Abstract
The 1,9-dimethylmethylene blue (DMMB) assay is widely used to quantify sulfated glycosaminoglycan (sGAG) contents of engineered tissues, culture media, tissue samples and bodily fluids, but is subject to interference from polyanions such as hyaluronic acid (HA), DNA and RNA. We examined whether specific combinations of dye pH and absorbance wavelength could minimize non-sGAG artifacts without compromising DMMB assay sensitivity. HA and DNA solutions generated substantial signal at pH 3 but not at pH 1.5. Reducing dye pH did not significantly alter sGAG measurements for normal cartilage and meniscus tissues, but eliminated anomalously high apparent sGAG contents for enzymatically isolated chondrocytes, adipose-derived stem cell (ADSC)-agarose constructs and ADSC pellets. In a cartilage tissue engineering case study, pH 3 dye indicated high apparent sGAG readings throughout culture in both basal and chondrogenic media, with a marked decline between day 14 and 21 for chondrogenic constructs. The pH 1.5 dye, however, indicated minimal sGAG accumulation in basal medium and stable sGAG content throughout culture in chondrogenic medium. As it is often difficult to know a priori whether all groups in a study will have sGAG contents high enough to overwhelm artifacts, we recommend modifying the standard DMMB assay to reduce the risk of spurious findings in tissue engineering and clinical research. Specifically, we recommend shifting to a pH 1.5 DMMB dye and basing quantification on the absorbance difference between 525 nm (μ peak) and 595 nm (β peak) to compensate for the moderate loss of sensitivity associated with reducing the dye pH.
Keywords: dimethylmethylene blue assay (DMMB), glycosaminoglycan, tissue engineering, chondrogenesis, mesenchymal stem cells, cartilage, extracellular matrix
Introduction
Proteoglycan (PG) production is an important marker in cartilage tissue engineering because PGs comprise a large fraction of the cartilage extracellular matrix, where they serve important structural and biological functions such as lubricating articulating surfaces and modulating cell signaling by selectively sequestering and trafficking growth factors (Knudson and Knudson, 2001). The predominant PG found in cartilage is aggrecan (Heinegård and Axelsson, 1977), a high molecular weight, aggregating PG comprised of a protein core bearing many unbranched, negatively charged sulfated glycosaminoglycan (sGAG) side chains, mainly chondroitin sulfate (CS) and keratan sulfate (KS) (Meyer et al., 1958). The high negative charge density associated primarily with aggrecan contributes to the tissue’s structural stiffness, in part through electrochemical interactions with the ionic interstitial fluid (Eisenberg and Grodzinsky, 1985; Maroudas, 1968). Other sGAGs found in cartilage include heparan sulfate (HS) and dermatan sulfate (DS) (Heinegård and Axelsson, 1977), while hyaluronic acid (HA) is a polyanionic non-sulfated GAG found in cartilage.
The most common approaches to quantifying sGAG levels in tissues and tissue engineered constructs and culture media involve variations on the 1,9-dimethylmethylene blue (DMMB) dye binding spectrophotometric assay (Farndale et al., 1986; Farndale et al., 1982). The DMMB assay has largely replaced the uronic acid carbazole assay (Bitter and Muir, 1962), which is less sensitive to GAG content and may be inaccurate in the presence of glucose or salts (Frazier et al., 2008; Mort and Roughley, 2007). Assays based on binding of other cationic dyes such as Alcian blue (Whiteman, 1973) or toluidine blue (Terry et al., 2000) see some use, but DMMB has been reported to be more sensitive and reliable (Chandrasekhar et al., 1987; de Jong et al., 1994). GAG analyses via high performance liquid chromatography (Hjerpe et al., 1979) or capillary electrophoresis (Hjerpe et al., 1979) are highly sensitive and specific (Frazier et al., 2008) and can reveal detailed sulfation patterns, but these more specialized methods are not amenable to routine analysis of large numbers of samples.
The ability of the DMMB assay to detect sGAG is based on the phenomenon of metachromasia, with the characteristic blue of the cationic DMMB dye shifting to a violet hue when the dye binds to polyanionic substrates such as sGAG (Templeton, 1988). This color shift is characterized by decreased absorbance of spectral peaks centered at approximately 590 nm (β band) and 650 nm (α band) and increased absorbance at approximately 525 nm (μ band) (Figure 1a). The higher energy β band is associated with the presence of dye dimer and the α band is associated with dye monomer (Nath et al., 2005; Templeton, 1988). When the levels of dye dimer and monomer are altered, such as by the interaction of DMMB dye with a polyanionic substrate, the α and β peaks are depressed and a third peak, the μ band, emerges. The degree of metachromasia is positively correlated with substrate-to-dye ratio and is affected by parameters such as duration of reaction, pH, salt content and the presence of interfering polyanions.
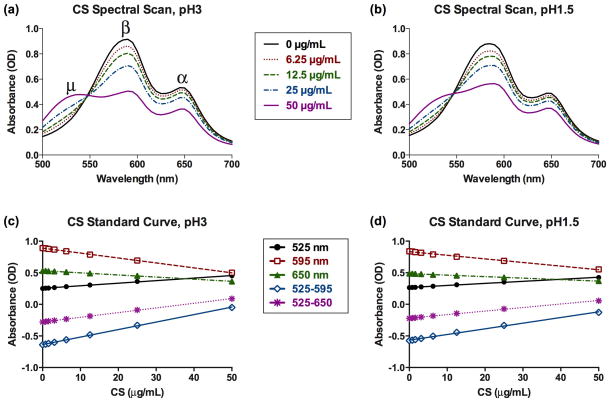
Figure 1.
(a) pH 3 and (b) pH 1.5 DMMB dyes react to increasing concentrations of chondroitin sulfate by shifting their spectral profiles; the β (590 nm) and α (650 nm) peaks decline while the μ (525 nm) peak emerges. Assay sensitivity, defined as slope of the standard curve, is greater for (c) pH 3 than for (d) pH 1.5 dye and is enhanced by using the difference between two wavelengths. The slopes of the standard curves for CS standards are significantly different from zero for all wavelengths, with P <0.0001 and R2> 0.99.
Numerous modifications to the original DMMB assay have been adopted over time to enhance assay stability, repeatability, accuracy and sensitivity. The dye-substrate metachromatic reaction occurs immediately after mixing but a dye-substrate precipitate can form within minutes, preventing accurate assessment using a spectrophotometer. Precipitation of the dye-substrate complex is hastened by physical agitation and increased sGAG content. An early attempt to address the temporal instability of the DMMB assay involved changing the DMMB dye buffer from citrate-phosphate to formate (Farndale et al., 1982). Rapid but consistently timed (e.g., 15 seconds after mixing) analysis of the reaction mixture was recommended to minimize precipitation and improve assay repeatability (Farndale et al., 1986). The addition of bovine serum albumin (BSA) to the reaction mixture to delay dye-substrate precipitation was introduced to prolong the window of time during which the reaction could be analyzed (Goldberg and Kolibas, 1990). Delaying precipitation was particularly useful in adapting the assay for use in multi-well plates.
Other protocols intentionally precipitate the dye-substrate complex as an alternate approach to quantify sGAG content. Following complex precipitation, the sGAG content can be inferred from the absorption of the remaining unbound dye, with sample sGAG content being inversely related to the amount of unbound dye (Müller and Hanschke, 1996). Alternatively, the dye-substrate precipitate can be collected and dissolved, with absorption of the resuspended dye used to determine sGAG content (Barbosa et al., 2003). A widely used commercial assay kit (Blyscan Glycosaminoglycan Assay, Biocolor Ltd., Carrickfergus, Northern Ireland) is based on a similar approach involving complex precipitation and resuspension. Problems have been noted, however, with the use of precipitation methods. Over-estimation of sGAG has been reported when analyzing unbound dye (Oke et al., 2003) and under-estimation of sGAG may result when analyzing the precipitate due to sGAG loss from the multiple processing steps (Huynh et al., 2012).
DMMB assay accuracy is also affected by polyanions and salts found in biological samples ranging from cartilage to muscle to urine. Farndale et al. (1986) proposed adding salt (NaCl) to the dye and decreasing the pH to 3.0 to decrease interference by polyanions such as HA, DNA, and RNA. Interference by HA (as evidenced by increased sample turbidity and/or presence of “stringy precipitate”) in HA-rich samples such as synovial fluid has been effectively minimized by using hyaluronidase (Dey et al., 1992; Goldberg and Kolibas, 1990; Oke et al., 2003). Other enzymes have been successfully used to selectively degrade and, by subtraction, quantify specific sGAG species (Farndale et al., 1986; Goldberg and Kolibas, 1990). Interference by DNA and RNA, of particular concern in cellularly dense tissue engineering applications, can likewise be avoided by enzymatic digestion using DNase and RNase (Dey et al., 1992) or by using DNA filters (Barbosa et al., 2003). These approaches can be procedurally cumbersome for large studies, and high levels of DNA can block the filter, resulting in low sGAG estimates (Huynh et al., 2012).
Aside from the common biological polyanions of HA, DNA and RNA, the DMMB assay can experience interference from polyanionic matrix or scaffold components. One commonly used polyanionic hydrogel is alginate, a polysaccharide whose negatively charged carboxyl groups react strongly with DMMB dye at pH 3. Enobakhare et al. (1996) adapted the DMMB assay for use with alginate samples by reducing the dye pH. At pH 1.5, the alginate carboxyl groups are protonated, allowing sulfate groups of sGAGs (with lower pKa values) to be detected. Further reduction of dye pH 1 rendered the assay unusable due to protonation of sulfate groups. Using the DMMB dye at pH 1.5 has since become standard for assessing samples that contain alginate, with some recommending use of pH 1.5 dye for alginate constructs and pH 3 dye for other scaffolds (Estes et al., 2010). As an additional measure to enhance assay sensitivity, some have measured sGAG based on the difference between two absorbance wavelengths (Hoemann, 2004; Seibel et al., 1992). By measuring increased signal at the μ peak and suppression in signal from either the α or β peaks, sensitivity can be enhanced when compared with using the μ peak alone.
Keeping in mind the many assay parameters that can be and have been altered, the goal of this study was to identify simple modifications to the standard DMMB assay that would minimize measurement artifacts due to non-sGAG polyanions for tissue samples, cell-seeded constructs and conditioned culture media. We examined the effects of dye pH and measurement wavelength on quantifying defined CS, DS, HS, HA and DNA standards and measured sGAG contents of native cartilage and meniscus tissues, enzymatically isolated chondrocytes and adipose-derived stem cell (ADSC) seeded agarose gels immediately after preparation. As tissue engineering case studies, we evaluated sGAG contents of ADSC pellets cultured for 28 days and of ADSC-seeded agarose gels and conditioned media over 21 days of chondrogenic induction. Our results indicate that caution should be exercised in interpreting reported sGAG contents in the current literature, but that simple modifications to the DMMB assay (pH and absorbance wavelength) can minimize non-sGAG interference without compromising sensitivity to sGAG content. These simple modifications can suppress a number of known artifacts and allow consistent reporting and interpretation of sGAG measurements in tissue engineering studies.
Materials and Methods
Dye Preparation
1,9-dimethylmethylene blue (DMMB; C18H22ClN3S · 0.5ZnCl2; CAS number 931418-92-7; molecular weight 416.05) dye (Sigma-Aldrich Ltd, St. Louis, MO) was prepared by dissolving 16 mg of DMMB in 5 mL ethanol (Fisher Scientific, Fair Lawn, NJ) overnight and volume adjusting to 1 L with deionized water containing 40 mM glycine (Sigma-Aldrich), 40 mM NaCl and sufficient HCl (EMD Chemicals, Gibbstown, NJ) to adjust the dye solution to the desired pH (Farndale et al., 1986). The dye was stored at room temperature in a bottle shielded from light and used within 1 month of preparation, within the recommended 3 months (Farndale et al., 1986). Hoechst 33258 dye (Sigma-Aldrich) solution was prepared by diluting in TNE buffer (50 mM Tris-HCL at pH 7.4 (Invitrogen, Carlsbad, CA), 100 mM NaCl (Fisher), 0.1mM ethylenediaminetetraacetic acid (EDTA, Sigma-Aldrich)) to a final working concentration of 0.1 μg dye/mL TNE buffer (Downs and Wilfinger, 1983).
Standards Preparation
Shark chondroitin sulfate (CS) (Sigma-Aldrich), calf thymus DNA (Invitrogen), bovine kidney heparan sulfate (Sigma-Aldrich), dermatan sulfate (Crescent Chemical Co., Islandia, NY) and sodium hyaluronate (Lifecore Biomedical, Chaska, MN) standards were prepared in 100 mM ammonium acetate (AA) (EMD). Media standards were prepared by serially diluting CS in one of four media formulations based on high glucose Dulbecco’s Modified Eagle’s Medium (DMEM) containing phenol red (Hyclone, South Logan, UT): (1) “DMEM” consisting of only DMEM, (2) “HEPES” consisting of high glucose DMEM and 10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulphonic acid buffer (HEPES) (Mediatech, Manassas, VA), (3) “FBS” consisting of high glucose DMEM and 10 % fetal bovine serum (FBS, Atlanta Biologicals, Lawrenceville, GA) and (4) “Total” consisting of high glucose DMEM, 10 mM HEPES, 50 μg/mL L-ascorbate 2-phosphate (Sigma-Aldrich), 1 % non-essential amino acids (NEAA) (Gibco, Grand Island, NY), 1 % insulin, transferrin, and selenous acid (ITS+) (BD Biosciences, Bedford, MA) and 0.4 mM L-proline (Sigma-Aldrich). Media standards were prepared in triplicate (n=3). CS standard curves within the linear range of sensitivity (0–50 μg/mL for cell and tissue assays, 0–25 μg/mL for media assays) were used to calculate apparent sGAG levels.
Enzyme Preparation
Proteinase K (PK) solution (Invitrogen) was prepared per manufacturer’s protocol by dissolving lyophilized PK powder in 10 mM TrisHCl, pH 7.5 (Invitrogen), 20 mM calcium chloride (Sigma-Aldrich), and 50 % glycerol (EMD Chemicals). Agarase solution was prepared per manufacturer’s protocol by dissolving lyophilized agarase (Sigma-Aldrich) to a final concentration of 1 U agarase/μL potassium phosphate buffer, pH 6.0 (0.2 mg/mL) (EMD). DNase stock solution was prepared by reconstituting lyophilized DNase I (Worthington Biosciences, Lakewood, NJ) into 10 KU DNase/mL stock solution (10 mM Tris-HCl, pH 7.5, 10 mM CaCl2, 10 mM MgCl2 and 50 % (v/v) glycerol). Prior to use, DNase stock was diluted to a 400 U DNase/mL working solution (200 mM Tris-HCl, pH 7.5, 50 mM MgCl2, 10 mM CaCl2). ChABC enzyme solution was prepared by reconstituting lyophilized chondroitinase ABC (Sigma-Aldrich) at 20 U/mL in a 0.01 % BSA aqueous solution (Invitrogen).
Micro-Plate Assays
The DMMB assays for CS, HS, DS, HA and DNA spectral scans and cell/tissue digests used a ratio of 20 μL sample of standard to 200 μL dye, with the samples plated first in triplicate in clear 96 well plates (Costar, Corning, NY) and later adding DMMB dye to the wells. In media assays, a higher sample- or standard-to-dye ratio of 20 μL to 150 μL was used to enhance signal due to the low sGAG contents of conditioned media. Samples were analyzed for absorbance at room temperature immediately after adding DMMB dye. Spectral scans were conducted in increments of 5 nm from 500 nm to 700 nm using a Synergy HT plate reader (Biotek Instruments, Winooski, VT). The Hoechst DNA assay (Downs and Wilfinger, 1983) used a sample-to-dye ratio of 10 μl to 200 μL, where standards and samples were plated in triplicate in black 96 well plates (Costar). DNA samples were analyzed for fluorescence (excitation wavelength 356 nm, emission wavelength 465 nm) at room temperature with a Synergy HT plate reader immediately after adding Hoechst dye.
Tissue & Cellular Assays
To determine the effects of DMMB dye pH on tissue and cellular sGAG measurements, cartilage and meniscus tissue samples, isolated chondrocytes and ADSC-seeded constructs were enzymatically digested and assayed at OD525-OD595 as indicated below.
Cartilage and Meniscus
Cartilage and meniscus tissues were extracted from 6 different juvenile bovine stifles (Research 87, Boylston, MA). Samples were cored with a 4 mm diameter biopsy punch (Miltex, York, PA) sliced to 2 mm thick discs, frozen, lyophilized and digested overnight at 65 °C in 1 mL PK-AA solution (150 μg PK per cartilage sample, 400 μg PK per meniscus sample) (n=6). For the assay, cartilage samples were diluted 1:100 and meniscus samples were diluted either 1:20 or 1:10 in AA to ensure that sGAG concentrations fell within the linear range of the CS standard curve.
Chondrocytes
Articular chondrocytes were enzymatically isolated from juvenile bovine femoral condylar cartilage (Research 87) via overnight collagenase digestion and cryopreserved in liquid nitrogen. Frozen cells were thawed, counted in an automated cell counter (Vi-Cell XR, Beckman Coulter, Brea, CA) and separated into individual aliquots of about one million chondrocytes (n=6). The cell aliquots were washed twice in PBS and once in 100 mM AA, frozen and lyophilized. Lyophilized samples were rehydrated with 100 mM AA and digested with 200 μg PK overnight at 65 °C for a final digest volume of 1 mL.
ADSC-Agarose Constructs
Human ADSCs (ASC-F Lot ADSC0036, Zen-Bio, Inc., Research Triangle Park, NC) were expanded in αMEM (Hyclone) and 16.5 % FBS until passage 6 per standard expansion protocol (Wolfe et al., 2008). As the focus here is on characterizing potentially artifactual readings in cell pellets and cell-seeded constructs, the use of pooled cells is not detrimental. At passage 6, cells were trypsinized (0.05 % trypsin/0.53 mM EDTA (Cellgro, Herndon, VA)) and washed 3 times with high glucose DMEM. ADSCs were seeded at 10e6 cells/mL in 3 % low gelling point agarose (Sigma-Aldrich) which was maintained at 40 °C and cast between glass plates spaced 3 mm apart. After solidification at room temperature for 20 minutes, 4 mm diameter discs were removed with a biopsy punch, immediately frozen and lyophilized (n=6). Samples were digested with 200 μg PK overnight at 65 °C, heated to 90 °C for 10–15 minutes to deactivate PK and cooled to 43 °C. After adding at least 2 U of agarase, each sample was incubated at 43 °C for at least 4 hours, followed by volume adjustment to 1 mL with 100 mM AA.
Enzymatic Treatments
To determine relative contributions of DNA and CS to assay results, samples were enzymatically treated with DNase and/or ChABC (Dey et al., 1992; Goldberg and Kolibas, 1990) using protocols based on preliminary studies using calf thymus DNA and shark CS (data not shown). Aliquots of tissue or cell digests were divided into four enzymatic treatment groups: (1) enzyme-free control, (2) DNase, (3) ChABC and (4) DNase + ChABC. All groups included 90 μL of PK-digested sample, 5 μL of DNase solvent and 5 μL of ChABC solvent, and groups with active enzyme included 2U DNAse and/or 0.1 U ChABC. All samples were incubated for 2 hours at 37 °C.
Tissue Engineering Case Studies
ADSC-Agarose Construct Culture
To examine the effect of DMMB assay parameters on results from a typical tissue engineering experiment, ADSCs were expanded to passage 4 and seeded at 15e6 cells/mL into 3 % agarose gels as described above to produce 4 mm diameter, 3 mm thick constructs. Constructs were cultured in either basal medium or chondrogenic medium as described above (1 mL per sample). Media were made fresh, changed every other day and conditioned media were collected at each change. At days 7, 14 and 21, ADSC constructs (n=6/condition) were frozen, lyophilized, digested with PK and agarase as described above and assayed for sGAG and DNA contents.
ADSC Pellet Culture
To examine the effect of DMMB assay parameters on cell-dense pellet culture, passage 5 ADSCs were centrifuged at 450 g for 10 minutes into conical tubes (2e5 cells per tube) and cultured in 1 mL of either defined basal medium (high glucose DMEM, 50 μg/mL L-ascorbate 2-phosphate, 1 % NEAA, 1 % ITS+, 10 μg/mL gentamicin (Gibco), 0.1 μM dexamethasone (MPBio, Solon, OH), 0.4 mM L-proline and 10 mM HEPES buffer) or chondrogenic medium (basal medium plus 10 ng/mL TGF-β3 (PeproTech, Rocky Hill, NJ)), with media changed twice per week. Pellets were cultured for 28 days (n=12/condition) at which time they were frozen, lyophilized, digested with PK and assayed for sGAG and DNA contents.
Data Analysis
Linear regressions for DMMB standard curves were performed with a least squares fit. Cell and tissue sGAG contents were compared across digestion groups and assay protocols using general linear models (GLMs) with main factors of enzymatic treatment, pH, and wavelength (Minitab 16, Minitab, Inc., State College, PA). ADSC case study data were analyzed using GLMs with main factors of pH and treatment (Minitab 16). Tukey’s post-hoc test was used for multiple pairwise comparisons and significance was set at p<0.05 (n=6). Differences between regression coefficients or elevations for media standard curves were analyzed using analysis of covariance with multiple comparison testing (Zar, 2010) (MatLab 7.11.0 R2010b, MathWorks, Inc., Natick MA). The pH for half-maximal change in assay sensitivity (EC50) was determined by fitting a logistic dose-response curve to sensitivity data at 525 nm, 595 nm and 650 nm, subject to the constraint of a common EC50 for all wavelengths (GraphPad Prism 6, GraphPad Software, Inc., La Jolla, CA). Data are presented as mean ± standard error of the mean (SEM).
RESULTS
Spectral Scan & Assay Sensitivity
DMMB was sensitive to CS at both pH 3 and pH 1.5 as evidenced by changes in spectral profile when reacted with different CS concentrations (Figure 1a, b). Increasing CS concentrations resulted in the decline of the β (590 nm) and α (650 nm) peaks and a rise of the μ (525 nm) peak. Plots of the DMMB-CS standard curves showed significant (p<0.0001) dependence of absorbance on CS concentration at both pH 3 and pH 1.5 for standard wavelengths (525 nm, 595 nm, 650 nm) and differences between wavelengths (525–595 nm and 525–650 nm) (Figure 1c, d). These patterns are all consistent with the well documented response of DMMB dye to sulfated glycosaminoglycans (Enobakhare et al., 1996; Farndale et al., 1982).
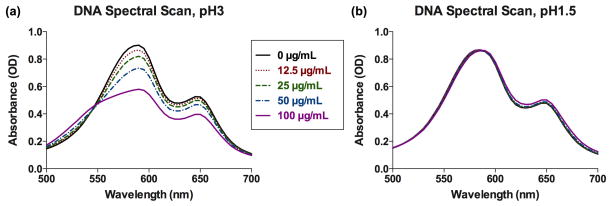
Reacting pH 3 DMMB dye with increasing concentrations of DNA likewise caused marked shifts in the α, β and μ bands (Figure 2a). However, when DNA standards were assayed with pH 1.5 DMMB dye, the spectral profile was essentially unaffected by DNA concentration (Figure 2b).
Figure 2.
(a) pH 3 DMMB dye is sensitive to the presence of DNA, as evidenced by altered spectral profiles with increasing DNA concentrations, but (b) pH 1.5 DMMB dye is minimally sensitive to DNA.
The sensitivity (slope of the standard curve) of DMMB dye to CS, DS, HS, HA and DNA all decreased when DMMB dye pH was decreased below 3 (Figure 3). The half maximal change in effective assay sensitivity (EC50) occurred between pH 2 and pH 2.5 (except for HA, for which EC50 was not calculated due to the steep transition of the assay sensitivity curve). Assay sensitivity for the sGAGs CS, DS and HS did not significantly vary for pH values between 3 and 6. Significant decreases in sensitivity resulted when switching from pH 3 to pH 1.5 dye for CS (−21 % at 525 nm), DS (−31 % at 525 nm) and HS (−36 % at 525 nm), with even greater decreases in sensitivity for HA (−80 % at 525 nm) and DNA (−74 % at 525 nm) (Table 1a). This decreased sensitivity to sGAGs at pH 1.5 was more than compensated for by the wavelength-difference method, which significantly enhanced sensitivity to CS (+186 %), DS (+188 %) and HS (+178 %) at 525–595 nm as compared with measurements at the single wavelength of 525 nm (Table 1b).
Figure 3.
Plot of DMMB assay sensitivity (standard curve slope) to (a) chondroitin sulfate (CS), (b) dermatan sulfate (DS), (c) heparan sulfate (HS), (d) hyaluronic acid, and (e) DNA as measured using dye ranging from pH 1 up to pH 6 at different absorbance wavelengths. Sensitivities were highest when using pH 3 or higher DMMB dye and when analyzed using 525–595 nm. Half maximal effective assay sensitivity (EC50) and standard error of EC50 are reported for all species but HA, whose transition was too abrupt to fit with the logistic dose-response model.
Table 1.
DMMB Assay Sensitivity
| (A) | Percent Decrease in Sensitivity | ||
|---|---|---|---|
| pH 1.5 vs pH 3 DMMB Dye | |||
| 525 nm | 595 nm | 650 nm | |
| CS | −21 % | −27 % | −29 % |
| DS | −31 % | −35 % | −38 % |
| HS | −36 % | −34 % | −34 % |
| DNA | −74 % | −97 % | −105 % |
| HA | −80 % | −78 % | −78 % |
| (B) | Percent Increase in Sensitivity | |
|---|---|---|
| pH 1.5 DMMB Dye | ||
| 525–595 nm vs 525 nm | 525–595 nm vs 595 nm | |
| CS | +186 % | +54 % |
| DS | +188 % | +53 % |
| HS | +178 % | +51 % |
(a) Sensitivity (quantified as absolute value of the slope of the standard curve for a given wavelength) of DMMB dye to the polyanions chondroitin sulfate (CS), heparan sulfate (HS), DNA and hyaluronan (HA) decreased when assayed at pH 1.5 compared with pH 3. The percentage decrease in sensitivity was greatest for the non sulfated polyanions DNA and HA. (b) Sensitivity of pH 1.5 DMMB dye to the sGAGs CS and HS were enhanced when assessed using the difference between two wavelengths (OD525-OD595) as compared with using a single wavelength (OD525 or OD595).
Media-CS Standard Curves
The slope of the CS standard curves varied significantly with solvent formulation, dye pH and absorbance wavelength. Although we found minimal differences at pH 3 or pH 1.5 among standard curves using AA solution, AA solution containing proteinase K, and phosphate buffered EDTA (data not shown), the presence of media components significantly influenced the standard curve. Standard curve slopes were significantly higher in AA solution than in media formulations containing DMEM (Table 2). For example, at 525 nm and pH 3, the standard curve slope in AA solution measured 4.03e-3 ± 3.9e-5 (OD/[μg/mL CS]) whereas in Total medium it measured 3.33e-3 ± 7.2e-5 (OD/[μg/mL CS]). At 525 nm and pH 1.5, the standard curve in AA measured 3.36e-3 ± 4.5e-5 (OD/[μg/mL CS]) whereas in Total medium, it measured 3.14 ± 6.0e-5 (OD/[μg/mL CS]). Use of a standard solvent differing in composition from the media could thus introduce substantial errors in the estimated sGAG content of conditioned media samples.
Table 2.
Media-CS Standard Curves
| pH 3 | pH 1.5 | |||||||
|---|---|---|---|---|---|---|---|---|
| Slope [OD/(μg/ml CS)] | S* | R2 | Y-intercept [OD] | Slope [OD/(μg/ml CS)] | S* | R2 | Y-intercept [OD] | |
| AA | 4.03e-3 ± 3.9e-5 | H,T,F | 0.994 | 0.201 ± 4.2e-4 | 3.36e-3 ± 4.5e-5 | d,h,t,f | 0.989 | 0.205 ± 4.9e-4 |
| DMEM | 3.81e-3 ± 6.2e-5 | T | 0.984 | 0.202 ± 6.8e-4 | 3.13e-3 ± 5.6e-5 | a,f | 0.981 | 0.219 ± 6.1e-4 |
| Hepes | 3.60e-3 ± 7.9e-5 | A,T | 0.971 | 0.200 ± 8.7e-4 | 3.01e-3 ± 6.0e-5 | a | 0.977 | 0.219 ± 6.5e-4 |
| Total | 3.33e-3 ± 7.2e-5 | A,D,H,F | 0.972 | 0.194 ± 7.8e-4 | 3.14e-3 ± 6.0e-5 | a,f | 0.978 | 0.212 ± 6.5e-4 |
| FBS | 3.60e-3 ± 6.3e-5 | A,T | 0.982 | 0.202 ± 6.8e-4 | 2.81e-3 ± 4.9e-5 | a,d,t | 0.982 | 0.210 ± 5.4e-4 |
Chondroitin sulfate (CS) standards formulated in different solvents: ammonium acetate (“AA”), in high glucose DMEM (“DMEM”), in DMEM plus HEPES buffer (“HEPES”), in Total medium (“Total” and in DMEM with 10% FBS (“FBS”). Slope and intercept of the standard curve at 525 nm depended significantly on pH of DMMB dye, solvent and wavelength. Letters A, D, H, T, F under the column S* indicate significant difference between the slopes when compared with CS formulated in AA, DMEM, HEPES, Total and FBS solvents, respectively, with upper case letters for pH 3 and lower case letters for pH 1.5. P<0.05
DMMB Assay for Tissues and Cells
For cartilage samples, dye pH did not significantly alter the estimated sGAG contents (44.24±2.40 μg sGAG/mg WW at pH 3 versus 43.87± 2.37 μg sGAG/mg WW at pH 1.5, analyzed using OD525-OD595 unless otherwise stated) (Figure 4a). Likewise, dye pH did not significantly affect estimated sGAG content of meniscus samples (8.80± 1.76 μg sGAG/mg WW at pH 3 versus 7.90± 1.68 μg sGAG/mg WW at pH 1.5) (Figure 4b). DNase digestion of both tissues had minimal effect on sGAG readings at either pH, whereas ChABC digestion substantially reduced readings at either pH.
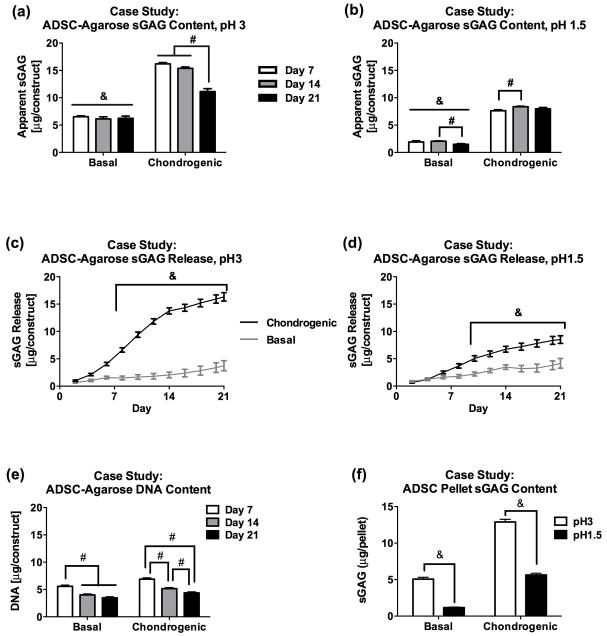
Figure 4.
DMMB measurements of sGAG content for (a) cartilage and (b) meniscus were not significantly different at pH 3 compared with pH 1.5. (c) Isolated chondrocytes have negligible sGAG content when assayed at pH 1.5 but not at pH 3. (d) Freshly seeded ADSC-agarose constructs had significantly higher apparent sGAG content at pH 3 than at pH 1.5, even after combined DNase and chondroitinase treatment. Analyses were performed using OD525-OD595. & Significantly different when assayed using pH 3 and pH 1.5 DMMB dye. (P <0.05)
Measurements of isolated chondrocytes (approximately one million cells per sample) at pH 3 indicated an unexpectedly high sGAG reading (3.43 ± 0.45 μg), whereas measurement of the same samples at pH 1.5 indicated low sGAG levels (0.21 ± 0.35 μg) (Figure 4c). DNase digestion significantly lowered the apparent sGAG levels at pH 3 (0.54 ± 0.29 μg), indicating that much of the signal at pH 3 was actually produced by DNA.
Similarly, sGAG estimates with pH 3 dye were significantly higher than estimates with pH 1.5 dye for freshly seeded ADSC-agarose gels (approximately 5.7e5 cells per construct) (Figure 4d). ADSC-agarose samples assayed with OD525-OD595 at pH 3 had absorbance readings equivalent to 8.15 ± 0.26 μg sGAG, whereas measurements of the same samples at pH 1.5 indicated minimal sGAG contents (0.21 ± 0.09 μg). The average apparent sGAG level at pH 3 decreased by 19 % with DNase treatment (6.58 ± 0.26 μg), by 29 % with ChABC treatment (5.81 ± 0.19 μg) and similarly with DNase+ChABC treatment (5.81 ± 0.37 μg), indicating that a substantial portion of the signal at pH 3 was due to species other than DNA and CS. At pH 1.5, apparent sGAG levels for ADSC samples were minimal in the DNase treated group (0.48 ± 0.06 μg). The apparent sGAG levels increased slightly after ChABC and DNase+ChABC treatments (1.25 ± 0.14 μg and 1.24 ± 0.15 μg respectively). This may be due to interaction with components of the digest solution, which was not added to the standards in this analysis. Additionally, we noted a difference in meniscus depth within the microplate wells after adding digest solution, which could alter the optical absorption path length in the microplate reader.
Tissue Engineering Case Studies
ADSC–Agarose Case Study
Measurements at either pH 3 or pH 1.5 indicated that construct sGAG contents were significantly greater for samples cultured in chondrogenic medium than basal medium, but both the levels and temporal patterns of apparent sGAG content varied with dye pH. The DMMB assay at pH 3 and OD525-OD595 indicated consistent sGAG levels throughout the 3 weeks of culture for constructs in basal medium (6.53 ± 0.14 μg, 6.12 ± 0.38 μg and 6.24±0.40 μg at days 7, 14 and 21) and indicated relatively high initial sGAG contents (16.19 ± 0.24 μg at day 7) that decreased significantly between days 14 (15.38 ± 0.25 μg) and 21 (11.12 ± 0.53 μg) for constructs in chondrogenic medium (Figure 5a). In contrast, the DMMB assay at pH 1.5 and OD525-OD595 indicated substantially lower sGAG contents for constructs cultured in both basal and chondrogenic media (Figure 5b). The pH 1.5 assay indicated low sGAG levels for the basal group that were consistent throughout culture (1.91 ± 0.21 μg, 2.05 ± 0.06 μg and 1.49 ± 0.12 μg on days 7, 14 and 21) and sGAG contents for the chondrogenic group that were stable throughout culture, with a small difference between day 7 and day 14 samples (7.62 ± 0.18 μg, 8.36 ± 0.09 μg and 7.98 ± 0.24 μg on days 7, 14 and 21). DNA content was lower at day 21 than at day 7 for both basal (3.47 ± 0.17 vs. 5.58 ± 0.18μg) and chondrogenic groups (4.38 ± 0.14 vs. 6.89 ± 0.21 μg). Measurements of cumulative sGAG release to the conditioned media also differed between pH 3 and pH 1.5 dyes (Figure 5c, d). Apparent sGAG levels of conditioned chondrogenic medium assayed at pH 1.5 were half of those assayed at pH 3 (8.50 ± 0.71 μg at pH 1.5 versus 16.33 ± 0.76 μg at pH 3 dye), with no significant difference between dye formulations for basal medium (4.17 ± 0.89 μg at pH 1.5 versus 3.74 ± 0.93 μg at pH 3). We speculate that other polyanionic ECM components (e.g., HA) were released in parallel with the sGAG and produced elevated readings for the pH 3 DMMB assay that were suppressed at pH 1.5.
Figure 5.
(a) Results from the pH 3 DMMB assay indicate that basal samples maintained approximately 6 μg sGAG throughout culture whereas the sGAG content of chondrogenic samples, although higher at day 7 (16.19 ± 0.24 μg), dropped by day 21 (11.12 + 0.53 μg). (b) Results from the pH 1.5 assay indicate that basal samples retained minimal sGAG while chondrogenic samples maintained stable sGAG content throughout culture. (c, d) Cumulative sGAG release measurements taken with pH 3 dye are almost double those assayed at pH 1.5 for chondrogenic samples. (e) DNA content for ADSC-agarose constructs cultured in either basal or chondrogenic medium decreased with time in culture, with day 21 DNA contents measuring lower than those on day 7. (f) ADSC pellets assayed after 28 days of culture in either basal or chondrogenic medium showed significantly lower sGAG measurements when assayed at pH 1.5 as compared with pH 3. Average DNA content per sample was 3.64 ± 0.13 μg for basal and 4.97 ± 0.16 μg for the chondrogenic group. Analyses were performed using OD525-OD595. & Significantly different between basal and chondrogenic groups. # Significantly different between time points. (P<0.05)
ADSC Pellet Case Study
Assayed at either pH, ADSC pellets cultured in chondrogenic medium for 28 days had higher sGAG contents than those cultured in basal medium, but the magnitude of the apparent sGAG contents varied significantly with dye pH (Figure 5f). The apparent sGAG content of basal samples analyzed at OD525-OD595 was 5.07 ± 0.25 μg at pH 3 vs. 1.17 ± 0.07 μg at pH 1.5, while the apparent sGAG content of chondrogenic samples was 12.89 ± 0.37 μg at pH 3 vs. 5.63 ± 0.23 μg at pH 1.5. The average DNA content per sample was 3.64 ± 0.13 μg for basal and 4.97 ± 0.16 μg for the chondrogenic group.
Discussion
The original spectrophotometric DMMB assay for measuring sGAG content (Farndale et al., 1982; Humbel and Etringer, 1974) has been modified in numerous ways to enhance accuracy, sensitivity and temporal stability for use with various specimens, equipment and processes. In this study, we explored how two assay parameters, dye pH and absorbance wavelength, affect sensitivity and accuracy of the assay. In native cartilaginous tissues, sGAGs are generally the predominant polyanions that are measured using the traditional DMMB assay. However, in tissue-engineered samples, conditioned media and bodily fluids, which have low levels of sGAG relative to non-sGAG polyanions signal from non-sGAG species can result in over-estimation of sGAG content using the DMMB assay.
Using DNA as an example of a specific polyanionic artifact, approaches for removing DNA contamination from the DMMB-sGAG assay include treating samples with DNase or DNA filtration (Barbosa et al., 2003; Dey et al., 1992; Goldberg and Kolibas, 1990), but these additional processing steps increase analysis time and cost and may introduce sources of error (Huynh et al., 2012). Although the use of pH 3 dye is purported to minimize DNA interference (Farndale et al., 1986), we found that pH 3 DMMB dye remains sensitive to DNA contamination, particularly when measuring about the β peak (590 nm). Further lowering the dye pH to 1.5 minimizes DNA-induced metachromasia, likely due to DNA protonation by the highly acidic dye. Similarly, interference from HA, which has an acid dissociation constant of 2.87 (Wishart et al., 2013), was significantly decreased by using dye below pH 2.5. While lowering dye pH to 1.5 moderately decreased sensitivity to sGAGs, the dye remains sensitive to sGAGs due to the presence of sulfate groups with very low acid dissociation constants (Wang et al., 1991). The pH 1.5 DMMB dye thus has the advantage of minimizing interference from contaminants such as DNA and HA while still being able to accurately detect sGAG levels.
Although not specifically intended to remove non-sGAG polyanionic contamination, pH 1.5 DMMB dye is commonly used in tissue engineering studies involving alginate scaffolds to remove anionic interference from alginate carboxyl groups, which are protonated at pH 1.5 (Enobakhare et al., 1996). Our results indicate that effects of a number of contaminants are also reduced or eliminated at pH 1.5. Consequently, use of different pH dyes for different scaffold materials (Estes et al., 2010) could bias comparisons between scaffolds due to differential elimination of artifacts. Although DMMB assay sensitivity to sGAG at pH 1.5 is lower than at pH 3, assay sensitivity can be enhanced by calculating sGAG levels based on the difference in OD between μ and β peaks (Figures 1 & 3). In addition to enhancing assay sensitivity, using the wavelength difference method also helps to correct for vertical shifts of the spectral curve that can be induced by contaminants (Templeton, 1988). If a single wavelength were to be used in calculating sGAG concentration, then a curve shift could be easily overlooked, resulting in inaccurate measurements.
Few published manuscripts explicitly address procedures for analysis of media samples, and even fewer describe procedures different from those for sample digests. Likewise, few published protocols explicitly address media samples. Mort and Roughley (2007) instruct that standards for media sGAG analysis should be prepared in the culture medium, but other protocols (Billington, 2001; Hughes et al., 2003; Shingleton, 2003) instruct that phosphate buffer or water should be used for standards and, if necessary, to dilute samples. Our results indicate the that the latter practice could lead to inaccurate results. Disparities among standard curves created using different solvents underscore the importance of matching the standards solvent with sample solvent as closely as possible (Table 2). Not only did we find significant differences between standard curves based on ammonium acetate and media as solvents, but we also found substantial differences among different media formulations. The ideal solvent would replicate the media formulation used in the experiment to the extent that it contains similar concentrations of ions, buffers, proteins, etc. Generating CS standards using a solvent that is dissimilar to the medium used in the experiment may result in inaccurate values of sGAG release values, and any dilution of media samples in a different solvent would introduce dilution-dependent inaccuracies. As for tissue or construct digests, the use of pH 1.5 DMMB dye and OD525-OD595 for media assays would reduce potential interference from polyanions and other species in the conditioned media.
The assayed sGAG content of cartilage and meniscus, tissues that contain high levels of sGAG, were not significantly different between pH 1.5 and pH 3 DMMB dye. Treatment with DNase did not significantly alter measured sGAG levels, confirming that DNA contamination was not substantial for cartilage or meniscus tissue. As expected, both tissue digests exhibited similarly low sGAG levels at either pH after chondroitinase digestion. Unlike native cartilage and meniscus tissues, isolated chondrocytes and freshly seeded, non-cultured ADSC-agarose constructs are expected to contain negligible levels of sGAG. It was therefore surprising that apparent sGAG levels for isolated chondrocytes and ADSC-agarose construct digests were significantly greater than zero when assayed with pH 3 dye. Treating chondrocyte digests with DNase eliminated interference from DNA and effectively decreased “apparent” sGAG levels to near zero at pH 3; however, simply reducing the dye pH to 1.5 (without DNase treatment) had a similar effect. Treating ADSC construct digests with chondroitinase and DNase reduced the apparent sGAG level but did not fully abolish it at pH 3, whereas assaying the same samples with pH 1.5 dye indicated negligible sGAG content (as expected for freshly-seeded, non-cultured constructs). The substantial sGAG measurement at pH 3 for DNase-treated ADSC samples indicates that artifacts other than DNA likely contributed to the reading. Interference from these artifacts was effectively minimized when dye pH was lowered to pH 1.5. Although not examined in this study, digesting samples with additional enzymes (e.g., hyaluronidase) could be used to identify the specific sources of DMMB metachromasia at pH 3.
As demonstrated by the case studies on ADSC chondrogenesis, accounting for the presence of interfering polyanions is particularly important due to the relatively low sGAG contents in many tissue engineering studies, especially at early time points. In the ADSC-agarose case study, not only was the amplitude of sGAG content different when assayed with dyes of pH 3 and pH 1.5, but also the temporal trend of sGAG accumulation within the constructs was also different. While results using pH 3 seem to indicate that ADSC sGAG production was enhanced by chondrogenic media early on but that constructs lost sGAG after day 14, results using pH 1.5 dye show a maintenance of sGAG content throughout the 21 days of culture in chondrogenic medium, suggesting accumulation over the initial week but no subsequent net change in content. This striking difference may partly be explained by a loss of non-sGAG polyanionic content within the constructs and a decrease in nucleic acid content, resulting in artifactual readings at pH 3 but not at pH 1.5 dye. The trend in sGAG content measured at pH 3 were consistent with safranin O staining for glycosaminoglycans and immunostaining for aggrecan-G1 (not shown), which indicated weak, pericellular staining in chondrogenic medium samples that was absent in basal medium samples. Cumulative sGAG release into media was significantly lower for chondrogenic samples when assayed with pH 1.5 dye then when assayed with pH 3 DMMB dye. As minimal DNA is expected in the conditioned culture media, this additional signal at pH 3 is likely due to concurrent release of other polyanionic extracellular matrix components. Similarly, the ADSC pellet culture case study confirms that results are highly dependent on assay pH, with the standard DMMB assay at pH 3 over-estimating the true sGAG content. These results are significant in that the pellet culture model is widely used to assess the degree of chondrogenic induction of various tissue engineering treatments and the DMMB assay is the main quantitative assay used to determine sGAG content, a measure of the degree of chondrogenesis.
The results of these analyses indicate that widely used methods have the potential to produce contaminated results. To broadly assess the potential impact on the literature, we individually examined manuscripts resulting from two separate PubMed searches. To examine literature involving pellet culture, we performed a search for “(chondrogenesis or chondrogenic) and (pellet or micromass)” and limited the search results to the five-year period from July 1, 2009 through June 30, 2014. After eliminating protocols and reviews, this yielded 294 primary research articles that we could obtain as full text English articles (an additional 16 were either published in another language or unavailable through our institutional subscriptions). Similarly, to examine the cartilage tissue engineering literature, we performed a search for “cartilage and (“tissue engineering” or “tissue engineered”)” and, due to the large number of manuscripts, limited the search results to the one-year period from July 1, 2013 through June 30, 2014. After eliminating protocols and reviews, we obtained full text English versions of 432 primary research articles (an additional 31 were either published in another language or unavailable through our institutional subscriptions). Some variant of the DMMB assay was employed by 111 (38%) of the 294 pellet culture articles and by 164 (38%) of the 432 cartilage tissue engineering articles (14 articles were captured in both PubMed searches). We further examined each manuscript to determine the cited source (if any) for the DMMB assay and the assay conditions.
The patterns of citation for the DMMB assay were similar for these two segments of the literature (Figure 6). Among the 111 pellet culture studies, 32 performed some variation of a precipitation-based assay (primarily using the Blyscan commercial kit), 40 cited Farndale et al. (1986); Farndale et al. (1982) or a close derivative, 2 used a modification of Farndale’s assay intended to minimize DNA signal, and 37 either cited multiple papers using inconsistent methods or provided no citation and few if any details of the assay conditions—we speculate that the majority of these manuscripts used some variation of Farndale’s assay (~pH 3), although this cannot be determined based on our review of the literature. Among the 164 cartilage tissue engineering studies, 44 performed some variation of a precipitation-based assay (primarily using the Blyscan commercial kit), 59 cited Farndale et al. (1986); Farndale et al. (1982) or a close derivative, 2 used a modification of the Farndale assay intended to minimize DNA signal (e.g., BSA), 9 used Enobakhare’s low-pH modification (intended for alginate culture), 2 cited relatively unusual implementations (pH 6.5, 520nm) and 48 either cited multiple papers using inconsistent methods or provided no citation—few of these provided assay details, and we again speculate that the majority of these manuscripts used some variation on Farndale’s assay (~pH 3). Thus, roughly 2/3 of the recent literature involving chondrogenic pellet culture or cartilage tissue engineering either used methods that may be susceptible to these avoidable artifacts or provided too few details (directly or via citation) to determine whether concerns exist. While it is virtually impossible for a reader to discern whether any individual study’s results were affected by these artifacts, even if using a method highly susceptible to artifact, it seems unlikely that the existing literature has not been affected to some extent.
Figure 6.

Citation patterns for the DMMB assay in (a) 111 research articles involving chondrogenic pellet culture over a 5 year period and (b) 164 research articles involving cartilage tissue engineering over a 1 year period, based on examination of articles identified through PubMed searches. In both literature segments, these articles employing some variant of the DMMB assay represent approximately 38% of the total number of identified articles.
Conclusion
Overall, results of this study indicate that artifacts in sGAG readings due to the presence of cell and matrix-associated polyanionic contaminants have the potential to substantially confound results and interpretations of tissue engineering studies. Presence of such artifacts may produce overly optimistic estimates of the actual extent of sGAG production in cartilage tissue engineering studies, particularly at early time points or for cells that actually produce relatively little sGAG. Simple modifications to the assay protocol, however, can substantially reduce or eliminate these artifacts without additional sample processing. Even in studies expected to have high sGAG contents (e.g., native tissue, highly mature engineered tissues), individual groups (e.g., degraded tissue, early stage constructs) may be susceptible to artifact. Unless specific circumstances require a different approach, we therefore recommend conducting the spectrophotometric DMMB assay using dye titrated to pH 1.5 and calculating concentrations based on the difference in absorption between μ and β peak wavelengths (i.e., OD525-OD595). This is particularly important for high density (e.g., pellet, micromass, monolayer) cultures and for tissue engineered constructs with low sGAG content and where polyanionic contamination (e.g. DNA, RNA, HA, etc.) may be of concern. We similarly recommend that conditioned media assays employ dye titrated to pH 1.5 and be based on the absorption difference between μ and β peaks, with standards dissolved in a solvent matching the full medium composition to account for any interference by medium components. By suppressing common but variable artifacts, these relatively simple modifications can reduce the impact of spurious results in the tissue engineering literature.
Acknowledgments
Supported by an Arthritis Foundation Innovative Research Grant, Grant R01AR054939 from NIAMS/NIH, and a National Science Foundation Graduate Research Fellowship.
References
- Barbosa I, Garcia S, Barbier-Chassefiere V, Caruelle JP, Martelly I, Papy-Garcia D. Improved and simple micro assay for sulfated glycosaminoglycans quantification in biological extracts and its use in skin and muscle tissue studies. Glycobiology. 2003;13:647–653. doi: 10.1093/glycob/cwg082. [DOI] [PubMed] [Google Scholar]
- Billington CJ. Cartilage proteoglycan release assay. Methods Mol Biol. 2001;151:451–456. doi: 10.1385/1-59259-046-2:451. [DOI] [PubMed] [Google Scholar]
- Bitter T, Muir HM. A modified uronic acid carbazole reaction. Anal Biochem. 1962;4:330–334. doi: 10.1016/0003-2697(62)90095-7. [DOI] [PubMed] [Google Scholar]
- Chandrasekhar S, Esterman MA, Hoffman HA. Microdetermination of proteoglycans and glycosaminoglycans in the presence of guanidine hydrochloride. Anal Biochem. 1987;161:103–108. doi: 10.1016/0003-2697(87)90658-0. [DOI] [PubMed] [Google Scholar]
- de Jong JG, Heijs WM, Wevers RA. Mucopolysaccharidoses screening: dimethylmethylene blue versus alcian blue. Ann Clin Biochem. 1994;31 (Pt 3):267–271. doi: 10.1177/000456329403100309. [DOI] [PubMed] [Google Scholar]
- Dey P, Saphos CA, McDonnell J, Moore VL. Studies on the quantification of proteoglycans by the dimethylmethylene blue dye-binding method. Specificity, quantitation in synovial lavage fluid, and automation. Connect Tissue Res. 1992;28:317–324. doi: 10.3109/03008209209016823. [DOI] [PubMed] [Google Scholar]
- Downs TR, Wilfinger WW. Fluorometric quantification of DNA in cells and tissue. Anal Biochem. 1983;131:538–547. doi: 10.1016/0003-2697(83)90212-9. [DOI] [PubMed] [Google Scholar]
- Eisenberg SR, Grodzinsky AJ. Swelling of articular cartilage and other connective tissues: electromechanochemical forces. J Orthop Res. 1985;3:148–159. doi: 10.1002/jor.1100030204. [DOI] [PubMed] [Google Scholar]
- Enobakhare BO, Bader DL, Lee DA. Quantification of sulfated glycosaminoglycans in chondrocyte/alginate cultures, by use of 1,9-dimethylmethylene blue. Anal Biochem. 1996;243:189–191. doi: 10.1006/abio.1996.0502. [DOI] [PubMed] [Google Scholar]
- Estes BT, Diekman BO, Gimble JM, Guilak F. Isolation of adipose-derived stem cells and their induction to a chondrogenic phenotype. Nat Protoc. 2010;5:1294–1311. doi: 10.1038/nprot.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farndale RW, Buttle DJ, Barrett AJ. Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim Biophys Acta. 1986;883:173–177. doi: 10.1016/0304-4165(86)90306-5. [DOI] [PubMed] [Google Scholar]
- Farndale RW, Sayers CA, Barrett AJ. A direct spectrophotometric microassay for sulfated glycosaminoglycans in cartilage cultures. Connect Tissue Res. 1982;9:247–248. doi: 10.3109/03008208209160269. [DOI] [PubMed] [Google Scholar]
- Frazier SB, Roodhouse KA, Hourcade DE, Zhang L. The Quantification of Glycosaminoglycans: A Comparison of HPLC, Carbazole, and Alcian Blue Methods. Open Glycosci. 2008;1:31–39. doi: 10.2174/1875398100801010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RL, Kolibas LM. An improved method for determining proteoglycans synthesized by chondrocytes in culture. Connect Tissue Res. 1990;24:265–275. doi: 10.3109/03008209009152154. [DOI] [PubMed] [Google Scholar]
- Heinegård D, Axelsson I. Distribution of keratan sulfate in cartilage proteoglycans. J Biol Chem. 1977;252:1971–1979. [PubMed] [Google Scholar]
- Hjerpe A, Antonopoulos CA, Engfeldt B. Determination of sulphated disaccharides from chondroitin sulphates by high-performance liquid chromatography. J Chromatogr. 1979;171:339–344. doi: 10.1016/s0021-9673(01)95313-0. [DOI] [PubMed] [Google Scholar]
- Hoemann CD. Molecular and biochemical assays of cartilage components. Methods Mol Med. 2004;101:127–156. doi: 10.1385/1-59259-821-8:127. [DOI] [PubMed] [Google Scholar]
- Hughes CE, Little CB, Caterson B. Measurement of aggrecanase-generated interglobular domain catabolites in the medium and extracts of cartilage explants using Western blot analysis. Methods Mol Biol. 2003;225:89–98. doi: 10.1385/1-59259-374-7:89. [DOI] [PubMed] [Google Scholar]
- Humbel R, Etringer S. A colorimetric method for the determination of sulphated glycosaminoglycans. Rev Roum Biochim. 1974;11:21–24. [Google Scholar]
- Huynh MB, Morin C, Carpentier G, Garcia-Filipe S, Talhas-Perret S, Barbier-Chassefiere V, van Kuppevelt TH, Martelly I, Albanese P, Papy-Garcia D. Age-related changes in rat myocardium involve altered capacities of glycosaminoglycans to potentiate growth factor functions and heparan sulfate-altered sulfation. J Biol Chem. 2012;287:11363–11373. doi: 10.1074/jbc.M111.335901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson CB, Knudson W. Cartilage proteoglycans. Semin Cell Dev Biol. 2001;12:69–78. doi: 10.1006/scdb.2000.0243. [DOI] [PubMed] [Google Scholar]
- Maroudas A. Physicochemical properties of cartilage in the light of ion exchange theory. Biophys J. 1968;8:575–595. doi: 10.1016/S0006-3495(68)86509-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Hoffman P, Linker A. Mucopolysaccharides of Costal Cartilage. Science. 1958;128:896–896. doi: 10.1126/science.128.3329.896. [DOI] [PubMed] [Google Scholar]
- Mort JS, Roughley PJ. Measurement of glycosaminoglycan release from cartilage explants. Methods Mol Med. 2007;135:201–209. doi: 10.1007/978-1-59745-401-8_12. [DOI] [PubMed] [Google Scholar]
- Müller G, Hanschke M. Quantitative and qualitative analyses of proteoglycans in cartilage extracts by precipitation with 1,9-dimethylmethylene blue. Connect Tissue Res. 1996;33:243–248. doi: 10.3109/03008209609028881. [DOI] [PubMed] [Google Scholar]
- Nath S, Ghosh SK, Panigrahi S, Pal T. Photo-induced decolorization of dimethylmethylene blue with selenious acid: a novel method to examine selective monomer-dimer distribution of the dye in micelle. Spectrochim Acta A Mol Biomol Spectrosc. 2005;61:2145–2151. doi: 10.1016/j.saa.2004.08.013. [DOI] [PubMed] [Google Scholar]
- Oke SL, Hurtig MB, Keates RA, Wright JR, Lumsden JH. Assessment of three variations of the 1,9-dimethylmethylene blue assay for measurement of sulfated glycosaminoglycan concentrations in equine synovial fluid. Am J Vet Res. 2003;64:900–906. doi: 10.2460/ajvr.2003.64.900. [DOI] [PubMed] [Google Scholar]
- Seibel MJ, Macaulay W, Jelsma R, Saed-Nejad F, Ratcliffe A. Antigenic properties of keratan sulfate: influence of antigen structure, monoclonal antibodies, and antibody valency. Arch Biochem Biophys. 1992;296:410–418. doi: 10.1016/0003-9861(92)90591-j. [DOI] [PubMed] [Google Scholar]
- Shingleton WD. In vitro model of human articular cartilage degradation. Methods Mol Biol. 2003;225:99–106. doi: 10.1385/1-59259-374-7:99. [DOI] [PubMed] [Google Scholar]
- Templeton DM. The basis and applicability of the dimethylmethylene blue binding assay for sulfated glycosaminoglycans. Connect Tissue Res. 1988;17:23–32. doi: 10.3109/03008208808992791. [DOI] [PubMed] [Google Scholar]
- Terry DE, Chopra RK, Ovenden J, Anastassiades TP. Differential use of Alcian blue and toluidine blue dyes for the quantification and isolation of anionic glycoconjugates from cell cultures: application to proteoglycans and a high-molecular-weight glycoprotein synthesized by articular chondrocytes. Anal Biochem. 2000;285:211–219. doi: 10.1006/abio.2000.4761. [DOI] [PubMed] [Google Scholar]
- Wang HM, Loganathan D, Linhardt RJ. Determination of the pKa of glucuronic acid and the carboxy groups of heparin by 13C-nuclear-magnetic-resonance spectroscopy. Biochem J. 1991;278 (Pt 3):689–695. doi: 10.1042/bj2780689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman P. The quantitative measurement of Alcian Blue-glycosaminoglycan complexes. Biochem J. 1973;131:343–350. doi: 10.1042/bj1310343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelnikov I, Arndt D, Xia J, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R, Scalbert A. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe M, Pochampally R, Swaney W, Reger RL. Isolation and culture of bone marrow-derived human multipotent stromal cells (hMSCs) Methods Mol Biol. 2008;449:3–25. doi: 10.1007/978-1-60327-169-1_1. [DOI] [PubMed] [Google Scholar]
- Zar JH. Biostatistical Analysis. Pearson Prentice-Hall; Upper Saddle River, NJ: 2010. Comparing simple linear regression equations; pp. 362–378. [Google Scholar]