Abstract
The role of Rad51 in an unperturbed cell cycle has been difficult to dissect from its DNA repair function. Here, using electron microscopy (EM) to visualize replication intermediates (RIs) assembled in Xenopus laevis egg extract we show that Rad51 is required to prevent the accumulation of ssDNA gaps at replication forks and behind them. ssDNA gaps at forks arise from extended uncoupling of leading and lagging strand DNA synthesis. Instead, ssDNA gaps behind forks, which are exacerbated on damaged templates, result from Mre11 dependent degradation of newly synthesized DNA strands as they can be suppressed by inhibition of Mre11 nuclease activity. These findings reveal direct and unanticipated roles for Rad51 at replication forks demonstrating that Rad51 protects newly synthesised DNA from Mre11 dependent degradation and promotes continuous DNA synthesis.
Introduction
Genomic DNA is highly vulnerable to mutagenesis during DNA replication as replication fork progression is frequently impaired by DNA lesions caused by exogenous or endogenous factors such as UV light and reactive oxygen species. Many redundant pathways preserve fork integrity in the presence of DNA damage 1,2. This prevents the lethal effects deriving from the complete collapse of replication forks leading to double stranded breaks (DSBs). DNA lesions can be bypassed by error-prone trans-lesion (TLS) polymerases such as Pol eta or Pol zeta 3. This polymerase switching requires mono-ubiquitilation of PCNA at Lys164 mediated by Rad6-Rad18 complex 4.
Another pathway called template switching (TS) ensures continuous replication across DNA lesions in an error-free mode using newly synthesized undamaged daughter strand as a template instead of the damaged parental one to bypass the lesion. TS was proposed to entail fork regression by annealing of nascent strands at the fork 1,2.
Strand invasion of the paused nascent strand into sister chromatid to continue replication is also possible. This pathway requires homologous recombination (HR) proteins such as Rad51, the eukaryotic ortholog of RecA in E. coli, which plays a central role in HR during meiosis as well as during DSB repair 5.
Rad51 is not essential in yeast, but is required for cell proliferation in vertebrates 6,7. This suggests that in vertebrates Rad51 plays indispensable roles not only in meiotic chromosomal recombination and segregation but also in normal cell cycle. A role for Rad51 in S-phase has been postulated 8-10. However, it is unclear whether Rad51 is solely required to repair DSBs that spontaneously arise during normal cell cycle, or whether Rad51 plays any additional replicative role beyond the conventional DSBs repair function.
The above-described pathways (TLS, TS and HR), known to be involved in post-replication-repair (PRR) could operate at the fork to ensure its progression through the damage. However, it is also possible that these pathways are not necessary for the fork progression itself and rather deployed to repair gaps behind the fork 11. This issue is only poorly understood due to the lack of enough structural information about replication forks and their surrounding regions. A few studies have highlighted the presence of DNA gaps present behind forks in the presence of obstacles to replication fork progression 12,13. A recent study suggests that Rad51 mediates two distinctive pathways; one promotes replication restart after short exposure to HU, while the other promotes repair of forks completely collapsed by prolonged exposure to HU 14. The former pathway is also supported by evidence showing that UV-irradiated nucleotide excision repair (NER) mutant cells accumulate collapsed forks, which are mainly rescued by Rad51-dependent pathway to enable restart 15. These results imply that Rad51 functions both at forks and behind them.
Here, we have established a cell-free system based on X. laevis egg extract to study the role of Rad51 during DNA replication. Taking advantage of EM based analysis to directly observe replication fork structures and a biochemical assay to detect DNA gaps we have discovered that Rad51 is required to prevent formation of DNA gaps at forks and behind them. DNA gaps behind forks are suppressed by inhibition of Mre11 nuclease activity indicating that Rad51 protects nascent DNA from nuclease-mediated degradation.
Results
Rad51 associates to replicating chromatin
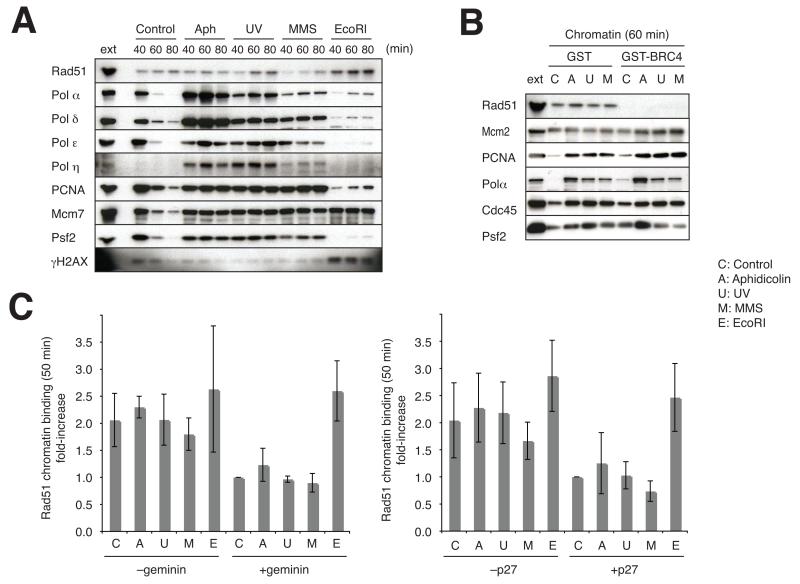
X. laevis egg extracts permit the biochemical characterization of essential DNA repair proteins involved in DNA replication 16-18,47. To verify whether Rad51 has a role in DNA replication we monitored chromatin binding of X. laevis Rad51, which is highly conserved among vertebrates and is present at the concentration of 20 nM in X. laevis egg extract (data not shown). We also monitored the binding of other replication factors during DNA replication on undamaged and damaged templates. We found that Rad51 binds to chromatin during DNA replication (Fig 1a). Its binding is impaired by inhibition of replication origin assembly, achieved by supplementing extract with geminin, which prevents MCM helicase loading 19, (Fig 1 and Supplementary Fig 1a) and by inhibition of origin firing, obtained by treating extracts with CDK inhibitor p27 20 (Fig 1 and Supplementary Fig 1b). Rad51 binding in the presence of agents that stall replication forks such as aphidicolin, UV and MMS was also sensitive to geminin and p27 (Fig 1 and Supplementary Fig 1b). In contrast, EcoRI endonuclease mediated induction of DSBs, revealed by the presence of γH2AX, was resistant to geminin and p27 treatments (Fig 1 and Supplementary Fig 1a and 1b). These data indicate that a fraction of Rad51 binding to chromatin takes place after replication forks have been established and depends at least in part on the number of active replication forks. Consistent with this the amount of Rad51 bound to chromatin was linearly correlated with the levels of Psf2 and therefore to the number of active forks (Fig 1a). Overall these data suggest that in addition to its well-known role in DSB repair, Rad51 is involved in DNA replication.
Figure 1.
Rad51 binding to undamaged and damaged chromatin during DNA replication. (A) The time course of chromatin association of Rad51 and the indicated replication proteins. Immunoblotting was performed for the chromatin fractions that were incubated in 30 μl of egg extract for the indicated times in the presence or absence of aphidicolin (10 μg ml−1) or EcoRl (0.1 unit μl−1). Where indicated sperm nuclei were treated with 1,000 J m −2 UV and 1% (v/v) MMS, respectively. As a control, 0.5 μl egg extract was also immunoblotted (ext). To measure the relative amount of Rad51 per fork we calculated the ratio between Rad51 and Psf2 signal intensity at 40 min, which was 0.35 for untreated extracts, 0.57 for aphidicolin, 0.55 for UV and 0.51 for MMS treated extracts. (B) The effect of BRC4 on the chromatin association of Rad51 and replication fork proteins. Immunoblotting was performed for the chromatin fractions that were incubated in 25 μl of egg extract for 60 min in the presence of 0.5 mg ml−1 GST or 0.5 mg ml−1 GST-BRC4. Sperm nuclei were incubated in extracts that were unreated (−) or incubated with 50 μg ml−1 aphidicolin. Where indicated sperm nuclei were irradiated with UV at 1,000 J m−2 or treated with 1% (w/v) MMS before the incubation in egg extract. As a control, 1 μl egg extract was also immunoblotted (ext). (C) Quantification of Rad51 bound to damaged and undamaged chromatin in the presence (+ geminin) or in the absence (− geminin) of 160 nM geminin, and in the presence (+p27) or absence (−p27) of 40 μg ml−1 p27 recombinant protein. The graph shows the average relative values of several repeated experiments taking as reference the amount of Rad51 bound to undamaged chromatin in the presence of geminin or p27 (C; control). Error bars indicate standard deviations. Representative immunoblots are shown in Supplementary S1A and S1B.
Effects of impaired Rad51 chromatin binding
To study Rad51 replication function we inhibited Rad51 binding to chromatin using recombinant human BRC4 (one of eight BRC motifs of BRCA2 that has a strong affinity for Rad5121) fused to GST (GST-BRC4), which efficiently binds X. laevis Rad51 even at high salt concentrations (Supplementary Fig 1c). The sole presence of GST-BRC4 completely suppressed Rad51 chromatin binding but did not impair the binding of replication proteins such as Mcm2, PCNA, Pol α, Cdc45 and Psf2 of the GINS complex (Fig 1b). This indicates that Rad51 is not required for the assembly of replication proteins onto chromatin. The BRC4 peptide used in this manuscript (aminoacid 1511–1579 of BRCA2) is different from the one used by Carreira et al 22, which has been shown to promote Rad51 binding to ssDNA in vitro. We used here the minimal GST-BRC4 concentration required to effectively suppress Rad51 binding to chromatin (data not shown). As at this concentration GST-BRC4 was able to suppress both ssDNA and dsDNA binding of Rad51 (Supplementary Fig 1d), we could not discriminate whether the effects that we observe on chromatin derive prevalently from the inhibition of Rad51 binding to ss or dsDNA.
To uncover the direct role of Rad51 in DNA synthesis we analyzed nascent ssDNA molecules recovered from X. laevis egg extracts in which Rad51 chromatin binding was inhibited by GST-BRC4. As redundant PRR pathways such as TLS could mask the role of HR in replication fork progression we also attenuated TLS using a recombinant mutant PCNA (PCNA-K164R) that cannot be ubiquitinated 23 and therefore does not fully support binding of TLS polymerase Pol η to chromatin (Supplementary Fig 1e). DNA replication efficiency was not affected by inhibition of Rad51 chromatin binding and/or impairment of TLS (Fig 2a and 2b, 1–4). Furthermore, following DNA damage induced by MMS, DNA replication efficiency decreased (Fig 2a and 2b, 5-10) due to hindrance of fork progression and checkpoint activation that inhibits further origin firing 24. However, residual DNA replication was not affected and nascent DNA strands matured with similar kinetics in the absence of Rad51 and/or PCNA ubiquitination (Fig 2a, 5–10). Collectively, these data show that Rad51 is dispensable for fork progression even with impaired TLS.
Figure 2.
Rad51 and PCNA modifications in DNA replication and ssDNA gap accumulation. (A) Rad51 and PCNA requirement for replication of untreated and MMS-treated DNA. Sperm nuclei were incubated in 10 μl egg extract with α32P-dATP for the indicated times in the presence or absence of 0.7 mg ml−1 GST or GST-BRC4 and MMS (− or +), and 0.2 mg ml−1 of recombinant wild type PCNA (WT) or mutated PCNA (K164R). Replication products were resolved on 1% (w/v) alkaline agarose gel and subjected to autoradiography. (B) The signal intensities obtained in (A) were quantified and reported on the graph. The experiments shown represent a typical result. (C) Gap labelling procedure using T4 DNA polymerase. Replicating genomic DNA was isolated and used as a template for gap-filling assay using T4 DNA polymerase. The labelled nascent molecules extended by T4 were then resolved on alkaline agarose gel. (D) Untreated (−MMS) and MMS treated (+MMS) sperm nuclei were incubated in 10 μl of egg extract in the presence of GST or GST-BRC4 for 60 min (1–4). Untreated sperm nuclei were incubated for 40, 60 or 80 min in the presence of PCNA-WT or PCNA-K164R (5–10). Genomic DNA was isolated and subjected to the gap labelling reaction followed by autoradiography. Exposure times are equivalent for the 2 gels although kinetic profile starts at 40 minutes in 5–10. The graph shows the relative fold increase in optical density measured for each lane taking as reference untreated chromatin recovered at 60 minutes. The experiment shows a typical result.
Accumulation of ssDNA gaps in the absence of Rad51
Having ruled out a role in replication fork progression we set out to uncover subtler genomic defects that could highlight Rad51 function in DNA replication. Chicken DT40 cells deficient for Rad51 accumulate ssDNA gaps 25 and DSBs 7 after one or few cell cycles, respectively. However, it is unclear whether such lesions arise directly from defects in the DNA replication process or in DNA repair. We did not detect formation of DSBs following one round of DNA replication in the absence of Rad51 bound to chromatin (data not shown). However, using a gap-filling assay 26 based on T4 DNA polymerase, which has primer extension and TLS 27 but not strand displacement activities (Fig 2c and 2d) we observed a five-fold increase of labelled ssDNA molecules on undamaged (above 10 Kb in size) and MMS damaged templates (between 0.5 and 10 Kb) in GST-BRC4-treated extracts, confirming that although DNA replication is not inhibited, ssDNA gaps accumulate in the absence of Rad51 bound to chromatin (Fig 2d, 1–4).
We also monitored ssDNA gaps accumulation in extracts deficient for TLS, which is involved in preventing ssDNA gaps accumulation following UV damage. As expected, increased ssDNA gaps were observed on damaged templates in the absence of TLS (Supplementary Fig S2), and no additive effects were observed in the presence of both GST-BRC4 and PCNA-K164R, suggesting that Rad51 and TLS operate in the same gapped regions. These observations are consistent with the PRR model, in which replication forks proceed past DNA damage leaving un-replicated single-stranded DNA gaps that are subsequently sealed by TLS and/or HR 11,28.
However, in contrast to Rad51, TLS impairment alone did not induce noticeable accumulation of ssDNA gaps on undamaged templates (Fig 2d, 5–10). This indicates that Rad51 but not TLS prevents the accumulation of such lesions on undamaged templates and suggest that Rad51 plays a specific role in preventing replication associated DNA lesions in addition to its known role in DNA repair.
Importantly, an excess of recombinant X. laevis Rad51 added back to egg extract containing GST-BRC4 suppressed the accumulation of ssDNA gaps (Supplementary Fig S3). Similar results were obtained by adding recombinant human Rad51 and not an irrelevant protein such as GST (data not shown). These control experiments confirmed the specificity of GST-BRC4 effects on Rad51.
Replication intermediate structure in the absence of Rad51
To uncover the nature Rad51 function during DNA replication we performed in vivo electron microscopic (EM) analyses of genomic RIs coupled to psoralen-crosslinking, taking advantage of established methods and procedures 29 that we adapted to sperm nuclei replicated in X. laevis egg extracts, in the presence and in the absence of DNA damage (see Methods). Upon standard enrichment procedures used for analogous analysis in yeast and mammalian cells 12,29,30, EM samples showed a high frequency of RIs. After identification of RIs (see Methods), we assessed frequency and length of ssDNA regions by detecting local differences in filament thickness. Although ssDNA stretches can also be revealed by EM upon binding of single-strand binding proteins, short ssDNA stretches fail to consistently assemble nucleoprotein complexes and may escape EM detection 12. In this respect, assessment of DNA thickness along replicating molecules proved a powerful and reliable tool to score number and size of ssDNA gaps, focusing on relative differences with control samples where similar criteria of assignment are applied 12. We found that, during DNA replication, 60% of RIs isolated from extracts in which Rad51 chromatin binding was inhibited showed at least one ssDNA gap behind the replication fork (internal gaps, Fig 3a-b), a rare event in control extracts. While MMS treatment led to minor accumulation of internal gaps in control extracts, MMS-treated Rad51-depleted ones showed 80% of gapped RIs, with 30% of RIs having more than two gaps on the same fork (Fig 3b). The size of the internal gaps is rather heterogeneous, but in most cases lower than 300 nucleotides (nt). Even though the frequency of the gaps increases in Rad51-depleted extracts, their size distribution is overall unchanged (Supplementary Fig S4). Such ssDNA gaps behind replication forks have been previously observed and related to re-priming events downstream of lesions on the template 12. Consistently, their persistence in HR and TLS deficient cells has been attributed to defects in PRR.
Figure 3.
Rad51 is required to prevent replication fork uncoupling and ssDNA accumulation on damaged and undamaged templates. (A) and (C) Electron micrographs (and schematic drawings) of representative RIs isolated from sperm nuclei, incubated in GST-BRC4 treated extracts. Black arrows point to ssDNA regions at the replication fork. White arrows point to ssDNA gaps along the replicated duplexes (internal gaps). (B) Statistical distribution of internal gaps in the analyzed population of molecules. The total number of molecules analyzed is indicated in brackets. (D) Statistical distribution of ssDNA length at replication forks isolated in the indicated conditions. The total number of forks analyzed is indicated in brackets.
As re-priming has been shown to result from extended uncoupling of leading and lagging strand synthesis 12, we analyzed RIs for the presence of ssDNA regions directly at the fork. Small ssDNA regions (<200 nt) are often detectable by this assay at unperturbed replication forks, marking discontinuous lagging strand synthesis 12. Strikingly, even in the absence of exogenous DNA damage, almost 50% of RIs in Rad51-depleted extracts showed an abnormally long (>200 nt) ssDNA region at the fork (Fig 3c-d), suggesting frequent uncoupling of leading and lagging strand synthesis. In many cases ssDNA regions up to 800 nt long could be detected (Fig 3c). As already shown for yeast 12, MMS treatment - even at concentrations that dramatically affect fork progression (Fig 2) - had only limited effects on leading and lagging strand uncoupling, as 80% of control RIs showed ssDNA regions at the fork only smaller than 200 nt in the presence of MMS (Fig 3d). This suggests that the mechanism producing ssDNA at forks is distinct from the one responsible for ssDNA gaps formation behind them in the presence of MMS induced DNA damage. Intriguingly, we also observed an accumulation of ssDNA tracts during DNA replication at forks and behind them in yeast rad52 mutants (Fig 4), in which Rad51 chromatin loading is impaired 31,32. Similar results were obtained with yeast rad51 mutants, although the accumulation of post-replicative ssDNA gaps was less pronounced than in rad52 cells. This likely reflects the described contribution of S. cerevisiae Rad59, a Rad52-paralogue that mediates Rad51-independent recombination mechanisms 33. Overall, these data indicate that Rad51 function preventing the accumulation of ssDNA gaps is conserved across different species.
Figure 4.
Accumulation of ssDNA gaps in the absence of Rad52 and Rad51 in S. cerevisiae.
Upper panels: Electron micrographs of a representative RIs isolated from rad52 mutant S. cerevisiae growing cells. The black arrow points to an extended ssDNA regions at the replication fork. White arrows show ssDNA gap behind the fork. Lower panels: Statistical distribution of ssDNA gap length (left) and of the number of ssDNA gaps (right) observed on RIs isolated from wild type, Rad52 and Rad51 mutant S. cerevisiae growing cells.
Mre11 dependent formation of ssDNA gaps behind forks
We then tested whether ssDNA accumulation arises from nuclease dependent degradation of newly synthesized DNA. To this end we treated extracts with Mirin, which specifically inhibits the activity of Mre11 34, a major nuclease present at replication forks 17,35. Remarkably, Mirin was able to prevent accumulation of detectable ssDNA gaps behind forks formed upon suppression of Rad51 binding to DNA (Fig 5a). In contrast, Mre11 inhibition by Mirin did not suppress accumulation of ssDNA at forks (Fig 5b). EM analysis was more useful in assessing the effects of Mirin and Rad51 inhibition than the gap-filling assay, as this could not discriminate between ssDNA at forks and behind them. However, consistent with the EM analysis Mirin induced a significant decrease in the amount of gaps detected with the gap-filling assay (Supplementary Fig S5). By this assay, the effect is more noticeable upon MMS treatment, where the vast majority of the labelled fragments result from ssDNA gaps behind the fork. These observations indicate that ssDNA gaps behind forks are due to Mre11 dependent degradation of nascent DNA in the absence of Rad51. ssDNA gaps at forks, instead, arise independently from Mre11, either mediated by a different nuclease or solely resulting from a DNA synthesis defect.
Figure 5.

Rad51 protects nascent strand DNA from Mre11-dependent degradation. (A) Statistical distribution of internal gaps in the analyzed population of molecules isolated from extracts that were supplemented with buffer (Control) or 100 μM Mirin and treated as indicated. The total number of molecules analyzed is indicated in brackets. (B) Statistical distribution of ssDNA length at replication forks isolated from extracts that were supplemented with buffer (Control) or 100 μM Mirin and treated as indicated. The total number of molecules analyzed is indicated in brackets. in the indicated conditions.
Discussion
The role of recombination factors in DNA replication has been postulated in the past. However, direct demonstration of this function has been impeded by the lack of an experimental system to directly address the function of recombination proteins during DNA replication. A possible direct role for Rad51 in the replication process has been inferred from recent investigations on Rad51 inactivation in DT40 cells 25. Consistent with this we also observed ssDNA gap accumulation at forks and behind them in yeast cells lacking Rad51 and in Rad52 deficient cells in which Rad51 function is impaired 31,32. Rad51 foci can be observed during unperturbed S-phase progression in cultured mammalian cells 36,37. Rad51 was also recently shown to mediate restart of transiently stalled forks, but this function is not linked to foci formation nor to its standard role in DSB repair 33.
While all these observations suggest that a replicative function of recombination factors could be well conserved among eukaryotes, they cannot effectively distinguish between a replicative and DNA repair function of Rad51. It is indeed possible that accumulation of DNA lesions in Rad51-defective cells depends on defective repair of DNA lesions accumulated after one or few cell cycles, or upon short genotoxic treatments. However, our results on X. laevis egg extracts can now effectively discriminate Rad51 function during the process of DNA synthesis in the presence and the absence of exogenous DNA lesions, combining selective Rad51 depletion just before one round of DNA replication and direct visualization of replication intermediates.
Overall our data suggest a dual role for the recombination factor Rad51 during DNA replication: restoring coupling of uncoupled leading and lagging strand synthesis and protection of nascent DNA from nucleolytic degradation (Fig 6). Our observations indicate that Rad51 binding to chromatin during DNA replication might be required to limit the size of ssDNA stretches at replication forks (Fig 6, 1–3). We propose that Rad51 is recruited to replication forks upon transient uncoupling of the fork at natural impediments and consequent accumulation of longer stretches of ssDNA, similarly to what extensively described for DSB end resection 5. Presumably, Rad51 might be recruited to forks undergoing problematic progression. Indeed, transient replication fork stalling and uncoupling (Fig 6, 2) may be frequent even during DNA replication in the absence exogenous DNA damage and may result from endogenous lesions, multi-protein complexes obstructing fork progression or sequences prone to form secondary structures. This is reflected in the high frequency of pathological ssDNA regions at forks upon suppression of Rad51 binding to chromatin during DNA replication in the absence of exogenous damage. We envision three possible non-mutually exclusive scenarios for the function of Rad51 in this context (Fig 6, 3): I) Rad51 could bind extended ssDNA on the blocked leading strand and use its strand annealing activity to favour re-annealing with the unwound lagging strand, thus counteracting helicase activity and limiting further fork uncoupling; II) Rad51 binding to the transiently uncoupled fork may assist the processivity of the stalled polymerases that encounter obstacles to DNA synthesis such as ssDNA secondary structures; III) Rad51 binding may facilitate local recruitment of translesion polymerases to promote continuous synthesis across endogenous lesions. Intriguingly translesion polymerases were shown to assist DNA synthesis on Rad51-dependent recombination intermediates 38. In addition RecA, Rad51 related protein in E. Coli, has been shown to promote recruitment of translesion polymerases 39,40.
Figure 6.
A model for possible roles of Rad51 during DNA replication. See text for explanation.
At the same time, if persistent uncoupling at bulky lesions leads to DNA synthesis re-priming, especially frequent in the presence of exogenous DNA damage (Figure 6, 4-5), Rad51 binding to the resulting ssDNA gaps behind the forks may effectively engage them in PRR (Fig 6, 7–8). According to this model, Rad51 binding to replication forks should be transient and selective for temporary uncoupled forks, whereas it is likely to be more stable in the presence of permanent DNA lesions. Intriguingly, mammalian Rad51 paralogs - that are known to regulate Rad51 recruitment - were recently reported to bind fork structures with high affinity and specificity 41. Analogously, it is tempting to speculate that anti-recombinase helicases such as Srs2, Bloom or R-TEL 42,43 may prevent unscheduled HR events by counteracting inappropriate or permanent Rad51 fork association.
While a general role for Rad51 in PRR is well established, the present data show that post-replicative ssDNA gaps not bound to Rad51 are prone to extensive Mre11-dependent degradation (Fig 6, 9–11). This may result from a direct role of Rad51 in counteracting Mre11 on these ssDNA substrates. We propose that Mre11 and Rad51 are in a dynamic equilibrium at ssDNA and counteract each other activity in a sort of feedback mechanism. On one hand, Mre11-dependent controlled resection could be required for Rad51 binding on ssDNA (Fig 6, 6), similarly to what extensively described for DSB repair and for RecA ssDNA-binding in E. coli 44. On the other hand, the engagement of these gaps in Rad51-dependent repair could avoid excessive nucleolytic degradation, sequestering the substrates once optimal Mre11 dependent resection is achieved (Fig 6, 7). In this view, the accumulation of Mre11 dependent ssDNA gaps behind forks in absence of Rad51 may reflect the accumulation of ssDNA intermediates unproductive for strand invasion, which may in turn become particularly susceptible to the resection apparatus. The absence of detectable post-replicative ssDNA gaps upon Mirin treatment and Rad51 depletion may indeed suggest that, in absence of Mre11 activity, non-resected post-replicative ssDNA gaps may be below the resolution limit of EM (50–100 nt) and escape detection even in the absence of Rad51.
Intriguingly, mutations in SbcD, the putative ortholog of Mre11 in E. coli, suppress lethality of RecBCD recombination defective cells in the presence of repetitive palindromic sequences 45. The suppression is due to the inability of SbcD mutant cells to degrade secondary structures formed at or behind replication forks 46. However, differently from Mre11 SbcD processes secondary structures formed on the template whereas the gaps we observe result from the degradation of nascent DNA strands. It is likely that the nuclease activity of Mre11 does not target parental DNA in eukaryotes. In any case these observations suggest that Mre11 dependent processing of replication dependent secondary structures arising in the context of recombination dependent events is conserved across species.
Methods
Recombinant proteins and antibodies
Recombinant human Rad51 proteins and the cDNA fragment encoding human BRC4 (amino acid 1511–1579 of BRCA2) cloned into pDONR221 (Invitrogen) were kindly provided by Dr F. Esashi (Oxford University). The fragment was then cloned into DEST15, an expression vector for GST-tagged recombinant proteins, using Gateway system (Invitrogen). The BRC4-DEST15 plasmid was transformed to BL21-Al cells, and recombinant GST-BRC4 protein production was induced by 0.2% (w/v) L-arabinose and purified with Glutathione Sepharose 4B according to standard procedures (GE Healthcare). Control GST protein was prepared using pGEX 6P-1 empty vector (GE Healthcare). The cDNA encoding full length X. laevis Rad51 was amplified by PCR using as 5′-primer (ATGGATCCATGGCCATGCAAGCTCACTATC) and 3′-primer (AGAATTCTCAGTCCTTGGCATCTCCCAC) using a X. laevis oocyte cDNA library and cloned into pGEX 6P-1. The GST-tagged recombinant protein was expressed and purified with Glutathione Sepharose 4B, and the GST-tag was removed by Prescission Protease (GE Healthcare) treatment to obtain an untagged version of Rad51. The pET28-based expression vectors of wild type and mutant (K164R) X. laevis PCNA and the pET21-based expression vector of human p27 were kindly provided from Dr H. Ulrich (Cancer Research UK) and Dr T. Hunt (Cancer Research UK) respectively, and the recombinant 6His-tagged proteins were purified with Ni-NTA agarose (QIAGEN). 6His-tagged geminin was prepared as described 47.
Antibodies against Rad51 (14B4, abcam), Pol alpha p180 subunit (ab31777, abcam), PCNA (MCA1558, serotec), Mcm7 (sc-9966, Santa Cruz), gamma-H2AX (JW301, upstate), RPA32 (ab10359, abcam) were obtained from the indicated providers. Antibodies against Mcm2, Cdc45, Psf2, Pol epsilon p60 subunit were provided by Dr H. Takisawa (Osaka University), Pol delta p125 subunit and Pol eta by Dr S. Waga (Japan Women’s University) and Dr M. Akiyama (Nara Institute of Technology).
X. laevis egg extracts, chromatin fractions, replication assay, gap-filling assay
Interphase egg extracts were prepared as described 47. To isolate chromatin fractions, usually 4,000 demembranated sperm nuclei per μl were incubated in egg extract and diluted with 20 volumes of EB-buffer (100 mM KCl, 50 mM Hepes-KOH/pH 7.5, 2.5 mM MgCl2) containing 0.2% (v/v) Triton X-100 and layered onto 200 μl of a 30% (w/v) sucrose cushion made with the same buffer. The chromatin was spun at 10,000 × g for 5 min at 4°C, washed with 300 μl of EB-buffer and spun again at 16,100 × g for 1 min. The pellet was suspended with SDS-PAGE sample buffer and analysed by immunoblotting. DNA replication assay with neutral 47 and alkaline agarose gel 26 and gap-labelling assay 26 were previously described. Mirin was kindly provided by Dr J. Gautier. Single- and double-stranded DNA celluloses were obtained from Worthington.
Electron microscopy
Demembranated sperm nuclei (5,000 μl−1) were incubated in 1.2-1.5 ml of egg extracts for 45 min (untreated sperm) or for 60 min (0.2% (v/v) MMS treated sperm), diluted with 5 ml of EB-buffer, layered onto 2 ml of EB-buffer + 30% (w/v) Sucrose and spun at 3,000×g for 10 min at 4°C. The pellets were re-suspended in 600 μl of EB-buffer and transferred to a 96 well plate (each 100 μl per well). 4, 5′, 8-Trimethylpsoralen (TMP) was added at 10 μg ml−1 to each well. Samples were incubated on ice for 5 min in the dark, irradiated with 366 nm UV light for 7 min on a pre-cooled metal block. The procedure from TMP addition to UV irradiation was repeated three more times. Then, the genomic DNA was purified through proteinase K (1 mg ml−1) and RNase A (167 μg ml−1) treatment, phenol/chloroform extraction, and isopropanol precipitation. The purified DNA (20 μg) was digested with NdeI endonuclease (100 units) for 5 hr, and the replication intermediates (RIs) were further purified on BND cellulose column 29 and were processed for the observation with electron microscopy as previously described 29. Upon length measurements (ImageJ) of the resulting micrographs, DNA replication intermediates are identified by two parameters: 1) the presence of at least one fork (three-way junction) and 2) the presence of at least two “legs” of equal length, as expected after restriction digestion of a genomic fragment containing a replication fork. The analysis of replication forks derived from rad52 mutant cells has been performed as previously described 12. Rad52 and Rad51 strains were previously described 12.
Supplementary Material
Acknowledgements
We thank S. West, S. Boulton and members of the genome stability lab for their insightful comments. We thank H. Mahbubani and J. Kirk for technical support with X. laevis. This work was funded by Cancer Research UK. V.C. is also supported by the European Research Council (ERC) start up grant (206281), the Lister Institute of Preventive Medicine and the EMBO Young Investigator Program (YIP). M. L. and A.R.C supported by the SNF grant (PP00A—114922). A.R.C. was supported by the EMBO short term fellowship.
References
- 1.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–19. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 2.Lambert S, Froget B, Carr AM. Arrested replication fork processing: interplay between checkpoints and recombination. DNA Repair (Amst) 2007;6:1042–61. doi: 10.1016/j.dnarep.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 3.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–53. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 4.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–79. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 5.West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:435–45. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 6.Tsuzuki T, et al. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci U S A. 1996;93:6236–40. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonoda E, et al. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. Embo J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–17. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 9.Lambert S, Watson A, Sheedy DM, Martin B, Carr AM. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell. 2005;121:689–702. doi: 10.1016/j.cell.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 10.Alabert C, Bianco JN, Pasero P. Differential regulation of homologous recombination at DNA breaks and replication forks by the Mrc1 branch of the S-phase checkpoint. EMBO J. 2009;28:1131–41. doi: 10.1038/emboj.2009.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehmann AR, Fuchs RP. Gaps and forks in DNA replication: Rediscovering old models. DNA Repair (Amst) 2006;5:1495–8. doi: 10.1016/j.dnarep.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 13.Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol. 2006;7:932–43. doi: 10.1038/nrm2058. [DOI] [PubMed] [Google Scholar]
- 14.Petermann E, Caldecott KW. Evidence that the ATR/Chk1 pathway maintains normal replication fork progression during unperturbed S phase. Cell Cycle. 2006;5:2203–9. doi: 10.4161/cc.5.19.3256. [DOI] [PubMed] [Google Scholar]
- 15.Moriel-Carretero M, Aguilera A. A postincision-deficient TFIIH causes replication fork breakage and uncovers alternative Rad51- or Pol32-mediated restart mechanisms. Mol Cell. 2010;37:690–701. doi: 10.1016/j.molcel.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 16.Hekmat-Nejad M, You Z, Yee MC, Newport JW, Cimprich KA. X. laevisATR is a replication-dependent chromatin-binding protein required for the DNA replication checkpoint. Curr Biol. 2000;10:1565–73. doi: 10.1016/s0960-9822(00)00855-1. [DOI] [PubMed] [Google Scholar]
- 17.Trenz K, Smith E, Smith S, Costanzo V. ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. Embo J. 2006;25:1764–74. doi: 10.1038/sj.emboj.7601045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costanzo V, et al. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol Cell. 2001;8:137–47. doi: 10.1016/s1097-2765(01)00294-5. [DOI] [PubMed] [Google Scholar]
- 19.McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–53. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- 20.Hashimoto Y, Takisawa HX. laevisCut5 is essential for a CDK-dependent process in the initiation of DNA replication. EMBO J. 2003;22:2526–35. doi: 10.1093/emboj/cdg238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thorslund T, West SC. BRCA2: a universal recombinase regulator. Oncogene. 2007;26:7720–30. doi: 10.1038/sj.onc.1210870. [DOI] [PubMed] [Google Scholar]
- 22.Carreira A, et al. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell. 2009;136:1032–43. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leach CA, Michael WM. Ubiquitin/SUMO modification of PCNA promotes replication fork progression in X. laevis egg extracts. J Cell Biol. 2005;171:947–54. doi: 10.1083/jcb.200508100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stokes MP, Michael WM. DNA damage-induced replication arrest in X. laevisegg extracts. J Cell Biol. 2003;163:245–55. doi: 10.1083/jcb.200306006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su X, Bernal JA, Venkitaraman AR. Cell-cycle coordination between DNA replication and recombination revealed by a vertebrate N-end rule degron-Rad51. Nat Struct Mol Biol. 2008;15:1049–58. doi: 10.1038/nsmb.1490. [DOI] [PubMed] [Google Scholar]
- 26.Fukui T, et al. Distinct roles of DNA polymerases delta and epsilon at the replication fork in X. laevisegg extracts. Genes Cells. 2004;9:179–91. doi: 10.1111/j.1356-9597.2004.00716.x. [DOI] [PubMed] [Google Scholar]
- 27.Tsujikawa L, Weinfield M, Reha-Krantz LJ. Differences in replication of a DNA template containing an ethyl phosphotriester by T4 DNA polymerase and Escherichia coli DNA polymerase I. Nucleic Acids Res. 2003;31:4965–72. doi: 10.1093/nar/gkg722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagaraju G, Scully R. Minding the gap: the underground functions of BRCA1 and BRCA2 at stalled replication forks. DNA Repair (Amst) 2007;6:1018–31. doi: 10.1016/j.dnarep.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopes M. Electron microscopy methods for studying in vivo DNA replication intermediates. Methods Mol Biol. 2009;521:605–31. doi: 10.1007/978-1-60327-815-7_34. [DOI] [PubMed] [Google Scholar]
- 30.Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- 31.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–70. doi: 10.1128/MMBR.66.4.630-670.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrini JH, Bressan DA, Yao MS. The RAD52 epistasis group in mammalian double strand break repair. Semin Immunol. 1997;9:181–8. doi: 10.1006/smim.1997.0067. [DOI] [PubMed] [Google Scholar]
- 33.Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–71. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 34.Dupre A, et al. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat Chem Biol. 2008;4:119–125. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petrini JH. S-phase functions of the Mre11 complex. Cold Spring Harb Symp Quant Biol. 2000;65:405–11. doi: 10.1101/sqb.2000.65.405. [DOI] [PubMed] [Google Scholar]
- 36.Tashiro S, et al. S phase specific formation of the human Rad51 protein nuclear foci in lymphocytes. Oncogene. 1996;12:2165–70. [PubMed] [Google Scholar]
- 37.Tarsounas M, Davies D, West SC. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene. 2003;22:1115–23. doi: 10.1038/sj.onc.1206263. [DOI] [PubMed] [Google Scholar]
- 38.McIlwraith MJ, et al. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol Cell. 2005;20:783–92. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 39.Schlacher K, Cox MM, Woodgate R, Goodman MF. RecA acts in trans to allow replication of damaged DNA by DNA polymerase V. Nature. 2006;442:883–7. doi: 10.1038/nature05042. [DOI] [PubMed] [Google Scholar]
- 40.Reuven NB, Arad G, Stasiak AZ, Stasiak A, Livneh Z. Lesion bypass by the Escherichia coli DNA polymerase V requires assembly of a RecA nucleoprotein filament. J Biol Chem. 2001;276:5511–7. doi: 10.1074/jbc.M006828200. [DOI] [PubMed] [Google Scholar]
- 41.Compton SA, Ozgur S, Griffith JD. Ring-shaped Rad51 paralog protein complexes bind Holliday junctions and replication forks as visualized by electron microscopy. J Biol Chem. 2010;285:13349–56. doi: 10.1074/jbc.M109.074286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–50. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 43.Barber LJ, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–71. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lisby M, Rothstein R. Choreography of recombination proteins during the DNA damage response. DNA Repair (Amst) 2009;8:1068–76. doi: 10.1016/j.dnarep.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibson FP, Leach DR, Lloyd RG. Identification of sbcD mutations as cosuppressors of recBC that allow propagation of DNA palindromes in Escherichia coli K-12. J Bacteriol. 1992;174:1222–8. doi: 10.1128/jb.174.4.1222-1228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eykelenboom JK, Blackwood JK, Okely E, Leach DR. SbcCD causes a double-strand break at a DNA palindrome in the Escherichia coli chromosome. Mol Cell. 2008;29:644–51. doi: 10.1016/j.molcel.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 47.Trenz K, Errico A, Costanzo V. Plx1 is required for chromosomal DNA replication under stressful conditions. EMBO J. 2008;27:876–85. doi: 10.1038/emboj.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.