Abstract

Polluxochrin (1) and dioschrin (2), two new dimers of sulochrin linked by thioether bonds, were purified from an Alternaria sp. isolate obtained from a Hawaiian soil sample. The structures of the two metabolites were established by NMR, mass spectrometry data, and X-ray analysis. Metabolite 1 was determined to be susceptible to intramolecular cyclization under aqueous conditions, resulting in the generation of 2 as well as another dimeric compound, castochrin (3). An additional nine new metabolites were also obtained, including four new pyrenochaetic acid derivatives (8–11), one new asterric acid analogue (13), and four new secalonic acid analogues (14–17). Bioassay analysis of these compounds revealed 1–3 displayed antimicrobial and weak cytotoxic activities.
The Hawaiian Islands consist of a patchwork of biodiverse habitats exhibiting a high degree of species endemism.1,2 Recent reports in the field of natural products have highlighted the impact of Hawaii’s biodiversity on the likelihood for discovering new bioactive metabolites, especially from microbial sources.3−7 For example, our research group reported the discovery of several new bioactive metabolites from a biosynthetically talented fungal (Aspergillus sp.) isolate obtained from soil collected near Waikiki Beach, Honolulu, Hawaii.8 Among the compounds obtained from this fungus, the new prenylated indole alkaloid waikialoid A was notable because of the potent inhibition it afforded against Candida albicans biofilm formation (IC50 1.4 μM).8
An examination of additional samples from the Hawaiian Islands has provided a second biosynthetically talented fungal isolate, Wailua PDA-3, from a soil sample collected in the vicinity of Wailua Falls. An ethyl acetate extract prepared from a small-scale solid-phase fermentation of this fungus exhibited antifungal activity against both C. albicans and Aspergillus fumigatus. Subsequent LC-PDA-ESIMS profiling of the isolate’s secondary metabolome revealed an assortment of chemically diverse metabolites with masses ranging from approximately 200 to 700 Da. Sequencing of the isolate’s ITS region suggested that the fungus was taxonomically similar to Alternaria longissima. LC-PDA-ESIMS-based dereplication of natural products reported from this genus revealed that none of the putative metabolites we observed could be readily attributed to those ascribed to an Alternaria sp.
In this report, we describe 12 new natural products from this Alternaria sp. isolate. All of the compounds were tested in bioassays targeting different cell types and bioactivity end points (including antibacterial, antifungal, cancer cell cytotoxicity, and disruption of Candida biofilm formation). Compounds 1–3, which are new sulochrin dimers linked via a thioether bridge, showed antimicrobial and weak cytotoxic activities.
Results and Discussion
The Alternaria sp. isolate was prepared for scale-up chemical analysis by culturing it for 4 weeks on a solid-state medium composed of Cheerios breakfast cereal supplemented with a 0.3% sucrose solution.9−11 The fungal biomass was extracted with EtOAc, yielding a crude extract that was subsequently processed over several types of sorbents (including silica gel, Sephadex LH20, and C18) to provide purified metabolites 1–17.
Compound 1 was obtained as a pale yellow, amorphous powder, and its molecular formula was established as C34H30O14S based on HRESIMS data ([M + Na]+ ion at m/z 717.1253, calcd 717.1248) indicating 20 degrees of unsaturation. The 1H and 13C NMR spectra of 1 provided evidence for approximately half of the expected proton and carbon resonances, which were assigned as belonging to 11 nonprotonated carbons, three methines, three methyls, and three exchangeable protons. The initial analysis of the data set led us to suspect that 1 might be a dimeric compound. Further examination of the 1H and 13C NMR data (Table 1) indicated that the resonances observed for 1 were similar to those for the known metabolite sulochrin (4),12 which we also purified from the same fungal extract. The major differences between 1 and 4 as revealed by NMR were the loss of signals for a methine unit (δH 6.91; δC 107.6) and the gain of a new nonprotonated sp2 carbon resonance (δC 109.1). The necessity of including a sulfur atom in 1 as required by the molecular formula led us to propose that the sulfur served as a thioether bridge linking the C-2 and C-2′ carbons. Thus, the structure of the new sulochrin dimer was proposed as illustrated (1), and the metabolite was given the trivial name polluxochrin. Metabolite 1 shares structural similarities to other sulochrin dimers including, disulochrin,9 as well as guignasulfide,13 which is a demethoxy analogue of 1.
Table 1. 1H (400 MHz) and 13C (100 MHz) NMR and HMBC Data for Compound 1 (DMSO-d6).
| no. | δC, mult. | δH, mult. | HMBC |
|---|---|---|---|
| 1/1′ | 136.2 C | ||
| 2/2′ | 109.1 C | ||
| 3/3′ | 160.4 C | ||
| 4/4′ | 100.9 CH | 6.52 s | 2/2′, 3/3′, 1a/1a′, 4a/4a′ |
| 4a/4a′ | 157.7 C | ||
| 1a/1a′ | 124.3 C | ||
| 5/5′ | 107.9 CH | 6.10 s | 7/7′, 11/11′, 8a/8a′ |
| 5a/5a′ | 161.9 C | ||
| 6/6′ | 148.3 C | ||
| 7/7′ | 107.9 CH | 6.10 s | 5/5′, 11/11′, 8a/8a′ |
| 8/8′ | 161.9 C | ||
| 8a/8a′ | 109.6 C | ||
| 9/9′ | 197.8 C | ||
| 10/10′ | 56.3 CH3 | 3.61 s | 4a/4a′ |
| 11/11′ | 22.1 CH3 | 2.16 s | 5/5′, 6/6′, 7/7′ |
| 12/12′ | 167.3 C | ||
| 13/13′ | 52.3 CH3 | 3.48 s | 12/12′ |
| 3/3′-OH | 10.3 br s | ||
| 8/8′-OH | 11.2 s | 7/7′, 8/8′, 8a/8a′ | |
| 5a/5a′-OH | 11.2 s | 5/5′, 5a/5a′, 8a/8a′ |
Chart 1.
Compound 2 was purified as pale yellow, block-like crystals. HRESIMS provided a pseudomolecular ion at m/z 685.0981 [M + Na]+ (calcd 685.0986) that supported the molecular formula C33H26O13S. This corresponded to 21 degrees of unsaturation. Inspection of the 1H and 13C NMR data for 2 revealed that the spectra superficially appeared as the superimposition of two sets of signals (Table 2) with one set virtually identical to those generated for compound 1 and the other set bearing substantial similarity to methyl-(1,6-dihydroxy-3-methylxanthone)-8-carboxylate (5),14 which was also purified from the fungal extract. The additional unit of unsaturation afforded via the incorporation of 5 in this metabolite readily accounted for the overall difference of one unsaturation unit between metabolites 1 and 2. The change from a single C-9 diaryl ketone signal in 1 (δC 197.8) (Table 1) to pyrone (δC 179.2) and diaryl (δC 197.7) ketone signals in 2 (Table 2) further supported the asymmetric dimeric nature of the new metabolite. This assertion was subsequently confirmed by HSQC and HMBC experiments (Figures S9 and S10), as well as data generated from a single-crystal X-ray diffraction experiment (Figure 1). Compound 2 has been given the trivial name dioschrin.
Table 2. 1H (400 MHz) and 13C (100 MHz) NMR and HMBC Data for Compound 2 (DMSO-d6).
| no. | δC, mult. | δH, mult. | HMBC | no. | δC, mult. | δH, mult. | HMBC |
|---|---|---|---|---|---|---|---|
| 1 | 137.0 C | 1′ | 136.2 C | ||||
| 2 | 109.6 C | 2′ | 107.9 C | ||||
| 3 | 164.9 C | 3′ | 159.9 C | ||||
| 4 | 102.8 CH | 6.80 s | 2, 3, 1a, 4a | 4′ | 100.8 CH | 6.50 s | 2′, 3′, 1a′, 4a′ |
| 4a | 157.2 C | 4a′ | 157.5 C | ||||
| 1a | 118.5 C | 1a′ | 124.4 C | ||||
| 5 | 107.8 CH | 6.84 s | 7, 10, 5a, 8a | 5′ | 107.9 CH | 6.11 s | 7′, 11′, 8a′ |
| 5a | 155.4 C | 5a′ | 161.9 C | ||||
| 6 | 149.1 C | 6′ | 148.3 C | ||||
| 7 | 111.7 CH | 6.63 s | 5, 8, 10, 8a | 7′ | 107.9 CH | 6.11 s | 5′, 11′, 8a′ |
| 8 | 161.0 C | 8′ | 161.9 C | ||||
| 8a | 106.1 C | 8a′ | 109.6 C | ||||
| 9 | 179.2 C | 9′ | 197.7 C | ||||
| 10 | 22.4 CH3 | 2.38 s | 5, 6, 7 | 10′ | 56.3 CH3 | 3.62 s | 4a′ |
| 11 | 167.4 C | 11′ | 22.1 CH3 | 2.17 s | 5′, 6′, 7′ | ||
| 12 | 53.0 CH3 | 3.88 s | 11 | 12′ | 166.8 C | ||
| 8-OH | 12.3 br s | 7, 8, 8a | 13′ | 52.2 CH3 | 3.48 s | 12′ | |
| 8′-OH | 11.2 s | 7′, 8′, 8a′ | |||||
| 5a′-OH | 11.2 s | 5′, 5a′, 8a′ |
Figure 1.

ORTEP structure generated from the X-ray diffraction data for a single crystal of 2.
Compounds 1 and 2 were determined to be susceptible to transformation under aqueous conditions. Upon incubation for 48 h in various aqueous solvent mixtures, compound 1 was observed by LC-PDA-ESIMS to transform into 2, which subsequently converted to another compound (3) (Figure S1). The transformation process was determined to be temperature dependent, and even mild heating for brief periods led to the rapid loss of 1 and generation of 2 and 3. To obtain material for structure characterization, 5.0 mg of 1 in aqueous MeOH was held at 40 °C for 6 h, generating 3.5 mg of 3. The purified product was examined by HRESIMS, which yielded a pseudomolecular ion at m/z 629.0757 [M – H]− (calcd 629.0754) corresponding to the molecular formula C32H22O12S. An analysis of the 1H and 13C NMR data for the product (Table S1) indicated that 3 was structurally similar to compounds 1 and 2. Immediately apparent was that the 13C NMR diaryl ketone signal in 1 (C-9/9′, δC 197.8) had been replaced by a pyrone ketone signal in 3 (C-9/9′, δC 179.1) (Tables 1 and S1). Subsequent 2D NMR experiments (Figures S14 and S15) confirmed that these changes resulted in the formation of a new compound that consisted of a thioether-linked dimer of 5 connected to the sulfur atom through C-2 and C-2′. In contrast to metabolite 1, compound 3 exhibited two nearly superimposable sets of NMR signals. The doubling of the signals is proposed to result from the hindered rotation of the S–C-2 and/or S–C-2′ bonds. Compound 3 has been given the trivial name castochrin. A similar cyclization phenomenon has been observed for the transformation of metabolite 4 to 5, but in this case, the reaction rate was much slower, taking upward of a week to occur. These results imply that the sulfur atom might serve to accelerate the intramolecular cyclization of 1 into 2 and 3.
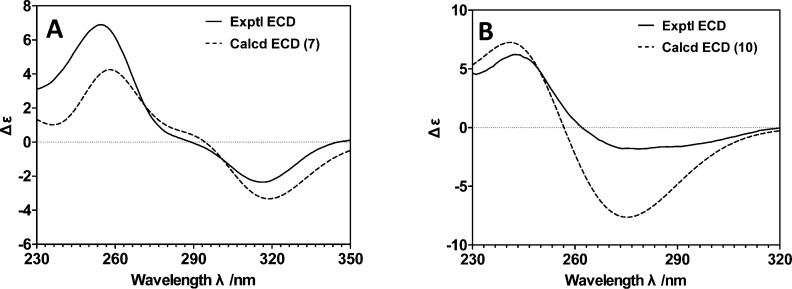
The molecular formula for compound 7 was determined by HRESIMS to be C13H16O5. An analysis of the 1D and 2D NMR (in acetone-d6 and DMSO-d6) for 7 indicated that this metabolite had the same planar structure as pyrenochaetic acid D,15 but the published account of this metabolite did not address the compound’s absolute configuration. To address this problem, we prepared crystals of 7 for X-ray diffraction analysis (Figure 3). The metabolite’s absolute configuration was determined to be 8S by the Hooft method.16 This was further supported by comparing the ECD experimental data for 7 with a computationally derived theoretical spectrum (Figure 4A, refer to the Supporting Information for a discussion of how the theoretical ECD data for 7 were prepared).
Figure 3.

ORTEP structure generated from the X-ray diffraction data for a single crystal of 7.
Figure 4.
Calculated and experimental ECD data for compounds 7 (A) and 10 (B).
Compound 8 was determined by HRESIMS to have the same molecular formula as 7. Analysis of the 1H and 13C NMR data for 8 (Tables 3 and 4) revealed that it possessed the same carbon skeleton as the co-occurring metabolite pyrenochaetic acid C (6).17 However, the carbon and proton signals attributable to an oxidized methylene (CH2-10, δH 3.42; δC 60.4) were replaced by signals for an aliphatic methyl. Taking into account the HRESIMS data, it was proposed that compound 8 contained a new C-10 hydroxyl group relative to 6. Combining the proton and carbon chemical shift patterns for 8 with the 1H–1H COSY correlations among H-8 and H-9 and H-10, as well as the HMBC correlations between H-8 and H-9 with C-7 (Figure 2), the position of the new hydroxyl group was unambiguously assigned. This new metabolite has been given the name pyrenochaetic acid E (8).
Table 3. 1H NMR Data for Compounds 7–11 (400 MHz, DMSO-d6).
| 7 | 8 | 9 | 10 | 11 | |
|---|---|---|---|---|---|
| no. | δH, mult. (J in Hz) | δH, mult. (J in Hz) | δH, mult. (J in Hz) | δH, mult. (J in Hz) | δH, mult. (J in Hz) |
| 2 | 7.36 s | 7.39 s | 7.40 s | 7.31 s | 7.42 s |
| 6 | 7.42 s | 7.44 s | 7.43 s | 7.38 s | 7.49 s |
| 7 | 5.31 s | ||||
| 8 | 4.25 dd (8.6, 3.9) | 2.76 t (7.4) | 2.81 t (5.7) | ||
| 9 | 1.67 m | 1.72 m | 1.89 m | 2.58 dq (18.0, 7.3) | 2.82 q (7.2) |
| 1.45 m | 2.31 dq (18.0, 7.3) | ||||
| 10 | 0.90 t (7.4) | 3.42 t (7.4) | 4.03 t (5.4) | 0.91 t (7.3) | 1.06 t (7.2) |
| 11 | 3.79 s | 3.83 s | 3.82 s | 3.73 s | 3.78 s |
| 12 | 2.15 s | 2.16 s | 2.15 s | 2.29 s | 2.29 s |
| 15 | 2.00 s | ||||
| 7-OH | 5.52 br s | ||||
| 13-OH | 11.43 br s | 13.32 br s |
Table 4. 13C NMR Data for Compounds 7–11 (100 MHz, DMSO-d6).
| 7 | 8 | 9 | 10 | 11 | |
|---|---|---|---|---|---|
| no. | δC, mult. | δC, mult. | δC, mult. | δC, mult. | δC, mult. |
| 1 | 133.1 C | 132.7 C | not detected | 131.7 C | 135.3 C |
| 2 | 109.5 CH | 109.7 CH | 109.8 CH | 109.9 CH | 110.3 CH |
| 3 | 156.4 C | 156.0 C | 156.0 C | 157.4 C | 159.3 C |
| 4 | 133.6 C | 135.2 C | 134.0 C | 132.7 C | 127.9 C |
| 5 | 136.3 C | 135.2 C | 134.9 C | 139.1 C | 140.1 C |
| 6 | 124.1 CH | 124.2 CH | 124.2 CH | 124.5 CH | 124.8 CH |
| 7 | 209.5 C | 207.1 C | 206.6 C | 72.5 CH | 196.3 C |
| 8 | 78.0 CH | 40.8 CH2 | 40.5 CH2 | 211.9 C | 201.7 C |
| 9 | 26.0 CH2 | 26.9 CH2 | 22.7 CH2 | 31.1 CH2 | 29.7 CH2 |
| 10 | 10.7 CH3 | 60.4 CH2 | 63.5 CH2 | 8.0 CH3 | 7.3 CH3 |
| 11 | 56.2 CH3 | 56.3 CH3 | 56.2 CH3 | 56.2 CH3 | 56.8 CH3 |
| 12 | 19.1 CH3 | 18.7 CH3 | 18.8 CH3 | 19.7 CH3 | 19.5 CH3 |
| 13 | 167.4 C | 167.3 C | 167.9 C | 167.5 C | 166.9 C |
| 14 | 170.8 C | ||||
| 15 | 21.1 CH3 |
Figure 2.
Selected 1H–1H COSY and HMBC correlations for compounds 7–11.
Pyrenochaetic acid F (9) was purified as a colorless, amorphous solid, and its molecular formula was established to be C15H18O6 based on HRESIMS data ([M – H]− ion at m/z 293.1034, calcd 293.1031). The compound exhibited NMR resonances similar to 8 with the addition of proton and carbon signals corresponding to an acetoxy group (Tables 3 and 4). The inclusion of the acetoxy group in the structure was confirmed on the basis of an HMBC correlation from H-10 (δH 4.03) to C-14 (δC 170.8) (Figure 2).
Pyrenochaetic acid G (10) was obtained as a colorless, amorphous solid. An [M – H]− molecular ion at m/z 251.0932 (calcd 251.0925) in the HRESIMS spectrum was in agreement with the molecular formula C13H16O5. Compound 10 displayed 13C NMR resonances that were similar to those for 7. The HMBC correlation data (H-7 to C-3, C-4, C-5, and C-8, as well as H-9 and H-10 to C-8) indicated that the oxidation pattern of C-7 and C-8 in 7 was reversed in 10 (i.e., C-7 bore the hydroxyl group and C-8 was a ketone) (Tables 3 and 4, Figure 2). The metabolite’s absolute configuration was determined to be 7R by comparing experimental ECD data with a theoretical spectrum of the compound (Figure 4B, refer to the Supporting Information for a discussion of how the theoretical ECD data for 10 were prepared). Thus, the structure of the new metabolite was confirmed as illustrated for 10 and was named pyrenochaetic acid G.
The molecular formula of 11 was determined to be C13H14O5 based on HRESIMS, which provided an [M – H]− ion at m/z 249.0776 (calcd 249.0768). A comparison of the 1D NMR data for 11 with 10 (Tables 3 and 4) revealed that the carbon and proton signals attributable to the C-7 methine were missing and had been replaced by a downfield ketone resonance (δC 196.3). Compound 11 was named pyrenochaetic acid H.
Dimethylamide asterrate (13) possessed the molecular formula C19H21NO7 as determined by HRESIMS, indicating that the metabolite possessed 10 degrees of unsaturation. The compound exhibited 1H and 13C NMR resonances (Table 5) that were similar to the known, co-occurring metabolite asterric acid (12),18 with the major exceptions being the addition of two new methyl groups (C-9′: δC 34.3, δH 2.91; C-10′: δC 37.5, δH 2.85). Considering the chemical shift and HMBC correlation data for 13 (δH 2.91 and 2.85 to δC 166.2), the new methyl groups were rationalized to be part of a tertiary amide that had replaced the carboxylic acid in 12.
Table 5. 1H (400 MHz) and 13C (100 MHz) NMR and HMBC Data for Compound 13 (DMSO-d6).
| no. | δC, mult. | δH, mult. | HMBC | no. | δC, mult. | δH, mult. | HMBC |
|---|---|---|---|---|---|---|---|
| 1 | 134.2 C | 1′ | 156.1 C | ||||
| 2 | 126.5 C | 2′ | 111.4 C | ||||
| 3 | 107.6 CH | 6.71 s | 1, 4, 5, 8 | 3′ | 154.7 C | ||
| 4 | 155.4 C | 4′ | 109.6 CH | 6.25 s | 2′, 3′, 6′, 7′ | ||
| 5 | 105.3 CH | 6.71 s | 1, 3, 6 | 5′ | 138.9 C | ||
| 6 | 153.7 C | 6′ | 104.3 CH | 5.57 s | 1′, 2′, 4′, 7′ | ||
| 7 | 56.5 CH3 | 3.62 s | 6 | 7′ | 21.7 CH3 | 2.02 s | 4′, 5′, 6′ |
| 8 | 165.6 C | 8′ | 166.2 C | ||||
| 9 | 52.5 CH3 | 3.62 s | 8 | 9′ | 34.3 CH3 | 2.91 s | 8′, 10′ |
| 4-OH | 9.47 br s | 10′ | 37.5 CH3 | 2.85 s | 8′, 9′ | ||
| 3′-OH | 9.81 br s |
Compound 14 was obtained as a pale yellow, amorphous solid. On the basis of the HRESIMS data ([M + H]+ ion at m/z 639.1708, calcd 639.1714), the molecular formula was established as C32H30O14 requiring 18 degrees of unsaturation. Analysis of 1H NMR, 13C NMR, and HSQC data (Table 6) revealed that this metabolite possessed 17 nonprotonated carbons, eight methines, three methylenes, and four methyl groups. Further analysis of the 2D NMR data (Figure 5) revealed that 14 shared the same planar structure as blennolide G.19 When the NMR solvent was changed from DMSO-d6 to CDCl3 (as reported in the literature), compound 14 provided 1H and 13C NMR spectra that were identical to blennolide G; however, their specific rotation values were in essence the opposite of one another {14: [α]25D −68.8 (c 0.125, CHCl3); blennolide G: [α]25D +81.1 (c 0.29, CHCl3)}. Further analysis of the ECD data for 14 (Figure S80) revealed a Cotton effect pattern that was the inverse of that generated for blennolide G. Thus, metabolite 14 was determined to be the antipode of blennolide G.
Table 6. 1H (400 MHz) and 13C (100 MHz) NMR Data for Compounds 14–17 (DMSO-d6).
|
14 |
15 |
16 |
17 |
|||||
|---|---|---|---|---|---|---|---|---|
| no. | δC, mult. | δH, mult. (J in Hz) | δC, mult. | δH, mult. (J in Hz) | δC, mult. | δH, mult. (J in Hz) | δC, mult. | δH, mult. (J in Hz) |
| 2 | 85.3 C | 84.9 C | 84.1 C | 84.6 C | ||||
| 3 | 101.0 C | 39.3 CH2 | 3.60 d (17.2) | 39.3 CH2 | 3.58 d (17.2) | 40.4 CH2 | 3.53 d (17.3) | |
| 3.11 d (17.2) | 3.08 d (17.2) | 3.03 d (17.3) | ||||||
| 4 | 187.4 C | 196.1 C | 196.2 C | 196.0 C | ||||
| 4a | 106.5 C | 107.5 C | 107.3 C | 107.6 C | ||||
| 5 | 158.9 C | 158.7 C | 158.6 C | 160.8 C | ||||
| 6 | 118.1 C | 117.5 C | 117.4 C | 109.9 CH | 6.58 d (8.6) | |||
| 7 | 140.3 CH | 7.39 d (8.5) | 141.0 CH | 7.49 d (8.5) | 141.1 CH | 7.47 d (8.6) | 141.1 CH | 7.47 d (8.6) |
| 8 | 107.9 CH | 6.55 d (8.5) | 107.7 CH | 6.67 d (8.5) | 107.6 CH | 6.65 d (8.6) | 115.4 C | |
| 8a | 157.8 C | 159.1 C | 158.8 C | 156.2 C | ||||
| 9 | 70.4 CH | 3.95 br s | 87.0 CH | 4.60 d (4.0) | 86.1 CH | 4.55 d (5.1) | 85.8 CH | 4.42 d (4.0) |
| 10 | 28.8 CH | 1.91 m | 29.8 CH | 2.89 m | 29.8 CH | 2.89 m | 29.6 CH | 2.42 m |
| 11 | 33.0 CH2 | 2.35 m | 36.1 CH2 | 2.87 dd (21.6, 9.1) | 36.2 CH2 | 2.89 dd (17.6, 9.3) | 35.5 CH2 | 2.13 dd (18.0, 8.9) |
| 2.28 dd (21.6, 9.1) | 2.31 dd (17.6, 6.0) | 2.05 dd (18.0, 5.4) | ||||||
| 12 | 180.5 C | 176.1 C | 175.8 C | 175.9 C | ||||
| 13 | 17.8 CH3 | 1.00 d (6.6) | 20.3 CH3 | 1.17 d (6.5) | 19.7 CH3 | 1.09 d (6.8) | 20.2 CH3 | 0.97 d (7.0) |
| 14 | 171.8 C | 169.3 C | 169.2 C | 169.1 C | ||||
| 15 | 53.9 CH3 | 3.64 s | 53.9 CH3 | 3.70 s | 54.1 CH3 | 3.71 s | 54.1 CH3 | 3.68 s |
| 5-OH | 11.67 s | 11.82 s | 11.79 s | 11.53 s | ||||
| 9-OH | 5.80 br s | |||||||
| 12-OH | 13.8 br s | |||||||
| 2′ | 84.8 C | 84.9 C | 84.9 C | 85.0 C | ||||
| 3′ | 39.4 CH2 | 3.58 d (17.2) | 39.3 CH2 | 3.60 d (17.2) | 39.3 CH2 | 3.59 d (17.2) | 39.3 CH2 | 3.60 d (17.2) |
| 3.09 d (17.2) | 3.11 d (17.2) | 3.09 d (17.2) | 3.12 d (17.2) | |||||
| 4′ | 196.1 C | 196.1 C | 196.1 C | 196.2 C | ||||
| 4a′ | 107.4 C | 107.5 C | 107.5 C | 107.5 C | ||||
| 5′ | 158.6 C | 158.7 C | 158.6 C | 158.6 C | ||||
| 6′ | 117.7 C | 117.5 C | 117.5 C | 117.3 C | ||||
| 7′ | 141.2 CH | 7.48 d (8.6) | 141.0 CH | 7.49 d (8.5) | 141.0 CH | 7.47 d (8.6) | 140.9 CH | 7.57 d (8.6) |
| 8′ | 107.6 CH | 6.64 d (8.6) | 107.7 CH | 6.67 d (8.5) | 107.7 CH | 6.67 d (8.6) | 107.7 CH | 6.67 d (8.6) |
| 8a′ | 159.1 C | 159.1 C | 159.1 C | 159.1 C | ||||
| 9′ | 87.0 CH | 4.58 d (4.0) | 87.0 CH | 4.60 d (4.0) | 87.0 CH | 4.58 d (4.0) | 86.9 CH | 4.59 d (4.2) |
| 10′ | 29.8 CH | 2.86 m | 29.8 CH | 2.89 m | 29.8 CH | 2.89 m | 29.8 CH | 2.86 m |
| 11′ | 36.1 CH2 | 2.84 m | 36.1 CH2 | 2.87 dd (21.6, 9.1) | 36.1 CH2 | 2.87 dd (21.8, 8.9) | 36.1 CH2 | 2.86 dd (21.0, 9.3) |
| 2.27 m | 2.28 dd (21.6, 9.1) | 2.26 dd (21.7, 8.8) | 2.28 dd (21.5, 9.2) | |||||
| 12′ | 176.1 C | 176.1 C | 176.1 C | 176.0 C | ||||
| 13′ | 20.3 CH3 | 1.15 d (6.4) | 20.3 CH3 | 1.17 d (6.5) | 20.3 CH3 | 1.15 d (6.6) | 20.2 CH3 | 1.15 d (6.3) |
| 14′ | 169.3 C | 169.3 C | 169.3 C | 169.2 C | ||||
| 15′ | 53.9 CH3 | 3.68 s | 53.9 CH3 | 3.70 s | 53.9 CH3 | 3.68 s | 53.9 CH3 | 3.69 s |
| 5′-OH | 11.81 s | 11.82 s | 11.81 s | 11.80 s | ||||
Figure 5.
Selected 1H–1H COSY and HMBC correlations for compounds 14–17.
Like compound 14, metabolites 15, 16, and 17 shared the same molecular formula (C32H30O14), as well as many other similarities among their 1H and 13C NMR spectral data (Table 6). In spite of this, compound 15 stood out as displaying half of the expected proton and carbon resonances, which indicated that it was a symmetrical dimer. HMBC correlation data (Figure 5) were essential for establishing the C-6–C-6′ linkage and overall planar structure of 15. It was subsequently determined that metabolite 16 shared the same planar structure as 15 (Figure 5). However, the presence of two distinct sets of resonances representing the two monomeric portions of 16 denoted that this metabolite was an asymmetric diastereomer of 15. In contrast to 15 and 16, HMBC correlations from H-7′ to C-8 and H-7 to C-6′ (Figure 5) indicated that metabolite 17 was covalently bonded via a C-8–C-6′ linkage. Since compounds 15–17 were not active in our panel of assays and were prone to consistent degradation during both freezer storage and room-temperature handling, our investigation of their relative configurations was discontinued.
All the purified compounds were screened for cancer cell cytotoxicity,20 antibacterial21 and antifungal22 activities, and inhibition of fungal biofilm formation.8 Only compounds 1–3 exhibited anti-MRSA activity, with MIC values of 4.1, 4.9, and 3.2 μM (2.9, 3.2, and 2.0 μg/mL), respectively, whereas the MIC for chloramphenicol was 5 μM (1.6 μg/mL). Metabolites 1–3 also showed weak mammalian cell cytotoxicity effects against pancreatic cancer cells (MIA PaCa-2) with IC50 values of 50.8, 30.3, and 29.3 μM, respectively.
Experimental Section
General Experimental Procedures
UV data were collected on a Hewlett-Packard 8452A diode array spectrophotometer. IR data were collected on a Shimadzu IR Affinity FTIR. Optical rotation data were determined on a Rudolph Research AUTOPOL III automatic polarimeter. NMR data were collected on a Varian 400 MHz NMR instrument. Accurate mass data were collected on an Agilent 6538 HRESI QTOF MS coupled to an Agilent 1290 HPLC. LC-MS data were obtained on a Shimadzu LC-MS 2020 system (ESI quadrupole) coupled to a photodiode array detector, with a Phenomenex Kinetex column (2.6 μm C18 column, 100 Å, 75 × 3.0 mm). The HPLC system utilized SCL-10A VP pumps and a system controller with Luna 5 μm C18 columns (110 Å, 250 × 21.2 mm, 10 mL/min and 110 Å, 250 × 10 mm, 4 mL/min). X-ray data were collected using a diffractometer with a Bruker APEX CCD area detector and graphite-monochromated Mo Kα radiation (λ = 0.710 73 Å). All solvents were of ACS grade or better.
Fungal Isolate Procurement and Culture Conditions
The fungal isolate (internal strain designation Wailua PDA-3) was obtained from a soil sample collected in the vicinity of Wailua Falls, Hawaii. The isolate was identified as similar to Alternaria longissima based on ITS sequence analysis (99% sequence homology). Sequence data for the isolate (GenBank accession no. KM088044) was compared by BLAST analysis to sequences publicly available through the NCBI database. The isolate was deposited in our laboratory’s fungal library. The fungal isolate was cultured under solid-state conditions on a medium composed of Cheerios breakfast cereal supplemented with a 0.3% sucrose solution with 0.005% chloramphenicol. The fungus was grown for 4 weeks at room temperature (25 °C).
Extraction and Purification
The scale-up solid-phase cultures were pooled and extracted twice overnight with EtOAc, and the organic solvent was partitioned against water. The resultant organic layers were concentrated under vacuum. The resulting extract (71 g) was separated by silica gel VLC (gradient mobile phase consisting of hexane–CH2Cl2–MeOH), which yielded four fractions (Fr.1–Fr.4). Fr.2 (eluted with 9:1 MeOH–CH2Cl2) was subjected to C18 VLC (H2O–MeOH) to give four subfractions. Subfraction 1 (eluted with 4:6 H2O–MeOH) was further purified by semipreparative HPLC (mobile phase 1:1 H2O–MeOH with 0.1% formic acid in water) to afford compounds 6 (2.0 mg), 7 (15.0 mg), 8 (2.5 mg), 9 (2.8 mg), 10 (3.0 mg), 11 (2.3 mg), and 12 (6.0 mg). Subfraction 3 (eluted with 8:2 H2O–MeOH) was applied to a Sephadex LH20 column and eluted with 1:1 CH2Cl2–MeOH. Further purification by semipreparative HPLC (mobile phase 6:4 to 9:1 MeOH–H2O with 0.1% formic acid in water) yielded 1 (8.0 mg), 2 (7.5 mg), 4 (25 mg), 5 (14 mg), 13 (6.5 mg), 14 (4.0 mg), 15 (5.1 mg), 16 (4.7 mg), and 17 (5.0 mg).
Polluxochrin (1):
pale yellow, amorphous solid; UV (MeOH) λmax (log ε) 210 (4.27), 286 (3.81), 346 (3.54) nm; IR (film) νmax 3600, 2953, 1712, 1641, 1514, 1454, 1344, 1211, 1091, 1022, 831 cm–1; 1H and 13C NMR see Table 1; HRESIMS [M + Na]+m/z 717.1253 (calcd for C34H30O14SNa, 717.1248).
Dioschrin (2):
pale yellow, block-like crystals; UV (MeOH) λmax (log ε) 208 (4.37), 238 (4.34), 364 (4.05) nm; IR (film) νmax 3600, 2930, 1712, 1645, 1585, 1514, 1365, 1209, 1089, 1020, 831 cm–1; 1H and 13C NMR see Table 2; HRESIMS [M + Na]+m/z 685.0981 (calcd for C33H26O13SNa, 685.0986).
Castochrin (3):
pale yellow powder; UV (MeOH) λmax (log ε) 210 (4.39), 246 (4.12), 298 (3.56), 364 (3.90) nm; IR (film) νmax 3610, 2935, 1739, 1699, 1651, 1514, 1365, 1269, 1211, 1008, 827 cm–1; 1H and 13C NMR see Table S1; HRESIMS [M – H]−m/z 629.0757 (calcd for C32H21O12S, 629.0754).
Pyrenochaetic acid D (7):
colorless cubic crystals; [α]25D −5.0 (c 0.18, EtOH); UV (MeOH) λmax (log ε) 212 (4.14), 252 (3.68), 302 (3.32) nm; CD (MeOH) λmax (Δε) 254 (6.82) 317 (−2.50); IR (film) νmax 2972, 1699, 1541, 1456, 1413, 1315, 1226, 1097, 974 cm–1; 1H and 13C NMR see Tables 3 and 4; HRESIMS [M – H]−m/z 251.0931 (calcd for C13H15O5, 251.0925).
Pyrenochaetic acid E (8):
colorless, amorphous solid; UV (MeOH) λmax (log ε) 212 (4.12), 246 (3.61), 298 (3.23) nm; IR (film) νmax 2941, 1697, 1541, 1456, 1413, 1315, 1234, 1095 cm–1; 1H and 13C NMR see Tables 3 and 4; HRESIMS [M – H]−m/z 251.0931 (calcd for C13H15O5, 251.0925).
Pyrenochaetic acid F (9):
colorless, amorphous solid; UV (MeOH) λmax (log ε) 210 (4.19), 298 (3.34) nm; IR (film) νmax 2966, 1737, 1687, 1546, 1454, 1413, 1238, 1097, 1039 cm–1; 1H and 13C NMR see Tables 3 and 4; HRESIMS [M – H]−m/z 293.1034 (calcd for C15H17O6, 293.1031).
Pyrenochaetic acid G (10):
colorless, amorphous solid; [α]25D −11.4 (c 0.18, MeOH); UV (MeOH) λmax (log ε) 216 (4.20), 248 (3.79), 296 (3.37) nm; CD (MeOH) λmax (Δε) 244 (6.48), 275 (−1.85); IR (film) νmax 2965, 1699, 1577, 1456, 1413, 1313, 1226, 1091 cm–1; 1H and 13C NMR see Tables 3 and 4; HRESIMS [M – H]−m/z 251.0932 (calcd for C13H15O5, 251.0925).
Pyrenochaetic acid H (11):
colorless, amorphous solid; UV (MeOH) λmax (log ε) 212 (4.19), 270 (3.88), 326 (3.46) nm; IR (film) νmax 2981, 1685, 1548, 1460, 1311, 1255, 1096 cm–1; 1H and 13C NMR see Tables 3 and 4; HRESIMS [M – H]−m/z 249.0776 (calcd for C13H13O5, 249.0768).
Dimethylamide asterrate (13):
pale yellow, amorphous solid; UV (MeOH) λmax (log ε) 212 (4.33), 284 (3.32), 310 (3.34) nm; IR (film) νmax 3439, 1625, 1469, 1357, 1249, 1205, 1149, 1060 cm–1; 1H and 13C NMR see Table 5; HRESIMS [M + H]+m/z 376.1392 (calcd for C19H22NO7, 376.1391).
(−)-Blennolide G (14):
pale yellow, amorphous solid; [α]25D −68.8 (c 0.125, CHCl3); UV (MeOH) λmax (log ε): 206 (4.26), 260 (3.97), 340 (3.85) nm; CD (MeOH) λmax (Δε) 211 (8.37), 223 (14.2), 328 (−3.35); IR (film) νmax 2967, 1788, 1739, 1612, 1435, 1361, 1213, 1047 cm–1; 1H and 13C NMR see Table 6; HRESIMS [M + H]+m/z 639.1708 (calcd for C32H31O14, 639.1714).
Blennolide H (15):
pale yellow, amorphous solid; [α]25D −75.3 (c 0.473, CHCl3); UV (MeOH) λmax (log ε) 208 (4.36), 258 (4.30), 364 (3.84) nm; IR (film) νmax 3439, 2965, 1788, 1645, 1433, 1359, 1273, 1207, 1056 cm–1; 1H and 13C NMR see Table 6; HRESIMS [M + Na]+m/z 661.1530 (calcd for C32H30O14Na, 661.1528).
Blennolide I (16):
pale yellow, amorphous solid; [α]25D −35.0 (c 0.035, CHCl3); UV (MeOH) λmax (log ε) 206 (4.32), 256 (4.25), 360 (3.79) nm; IR (film) νmax 3458, 2962, 1791, 1647, 1433, 1359, 1286, 1207, 1012 cm–1; 1H and 13C NMR see Table 6; HRESIMS [M + Na]+m/z 661.1531 (calcd for C32H30O14Na, 661.1528).
Blennolide J (17):
pale yellow, amorphous solid; [α]25D −28.8 (c 0.125, CHCl3); UV (MeOH) λmax (log ε) 210 (4.50), 256 (4.43), 360 (3.93) nm; IR (film) νmax 3441, 1788, 1745, 1647, 1469, 1354, 1280, 1172, 1055 cm–1; 1H and 13C NMR see Table 6; HRESIMS [M + Na]+m/z 661.1527 (calcd for C32H30O14Na, 661.1528).
X-ray Crystal Structure Analysis of 2 and 7
A yellow plate-shaped crystal of dimensions 0.46 × 0.26 × 0.08 mm of 2 and a colorless prism-shaped crystal of dimensions 0.44 × 0.32 × 0.18 mm of 7 were selected for structural analysis. Intensity data for these two compounds were collected using a diffractometer with a Bruker APEX ccd area detector and graphite-monochromated Mo K radiation (0.710 73 Å). Details of the X-ray crystal structure analysis for 2 and 7 are available in the Supporting Information. The X-ray crystallographic data for 2 and 7 have been deposited with the Cambridge Crystallographic Data Center under accession numbers CCDC 993668 and 993669, respectively. These data can be accessed free of charge at http://www.ccdc.cam.ac.uk/.
ECD Calculation Details of 7 and 10
Conformational analyses were carried out using Spartan’10.23 Geometry, frequency, and ECD calculations were applied at the DFT level with Gaussian 09.24 SpecDis 1.60 was used to average single ECD or UV–vis spectra after Boltzmann statistical weighting.25
Cytotoxicity Assay
Mammalian cell cytotoxicity assays were performed on pancreatic cancer (MIA PaCa-2) cells by adding 5000 cells per well into 96-well plates. The cells were allowed to adhere overnight at 37 °C in a humidified incubator (5% CO2 atmosphere). Test compounds were diluted in DMSO and added to the wells so that the final concentration of DMSO per well did not exceed 1% by volume. The plates containing treated and control cells were incubated for 48 h, and cell viability was determined by MTT assay.26
Antibacterial Assay
Compounds were tested against methicillin-resistant Staphylococcus aureus (MRSA) strain ATCC 700787, which also exhibits reduced susceptibility to vancomycin. A stock culture was diluted in brain heart infusion broth and delivered into presterilized 96-well plates. Stocks prepared in DMSO were added to each well of the 96-well plate (1% final DMSO concentration). Optical density measurements (OD600) of the cultures were determined on a Tecan Infinite M200 plate reader. The plates were incubated for 17 h in a humidified incubator at 37 °C. Plates were removed and orbitally shaken, and a final OD600 value was recorded. The final OD600 reading was subtracted from the initial OD600 reading to obtain the change in OD600 (antimicrobial activity was determined based on the change in optical density).
Acknowledgments
Research reported in this publication was supported by the National Institutes of Health (1R01AI085161), NSF (CHE-0130835), a Challenge Grant from the Office of the Vice President for Research at the University of Oklahoma, and an award through the Shimadzu Equipment Grant Program.
Supporting Information Available
NMR (1H, 13C, 1H–1H COSY, HSQC, and HMBC) and HRESIMS data of 1–3, 7–11, and 13–17, X-ray crystal structure analysis for 2 and 7, ECD data for compound 14, and detailed analyses of calculated and experimental ECD data for compounds 7 and 10. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Conry P. J.; Cannarella R.. Hawaii Statewide Assessment of Forest Conditions and Trends; Department of Land and Natural Resources, Division of Forestry and Wildlife, Honolulu, HI, 2010; pp 154–178. [Google Scholar]
- Lowery P. P.Ocean and Aquatic Ecosystems - Vol. II - Patterns of Species Richness, Endemism, and Diversification in Oceanic Island Floras; United Nations Educational, Scientific and Cultural Organization & Encyclopedia of Life Support Systems, 2009; pp 201–220. [Google Scholar]
- Greshock T. J.; Grubbs A. W.; Jiao P.; Wicklow D. T.; Gloer J. B.; Williams R. M. Angew. Chem., Int. Ed. 2008, 47, 3573–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy A. A.; Swenson D. C.; Gloer J. B.; Wicklow D. T. J. Nat. Prod. 2008, 71, 415–419. [DOI] [PubMed] [Google Scholar]
- Shim S. H.; Swenson D. C.; Gloer J. B.; Dowd P. F.; Wicklow D. T. Org. Lett. 2006, 8, 1225–1228. [DOI] [PubMed] [Google Scholar]
- Neff S. A.; Lee S. U.; Asami Y.; Ahn J. S.; Oh H.; Baltrusaitis J.; Gloer J. B.; Wicklow D. T. J. Nat. Prod. 2012, 75, 464–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q.-X.; Crews M. S.; Draskovic M.; Sohn J.; John T. A.; Tenney K.; Valeriote F. A.; Yao X.-J.; Bjeldanes L. F.; Crews P. Org. Lett. 2010, 12, 4458–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; You J.; King J. B.; Powell D. R.; Cichewicz R. H. J. Nat. Prod. 2012, 75, 707–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerea A. L.; Branscum K. M.; King J. B.; You J.; Powell D. R.; Miller A. N.; Spear J. R.; Cichewicz R. H. Tetrahedron Lett. 2012, 53, 4202–4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L.; King J. B.; Morrow B. H.; Shen J. K.; Miller A. N.; Cichewicz R. H. J. Nat. Prod. 2012, 75, 1819–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S.; Du L.; Gerea A. L.; King J. B.; You J.; Cichewicz R. H. Org. Lett. 2013, 15, 4186–4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J.; Lee J. H.; Hwang B. Y.; Kim H. S.; Lee J. J. J. Antibiot. 2002, 55, 552–556. [DOI] [PubMed] [Google Scholar]
- Wang F.-W.; Ye Y.-H.; Ding H.; Chen Y.-X.; Tan R.-X.; Sun Y.-C. Chem. Biodiversity 2010, 7, 216–220. [DOI] [PubMed] [Google Scholar]
- Hamasaki T.; Kimura Y. Agric. Biol. Chem. 1983, 47, 163–165. [Google Scholar]
- Zhang H.; Mao L.-L.; Qian P.-T.; Shan W.-G.; Wang J.-D.; Bai H. J. Asian Nat. Prod. Res. 2012, 11, 1078–1083. [DOI] [PubMed] [Google Scholar]
- Hooft R. W. W.; Straver L. H.; Spek A. L. J. Appl. Crystallogr. 2008, 41, 96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H.; Konoma K.; Sakamura S. Agric. Biol. Chem. 1981, 45, 1675–1679. [Google Scholar]
- Ohashi H.; Akiyama H.; Nishikori K.; Mochizuki J. J. Antibiot. 1992, 45, 1684–1685. [DOI] [PubMed] [Google Scholar]
- Zhang W.; Krohn K.; Zia-U; Flörke U.; Pescitelli G.; Bari L. D.; Antus S.; Kurtán T.; Rheinheimer J.; Draeger S.; Schulz B. Chem.—Eur. J. 2008, 14, 4913–4923. [DOI] [PubMed] [Google Scholar]
- Joyner P. M.; Waters A. L.; Williams R. B.; Powell D. R.; Janakiram N. B.; Rao C. V.; Cichewicz R. H. J. Nat. Prod. 2011, 74, 857–861. [DOI] [PubMed] [Google Scholar]
- Wang X.; Filho J. G. S.; Hoover A. R.; King J. B.; Ellis T. K.; Powell D. R.; Cichewicz R. H. J. Nat. Prod. 2010, 73, 942–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrikson J. C.; Ellis T. K.; King J. B.; Cichewicz R. H. J. Nat. Prod. 2011, 74, 1959–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y.; Molnar L. F.; Jung Y.; Kussmann J.; Ochsenfeld C.; Brown S. T.; Gilbert A. T.; Slipchenko L. V.; Levchenko S. V.; O’Neill D. P.; DiStasio R. A. Jr.; Lochan R. C.; Wang T.; Beran G. J.; Besley N. A.; Herbert J. M.; Lin C. Y.; Van Voorhis T.; Chien S. H.; Sodt A.; Steele R. P.; Rassolov V. A.; Maslen P. E.; Korambath P. P.; Adamson R. D.; Austin B.; Baker J.; Byrd E. F.; Dachsel H.; Doerksen R. J.; Dreuw A.; Dunietz B. D.; Dutoi A. D.; Furlani T. R.; Gwaltney S. R.; Heyden A.; Hirata S.; Hsu C. P.; Kedziora G.; Khalliulin R. Z.; Klunzinger P.; Lee A. M.; Lee M. S.; Liang W.; Lotan I.; Nair N.; Peters B.; Proynov E. I.; Pieniazek P. A.; Rhee Y. M.; Ritchie J.; Rosta E.; Sherrill C. D.; Simmonett A. C.; Subotnik J. E.; Woodcock H. L. 3rd; Zhang W.; Bell A. T.; Chakraborty A. K.; Chipman D. M.; Keil F. J.; Warshel A.; Hehre W. J.; Schaefer H. F. 3rd; Kong J.; Krylov A. I.; Gill P. M.; Head-Gordon M. Phys. Chem. Chem. Phys. 2006, 8, 3172–3191. [DOI] [PubMed] [Google Scholar]
- Frisch M. J.; Trucks G. W.; Schlegel H. B.; Scuseria G. E.; Robb M. A.; Cheeseman J. R.; Scalmani G.; Barone V.; Mennucci B.; Petersson G. A.; Nakatsuji H.; Caricato M.; Li X.; Hratchian H. P.; Izmaylov A. F.; Bloino J.; Zheng G.; Sonnenberg J. L.; Hada M.; Ehara M.; Toyota K.; Fukuda R.; Hasegawa J.; Ishida M.; Nakajima T.; Honda Y.; Kitao O.; Nakai H.; Vreven T.; Montgomery J. A. Jr.; Peralta J. E.; Ogliaro F.; Bearpark M.; Heyd J. J.; Brothers E.; Kudin K. N.; Staroverov V. N.; Kobayashi R.; Normand J.; Raghavachari K.; Rendell A.; Burant J. C.; Iyengar S. S.; Tomasi J.; Cossi M.; Rega N.; Millam J. M.; Klene M.; Knox J. E.; Cross J. B.; Bakken V.; Adamo C.; Jaramillo J.; Gomperts R.; Stratmann R. E.; Yazyev O.; Austin A. J.; Cammi R.; Pomelli C.; Ochterski J. W.; Martin R. L.; Morokuma K.; Zakrzewski V. G.; Voth G. A.; Salvador P.; Dannenberg J. J.; Dapprich S.; Daniels A. D.; Farkas Ö.; Foresman J. B.; Ortiz J. V.; Cioslowski J.; Fox D. J.. Gaussian 09, Revision A.1; Gaussian, Inc.: Wallingford, CT, 2009.
- Bruhn T.; Schaumlöffel A.; Hemberger Y.; Bringmann G.. SpecDis, Version 1.60; University of Wuerzburg: Germany, 2012.
- Hansen M. B.; Nielsen S. E.; Berg K. J. Immunol. Methods 1989, 119, 203–210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.