas AlFx is more similar to the spectrum of k1 than is BeF3−. Because several tyrosine vibrations were found in this region (Fig. S6A), the spectra might represent a change in the α5T/α6T glutamine finger loop. Furthermore, the β-phosphate band in the AlFx complex is shifted to lower wave numbers relative to the BeF3− complex (Fig. S8B), which indicates a different charge distribution, in line with a more product state-like charge distribution. However, the charge shift might be of artificial nature due to the analogs used.
The catalytic mechanism for the mammalian Rab⋅RabGAP pair elucidated here agrees well with the proposed mechanism based on the hybrid Rab⋅RabGAP complex with yeast Gyp1p and mammalian Rab33. Gyp1p also employs an arginine finger, and its glutamine finger takes the role played by the cis-glutamine in most other GTPases (11).
Discussion
Our results are summarized in Fig. 5 and show the dynamics of the active center in RabGAP. With the rate constant k1, Q67R of Rab is pulled out of the active site by the YxQ motif of GAP by interacting with the YxQ backbone, leaving the active site accessible for the catalytic GAP residues; this allows Q144T of GAP to move into the active site. In contrast to the intrinsic case, the attacking water molecule can now be stabilized in the correct position for nucleophilic attack, which is probably the key contribution of RabGAP to catalysis. Furthermore, R105T of GAP is pulled into the active site, probably by coulomb interactions with the negatively charged GTP with k1 as well; in contrast to RasGAP, this does not immediately induce bond breakage, which occurs with the final rate k2. Simultaneously, the conformational changes of Q67R, Q144T, and R105T are reversed as Pi is released. There is no observable accumulation of a protein-bound phosphate intermediate as in the RasGAP catalysis, because bond breakage is the rate-limiting step.
Fig. 5.
Summary of the reaction mechanism. (A) Illustration of the observed processes. With k1, the arginine and glutamine fingers of GAP move into the active site, whereas the intrinsic glutamine of the GTPase moves out to interact with the backbone of GAP. Thus, the catalytically active conformation is formed. With k2, these conformational changes are reversed, and, simultaneously, bond cleavage occurs and Pi is released. (B) Reaction scheme of the Rab·RabGAP reaction.
We have demonstrated here a bona fide human Rab·RabGAP pair, that RabGAP proteins with a TBC domain catalyze GTP hydrolysis using a dual-finger mechanism, as first proposed for a mouse/yeast hybrid pair (11). It is therefore highly likely that all TBC domain-containing RabGAPs use this basic mechanism. We have provided detailed insights into the reaction mechanism using time-resolved FTIR, which revealed a coordinated removal of the cis-glutamine from the region of the γ-phosphate group of GTP and the recruitment of a trans-glutamine from the GAP molecule. The cis-glutamine is essential for the intrinsic reaction but not for GAP activity, in which case the mutant is almost as strongly accelerated as wild type. Importantly, mutation of the cis-glutamine is still widely used to generate GTP-locked Rab GTPases for in vivo studies. However, it needs to be recognized that these mutants can be activated by GAPs present in vivo.
There are now several different mechanisms of GAP activation of GTPases that have been recognized, some (but not all) of which involve an arginine from the GAP molecule. However, there appear to be no exceptions to the observation that a carboxamide function from a glutamine or an asparagine residue is essential for catalysis. This carboxamide function is contributed either by the GTPase itself or by the GAP molecule, as in the case described here. The significance of these differences in the physiological context is not clear, and it is also not yet clear whether restrictions imposed by the GTPase structure itself determine the need for a different carboxamide, or specific type of GAP activation mechanism, except in the case of Rap, where the lack of a correctly positioned cis-glutamine necessitates this. Interestingly, in the case of dual-specificity GAPs, there is indeed an involvement of a cis-glutamine of Rap, which occurs two residues removed from the glutamine in other small GTPases. This finding suggests that different GAP mechanisms can operate on a single type of GTPase, and it will be of interest to search for other examples of this nature. The characterization of the different mechanisms at the level described here will help to define the significance of the differences and lead to approaches for manipulating or emulating GAP activity in specific cases.
Materials and Methods
Protein Isolation.
Rab1b3–174 and TBC1D201–305/1–362/14–305 were expressed and purified as described previously (27, 39) and in more detail in SI Materials and Methods. SeMet labeling of TBC1D2014–305 was achieved during protein overexpression in M9 media by inhibition of methionine biosynthesis (40). For isotopic labeling, we used optimized M9 media (composition as described in ref. 41; amino acids raised to 1.2 mM, NH4Cl to 46 mM) in which the amino acid of interest was replaced by its isotopically labeled form (166 mg/L in all cases). For glutamine labeling of proteins, the E. coli strain TH16#4 (42) was used, and concentrations of glutamate, asparagine, and aspartate in the media were raised to 2.4 mM. η-[15N2]Arginine, [5-13C]glutamine, and [13C915N]tyrosine were purchased from Cambridge Isotope Laboratories. The degree of incorporation and possible spreading were checked by mass spectrometry (41) (Table S2).
Structure Determination.
Details of crystallization, structure determination, and synthesis of caged nucleotides are provided in SI Materials and Methods.
FTIR Measurements.
For FTIR measurements, Rab1b was loaded with the nonhydrolysable, photoactivatable caged nucleotide (43), and the buffer was exchanged to 1 mM Hepes (pH 7.0), 2 mM NaCl, 0.05 mM DTT, and 0.05 mM MgCl2. TBC1D201–362 used for FTIR measurements was kept in a higher concentrated buffer to avoid precipitation: 5 mM Hepes (pH 7.0), 20 mM NaCl, 1 mM DTT. The sample was prepared between two CaF2 windows as described (44). The final sample composition was 5.6 mM Rab1b, 6.1 mM TBC1D20, 20 mM MgCl2, 20 mM DTT, and 200 mM Hepes (pH 7.0) for the 1:1 complex measurements. Photolysis of the caged compounds was performed by an LPX 240 XeCl excimer laser (308 nm; Lambda Physics) by 12 flashes within 24 ms. A modified Bruker IFS 66v/S spectrometer in the fast-scan mode was used for the measurement (45). The data were analyzed between 1,800 and 950 cm−1 with a global fit method (46). Further details of sample composition, measurement conditions, and fit equations are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank H. E. Meyer and K. Kuhlmann for the mass spectrometric analysis of isotopically labeled proteins; Y. Suveyzdis for the labeled caged nucleotides; and F. Großerüschkamp for improvement of the fitting routine. This work was supported by the Ruhr University Research School funded by the Deutsche Forschungsgemeinschaft in the framework of the Excellence Initiative; the Deutsche Forschungsgemeinschaft within Sonderforschungsbereich 642; and the Max Planck Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes: 4HL4 for TBC1D20_14-305 and 4HLQ for Rab1b:TBC1D20_1-305).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214431110/-/DCSupplemental.
References
- 1.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10(8):513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 2.Frasa MAM, Koessmeier KT, Ahmadian MR, Braga VMM. Illuminating the functional and structural repertoire of human TBC/RABGAPs. Nat Rev Mol Cell Biol. 2012;13(2):67–73. doi: 10.1038/nrm3267. [DOI] [PubMed] [Google Scholar]
- 3.Albert S, Will E, Gallwitz D. Identification of the catalytic domains and their functionally critical arginine residues of two yeast GTPase-activating proteins specific for Ypt/Rab transport GTPases. EMBO J. 1999;18(19):5216–5225. doi: 10.1093/emboj/18.19.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wittinghofer A, Vetter IR. Structure-function relationships of the G domain, a canonical switch motif. Annu Rev Biochem. 2011;80:943–971. doi: 10.1146/annurev-biochem-062708-134043. [DOI] [PubMed] [Google Scholar]
- 5.Scheffzek K, et al. The Ras-RasGAP complex: Structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277(5324):333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 6.Seewald MJ, Körner C, Wittinghofer A, Vetter IR. RanGAP mediates GTP hydrolysis without an arginine finger. Nature. 2002;415(6872):662–666. doi: 10.1038/415662a. [DOI] [PubMed] [Google Scholar]
- 7.Rittinger K, et al. Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP. Nature. 1997;388(6643):693–697. doi: 10.1038/41805. [DOI] [PubMed] [Google Scholar]
- 8.Daumke O, Weyand M, Chakrabarti PP, Vetter IR, Wittinghofer A. The GTPase-activating protein Rap1GAP uses a catalytic asparagine. Nature. 2004;429(6988):197–201. doi: 10.1038/nature02505. [DOI] [PubMed] [Google Scholar]
- 9.Scrima A, Thomas C, Deaconescu D, Wittinghofer A. The Rap-RapGAP complex: GTP hydrolysis without catalytic glutamine and arginine residues. EMBO J. 2008;27(7):1145–1153. doi: 10.1038/emboj.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sot B, et al. Unravelling the mechanism of dual-specificity GAPs. EMBO J. 2010;29(7):1205–1214. doi: 10.1038/emboj.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan X, Eathiraj S, Munson M, Lambright DG. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature. 2006;442(7100):303–306. doi: 10.1038/nature04847. [DOI] [PubMed] [Google Scholar]
- 12.Scheidig AJ, Burmester C, Goody RS. The pre-hydrolysis state of p21(ras) in complex with GTP: New insights into the role of water molecules in the GTP hydrolysis reaction of ras-like proteins. Structure. 1999;7(11):1311–1324. doi: 10.1016/s0969-2126(00)80021-0. [DOI] [PubMed] [Google Scholar]
- 13.Pai EF, et al. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: Implications for the mechanism of GTP hydrolysis. EMBO J. 1990;9(8):2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosnizeck IC, et al. Stabilizing a weak binding state for effectors in the human Ras protein by cyclen complexes. Angew Chem Int Ed Engl. 2010;49(22):3830–3833. doi: 10.1002/anie.200907002. [DOI] [PubMed] [Google Scholar]
- 15.Rudack T, Xia F, Schlitter J, Kötting C, Gerwert K. Ras and GTPase-activating protein (GAP) drive GTP into a precatalytic state as revealed by combining FTIR and biomolecular simulations. Proc Natl Acad Sci USA. 2012;109(38):15295–15300. doi: 10.1073/pnas.1204333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia F, Rudack T, Kötting C, Schlitter J, Gerwert K. The specific vibrational modes of GTP in solution and bound to Ras: A detailed theoretical analysis by QM/MM simulations. Phys Chem Chem Phys. 2011;13(48):21451–21460. doi: 10.1039/c1cp22741f. [DOI] [PubMed] [Google Scholar]
- 17.Martín-García F, Mendieta-Moreno JI, López-Viñas E, Gómez-Puertas P, Mendieta J. The Role of Gln61 in HRas GTP hydrolysis: A quantum mechanics/molecular mechanics study. Biophys J. 2012;102(1):152–157. doi: 10.1016/j.bpj.2011.11.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rohrer M, et al. Structure of the metal-water complex in Ras x GDP studied by high-field EPR spectroscopy and 31P NMR spectroscopy. Biochemistry. 2001;40(7):1884–1889. doi: 10.1021/bi002164y. [DOI] [PubMed] [Google Scholar]
- 19.Kötting C, et al. A phosphoryl transfer intermediate in the GTPase reaction of Ras in complex with its GTPase-activating protein. Proc Natl Acad Sci USA. 2006;103(38):13911–13916. doi: 10.1073/pnas.0604128103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kötting C, Gerwert K. Time-resolved FTIR studies provide activation free energy, activation enthalpy and activation entropy for GTPase reactions. Chem Phys. 2004;307:227–232. [Google Scholar]
- 21.Kötting C, Kallenbach A, Suveyzdis Y, Wittinghofer A, Gerwert K. The GAP arginine finger movement into the catalytic site of Ras increases the activation entropy. Proc Natl Acad Sci USA. 2008;105(17):6260–6265. doi: 10.1073/pnas.0712095105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allin C, Ahmadian MR, Wittinghofer A, Gerwert K. Monitoring the GAP catalyzed H-Ras GTPase reaction at atomic resolution in real time. Proc Natl Acad Sci USA. 2001;98(14):7754–7759. doi: 10.1073/pnas.131549798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sklan EH, et al. TBC1D20 is a Rab1 GTPase-activating protein that mediates hepatitis C virus replication. J Biol Chem. 2007;282(50):36354–36361. doi: 10.1074/jbc.M705221200. [DOI] [PubMed] [Google Scholar]
- 24.Haas AK, et al. Analysis of GTPase-activating proteins: Rab1 and Rab43 are key Rabs required to maintain a functional Golgi complex in human cells. J Cell Sci. 2007;120(Pt 17):2997–3010. doi: 10.1242/jcs.014225. [DOI] [PubMed] [Google Scholar]
- 25.Isberg RR, O’Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol. 2009;7(1):13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukherjee S, et al. Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature. 2011;477(7362):103–106. doi: 10.1038/nature10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Müller MP, et al. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science. 2010;329(5994):946–949. doi: 10.1126/science.1192276. [DOI] [PubMed] [Google Scholar]
- 28.Goody PR, et al. Reversible phosphocholination of Rab proteins by Legionella pneumophila effector proteins. EMBO J. 2012;31(7):1774–1784. doi: 10.1038/emboj.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Antoni A, Schmitzová J, Trepte HH, Gallwitz D, Albert S. Significance of GTP hydrolysis in Ypt1p-regulated endoplasmic reticulum to Golgi transport revealed by the analysis of two novel Ypt1-GAPs. J Biol Chem. 2002;277(43):41023–41031. doi: 10.1074/jbc.M205783200. [DOI] [PubMed] [Google Scholar]
- 30.Chakrabarti PP, Suveyzdis Y, Wittinghofer A, Gerwert K. Fourier transform infrared spectroscopy on the Rap.RapGAP reaction, GTPase activation without an arginine finger. J Biol Chem. 2004;279(44):46226–46233. doi: 10.1074/jbc.M405603200. [DOI] [PubMed] [Google Scholar]
- 31.Chakrabarti PP, et al. Insight into catalysis of a unique GTPase reaction by a combined biochemical and FTIR approach. J Mol Biol. 2007;367(4):983–995. doi: 10.1016/j.jmb.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 32.Brucker S, Gerwert K, Kötting C. Tyr39 of ran preserves the Ran.GTP gradient by inhibiting GTP hydrolysis. J Mol Biol. 2010;401(1):1–6. doi: 10.1016/j.jmb.2010.05.068. [DOI] [PubMed] [Google Scholar]
- 33.Wellner N, Belton PS, Tatham AS. Fourier transform IR spectroscopic study of hydration-induced structure changes in the solid state of ω-gliadins. Biochem J. 1996;319(Pt 3):741–747. doi: 10.1042/bj3190741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brauner JW, Dugan C, Mendelsohn R. 13C isotope labeling of hydrophobic peptides. Origin of the anomalous intensity distribution in the infrared Amide I spectral region of β-sheet structures. J Am Chem Soc. 2000;122(4):677–683. [Google Scholar]
- 35.Kötting C, Kallenbach A, Suveyzdis Y, Eichholz C, Gerwert K. Surface change of Ras enabling effector binding monitored in real time at atomic resolution. ChemBioChem. 2007;8(7):781–787. doi: 10.1002/cbic.200600552. [DOI] [PubMed] [Google Scholar]
- 36.Díaz JF, Sillen A, Engelborghs Y. Equilibrium and kinetic study of the conformational transition toward the active state of p21Ha-ras, induced by the binding of BeF3- to the GDP-bound state, in the absence of GTPase-activating proteins. J Biol Chem. 1997;272(37):23138–23143. doi: 10.1074/jbc.272.37.23138. [DOI] [PubMed] [Google Scholar]
- 37.Bergbrede T, Pylypenko O, Rak A, Alexandrov K. Structure of the extremely slow GTPase Rab6A in the GTP bound form at 1.8A resolution. J Struct Biol. 2005;152(3):235–238. doi: 10.1016/j.jsb.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 38.Garczarek F, Gerwert K. Functional waters in intraprotein proton transfer monitored by FTIR difference spectroscopy. Nature. 2006;439(7072):109–112. doi: 10.1038/nature04231. [DOI] [PubMed] [Google Scholar]
- 39.Schoebel S, Oesterlin LK, Blankenfeldt W, Goody RS, Itzen A. RabGDI displacement by DrrA from Legionella is a consequence of its guanine nucleotide exchange activity. Mol Cell. 2009;36(6):1060–1072. doi: 10.1016/j.molcel.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 40.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229(1):105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 41.Warscheid B, et al. Systematic approach to group-specific isotopic labeling of proteins for vibrational spectroscopy. Vib Spectrosc. 2008;48(1):24–36. [Google Scholar]
- 42.Ueno-Nishio S, Backman KC, Magasanik B. Regulation at the glnL-operator promoter of the complex glnALG operon of Escherichia coli. J Bacteriol. 1983;153(3):1247–1251. doi: 10.1128/jb.153.3.1247-1251.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.John J, et al. Kinetics of interaction of nucleotides with nucleotide-free H-ras p21. Biochemistry. 1990;29(25):6058–6065. doi: 10.1021/bi00477a025. [DOI] [PubMed] [Google Scholar]
- 44.Cepus V, Scheidig AJ, Goody RS, Gerwert K. Time-resolved FTIR studies of the GTPase reaction of H-ras p21 reveal a key role for the β-phosphate. Biochemistry. 1998;37(28):10263–10271. doi: 10.1021/bi973183j. [DOI] [PubMed] [Google Scholar]
- 45.Gerwert K, Souvignier G, Hess B. Simultaneous monitoring of light-induced changes in protein side-group protonation, chromophore isomerization, and backbone motion of bacteriorhodopsin by time-resolved Fourier-transform infrared spectroscopy. Proc Natl Acad Sci USA. 1990;87(24):9774–9778. doi: 10.1073/pnas.87.24.9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hessling B, Souvignier G, Gerwert K. A model-independent approach to assigning bacteriorhodopsin’s intramolecular reactions to photocycle intermediates. Biophys J. 1993;65(5):1929–1941. doi: 10.1016/S0006-3495(93)81264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.