Abstract
Objective. The goal of this study was to define viral kinetics after initiation of raltegravir (RAL)–based antiretroviral therapy (ART).
Methods. ART-naive patients received RAL, tenofovir disoproxil fumarate, and emtricitabine for 72 weeks. Human immunodeficiency virus type 1 (HIV-1) RNA were measured by ultrasensitive and single-copy assays, and first (d1)–, second (d2)–, and, third (d3)–phase decay rates were estimated by mixed-effects models. Decay data were compared to historical estimates for efavirenz (EFV)– and ritonavir/lopinavir (LPV/r)–based regimens.
Results. Bi- and tri-exponential models for ultrasensitive assay (n = 38) and single-copy assay (n = 8) data, respectively, provided the best fits over 8 and 72 weeks. The median d1 with ultrasensitive data was 0.563/day (interquartile range [IQR], 0.501–0.610/day), significantly slower than d1 for EFV-based regimens [P < .001]). The median duration of d1 was 15.1 days, transitioning to d2 at an HIV-1 RNA of 91 copies/mL, indicating a longer duration of d1 and a d2 transition at lower viremia levels than with EFV. Median patient-specific decay estimates with the single-copy assay were 0.607/day (IQR, 0.582–0.653) for d1, 0.070/day (IQR, 0.042–0.079) for d2, and 0.0016/day (IQR, 0.0005–0.0022) for d3; the median d1 duration was 16.1 days, transitioning to d2 at 69 copies/mL. d3 transition occurred at 110 days, at 2.6 copies/mL, similar to values for LPV/r-based regimens.
Conclusions. Models using single-copy assay data revealed 3 phases of decay with RAL-containing ART, with a longer duration of first-phase decay consistent with RAL-mediated blockade of productive infection from preintegration complexes.
Clinical Trials Registration. NCT00660972.
Keywords: raltegravir, viral decay, single copy assay, Roche ultrasensitive assay, ACTG A5248, ACTG A5160s, ACTG A5166s
Infection with human immunodeficiency virus type 1 (HIV-1) is characterized by persistent high levels of viremia resulting from a dynamic equilibrium between virus production and clearance and the associated rapid and continuous turnover of the virus population (approximately half of the circulating virus population is replaced with newly produced virus daily) [1, 2]. When this quasi–steady state is perturbed by initiation of potent antiretroviral therapy (ART), the resultant decrease in plasma viremia proceeds through several phases [3]. Decay during the first 7–10 days, which is dominated by loss of short-lived productively infected cells (eg, activated CD4+ T lymphocytes), is characterized by the parameter estimate d1, with a half-life of approximately 1 day. A slower second phase (d2), with an average half-life of 14 days, is believed to represent the loss of longer-lived, productively infected cells. An even slower third phase (d3) has been revealed by HIV-1 RNA assays with single-copy sensitivity and may represent decay of latently infected resting CD4+ T cells that become activated stochastically to produce HIV-1 but go unobserved because plasma HIV-1 RNA levels have usually already fallen below the lower limit of quantification for standard assays [4, 5]. A study using an HIV-1 RNA assay with single-copy sensitivity estimated the half-life for d3 to be 39–63 weeks, similar to that of latently infected CD4+ T cells (44 weeks), in patients receiving 2 nucleoside analog reverse-transcriptase inhibitors (NRTIs) and ritonavir-boosted lopinavir (LPV/r) [6, 7]. In that same study [6], a fourth phase (d4) was described as occurring 4–5 years after ART initiation and extending through 7 years of follow-up, during which there was no further decline in viremia.
Raltegravir (RAL) is an integrase strand transfer inhibitor [8]. Clinical trials show that treatment-naive patients who initiate ART with a RAL-containing regimen achieve plasma HIV-1 RNA levels below the limit of detection by conventional assays (≤50 copies/mL) faster than patients treated with an efavirenz (EFV)–containing regimen [9]. The reasons for the apparently faster reduction in viremia are unclear. One study found no significant difference in d1 between patients receiving RAL and EFV-containing regimens but noted that the transition from d1 to d2 occurred later and at lower plasma HIV-1 RNA levels in the RAL recipients [10]. That analysis, however, was based on data from a 10-day RAL monotherapy trial with frequent quantification of plasma HIV-1 RNA and from a phase 2 trial of tenofovir disoproxil fumarate plus emtricitabine (TDF/FTC) plus either RAL or EFV in which there was sparse sampling after ART initiation, making it difficult to estimate accurately d1 and the time of transition to d2. Mathematical models of viral decay suggest that the rapid suppression of plasma HIV-1 RNA observed with RAL-containing ART reflects the stage of the viral life cycle at which the drug acts [11].
To estimate viral decay parameters more precisely and to explore the underlying mechanism for more-rapid suppression of viremia to <50 copies/mL after initiation of RAL-containing ART, we conducted a prospective, single-arm study to estimate the various phases of viral decay in treatment-naive subjects initiating ART with RAL and TDF/FTC.
METHODS
Study Design
AIDS Clinical Trials Group (ACTG) A5248 was a prospective, open-label, multicenter, single-arm, 72-week pilot study designed to estimate viral decay kinetic parameters in ART-naive subjects initiating treatment with RAL and FTC/TDF. Eligible HIV-1–infected patients were ≥18 years of age with plasma HIV-1 RNA levels of >10 000 to ≤300 000 copies/mL. Patients were ineligible if their screening genotype revealed evidence of major mutation(s) associated with resistance to NRTIs, nonnucleoside reverse-transcriptase inhibitors (NNRTIs), or protease inhibitors (PIs). The study protocol was approved by the institutional review board at each of the 15 participating ACTG clinical research sites, and all subjects provided signed informed consent.
Patients meeting entry criteria underwent HIV-1 RNA testing before entry, at entry, and on days 2, 7, 10, 14, 21, 28, and 56 and weeks 12, 16, 20, 24, 36, 48, 60, and 72 after entry. Plasma was assayed for HIV-1 RNA by using the Amplicor HIV-1 Monitor, version 1.5, UltraSensitive protocol (≤50 copies/mL; Roche Molecular Systems, Branchburg, NJ; hereafter, the ultrasensitive assay), and, in a subset of patients, by a quantitative real-time reverse-transcriptase–initiated polymerase chain reaction assay that can detect a single copy of HIV-1 RNA in a plasma sample (hereafter, the single-copy assay) [6]. Of all 11 subjects in ACTG substudy A5249s (which was designed to estimate the time of onset of virologic decay) and an additional random sample of 8 subjects who did not exhibit virologic failure (defined as an HIV-1 RNA level of ≥1000 copies/mL 16–24 weeks after ART initiation or ≥200 copies/mL on or after week 24), 11 were eligible for inclusion in single-copy assay models (ie, they exhibited neither inefficient amplification of virus in day 0 samples nor virologic failure).
Because accurate estimation of viral decay rates requires that subjects take study drugs as prescribed, plasma HIV-1 RNA values were excluded from modeling if they were collected after a single missed dose on or before day 14, if subjects had a second missed dose during days 15–56, if they had a seventh cumulative missed dose after week 8, or if their viral load underwent an analytic rebound (defined as an increase of >0.3 log10 copies/mL from the previous observation, excluding oscillations below 1 copy/mL), to avoid model-fitting difficulties due to nonmonotonicity. Day 0 HIV-1 RNA values (geometric mean of values before entry and at entry) were excluded from ultrasensitive assay models if they were below the day 2 value. One of 39 subjects with ultrasensitive assay HIV-1 RNA data was excluded from ultrasensitive assay models because of a missed dose before day 2. Exclusions for missed doses resulted in too few single-copy assay values from weeks 8 through 72 for 3 subjects; thus, models to week 72 are based on data from 8 subjects. Calculated day/week was used as the independent variable; results collected outside protocol-specified windows were retained.
Adherence was assessed by patient self-report; participants were given medication diaries to facilitate accurate reporting of doses taken during the first 56 days of treatment.
Statistical Methods
Statistical Power
The original accrual target was 34 subjects, a sample size expected to yield plasma HIV-1 RNA data from 30 subjects for model fitting and to provide a 95% confidence interval (CI) around the estimated d1 no wider than ±0.09/day; expected precision was ±0.06/day. If a subject changed their regimen or missed any dose in the first 14 days, an additional subject was accrued. For every 3 subjects who changed their regimen or missed ≥2 doses between days 15 and 56, an additional subject was accrued. Subjects altering their treatment regimen or missing doses between weeks 12 and 72 were not replaced.
Estimating and Comparing Viral Decay Rates
To estimate virus decay rates, mono-, bi- and tri-exponential nonlinear mixed-effects models were fitted using R software (version 2.9.2). Several approaches were taken for handling plasma HIV-1 RNA levels below the assay lower limit of detection (50 copies/mL for the ultrasensitive assay and 1 copy/mL for the single-copy assay). A multiple imputation approach, in which values below the detection limit are replaced with values imputed from sequential model fits, exhibits minimal bias [12]. For fits to ultrasensitive assay data, we also considered (1) a fit excluding all values after the first value below detection and replacing the first such value with 50 (L1–50) or 25 (L1–25) copies/mL and (2) a fit excluding the first value below detection (L0). When fitting models to single-copy assay data, we estimated parameters in 2 ways: (1) via multiple imputation and (2) for consistency with other studies, by retaining all values and setting those reported below detection to 0.5 copies/mL.
Model fits were compared via log-likelihood values (for which higher values indicate a better fit), Akaike and Bayesian information criteria (for which smaller values indicate a better fit), and estimated error variance (for which a lower value indicates a better fit); exploratory plots and qualitative results were also considered in selecting the best model.
Subject-specific empirical Bayesian estimates of d1 and d2 from the A5248 RAL-based regimen were compared to corresponding subject-specific decay-rate estimates from the EFV plus 2 NRTI arms of ACTG A5166s [13] and A5160s [14], using a 2-sided Wilcoxon rank sum test unadjusted for multiple comparisons. Subject-specific measurements derived from empirical Bayesian estimates included transition time (defined as the time at which production of HIV-1 RNA decay originating from short- and longer-lived cells is equal), the predicted HIV-1 RNA level at these transition times, and the times at which the predicted HIV-1 RNA level was <50 copies/mL. Subject-specific empirical Bayes estimates were summarized by medians and interquartile ranges.
Type 1 Error Rates and Confidence Intervals (CIs)
Type I error rates were set at 5%; CIs were constructed to have 95% coverage. For inference about dichotomous end points, Clopper-Pearson exact binomial CIs were reported.
RESULTS
Baseline Characteristics
A total of 39 adult subjects were enrolled in A5248. Data from 1 subject were excluded because of a missed dose prior to day 2; 11% were females, 47% were non-Hispanic white, 26% were non-Hispanic black, and were 18% were Hispanic. Data for the single-copy assay, from a subset of 8 patients (1 female, 5 non-Hispanic white individuals, and 1 non-Hispanic black individual, and 2 Hispanic individuals), were also analyzed. Of the 25 subjects enrolled in the previously reported 3-drug EFV arm of A5160s, 13 were female, 11 were non-Hispanic white, 9 were non-Hispanic black, and 4 were Hispanic. Of the 18 subjects enrolled in A5166s, 7 were female, 4 were non-Hispanic white, 5 were non-Hispanic black, and 8 were Hispanic (Table 1).
Table 1.
Baseline Characteristics for 3 ACTG Viral Decay Studies (A5248, A5160s, and A5166s)
| Characteristic | RAL + FTC/TDF A5248 (n = 38) | RAL + FTC/TDF A5248 SCA (n = 8) | EFV + 2NRTIs A5160Ss (n = 25) | ZDV/3TC/EFV A5166s (n = 18) |
|---|---|---|---|---|
| Female sex | 4 (11) | 1 (13) | 13 (52) | 7 (39) |
| Age, y | 44 (32–50) | 49 (43–52) | 40 (37–47) | 36 (29–44) |
| Race/ethnicitya | ||||
| White | 18 (47) | 5 (63) | 11 (44) | 4 (22) |
| Black | 10 (26) | 1 (13) | 9 (36) | 5 (28) |
| Hispanic | 7 (18) | 2 (25) | 4 (16) | 8 (44) |
| Other | 3 (8) | 1 (4) | 1 (6) | |
| Plasma HIV-1 RNA load, log10 copies/mL | 4.6 (4.4–4.8) | 4.7 (4.4–5.0) | 4.8 (4.5–4.9) | 4.7 (4.5–5.2) |
| Plasma HIV-1 RNA load ≥ 100 000 copies/mL | 4 (11) | 2 (25) | 8 (32) | 6 (33) |
| CD4+ T-cell count, cells/mm3 | 258 (196–348) | 200 (58–297) | 261 (148–352) | 203 (68–335) |
Data are no. (%) of subjects or median (interquartile range). Data for A5160s and A5166s are from Haubrich et al [14] and Kuritzkes et al [13], respectively.
Abbreviations: EFV, efavirenz; FTC, emtricitabine; HIV-1, human immunodeficiency virus type 1; NRTIs, nucleoside reverse-transcriptase inhibitors; RAL, raltegravir; TDF, tenofovir disoproxil fumarate; ZDV, zidovudine; 3TC, lamivudine.
a Data in some columns fail to sum to 100% because of rounding.
The median baseline plasma ultrasensitive assay HIV-1 RNA level was 4.6 log10 copies/mL for subjects in A5248 (4.7 log10 copies/mL for the single-copy assay subset) and 4.8 log10 copies/mL and 4.7 log10 copies/mL for subjects in A5160s and A5166s, respectively. The median baseline CD4+ T-cell count was 258 cells/mm3 for A5248 (200 cells/mm3 for the single-copy assay subset) and 261 cells/mm3 and 203 cells/mm3, respectively, for A5160s and A5166s.
Overall, the baseline characteristics of subjects enrolled into the current study were similar to those of subjects enrolled in A5160s and A5166s, except for a greater proportion of female subjects in the latter 2 studies.
Virologic Suppression
By day 56, 85% of subjects in ACTG A5248 had plasma HIV-1 RNA levels of <50 copies/mL, compared with 17% and 57% of subjects in A5160s and A5166s, respectively (Supplementary Table 1). These differences persisted after stratification by screening plasma HIV-1 RNA level (data not shown). These results are consistent with those of previous studies reporting that subjects receiving RAL-based ART reach undetectable HIV-1 RNA levels earlier than those receiving 2 NRTIs plus EFV [9]. As expected, at each time point, the plasma HIV-1 RNA was detectable in a greater proportion of subjects by the single-copy assay than by the ultrasensitive assay.
Assessment of Viral Dynamics by the Ultrasensitive Assay, Weeks 0–8
The decay in HIV-1 RNA over weeks 0–8 was best described by a bi-exponential model (data not shown). Due to the rapid virologic suppression by RAL plus FTC/TDF seen in previous studies, we hypothesized that first phase decay would be steeper (d1 larger) for RAL- than for EFV-based regimens. However, d1 estimates for EFV-based regimens were significantly larger than those for RAL: median d1 values for A5160s and A5248 were 0.652/day and 0.590/day, respectively (P = .015, by the L1–25 approach); median d1 values for A5166s and A5248 were 0.677/day and 0.563/day, respectively (P < .001, by the multiple imputation approach; Table 2). (The model using multiple imputations for A5160s did not converge.) However, d1 estimates for EFV-based regimens were significantly larger than those for RAL-based regimens (Figure 1): median times to suppression for A5160s and A5248 were 74 and 22 days, respectively (P < .001, by the L1–25 approach); for A5166s and A5248, median times to suppression were 55 days and 28 days (P < .001, by the multiple imputation approach).
Table 2.
Estimates of d1 and d2 per Day Based on Ultrasensitive Plasma Human Immunodeficiency Virus Type 1 (HIV-1) RNA Measurements After Initiating Antiretroviral Therapy, Weeks 0–8
| Handling of VLs Below LLD | ACTG Study | VLs/ Subjects, No. |
d1 |
d2 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
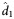 (95% CI) (95% CI) |
Median (IQR) | t1/2, Days, Median (IQR) | Pa |
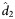 (95% CI) (95% CI) |
Median (IQR) | t1/2, Days, Median (IQR) | Pa | |||
| MIb | A5248 | 276/38 | 0.561 (.521–.060) | 0.563 (0.501–0.610) | 1.23 (1.14–1.39) | 0.042 (.037–.047) | 0.043 (0.041–0.044) | 16.09 (15.59–16.91) | ||
| A5166s | 103/18 | 0.684 (.594–.774) | 0.677 (0.630–0.769) | 1.02 (0.90–1.10) | <.001 | 0.055 (.041–.069) | 0.052 (0.040–0.072) | 13.46 (19.65–17.28) | .026 | |
| L1–25c | A5248 | 244/38 | 0.587 (.543–.631) | 0.590 (0.521–0.640) | 1.17 (1.08–1.33) | 0.082 (.062–.102) | 0.084 (0.055–0.105) | 8.27 (6.57–12.50) | ||
| A5160s | 142/25 | 0.661 (.564–.759) | 0.652 (0.592–0.728) | 1.06 (0.95–1.17) | .015 | 0.037 (.027–.048) | 0.033 (0.028–0.045) | 21.30 (15.47–24.60) | <.001 | |
Abbreviations: ACTG, AIDS Clinical Trials Group; d1, first phase of viral decay; d2, second phase of viral decay; 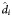 , population based fixed-effects estimate; IQR, interquartile range; LLD, lower limit of detection; t1/2, half-life; VL, viral load.
, population based fixed-effects estimate; IQR, interquartile range; LLD, lower limit of detection; t1/2, half-life; VL, viral load.
a By the Wilcoxon rank sum test.
b VLs below the LLD (50 copies/mL) were handled by multiple imputation (MI).
c The first VL below the LLD was replaced with a value of 25 copies/mL (half the LLD), and all subsequent VLs were excluded from model fits. (The model using multiple imputation for A5160s did not converge.)
Figure 1.
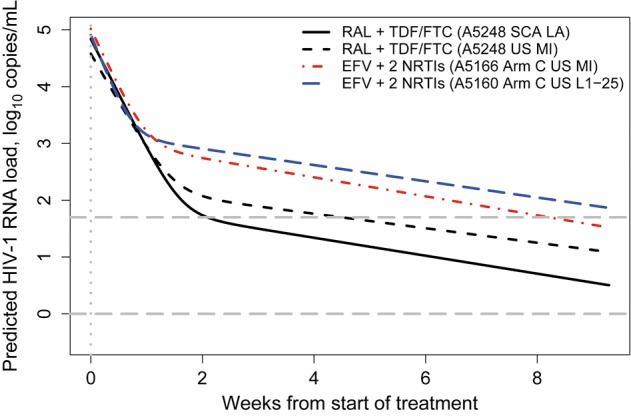
Predicted viral decay curves (weeks 0–8) across 3 AIDS Clinical Trials Group studies, using a single-copy assay (SCA) and an ultrasensitive assay (US). Plasma human immunodeficiency virus type 1 (HIV-1) RNA load, in log10 copies/mL, in treatment-naive subjects initiating highly active antiretroviral therapy, predicted by population-average bi-exponential mixed-effects models of data from weeks 0–8. Solid black line, SCA results for A5248 subjects initiating raltegravir (RAL) plus emtricitabine/tenofovir disoproxil fumarate (FTC/TDF); all plasma HIV-1 RNA values were retained; values of 0.5 copies/mL were imputed for those below the lower limit of detection (LLD) of 1 copy/mL. Dashed black line, US results for A5248 subjects initiating RAL plus FTC/TDF; multiple imputation of plasma HIV-1 RNA values below the LLD of 50 copies/mL. Dashed red line, US results for A5166s subjects in arm C, initiating efavirenz (EFV) plus 2 nucleoside reverse-transcriptase inhibitors (NRTIs); multiple imputation of plasma HIV-1 RNA values below the LLD of 50 copies/mL. Dashed blue line, US results for A5160s subjects in arm C, initiating EFV plus 2 NRTIs; only the first plasma HIV-1 RNA values below the LLD of 50 copies/mL was retained (values of 25 copies/mL were imputed), with all other data excluded from model fit (multiple imputation fit failed to converge). Dashed gray lines show US and SCA LLDs (log10 50 and log10 1 copies/mL, respectively). Dotted gray line shows time of treatment initiation. Rate constants for the first and second phases of decay are reported in Table 2 (for the models using data from the UltraSensitive HIV-1 Monitor Assay) and Table 3 (for the model using data from the SCA). Abbreviation: MI, multiple imputation.
Estimates of d2 were highly sensitive to the method of handling HIV-1 RNA values below the limit of quantification. By using the multiple imputation approach, which is preferred because it introduces least bias, d2 for RAL plus FTC/TDF (median, 0.043/day) was significantly slower than for the EFV-based regimen in A5166s (median, 0.052/day; P = .026). However, by using the L1–25 approach, d2 for RAL plus FTC/TDF (median, 0.084/day) was significantly faster than that for the EFV-based regimen in A5160s (median, 0.033/day; P < .001; Table 2). We believe the estimates from multiple imputation are stronger; in the presence of so many A5248 values below the detection limit, the L1–25 approach overestimates the second-phase decay rate. Additional model fit statistics for A5248 data are shown in Supplementary Table 2.
Because differences in d1 did not explain the more rapid virologic suppression observed for subjects receiving RAL-based therapy, we asked whether the transition time from d1 to d2 differed across studies. We found that the estimated time of transition from d1 to d2 was later for RAL plus FTC/TDF (median, 13.2 days [interquartile range {IQR}, 12.5–15.6 days], by the L1–25 approach; median, 15.1 days [IQR, 13.8–17.2 days], by the multiple imputation approach), compared with EFV plus 2 NRTI (A5160s: median, 9.3 days [IQR, 7.9–10.4 days], by the L1–25 approach; A5166s: median, 11.2 days [IQR, 9.7–13.3 days], by the multiple imputation approach). The d1 to d2 transition also occurred at a lower HIV-1 RNA level in subjects receiving RAL-based therapy (2.07 log10 copies/mL, by the L1–25 approach, and 1.96 log10 copies/mL, by the multiple imputation approach [this study] vs 2.99 log10 copies/mL [A5160s] and 2.77 log10 copies/mL [A5166s]). These findings are consistent with RAL blocking the transition of cells from a state of “preintegration latency” to productively infected, longer-lived cells, thereby reducing the pool of cells contributing to d2.
Assessment of Viral Dynamics by the Single-Copy Assay, Weeks 0–8 and 0–72
The rapid decline in plasma HIV-1 RNA load among subjects receiving a RAL-based regimen resulted in a large proportion of HIV-1 RNA determinations that were below the limit of detection. As noted above, the approach taken to handle censored data had a large effect on estimates of the parameters of viral decay kinetics. We therefore applied the more sensitive single-copy assay to samples from a subset of patients to obtain more accurate estimates of viral decay parameters. Baseline characteristics of patients with single-copy assay data were similar to those of the study population overall (Table 1). For single-copy assay data from weeks 0–8 and weeks 0–72, bi- and tri-exponential models, respectively, proved the best fit (Table 3). Median patient-specific decay estimates were 0.607/day (IQR, 0.582–0.653) for d1, 0.070/day (IQR, 0.042–0.079) for d2, and 0.0016/day (IQR, 0.0005–0.0022) for d3. d1 estimates based on single-copy assay data were larger than those based on ultrasensitive assay data (0.563/day [IQR, 0.501–0.610]; P = .06, by the Wilcoxon rank sum test). The estimated transition from d1 to d2 occurred 16.1 days after initiating ART, at a median predicted HIV-1 RNA level of 69 copies/mL (1.8 log10 copies/mL), compared with 15.1 days and 91 copies/mL (1.96 log10 copies/mL) for ultrasensitive assay viral load data (Figure 1). The estimated transition from d2 to d3 occurred 110 days (15.7 weeks) after ART initiation, at a median predicted HIV-1 RNA level of 2.6 copies/mL (IQR, 2.5–3.5; Figure 2). The (bootstrap) mean third-phase half-life was 497.84 days (71 weeks), which falls near the point estimate of 69 weeks for the LPV/r-based regimen of M98–863 [6], and our bootstrap-based CIs exhibit substantial overlap with the CIs reported for M98–863 and M97–720 (Table 3). No fourth decay phase (d4) was evident by week 72.
Table 3.
Fixed-Effects Estimates of d1, d2, and d3 With Tri- and Bi-Exponential Decay Models Based on Single-Copy Assay Viral Load (VL) Measurements
| Period | Phases, No. | First-Phase Decay, per Day |
Second-Phase Decay, per Day |
Third-Phase Decay, per Day |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
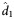 (95% CI) (95% CI) |
Subject-Specific Estimates |
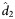 (95% CI) (95% CI) |
Subject-Specific Estimates |
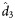 (95% CI) (95% CI) |
Subject-Specific Estimates |
||||||||
| d1, Median (IQR) |
t1/2, Days |
d2, Median (IQR) |
t1/2, Days |
d3, Median (IQR) |
t1/2, Days |
||||||||
| Median (IQR) | Mean (95% CI) | Median (IQR) | Mean (95% CI) | Median (IQR) | Mean (95% CI) | ||||||||
| Weeks 0–72 | 3 | 0.615 (−.444 to 1.674) | 0.607 (0.582–0.653) | 1.14 (1.07–1.19) | 1.16 (1.11–1.32) | 0.065 (−.994 to 1.124) | 0.070 (0.042–0.079) | 9.19 (7.43–11.31) | 12.19 (9.92–18.49) | 0.0016 (−1.0574 to 1.0606) | 0.0016 (0.0005–0.0022) | 340.53 (243.53–757.92) | 497.84 (376.00–843.49) |
| Weeks 0–8 | 2 | 0.637 (−.366 to 1.639) | 0.614 (0.580–0.748) | 1.13 (0.93–1.20) | 1.13 (1.08–1.30) | 0.052 (−.951 to 1.054) | 0.056 (0.038–0.057) | 12.44 (12.09–18.38) | 15.12 (14.01–18.81) | … | … | … | … |
All VLs were retained for model fits; those below the lower limit of detection of 1 copy/mL were set to 0.5 c/mL. The wide CIs around the population-based fixed-effect parameter estimates likely reflect the small sample size (8 subjects) for which models covering weeks 0–72 could be fit. Means and 95% CIs around subject-specific t1/2 values are bootstrap estimates (10 000 bootstrap samples). The bootstrap mean for the third-phase half-life for the raltegravir-based regimen of A5248 is 498 days, or 71 weeks, a half-life that is near the mean of 69 weeks reported for the boosted lopinavir-based regimen of M98–863. The 95% CI for A5248's third-phase half-life of 376–843 days (54–120 weeks) exhibits substantial overlap with the corresponding CIs of 38–408 weeks, for M98–863, and 25–90 weeks, for M97–720 [6].
Abbreviations: CI, confidence interval; d1, first phase of viral decay; d2, second phase of viral decay; d3, third phase of viral decay; 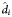 , population based fixed-effects estimate with model-based CI; IQR, interquartile range; t1/2, half-life.
, population based fixed-effects estimate with model-based CI; IQR, interquartile range; t1/2, half-life.
Figure 2.
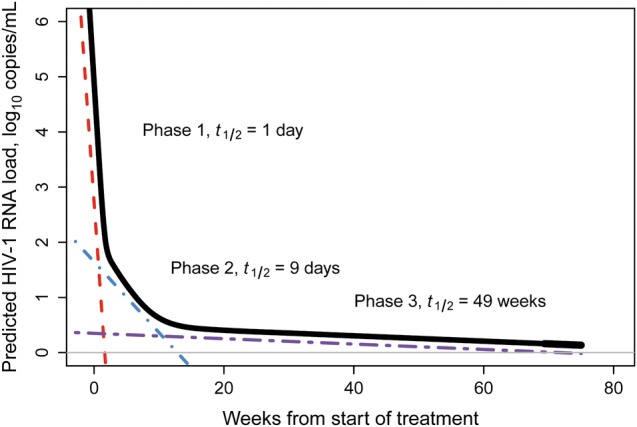
Tri-exponential model of predicted viral decay (weeks 0–72), using single-copy assay data. Plasma human immunodeficiency virus type 1 (HIV-1) RNA decay (log10 copies/mL) in treatment-naive subjects initiating raltegravir plus emtricitabine/tenofovir disoproxil fumarate, predicted by population-average tri-exponential mixed-effects models of single-copy assay data for weeks 0–72 (solid black line), using all values obtained (those below the lower limit of detection of 1 copy/mL were imputed at 0.5 copies/mL). The colored dotted lines show the slopes (tangent lines) for each of the 3 phases. Half-lives for each phase are also shown.
DISCUSSION
Viral decay kinetics following the initiation of ART reveal the dynamic nature of HIV-1 infection and provide important information regarding the mechanisms of drug action. The various phases of viral clearance also provide insights into the cellular reservoirs that contribute to HIV-1 persistence. The rapid reduction of plasma HIV-1 RNA to levels below the limits of detection by conventional assays in patients initiating treatment with a RAL-containing regimen raises 2 important questions: what accounts for the apparently faster initial clearance of plasma virus in patients receiving RAL, and are the kinetics of subsequent phases of viral decay different in patients receiving an integrase strand transfer inhibitor versus inhibitors of reverse transcriptase or protease? Sparse sampling and/or constraints imposed by the limitation of detection of standard assays have hampered previous attempts to answer these questions.
We used a combination of frequent sampling and 2 different plasma HIV-1 RNA assays to estimate clearance rates for plasma virus in treatment-naive patients initiating RAL plus FTC/TDF. Using ultrasensitive assay data, we found that d1 lasted longer in RAL-treated patients (15 days) than in EFV-treated patients (9 days in A5166s; 11 days in A5160s); at the time of transition, the HIV-1 RNA level was lower (91 copies/mL vs 589 copies/mL in A5166s and 977 copies/mL in A5160s). Models incorporating single-copy assay data showed that d1 extends for 16 days, with the d1 to d2 transition occurring at a plasma HIV-1 RNA level of 69 copies/mL [10]. Although d1 lasted longer in RAL-treated patients than in EFV-treated patients, the rate of d1 decay was significantly faster in EFV-treated patients. The reason for this difference is not clear; it may reflect (1) a more effective initial inhibition of viral replication by EFV-containing regimens or (2) a cytotoxic effect of EFV on HIV-1–producing cells, mediated through promotion of gag-pol dimerization, proteolytic processing of gag-pol, and intracellular release of active HIV-1 protease, resulting in cell death [15].
The proportion of subjects with plasma HIV-1 RNA levels below the ultrasensitive assay limit of detection increased from 10% at the start of d2 to 85% by day 56. Given the increasing proportion over time of ultrasensitive assay values of ≤50 copies/mL, it is not surprising that d2 estimates were dependent on the method of handling censored data. This problem was minimized in models that relied on single-copy assay data, because a majority of subjects (46%) still had detectable viremia at week 60, as measured by this assay. More importantly, use of single-copy assay data revealed a third phase of decay, with a corresponding median half-life of 71 weeks, similar to that observed in patients treated with a ritonavir-boosted PI [6].
Our results demonstrate convincingly that the rapid reduction in plasma HIV-1 RNA load to below the detection limits of standard HIV-1 RNA assays with RAL-containing regimens is due to the longer duration of d1 when compared to EFV-based regimens and not to faster initial clearance. This longer duration is most likely explained by RAL's mechanism of action, which may prevent the transition of cells from a state of “preintegration latency” to productively infected cells by blocking the integration of linear double-stranded viral DNA products of reverse transcription from previously infected cells, thereby reducing the pool of cells that contribute to d2 [10] and that would otherwise obscure d1. Although our results are qualitatively similar to those previously reported [10], the design of the current study enabled us to estimate d1 and the time of transition to d2 more accurately because of more frequent plasma HIV-1 RNA measurements at early and late time points of the viral decay curve and because of the use of the more sensitive single-copy assay.
The single-copy assay also revealed a third phase of decay in RAL-treated patients, but the estimated decay rate did not differ from that observed in patients receiving LPV/r. This finding suggests that cells harboring long-lived unintegrated forms of HIV-1 DNA do not contribute significantly to d3, and it is consistent with the hypothesis that this third phase of decay results from long-lived reservoirs of either latently infected cells that are stochastically activated or from long-lived productively infected cells. These results also suggest that integrase strand-transfer inhibitors, which, like all other approved antiretrovirals, block new cycles of HIV-1 replication, do not have an effect on latently infected cells. Our results are also consistent with the outcome of intensification studies that showed that addition of RAL to suppressive ART had no effect on residual plasma viremia [16–18]. In our study, we did not detect a plateau of HIV-1 RNA load decline, which was previously reported for LPV/r-containing ART [6]. However, the plateau was observed 4–5 years after initiation of LPV/r-containing ART, much later than the 72-week observation period for the current study.
This study has several limitations. Because it was not randomized, comparisons with historical data from EFV-treated patients must be interpreted with caution. Nevertheless, our population was similar overall to populations in the earlier studies. Moreover, the time course of virologic suppression in the current study was consistent with that observed in randomized trials of RAL in treatment-naive patients [9, 19, 20]. Another potential limitation is that the single-copy assay was performed on samples from only a subset of patients, but this subset was similar to the study population as a whole. We did not examine the levels of virus in tissues such as the gastrointestinal mucosa, genital tract, or central nervous system. Such analyses may provide additional insights into the effects of RAL on viral replication but are not required to assess viremia decay kinetics. Another limitation is that although multiple imputation is the preferred approach to handling viral loads reported as below detection, problems with convergence prevented us from using it in all cases. Finally, the viral dynamics models we used assumed perfect drug efficacy [21]. Other models that account for variation in adherence to antiretroviral drugs might have produced different results.
In conclusion, using frequent sampling and a single-copy assay for measuring the plasma HIV-1 RNA load, we describe a more complete model of viral decay in patients initiating ART with RAL plus TDF/FTC. This model revealed 3 phases of decay characterized by a significantly extended d1 and d2 that began at lower levels of plasma HIV-1 RNA than observed in prior viral dynamics studies involving patients receiving EFV-based regimens. These results are consistent with a reduction in the size of the pool of cells contributing to d2 resulting from RAL's ability to prevent cells in a state of “preintegration latency” from becoming productively infected cells. Additional insights may be gained by analyzing the decay in integrated and unintegrated forms of viral DNA, including 2 long terminal repeat circles, in peripheral blood mononuclear cells, and in lymphoid tissues. It will be of interest to explore associations between the rapid clearance of HIV-1 RNA and changes in immunological parameters, including the kinetics of CD4+ T-cell count recovery and changes in immune activation.
STUDY TEAM MEMBERS AND PARTICIPATING INVESTIGATORS
Additional ACTG A5248 Protocol Team Members
Laboratory technologists: Dan Eggers (Massachusetts General Hospital) and David Shugarts (University of Colorado Health Sciences Center); laboratory data coordinators: Ken Braun, Travis Behm, and Amy Jennings; field representative: Karen Cavanaugh (New York University Medical Center); DAIDS pharmacists: Ana Martinez and Debra Meres; NCAB representatives: Louis Zimmerman and Patrick Kramme; industry representative: Randi Leavitt (Merck); and protocol specialist: Katherine Bergstrom (ACTG Operations Center).
Additional Participating Investigators and AIDS Clinical Trials Units
Constance Benson, MD, University of California, San Diego; Michael Keefer, MD, University of Rochester; David Clifford, MD, University of Washington; Susan Koletar, MD, Ohio State University; Robert Murphy, MD, Northwestern University; Richard D'Aquila, MD, Vanderbilt University; Charles Davis, MD, University of Maryland; Roberto Arduino, MD, Houston AIDS Research Team; and Scott Hammer, MD, Harlem ACTG CRS.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank Merck for donating drug and providing funds to support the intensive viral sampling portion of this study.
Financial support. This work was supported by grants to the AIDS Clinical Trials Group (U01 AI068636, AI069472, AI68634, AI069465, AI069494, and AI69450), including ACTG support of the Harvard and University of Pittsburgh Virology Specialty Laboratories, the Harvard University Center for AIDS Research (P30 AI060354), and the Statistical and Data Management Center (UM1 AI068634), funded by the National Institute of Allergy and Infectious Diseases; and by Clinical and Translational Science Centers (UL1 RR 025005, RR025780, and RR025780), funded by the National Center for Research Resources. Merck provided drug for this study, as well as financial support for plasma HIV-1 RNA determinations.
Potential conflicts of interest. A. A. received research grant support from GlaxoSmithKline. E. S. D. received research support from Abbott, Gilead, Merck, Pfizer, and ViiV Healthcare and is a consultant/advisor for Bristol Myers Squibb, Merck, ViiV Healthcare, and Gilead. M. L. is a consultant for Merck. C. F. has received grant support from GlaxoSmithKline; has served on scientific advisory boards for Bristol-Myers Squibb, GlaxoSmithKline, Merck, Roche, Tibotec Pharmaceuticals, Vertex, ViiV Healthcare, Virostatics, and Inhibitex; and has received honoraria from Abbott Laboratories. J. M. is a consultant to and has received honoraria from Gilead Sciences and Johnson & Johnson and owns shares of RFS Pharmaceuticals. D. R. K. is a consultant to and has received honoraria and/or research grant support from Abbott, Bristol-Myers Squibb, Gilead, GlaxoSmithKline, Merck, Roche, and ViiV Healthcare. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–6. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 2.Wei X, Ghosh SK, Taylor ME, et al. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–22. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 3.Perelson AS, Essunger P, Cao Y, et al. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387:188–91. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 4.Ramratnam B, Mittler J, Zhang L, et al. The decay of the latent reservoir of replication-competent HIV-1 is inversely correlated with the extent of residual viral replication during prolonged anti-retroviral therapy. Nat Med. 2000;6:82–5. doi: 10.1038/71577. [DOI] [PubMed] [Google Scholar]
- 5.Finzi D, Blankson J, Siliciano JD, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5:512–25. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 6.Palmer S, Maldarelli F, Wiegand A, et al. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc Natl Acad Sci U S A. 2008;105:3879–84. doi: 10.1073/pnas.0800050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siliciano JD, Kajdas J, Finzi D, et al. Long term follow-up studies confirm the extraordinary stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nature Med. 2003;9:727–8. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 8.Hazuda DJ, Felock P, Witmer M, et al. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science. 2000;287:646–50. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 9.Lennox JL, DeJesus E, Lazzarin A, et al. Safety and efficacy of raltegravir-based versus efavirenz-based combination therapy in treatment-naive patients with HIV-1 infection: a multicentre, double-blind randomised controlled trial. Lancet. 2009;374:796–806. doi: 10.1016/S0140-6736(09)60918-1. [DOI] [PubMed] [Google Scholar]
- 10.Murray JM, Emery S, Kelleher AD, et al. Antiretroviral therapy with the integrase inhibitor raltegravir alters decay kinetics of HIV, significantly reducing the second phase. AIDS. 2007;21:2315–21. doi: 10.1097/QAD.0b013e3282f12377. [DOI] [PubMed] [Google Scholar]
- 11.Sedaghat AR, Dinoso JB, Shen L, et al. Decay dynamics of HIV-1 depend on the inhibited stages of the viral life cycle. Proc Natl Acad Sci U S A. 2008;105:4832–7. doi: 10.1073/pnas.0711372105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fitzgerald AP, DeGruttola VG, Vaida F. Modeling HIV viral rebound using non-linear mixed effects models. Stat Med. 2002;21:2093–108. doi: 10.1002/sim.1155. [DOI] [PubMed] [Google Scholar]
- 13.Kuritzkes DR, Ribaudo HJ, Squires KE, et al. Plasma HIV-1 RNA dynamics in antiretroviral-naive subjects receiving either triple-nucleoside or efavirenz-containing regimens: ACTG A5166s. J Infect Dis. 2007;195:1169–76. doi: 10.1086/512619. [DOI] [PubMed] [Google Scholar]
- 14.Haubrich RH, Riddler SA, Ribaudo H, et al. Initial viral decay to assess the relative antiretroviral potency of protease inhibitor-sparing, nonnucleoside reverse transcriptase inhibitor-sparing, and nucleoside reverse transcriptase inhibitor-sparing regimens for first-line therapy of HIV infection. AIDS. 2001;25:2269–78. doi: 10.1097/QAD.0b013e32834d0c20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jochmans D, Anders M, Keuleers I, et al. Selective killing of human immunodeficiency virus infected cells by non-nucleoside reverse transcriptase inhibitor-induced activation of HIV protease. Retrovirology. 2010;7:89. doi: 10.1186/1742-4690-7-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gandhi RT, Zheng L, Bosch RJ, et al. The effect of raltegravir intensification on low-level residual viremia in HIV-infected patients on antiretroviral therapy: a randomized controlled trial. PLoS Med. 2010;7 doi: 10.1371/journal.pmed.1000321. pii: e1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinoso JB, Kim SY, Wiegand AM, et al. Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 2009;106:9403–8. doi: 10.1073/pnas.0903107106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dahl V, Sinclair E, et al. A randomized, controlled trial of raltegravir intensification in antiretroviral-treated, HIV-infected patients with a suboptimal CD4+ T cell response. J Infect Dis. 2011;203:960–8. doi: 10.1093/infdis/jiq138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Markowitz M, Morales-Ramirez JO, Nguyen BY, et al. Antiretroviral activity, pharmacokinetics, and tolerability of MK-0518, a novel inhibitor of HIV-1 integrase, dosed as monotherapy for 10 days in treatment-naive HIV-1-infected individuals. J Acquir Immune Defic Syndr. 2006;43:509–15. doi: 10.1097/QAI.0b013e31802b4956. [DOI] [PubMed] [Google Scholar]
- 20.Markowitz M, Nguyen BY, Gotuzzo E, et al. Rapid and durable antiretroviral effect of the HIV-1 Integrase inhibitor raltegravir as part of combination therapy in treatment-naive patients with HIV-1 infection: results of a 48-week controlled study. J Acquir Immune Defic Syndr. 2007;46:125–33. doi: 10.1097/QAI.0b013e318157131c. [DOI] [PubMed] [Google Scholar]
- 21.Perelson AS, Neumann AU, Markowitz M, et al. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–6. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


