Abstract
The elucidation of genetic causes of cholestasis has proved to be important in understanding the physiology and pathophysiology of the liver. Protein-truncating mutations in the tight junction protein 2 gene (TJP2) are shown to cause failure of protein localisation, with disruption of tight-junction structure leading to severe cholestatic liver disease. This contrasts with the embryonic-lethal knockout mouse, highlighting differences in redundancy in junctional complexes between organs and species.
Progressive familial intrahepatic cholestasis (PFIC) was described in 19691. Common to all patients is early-onset progressive liver disease. PFIC may be subdivided based on levels of serum γ-glutamyl transferase activity (GGT)2-4. While most forms of cholestasis manifest elevated levels of GGT, most patients with PFIC have normal or near-normal GGT. In 1998, 2 genes were identified as bearing recessive mutation in different subsets of patients with normal-GGT PFIC2, 4. FIC1 (familial intrahepatic cholestasis protein 1) deficiency is caused by mutations in ATP8B1, and BSEP (bile salt export pump) deficiency by mutations in ABCB11. Studies have since shown that approximately one third of patients with normal-GGT PFIC do not have mutations in either gene5.
Thirty-three children from 29 families were studied. All children had chronic cholestatic liver disease, with low GGT for the degree of cholestasis, and were free of mutations in ABCB11 and ATP8B1. Eighteen of the 29 families were consanguineous. Samples were examined by targeted resequencing (TRS), whole-exome sequencing (WES), or both. Four separate sequencing projects contributed to this work (Supplementary Fig. 1). The TRS assay was designed to capture 21 genes known to be associated with cholestasis. In addition to the PFIC genes, the list included genes responsible for other inherited disorders in which cholestasis features (Supplementary Table 1).
For both next-generation sequencing methods, reads were mapped and variants called using NextGene® software or exome sequencing pipelines with the ExomeDepth program®, a copy number variation (CNV) detection tool. Common variants were removed by comparison with dbSNP, 1000 Genome Project, and NHLBI Exome Sequencing Project databases. Synonymous mutations and non-synonymous mutations that are not predicted to have a deleterious effect on the function of the protein by PolyPhen, SIFT, or MutationTaster tools were filtered out.
TRS of 18 patients identified 5 individuals, each bearing a distinct homozygous mutation in TJP2 (patients 1-5). Independently performed, WES identified a disease-causing mutation in TJP2 in 2 siblings from a consanguineous family. One of these was also amongst the 5 identified by TRS (patient 2a). In light of this knowledge homozygous mutations in TJP2 were discovered in 3 further individuals by examination of WES data. No other disease-causing variants were found in these families (Supplementary Tables 2, 3 and 4a-e). Three affected siblings of patients identified through TRS were confirmed by Sanger sequencing (SS) to have the same mutations (Supplementary Fig. 1). Clinical and genetics features of the 12 affected patients are described in Table 1 and Supplementary Fig.2. A 4-base deletion in family 1 and 2 one-base deletions in families 2 and 3 were located in exon 5, causing frameshifts and generation of novel protein-coding sequences ending with the same premature termination codon (PTC). An additional one-base deletion was identified in exon 9 in family 4, causing a frameshift and a consequent downstream PTC. Family 5 harboured a mutation in the splice acceptor site of intron 13; cDNA sequencing confirmed that this mutation resulted in alternative splicing utilizing the contiguous AG bases on the 5′ end of exon 14, causing a 2-base deletion in the cDNA, introducing a frameshift and consequent PTC. Structural variations were identified with the CNV tool and examination of the WES data. A deletion of 11 exons (exons 6-16) was found in family 6; splicing was shown to occur between the 2 exons adjacent to the intronic breakpoints. The novel mRNA was confirmed, by cDNA sequencing, to contain a PTC. An additional rearrangement was identified in family 7 on inspection of WES data, with the final exon of the gene completely deleted. Last, in WES data a nonsense mutation was identified in family 8. All mutations were confirmed by PCR/SS, although in family 7 the exact breakpoints have not been found. All are predicted to abolish protein translation, and none has been identified previously.
Table 1. Clinical and genetic characteristics of the 12 patients found to harbour TJP2 homozygous mutations.
All presented in the first 3 months of life. Eight of 12 undergone OLT. None manifested elevated GGT, after the post-natal period. Normal range for GGT at 2 months is <200 IU/1; by 9 months the upper limit of normal is 55 IU/1. Mutations were described using the transcript NM_004817 as a reference sequence.
| Family number | Patient number | Age at presentation (months) |
Consanguinity | Mutation discovery | Exon/intron number | Nucleotide change | Predicted amino acid change |
Age at OLT (years) | Early, post-neonatal GGT (IU/l) |
Age GGT measured (months) |
Notes |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 3 | Y | TRS | Exon 5 | c.766_769delGCCT | p.Ala256ThrfsTer54 | 2.6 | 15 | 6 | |
| 2 | 2a | 0.5 | Y | WES | Exon 5 | c.885delC | p.Ser296AlafsTer15 | ND | 58 | 4 | Stable age 7 |
| 2b | 2 | Y | TRS and WES | 10 | 74 | 2 | Subdural hematomas | ||||
| 3 | 3 | 2 | Y | TRS | Exon 5 | c.782delA | p.Tyr261SerfsTer50 | 2 | 44 | 3 | |
| 4 | 4a | 1 | Y | TRS | Exon 9 | c.1361delC | p.Ala454GlyfsTer60 | 2.5 | 67 | 27 | |
| 4b | 0.25 | Y | SS | 8.5 | 75 | 24 | |||||
| 5 | 5a | 2 | Y | TRS | Intron 13 | c.1992-2A>G | p.Arg664SerfsTer2 | 6 | 22 | 23 | |
| 5b | 2 | Y | SS | 4 | 27 | 23 | |||||
| 6 | 6a | 3 | Y | WES | Exon 6-16 | c.953-735_2356- 249del |
p.Glu318GlyfsTer2 | 4 | 63 | 24 | Chronic respiratory disease |
| 6b | 2 | Y | SS | ND | 109 | 2 | Died at 13 months | ||||
| 7 | 7 | 1 | Y | WES | Exon 23 | c.3408-?_3573+?del | p.Ser1136ArgTer2 | ND | 37 | 24 | Stable age 4, PEBD |
| 8 | 8 | 1 | N | WES | Exon 13 | c.1894C>T | p.Arg632Ter | 1.5 | 48 | 2.5 |
Abbreviations: OLT, Orthotopic liver transplantation; GGT, serum γ-glutamyl transferase; ND, not done, PEBD, Partial external biliary diversion.
The effect of mutations on gene expression within the liver was investigated using qRT-PCR. Total RNA was isolated from liver biopsy specimens from 5 affected individuals (1, 4a, 5a, 5b, 6a) and from liver of 6 allograft donors. GAPDH, encoding glyceraldehyde 3-phosphate dehydrogenase, served as internal control. A mean reduction of 50% of TJP2 mRNA levels was found in patients compared with controls, suggesting activation of nonsense-mediated decay (Supplementary Fig. 3).
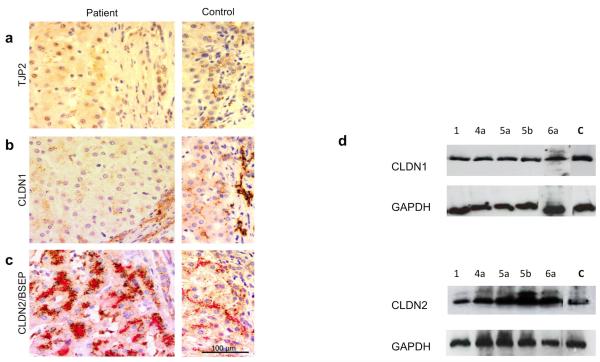
Using liver tissue, immunohistochemical studies (patients 1, 3, 4a, 4b, 5a, 5b, 6a) and western blotting (patients 1, 4a, 5a, 5b, 6a) performed with an antibody against a C-terminal TJP2 epitope unsurprisingly detected no protein (Fig. 1a). Claudins are integral to tight-junction structure6. They have been demonstrated to bind to the PDZ1 domain of TJPs in vitro7. Claudin-1 (CLDN1) and claudin-2 (CLDN2) are known to be expressed in the liver6 and to manifest different sub-cellular localisations8. Immunohistochemical analysis of CLDN1 expression showed tight-junction marking in controls. Staining was substantially reduced in liver tissue from patients (Fig. 1b). Normal expression of CLDN1 was, however, identified by western blotting analysis (Fig. 1d). As expected, CLDN2 showed a pericanalicular staining pattern in controls8. This distribution was preserved in patient material with an apparent increase in abundance (Figure 1c). However, western blotting analysis showed normal levels of CLDN2 protein (Fig. 1d). These findings suggest that CLDN1, although expressed, fails to localise in the absence of TJP2, whilst CLDN2 localisation is maintained. At transmission electron microscopy tight junctions appeared elongated and lacked the densest part of the zona occludens, as seen in ZO-2 (Tjp2) knockout mouse embryos9 (Supplementary Fig. 4).
Figure 1. rotein consequences of TJP2 mutation.
Immunohistochemical staining of patient 5a (left panels), with control (right panels) for TJP2 (a), CLDN1 (b) and CLDN2 / BSEP (c) proteins. TJP2 expression (brown) is absent from cholangiocytes and canalicular margins. CLDN1 expression (brown) is markedly reduced at both sites. CLDN2 expression (brown), clustered within cytoplasm adjoining bile canaliculi (highlighted by BSEP, red), as has been shown previously. Hematoxylin counterstain; scale bar= 100 μm. (d) Western blot for CLDN1 and CLDN2 proteins isolated from liver biopsy tissues (1 control and 5 patients). Normalization was assessed using GAPDH as a loading control.
TJP2 is a cytosolic component of several classes of cell-cell junctions10. Through interaction with cytoskeletal proteins and integral membrane proteins, members of the TJP family are important in the localisation of components of these paracellular structures. Human TJP2 generates 2 isoforms (A and C), both widely expressed11. A single incompletely penetrant homozygous missense mutation (affecting both isoforms) was previously identified in TJP2 in some Amish patients with familial hypercholanaemia, a disorder manifesting pruritus and fat malabsorption but not progressive liver disease. This mutation impaired binding of TJP2 to claudins in vitro12. In contrast, the mutations identified in the patients presented here are predicted to abolish translation of both isoforms of TJP2. Long-term survival in these patients contrasts with the embryonic lethality in the homozygous mouse ortholog knockout9 and suggests important inter-species differences.
All patients investigated had severe liver disease. Nine of 12 have required liver transplantation, 2 have stable liver disease with mild portal hypertension aged 4 and 7 years, and one died at 13 months. In addition, one had recurrent unexplained hematomas, despite normal clotting at 8 years and one has poorly characterised lung disease. Immunohistochemical study finds that CLDN1 fails to localise normally to cholangiocyte-cholangiocyte borders and to biliary canaliculus margins, despite normal protein levels. However, localisation of CLDN2 does not seem similarly dependent on TJP2 binding. We hypothesise that, in humans, the lack of TJP2 alone is insufficient to cause substantial disruption to tight junctions at epithelia outside the liver. In the unusually hostile environment of the canalicular and cholangiocytic membranes, exposed as these are to high concentrations of detergent bile acids, expression of TJP2 may be essential for hepatobiliary integrity13. In mice, Tjp2 has been shown to be non-redundant. We have now shown that this lack of redundancy is, for the human ortholog, probably restricted to the liver and that a complete absence of TJP2 causes severe liver disease.
Online Methods
Patients
Patients were selected from clinical databases in each centre, or from the Childhood Liver Disease Research and Education Network (ChiLDREN) longitudinal study of genetic causes of intrahepatic cholestasis (LOGIC; ClinicalTrials.gov Identifier: NCT00571272). All had chronic cholestatic liver disease, with GGT low for the degree of cholestasis and were known to be free of mutations in ABCB11 and ATP8B1. All samples had previously been collected for research purposes, after IRB/Ethics approval and parental consent. Samples used were all de-identified and coded. In the case of local databases this was performed after collection of clinical data; within the ChiLDREN network this was done prospectively.
Next-generation sequencing
For the Target Enrichment Sequencing method, the probes library was designed using the Agilent eArray web portal (https://earray.chem.agilent.com/earray/). The 176 kilobase (kb) of targeted regions included 364 translated and untranslated exons of 21 genes involved in the major inherited cholestatic disorders (Supplementary Table 1), and from 1 kb to 3 kb upstream of transcriptional start site of gene promoters. The bait library was optimized for 120-mer bait length centred on the region of interest with 4X coverage, with 40 base pairs overlapping the target region boundaries; masking was enabled to avoid repeat regions. To increase the coverage and consequently the adequacy of base-calling detection, the designed bait library was divided into 6 different groups according to the amount of GC content using the SureSelect Bin Orphan and High GC Baits Workflow from the Galaxy web-based platform; each group of baits was then boosted. Samples were prepared with the SureSelectXT Target Enrichment Kit (Agilent Technologies). Whole-exome sequencing was3undertaken using the SureSelectXT Human All Exon V4 kit (Agilent Technologies) and the SeqCap EZ Human exome Library v3.0 (Roche NimblGen), according to manufacturers’ protocols. In both cases cluster generation was carried on with C-Bot cluster generation automated system and multiplexed sequencing on HiSeq 2000/2500 sequencers with 2X100 paired end module (Illumina).
NGS Data analysis
NGS data were processed and analyzed using NextGENe® software (Softgenetics) and exome sequencing pipelines. Sequences were all aligned to hg19/GRCh37 human reference genome.
NextGENe® Software
Reads were converted in FASTA format using a Phred score ≥15 and a minimum coverage of 25X as a quality filter. Reads were mapped using a preloaded BWT alignment algorithm and mutations were called only when meeting the following values: mutation frequency at a given position ≥20% and mutation coverage ≥5×.
Exome sequencing analysis
Slight differences were employed by different laboratories. However, sequence alignment was performed using Novalign or BWA. Alignments were processed in the SAM/BAM format14 using SAMtools14 and Picard tool, and SNVs and InDels were called using GATK Unified Genotyper15, 16. Afterward, genetic variations were annotated using ANNOVAR. ExomeDepth was used for CNV analysis 17.
Subsequently, variations were compared to several different databases: NCBI dbSNP build 129/132, 1000 Genome Project, NHLBI Exome Sequencing Project (ESP), Exome Variant Server, and internal databases. Variations occurring with allele frequency greater than 2% in the population were then filtered out. Potential disease-causing mutations were predicted using PolyPhen, SIFT, MutationTaster, and Ensembl’s SNP Effect Predictor18. A bespoke Perl script was used to identify biallelic variants compatible with autosomal recessive inheritance (either homozygous or potential compound heterozygous variants). Perl scripts are available on request from DAP (d.a.parry@leeds.ac.uk).
CNVs were removed if the read ratio was between 0.2 and 1.5, if they were known19 or if the Bayes Factor was ≤10.
Sanger sequencing validation
Sanger sequencing validation was performed for all patients found to harbour a TJP2 mutation and for their affected siblings. Forward and reverse primers were manually designed on the flanking mutated regions (listed with annealing temperature conditions in Supplementary Table 5a). PCR amplification was optimized in accordance to the standard PCR protocol using FastStart Taq DNA Polymerase, dNTPack (Roche Applied Science). Sequencing reaction was performed using the BigDye® v.1.1 Terminator cycle sequencing kit and the ABI Prism® 3130xl Genetic Analyzer (Life Technologies). To investigate the effect of splice site mutation and frameshift deletions on mRNA splicing, cDNA was also Sanger sequenced. Forward and reverse primers were manually designed, some within exons and others across boundaries (listed with annealing temperature conditions in Supplementary Table 5b).
TJP2 mRNA expression
Frozen liver from 5 liver biopsy specimens and 6 liver-donor controls was used. Total RNA was isolated from 30 mg of liver tissue using Trizol® (Invitrogen); cDNA was synthesized using 1 μg/μl RNA and the high capacity RNA-to-cDNA kit (Life Technologies) following the manufacturers’ protocols. To evaluate TJP2 mRNA expression, quantitative RT-PCR was carried out using a TaqMan® Gene expression assay with glyceraldehyde 3-phosphate dehydrogenase (GADPH) as endogenous control. Primers and probe sets for TJP2 and GAPDH were purchased from Life Technologies. Reactions were performed in triplicate, with an ABI Prism® 7000 Sequence Detection System following manufacturer’s instructions. The data were analyzed by comparative Ct method (2 −ΔΔCt).
Western blot analysis
Proteins were extracted from 50 mg of frozen liver tissues, homogenized with 200 μl of radioimmunoprecipitation assay buffer containing 1% nonyl phenoxypolyethoxylethanol, 0.5% sodium deoxycholate and 0.1% sodium dodecyl sulphate (SDS) with protease inhibitors added just before use (10 μl/ml phenylmethylsulfonyl fluoride, 30 μl/ml aprotinin, 10 μl/ml sodium orthovanadate). Protein concentrations were determined using Lowry’s method with bovine serum albumin protein standard. Protein samples were normalized to a concentration of 1 μg/μl and heated with 10% of western blot buffer (0.5 M Tris pH 6.8, 0.35 M SDS, 30% glycerol, 0.6 M dithiothreitol, and 0.175 M bromphenol blue) and 10% of β-mercaptoethanol for 5 minutes at 100°C. They then were submitted to electrophoretic separation on 8% and 15% SDS-polyacrylamide gels for CLDN-1/CLDN-2 and TJP2 respectively. Proteins were transferred onto Hybond enhanced-chemiluminescence nitrocellulose membranes (GE Healthcare) and blocked with 10% of non-fat milk diluted in 0.1% of Tris-buffered saline and 0.05% Tween 20 (TBST) for 1 hour. Primary immunoblotting was carried out using antibodies to CLDN-1 (1:5,000; 18-7362, Zymed Laboratories, Invitrogen), CLDN-2 (1:5,000; 32-5600, Invitrogen), TJP2 (1:5,000; LS-B2185, Source BioScience) and GAPDH as loading control (1:20,000; MAB374, Millipore), and incubated overnight at 4 °C. The membranes were washed with TBST 0.1% for 3 × 10 minutes. Secondary immunoblotting was carried out with horseradish peroxidase (HRP)-conjugated rabbit-specific antibody (1:10,000; sc-3837, Santa Cruz Biotech) or HRP-conjugated mouse-specific antibody (1:20,000; NA931V, GE Healthcare) for 1 hour incubation at room temperature. The membranes were washed with TBST 0.1% for 3 × 10 minutes. The Amersham ECL Prime chemiluminescence detection system was used to visualize protein bands (GE Healthcare).
Immunohistochemistry
Archival material obtained for purposes of clinical diagnosis and treatment, fixed in formalin and processed into paraffin wax by routine methods, from patients with mutation in TJP2 and from patients with cholestatic liver disease ascribed to known mutation in other genes (ATP8B1, ABCB11) or considered idiopathic (extrahepatic biliary atresia) was sectioned at 4 μl and routinely immunostained (BondMax, Leica Biosystems) with the antibodies used for western blotting (see above) against TJP2, CLDN1 and CLDN2 proteins, with hematoxylin counterstaining. The antibody to BSEP was rabbit polyclonal HPA019035, Sigma-Aldrich.
Transmission electron microscopy
Tissue specimens were primarily fixed in paraformaldehyde / glutaraldehyde, post-fixed in osmium tetroxide (OsO4), and embedded in resin. Ultrathin sections were stained with uranyl acetate / lead citrate.
Supplementary Material
Acknowledgments
M.S. was funded by Alex Mowat PhD Studentship. R.J.T. and B.E.C. received funding for this work from a consumables grant awarded by King’s College Hospital Department of Research and Development. TRS and WES were supported by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health. C.A.J. was supported by a Sir Jules Thorn Award for Biomedical Research (JTA/09). Additional funding for this project included NIH R56 DK094828 to L.N.B. and R.J.T., the UCSF-King’s College Health Partners Faculty Fellowship Travel Grant (UCSF Academic Senate) to L.N.B., NIH U01 DK062500 to P. Rosenthal, NIH U01 DK062453 and UL1 TR000154 to R.J.S. and NIH U01 DK062456 to J.C.M. Further WES was undertaken by the University of Washington Center for Mendelian Genomics (UW CMG) and was funded by the National Human Genome Research Institute and the National Heart, Lung and Blood Institute grant 1U54HG006493 to Drs. Debbie Nickerson, Jay Shendure, and Michael Bamshad.
Footnotes
AUTHOR CONTRIBUTIONS
M.S. performed the majority of the experimental work, analysed and interpreted data, and wrote the manuscript. S.S., E.P., C.V.L. performed experiments. P.R. and B.C. helped design the targeted capture. D.A.P., J.D.S and M.A.S. analysed WES data. L.J.N. helped with Western blotting. B.E.W. performed electron microscopy. B.M.K., S.L. and P.Mc provided patients and sequence data. G.M.V. and T.G. provided patients. J.C.M. and R.J.S. run ChiLDREN and provided samples and clinical data. C.M.G. at UW performed WES for ChiLDREN patients. A.S.K. undertook histopathological analysis. C.A.J. and L.N.B. directed WES projects. R.J.T. initiated the project, analysed the data and wrote the manuscript. All authors commented on and edited the final manuscript.
References
- 1.Clayton RJ, et al. Am J Dis Child. 1969;117:112–24. [PubMed] [Google Scholar]
- 2.Bull LN, et al. Nat Genet. 1998;18:219–24. doi: 10.1038/ng0398-219. [DOI] [PubMed] [Google Scholar]
- 3.de Vree JM, et al. Proc Natl Acad Sci U S A. 1998;95:282–7. doi: 10.1073/pnas.95.1.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strautnieks SS, et al. Nat Genet. 1998;20:233–8. doi: 10.1038/3034. [DOI] [PubMed] [Google Scholar]
- 5.Davit-Spraul A, et al. Hepatology. 2010;51:1645–55. doi: 10.1002/hep.23539. [DOI] [PubMed] [Google Scholar]
- 6.Mitic LL, et al. Am J Physiol Gastrointest Liver Physiol. 2000;279:G250–4. doi: 10.1152/ajpgi.2000.279.2.G250. [DOI] [PubMed] [Google Scholar]
- 7.Itoh M, et al. J Cell Biol. 1999;147:1351–63. doi: 10.1083/jcb.147.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holczbauer A, et al. J Histochem Cytochem. 2013;61:294–305. doi: 10.1369/0022155413479123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu J, et al. Mol Cell Biol. 2008;28:1669–78. doi: 10.1128/MCB.00891-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fanning AS, et al. Mol Biol Cell. 2012;23:577–90. doi: 10.1091/mbc.E11-09-0791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chlenski A, et al. Biochim Biophys Acta. 2000;1493:319–24. doi: 10.1016/s0167-4781(00)00185-8. [DOI] [PubMed] [Google Scholar]
- 12.Carlton VE, et al. Nat Genet. 2003;34:91–6. doi: 10.1038/ng1147. [DOI] [PubMed] [Google Scholar]
- 13.Grosse B, et al. Hepatology. 2012;55:1249–59. doi: 10.1002/hep.24761. [DOI] [PubMed] [Google Scholar]
- 14.Li H, et al. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DePristo MA, et al. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKenna A, et al. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plagnol V, et al. Bioinformatics. 2012;28:2747–54. doi: 10.1093/bioinformatics/bts526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLaren W, et al. Bioinformatics. 2010;26:2069–70. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conrad DF, et al. Nature. 2010;464:704–12. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.