Abstract
Delineation of antibody epitopes at the residue level is key to understanding antigen resistance mutations, designing epitope-specific probes for antibody isolation, and developing epitope-based vaccines. Ideally, epitope residues are determined in the context of the atomic-level structure of the antibody-antigen complex, though structure determination may in many cases be impractical. Here we describe an efficient computational method to predict antibody-specific HIV-1 envelope (Env) epitopes at the residue level, based on neutralization panels of diverse viral strains. The method primarily utilizes neutralization potency data over a set of diverse viral strains representing the antigen, and enhanced accuracy could be achieved by incorporating information from the unbound structure of the antigen. The method was evaluated on 19 HIV-1 Env antibodies with neutralization panels comprising 181 diverse viral strains and with available antibody-antigen complex structures. Prediction accuracy was shown to improve significantly over random selection, with an average of greater-than-8-fold enrichment of true positives at the 0.05 false-positive rate level. The method was used to prospectively predict epitope residues for two HIV-1 antibodies, 8ANC131 and 8ANC195, for which we experimentally validated the predictions. The method is inherently applicable to antigens that exhibit sequence diversity, and its accuracy was found to correlate inversely with sequence conservation of the epitope. Together the results show how knowledge inherent to a neutralization panel and unbound antigen structure can be utilized for residue-level prediction of antibody epitopes.
INTRODUCTION
Broadly neutralizing antibodies (bNAbs) against various antigens, such as HIV-1 envelope glycoprotein (Env) (1–4) and influenza virus hemagglutinin (HA) (5, 6), can have utility as therapeutics in the context of passive transfer (7) and as templates for the design of epitope-specific vaccines (8). The determination of the epitope targeted by an antibody of interest can assist in understanding virus resistance and escape mutations (9), offer clues for antibody affinity improvement (10, 11), and guide immunogen design for focusing the immune response toward neutralizing epitopes (12). Structure determination, by X-ray crystallography or nuclear magnetic resonance spectroscopy, can provide atomic-level resolution of epitopes and interactions in antibody-antigen complexes, but structures for many such complexes can be difficult or even infeasible to obtain (13). Cryo-electron microscopy could also be used to identify general epitope regions, but this method is typically associated with lower-resolution structures and in most cases cannot provide atomic-level information (14). A variety of other experimental methods can also be applied to epitope residue mapping, though these are typically laborious and can be limited by different factors, such as sensitivity to effects from distal residues not part of the direct antibody-antigen interactions (e.g., alanine scanning) or dependence on the presence of substantial antibody interactions in a sequentially continuous region of the antigen (e.g., pepscan) (15). The latter case is particularly limiting since most antibody epitopes are discontinuous (i.e., involving multiple sequentially noncontiguous regions) (13). In silico methods for epitope prediction are also available, but the majority focus on predicting protein residues that can be part of any epitope and are thus not antibody specific (16–19). Only a limited number of antibody-specific epitope prediction methods have been proposed thus far (20, 21). Computational docking can also be used to predict epitope residues by means of generating a structural model of the antibody-antigen complex. However, docking depends on the existence of separate antigen and antibody structures (or accurate structural models), and docking scoring functions are, in general, not optimal (22, 23) and in many cases unable to accurately predict the epitope of interest (24). Recently, a computational method was proposed for predicting the epitopes of query antibodies based on the similarity of their neutralization fingerprints to the fingerprints of antibodies with known epitopes (25). This method, however, does not provide residue-level information and is not applicable to antibodies that bind to novel epitopes. Another recent study utilized HIV-1 antibody neutralization panels to identify antigen residues functionally important for binding to specific antibodies (26). The method used in the study, however, aims at predicting a limited number of antigen residues of functional importance for a given antibody, rather than identifying the antibody epitope.
Here we present a computational method for antibody-specific prediction of epitope residues based on neutralization data from a panel of diverse viral strains. The method is applicable to viruses that exhibit strain diversity, such as HIV-1 and influenza virus, and relies on the hypothesis that antibody neutralization potency can be expected to be affected more substantially by the choice of residue type at an epitope position but to a lesser extent by the choice of residue type at a nonepitope position. Conversely, residue positions whose variation is found to affect neutralization potency can be expected to be part of the respective antibody epitope (Fig. 1), although distal effects and residue covariation may also play a role. To explore this relationship between neutralization potency and antigen sequence variation, we applied a neutralization-based method to a set of 19 HIV-1 Env antibodies with published antibody-antigen complex structures and neutralization panels of diverse viral strains. The incorporation of data from diverse strains for the same virus can increase the likelihood of observing an association between changes in neutralization potency and variations in sequence. Variants of the method that further incorporated antigen structural information were also evaluated. Finally, we prospectively applied our method to the prediction of epitope residues for two HIV-1 antibodies and experimentally validated the computational predictions. Overall, our results indicate that a neutralization-based method in combination with structural information could be a useful tool for the accurate prediction of antibody epitopes of genetically diverse antigens.
Fig 1.
Schematic overview of utilizing the mutual variation between amino acid types at each position and neutralization potency for the prediction of epitope residues. For a given antibody, antigen sequence positions are colored according to amino acid type, while each strain is further colored according to neutralization potency.
MATERIALS AND METHODS
Data set.
A neutralization panel of 181 HIV-1 strains and antibody-antigen complex crystal structures for 19 HIV-1 broadly neutralizing antibodies downloaded from the Protein Data Bank (PDB) (27) were used to assess the performance of the computational methods (Table 1; see Table S1 in the supplemental material) (10, 25, 28–39). Antibody-mediated neutralization of 181 pseudotyped HIV-1 isolates was measured using TZM-bl cells and a luciferase reporter gene assay as described previously (40–42). Neutralization curves were fit by nonlinear regression using a 5-parameter hill slope equation as described previously (41) to determine the antibody concentrations required to inhibit infection by 50% (IC50). Some of the neutralization data have been previously published (4, 25, 30, 33, 36, 43, 44). The gp160 amino acid sequences for the 181 HIV-1 strains were aligned using HIVAlign (http://www.hiv.lanl.gov/content/sequence/VIRALIGN/viralign.html). The antigen residues for the CD4-binding-site antibodies were defined as the residues in a ligand-free gp120 core structure (PDB ID 3TGT) (residues 44 to 124, 198 to 301, 324 to 396, and 408 to 492); for PG9 and PG16, the antigen residues were, respectively, defined as the residues in the V1/V2 loop present in the crystal structures (residues 126 to 196, except for the missing residues in the respective structures); and for MPER antibodies, the antigen residues were defined as residues in the MPER region (residues 659 to 685). Epitope residues were defined as antigen residues for which any heavy atom was within 5 Å of any heavy atom in the given antibody, as observed in the crystal structures.
Table 1.
The 19 HIV-1 antibodies used in the analysis
| Antibody | PDB ID | Site of recognition |
|---|---|---|
| VRC01 | 3NGB | CD4-binding site |
| VRC-PG04 | 3SE9 | CD4-binding site |
| b12 | 2NY7 | CD4-binding site |
| b13 | 3IDX | CD4-binding site |
| F105 | 3HI1 | CD4-binding site |
| NIH45-46 | 3U7Y | CD4-binding site |
| 3BNC117 | 4JPV | CD4-binding site |
| VRC-PG20 | 4LSUP | CD4-binding site |
| VRC23 | 4J6R | CD4-binding site |
| CH103 | 4JAN | CD4-binding site |
| VRC03 | 3SE8 | CD4-binding site |
| VRC06 | 4JB9 | CD4-binding site |
| VRC-CH31 | 4LSP | CD4-binding site |
| 17b | 2NXY | CD4i |
| 10E8 | 4G6F | MPER |
| 2F5 | 1TJI | MPER |
| 4E10 | 2FX7 | MPER |
| PG9 | 3U4E | V1/V2 |
| PG16 | 4DQO | V1/V2 |
Epitope prediction algorithm.
Six different method variants were evaluated using the set of 19 HIV-1 antibodies. We used the true-positive (TP) rate and false-positive (FP) rate to evaluate the prediction accuracy, where the TP rate was defined as the ratio of the number of correctly predicted epitope residues to the number of real (as defined by the antibody-antigen structure [see above]) epitope residues, while the FP rate was defined as the ratio of the number of incorrectly predicted epitope residues to the number of real nonepitope residues. For methods where a set of optimal parameters (weight or radius [see below]) had to be determined, the prediction accuracy of the method was evaluated with leave-one-out cross-validation, where the optimal parameter value that maximized the average TP rate at the 0.05 FP rate level for 18 of the antibodies was used to evaluate the performance on the remaining antibody.
For each method variant, the TP rates for the predictions at FP rates of 0.05, 0.10, 0.15, 0.20, and 0.25 were evaluated. The parameters selected for each method variant were the optimal parameters to maximize the average TP rate at the 0.05 FP rate level for the epitope prediction of the 19 HIV-1 antibodies. The six variants were as follows.
- Normalized mutual information (nMI). Each antigen residue i was ranked by the normalized mutual information between amino acid types and neutralization IC50. The score for residue i was then defined as
where Xi is a variable that encompasses the possible amino acid types at position i (for a total of 21 possible values, i.e., the 20 natural amino acid types and a gap in the sequence alignment), while Y is a binary variable defined by the respective neutralization IC50 (for a total of 2 possible values, i.e., neutralizing [IC50 < 50 μg/ml] and nonneutralizing [IC50 ≥ 50 μg/ml]). MI(Xi;Y) is the standard mutual information defined between Xi and Y (45). Since the mutual information is dependent on the Shannon entropy of each variable, here we normalized the mutual information by the Shannon entropy of the amino acid types at each position [H(Xi)] (the Shannon entropy of the amino acid types at each position is a function of the selected input sequences); the mutual information was not normalized by H(Y) since, for a given antibody, H(Y) was constant at each position.
- Normalized mutual information with window smoothing (nMIwindow). The score for each residue was the normalized mutual information calculated as in method variant 1 averaged with the normalized mutual information of n upstream and downstream (in sequence) residues:
Different n values (1 to 5) were evaluated, and a value of 4 was selected from the leave-one-out analysis on the 19 HIV-1 antibodies.
Normalized mutual information with prior filtering of non-solvent-accessible residues (SA/nMI). From the antibody-free antigen structure (PDB ID 3TGT for the gp120 core structure, PDB ID 3U4E for PG9 and PDB ID 4DQO for PG16 for the V1/V2 region, and PDB ID 4G6F for MPER), the solvent-accessible area of each residue was calculated using NACCESS (46), and the residues with a solvent-accessible area of less than 5 Å2 were considered non-solvent-accessible residues and were excluded from further analysis. The remaining antigen residues were ranked by the normalized mutual information as in method variant 1.
- Prior filtering of nonaccessible residues and summation of the normalized mutual information of residues in spatial proximity (SA/nMIproxsum). The non-solvent-accessible residues were filtered out as in method variant 3. The score of each of the remaining residues was the normalized mutual information of the residue as calculated in method variant 1 plus a distance-dependent sum of the normalized mutual information of surrounding (in space) residues:
where A is the set containing all solvent-accessible residues of the antigen, rij is the shortest distance between any side chain heavy atom of residues i and j (for glycine, Cα was used instead of a side chain atom), and R is a normalization distance. A number of different values for R (5.5 to 15.0 Å in increments of 0.5) were evaluated, and a value of 9 Å was selected from the leave-one-out analysis on the 19 HIV-1 antibodies. Such a simple scoring function that gradually decreases the weight of the score as the distance increases has also been used elsewhere (47, 48).
The last two method variants add an epitope propensity score (PS) to two of the previous methods (nMIwindow and SA/nMIproxsum). An epitope log odds ratio for each residue type was proposed previously (see Table 1 in reference 16) as an estimate of the likelihood of each type of amino acid being an epitope residue. Since an ensemble of antigen sequences was used as input, the PS for each antigen position i was defined as the average of epitope log odds ratios of position i from all input sequences.
- Normalized mutual information plus a weighted epitope propensity score with window smoothing (nMIwindow/PS). The score composed of a window-smoothed normalized mutual information plus a weighted epitope propensity (we used n = 4 as determined in method variant 2):
Different weights (w1 = 0.01 to 0.20 in increments of 0.01) were evaluated, and a weight of 0.039 ± 0.002 (mean ± standard error) was selected from the leave-one-out analysis.
- Prior filtering of nonaccessible residues and summation of normalized mutual information and epitope propensity of residues in spatial proximity (SA/nMIproxsum/PS). This was similar to method variant 4, but a weighted epitope propensity score (PS) was added to the normalized mutual information score (we used R = 9.0 as determined in method variant 4):
Different weights (w2 = 0.01 to 0.20 in increments of 0.01) were evaluated, and a weight of 0.031 ± 0.005 was selected from the leave-one-out analysis.
All calculations were performed using a combination of Java and Perl code.
Panel size and strain diversity analysis.
The effect of neutralization panel size on the prediction accuracy was evaluated by removing nonredundant strains from the full panel. In this analysis, strains with missing neutralization data for any antibody (see Table S1 in the supplemental material) were removed from the panel, resulting in a panel of 171 strains. The redundancy of the 171 strains was reduced by the “decrease redundancy” tool on the ExPASy server (49) based on the gp160 amino acid sequences, with maximum similarities set at 97.5%, 95%, 92.5%, 90%, 87.5%, 85%, 82.5%, and 80%, resulting in panels with sizes of 166, 165, 158, 152, 129, 96, 34, and 16 strains, respectively. The average performance of 100 randomly selected panels with the same panel sizes was also evaluated for comparison. Two clade B-specific panels were also created for 16- and 34-strain panel sizes by randomly selecting strains from the total of 39 clade B strains in the data set.
Residue conservation analysis.
The conservation level of the epitopes was defined by the number of epitope residues that are fully conserved among all tested strains divided by the total number of epitope residues.
Statistical analysis.
A two-tailed paired t test was used to evaluate the significance of the differences in prediction accuracy (based on the TP rates at the 0.05 FP rate level) between the different method variants and random selection (TP rate equals FP rate) evaluated on the 19 HIV-1 antibodies; computed P values were further adjusted using the false-discovery rate (FDR) method (50).
Prospective epitope prediction for HIV-1 antibodies 8ANC131 and 8ANC195.
The SA/nMIproxsum/PS method was applied to predict epitope residues for HIV-1 antibodies 8ANC131 and 8ANC195, using a neutralization panel of 181 HIV-1 strains (see Table S2 in the supplemental material) and the ligand-free gp120 core structure (PDB ID 3TGT) as structural input (0.031 was used for w2 in the SA/nMIproxsum/PS method). Binding of 8ANC131 to full-length gp120 (YU2) variants (wild type, R456S, K282N, I326T, and D78N) and binding of 8ANC195 to 2CC (a recombinant HXB2 gp120 core with stabilizing mutations W96C/V275C/I109C/Q428C/M95W/T257S/S375W/A433M) (38, 51) variants (wild type, deglycosylated wild type, N234S, T236K, and N276D) were determined using a previously described enzyme-linked immunosorbent assay (ELISA) method (4). Briefly, proteins were expressed by transient transfection in Freestyle 293-F cells (Invitrogen, Carlsbad, CA). gp120 proteins were purified either using DEAE-agarose resin (GE Healthcare, Little Chalfont, United Kingdom) followed by nickel chelation resin (Sigma, St. Louis, MO) or by affinity chromatography using monoclonal antibody 17b. Antibodies 8ANC131 and 8ANC195 were purified by protein A-affinity chromatography. Deglycosylated 2CC was prepared using methods described previously (38). Briefly, 2CC was expressed by transient transfection in HEK293S GnTi− cells, purified by 17b affinity chromatography, deglycosylated with endoglycosidase H, and purified with concanavalin A (ConA)-Sepharose (Sigma) and Superdex 200 (GE Healthcare) chromatography. Purified recombinant gp120 proteins were diluted in phosphate-buffered saline, and 200 ng per well was adsorbed onto Reacti-Bind 96-well plates (Pierce, Rockford, IL). Plates were blocked and incubated with a 5-fold dilution series of antibody with a starting concentration of 5 μg/ml. Horseradish peroxidase-conjugated goat anti-human IgG Fc antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) was used to detect bound antibody, and the color reaction was carried out using SureBlue 3,3′,5,5′-tetramethylbenzidine (Kirkegaard & Perry Laboratories, Gaithersburg, MD). For 8ANC131, the epitope predictions were further compared to a recently determined crystal structure of the antibody-gp120 complex (52). For 8ANC195, the neutralization of the 3337.V2.C6 virus variants (wild type, N234S, T236K, and N276D) was determined as described in “Data set” above. Site-directed mutagenesis was performed by GeneImmune Biotechnology (Rockville, MD).
Source code availability.
Source code is freely available to noncommercial users upon request.
RESULTS
Epitope prediction for 19 HIV-1 antibodies.
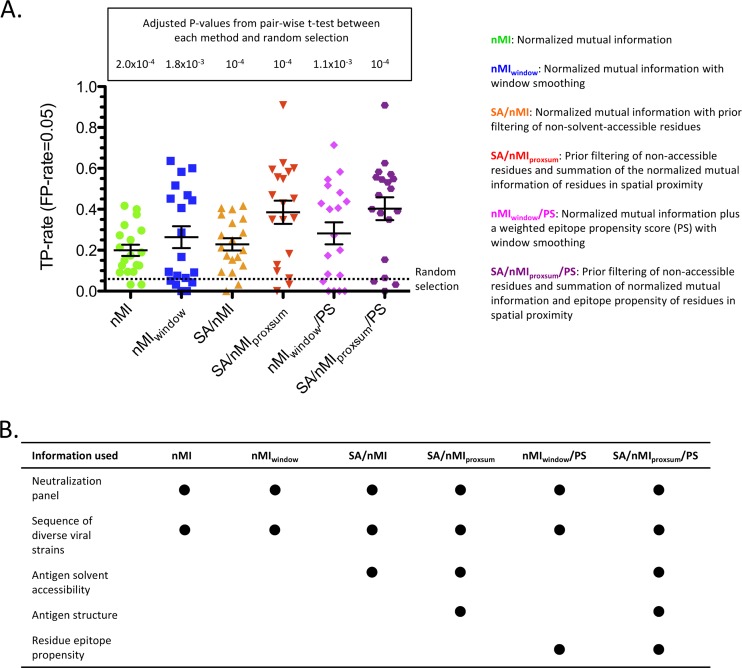
To interrogate whether, for the purposes of epitope prediction, there is sufficient association between variation in strain sequence and changes in antibody neutralization potency on a panel of diverse viral strains, we applied a neutralization-based approach to a set of 19 HIV-1 Env antibodies for which antibody-antigen complex crystal structures were available (Table 1; see Table S1 in the supplemental material). Specifically, putative epitope residues were selected based on the normalized mutual information (nMI) scores between neutralization potency for a given antibody and the sequence variation at a given antigen residue for the given set of strains (see Materials and Methods). A higher nMI score for a given residue position was taken to indicate a possible epitope residue. The prediction accuracy using the nMI approach was significantly better than random (P < 0.0005) (see Table S3 in the supplemental material), with an average true-positive (TP) rate of 0.199 at the 0.05 false-positive (FP) rate level (Fig. 2; see Fig. S1 in the supplemental material). Window smoothing of the nMI scores (nMIwindow; see Materials and Methods) further improved the prediction (average TP rate of 0.264 at the 0.05 FP level).
Fig 2.
Six different variants of the neutralization-based epitope prediction method. (A) TP rate at an FP rate level of 0.05 for each of the 19 antibodies, with the average (long black horizontal bar) and standard error (shorter black horizontal bar), for each method variant. The adjusted P value of the pairwise t test between each method and random selection based on the TP rate at an FP rate of 0.05 is displayed. (B) The different types of information that each method variant uses for prediction.
Next, we evaluated the effects of augmenting the epitope prediction of the neutralization-based method with antigen structural information. Filtering out non-solvent-accessible residues based on the antigen structure before scoring the remaining residues by nMI (SA/nMI; see Materials and Methods) gave minimal improvement to the prediction accuracy (Fig. 2; see Fig. S1 in the supplemental material). However, averaging in the nMI scores of structure-proximal residues in conjunction with filtering out non-solvent-accessible residues (SA/nMIproxsum; see Materials and Methods) significantly improved the performance compared to those of the nMI and nMIwindow methods (P < 0.0005 and P < 0.005, respectively) (see Table S3 in the supplemental material). Specifically, the average TP rate at the 0.05 FP rate level was 0.385, an almost 8-fold improvement over random and a 1.75-fold improvement over using only nMI as a predictor. For 17 out of the 19 HIV-1 antibodies, SA/nMIproxsum gave predictions equal to or better than those by nMI at the 0.05 FP rate level (see Table S4 in the supplemental material).
The effect of adding an epitope propensity score (see Materials and Methods) to two of the method variants, nMIwindow (nMIwindow/PS) and SA/nMIproxsum (SA/nMIproxsum/PS), was also evaluated (Fig. 2; see Fig. S1 in the supplemental material). Adding an epitope propensity score gave modest improvements to the two methods (TP rate increases of 0.264 to 0.283 and 0.385 to 0.403, respectively, at the 0.05 FP rate level). The prediction accuracy of SA/nMIproxsum/PS was significantly better than that of nMIwindow/PS (P < 0.01), and this was the best method among the six variants in terms of average prediction accuracy, although the improvement over the prediction accuracy of SA/nMIproxsum was not significant.
Table 2 displays the prediction accuracy of the SA/nMIproxsum/PS method on the 19 HIV-1 antibodies. For 14 of the antibodies, the SA/nMIproxsum/PS method resulted in an enrichment of TP rate over random of greater than 5-fold at the 0.05 FP rate level; for 2 of the antibodies, the enrichment over random was between 1- and 5-fold; and for the remaining 3 antibodies, the SA/nMIproxsum/PS method performed worse than random. The 14 antibodies with greater-than-5-fold enrichment by the SA/nMIproxsum/PS method included all 3 of the MPER and all 2 of the V1/V2 antibodies as well as 9 of the 13 CD4-binding-site antibodies, but not the sole CD4i antibody.
Table 2.
Prediction accuracy of the SA/nMIproxsum/PS method on 19 HIV-1 antibodies at different false-positive rates
| Antibody | No. of: |
TP rate at an FP rate of: |
|||||
|---|---|---|---|---|---|---|---|
| Antigen residues | Epitope residues | 0.05 | 0.10 | 0.15 | 0.20 | 0.25 | |
| VRC01 | 342 | 31 | 0.548 | 0.613 | 0.645 | 0.677 | 0.742 |
| VRC-PG04 | 342 | 27 | 0.556 | 0.556 | 0.593 | 0.667 | 0.667 |
| b12 | 342 | 31 | 0.032 | 0.097 | 0.161 | 0.161 | 0.226 |
| b13 | 342 | 21 | 0.048 | 0.143 | 0.190 | 0.238 | 0.238 |
| F105 | 342 | 31 | 0.065 | 0.129 | 0.161 | 0.194 | 0.258 |
| NIH45-46 | 342 | 42 | 0.500 | 0.643 | 0.690 | 0.786 | 0.833 |
| 3BNC117 | 342 | 32 | 0.531 | 0.562 | 0.594 | 0.625 | 0.656 |
| VRC-PG20 | 342 | 42 | 0.381 | 0.571 | 0.667 | 0.738 | 0.762 |
| VRC23 | 342 | 32 | 0.469 | 0.562 | 0.656 | 0.688 | 0.719 |
| CH103 | 342 | 23 | 0.391 | 0.478 | 0.478 | 0.478 | 0.522 |
| VRC03 | 342 | 40 | 0.350 | 0.525 | 0.525 | 0.625 | 0.675 |
| VRC06 | 342 | 39 | 0.154 | 0.410 | 0.462 | 0.538 | 0.615 |
| VRC-CH31 | 342 | 24 | 0.625 | 0.625 | 0.625 | 0.708 | 0.750 |
| 17b | 342 | 16 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 10E8 | 27 | 12 | 0.583 | 0.583 | 0.667 | 0.667 | 0.667 |
| 2F5 | 27 | 10 | 0.400 | 0.900 | 0.900 | 0.900 | 0.900 |
| 4E10 | 27 | 11 | 0.909 | 0.909 | 1.000 | 1.000 | 1.000 |
| PG9 | 52 | 14 | 0.571 | 0.643 | 0.643 | 0.643 | 0.643 |
| PG16 | 41 | 11 | 0.545 | 0.636 | 0.636 | 0.636 | 0.636 |
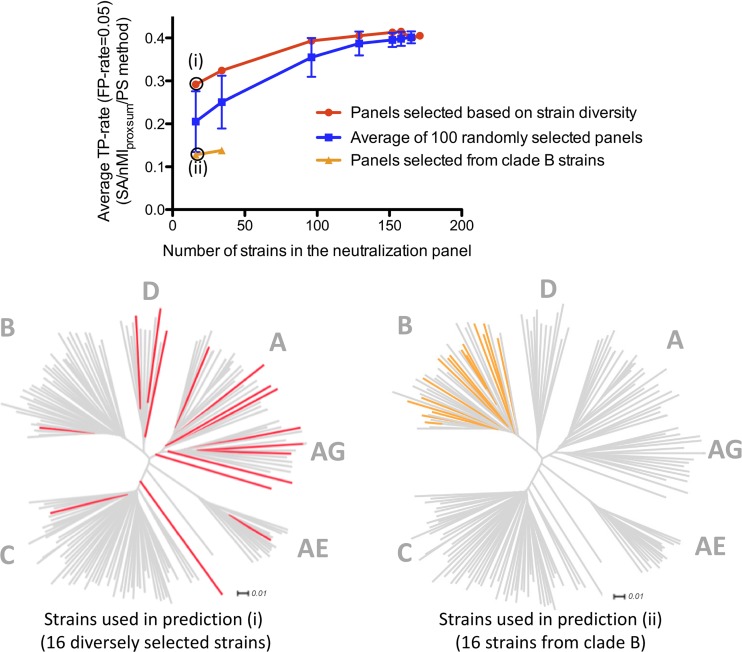
Prediction accuracy versus neutralization panel size and strain diversity.
Since larger panels of viral strains can be expected to contain more information with respect to sequence diversity, we investigated how the size of the neutralization panels may affect the prediction accuracy of our method. To that end, we generated subsets of the 181-strain panel at different cutoffs for sequence identity among included strains (see Materials and Methods). The effect of reducing the size of the neutralization panel by removing sequence-redundant strains is shown in Fig. 3 (red line). The average TP rate for the SA/nMIproxsum/PS method at the 0.05 FP rate level decreased 31.6% for the panel of 16 strains (corresponding to a sequence identity cutoff of 80.0%) and 20% for the panel of 34 strains (82.5% cutoff), while the prediction accuracy was similar (within 10%) to that for the full-size panel for the panels of 96 strains or larger (≥85% cutoff).
Fig 3.
Effect of the number of viral strains and strain diversity on epitope prediction. The average epitope prediction accuracy for 19 HIV-1 antibodies by the SA/nMIproxsum/PS method for panels of different sizes and different degrees of diversity is shown as the average TP rate at the 0.05 FP rate level. The red line represents predictions using different smaller panels constructed by removing sequence-redundant strains with ExPASy. The blue line represents the average prediction accuracy of 100 randomly generated panels for the same panel sizes (error bars indicate standard errors). The orange line represents the prediction accuracy using strains selected only from clade B (16- and 34-strain panels only). The dendrograms display the distribution of the viral strains used in the nonredundant 16-strain panel selected from ExPASy (red) and the clade B 16-strain panel (orange) among all 171 strains (gray).
The effect of reducing the size of the neutralization panel by random selection was also examined (Fig. 3). When the prediction accuracies from 100 randomly selected panels were averaged for each panel size, the average TP rate for the SA/nMIproxsum/PS method at the 0.05 FP rate level decreased 49.33% for random panels of 16 strains, 38.13% for random panels of 34 strains, and 12.40% for random panels of 96 strains, while the prediction accuracy was similar (within 10%) to that for the full-size panel for random panels of 129 strains or larger. Smaller panels performed significantly worse than the full-size panel when comparing the average prediction accuracies of randomly selected panels (see Table S5 in the supplemental material).
The effect of strain diversity on prediction accuracy of the neutralization-based algorithm was seen by comparing the nonredundant panels with clade B-specific panels, which are composed of sequences with lower sequence variation. The prediction by the SA/nMIproxsum/PS method using the panel of 16 clade B strains was significantly worse than the prediction using the panel of 16 nonredundant strains (P < 0.05), with a 56.16% decrease in terms of the average TP rate at the 0.05 FP rate level (Fig. 3). This result indicates that prediction accuracy can be improved when using diverse strains compared to using strains with lower sequence variation.
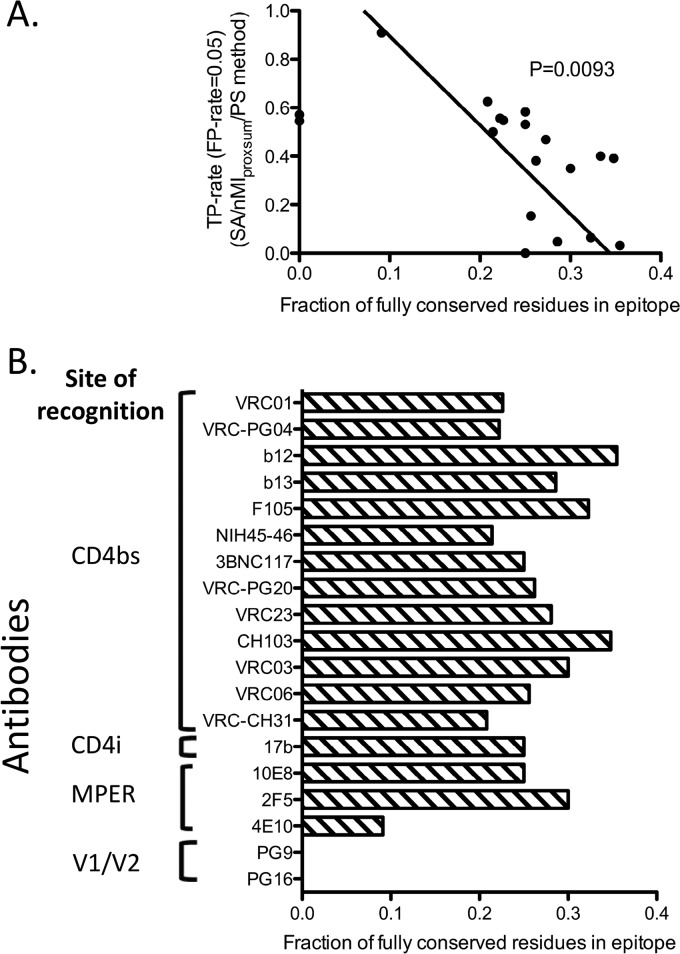
Prediction accuracy versus epitope sequence conservation.
The prediction accuracy of the SA/nMIproxsum/PS method described here was highly dependent on the variability of each residue among the different strains. Of note, the prediction accuracy was inversely correlated with the conservation level of the epitope residues (Fig. 4; see Table S6 in the supplemental material). The residues in the epitopes for antibodies b12 and b13, for example, had higher levels of sequence conservation and were less accurately predicted than other CD4-binding-site antibodies (see Fig. S2 in the supplemental material).
Fig 4.
Relationship between prediction accuracy of the SA/nMIproxsum/PS method and epitope sequence conservation for the 19 HIV-1 antibodies. (A) Fraction of fully conserved residues in the antibody epitopes (horizontal axis) versus TP rate at the 0.05 FP rate level (vertical axis). Two antibodies, PG9 and PG16, were labeled on the y axis since there were no fully conserved residues in their epitopes. A negative correlation was observed between the fraction of fully conserved residues in epitope and the prediction accuracy for a given antibody (P = 0.0093, Deming linear regression). (B) Fraction of fully conserved residues for each of the 19 HIV-1 antibodies. A residue is defined as conserved only if it is 100% conserved among all viral strains in the data set. For example, although 54.55% of the 4E10 epitope residues have a conservation of greater than 98%, only 9.09% of the 4E10 epitope residues are 100% conserved.
Prospective prediction of epitope residues for HIV-1 antibody 8ANC131.
The SA/nMIproxsum/PS method was applied to prospectively predict the epitope for gp120 antibody 8ANC131, a CD4-binding-site antibody (2). The top 10 predicted residues (see Table S7 in the supplemental material) were located in three distinct regions of the gp120 structure (Fig. 5A and B): region 1 (residues 456, 466, 280, 96, 282, and 461), region 2 (residues 78, 79 and 80), and region 3 (residue 326). We selected residues from each region for mutation analysis (residues 456 and 282 from region 1, residue 78 from region 2, and residue 326 from region 3); for each position, the YU2 gp120 wild-type residue was mutated to an amino acid type preferentially found in 8ANC131-resistant strains (53). The ELISA binding analysis demonstrated that only R456S and K282N resulted in a substantial decrease in binding of 8ANC131 to YU2 gp120 (Fig. 5C; see Fig. S3 in the supplemental material). These results and the fact that region 1 had the largest number of residues in the top 10 predictions suggested that the predicted residues from region 1 are likely part of the 8ANC131 epitope. As a further validation, the computational predictions were further compared to the actual epitope residues defined by a recently determined crystal structure of the antibody-gp120 complex (52). The SA/nMIproxsum/PS method demonstrated multiple fold increases of true positive rates at different FP rate levels compared to random selection (Fig. 5D). In particular, TP rates were improved by 6.98-fold at the 0.05 FP rate level compared to random selection. Among the top 10 predicted residues, five of the six region 1 residues (456, 280, 96, 282, and 461) but none of the region 2 and region 3 residues are part of the actual antibody epitope.
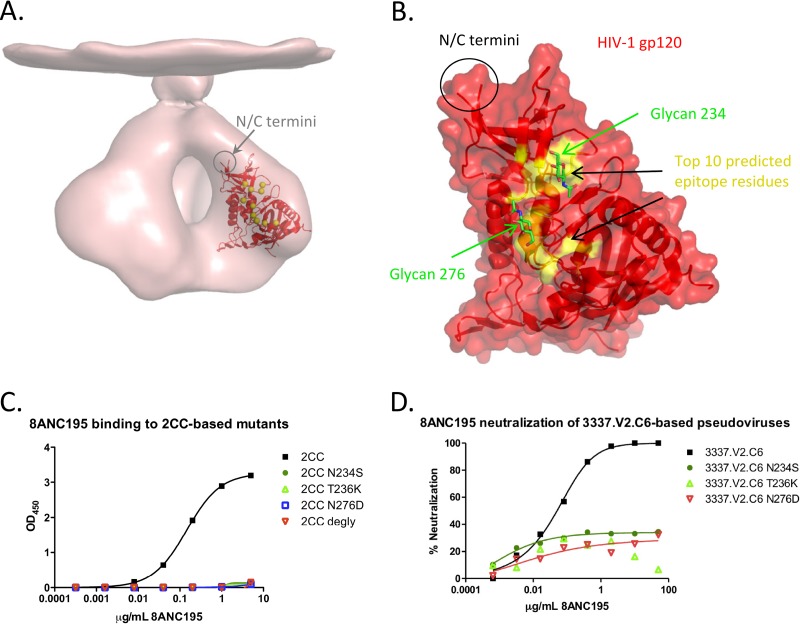
Fig 5.
Epitope prediction for HIV-1 antibody 8ANC131 using the SA/nMIproxsum/PS method. (A) The top 10 predicted residues (yellow spheres) mapped on a gp120 core crystal structure (red) (PDB ID 3TGT), fitted in an unliganded HIV-1 Env trimer cryo-EM map (EMD accession code 5019, pink). (B) Outline of the top 10 predicted epitope residues (yellow) for 8ANC131 shown on a transparent surface and cartoon representation of gp120 (red). The top 10 predicted epitope residues can be clustered into three distinct sites outlined by yellow dashed circles. (C) ELISA binding of 8ANC131 to YU2 gp120, YU2 gp120 R456S, YU2 gp120 K282N, YU2 gp120 D78N, and YU2 gp120 I326T. Mutants R456S and K282N had a substantially decreased binding to 8ANC131. (D) Prediction accuracy determined by comparison to the complex structure. Shown are the TP rates at different FP rate levels (0.05, 0.10, 0.15, 0.20, and 0.25).
Prospective prediction of epitope residues for HIV-1 antibody 8ANC195.
The SA/nMIproxsum/PS method was applied to prospectively predict the epitope for gp120 antibody 8ANC195, which was isolated using 2CC, a gp120-based probe stabilized in the CD4-bound conformation (38, 51). This antibody does not possess the typical characteristics of a CD4-binding-site antibody, and its precise epitope is currently unknown (2). The 10 top-scoring residues were focused on a patch of the gp120 surface encompassing a loop preceding the α1 helix, loops A and D, and the β23/V5/β24 strand/loop/strand region (Fig. 6; see Table S8 in the supplemental material). Three of the top 10 residues (234, 236, and 276) affected the presence of an N-linked glycosylation sequon (for glycans at positions 234 and 276), and since enzymatic deglycosylation of 2CC knocked out its binding to 8ANC195 (Fig. 6C), we selected these three residues for mutation analysis: for each position, the 2CC residue was mutated to an amino acid type preferentially found in 8ANC195-resistant strains (53). The selected mutations (N234S, T236K, and N276D) each removed a single N-linked glycosylation site (Fig. 6C; see Fig. S4 in the supplemental material). Each of the three mutations knocked out 8ANC195 binding (Fig. 6C), in agreement with the computational predictions, suggesting that 8ANC195 is a glycan-reactive antibody targeting a novel epitope on gp120. Additionally, we tested the effects of N234S, T236K, and N276D on neutralization by 8ANC195 by introducing each of these three mutations into 3337.V2.C6, the strain most sensitive to 8ANC195 in the 181-strain neutralization panel. All three mutant viruses were resistant (IC50 of >50 μg/ml) to neutralization by 8ANC195 (Fig. 6D; see Fig. S5 in the supplemental material), further confirming that the N-linked glycans at positions 234 and 276 are important for viral recognition by this antibody.
Fig 6.
Epitope prediction for HIV-1 antibody 8ANC195 using the SA/nMIproxsum/PS method. (A) The top 10 predicted residues (yellow spheres) mapped on a gp120 core crystal structure (red) (PDB ID 3TGT), fitted in an unliganded HIV-1 Env trimer cryo-EM map (EMD accession code 5019, pink). (B) Outline of the top 10 predicted epitope residues (yellow) for 8ANC195 shown on a transparent surface and cartoon representation of gp120 (red). Two N-linked glycosylation sites, at residues 234 and 276, were among the selected residues; the N-acetylglucosamine moieties linked directly to the two residues are shown in green sticks to highlight their location. (C) ELISA binding of 8ANC195 to the stabilized gp120 core 2CC, 2CC N234S, 2CC T236K, 2CC N276D, and deglycosylated 2CC (2CC degly). 8ANC195 showed good binding to the wild type but did not bind deglycosylated 2CC. The mutations N234S, T236K, and N276D, each of which resulted in the removal of an N-linked glycan, knocked out binding of 2CC to 8ANC195. (D) 8ANC195 neutralization of wild-type, N234S, T236K, and N276D 3337.V2.C6. Wild-type 3337.V2.C6 was potently neutralized by 8ANC195, while all of the N234S, T236K, and N276D mutant viruses were resistant to neutralization by that antibody.
DISCUSSION
Most of the currently available computational methods for epitope prediction are based on analyzing sequence or biochemical properties of the general protein surfaces and are not antibody specific. Based on the hypothesis that sequence variation of epitope residues likely affects antibody neutralization, the neutralization-based method described here provides an efficient way to identify epitope residues in cases where antibody neutralization panels of diverse viral strains are available. Neutralization information has been used elsewhere to classify broadly neutralizing antibodies and to understand the antigenic signature associated with these antibodies (25, 54). We have demonstrated that this method can predict the epitopes of HIV-1 antibodies with high accuracy. Experimental validation of the prospective predictions for antibody 8ANC195 revealed dependence of antigen recognition by this antibody on the presence of N-linked glycans at residues 234 and 276, indicating that these residues might indeed be part of the antibody epitope. To our knowledge, there is no other known HIV-1 broadly neutralizing antibody reported to target this epitope, making 8ANC195 an interesting target for further structural characterization. Similar findings with respect to glycan reactivity of antibody 8ANC195 were also reported recently (26).
Although the availability of neutralization panels over diverse viral strains is a prerequisite for neutralization-based methods, such panels are often performed as one of the first steps in the evaluation of an antibody of interest, in order to define its neutralization breadth and potency. Furthermore, although some variability in the measurements of neutralization potency can be observed upon multiple repeats of the experiment for the same antibody against the same virus, such variability is expected to have minimal effects on the prediction accuracy, since neutralization potency was treated as a binary value. Indeed, this was the case when our method was applied to predict the epitopes of antibodies VRC01 and VRC-PG04 based on two neutralization panels with an overlapping set of strains (see Fig. S6 and Table S9 in the supplemental material). We further note that, in addition to neutralization panels, the computational framework described here can also be used with other experimental measurements, such as binding affinity panels.
A number of factors that could affect the performance of the methods were explored further. Although the neutralization panel used in this study contained 181 viral strains, a panel of around 100 non-sequence-redundant viral strains could be sufficient to achieve similar prediction accuracy, and panels a third that size did not perform significantly worse. Hence, while smaller panels may not fully capture the true diversity of the viral protein, they nevertheless seem to possess sufficient information for the prediction of epitope residues. Since the variability of the residue types at each position is dependent on the input sequences, the mutual information scores were normalized with the Shannon entropy of the residue type at each position (see Materials and Methods) instead of using mutual information directly as the scoring function. The prediction accuracy using this normalized mutual information was slightly better than that of its unnormalized counterpart in the low-FP-rate region (see Fig. S7 in the supplemental material). Furthermore, in our computational protocol, the IC50 cutoff for neutralization potency was set as a binary variable at 50 μg/ml, which was the detection limit of the neutralization assay. Although alternative cutoffs may also be used, we note that, for the current set of antibodies, the prediction accuracy with an IC50 cutoff at 50 μg/ml was higher than that using other IC50 cutoffs with lower values (see Fig. S8 in the supplemental material). A potential explanation for this observation may be that a cutoff at the detection limit of the assay (50 μg/ml) may give a more robust classification of neutralized versus nonneutralized strains for each antibody, compared to using a lower IC50 as a cutoff. Nevertheless, we note that the cutoff must be lowered for pan-neutralizing antibodies (for which the neutralization IC50s are less than 50 μg/ml for all viral strains), since otherwise the mutual information scores between residue types at each position and neutralization potency will always be zero. We also note that the computation of mutual information scores dictates the use of discrete classes, since classes for sequence variation were formed based on amino acid types. A binary variable was selected for the neutralization potency, although discretization into a larger number of classes could also be performed; however, using a ternary variable reduced the prediction accuracy for the current set of antibodies (see Fig. S9 in the supplemental material). Finally, while an inverse trend between the number of epitope residues and the prediction accuracy was observed, the correlation was not statistically significant (see Fig. S10 in the supplemental material). It is interesting to speculate that a possible explanation for such a trend may be that, in general, there exists only a small number of epitope residues whose variation has a more substantial effect on antibody neutralization and that can thus be more easily picked up by the neutralization-based epitope prediction method, and this may have a more pronounced effect within the context of a smaller versus a larger epitope. Nevertheless, Fig. S10 in the supplemental material shows that, with few exceptions, the TP rates for most antibodies fall within the range of approximately 0.4 to 0.6, indicating that epitope size is not a major determinant for prediction accuracy.
Incorporating information from unbound antigen structures, which are available for many antigens (such as for different regions of HIV-1 Env and for influenza virus HA), was found to enhance the neutralization-based epitope prediction. In addition to removing surface-inaccessible residues, we conjectured that residues with lower nMI scores could nevertheless be a part of the epitope if they are surface proximal to patches that contained residues with higher nMI scores, since antibody epitopes are generally formed by one or more contiguous surface patches. Indeed, scoring each residue not by its nMI score alone but by adding a weighted sum of the nMI scores for structure-proximal residues improved the predictions in most of the test cases. On the other hand, if a residue with a high nMI score is in an isolated patch surrounded only by residues with low nMI scores, it may actually not be part of the epitope and its score may be due either to some distal effects responsible for antibody resistance or to an artifact of, e.g., sequence covariation with epitope residues. For the method variants without antigen structure incorporation, averaging in the nMI scores of sequence-neighboring residues achieved similar but lesser effects. Adding the epitope propensity score developed by Haste Andersen et al. (16), which reflects the tendency of a particular amino acid type to be an epitope residue, into the scoring function also slightly improved the prediction accuracy. The score is statistically derived and scores amino acid types based on the frequency with which they are observed in epitope regions compared to nonepitope regions (e.g., asparagine, arginine, proline, and lysine have higher propensity scores since they are overrepresented in epitope regions, while cysteine, alanine, leucine, valine, and phenylalanine have lower scores since they are underrepresented in epitope regions). Hence, the inclusion of the precomputed propensity scores does not substantially increase computational complexity. Since the SA/nMIproxsum/PS method had the highest average prediction accuracy at the 0.05 FP rate level for the 19 HIV-1 antibodies in our data set, we selected it as the default method variant. However, it should be noted that, statistically, this method does not perform significantly better than SA/nMIproxsum (see Table S3 in the supplemental material), indicating that the prediction power of SA/nMIproxsum/PS still derives mainly from summing the mutual information scores of spatially proximal residues. Further improvements in prediction accuracy may be achieved by incorporating additional sequence and structural information from mining known antibody-antigen interfaces.
The neutralization-based methods described here are limited to observing the direct effects on neutralization of sequence variation within an antibody epitope. For example, the methods failed to predict epitope residues for the CD4i antibody 17b, possibly due to the dependence of neutralization by 17b on the requirement for conformational change of the viral protein (55), a secondary effect that is not taken into account by the neutralization-based methods and that can misguide the epitope predictions. Another major caveat of using mutual information between residue types and neutralization potency to predict epitope residues is the pronounced inverse dependence of prediction accuracy on the conservation level of epitope residues. For example, residue D368 of HIV-1 gp120, which is important for the binding of various VRC01 class antibodies, could not be predicted as an epitope residue by calculating the mutual information alone, since that residue was conserved among all viral strains in our data set. Thus, the neutralization-based epitope prediction methods described here are more applicable for antigens (and epitopes) showing larger diversity.
We further note that approximately half of the residues incorrectly predicted (FP rate = 0.05) by the SA/nMIproxsum/PS method to be part of the epitope for the set of 19 antibodies were found within 5 Å within an epitope residue, and ∼65% were within 10 Å (see Fig. S11 in the supplemental material). This observation could indicate that some of these false-positive residues may nevertheless be important for interactions with the given antibody, either through direct interactions explained by strain-to-strain epitope variation, or in a supporting role for other neighboring epitope residues. Indeed, although we evaluated the prediction accuracy of the algorithm relative to antibody-antigen complex structures, the nature of the neutralization-based methods should also allow the identification of residues that are not necessarily part of, or even in proximity to, the epitope defined by the antibody-antigen complex structures but that may still, through distal effects, be of functional importance for antibody recognition.
Despite their limitations, the neutralization-based methods generated promising results, with significant enrichment and multiple-fold improvement over random epitope selection for the majority of the tested antibodies. It would also be of utility if these methods could be associated with a measure of the reliability for the epitope predictions for a given antibody. To that end, the neutralization-based methods can be augmented with the epitope-specific antibody clustering method described previously (25), which is applicable to both conserved and diverse antigens but is dependent on the existence of a reference set of antibodies with known epitopes and cannot make predictions at the residue level since no antigen sequence or structural information is incorporated. Where applicable, the antibody clustering method can be applied to predict the general epitope region for a query antibody, possibly also providing an indication of the level of conservation of the epitope. In turn, this can serve as a measure of the reliability of the neutralization-based residue-level epitope predictions. Alternatively, in the general case of a novel epitope where prior knowledge of the sequence conservation level of the epitope residues is not available, it may be beneficial to examine other factors that may affect the reliability of the predictions. Hence, we determined that there is a significant relationship between residue score distribution and prediction accuracy for a given epitope: the larger the ratio of the median of the top residue scores to the median of all residue scores, the higher the prediction accuracy (see Fig. S12 in the supplemental material). Further work will be necessary to evaluate variants of the residue score distribution and other factors as measures of reliability for the epitope predictions for a given antibody.
The epitope prediction methods described here show that it is possible to obtain detailed structural information on HIV-1 Env antibody recognition through analysis of functional readouts such as antibody-antigen neutralization data. It is interesting to speculate that the inverse information flow could also be exploited: for example, given a target epitope region for which currently no known neutralizing antibodies exist, it may be possible to devise a (hypothetical) antibody neutralization fingerprint that matches that epitope. Such epitope-specific neutralization fingerprints (25) can then be used to analyze donor sera, as a way to identify new broad and potent antibodies targeting the epitopes of interest. Similarly, one can start by first devising hypothetical antibody neutralization fingerprints, followed by prediction of the epitopes that may match such fingerprints. It is likely that not all generated fingerprints could represent real antibodies, so a search over the neutralization fingerprint space may be necessary. This process could give insight into the susceptibility of a specific virus to broad neutralization and can help identify novel broadly neutralizing epitopes as new targets for immunogen design.
The identification of neutralizing antibody epitopes plays an important role in understanding how these antibodies bind to their antigen and is a key step to designing epitope-specific immunogens. Since structure determination of antibody-antigen complexes is not always feasible, the wealth of information hidden in antibody neutralization panels can be used to efficiently extract antibody epitope information at the residue level and can be augmented with other experimental methods (such as cryo-electron microscopy and alanine or peptide scanning) for confirmation or more complete delineation of the epitopes for antibodies of interest. The identification of a novel epitope for a highly potent and broad neutralizing antibody can also be used as a starting point for designing immunogens to elicit similar antibodies. The computational framework described here therefore provides an efficient way to identify epitope residues on antigens characterized by sequence diversity and could further improve the understanding of strain-specific antibody resistance and facilitate the development of epitope-based vaccines.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program of the Vaccine Research Center, NIAID, the International AIDS Vaccine Initiative's Neutralizing Antibody Consortium, the Office of AIDS Research, NIH, and the following grants to M.C.N.: Bill and Melinda Gates Foundation grant 38619s and NIH grants AI 100663-01 and AI 100148-01. M.C.N. is an HHMI investigator.
We thank J. Stuckey for assistance with graphics and the Structural Biology Section, Structural Bioinformatics Core, Humoral Immunology Section, and Humoral Immunology Core at the NIH Vaccine Research Center for helpful discussions or comments on the manuscript. We thank J. Baalwa, D. Ellenberger, D. Gabuzda, F. Gao, B. Hahn, K. Hong, J. Kim, F. McCutchan, D. Montefiori, L. Morris, J. Overbaugh, E. Sanders-Buell, G. Shaw, R. Swanstrom, M. Thomson, S. Tovanabutra, C. Williamson, and L. Zhang for contributing the HIV-1 envelope plasmids used in our neutralization panels.
Footnotes
Published ahead of print 10 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00984-13.
REFERENCES
- 1. Corti D, Langedijk JP, Hinz A, Seaman MS, Vanzetta F, Fernandez-Rodriguez BM, Silacci C, Pinna D, Jarrossay D, Balla-Jhagjhoorsingh S, Willems B, Zekveld MJ, Dreja H, O'Sullivan E, Pade C, Orkin C, Jeffs SA, Montefiori DC, Davis D, Weissenhorn W, McKnight A, Heeney JL, Sallusto F, Sattentau QJ, Weiss RA, Lanzavecchia A. 2010. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One 5:e8805. 10.1371/journal.pone.0008805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. 2011. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333:1633–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Protocol GPI, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, Lee JH, Dillon MA, O'Neil RE, Faynboym AM, Horowitz M, Horowitz L, Ward AB, Palese P, Webby R, Lerner RA, Bhatt RR, Wilson IA. 2012. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489:526–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, Kepler TB, Liao HX, Harrison SC. 2011. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. U. S. A. 108:14216–14221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, Bournazos S, Mouquet H, Spatz LA, Diskin R, Abadir A, Zang T, Dorner M, Billerbeck E, Labitt RN, Gaebler C, Marcovecchio PM, Incesu RB, Eisenreich TR, Bieniasz PD, Seaman MS, Bjorkman PJ, Ravetch JV, Ploss A, Nussenzweig MC. 2012. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature 492:118–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sette A, Fikes J. 2003. Epitope-based vaccines: an update on epitope identification, vaccine design and delivery. Curr. Opin. Immunol. 15:461–470 [DOI] [PubMed] [Google Scholar]
- 9. Adams O, Bonzel L, Kovacevic A, Mayatepek E, Hoehn T, Vogel M. 2010. Palivizumab-resistant human respiratory syncytial virus infection in infancy. Clin. Infect. Dis. 51:185–188 [DOI] [PubMed] [Google Scholar]
- 10. Diskin R, Scheid JF, Marcovecchio PM, West AP, Jr, Klein F, Gao H, Gnanapragasam PN, Abadir A, Seaman MS, Nussenzweig MC, Bjorkman PJ. 2011. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science 334:1289–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yu CM, Peng HP, Chen IC, Lee YC, Chen JB, Tsai KC, Chen CT, Chang JY, Yang EW, Hsu PC, Jian JW, Hsu HJ, Chang HJ, Hsu WL, Huang KF, Ma AC, Yang AS. 2012. Rationalization and design of the complementarity determining region sequences in an antibody-antigen recognition interface. PLoS One 7:e33340. 10.1371/journal.pone.0033340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ofek G, Guenaga FJ, Schief WR, Skinner J, Baker D, Wyatt R, Kwong PD. 2010. Elicitation of structure-specific antibodies by epitope scaffolds. Proc. Natl. Acad. Sci. U. S. A. 107:17880–17887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gershoni JM, Roitburd-Berman A, Siman-Tov DD, Tarnovitski Freund N, Weiss Y. 2007. Epitope mapping: the first step in developing epitope-based vaccines. BioDrugs 21:145–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Glaeser RM, Hall RJ. 2011. Reaching the information limit in cryo-EM of biological macromolecules: experimental aspects. Biophys. J. 100:2331–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Geysen HM, Meloen RH, Barteling SJ. 1984. Use of peptide synthesis to probe viral antigens for epitopes to a resolution of a single amino acid. Proc. Natl. Acad. Sci. U. S. A. 81:3998–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haste Andersen P, Nielsen M, Lund O. 2006. Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci. 15:2558–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kulkarni-Kale U, Bhosle S, Kolaskar AS. 2005. CEP: a conformational epitope prediction server. Nucleic Acids Res. 33:W168–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Parker JM, Guo D, Hodges RS. 1986. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry 25:5425–5432 [DOI] [PubMed] [Google Scholar]
- 19. Ponomarenko J, Bui HH, Li W, Fusseder N, Bourne PE, Sette A, Peters B. 2008. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics 9:514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ahmad S, Mizuguchi K. 2011. Partner-aware prediction of interacting residues in protein-protein complexes from sequence data. PLoS One 6:e29104. 10.1371/journal.pone.0029104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhao L, Wong L, Li J. 2011. Antibody-specified B-cell epitope prediction in line with the principle of context-awareness. IEEE/ACM Trans. Comput. Biol. Bioinform. 8:1483–1494 [DOI] [PubMed] [Google Scholar]
- 22. Halperin I, Ma B, Wolfson H, Nussinov R. 2002. Principles of docking: An overview of search algorithms and a guide to scoring functions. Proteins 47:409–443 [DOI] [PubMed] [Google Scholar]
- 23. Vajda S, Camacho CJ. 2004. Protein-protein docking: is the glass half-full or half-empty? Trends Biotechnol. 22:110–116 [DOI] [PubMed] [Google Scholar]
- 24. Wodak SJ, Mendez R. 2004. Prediction of protein-protein interactions: the CAPRI experiment, its evaluation and implications. Curr. Opin. Struct. Biol. 14:242–249 [DOI] [PubMed] [Google Scholar]
- 25. Georgiev IS, Doria-Rose NA, Zhou T, Kwon YD, Staupe RP, Moquin S, Chuang GY, Louder MK, Schmidt SD, Altae-Tran HR, Bailer RT, McKee K, Nason M, O'Dell S, Ofek G, Pancera M, Srivatsan S, Shapiro L, Connors M, Migueles SA, Morris L, Nishimura Y, Martin MA, Mascola JR, Kwong PD. 2013. Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science 340:751–756 [DOI] [PubMed] [Google Scholar]
- 26. West AP, Jr, Scharf L, Horwitz J, Klein F, Nussenzweig MC, Bjorkman PJ. 2013. Computational analysis of anti-HIV-1 antibody neutralization panel data to identify potential functional epitope residues. Proc. Natl. Acad. Sci. U. S. A. 110:10598–10603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cardoso RM, Brunel FM, Ferguson S, Zwick M, Burton DR, Dawson PE, Wilson IA. 2007. Structural basis of enhanced binding of extended and helically constrained peptide epitopes of the broadly neutralizing HIV-1 antibody 4E10. J. Mol. Biol. 365:1533–1544 [DOI] [PubMed] [Google Scholar]
- 29. Chen L, Kwon YD, Zhou T, Wu X, O'Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, Yang ZY, Zhang MY, Arthos J, Burton DR, Dimitrov DS, Nabel GJ, Posner MR, Sodroski J, Wyatt R, Mascola JR, Kwong PD. 2009. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science 326:1123–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. 2012. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491:406–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu RB, Fu BZ, Gnanapragasam PN, Oliveira TY, Seaman MS, Kwong PD, Bjorkman PJ, Nussenzweig MC. 2013. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 153:126–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, Comparative Sequencing Program NISC. Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia SM, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BT, Kwong PD, Mascola JR, Haynes BF. 2013. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496:469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O'Dell S, Patel N, Shahzad-ul-Hussan Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang GY, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang ZY, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang LX, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ofek G, Tang M, Sambor A, Katinger H, Mascola JR, Wyatt R, Kwong PD. 2004. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J. Virol. 78:10724–10737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pancera M, Shahzad-Ul-Hussan S, Doria-Rose NA, McLellan JS, Bailer RT, Dai K, Loesgen S, Louder MK, Staupe RP, Yang Y, Zhang B, Parks R, Eudailey J, Lloyd KE, Blinn J, Alam SM, Haynes BF, Amin MN, Wang LX, Burton DR, Koff WC, Nabel GJ, Mascola JR, Bewley CA, Kwong PD. 2013. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nat. Struct. Mol. Biol. 20:804–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O'Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, Program NCS, Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. 2011. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333:1593–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P, Georgiev I, Altae-Tran H, Chuang GY, Joyce MG, Kwon YD, Longo NS, Louder MK, Luongo T, McKee K, Schramm CA, Skinner J, Yang Y, Yang ZY, Zhang Z, Zheng A, Bonsignori M, Haynes BF, Scheid JF, Nussenzweig MC, Simek M, Burton DR, Koff WC, Program NCS, Mullikin JC, Connors M, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. B cell ontogeny of the VRC01 class of HIV-1-neutralizing antibodies. Immunity, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 84:1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shu Y, Winfrey S, Yang ZY, Xu L, Rao SS, Srivastava I, Barnett SW, Nabel GJ, Mascola JR. 2007. Efficient protein boosting after plasmid DNA or recombinant adenovirus immunization with HIV-1 vaccine constructs. Vaccine 25:1398–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Doria-Rose NA, Louder MK, Yang Z, O'Dell S, Nason M, Schmidt SD, McKee K, Seaman MS, Bailer RT, Mascola JR. 2012. HIV-1 neutralization coverage is improved by combining monoclonal antibodies that target independent epitopes. J. Virol. 86:3393–3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li Y, O'Dell S, Wilson R, Wu X, Schmidt SD, Hogerkorp CM, Louder MK, Longo NS, Poulsen C, Guenaga J, Chakrabarti BK, Doria-Rose N, Roederer M, Connors M, Mascola JR, Wyatt RT. 2012. HIV-1 neutralizing antibodies display dual recognition of the primary and coreceptor binding sites and preferential binding to fully cleaved envelope glycoproteins. J. Virol. 86:11231–11241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Steuer R, Kurths J, Daub CO, Weise J, Selbig J. 2002. The mutual information: detecting and evaluating dependencies between variables. Bioinformatics 18(Suppl 2):S231–S240 [DOI] [PubMed] [Google Scholar]
- 46. Hubbard SJT, Thornton JM. 1993. NACCESS. Department of Biochemistry and Molecular Biology, University College London, London, United Kingdom [Google Scholar]
- 47. Kringelum JV, Lundegaard C, Lund O, Nielsen M. 2012. Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLoS Comput. Biol. 8:e1002829. 10.1371/journal.pcbi.1002829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sweredoski MJ, Baldi P. 2008. PEPITO: improved discontinuous B-cell epitope prediction using multiple distance thresholds and half sphere exposure. Bioinformatics 24:1459–1460 [DOI] [PubMed] [Google Scholar]
- 49. Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, Grosdidier A, Hernandez C, Ioannidis V, Kuznetsov D, Liechti R, Moretti S, Mostaguir K, Redaschi N, Rossier G, Xenarios I, Stockinger H. 2012. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40:W597–W603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57:289–300 [Google Scholar]
- 51. Dey B, Svehla K, Xu L, Wycuff D, Zhou T, Voss G, Phogat A, Chakrabarti BK, Li Y, Shaw G, Kwong PD, Nabel GJ, Mascola JR, Wyatt RT. 2009. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog. 5:e1000445. 10.1371/journal.ppat.1000445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Acharya P, Zhou T, Luongo T, Moquin S, Louder MK, Schmidt SD, Zhang B, Yang Y, Scheid JF, Nussenzweig MC, Mascola JR, Kwong PD. 2013. CD4-binding-site recognition by VH1-46-derived HIV-1 neutralizers, p 136 In Proceedings of the 2013 Keystone Symposia Meeting on HIV Vaccines, Keystone Symposia on Molecular and Cellular Biology, Keystone, CO [Google Scholar]
- 53. Doria-Rose NA, Georgiev I, O'Dell S, Chuang GY, Staupe RP, McLellan JS, Gorman J, Pancera M, Bonsignori M, Haynes BF, Burton DR, Koff WC, Kwong PD, Mascola JR. 2012. A short segment of the HIV-1 gp120 V1/V2 region is a major determinant of resistance to V1/V2 neutralizing antibodies. J. Virol. 86:8319–8323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gnanakaran S, Daniels MG, Bhattacharya T, Lapedes AS, Sethi A, Li M, Tang H, Greene K, Gao H, Haynes BF, Cohen MS, Shaw GM, Seaman MS, Kumar A, Gao F, Montefiori DC, Korber B. 2010. Genetic signatures in the envelope glycoproteins of HIV-1 that associate with broadly neutralizing antibodies. PLoS Comput. Biol. 6:e1000955. 10.1371/journal.pcbi.1000955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang W, Canziani G, Plugariu C, Wyatt R, Sodroski J, Sweet R, Kwong P, Hendrickson W, Chaiken I. 1999. Conformational changes of gp120 in epitopes near the CCR5 binding site are induced by CD4 and a CD4 miniprotein mimetic. Biochemistry 38:9405–9416 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.