Summary
Dynamic interactions between membrane-bound organelles and the microtubule cytoskeleton are crucial to establish, maintain and remodel the internal organization of cells throughout the cell cycle. However, the molecular nature of these interactions remains poorly understood. We performed a biochemical screen for microtubule-membrane linkers and identified REEP4, a previously uncharacterized endoplasmic reticulum (ER) protein. Depletion of REEP4 and the closely related REEP3 from HeLa cells causes defects in cell division and a proliferation of intranuclear membranes derived from the nuclear envelope. This phenotype originates in mitosis, when ER membranes accumulate on metaphase chromosomes. Microtubule binding and mitotic ER clearance from chromosomes both depend on a short, positively charged amino acid sequence connecting the two hydrophobic domains of REEP4. Our results show that REEP3/4 function redundantly to clear the ER from metaphase chromatin, thereby ensuring correct progression through mitosis and proper nuclear envelope architecture.
Introduction
The internal spatial organization of cells is critically important for their functions. This is illustrated by the characteristic positioning of organelles in many differentiated cell types and by the re-organization of the cellular interior upon polarization and division. In recent years, progress has been made towards elucidating how the positioning of lysosomes, Golgi complex and the nucleus is achieved (Rosa-Ferreira and Munro, 2011; Yadav et al., 2012; Starr, 2007). Furthermore, organelle-shaping proteins have been identified, most notably reticulon and DP1/REEP5 proteins that generate high-curvature ER membranes (Voeltz et al., 2006). The microtubule cytoskeleton acts as a crucial organizing element, and a number of organelle-microtubule linker proteins have been identified. These include proteins that link organelles to microtubule motors such as golgin160 (Yadav et al., 2012), as well as proteins that link to growing microtubule plus ends, like CLIP-170 and STIM1 (Pierre et al. 1992; Grigoriev et al., 2008). However, much is unknown about how the microtubule cytoskeleton organizes membrane-bound organelles, in particular how cell cycle-dependent changes in organelle morphology and positioning are achieved and how these changes contribute to proper cell division and organelle inheritance.
Dramatic membrane restructuring occurs upon nuclear envelope breakdown (NEBD) during metazoan mitosis (Hetzer, 2010; Puhka et al., 2007; Lu et al., 2009; Puhka et al., 2012). Microtubules support this process and promote the removal of nuclear envelope components from chromatin (Beaudouin et al., 2002; Salina et al., 2002; Muhlhausser and Kutay, 2007). After NEBD the nuclear membrane is resorbed into the ER, which is absent from chromosomes and the area between the spindle poles in early mitosis (Puhka et al., 2007; Anderson and Hetzer, 2008). Only in late anaphase do ER membranes establish contact with the separated daughter chromatin masses to initiate nuclear envelope reassembly. One mechanism that helps prevent ER association with the spindle relies on mitotic phosphorylation of the ER membrane protein STIM1, which inhibits binding of STIM1 to microtubule plus ends (Smyth et al., 2012). But how mitotic chromosomes are maintained clear of ER membrane until the onset of nuclear envelope reformation is unknown.
We have used a biochemical approach to identify previously uncharacterized proteins capable of linking organelles and microtubules. Among the candidates obtained was REEP4, a protein related to DP1/REEP5. Here we show that REEP4 and the closely related REEP3 are essential for sequestering nuclear envelope components away from chromatin during metaphase, thereby contributing to the fidelity of chromosome segregation and to proper formation and architecture of the nuclear envelope.
Results
Identification of REEP4 as a microtubule-binding ER protein
To identify novel proteins that link cell organelles and microtubules, we isolated total membranes from Xenopus laevis cytoplasmic egg extracts by pelleting and flotation, extracted membrane proteins by treatment with the detergent CHAPS, and incubated the extracted proteins with taxol-stabilized microtubules. Microtubule-bound proteins were eluted with a high-salt buffer and analyzed by mass spectrometry. We isolated a number of known organelle-microtubule linker proteins including STIM1, CLIMP63 and p22, validating our purification approach. Among the candidates isolated was REEP4, an uncharacterized member of the DP1/REEP5 family of ER morphogenic proteins whose closest studied relative, REEP1, is a neuron-specific ER-microtubule linker (Park et al., 2010).
We first determined REEP4’s subcellular localization. In agreement with its similarity to the ER protein DP1/REEP5, REEP4 tagged with a hemagglutinin (HA) epitope at its amino- or carboxy-terminus localized to the ER in HeLa and COS-7 cells (Figures 1A, S1A, S1B and data not shown). Consistent with our original screening approach, REEP4 co-pelleted with taxol-stabilized microtubules added to tissue culture cell lysates (Figure S1C). Thus, REEP4 is an ER protein that binds to microtubules.
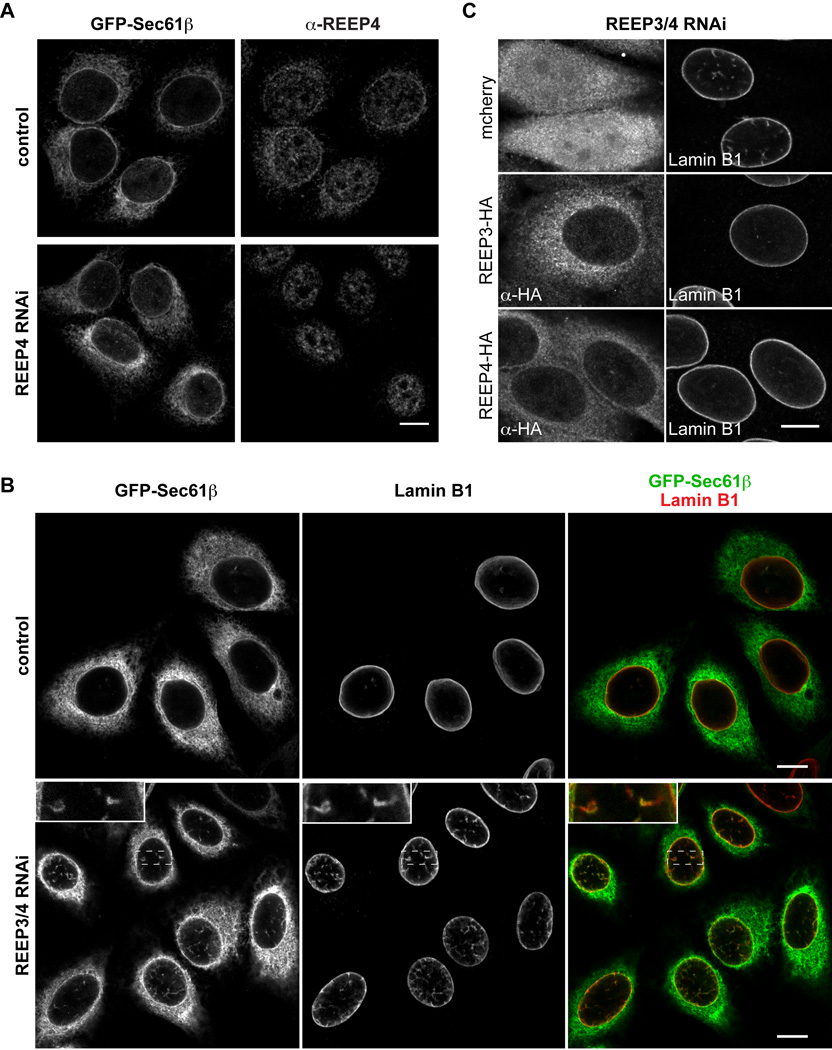
Figure 1. Depletion of REEP3 and REEP4 causes interphase nuclear envelope defects.
(A) Endogenous REEP4 stained with a specific antibody in HeLa cells stably expressing the ER-marker GFP-Sec61β. REEP4 nuclear envelope and ER staining are apparent in control cells but not in REEP4-depleted cells. Intranuclear background staining of the antibody is present in both.
(B) HeLa cells expressing GFP-Sec61β stained for Lamin B1. In control cells, GFP-Sec61β and Lamin B1 label is restricted to the nuclear rim, whereas in REEP3/4 double knockdown cells numerous GFP-Sec61β- and Lamin B1-positive structures are visible within the nucleus.
(C) HeLa cells co-transfected with siRNA targeting REEP3/4 and plasmids encoding either mcherry or RNAi-resistant versions of HA-tagged REEP3 or REEP4 and stained for HA and Lamin B1. Normal nuclear morphology is restored in cells expressing either REEP3 or REEP4. Scale bars are 10 µm. See Figure S1 and Movie S1.
REEP3 and REEP4 are required for proper nuclear envelope architecture
The fact that REEP4 binds microtubules while the more divergent tubule-shaping protein REEP5 does not (Figure S1C and Park et al., 2010), raised the possibility that REEP4 and the closely related REEP2 and REEP3 perform distinct functions. REEP2 mRNA could not be detected in HeLa cells and this protein was not studied further (data not shown). When REEP4 and REEP3 were co-depleted from HeLa cells by RNA interference (RNAi), defects in nuclear envelope structure were observed. In control cells, markers for the ER (GFP-Sec61β) and the nuclear envelope (Lamin B1, POM121-GFP) were almost exclusively restricted to the nuclear rim, as expected, whereas REEP3/4-depleted cells contained numerous intranuclear membrane structures positive for these markers (Figures 1B and S1E, Movie S1). We analyzed REEP3/4 RNAi cells by electron microscopy and confirmed the abundant presence of membrane structures in the nuclear interior (Figures S1F and S1G). These structures contained nuclear pores and are reminiscent of a previously described nuclear envelope feature termed the nucleoplasmic reticulum (Malhas et al., 2011).
We examined single confocal sections at the nuclear center and found that in controls 45% of them were free of internal Lamin B1 label, 45% contained one to three Lamin B1-positive intranuclear structures, and 10% contained four or more such structures. In contrast, 76% of confocal sections of nuclei in REEP3/4-depleted cells contained four or more Lamin B1-positive intranuclear structures (Figure S2B). This phenotype required depletion of both REEP3 and REEP4 (Figure S1H) and could be rescued by expression of RNAi-resistant HA-tagged versions of either protein, indicating that the two proteins function redundantly (Figure 1C). Depletion of REEP2/3/4 also caused nuclear envelope defects in ARPE-19 and U2OS cells, indicating their conserved activity in establishing proper nuclear envelope structure (Figure S1I).
REEP3/4 clear the ER from chromosomes during mitosis
In metazoans undergoing open mitosis, the nuclear envelope reforms from ER membranes following chromosome segregation (Hetzer, 2010). To determine how REEP3/4 depletion caused nuclear envelope defects we therefore examined ER distribution in mitotic cells by live imaging. The ER was excluded from chromosomes and the central spindle area in control metaphase cells, but, strikingly, was closely associated with mitotic chromosomes and the spindle in REEP3/4 knockdown cells (Figure 2A). Moreover, the characteristic ER enrichment at spindle poles was abolished in REEP3/4-depleted cells (Figure 2A, C). Observation of nuclear envelope reformation in live cells expressing the ER marker GFP-Sec61β revealed that control cells excluded ER from the segregating chromosomes throughout anaphase. A distinct boundary between the nuclear interior and the cytosolic ER network was created by the emerging nuclear envelope late in anaphase (Figure 2B, see Movie S2 for an overlay with the chromatin marker histone H2B-mcherry). In REEP3/4 knockdown cells, however, ER membranes were tightly associated with chromatin throughout anaphase and during the course of nuclear envelope assembly. After nuclear envelope reformation, ER membranes were gradually cleared from the nuclear interior, but features identical to the interphase intranuclear membrane structures persisted (Figure 2B, Movie S2). These observations indicate that interphase defects in nuclear envelope structure following REEP3/4 depletion arise during mitosis. In support of this notion, the interphase phenotype was largely rescued when cell cycle progression through S-phase and mitosis was inhibited in REEP3/4-depleted cells by addition of thymidine (Figure S2A, B). REEP3/4 mRNAs were depleted with equal efficiency in the presence or absence of thymidine as judged by quantitative real-time PCR (data not shown). REEP4 continued to co-localize with the ER in mitotic cells (Figure S2C, D) and endogenous REEP4 co-pelleted with microtubules in extracts from cells arrested in metaphase (Figure S2E), consistent with a spindle-related function for REEP3/4.
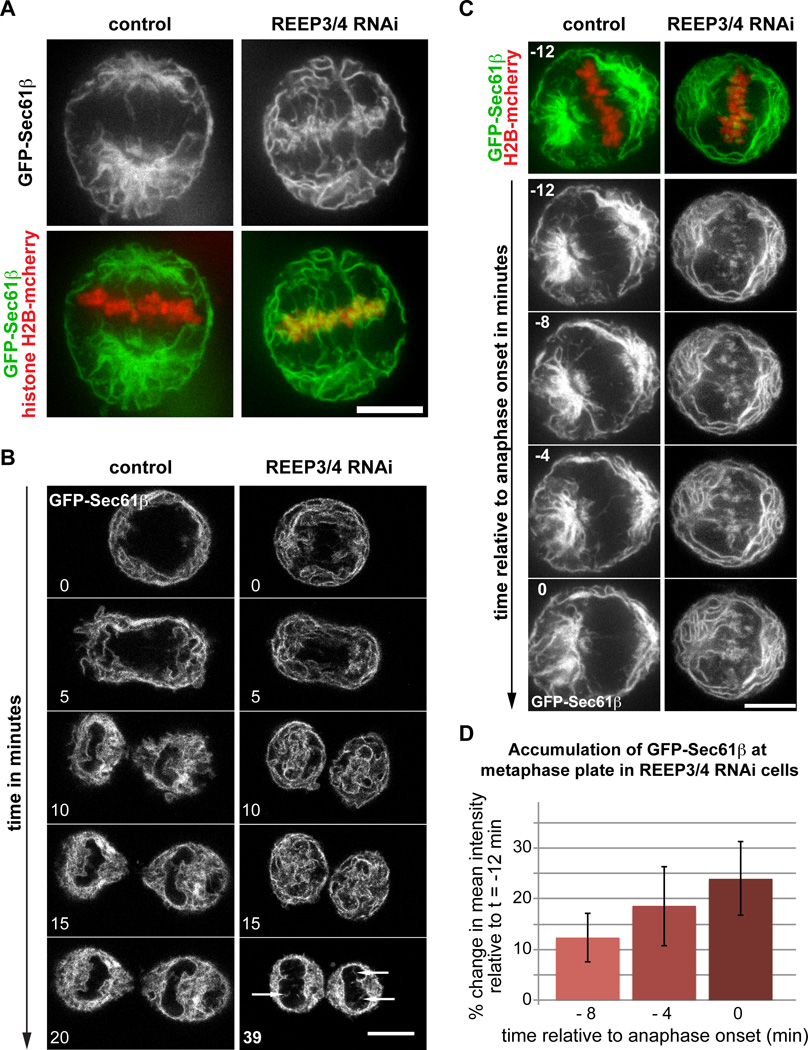
Figure 2. Nuclear envelope defects arise in mitosis.
(A) HeLa cells expressing GFP-Sec61β and histone H2B-mcherry imaged live at metaphase. GFP-Sec61β is mostly excluded from the chromosome area in control cells but shows extensive association with chromatin in REEP3/4 RNAi cells.
(B) Still images from time-lapse acquisitions of HeLa cells expressing GFP-Sec61β in mitosis. In control cells, GFP-Sec61β is excluded from chromatin until the onset of nuclear envelope reformation. In REEP3/4 RNAi cells GFP-Sec61β associates with chromosomes throughout anaphase and during nuclear envelope reformation. It is subsequently gradually and incompletely cleared from the inside of the nucleus after nuclear envelope reformation, with some remnants persisting (arrows). For REEP3/4 RNAi a later final time point (39 minutes instead of 20 minutes for control) is shown to illustrate clearing of ER from the nuclear interior and the persistence of intranuclear membranes. See Movie S2.
(C) Still images from a time-lapse acquisition of control and REEP3/4 RNAi HeLa cells expressing GFP-Sec61β and H2B-mcherry. The initial frame is shown with both, GFP-Sec61β and H2B-mcherry signal. Below, the time series is shown with only the GFP-Sec61β signal to make ER accumulation discernable. In control cells, the chromatin area remains free of ER over the course of metaphase but in REEP3/4 RNAi cells, ER accumulates at the metaphase plate.
(D) Quantification of ER accumulation at the metaphase plate in REEP3/4 RNAi cells as % change compared to the initial frame taken 12 minutes before anaphase onset. Mean pixel intensities for GFP-Sec61β fluorescence at the metaphase plate were measured. Data are mean ± SEM from 10 cells analyzed. Means for each time point are significantly different from t = −12 min to p<0.05 (t = −8 min and t = −4 min), and to p<0.005 for t = 0 min. p-values were obtained using a two-tailed, unpaired student’s t-test. Scale bars represent 10 µm. See Figure S2.
Therefore, intranuclear membranes in REEP3/4 knockdown cells arise during mitosis and are caused by perturbations in nuclear envelope reformation that result from ER mislocalization during metaphase.
ER accumulates on metaphase chromatin in REEP3/4-depleted cells
ER is targeted to chromosomes at the end of anaphase to initiate nuclear envelope reformation. We hypothesized that association of ER membrane with metaphase chromosomes in REEP3/4-depleted cells could be caused by premature targeting of ER to chromosomes. To evaluate this scenario, we imaged live cells expressing GFP-Sec61β during metaphase. In control cells, ER was excluded from the chromatin throughout metaphase (Figure 2C). In REEP3/4 knockdown cells, however, ER invaded the chromatin region and accumulated in the central spindle area over time. Quantification of mean GFP-Sec61β fluorescence intensity at metaphase chromosomes revealed that REEP3/4-depleted cells displayed an increase in ER fluorescence at the metaphase plate in the period leading up to sister chromatid separation in anaphase (Figure 2D). The 24% increase observed likely underestimates total ER accumulation, since in many cells the ER was already associated with mitotic chromosomes at the beginning of the analysis, 12 minutes before anaphase onset. These observations suggest that REEP3/4-mediated sequestration of ER away from chromatin prevents premature targeting of the ER to metaphase chromosomes.
REEP3/4-depleted cells exhibit mitotic defects
Next, we asked whether aberrant chromatin-association of ER in REEP3/4-depleted cells affected mitotic progression. We did not observe obvious changes in cell proliferation, and mitotic spindles formed normally following REEP3/4 RNAi. However, daughter cells were often abnormally close to one another at telophase (Figure 3A). In late mitosis, by the time the cleavage furrow had constricted to 1 µm or less, REEP3/4-depleted cells had separated their nuclei to only ~70 % of the distance in control cells (Figure 3B). Furthermore, the fraction of cells showing anaphase bridges and lagging chromosomes in late anaphase and telophase increased 1.5-fold in REEP3/4 RNAi cells (Figure 3A, C). These observations suggest that chromosomes need to be kept clear of membranes in mitosis for spindle microtubules to properly attach to and segregate sister chromatids.
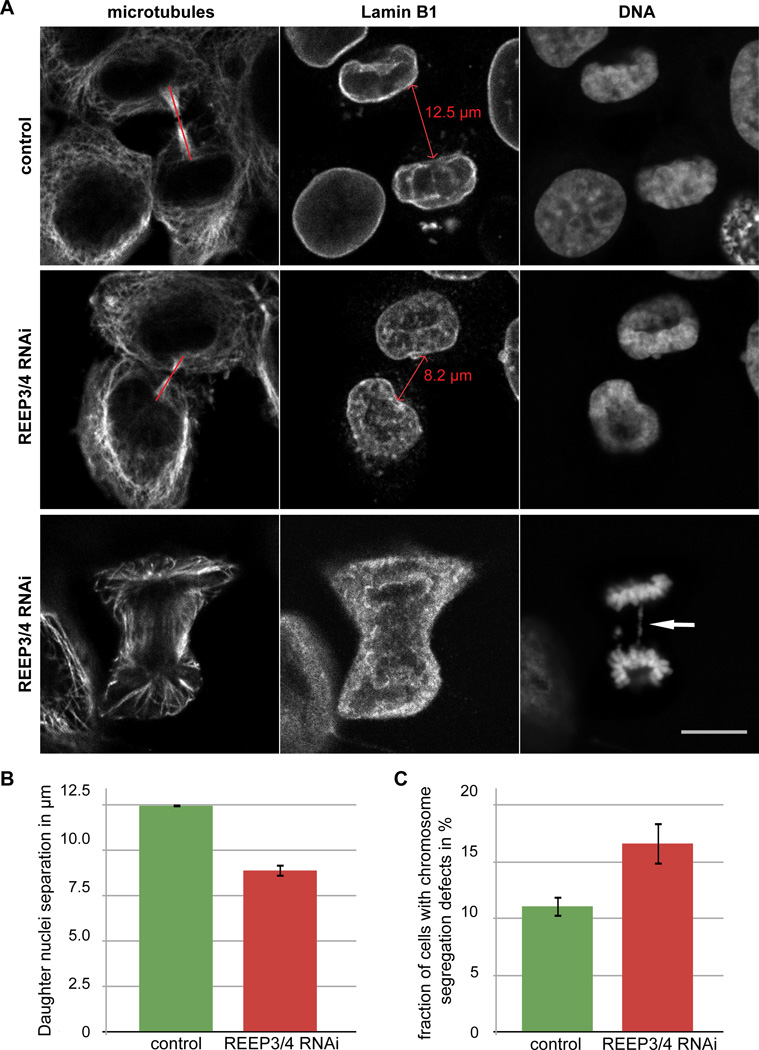
Figure 3. Depletion of REEP3/4 impairs daughter nuclei separation and leads to an increase in chromosome segregation defects.
(A) Top panels: Control and REEP3/4 depleted cells in late stages of cell division. Separation between daughter cell nuclei in REEP3/4 RNAi is reduced compared to controls. Bottom panel: Example for chromosome segregation defect in REEP3/4 RNAi cells. Scale bar is 10 µm.
(B) Quantification of daughter nuclei separation. When the cleavage furrow had narrowed to ≤ 1 µm, nuclei were separated by 12.5 µm in control cells but only 8.9 µm in REEP3/4 RNAi cells. Data are mean +/− SEM, from three independent experiments. In each experiment, at least 14 control and 22 REEP3/4 RNAi cells were analyzed. Control and REEP3/4 RNAi were significantly different to p<0.001 in each experiment using a two-tailed, unpaired Welch’s t-test.
(C) Quantification of chromosome segregation defects (anaphase bridges and lagging chromosomes). 11.1% of control cells and 16.6% of REEP3/4 RNAi cells show chromosome segregation defects. Data shown are mean +/− SEM from seven experiments. At least 50 cells were analyzed per condition and experiment. Control and REEP3/4 RNAi are significantly different to p<0.03 using a two-tailed, unpaired student’s t-test..
Microtubule-association is important for REEP3/4 function
Given that we identified REEP4 as an ER-microtubule linker protein, we asked whether its mitotic membrane-clearing function required association with microtubules. To identify domains potentially important for REEP4 interaction with microtubules, we compared the sequences of the microtubule-binding REEP1-REEP4 subfamily and the non-microtubule binding REEP5/6 subfamily. While REEP C-termini are divergent and longer in REEP1–4 than in REEP5/6, we found that the C-terminal 134 amino acids of REEP4 were not essential for function (data not shown). The more conserved N-terminal regions of REEP proteins contain two hydrophobic domains required for membrane association (Voeltz et al., 2006). Interestingly, the 17 amino acid cytoplasmic stretch between these hydrophobic domains is positively charged for the REEP1-REEP4 subfamily but negatively charged for REEP5/6 (Figure 4A). Since positively charged residues frequently contribute to microtubule binding, we replaced this putative microtubule-binding sequence in REEP4 with the corresponding sequence of REEP5. Given that REEP3 and REEP4 are highly similar, especially at their amino termini (67% similarity over the entire sequence; 86% similarity and 74% identity over the N-terminal 130 amino acids), we expect similar results for both REEP4 and REEP3. We first assayed the C-terminally HA-tagged REEP4/5 chimera for association with microtubules and found that only 13% co-sedimented with microtubules, compared to 85% of wild type REEP4-HA (Figure 4B). This result shows that the short and charged cytoplasmic region between the hydrophobic domains contributes significantly to microtubule binding, and we therefore termed the chimeric protein REEP4dMT to indicate its deficiency in microtubule association.
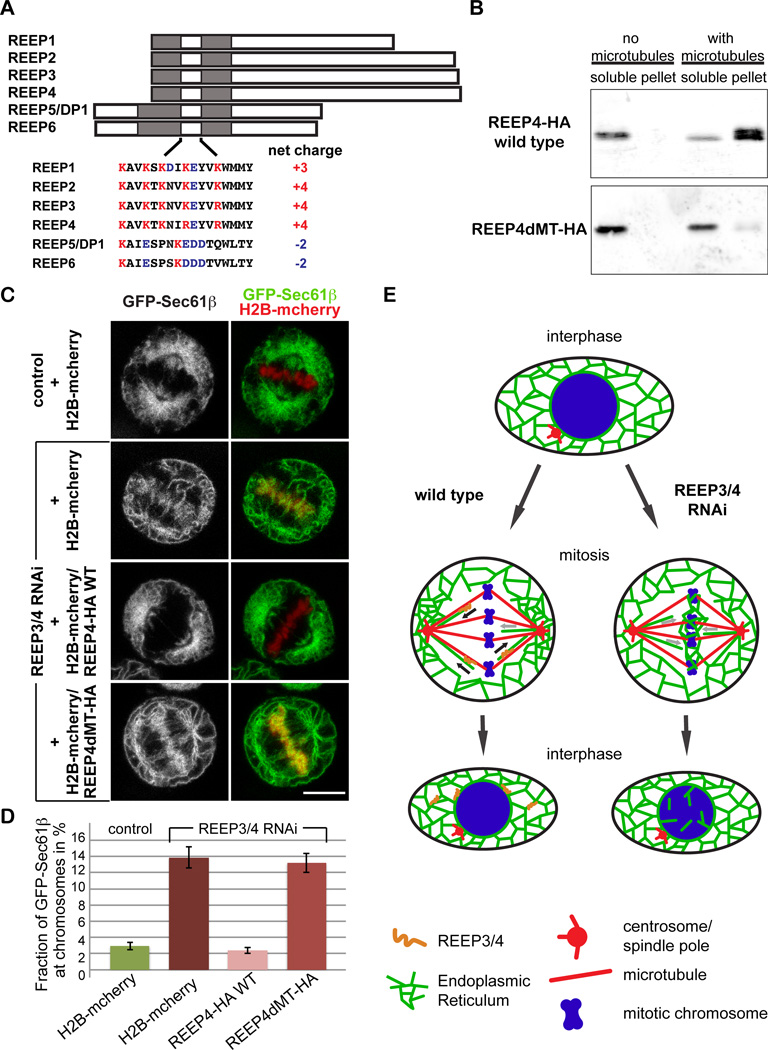
Figure 4. Microtubule binding is important for REEP3/4 function.
(A) Schematic representation of the REEP protein family. Shaded regions represent the hydrophobic domains. The sequences for the cytoplasmic stretches connecting the hydrophobic domains are shown.
(B) Western blot of samples from microtubule co-pelleting assay performed with HA-tagged REEP4 and the REEP4/5 chimeric protein (REEP4dMT). 85% of wild type REEP4-HA but only 13% of REEP4dMT-HA co-pellet with microtubules.
(C) HeLa cells expressing GFP-Sec61β and histone H2B-mcherry were imaged live at metaphase. Expression of RNAi-resistant wild type REEP4 restores ER clearance from chromosomes in REEP3/4 RNAi cells whereas expression of RNAi-resistant REEP4dMT does not rescue. Scale bar is 10 µm.
(D) Total and metaphase plate GFP-Sec61β fluorescence were quantified from images as in (C). The fraction of cellular GFP-Sec61β fluorescence at the metaphase plate is shown. Data are mean ± SEM from three independent experiments. At least 10 cells were analyzed per experiment and condition. Means for control and REEP3/4 RNAi (p<0.002) as well as control and REEP3/4 RNAi + REEP4dMT-HA (p<0.002) are significantly different. The means for control versus REEP3/4 RNAi + REEP4-HA WT are not significantly different (p>0.4). The means for REEP3/4 RNAi versus REEP3/4 RNAi + REEP4dMT-HA are not significantly different (p>0.7). pvalues were obtained using a two-tailed, unpaired student’s t-test.
(E) Model depicting REEP3/4 function. In wild type cells, REEP3/4 transport ER towards microtubule minus ends, thus clearing chromosomes, opposing ER movement towards chromatin and clustering ER at the spindle poles. Without REEP3/4, ER clearance from chromosomes fails and ER invasion of the chromosome area causes accumulation at the metaphase plate.
WT: wild type. See Figure S3.
Next, we tested whether REEP4dMT was functional. In a REEP3/4 RNAi background, HA-tagged, RNAi-resistant REEP4dMT showed the same localization as RNAi-resistant REEP4-HA and was expressed at similar levels. In contrast to wild type REEP4, however, REEP4dMT could not rescue the interphase nuclear envelope defects caused by REEP3/4 depletion (Figure S3A). To examine mitotic ER distribution, we co-transfected REEP3/4 RNAi-treated cells expressing GFP-Sec61β with the chromatin marker H2B-mcherry and the respective RNAi-resistant HA-tagged rescue constructs. Whereas 11% of control mitotic cells showed association of ER with the metaphase plate, this fraction increased to 88% upon REEP3/4 depletion. Remarkably, this defect could be rescued to 14% by expression of wild type REEP4, but not by REEP4dMT, which yielded 84% of cells displaying ER association with mitotic chromosomes (Figure 4C). Expression of REEP4dMT alone did not lead to ER accumulation on metaphase chromosomes (Figure S3B). As an alternative means to quantify rescue efficiency, we plotted GFP-Sec61β fluorescence intensity at the metaphase plate as a fraction of total cellular GFP-Sec61β fluorescence (Figure 4D). In control cells, GFP-Sec61β intensity at the metaphase plate accounted for 2.9% of total cellular fluorescence. In REEP3/4 RNAi cells, this fraction was 13.9% but could be rescued to 2.4% by expression of wild type REEP4. In contrast, expression of REEP4dMT did not rescue, as 13.2% of total GFP-Sec61β fluorescence was associated with metaphase chromosomes (Figure 4D), corroborating the finding that wild type REEP4-HA was functional whereas REEP4dMT was not.
These results show that a short cytoplasmic region that connects the two hydrophobic domains of REEP4 is critical for microtubule binding and essential for the function of REEP4 in clearing ER from metaphase chromosomes, and indicate that REEP3/4 function through linking ER to microtubules.
Discussion
Our findings indicate that REEP3/4 ensure proper cell division and nuclear envelope reassembly by sequestering ER away from chromosomes during mitotic metaphase (Figure 4E). Previous models for nuclear envelope reformation proposed that cell cycle-dependent phosphorylation/dephosphorylation events regulate the association of nuclear envelope membrane with chromosomes throughout mitosis (Foisner and Gerace, 1993). We show here that an additional, microtubule-dependent mechanism acts during metaphase to ensure clearance of ER from chromosomes before the onset of nuclear envelope formation. Our results further indicate that clearance of ER from chromosomes is required for (1) proper chromosome segregation during anaphase, potentially because membranes in the chromosome area interfere with microtubule-kinetochore interactions and chromosome movements, (2) correct separation of daughter cells, and (3) formation of an interphase nucleus free of aberrant membranes.
Even though REEP3/4 RNAi had no obvious effect on the growth and viability of HeLa cells grown in culture, it is conceivable that insufficient separation of daughter cells could be detrimental to the generation and maintenance of a well-organized tissue. In addition, the excessive accumulation of nuclear envelope proteins in the nuclear interior could impair nuclear transport and other nuclear processes, which again may affect cells in an organism more than cultured cells. Interestingly, REEP4 was identified in the Mitocheck genome-wide RNAi screen as a factor whose depletion caused an increase in binucleate cells (Neumann et al., 2010). While we did not observe more binucleate cells upon REEP3/4 knockdown, perhaps due to differences in culture conditions, the phenotype reported by Neumann et al. is consistent with our results and might be a manifestation of the daughter nuclei separation defect we observed.
Our data strongly suggest that microtubule binding of REEP3/4 is important for their role in clearing ER from chromosomes. Presumably, microtubule minus end-directed transport is needed for REEP3/4 to remove ER from the chromosomes and to support ER focusing at spindle poles, where microtubule minus-ends are clustered. We do not yet know whether microtubule binding of REEP3/4 is direct or mediated by interaction partners, e.g. microtubule motors. If REEP3/4 bound directly, they might harness microtubule flux toward the spindle poles to transport ER to minus ends. If REEP3/4 associated with microtubules indirectly through binding to other proteins, the microtubule minus-end directed motor dynein would be a likely candidate. We have not been able to detect interactions of REEP4 and members of the dynein complex (our unpublished observations), but further studies on REEP3/4 binding partners and specifically the differences between REEP3/4 and REEP4dMT will shed light on these questions.
In a recent paper, Smyth et al. showed that ER targets the mitotic spindle when microtubule plus tip association of the ER protein STIM1 persists during mitosis (Smyth et al., 2012). Importantly, no accumulation of ER membrane at the metaphase plate was observed under these conditions, indicating that solely the aberrant presence of ER in the spindle region does not lead to its premature association with chromatin. This supports a specific function for REEP3/4 in the timing of ER association with chromosomes and nuclear envelope reformation.
While it seems intuitive that membrane-bound organelles should be excluded from the spindle and chromosomes during mitosis so that they do not interfere with chromosome segregation or are incorporated into newly forming nuclei, the exclusion mechanism(s) involved have remained unknown. REEP3 and REEP4 play a key role in clearing organelle membrane from metaphase chromosomes and we predict that further study of proteins that link organelles to microtubules will considerably advance our understanding of the dramatic cellular reorganization that occurs during mitosis.
Experimental procedures
Isolation of REEP4
CSF-arrested cytoplasmic extracts from Xenopus laevis eggs were prepared as described (Hannak and Heald, 2006). Eight to ten milliliters of egg extract were used per purification. Membranes were recovered by a high-speed spin (200,000 × g, 1.5 hours at 4°C), re-suspended in extract buffer and purified by centrifugation through a sucrose cushion. The membrane pellet was re-suspended and further purified by flotation over high sucrose buffer at 280,000 × g, for 18 hours at 4°C in a SW40 rotor. The purified membrane fraction was treated with 25 mM CHAPS for 30 minutes at 4°C to solubilize membranes, followed by a clarifying spin at 200,000 × g for 1 hour at 4°C. The supernatant was incubated with taxol-stabilized microtubules for 30 minutes at room temperature and the sample was centrifuged consecutively through three sucrose cushions to remove contaminants. The final pellet was treated with elution buffer containing 1 M KCl. The eluate was precipitated by methanol-chloroform and the pellet analyzed by MudPIT mass spectrometry.
REEP4 was isolated in 4 out of 5 purifications from CSF (meiotic arrested) egg extracts. Sequence coverage for REEP4 in these purifications was between 19% and 39%.
For details on the mass spectrometry analysis see the supplementary experimental procedures.
Cell culture and siRNAs
HeLa and HEK293T cells were cultured in DMEM supplemented with 10% fetal bovine serum at 37° C in a humidified 5% CO2 incubator. For REEP3/4 depletion, Ambion siRNA s37271 was used. By quantitative PCR this siRNA reduced REEP3 mRNA by >95% and REEP4 mRNA by >90% within 72 hours after transfection. Protein levels of REEP4 were reduced by 70 % after 48 hours (when most experiments were performed) as determined by Western blot (Figure S1D). Control cells were transfected with Silencer Negative Control No. 1 siRNA (AM4611). For a description of plasmids and transfection procedures used in this study see the Supplementary Experimental Procedures.
Antibodies, immunofluorescence, and light microscopy
Rabbit anti-REEP4 antibody was generated using a 6×His-fusion protein of the C-terminal 100 amino acids of REEP4 (aa 158–257) as antigen and the serum obtained was affinity-purified using the same fragment. Mouse anti-HA antibody HA.11 (Covance) was used 1:100. Anti-LaminB1 antibody ab16048 (Abcam) was used 1:500. Microtubules were stained with DM1α antibody from Sigma (T9026) diluted 1:500. For immunofluorescence, cells grown on coverslips were fixed 48 hours after siRNA-transfection with 3% formaldehyde/0.02 % glutaraldehyde, quenched with 0.5 mg/ml NaBH4 and permeabilized with 0.1% Triton X-100. For staining of endogenous REEP4, cells were fixed in ice-cold methanol for 10 minutes. Live cell imaging was performed 36 to 42 hours after siRNA transfections. Imaging was performed on either a Zeiss LSM 710 microscope or a Nikon eclipse Ti microscope equipped with an Andor Clara CCD camera, Yokogawa spinning disk head and diode lasers with 488 nm and 561 nm laser lines. Both microscopes were equipped with an environmental chamber.
Image analysis and quantifications
For quantification of daughter nuclei separation, cells late in cell division were chosen which had their cytokinetic furrows constricted to 1.0 µm or less, a straight line was aligned along the cytoplasmic bridge between the daughter cells and the distance between the daughter nuclei along this line was measured using Lamin B1 staining as the border of each daughter nucleus. Statistical significance was determined using a two-sided, unpaired Welch’s t-test.
For figures 2D and 4D, GFP-Sec61β fluorescence intensity at the metaphase plate was quantified using the software ImageJ by thresholding the H2B-mcherry signal and thus identifying the region occupied by the chromosomes. GFP-Sec61β signal in the metaphase plate region was measured in the corresponding channel.
For the quantification of changes in ER amounts during metaphase (Figure 2D), mean pixel intensities for GFP-Sec61β fluorescence were determined. Images were corrected for background and for bleaching that occurred over the time frame of the acquisition. The initial frame (12 minutes before anaphase onset) was chosen as a reference and the percent change in total GFP-fluorescence at the metaphase plate in the following frames relative to the initial frame was calculated.
For the quantification of rescue efficiencies of RNAi-resistant REEP4 and REEP4dMT (Figure 4D), total GFP-Sec61β fluorescence was measured in addition to the fluorescence intensity at the metaphase plate and the ratio of fluorescence intensity at the chromosomes to total cellular intensity was calculated.
To determine statistical significance, two-tailed, unpaired student’s t-tests were performed on these data sets.
For the quantification of Western blot signal to determine the fraction of microtubule-bound REEP4-HA and REEPdMT-HA (Figure 4B) the digital image of the developed blot obtained from a LI-COR (LI-COR Biosciences) scan was thresholded, the regions with signal identified and the integrated intensity measured. Intensity directly above and below the band of interest was also measured and the average of these two values was used to subtract the background intensity from the measured signal.
Microtubule co-pelleting assays
For microtubule co-pelleting assays, HEK293T cells or HeLa cells were transfected with the respective constructs 48 hours before the experiment, lysed by passing twenty times through a 30-gauge needle and centrifuged for 15 minutes at 400 × g. The postnuclear supernatant was treated with 0.5% Triton X-100 for 30 minutes at room temperature and centrifuged for 1 h at 200,000 × g at 25 °C. The supernatant was supplemented with 1 mM GTP, split in two and incubated with either buffer or with taxol-stabilized microtubules for 30 minutes at room temperature. Samples were centrifuged through a sucrose cushion for 30 minutes at 80,000 × g at 25 °C. Supernatants and pellets were adjusted to equal volumes and equivalent amounts were analyzed for presence of REEP3, REEP4, REEP4dMT or REEP5 by immunoblotting and detection of the HA-tag or endogenous REEP4.
Supplementary Material
Acknowledgements
We are grateful to Elisa Dultz, Sebastian Schuck and Daniel Levy for critical reading of the manuscript as well as for discussions and advice throughout this study.
This work was supported by postdoctoral fellowships from the Jane Coffin Childs Memorial Fund for Medical Research and the American Heart Association to A. S. and NIH grant R01 GM057839 to R.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson DJ, Hetzer MW. Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J. Cell Biol. 2008;182:911–924. doi: 10.1083/jcb.200805140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudouin J, Gerlich D, Daigle N, Eils R, Ellenberg J. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell. 2002;108:83–96. doi: 10.1016/s0092-8674(01)00627-4. [DOI] [PubMed] [Google Scholar]
- Foisner R, Gerace L. Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell. 1993;73:1267–1279. doi: 10.1016/0092-8674(93)90355-t. [DOI] [PubMed] [Google Scholar]
- Grigoriev I, Gouveia SM, van der Vaart B, Demmers J, Smyth JT, Honnappa S, Splinter D, Steinmetz MO, Putney JW, Jr, Hoogenraad CC, et al. STIM1 is a MT-plus-end-tracking protein involved in remodeling of the ER. Curr. Biol. 2008;18:177–182. doi: 10.1016/j.cub.2007.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannak E, Heald R. Investigating mitotic spindle assembly and function using Xenopus laevis egg extracts. Nat. Protoc. 2006;1:2305–2314. doi: 10.1038/nprot.2006.396. [DOI] [PubMed] [Google Scholar]
- Hetzer MW. The nuclear envelope. Cold Spring Harb. Perspect. Biol. 2010;2(3):a000539. doi: 10.1101/cshperspect.a000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Ladinsky MS, Kirchhausen T. Cisternal organization of the endoplasmic reticulum during mitosis. Mol. Biol. Cell. 2009;20:3471–3480. doi: 10.1091/mbc.E09-04-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhas A, Goulbourne C, Vaux DJ. The nucleoplasmic reticulum: form and function. Trends Cell Biol. 2011;21:362–373. doi: 10.1016/j.tcb.2011.03.008. [DOI] [PubMed] [Google Scholar]
- Muhlhausser P, Kutay U. An in vitro nuclear disassembly system reveals a role for the RanGTPase system and microtubule-dependent steps in nuclear envelope breakdown. J. Cell Biol. 2007;178:595–610. doi: 10.1083/jcb.200703002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Zhu PP, Parker RL, Blackstone C. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J. Clin. Invest. 2010;120:1097–1110. doi: 10.1172/JCI40979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre P, Scheel J, Rickard JE, Kreis TE. CLIP-170 links endocytic vesicles to microtubules. Cell. 1992;70:887–900. doi: 10.1016/0092-8674(92)90240-d. [DOI] [PubMed] [Google Scholar]
- Puhka M, Joensuu M, Vihinen H, Belevich I, Jokitalo E. Progressive sheet-to-tubule transformation is a general mechanism for endoplasmic reticulum partitioning in dividing mammalian cells. Mol. Biol. Cell. 2012;23:2424–2432. doi: 10.1091/mbc.E10-12-0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhka M, Vihinen H, Joensuu M, Jokitalo E. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J. Cell Biol. 2007;179:895–909. doi: 10.1083/jcb.200705112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa-Ferreira C, Munro S. Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev. Cell. 2011;21:1171–1178. doi: 10.1016/j.devcel.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salina D, Bodoor K, Eckley DM, Schroer TA, Rattner JB, Burke B. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 2002;108:97–107. doi: 10.1016/s0092-8674(01)00628-6. [DOI] [PubMed] [Google Scholar]
- Smyth JT, Beg AM, Wu S, Putney JW, Jr, Rusan NM. Phosphoregulation of STIM1 leads to exclusion of the endoplasmic reticulum from the mitotic spindle. Curr. Biol. 2012;22:1487–1493. doi: 10.1016/j.cub.2012.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA. Communication between the cytoskeleton and the nuclear envelope to position the nucleus. Mol. Biosyst. 2007;3:583–589. doi: 10.1039/b703878j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- Yadav S, Puthenveedu MA, Linstedt AD. Golgin160 recruits the dynein motor to position the Golgi apparatus. Dev. Cell. 2012;23:153–165. doi: 10.1016/j.devcel.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.