Abstract
Malignancy is associated with altered expression of glycans and glycoproteins that contribute to the cellular glycocalyx. We constructed a glycoprotein expression signature, which revealed that metastatic tumours upregulate expression of bulky glycoproteins. A computational model predicted that these glycoproteins would influence transmembrane receptor spatial organization and function. We tested this prediction by investigating whether bulky glycoproteins in the glycocalyx promote a tumour phenotype in human cells by increasing integrin adhesion and signalling. Our data revealed that a bulky glycocalyx facilitates integrin clustering by funnelling active integrins into adhesions and altering integrin state by applying tension to matrix-bound integrins, independent of actomyosin contractility. Expression of large tumour-associated glycoproteins in non-transformed mammary cells promoted focal adhesion assembly and facilitated integrin-dependent growth factor signalling to support cell growth and survival. Clinical studies revealed that large glycoproteins are abundantly expressed on circulating tumour cells from patients with advanced disease. Thus, a bulky glycocalyx is a feature of tumour cells that could foster metastasis by mechanically enhancing cell-surface receptor function.
The composition of cell surface glycans and glycoproteins changes markedly and in tandem with cell fate transitions occurring in embryogenesis, tissue development, stem-cell differentiation and diseases such as cancer1–3. Nevertheless, our understanding of the biochemical functions of glycans fails to explain fully why broad changes in glycosylation and glycoprotein expression are critical to cell fate specification and in what ways are they linked to disease. It is currently unclear whether changes in glycan and glycoprotein expression reflect a global and more general mechanism that directs cell and tissue behaviour.
From a materials perspective, glycan and glycoprotein expression dictates the bulk physical properties of the glycocalyx—the exterior cell surface layer across which information flows from the microenvironment to signal transduction pathways originating at the plasma membrane. Although the biophysical functions of the glycocalyx are largely untested, computational models predict that bulky glycoproteins can promote transmembrane receptor organization, including the clustering of integrins at adhesion sites4. These models suggest that glycocalyx-mediated integrin clustering would promote the assembly of mature adhesion complexes and collaborate to enhance growth factor signalling5—phenotypes that are associated with cancer6,7. We demonstrate that a global modulation of the physical properties of the glycocalyx alters integrin organization and function, and present evidence for how the glycocalyx can be co-opted in malignancy to support tumour cell growth and survival.
Regulation of integrin assembly by bulky glycoproteins
To determine whether glycocalyx bulk contributes to a cancer phenotype, we used gene expression microarray data to relate metastasis to expression of genes for which protein products contribute to the glycocalyx. The likely contribution of gene product to glycocalyx bulk was estimated based on the protein’s extracellular domain structure and predicted number of glycosylation sites (Extended Data Fig. 1). Using these estimates we obtained evidence for upregulation of transcripts encoding bulky glycoproteins and some classes of glycosyltransferases, which catalyse the glycosylation of cell surface proteins, in primary tumours of patients with distant metastases relative to those with localized tumour growth (P = 0.032 for bulky transmembrane proteins, Kolmogorov–Smirnov test; Fig. 1a and Extended Data Fig. 1).
Figure 1. The cancer glycocalyx drives integrin clustering.
a, Violin plots showing increased expression of genes encoding bulky transmembrane proteins in primary tumours of patients with distant metastases relative to those with local invasion. White dots and thick black lines indicate the median and interquartile range of the P value distribution of all transcripts within each class: all genes, all membrane proteins (Mem.), and bulky transmembrane proteins (Bulky). b, Computed relative rate of integrin–ECM ligand bond formation as a function of distance from a pre-existing adhesion cluster. c, Model of proposed glycocalyx-mediated integrin clustering. Shorter distances between integrin–ligand pairs result in faster kinetic rates of binding. d, Cartoon showing structure of glycoprotein mimetics with lipid insertion domain. e, Fluorescence micrographs of MEC adhesion complexes (vinculin–mCherry) and glycomimetics of the indicated length (scale bar, 3 µm). f, SAIM images of DiI-labelled ventral plasma membrane topography in MECs incorporated with glycomimetics (scale bar, 2.5 µm). g, Rate of integrin–substrate adhesion measured using single cell force spectroscopy in MECs with incorporated glycomimetics. h, Quantification of the total adhesion complex area per cell in MECs with incorporated glycomimetics. All results are the mean ± s.e.m. of three separate experiments. Statistical significance is given by *P < 0.05; **P < 0.01; ***P < 0.001.
To understand whether bulky glycoproteins could promote tumour aggression by regulating integrin adhesions, we developed an integrated biochemical and mechanical model that incorporates integrins, the extracellular matrix (ECM), the cell membrane and the glycocalyx (Extended Data Fig. 2). The model revealed that the kinetic rates of integrin–ECM interactions are tightly coupled to the distances between receptor–ligand pairs and, thus, the physical constraints imposed by the glycocalyx. In the presence of bulky glycoproteins, the model predicted that integrin–ECM binding is most favourable at sites of pre-existing adhesive contact, where the membrane and ECM substrate are in closest proximity (Fig. 1b). Elsewhere, bulky glycoproteins sterically restrict efficient integrin–matrix engagement (Fig. 1b) by increasing the gap between the plasma membrane and ECM. Thus, the model predicted that whereas bulky glycoproteins reduce the overall integrin-binding rate, they enhance, rather than impede, integrin clustering and focal adhesion assembly by generating a physically based kinetic trap (Fig. 1c).
To test experimentally whether bulky glycoproteins could drive integrin clustering and focal adhesion assembly, we generated a series of synthetic mucin glycoprotein mimetics of increasing length that rapidly intercalate into the plasma membrane and project perpendicularly to the cell surface8,9. These glycopolymers consisted of a long-chain polymer backbone, pendant glycan chains that mimic the structures of natural mucin O-glycans, a phospholipid for membrane insertion, and a fluorophore for imaging (Fig. 1d and Extended Data Fig. 3a–c). We found that large glycopolymers with lengths of 80 nm, significantly longer than the reported integrin length of 20 nm10, are consistently excluded from sites of integrin adhesion on the surface of non-malignant mammary epithelial cells (MECs; Fig. 1e). Shorter polymers with lengths of 3 or 30 nm were not excluded (Fig. 1e). Because the mimetics possessed minimal biochemical interactivity with cell surface proteins (Extended Data Fig. 3d), the data suggest a physical interplay between bulky glycoproteins and integrin receptors.
To determine how the largest polymer mimetics influence the nanoscale spatial features of the cell–ECM interface, we measured the topography of the ventral cell membrane using scanning angle interference microscopy (SAIM), a fluorescence-based microscopy technique that enables imaging with 5–10-nm axial resolution and diffraction-limited (~250 nm) lateral resolution11. Polymers designed to mimic large native glycoproteins (~80 nm) expanded the membrane–ECM gap by 19 nm (Fig. 1f). Consistent with computational predictions, the large glycoprotein mimetics reduced the overall rate of integrin bond formation, but significantly enhanced clustering of integrins into focal adhesions (Fig. 1g, h). Shorter glycoprotein mimetics (3 and 30 nm) did not have an impact on integrin clustering, even when incorporated at higher surface densities (Fig. 1h).
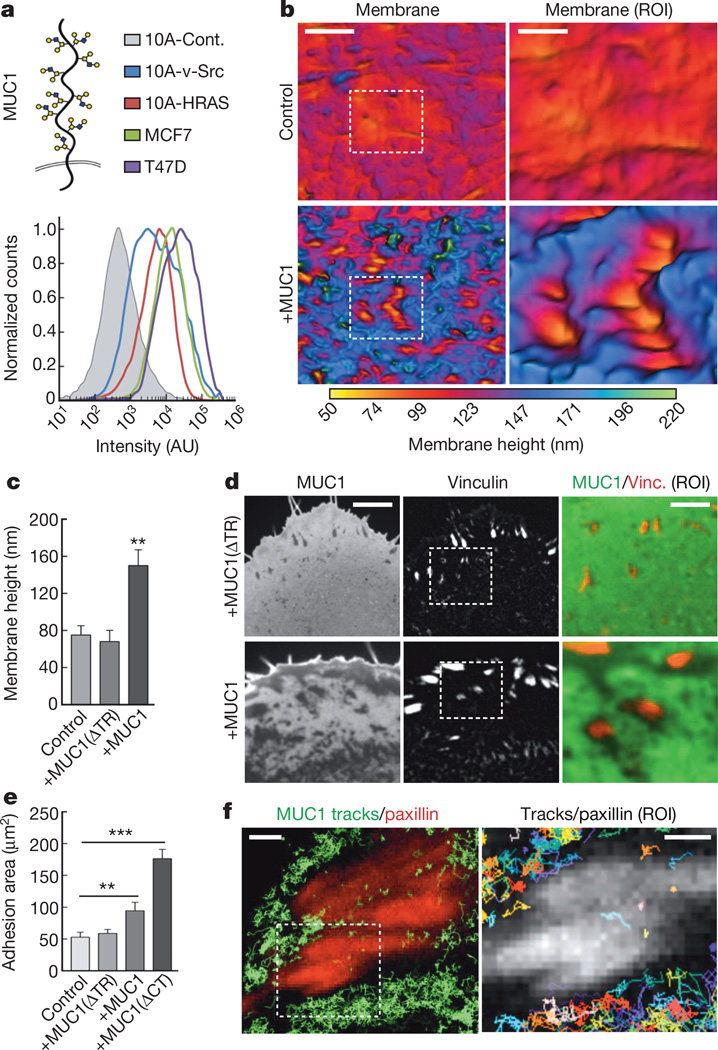
We next asked whether cancer-associated glycoproteins could similarly influence the spatial distribution of integrins and the assembly of focal adhesions. On the basis of our large-scale gene expression analysis, we determined that the transmembrane mucin glycoprotein, MUC1, which has a highly glycosylated ectodomain that projects out up to 200 nm from the cell surface12, was upregulated in metastatic tumours (nominal P = 0.0028 via one-sided t-test). In agreement with our analysis, we measured high levels of MUC1 on the surface of several breast cancer cell lines, as well as v-Src and HRAS-transformed MECs (Fig. 2a).
Figure 2. The bulky cancer-associated glycoprotein MUC1 drives integrin clustering.
a, Cartoon of MUC1 and quantification of MUC1 cell-surface levels on control (10A-Cont.), transformed (10A-v-Src, 10A-HRAS) and tumour (MCF7, T47D) cells. b, Topographical SAIM images of representative mCherry–CAAX-labelled ventral plasma membranes in control and MUC1–GFP-expressing (+MUC1) MECs (Scale bar, 5 µm; region of interest (ROI) scale bar, 2 µm). c, Quantification of mean plasma membrane height in control MECs and those ectopically expressing ectodomain-truncated MUC1–GFP (+MUC1(ΔTR)) and wild-type MUC1–GFP (+MUC1). Results are the mean ± s.e.m. of at least 15 cell measurements in duplicate experiments. d, Fluorescence micrographs of MUC1 (ΔTR) or wild-type MUC1 expressed in MECs and their focal adhesions labelled with vinculin–mCherry (scale bar, 3 µm; ROI scale bar, 1.5 µm). e, Quantification of the total adhesion complex area per cell in control non-malignant MECs (control) and those ectopically expressing MUC1 (ΔTR), wild-type MUC1, or cytoplasmic-tail-deleted MUC1 (+MUC1(ΔCT)). Results are the mean ± s.e.m. of three separate experiments. f, Left panel: trajectories of individual mEOS2-tagged MUC1 proteins recorded at 50 Hz using sptPALM (green) and focal adhesions visualized with paxillin–GFP (red) in MEFs (scale bar, 3 µm). Right panel: the ROI from the left panel with individual MUC1 tracks displayed in multiple colours (scale bar, 1 µm). Statistical significance is given by *P < 0.05; **P < 0.01; ***P < 0.001.
To assess the impact of MUC1 on focal adhesion assembly, we expressed MUC1 on the surface of non-malignant MECs, to levels comparable to those of transformed MECs and breast cancer lines. MUC1 expression induced striking membrane topographical features, which included regions of high curvature, and a significant expansion of the cell membrane–ECM gap (Fig. 2b, c and Extended Data Fig. 4a). Expression of an ectodomain-truncated MUC1 construct did not significantly change the gap compared to control MECs (Fig. 2c and Extended Data Fig. 4a). Our model predicted that the membrane topographies we observed in MUC1-expresing cells would facilitate integrin clustering through the kinetic trap. In agreement with these predictions, expression of full-length MUC1 significantly enhanced the size of adhesion clusters and the total adhesion area per cell (Fig. 2d, e and Extended Data Fig. 5a). The adhesion assembly phenotype did not require the MUC1 cytoplasmic tail, which mediates MUC1’s biochemical activity (Fig. 2e)13, or direct interactions between MUC1 and fibronectin (Extended Data Fig. 5b). Together, these results are consistent with a physically based mechanism of integrin clustering.
To gain additional insight into the coupled dynamics between integrins and MUC1, we conducted time-lapse imaging of fluorescently labelled MUC1 and the adhesion plaque protein vinculin. We observed that MUC1 and integrin adhesions spatially segregate on the cell surface in a temporally correlated manner (Fig. 2d, Extended Data Fig. 5c and Supplementary Movie 1), suggesting a physical communication between these molecules. Further evidence for a physical interplay between MUC1 and integrins was obtained in mouse embryonic fibroblasts (MEFs) using single-particle tracking photo-activation localization microscopy (sptPALM14,15) to track MUC1 diffusive trajectories. We noted that whereas MUC1 was mobile in the plasma membrane, it rarely crossed into integrin adhesion zones (Fig. 2f and Extended Data Fig. 6a).
We next tested our model’s prediction that MUC1 would favour integrin clustering by physically impeding integrin–ECM binding outside of adhesive contacts. We recorded the trajectories of individual β3 integrin molecules using sptPALM to determine the location and fraction of mobile (confined and freely diffusive) and matrix-bound, immobilized integrin15. Analysis of β3 integrin trajectories after manganese activation in MEFs revealed a significant increase in the total level of immobilized integrin at the plasma membrane, both inside and outside adhesive contacts (Fig. 3a and Extended Data Fig. 6b–e). By contrast, the immobilized β3 integrin in MEFs expressing high MUC1 was restricted to sites of adhesion (Fig. 3a–c and Extended Data Fig. 6e). These results are consistent with single-cell force spectroscopy measurements, which indicated that MUC1 expression reduces the net rate of integrin–ECM bond formation (Extended Data Fig. 5d). Mucin expression did not have a significant impact on the free diffusion of integrins (Extended Data Fig. 6b–d). Importantly, we observed that integrins frequently diffused across the mucin–adhesive zone boundary and could immobilize rapidly once in the adhesive zone (Fig. 3d, Extended Data Fig. 6f, g and Supplementary Movie 2). Together, our results indicate that large glycoproteins act as physical ‘steric’ barriers that impede integrin immobilization and thus funnel integrins into adhesive contacts.
Figure 3. Bulky glycoproteins spatially regulate immobilization of activated integrins.
a, Left panels: fluorescence micrographs displaying paxillin–GFP-labelled focal adhesions in control cells or MUC1-rich regions in MUC1–GFP-expressing MEFs, and positions of individual mEOS2-fused β3 integrins. Cells were treated without or with Mn2+ to activate integrins (scale bar, 3 µm). Right panels: magnified area of interest showing fluorescence micrographs of focal adhesions visualized with paxillin–GFP in control MEFs or MUC1 in MUC1–GFP-expressing MEFs, and individual β3 integrin trajectories recorded with sptPALM. Single molecule trajectories are colour-coded to indicate immobile and mobile (confined and freely diffusive) β3 integrins (scale bar, 1 µm). b, Distribution of β3 integrin diffusion coefficients recorded before or after Mn2+ treatment in control MEFs outside of adhesive contacts (left), MUC1-transfected MEFs inside MUC1-rich areas (middle), and MUC1-transfected MEFs outside MUC1-rich areas, including adhesive contacts (right). c, Fraction of immobilized, confined and freely diffusive β3 integrins outside of adhesive contacts in control MEFs (Ctrl) and MUC1-transfected cells (MUC1) before and after Mn2+ treatment. Results are the mean ± s.e.m. Statistical significance is given by *P < 0.05; **P < 0.01; ***P < 0.001. d, Fluorescence micrograph of MUC1–GFP and an illustrative single integrin trajectory in MEFs treated with Mn2+ (scale bar, 1 µm).
Bulky glycoproteins exert force on integrin bonds
Integrins switch between activity states by undergoing a conformational change that is facilitated by tensile force16,17. Given the order of magnitude difference in the size of MUC1 (~200 nm12) as compared to integrins (~20 nm10), and the close proximity of these molecules within the cell–ECM interface, we hypothesized that large glycoproteins, such as MUC1, modify integrin structure and function by applying force to matrix-bound receptors. Abiding by Newton’s third law, if large glycoproteins exert a tensile force on integrins, then we should detect a reciprocal strain on the glycoproteins. Consistent with this hypothesis, mucins imaged with SAIM appeared compressed or mechanically bent near integrin adhesive contacts (Fig. 4a and Extended Data Fig. 4a). Furthermore, single-cell force spectroscopy revealed that MECs expressing high levels of exogenous MUC1 required higher compressive force application at the ECM–substrate interface to promote integrin-mediated adhesion when compared to control MECs (Fig. 4b).
Figure 4. Integrins are mechanically loaded and re-enforced by bulky glycoproteins.
a, GFP-fluorescence and topographic SAIM images of MUC1–GFP (scale bars, 3 µm) and the corresponding focal adhesions visualized with vinculin–mCherry. b, Adhesion rate versus force of contact between cell and substrate (compressive force) measured with single-cell force spectroscopy for control and MUC1-expressing MECs. Results are the mean ± s.e.m. of at least 10 cell measurements per point. c, Schematic of FRET-based MUC1 compressive strain gauge. d, FRET efficiency maps of ectodomain-truncated (+MUC1(ΔTR) sensor) and wild-type (+MUC1 sensor) strain gauges measured at the ventral cell surface of MECs and the corresponding vinculin–mCherry-labelled focal adhesions (scale bar, 8 µm; ROI scale bar, 1 µm). e, Histogram of observed FRET efficiencies of wild-type MUC1 and MUC1 (ΔTR) strain gauges. f, Quantification of fibronectin-crosslinked α5 integrin in control and MUC1-expressing normal MECs treated with solvent alone (DMSO), myosin-II inhibitor (blebbistatin; 50 µM), or Rho kinase inhibitor (Y-27632; 10 µM) for 1 h followed by detergent-extraction to reveal the fibronectin bound integrin that is under mechanical tension. Results are the mean ± s.e.m. of three separate experiments. Statistical significance is given by *P < 0.05; **P < 0.01; ***P < 0.001.
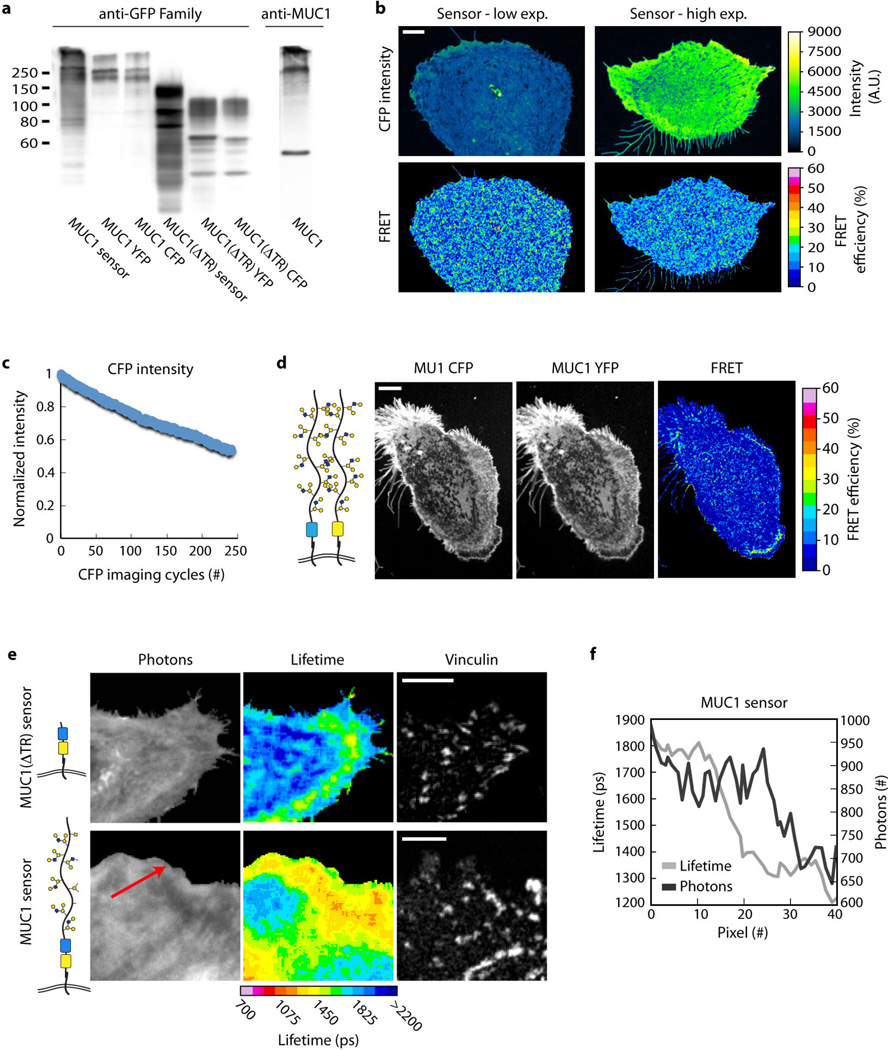
To test further whether integrin adhesions strain bulky transmembrane glycoproteins, we generated a genetically encoded construct conceptually similar to a strain gauge, consisting of a cysteine-free cyan and yellow fluorescent protein pair (CFP and YFP) separated by an elastic linker18, which we inserted into the ectodomain of full-length and truncated MUC1 proteins (Fig. 4c and Extended Data Fig. 4b). Fluorescence resonance energy transfer (FRET) served as the readout of distance between the CFP and YFP pair and, thus, functioned as a reporter of molecular strain. When the full-length reporter was expressed in MECs, we observed high FRET efficiencies in the cell–substrate interface (Fig. 4d, e and Extended Data Fig. 7). FRET efficiency was significantly lower in MECs expressing the ectodomain-truncated construct, indicative of lower molecular strain (Fig. 4d, e). The highest FRET efficiencies correlated spatially with sites of adhesive contact, consistent with integrin adhesions straining bulky transmembrane glycoproteins and glycoproteins exerting a reciprocal restoring force on integrins (Fig. 4d and Extended Data Fig. 7e, f).
We next examined whether the bulky glycoprotein MUC1 could induce conformational changes that would activate integrins independent of the contractile cytoskeleton. We used a bi-functional crosslinker that can specifically link extracellular fibronectin and bound α5β1 integrins that are in a tension-dependent conformation17. Inhibition of actomyosin contractility, using the myosin inhibitor blebbistatin or the Rho kinase inhibitor Y-27632, abrogated most of the fibronectin crosslinked integrins in MECs expressing empty vector (Fig. 4f and Extended Data Fig. 8a). By contrast, MUC1-expressing MECs formed tensioned bonds with the ECM substrate, even when cells were pre-treated with contractility inhibitors before plating (Fig. 4f and Extended Data Fig. 8a). Of note, the myosin-independent integrin clusters observed in the MUC1-expressing MECs recruited activated cytoplasmic adaptors typically associated with mature adhesion structures and nucleated actin (Extended Data Fig. 8b). These results suggest that large, cancer-associated glycoproteins not only facilitate integrin clustering but also physically alter integrin state and do so, at least in part, independently of cytoskeletal tension.
Bulky glycoproteins promote growth and survival
Tumour metastasis is a multi-step process that depends on the efficient dissemination of primary cancer cells and their subsequent colonization at distant metastatic sites19. Thus, the ability to survive, particularly within unfavourable microenvironments and under minimally adhesive conditions, is a prerequisite for efficient tumour cell metastasis19. Given their ability to promote integrin adhesion assembly, we hypothesized that bulky glycoproteins could facilitate metastasis by promoting focal adhesion signalling to enhance tumour cell growth and survival. Consistent with this notion, analysis of human data sets revealed that patients with aggressive breast cancers that presented with circulating tumour cells (CTCs) express disproportionately high amounts of bulky glycoproteins and have altered glycosyltransferase expression profiles (Fig. 5a and Extended Data Fig. 1d, e). Furthermore, analysis of genes expressed within CTCs isolated from a cohort of breast cancer patients with metastatic disease confirmed that several predicted bulky glycoproteins could be detected in these patient samples (Fig. 5b).
Figure 5. Bulky glycoproteins promote cell survival and are expressed in CTCs.
a, Violin plots showing that genes encoding bulky transmembrane proteins are more highly expressed in primary human tumours in patients with circulating tumour cells (CTCs). White dots and thick black lines indicate the median and interquartile range of the P-value distribution of transcripts of all cellular genes (all genes), all transmembrane proteins (membrane), and bulky transmembrane proteins (bulky). b, Heat map quantifying gene expression of bulky glycoproteins in CTCs isolated from 18 breast cancer patients (x axis; left), and representative immunofluorescence micrograph of MUC1 detected on human patient CTCs (right; scale bar, 5 µm). Quantification of the percentage of CTCs with detectable MUC1 is shown. c, Cell death in control non-malignant MECs and those with incorporated glycomimetics quantified 24 h after plating on a soft (140 Pa) fibronectin-conjugated hydrogel substrate. d, Cell death (left graph) and growth (right graph) of control MECs and those expressing cytoplasmic-tail-deleted MUC1 (+MUC1(ΔCT)) quantified 48 h after plating on a soft hydrogel. e, Quantification of the number of vehicle (DMSO), PI(3)K inhibitor, MEK inhibitor, or Src inhibitor-treated control and MUC1(ΔCT)-expressing MECs per colony 48 h after plating on a soft hydrogel. f, Proliferation of solvent (DMSO) or FAK-inhibitor-treated MUC1(ΔCT)-expressing MECs quantified at the indicated day after plating on soft hydrogels. g, Representative western blots showing phosphorylated and total ERK in control and MUC1(ΔCT)-expressing MECs plated on soft hydrogels unstimulated or stimulated with EGF. Cells were treated with solvent (control, +MUC1(ΔCT)) or FAK inhibitor (+MUC1(ΔCT) + FAKi) before stimulation. h, Bar graphs showing quantification of immunoblots probed for activated AKT in control and MUC1(ΔCT)-expressing non-malignant MECs 24 h after plating on soft versus stiff hydrogels. i, Model summarizing biophysical regulation of integrin-dependent growth and survival by bulky glycoproteins. In all bar graphs, results are the mean ± s.e.m. of at least 2–3 separate experiments (*P < 0.05; **P < 0.01; ***P < 0.001).
We next examined whether a bulky glycocalyx could promote growth and survival of non-malignant MECs. Using our glycoprotein mimetics, we observed that untreated MECs or MECs incorporated with short (3 nm) or medium (30 nm) length mimetics were not viable 24–48 h after they were plated on highly compliant hydrogel substrates that mimic the stiffness of soft sites of colonization, like lung or brain (Young’s modulus, E = 140 Pa; Fig. 5c). By contrast, MECs incorporated with long glycoprotein mimetics (80 nm) remained viable (Fig. 5c). Analysis of gene expression profiles and immunofluorescence analysis of freshly isolated CTCs in our human metastatic breast cancer cohort revealed that MUC1 could be detected in most of the samples examined (Fig. 5b). Similar to results with the synthetic mimetics, we observed that ectopic expression of either full-length or a tailless, signalling-defective MUC1 in non-malignant MECs permitted their growth and survival even when plated as single cells on compliant hydrogels (Fig. 5d and Extended Data Fig. 9a).
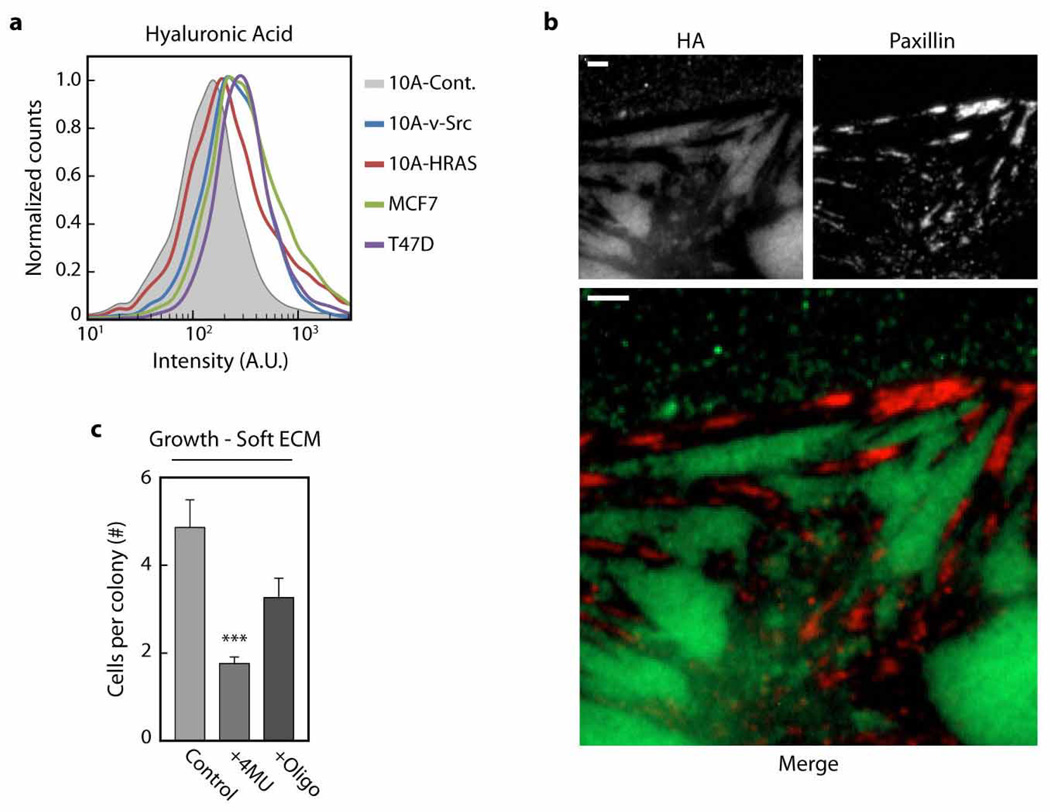
We noted that the CTCs in our cohort also expressed high levels of CD44, a receptor that binds and retains bulky hyaluronic acid (HA) glycan structures on the cell surface (Extended Data Fig. 10a)20. Similar to our observations with MUC1 and bulky glycoprotein mimetics, we observed that HA and integrins exhibit an anti-correlated spatial distribution on the surface of transformed MECs (Extended Data Fig. 10b). Inhibition of HA synthesis or HA cell-surface retention significantly reduced the growth of transformed MECs on compliant hydrogels, raising the possibility that bulky cell-surface constituents, in addition to MUC1, could similarly promote tumour aggression (Extended Data Fig. 10b). However, unlike the experiments with tailless MUC1 or the glycoprotein mimetics, which lack signalling capability, we cannot exclude that HA-induced growth and survival phenotypes are not also, at least in part, induced through HA’s direct biochemical signalling activity20,21.
We next addressed whether a bulky glycocalyx promotes MEC growth and survival by regulating focal adhesion assembly and crosstalk with growth factor signalling pathways5,7. We found that pharmacological inhibition of kinases linked to growth factor signalling, including phosphoinositide 3-kianse (PI(3)K), mitogen-activated kinase, and Src kinase, each independently inhibited the growth and survival of MUC1-expressing MECs on highly compliant substrates (Fig. 5e). We also noted that the MUC1 growth and survival phenotype requires integrin engagement and integrin signalling through focal adhesion kinase (FAK), which mediates crosstalk between integrin and growth factor signalling pathways (Fig. 5f and Extended Data Fig. 9b)5,6. Non-malignant MECs expressing the MUC1 ectodomain, but not control MECs, assembled distinct focal adhesion structures with activated Y397-phosphorylated FAK on compliant substrates (Extended Data Fig. 8c). Furthermore, MECs expressing wild-type or tailless, signalling defective MUC1, and plated on the compliant substrates showed enhanced Y118-phosphorylated paxillin, ERK and AKT activation in response to epidermal growth factor stimulation (Fig. 5g and Extended Data Fig. 8d). This response was attenuated by FAK inhibition (Fig. 5g, h and Extended Data Fig. 8d). Together, these findings indicate that a bulky glycocalyx can promote tumour aggression by enhancing integrin-dependent growth and survival (Fig. 5i).
Discussion
We present evidence to support a new paradigm for the biological function of cell surface glycans and glycoproteins. Independent of, and in addition to, their biochemical properties, we demonstrate how bulky constituents of the glycocalyx can physically influence receptor organization and activity. Although the current investigation focuses on integrins, a bulky glycocalyx could, in principle, regulate any transmembrane receptor that interacts with a tethered ligand. Candidate systems include neurological and immunological synapses22, cell–celladhesions23, and juxtacrine signalling complexes composed of receptors, like ephrin24. Membrane topographical features imprinted by large glycoproteins could also directly influence plasma membrane lipid organization, protein sorting and endocytosis25,26. The diversity of these processes suggests that the physiological relevance of the glycocalyx may be broad. For example, bulky glycoproteins and glycan structures, such as neuroligins, neurexins and polysialic acid, have a crucial role in neuronal development, maintenance and plasticity27,28. Thus, it is plausible that the glycocalyx has a prominent role in orchestrating multiple biological processes occurring at the plasma membrane.
Our observations provide a tractable explanation for why large glycan structures and glycoproteins, like HA and mucins, as well as regulatory enzymes, are so frequently elevated in many solid tumours13,20. Indeed, the growth and survival advantages afforded by these molecules may preferentially select for cancer cells with a prominent glycocalyx and favour tumour cell dissemination and metastasis. Mechanical perturbations to cell and tissue structure have a causal role in tumour development and progression29,30, and we now implicate the glycocalyx’s importance in the metastatic mechano-phenotype. Our results suggest that the glycocalyx and its molecular constituents are attractive targets for therapeutic interventions aimed at normalizing transmembrane receptor signalling.
METHODS
Bioinformatics
To estimate protein-level contributions to extracellular membrane bulkiness, we used TMHMM to identify extracellular domains within each isoform sequence (RefSeq v47) and counted the number of putative extracellular glycosylation sites predicted by NetOGlyc 3.1 and search of N-glycosylation motifs. Gene-wise enrichment of mRNA upregulation among bulkyproteins in clinical data (GEO accessions GSE12276 and GSE31364) was tested by permuting P values quantifying evidence for upregulation in the appropriate samples. Variance in mRNA upregulation explained by membrane bulkiness was estimated by regressing the negative log-transformed P values on the square root of the combined N- and O-glycosylation sites and comparing the residuals with the intercept model. Additional details of the analysis and models are provided in Supplementary Note 1.
Computational model
A mechanical model of the cell–ECM interface was constructed as described previously4. A summary of the model is described in Supplementary Note 2 and parameters are detailed in Supplementary Table 1.
Antibodies and reagents
Antibodies used include: mouse monoclonal antibody (mAb) vinculin (MAB674; Millipore), mouse mAb talin (8d4; Sigma), rat mAbβ1-integrin (AIIBII), rabbit mAb paxillin (Y113; Abcam), rabbit mAb FAK pY397 (141-9; Invitrogen), rabbit polyclonal antibody (pAb) α5-integrin (AB1928; Millipore), mouse mAb MUC1 (HMPV; BD Pharminigen), hamster mAb MUC1 (CT2; Thermo Scientific), rabbit mAb Src Family pY416 (D49G4; Cell Signaling), mouse mAb FAK (77; BD Transduction Laboratories), rabbit pAb paxillin pY118 (2541; Cell Signaling), rabbit mAb pan-AKT (C67E7; Cell Signaling), rabbit pAb AKT pS473 (9271; Cell Signaling); rabbit mAb ERK1/2 pT202/pT204 (197G2; Cell Signaling); rabbit pAb ERK1/2 (9102; Cell Signaling); rabbit mAb Gapdh (14C10; Cell Signaling); Alexa 488 and Alexa 568 conjugated goat anti-mouse and anti-rabbit mAbs (Invitrogen); FITC conjugated anti-hamster mAbs; Cy5-conjugated goat anti-mouse and rabbit mAbs (Jackson); and HRP conjugated anti-rabbit and anti-mouse mAbs. Chemical inhibitors used in these studies include ROCK inhibitor Y-27632 (Cayman Chemical), myosin-II inhibitor (−)-blebbistatin (Cayman Chemical), FAK inhibitor FAK inhibitor 14 (Tocris), MEK inhibitor U0126 (Cell Signaling), PI(3)K inhibitor Wortmannin (Cell Signaling), Src inhibitor Src I1 (Sigma), and DiI (Molecular Probes).
Cell culture conditions
All cells were maintained at 37 °C and 5%CO2. MCF10A human MECs (ATCC) were cultured in DMEMF12 (Invitrogen) supplemented with 5%donorhorse serum (Invitrogen), 20 ng ml−1 epidermal growth factor (Peprotech), 10 µg ml−1 insulin (Sigma), 0.5 µg ml−1 hydrocortisone (Sigma), 0.1 µg ml−1 cholera toxin (Sigma), and 100 units ml−1 penicillin/streptomycin. MCF7 and T47D breast tumour lines (ATCC) were grown in DMEM supplemented with 10% fetal bovine serum (Hyclone) and 100 units ml−1 penicillin/streptomycin. 293T cells (ATCC) were maintained in DMEM supplemented with 10% donor horse serum, 2 mM l-glutamine, and penicillin/streptomycin. Mouse embryonic fibroblasts (MEFs) were cultured in DMEM with 10% fetal bovine serum. Cell lines were tested routinely for mycoplasma contamination. For transient gene expression in MECs, constructs in pcDNA3.1 or Clonetech-style vectors were nucleofected with Kit V (Lonza) using program T-024 24 h before experimentation. Transient transfection in MEFs was conducted 48 h before experimentation using Fugene 6 (Roche) or nucleofection. For stable cell lines harbouring tetracycline inducible transgenes, expression was induced with 0.2 ng ml−1 doxycycline 24 h before experimentation. The conditional v-Src oestrogen receptor fusion (v-Src–ER) was activated with 1 µM 4-hydroxytamoxifen 48 h before experimentation to achieve transformation. For pERK, pY118paxillin, and pAKT studies, cells were plated on fibronectin-conjugated polyacrylamide hydrogels, serum-starved overnight, and stimulated with 20 ng ml−1 EGF before collecting protein lysates. Data are reported as the fold increase of phosphorylated protein relative to total protein, following EGF stimulation.
Preparation of cellular substrates
Glass and silicon substrates were prepared by glutaraldehyde activation followed by conjugation with 10 µg ml−1 (glass) or 20 µg ml−1 (silicon) fibronectin as described11. Compliant polyacrylamide hydrogel substrates (soft: 2.5% acrylamide, 0.03% Bis-acrylamide; stiff: 10% acrylamide, 0.5% Bisacrylamide) were prepared as previously described with one modification: functionalization with succinimidyl ester was with 0.01% N6, 0.01% Bis-acrylamide, 0.025% Irgacure 2959, and 0.002% Di(trimethylolpropane) tetraacrylate (Sigma)31. Following functionalization with succinimidyl ester, hydrogels were conjugated overnight with 20 µg ml−1 fibronectin at 4 °C and rinsed twice with PBS and DMEM before cell plating.
Generation of expression constructs
A description of cDNA constructs and their construction is provided in Supplementary Note 3.
Generation of stable cell lines
Stable transgene expression was achieved through retroviral or lentiviral transduction as previously described11,30.
Flow cytometry and cell sorting
Cell surface MUC1 was labelled directly with FITC-conjugated mAb MUC1 (clone HMPV). Cytometry and sorting were conducted on a FACSAria II (BD Biosciences).
Immunofluorescence and imaging
Cells were fixed and labelled as previously described and imaged at random on a Zeiss LSM 510 microscope system with a 100× Plan Apochromat NA 1.4 objective and 488 nm Argon, 543 nm HeNe, and 633 nm HeNe excitation lines30.
Live epithelial cell imaging and FRET
Normal growth media was exchanged for a similar formulation lacking phenol red and supplemented with 15 mM HEPES buffer, pH7.4. Cells were imaged on a Ti-E Perfect Focus System (Nikon) equipped with a CSU-X1 spinning disk confocal unit; 454 nm, 488 nm, 515 nm and 561 nm lasers; an Apo TIRF 60×NA 1.49 objective; electronic shutters; a charged-coupled device camera (Clara; Andor) and controlled by NIS-Elements software (Nikon).
For measurement of FRET efficiency, the acceptor photobleaching method pbFRET was implemented with live cells on the spinning disk confocal. Cyan fluorescent protein (CFP) was first imaged with 454 nm excitation and a 480/20 emission filter, yellow fluorescent protein (YFP) was subsequently bleached using a 100 mW 515 nm laser for 10 s, and CFP was imaged again following bleaching of YFP. Microscope Z-focus was maintained during image acquisition using the Perfect Focus System. Background images were constructed by imaging 10 unique cell-free regions on the coverslip and averaging the intensity at each pixel. The FRET efficiency was calculated on a pixel-by-pixel basis according to:
where Ipre is the CFP intensity before bleaching YFP, Bpre is the CFP-channel background intensity before bleaching YFP, Ipost is the CFP intensity after bleaching YFP, and Bpost is the CFP-channel background intensity after bleaching YFP. Appropriate controls were implemented to account for inadvertent CFP photobleaching, incomplete YFP photobleaching, and intermolecular FRET (see Supplementary Note 4).
Time-domain fluorescence lifetime imaging microscopy (FLIM) for additional FRET sensor characterization was implemented with an inverted Zeiss LSM510 Axiovert 200M microscope with a Plan NeoFLUAR 40×/1.3 NA DIC oil-immersion objective lens, equipped with a TCSPC controller (SPC-830 card; Becker & Hickl, Berlin, Germany) as described previously32. CFP was excited with 440 nm light generated by frequency doubling of 880 nm pulses from a mode-locked Ti:sapphire laser(Mai-Tai, Spectra-Physics, 120–150 fs pulse width, 80 MHz repetition rate, and Frequency Doubler and Pulse Selector, Spectra-Physics, Model 3980). The emission light was passed through a NFT 440 beamsplitter, directed to the fibre-out port of the confocal scan-head, filtered with a 480BP40 filter (ChromaTechnology, Rockingham, VT) and detected by a PMC-100 photomultiplier (Becker & Hickl). The pinhole was set to give an optical slice of <4.0 µm. Images of 386 × 386 pixels were averaged over <120 s. Data analysis to produce an intensity image and a FLIM image was done offline using the pixel-based fitting software SPCImage (Becker & Hickl), assuming double exponential decay during the first 8.5 ns of the 12.5 ns interval between laser pulses. Images were scaled to 256 × 256 pixels and no binning was used. Lifetime distributions were calculated for a masked portion of the FLIM image, generated with a triangle algorithm threshold of the photo count intensity image.
Scanning angle interference microscopy
Cells were plated overnight on reflective silicon substrates, fixed or roofed to remove the dorsal membrane (for MUC1–GFP imaging) and then fixed, and imaged randomly as previously described, scanning the incident angle of excitation light from 0° to 42° with a one-degree sampling rate11. Z-positions were localized with custom algorithms previously described and available on request11.
Single particle tracking photo-activation localization microscopy (sptPALM)
sptPALM experiments were performed and analysed as previously described15. Briefly, live MEFs were imaged at 37 °C in a Ludin chamber on a Ti Perfect Focus System equipped with a Plan Apo 100× NA 1.45 objective, and an electron multiplying charge-coupled device (Evolve; Photometrics). For photo-activation localization, cells expressing mEOS2-tagged constructs were activated using a 405 nm laser (Omicron) and the photo-activated fluorophores were excited simultaneously with a 561 nm laser (Cobolt Jive). The powers of the activation and excitation lasers were adjusted to keep the number of activated molecules constant and well separated. GFP fusions of paxillin or MUC1 were imaged in between each sptPALM sequence by imaging the GFP signal above the unconverted mEOS2 background. The acquisition was driven by Metamorph software (Molecular Devices) in streaming mode at 50 Hz. For tracking, single-molecules were localized and tracked over time using a combination of wavelet segmentation and simulated annealing algorithms. Trajectories lasting at least 20 frames were selected for further quantification, including calculation of immobile, confined and free-diffusing fraction (see Supplementary Note 5)15.
Preparation of glycopolymer-coated cell surfaces
Mucin mimetic glycopolymers with lipid insertion domains were synthesized and characterized as described in Supplementary Note 6. For incorporation into the plasma membrane, cells were suspended in DMEM and incubated with 2 µM glycopolymer for 1 h. Cells were pelleted by centrifugation and re-suspended in growth media to remove unincorporated polymer.
Quantification of adhesion complexes
Images of adhesions in fixed, immuno-labelled cells or cells expressing paxillin–mCherry were randomly acquired, smoothed with a median filter, and background subtracted (12 pixel diameter) in ImageJ. Adhesion sizes and the number of adhesions per cell were subsequently quantified in ImageJ with the ‘Analyze Particles’ tool.
Integrin crosslinking assay
Cells were incubated in suspension with inhibitor (Y-27632 or Blebbistatin) or control solvent for 1 h before plating on glass substrates. Integrin was crosslinked to fibronectin with 1 mM 3,3′-dithiobis(sulfosuccinimidylpropionate) (Pierce Chemical) and cells were extracted with SDS buffer as previously described17. Crosslinked α5 integrin was immuno-labelled and imaged at random with a Plan Apo VC 60× objective on a Nikon TE2000 epi-fluorescence microscope equipped with a charged-coupled device camera (HQ2; Photometrics).
Single cell force spectroscopy
Measurements were performed on an Asylum MFP-3D-BIO atomic force microscope as previously described33. Briefly, cells were attached to a streptavidin-coated, tipless cantilever using biotinylated jacalin (MUC1-expressing cells) or concanavalin A (all other cells) and pressed against the adhesive substrate with a calibrated force and duration before measuring the force required to detach the cell from the substrate. All measurements were conducted on fibronectin- or BSA-coated glass slides at room temperature. The relative rate of adhesion was calculated as the slope of a linear fit of cellular detachment force against contact time.
Assessment of fibronectin–fibrillogenesis
Human recombinant fibronectin was labelled with N-hydroxysuccinimide Alexa568 (Invitrogen) according to manufacturer’s protocol and dialysed extensively in PBS. Conversion of soluble, fluorescently labelled fibronectin from the growth media into insoluble fibrils was imaged according to published protocol34. Briefly, MCF10A complete growth media was prepared with donor horse serum that was depleted of fibronectin using gelatin Sepharose 4B (GE Healthcare). MCF10A cells were plated in the depleted media on fibronectin-conjugated glass coverslips and incubated the next day in 10 µg ml−1 labelled fibronectin for one hour. Cells were quickly rinsed in PBS, fixed in 4% paraformaldehyde, and imaged at random on a spinning disk confocal.
Isolation and gene expression profiling of CTCs
Twenty CTC samples were isolated from the blood of 18 metastatic breast cancer patients as previously described35. Briefly, whole blood was subjected to EpCAM-based immunomagnetic enrichment followed by fluorescence-activated cell sorting of CTCs defined as nucleated, EpCAM-positive, CD45-negative cells. CTCs were sorted directly onto lysis buffer (Taqman PreAmp Cells-to-Ct kit, Life Technologies). cDNA of target genes were pre-amplified (14 cycles) and measured via qPCR analysis. The mean Ct for ACTB and GAPDH was used for normalization to calculate relative gene expression (ΔCt). Studies involving CTCs were approved by the UCSF Committee on Human Research. Samples were obtained with IRB approved consent from all patients.
Immunofluorescence labelling of CTCs
CTC samples were isolated from the blood of three metastatic breast cancer patients as described for gene expression profiling. Isolated CTCs were mounted and fixed on poly-l-lysine-coated slides and labelled with FITC-conjugated MUC1 mAb (Clone HPMV). As a control, purified white blood cells from the same patients were prepared similarly, and their immunofluorescence was compared to CTC samples.
Statistics
Statistical significance of experimental data sets was determined by Student’s t-test after confirming that the data met appropriate assumptions (normality, homogenous variance and independent sampling). Statistical analyses of microarray gene expression data sets are described in detail in Supplementary Note 1. All public microarray data were downloaded from the NCBI Gene Expression Omnibus website and analysed using custom R scripts (all Perl, PHP and R scripts used in this work are available on request).
Extended Data
Extended Data Figure 1. Large-scale gene expression analysis reveals increased expression of genes encoding bulky glycoproteins and glycan-modifying enzymes in primary tumours of patients with disseminated disease.
a, Bioinformatics pipeline to estimate the extracellular bulkiness of a protein from its corresponding amino acid sequence. For each isoform sequence, the transmembrane and extramembrane domains were identified using a hidden Markov model (TMHMM). A combination of motif searches and neural network prediction then identified likely N- and O-glycosylation sites within each sequence. Isoform-level bulkiness estimates were generated by summing the number of predicted N- and O-glycosylation sites located within the extramembrane regions of the isoform. b, Heat map depicting the pairwise spearman correlation coefficients calculated by comparing all per-gene estimates of the total number of extra-membrane amino acids (AAoutside), N-glycosylation sites (Nglyc), O-glycosylation sites (Oglyc), and the overall bulkiness measure (total sites; for example, the sum of extra-membrane N- and O- glycosylation sites). Correlation coefficients relating the corresponding gene-wise measures are listed in the corresponding cells and depicted on a colour scale, where white corresponds to perfect correlation (rho = 1), and the dendrograms indicate the overall relationship between the parameters, estimated by Euclidean distance. High correlation coefficients indicate that gene-wise estimates of the compared parameters are similarly ranked (for example, genes with high values of X also tend to have high values of Y). The data indicate that the number of extracellular N-glycosylation sites and O-glycosylation sites identified within a gene are only weakly correlated, and neither dominates the total number of sites estimated per gene. c, Violin plots contrasting the distributions of gene-wise one-sided P values (y axis) quantifying evidence for transcriptional upregulation of glycosidases and glycosyltransferases, and subsets of glycosyltransferases (sialyltransferases and N-acetylgalactosaminyltransferases) with the full distribution. White dots and thick black lines indicate the median and interquartile range of the gene-wise P-value distribution among category members, and the width of the violin along the y axis indicates the density of the corresponding values. P values are derived from comparisons of expression levels in primary tumours of patients with or without distant metastases using a t-test. Indicated P values were estimated using a one-sided Kolmogorov–Smirnov test. d, Violin plots quantifying transcriptional upregulation of glycan-modifying enzymes in primary tumours of patients presenting with circulating tumour cells compared to tumours without detectable circulating tumour cells. e, Table of bulky glycoproteins and potential bulk-adding glycosyltransferases whose expression is upregulated in tumours that present with circulating tumour cells.
Extended Data Figure 2. Computational model of the cell–ECM interface.
Schematic of an integrated model that describes how the physical properties of the glycocalyx influence integrin–ECM interactions. The cell surface is modelled as a three-dimensional elastic plate; the ECM as a rigid substrate underneath the cell surface; and the glycocalyx as a repulsive potential between the plate and substrate. To compute stress–strain behaviour, the model is discretized using the three-dimensional lattice spring method, the cross-section of which is depicted above. Integrins are tethered to the cell surface and their distance-dependent binding to the ECM–substrate is calculated according to the Bell model. To calculate integrin-binding rate as a function of lateral distance from an adhesion cluster, an adhesion cluster is first constructed by assembling a 3 × 3 bond structure. The rates for additional integrin–ECM bonds then are computed at various distances from the cluster.
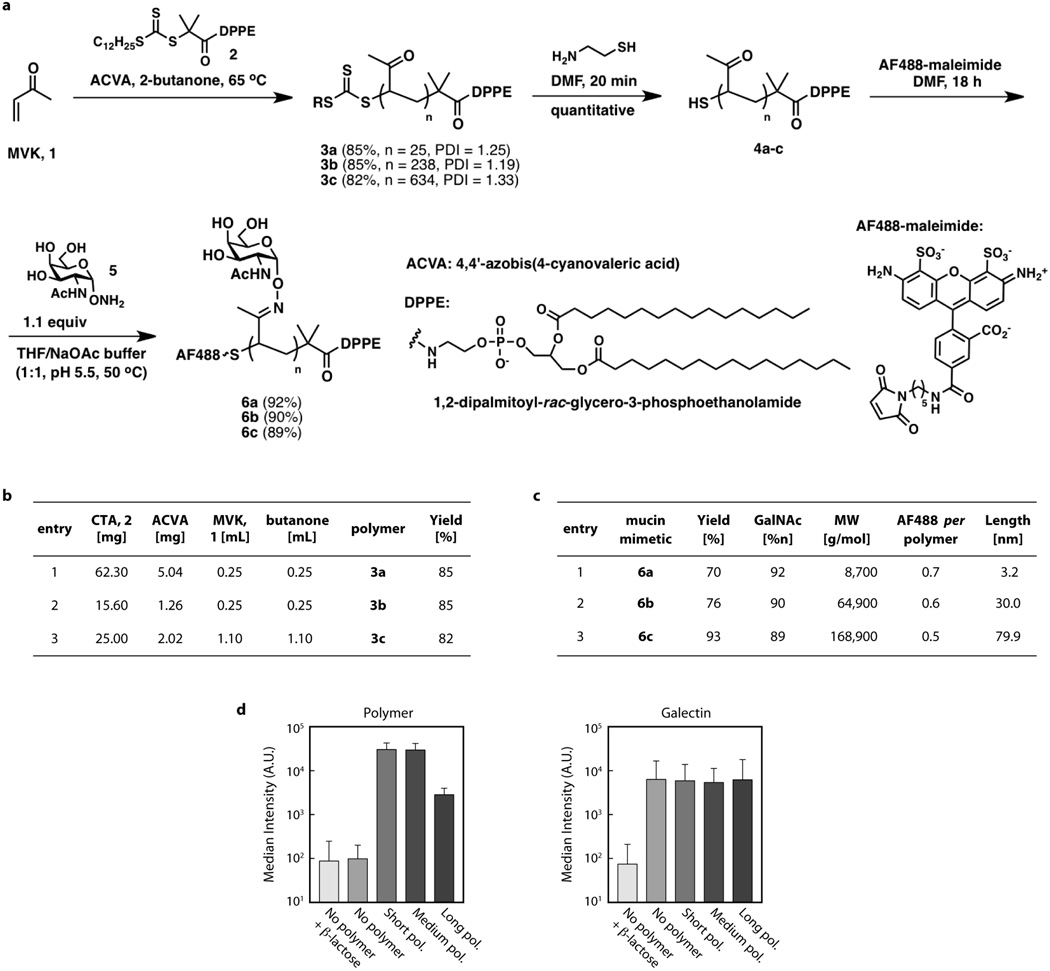
Extended Data Figure 3. Synthesis and characterization of glycoprotein mimetics.
a, Scheme for synthesis of lipid-terminated mucin mimetics labelled with Alexa Fluor 488 (AF488). b, Reagents and yields for the synthesis of polymers 3a–c. c, Characteristics of polymers 6a–c based on 1H NMR spectra. Glycoprotien mimetics were engineered to have minimal biochemical interactivity with cell surface lectins. d, Flow cytometry results quantifying incorporation of polymer on the surface of mammary epithelial cells (left) and binding with recombinant Alexa568-labelled galectin-3 with or without competitive inhibitor, β-lactose (right). Although a weak affinity between galectin-3 and the pendant N-acetylgalactosimes has previously been reported, the results suggest that incorporation of polymer does not significantly change the affinity of the cell surface for lectins.
Extended Data Figure 4. MUC1 expression constructs.
a, Schematic of MUC1 expression constructs. Full-length MUC1 consists of a large ectodomain with 42 mucin-type tandem repeats, a transmembrane domain, and short cytoplasmic tail. The tandem repeats and cytoplasmic tail are deleted in MUC1(ΔTR) and MUC1(ΔCT), respectively. For fluorescent protein fusions, mEmerald (GFP) and mEOS2 are fused to the C terminus of full-length MUC1 or MUC1(ΔCT). b, Schematic of MUC1 strain sensor and control constructs. Cysteine-free mTurqoiuse2 (CFP), Venus (YFP), or a FRET module consisting of the fluorescent proteins separated by an elastic linker (8 repeats of GPGGA) are inserted into the MUC1 ectodomain adjacent to the MUC1 tandem repeats. The mucin tandem repeats are deleted in ectodomain-truncated variants (ΔTR).
Extended Data Figure 5. MUC1-mediated adhesion formation.
a, Quantification of the average number of large adhesions, greater than 1 µm2, per area of cell in control epithelial cells (Control) and those ectopically expressing ectodomain-truncated MUC1 (+MUC1(ΔTR)), wild-type MUC1 (+MUC1), or cytoplasmic-tail-deleted MUC1 (+MUC1(ΔCT)). Results are the mean ± s.e.m. of three separate experiments. b, Fluorescence micrographs showing immuno-labelled MUC1 and fluorescently labelled fibronectin fibrils in control and MUC1-expressing epithelial cells. Soluble, labelled fibronectin in the growth media was deposited by cells at sites of cell–matrix adhesion. Binding of soluble fibronectin to MUC1 was not detected. Scale bar, 10 µm. c, Time lapse images of MUC1–YFP and vinculin–mCherry, showing the dynamics of adhesion assembly (Vinc.) and MUC1 patterning (MUC1). Scale bar, 1 µm. d, Rate of adhesion measured with single cell force spectroscopy of control (Cont.), α5 integrin-blocked (anti-α5), and MUC1-expressing cells (+MUC1) to fibronectin-coated surfaces and control cells to BSA-coated surfaces (BSA). Results are the mean ± s.e.m. of at least 15 cell measurements. Statistical significance is given by *P < 0.05; **P < 0.01; ***P < 0.001.
Extended Data Figure 6. β3 integrin mobility in MUC1-expressing cells.
a, Molecular diffusivity and adhesion enrichment measured with sptPALM in mouse embryonic fibroblasts (MEFs). Adhesion enrichment is reported as the ratio of the number of molecules detected inside focal adhesions per unit area to the number of molecules detected outside focal adhesions per unit area. b, Mean diffusion coefficients measured for freely diffusive β3 integrin tracks outside of adhesive contacts in control (Cont.) and MUC1-transfected (+MUC1) MEFs with and without Mn2+ to activate β3. c, Mean diffusion coefficients measured for confined β3 integrin tracks outside of adhesive contacts in MEFs with and without Mn2+. d, Mean radius of confinement measured for confined β3 integrin tracks outside of adhesive contacts in MEFs with and without Mn2+. e, Fraction of immobilized (Imm.), confined (Conf.), and freely diffusive (Free) β3 integrins inside of adhesive contacts in control and MUC1-transfected MEFs with and without Mn2+ treatment. f, From left to right, panels show GFP-tagged wild-type MUC1 (red) and positions of individual β3 integrins (green) in MEFs without Mn2+ treatment (left panel) and individual integrin trajectories recorded with sptPALM within MUC1-rich regions, outside MUC1-rich regions, and that cross MUC1 boundaries (scale bar, 2 µm). The ratio of integrins crossing out versus crossing in the MUC1 boundaries per cell is close to one (1.0 ± 0.1, n = 9 cells, 4,145 trajectories) showing that the flux of free diffusing integrins crossing in or out the mucin region is the same. g, From left to right, panels show integrin trajectories within an arbitrary region drawn in a MUC1-rich area (dashed white circles), outside of the circled region, and that cross the circled region (scale bar, 2 µm). The ratio of integrins crossing the MUC1-rich boundaries versus the fictive boundaries per cell is close to one (1.2 ± 0.2, n = 9 cells, 9,321 trajectories), showing that the MUC1–adhesive zone boundary does not affect the diffusive crossing of integrins. For all bar graphs, results are the mean ± s.e.m.
Extended Data Figure 7. MUC1 strain gauge.
a, Western blot of indicated construct expressed in HEK 293T cells and probed with anti-GFP family antibody, or full-length MUC1 construct expressed in HEK 293T cells and probed with an antibody against the MUC1 tandem repeats. b, Pseudocoloured images showing similar FRET efficiencies measured by the photobleaching FRET method for mammary epithelial cells (MECs) expressing low (Low) and high (High) levels of the sensor construct. Scale bar, 5 µm. c, Plot showing the level of CFP bleaching per CFP imaging cycle in MECs. d, Control images showing minimal intermolecular FRET in MECs expressing similar levels of bothMUC1 CFP and MUC1 YFP. e, Micrographs showing the emitted photons from CFP and their fluorescence lifetimes in MECs expressing ectodomain-truncated (MUC1(ΔTR) sensor) or full-length MUC1 strain sensors (MUC1 sensor). Shorter lifetimes are indicative of higher energy transfer between the CFP donor and YFP acceptor, and thus closer spatial proximity of the donor and acceptor (scale bar, 10 µm). f, Representative profile of CFP lifetimes and emitted photons of the full-length MUC1 sensor along the red line in panel e. Pixels 0 and 40 correspond to the base and tip of the arrow, respectively. A drop in fluorescence lifetime (Lifetime) is often observed before the drop in MUC1 molecular density (Photons) as an adhesive zone is approached.
Extended Data Figure 8. Tension-dependent integrin activation and focal adhesion assembly in MUC1-expressing cells.
a, Fluorescence micrographs of fibronectin-crosslinked α5 integrin in control and MUC1-expressing mammary epithelial cells (MECs) treated with solvent alone (DMSO), myosin-II inhibitor (blebbistatin; 50 µM), or Rho kinase inhibitor (Y-27632; 10 µM) for 1 h and detergent-extracted following crosslinking. Only fibronectin-bound integrins under mechanical tension are crosslinked and visualized following detergent extraction (scale bar, 15 µm). b, Fluorescence micrographs showing formation of myosin-independent adhesion complexes in MUC1-expressing MECs. Cells were pre-treated for 1 h and plated for 2 h in 50 µM blebbistatin (scale bar, 10 µm). c, Fluorescence micrographs of paxillin–mCherry and immuno-labelled activated FAK (pY397) in control and MUC1(ΔCT) expressing MECs plated on compliant fibronectin-conjugated hydrogels (E = 140 Pa; scale bar, 3 µm; ROI scale bar, 0.5 µm). d, Western blots showing phosphorylation of paxillin (pY118) in control and MUC1-expressing MECs on compliant substrates (E = 140 Pa) following overnight serum starvation and stimulation with EGF. MUC1-expressing cells treated with a pharmacological inhibitor of focal adhesion kinase (+FAKi) for 1 h before EGF stimulation did not exhibit robust paxillin phosphorylation.
Extended Data Figure 9. Cell proliferation on soft ECM.
a, Fluorescence micrographs showing DAPI-stained nuclei of control and MUC1(ΔCT)-expressing MECs after 24 h of plating on soft, fibronectin-conjugated hydrogels (E = 140 Pa; scale bar, 250 µm). The majority of cells plated as single cells, indicating that multi-cell colonies that formed at later time points were largely attributed to cell proliferation. b, Quantification of cell proliferation of MUC1(ΔCT)-expressing epithelial cells on soft hydrogels conjugated with bovine serum albumin (BSA) or fibronectin (Fn). Cells plated similarly on BSA– and Fn–hydrogels, but cell proliferation was significantly enhanced on Fn–hydrogels. Results are the mean ± s.e.m with statistical significance given by *P < 0.05; **P < 0.01; ***P < 0.001.
Extended Data Figure 10. Hyaluronic acid production by tumour cells promotes cellular growth.
a, Quantification of hyaluronic acid (HA) cell surface levels on control (10A-Cont.), transformed (10A-v-Src, 10A-HRAS) and malignant (MCF7, T47D) mammary epithelial cells (MECs). b, Fluorescence micrographs of HA and immuno-labelled paxillin on the v-Src transformed MECs (scale bars, 3 µm). c, Quantification of the number of v-Src-transformed MECs per colony 48 h after plating on soft polyacrylamide gels (fibronectin-conjugated) and treated with vehicle (DMSO), hyaluronic acid synthesis inhibitor 4-methylumbelliferone (+4MU; 0.3 µM), or competitive inhibitor HA oligonucleotides (+Oligo; 12-mer average oligonucleotide size; 100 mg ml−1). Results are the mean ± s.e.m with statistical significance is given by *P < 0.05; **P < 0.01; ***P < 0.001.
Supplementary Material
Acknowledgements
We thank S. Gendler, J. Goedhart and M. McMahon for cDNAs, as indicated in the Methods section. We thank A. Walker for bioinformatics support, L. Hauranieh for assistance in CTC analysis, H. Aaron for assistance with FLIM, J. B. Sibarita and M. Lagardère for support in sptPALM analysis, B. Hoffman in design of the FRET sensor, and T. Wittmann in assistance with pbFRET measurements. Image acquisition was partly performed at the Nikon Imaging Center and Biological Imaging Development Center at UCSF and the Berkeley Molecular Imaging Center. This work was supported by the Kavli Institute and UCSF Program for Biomedical Breakthrough postdoctoral fellowships to M.J.P.; DoD NDSEG Fellowship to M.G.R.; NIH GM59907 to C.R.B.; NIH Pathway to Independence Award K99 EB013446-02 to K.G.; French Ministry of Research, CNRS, ANR grant Nanomotility, INSERM, Fondation ARC pour la Recherche sur le Cancer, France BioImaging ANR-10-INBS-04-01, and Conseil Régional Aquitaine to O.R. and G.G.; NIH AI082292-03A1 to D.A.H.; The Breast Cancer Research Foundation to M.J.M., H.S.R. and J.W.P.; NIH 2R01GM059907-13 to C.R.B. and V.M.W.; and BCRP DOD Era of Hope Scholar Expansion grant BC122990, and NIH NCI grants U54CA163155-01, U54CA143836-01, 1U01 ES019458-01, and CA138818-01A1 to V.M.W.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions V.M.W. and M.J.P. conceived the project. V.M.W., M.J.P. and C.R.B. provided project management. M.J.P., A.C.W., M.W.D. and J.N.L. designed and constructed expression constructs. C.C.D. and M.J.P conducted single cell force spectroscopy measurements. M.J.P. and K.S.T. designed and implemented the interference microscope. M.J.P., L.C. and M.G.R. performed fluorescence, FRET and interferometric imaging and M.J.P. wrote the accompanying analysis software. O.R. and G.G. conducted sptPALM experiments and analysed the results. V.M.W., R.B. and M.J.P. designed the bioinformatics pipeline and R.B. implemented the pipeline and performed the large-scale gene expression analysis. J.K.M., A.C.W. and M.J.P. fabricated and conducted experiments on compliant hydrogels. M.J.M., H.S.R. and J.W.P. isolated CTCs and measured CTC gene and protein expression. K.G., J.E.H. and C.R.B. designed, synthesized and characterized the glycoprotein mimetics. M.J.P and D.A.H. constructed and implemented the computational model. M.J.P. and V.M.W wrote the manuscript with input from all authors.
The authors declare no competing financial interests.
References
- 1.Lanctot PM, Gage FH, Varki AP. The glycans of stem cells. Curr. Opin. Chem. Biol. 2007;11:373–380. doi: 10.1016/j.cbpa.2007.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haltiwanger RS, Lowe JB. Role of glycosylation in development. Annu. Rev. Biochem. 2004;73:491–537. doi: 10.1146/annurev.biochem.73.011303.074043. [DOI] [PubMed] [Google Scholar]
- 3.Varki A, Kannagi R, Toole BP. In: Essentials Glycobiol. Varki A, et al., editors. Cold Spring Harbor Laboratory Press; 2009. http://www.ncbi.nlm.nih.gov/books/NBK1963/ [Google Scholar]
- 4.Paszek MJ, Boettiger D, Weaver VM, Hammer DA. Integrin clustering is driven by mechanical resistance from the glycocalyx and the substrate. PLOS Comput. Biol. 2009;5:e1000604. doi: 10.1371/journal.pcbi.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker JL, Fournier AK, Assoian RK. Regulation of growth factor signaling and cell cycle progression by cell adhesion and adhesion-dependent changes in cellular tension. Cytokine Growth Factor Rev. 2005;16:395–405. doi: 10.1016/j.cytogfr.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nature Rev. Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shibue T, Weinberg RA. Integrin β1-focal adhesion kinase signaling directs the proliferation of metastatic cancer cells disseminated in the lungs. Proc. Natl Acad. Sci. USA. 2009;106:10290–10295. doi: 10.1073/pnas.0904227106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Godula K, et al. Control of the molecular orientation of membrane-anchored biomimetic glycopolymers. J. Am. Chem. Soc. 2009;131:10263–10268. doi: 10.1021/ja903114g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Godula K, Rabuka D, Nam KT, Bertozzi CR. Synthesis and microcontact printing of dual end-functionalized mucin-like glycopolymers for microarray applications. Angew. Chem. Int. Edn Engl. 2009;48:4973–4976. doi: 10.1002/anie.200805756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eng ET, Smagghe BJ, Walz T, Springer TA. Intact αIIbβ3 integrin is extended after activation as measured by solution X-ray scattering and electron microscopy. J. Biol. Chem. 2011;286:35218–35226. doi: 10.1074/jbc.M111.275107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paszek MJ, et al. Scanning angle interference microscopy reveals cell dynamics at the nanoscale. Nature Methods. 2012;9:825–827. doi: 10.1038/nmeth.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hattrup CL, Gendler SJ. Structure and function of the cell surface (tethered) mucins. Annu. Rev. Physiol. 2008;70:431–457. doi: 10.1146/annurev.physiol.70.113006.100659. [DOI] [PubMed] [Google Scholar]
- 13.Kufe DW. Mucins in cancer: function, prognosis and therapy. Nature Rev. Cancer. 2009;9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manley S, et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nature Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 15.Rossier O, et al. Integrins β1 and β3 exhibit distinct dynamic nanoscale organizations inside focal adhesions. Nature Cell Biol. 2012;14:1057–1067. doi: 10.1038/ncb2588. [DOI] [PubMed] [Google Scholar]
- 16.Chen W, Lou J, Evans EA, Zhu C. Observing force-regulated conformational changes and ligand dissociation from a single integrin on cells. J. Cell Biol. 2012;199:497–512. doi: 10.1083/jcb.201201091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedland JC, Lee MH, Boettiger D. Mechanically activated integrin switch controls alpha5beta1 function. Science. 2009;323:642–644. doi: 10.1126/science.1168441. [DOI] [PubMed] [Google Scholar]
- 18.Grashoff C, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen DX, Bos PD, Massagué J. Metastasis: from dissemination to organ-specific colonization. Nature Rev. Cancer. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 20.Toole BP. Hyaluronan: from extracellular glue to pericellular cue. Nature Rev. Cancer. 2004;4:528–539. doi: 10.1038/nrc1391. [DOI] [PubMed] [Google Scholar]
- 21.Bono P, Rubin K, Higgins JM, Hynes RO. Layilin, a novel integral membrane protein, is a hyaluronan receptor. Mol. Biol. Cell. 2001;12:891–900. doi: 10.1091/mbc.12.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dustin ML. Signaling at neuro/immune synapses. J. Clin. Invest. 2012;122:1149–1155. doi: 10.1172/JCI58705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coombs D, Dembo M, Wofsy C, Goldstein B. Equilibrium thermodynamics of cell-cell adhesion mediated by multiple ligand-receptor pairs. Biophys. J. 2004;86:1408–1423. doi: 10.1016/S0006-3495(04)74211-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salaita K, et al. Restriction of receptor movement alters cellular response: physical force sensing by EphA2. Science. 2010;327:1380–1385. doi: 10.1126/science.1181729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groves JT. Bending mechanics and molecular organization in biological membranes. Annu. Rev. Phys. Chem. 2007;58:697–717. doi: 10.1146/annurev.physchem.56.092503.141216. [DOI] [PubMed] [Google Scholar]
- 26.Lundmark R, Carlsson SR. Driving membrane curvature in clathrin-dependent and clathrin-independent endocytosis. Semin. Cell Dev. Biol. 2010;21:363–370. doi: 10.1016/j.semcdb.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 27.Comoletti D, et al. Synaptic arrangement of the neuroligin/β-neurexin complex revealed by X-ray and neutron scattering. Structure. 2007;15:693–705. doi: 10.1016/j.str.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hildebrandt H, Mühlenhoff M, Weinhold B, Gerardy-Schahn R. Dissecting polysialic acid and NCAM functions in brain development. J. Neurochem. 2007;103(Suppl. 1):56–64. doi: 10.1111/j.1471-4159.2007.04716.x. [DOI] [PubMed] [Google Scholar]
- 29.Levental KR, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paszek MJ, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 31.Lakins JN, Chin AR, Weaver VM. Exploring the link between human embryonic stem cell organization and fate using tension-calibrated extracellular matrix functionalized polyacrylamide gels. Methods Mol. Biol. 2012;916:317–350. doi: 10.1007/978-1-61779-980-8_24. [DOI] [PubMed] [Google Scholar]
- 32.Bruns N, Pustelny K, Bergeron LM, Whitehead TA, Clark DS. Mechanical nanosensor based on FRET within a thermosome: damage-reporting polymeric materials. Angew. Chem. Int. Ed. 2009;48:5666–5669. doi: 10.1002/anie.200900554. [DOI] [PubMed] [Google Scholar]
- 33.Friedrichs J, Helenius J, Muller DJ. Quantifying cellular adhesion to extracellular matrix components by single-cell force spectroscopy. Nature Protocols. 2010;5:1353–1361. doi: 10.1038/nprot.2010.89. [DOI] [PubMed] [Google Scholar]
- 34.Pankov R, Momchilova A. Fluorescent labeling techniques for investigation of fibronectin fibrillogenesis (labeling fibronectin fibrillogenesis) Methods Mol. Biol. 2009;522:261–274. doi: 10.1007/978-1-59745-413-1_18. [DOI] [PubMed] [Google Scholar]
- 35.Magbanua MJM, et al. Genomic profiling of isolated circulating tumor cells from metastatic breast cancer patients. Cancer Res. 2013;73:30–40. doi: 10.1158/0008-5472.CAN-11-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.