Abstract
Advances in genomics have allowed unbiased genetic studies of human disease with unexpected insights into the molecular mechanisms of cellular immunity and autoimmunity1. We performed whole exome sequencing (WES) and targeted sequencing in patients with an apparent Mendelian syndrome of autoimmune disease characterized by high-titer autoantibodies, inflammatory arthritis and interstitial lung disease (ILD). In five families, we identified four unique deleterious variants in the Coatomer subunit alpha (COPA) gene all located within the same functional domain. We hypothesized that mutant COPA leads to a defect in intracellular transport mediated by coat protein complex I (COPI)2–4. We show that COPA variants impair binding of proteins targeted for retrograde Golgi to ER transport and demonstrate that expression of mutant COPA leads to ER stress and the upregulation of Th17 priming cytokines. Consistent with this pattern of cytokine expression, patients demonstrated a significant skewing of CD4+ T cells toward a T helper 17 (Th17) phenotype, an effector T cell population implicated in autoimmunity5,6. Our findings uncover an unexpected molecular link between a vesicular transport protein and a syndrome of autoimmunity manifested by lung and joint disease. These findings provide a unique opportunity to understand how alterations in cellular homeostasis caused by a defect in the intracellular trafficking pathway leads to the generation of human autoimmune disease.
Monogenic disorders, while rare, have proven powerful in elucidating several fundamental biological mechanisms underlying autoimmunity7,8. Growing evidence from genomic studies and human immunology has demonstrated that autoimmunity can arise from perturbations in several nonclassical pathways. Indeed, components of inflammatory and innate immune machinery once thought to participate predominantly in anti-bacterial and anti-viral immunity have been implicated in the pathogenesis of autoimmune disease7. Defects in the sophisticated intracellular trafficking mechanisms employed by cells of the innate and adaptive immune response9 may also be anticipated to cause autoimmunity, since disruptions in protein trafficking lead to ER stress and activation of the unfolded protein response (UPR), both of which have been implicated in the pathogenesis of human autoimmune disease10.
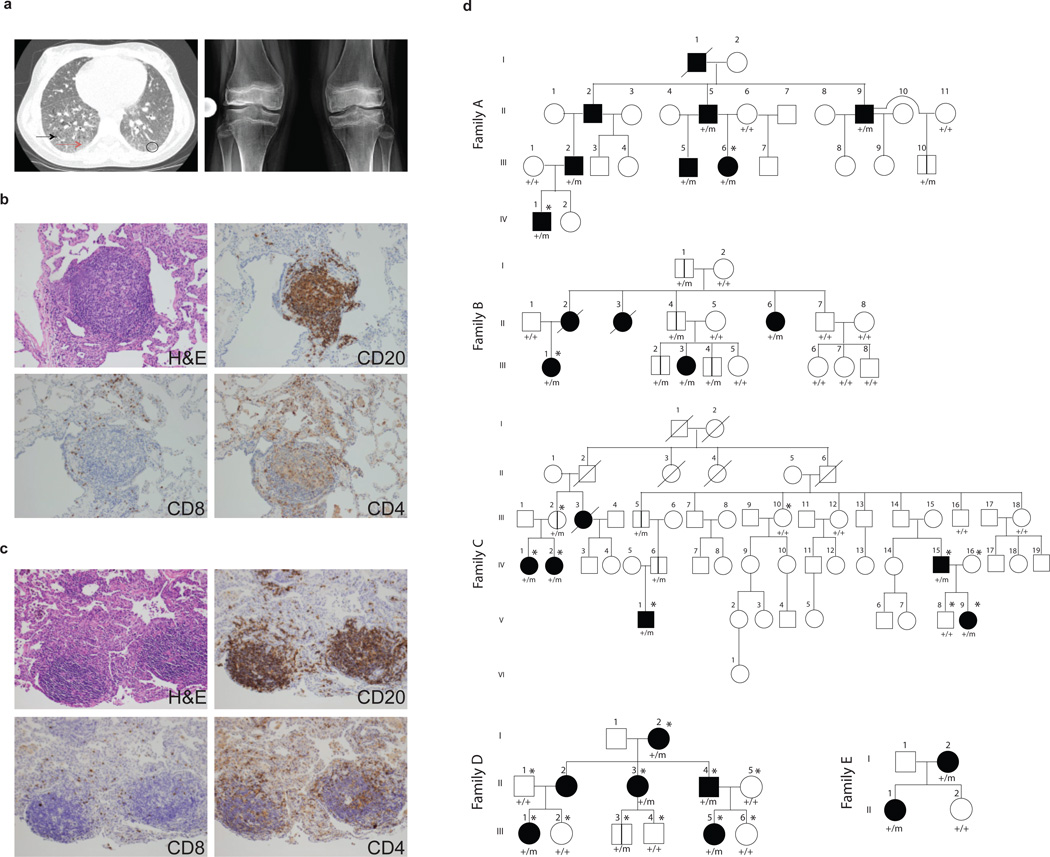
We identified five families with a novel Mendelian syndrome consisting of autoimmunity manifested by high-titer autoantibodies, ILD and inflammatory arthritis (Fig. 1a–d, Table 1, Supplementary Table 1). Among the five families there were twenty-seven affected patients, and we had access to clinical information and DNA for twenty-one. The average age of presentation was 3.5 years with a range of 6 months to 22 years. Sixteen of twenty-one subjects in the five families studied developed signs and symptoms of disease before age five. Several of the patients presented with acute pulmonary hemorrhage requiring high doses of immunosuppression. All of the patients have lung disease diagnosed as pulmonary hemorrhage, interstitial lung disease or both (Table 1, Supplementary Table 1). A comparison of lung biopsies from unrelated patients revealed significant interstitial infiltration by lymphocytes with germinal center formation (Fig. 1b–c), findings consistent with a pattern of ILD observed in systemic autoimmune syndromes11. Immunohistochemical staining of patient lung tissue revealed CD20+ B cells within the germinal centers and a significant number of CD4+ T cells infiltrating the lung interstitium (Fig. 1b–c). Autoantibodies were detected in 86% of affected patients, including antinuclear antibodies (ANA), anti-neutrophil cytoplasmic antibodies (ANCA) and rheumatoid factor (RF) (Table 1, Supplementary Table 2). Immunoglobulin levels, absolute lymphocyte counts and CD4 to CD8 cell ratios were largely normal (Supplementary Table 3). Ninety-five percent of patients have arthritis and some initially presented with joint pain. Four unrelated patients (A.III.6, B.III.1, D.III.5, E.I.2), all of who have circulating autoantibodies, underwent renal biopsies that demonstrate immune-mediated renal disease (Supplementary Table 1, Supplementary Fig. 1), providing further evidence of autoimmunity. All patients have required long-term immunosuppression to treat their disease.
Figure 1. Genetic segregation of ILD and arthritis with COPA mutations.
(a) On left, a chest CT scan of patient B.III.1 reveals basilar ground glass opacities (circle), reticulations (red arrow) and cystic formations (black arrow). Right panel is a bilateral knee radiograph from A.III.6 prior to restorative surgery. The radiograph shows generalized osteopenia with pathologic fracture of the distal femur. There is bilateral avascular necrosis of the metaphysis and diaphysis of the distal femur and proximal tibia with slackening of the condyles and articular cartilage destruction. (b,c) Lung biopsy sections from patients A.III.6, and B.III.1 stained with hematoxylin and eosin show cellular interstitial infiltrates and germinal center formation while immunohistochemical analysis demonstrates presence of CD20+ B cells and CD4+ and CD8+ T cells. (d) The pedigrees for Families A-E are shown with the presence of a wild type (+) or mutant (m) COPA allele. Family A (c.G698A) shows incomplete penetrance with one unaffected mutant carrier over four generations. Family B (c.A728G) shows incomplete penetrance of severe disease in females affected over three generations. Family C (c.G721A) shows incomplete penetrance of severe disease and segregation of the mutation within affected patients over six generations. For Family D (c.G690T) there is an unaffected mutation carrier who is 1 year old, which is below the age of onset. Family E (c.G698A) shows complete penetrance over two generations. Square=male; circle=female; black filled=affected with severe disease; unfilled=unaffected; /=deceased; *=submitted for WES; vertical lines=unaffected carrier. Roman numerals represent the generation within the family and Arabic numerals designate individuals within the generation.
Table 1.
Demographic and Clinical Characteristics of Patients*
| Patients No. (%) |
|
|---|---|
| Total | 21 |
| Age at presentation <5yrs | 16 (76%) |
| Sex | |
| Male | 8 (38%) |
| Female | 13 (62%) |
| Symptom at initial presentation | |
| Tachypnea, cough, hemoptysis | 14 (67%) |
| Joint pain | 5 (24%) |
| Arthritis | 20 (95%) |
| Pulmonary manifestations | |
| Hemorrhage or ILD | 21 (100%) |
| Autoantibodies | 18 (86%) |
| ANA | 14 (67%) |
| ANCA | 15 (71%) |
| RF | 9 (43%) |
| Response to immunosuppression | 21 (100%) |
Data shown are for patients with clinical records and DNA made available to investigators. The term ANA denotes anti-nuclear antibody, ANCA anti-neutrophil cytoplasmic antibodies, RF rheumatoid factor.
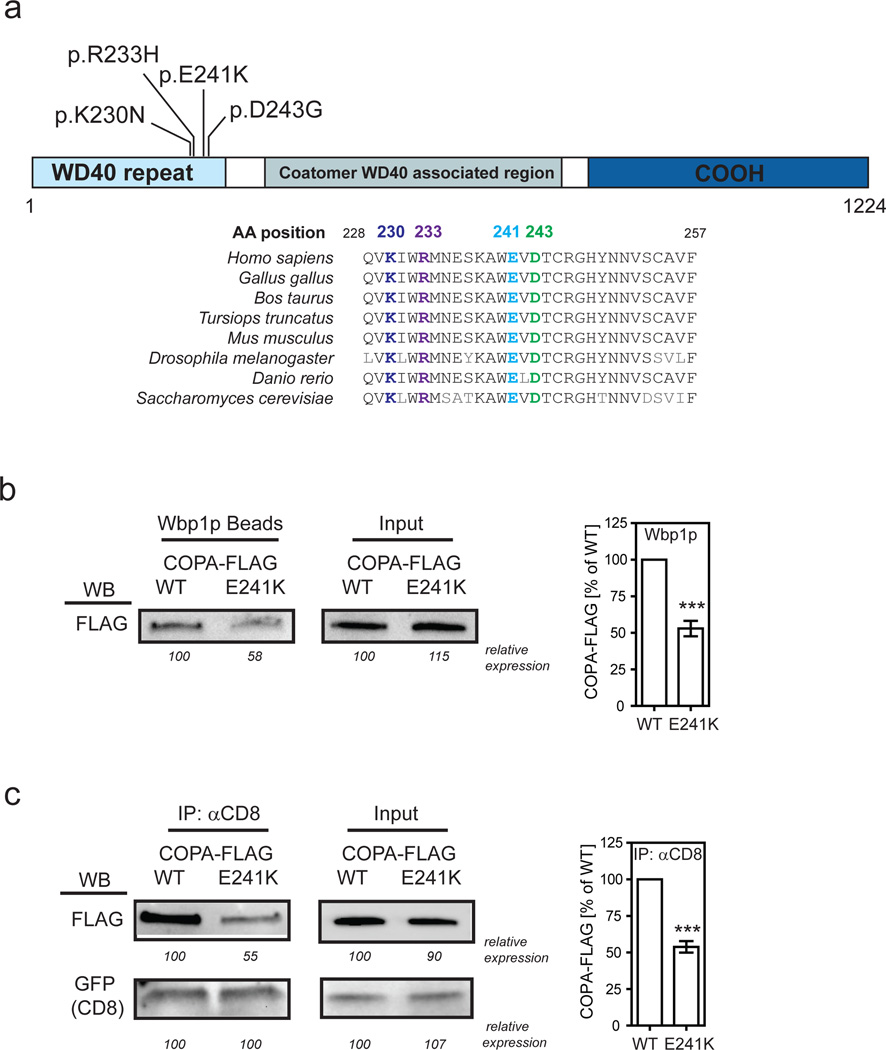
We enrolled patients for WES in order to identify potential genetic variants associated with disease. An examination of family pedigrees suggested autosomal dominant inheritance with incomplete penetrance (Fig. 1d). WES data were obtained for A.III.6, A.IV.1, and B.III.1 and analyzed for shared variants. There was only one gene observed, COPA, for which the two individuals in family A shared the c.G698A (p.R233H) (NM_004371.3) variant and individual III.1 from family B had c.A728G (p.D243G) (NM_004371.3). These variants have not been identified in various databases [NHLBI GO Exome Sequencing Project (ESP) server, 1000 Genomes, and two in-house generated databases that include WES data on ~4000 individuals from the Atherosclerosis Risk in Communities (ARIC) study and >2000 individuals who were sequenced as part of the Baylor College of Medicine/Johns Hopkins-Center for Mendelian Genomics (BH-CMG)]12. The variants co-segregated with the disease phenotype and were identified in all other affected family members (Supplementary Fig. 2). For family C we performed WES analysis and direct sequencing for 5 affected individuals (C.IV.1, C.IV.2, C.IV.15, C.V.1, C.V.9) and 4 unaffected pedigree members (C.III.2, C.III.10, C.IV.16, C.V.8) and identified the c.G721A (p.E241K) (NM_004371.3) COPA variant on chromosome 1 to be the only rare, non-synonymous variant that segregated with the disease phenotype (Table 2, Supplementary Fig. 2). In family D, WES of subjects D.I.2, D.II.1–5, D.III.1–6, identified another COPA variant c.G690T (p.K230N) (NM_004371.3) that co-segregated with the disease phenotype. In family E, given a clinical phenotype nearly identical to the other COPA patients, targeted Sanger sequencing of COPA exons 8 and 9 was performed to identify the c.G698A (p.R233H) (NM_004371.3) missense variant that was also discovered in family A (Table 2, Supplementary Fig. 2). Interestingly, the genotype analysis of family A and E revealed the presence of the same ancestral haplotype in COPA locus spanning approximately 140kb and 29 SNPs. The presence of a common haplotype provides a potential explanation for the co-occurrence of COPA p.R233H variant in these two apparently unrelated families (Supplementary Table 4). Overall, we employed WES and targeted sequencing at two institutions to independently identify four unique variants in five families. Notably, all variants identified are predicted to be deleterious and were located within a 14 amino acid region in the highly conserved, functionally important WD40 domain of COPA3 (Table 2, Fig. 2a).
Table 2.
Bioinformatic evaluation of segregating missense variants within COPA
| FAMILY ID | A & E | B | C | D |
|---|---|---|---|---|
| hg19 physical position | 160,293,229 | 160,283,894 | 160,283,901 | 160,293,237 |
| phyloP (100wayall) | 7.12 | 7.53 | 7.26 | 2.81 |
| phastCons | 1 | 1 | 1 | 1 |
| GERP | 4.73 | 5.62 | 5.62 | 2.87 |
| cDNA change | c.698G>A | c.728A>G | c.721G>A | c.690G>T |
| Amino acid change | p.Arg233His | p.Asp243Gly | p.Glu241Lys | p.Lys230Asn |
| PolyPhen-2 HVAR | Probably damaging | Probably damaging | Probably damaging | Probably damaging |
| SIFT | Damaging | Damaging | Damaging | Damaging |
| LRT | Damaging | Damaging | Damaging | Damaging |
| MutationAssessor | Low | Neutral | Low | Medium |
| MutationTaster | Disease-causing | Disease-causing | Disease-causing | Disease-causing |
| CADD (scaled) | 32 | 26.7 | 35 | 19.81 |
PhyloP, phastCons and GERP scores for nucleotide conservation were derived from the UCSC Genome Browser. PolyPhen-2, SIFT, LRT, MutationAssessor and MutationTaster provide functional prediction for each missense variant. For Combined Annotation Dependent Depletion (CADD), scaled C-scores ≥10 puts a variant within the top 10% of likely deleterious variants, while a score of ≥20 denotes the top 1% across the genome.
Figure 2. Binding of mutant COPA to dilysine motifs is impaired.
(a) Schematic of the COPA protein and its domains is shown. The amino acid position of the mutations identified and the conserved sequence homology at those sites in distantly related eukaryotic organisms are indicated. (b) Binding of mutant COPA to a dilysine motif was examined by taking either FLAG-tagged wild-type or mutant E241K COPA and incubating it with Wbp1p peptide-coupled agarose beads. The protein-bead complexes were analyzed by western blot (WB) using an anti-FLAG antibody. Numbers indicate the relative expression in percent of wild-type COPA. Statistical analysis of 6 independent experiments is shown. ***P<0.001 (c) Binding of mutant COPA to a dilysine motif was also assessed within T-Rex-293 cells stably expressing FLAG-tagged wild-type or mutant COPA by transiently transfecting cells with an expression construct for GFP-CD8-KKTN. Cell lysates were precipitated with anti-CD8 and immunoprecipitates were assayed for FLAG-COPA expression and GFP-CD8 enrichment. Numbers indicate the relative expression in percent of wild-type COPA or GFP-CD8 expressed in WT COPA cells. Statistical analysis of 5 independent experiments is shown. ***P<0.001.
To provide statistical evidence that variants in COPA are involved in disease etiology we performed an Affected-only parametric linkage analysis on the four families that are informative, analyzing COPA missense variants in each of the families. Although the pedigrees indicate an autosomal dominant mode of inheritance with reduced penetrance (21 of 30 COPA carriers were affected by disease while 9 were unaffected), no accurate estimate of the penetrance for this rare phenotype exists. Therefore an Affected-only parametric linkage analysis was performed where all unaffected individuals’ phenotypes were made unknown. Two-point LOD scores were calculated using SUPERLINK13 assuming an autosomal dominant mode of inheritance. The disease allele frequency and variant allele frequency were both assumed to be 0.001. The four informative pedigrees provided the following LOD scores: Family A: 1.5; Family B: 0.90; Family C: 2.7; Family D: 0.60 at theta=0 with the identified COPA variants. The combined statistically significant LOD score of 5.7 for the families provided statistical evidence that COPA is involved in disease etiology. Given that we observe four different variants in five families of the COPA gene, the rare variants co-segregate with the disease and the variation at this locus is linked to the autoimmunity phenotype, we conclude that it is highly likely that pathological variants in COPA are the underlying cause of disease etiology.
COPA encodes the COPA subunit of the seven member coat protein complex I (COPI). COPI and COPII are key components of the vesicular trafficking machinery in eukaryotic cells that serve as integral carrier complexes required for mediating bidirectional membrane traffic between the endoplasmic reticulum (ER) and the Golgi. COPII functions to facilitate the anterograde movement of newly created proteins or lipids from the ER toward the cell surface, while COPI is engaged in the retrograde movement of proteins being retrieved from the Golgi and returned to the ER14. To investigate for potential mechanisms by which the COPA variants lead to disease, we next asked whether patients exhibited a reduction in the amount of COPA protein produced. We assessed the expression level of the COPA transcript and protein by performing quantitative PCR and immunoblots. Using RNA and protein lysates from patient cells, we found no difference between COPA expression in patients versus controls (Supplementary Fig. 3a–c). Imaging analysis of COPA in patient cells demonstrated that the protein appeared to be distributed normally throughout the cell4, indicating that the variant did not cause aggregation or mislocalization of COPA to impair its function (Supplementary Fig. 3d).
Given that COPA was expressed at normal levels in patients, we sought to determine a functional consequence of the COPA variants. Importantly, point mutations in the COPA WD40 domain in yeast, analogous to the disease-associated damaging alleles we observed in patients, impair binding of dilysine-tagged proteins and cause a defect in retrograde transport2,3,15. Thus, we sought to directly test whether binding of dilysine proteins to the COPA mutants identified was similarly impaired. We generated a synthetic peptide for the cytoplasmic tail of yeast Wbp1 because this bears a dilysine retrieval signal that binds to mammalian COPA2,16. We conjugated the Wbp1 peptide to agarose beads and incubated them with recombinant mutant COPA containing the E241K amino acid substitution. We found that E241K mutant COPA does indeed have impaired binding to Wbp1 in comparison to wild-type COPA protein (Fig. 2b). To further experimentally verify this observation, we transfected cell lines stably expressing the E241K mutant COPA with a reporter protein bearing a dilysine retrieval signal. After performing an immunoprecipitation assay for the reporter, we also found impaired binding of the dilysine reporter to E241K mutant COPA (Fig. 2c). Comparable results were obtained by testing K230N mutant COPA (Supplementary Fig. 4). Taken together, these results demonstrate that mutant COPA bearing a WD40 variant identified in patients behaves in an analogous anomalous way to point mutations in yeast previously shown to cause a defect both in binding of dilysine-tagged proteins and retrograde transport2,3,15.
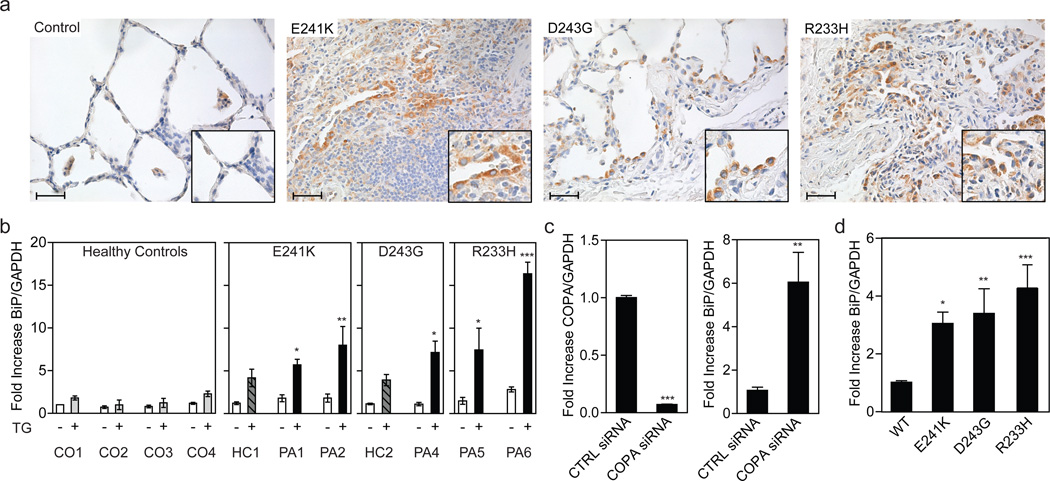
Having demonstrated a functional defect in mutant COPA, we next sought to identify physiologic effects of mutant COPA that might be linked mechanistically to autoimmune disease. Defects in the COPI pathway, including those caused by WD40 variants in COPA2,3,15, have been shown to affect not only retrograde trafficking but also COPII-mediated anterograde trafficking from the ER to Golgi4. Thus, we hypothesized that the COPA variants in patients likewise impair intracellular protein trafficking and as a result lead to ER stress and activation of the unfolded protein response (UPR)4,17. Importantly, ER stress and activation of the UPR have been linked to both autoimmunity and lung disease10,18,19. To determine whether patients exhibited evidence for increased ER stress, we examined lung biopsies for the molecular chaperone binding immunoglobulin protein (BiP), a well-established indicator of ER stress20. We found that BiP was markedly increased in the lung epithelium and alveolar macrophages of patients compared to controls (Fig. 3a and Supplementary Fig. 5a–b), consistent with our hypothesis.
Figure 3. Mutant COPA leads to an increase in ER stress.
(a) Immunohistochemistry stain for the ER stress component BiP in lung biopsy samples from patients with E241K, D243G, or R233H mutant COPA is shown. Biopsies from individuals not carrying a COPA mutation were used as a control. Hematoxylin was used for nuclear counterstain. Scale bars indicate 40µm. Enlarged inserts emphasize BiP staining in lung epithelial cells. (b) Quantitative PCR (qPCR) analysis of BiP expression in untreated (white bars; −) B lymphoblastoid cell-lines (BLCL) and treated (striped, gray and black bars; +) BLCL after stimulation with thapsigargin. Left graph indicates the results from 4 controls (CO1 (N=12), CO2 (N=3), CO3 (N=4), CO4 (N=9); CO3 is a related control of Family D, D.III.6, and CO4 of Family C, C.III.10). All results were normalized to values of untreated control 1. Data shown are from a healthy carrier (HC1, C.III.2, N=9) and 2 patients (PA1-2, C.IV.15 (N=12), C.V.1 (N=8)) of the E241K COPA family C, a healthy carrier (HC2, B.III.4, N=5) and one patient (PA4, B.III.3, N=5) of the D243G COPA family B, and 2 patients (PA 5–6, A.II.5 (N=5), A.III.6 (N=4)) of the R233H family A. One way ANOVA followed by Dunnett's Multiple Comparison Test was used to compare thapsigargin-treated experimental groups with thapsigargin-treated control 1 as a reference. Mean and SEM of pooled data from 2–4 independent experiments are shown. *P<0.05, **P<0.01, ***P<0.001. (c) COPA knockdown in HEK 293 cells. Left graph demonstrates greater than 90% reduction in COPA mRNA expression levels after treatment with COPA siRNA compared to cells treated with control siRNA. Right graph shows the induction of ER stress measured by BiP expression after COPA knockdown. Mean and SEM of pooled data from 3 independent experiments are shown (N=11). Statistical comparisons were made using Student’s unpaired t test with Welch's correction. **P<0.01, ***P<0.001. (d) HEK 293 cells were transfected with wild-type COPA (N=17) or mutant E241K (N=14), D243G (N=12), or R233H (N=12) COPA expression constructs. The graph shows BiP expression 48h after transfection as assessed by qPCR. Cells transfected with mutant COPA showed significantly increased BiP levels when normalized to cells transfected with wild-type COPA. Mean and SEM are shown for each column. One-way ANOVA followed by Dunnett's Multiple Comparison Test was use to compare experimental groups against wild-type transfected HEK293 cells as the control column. Results shown are pooled data from 3 independent experiments. *P<0.05, **P<0.01, ***P<0.001. CO denotes control, HC healthy carrier, PA patient, TG thapsigargin, WT wild-type.
We next performed dynamic functional assessments of patient cells for further evidence of enhanced ER stress caused by mutant COPA. B lymphoblastoid cell lines (BLCL) from patients were generated and treated with thapsigargin. Cells from patients with COPA variants had significantly elevated levels of BiP (Fig. 3b, Supplementary Fig. 5c) in comparison to related and unrelated control cells. Interestingly, the clinically asymptomatic carriers of COPA variants (i.e. apparent nonpenetrant individuals) also demonstrated an intermediate increase in ER stress compared to levels seen in cells from those severely affected (Fig. 5c), suggesting that the overall effect of the mutation on healthy carriers may be below a threshold required for the development of clinical disease.
To establish the specific role of mutant COPA in the induction of ER stress we performed siRNA knock down experiments in Human Embryonic Kidney (HEK) cells. Indeed, we found that reducing COPA expression resulted in higher levels of ER stress as indicated by a marked increase in BiP expression (Fig. 3c). To demonstrate that the increased ER stress observed in patient cell lines was specifically due to mutant COPA, we overexpressed wild-type or mutant COPA in HEK cells (Supplementary Fig. 5e–f). Consistent with a dominant-negative effect of the mutations, presence of any of the four patient-derived COPA mutations resulted in elevated BiP compared to cells transfected with wild-type COPA (Fig. 3d, Supplementary Fig. 5d). Levels of additional ER stress markers such as ATF4 and CHOP were also significantly increased when COPA expression was reduced or mutant COPA was present (Supplementary Fig. 5g–h). Given the role of autophagy in alleviating ER stress21 and in regulating immune responses22,23, we sought to evaluate alterations in this pathway. Interestingly, fluorescent imaging of cells revealed an increase in autophagosome and endo-lysosome size (Supplementary Figs. 6a–f and 7a–d) and patient BLCLs demonstrated a decrease in both the autophagy substrate p62 and torin-induced p62 catabolism (Supplementary Fig. 8a–b) This constellation of findings suggested a baseline defect in autophagic function that may be due to impaired early endosomal function in patients.24 Thus, it is possible that impaired autophagy further exacerbates the increase in ER stress caused by mutant COPA25. In aggregate, our findings demonstrate that the expression of mutant forms of COPA identified in patients is sufficient for the induction of ER stress.
Finally, to further investigate potential immunological links between mutant COPA and the clinical phenotype, we examined CD4+ T cells given their high numbers in diseased lungs (Fig. 1b–c) and the critical role these cells play in orchestrating autoimmune disease26. Interestingly, recent studies reveal a role for ER stress in the generation of CD4+ T helper cell subsets and demonstrate that ER stress results in cytokine secretion by antigen presenting cells (APCs) necessary for the induction and expansion of Th17 cells27–30, a CD4+ effector T cell population implicated in autoimmunity5,6. Thus, we hypothesized that mutant COPA might lead to ER stress and the generation of Th17 cells through a similar mechanism.
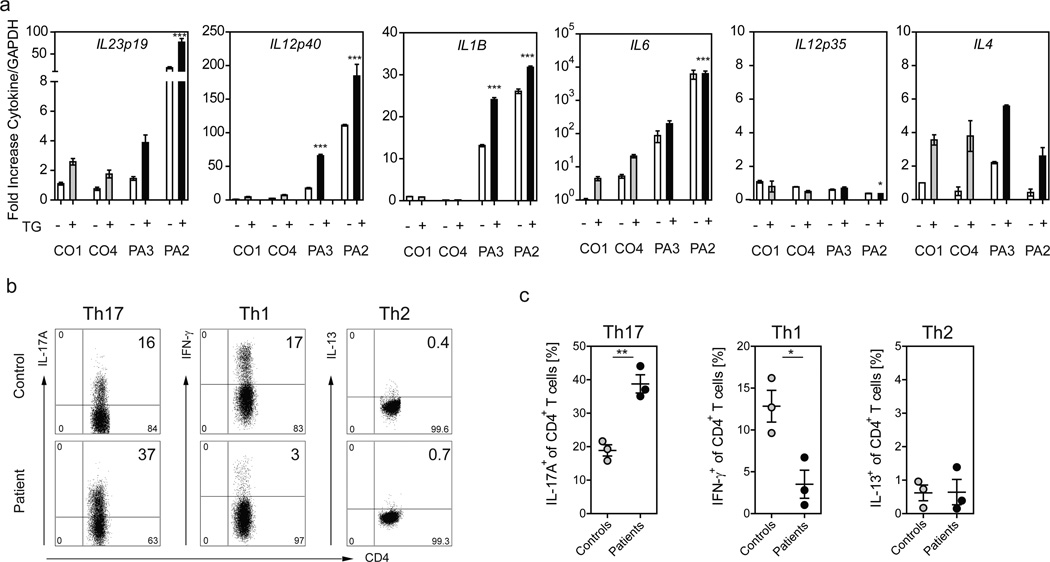
Given that B cells serve as professional antigen presenting cells, we tested our BLCLs for the expression of cytokines involved in the generation of the CD4+ helper T cell subsets Th1, Th2 and Th17. We discovered that patient BLCLs with mutant COPA showed a significant elevation of transcripts encoding for IL-1β, IL-6 and IL-23 (IL23p19 and IL12p40) (Fig. 4a), cytokines critical for the priming and expansion of Th17 cells31, and also found that this effect was further enhanced by an increase in ER stress. In contrast, transcripts for the Th2 priming cytokine IL-4 in patients were similar to controls while the Th1 priming cytokine IL-12 (IL12p35) was expressed at low levels in either group (Fig. 4a). Levels of IL-10 or TGFB were not different between patients and controls and IFNA2 levels in patients were increased and decreased in comparison to controls (Supplementary Fig. 9). To determine whether CD4+ T cells in patients demonstrated an immunological phenotype consistent with this pattern of cytokine expression, we profiled peripheral blood mononuclear cells. Intracellular cytokine staining analysis revealed equal frequencies of IL-13 secreting Th2 cells in patients and controls (Fig. 4b–c). By contrast, patient CD4+ T cells demonstrated a marked skewing away from IFN-γ secreting Th1 cells and a significant increase in the frequency of IL-17A secreting Th17 cells (Fig. 4b–c, Supplementary Fig. 10). Thus, mutant COPA leads both to higher levels of ER stress in patient BLCLs and the production of cytokines that promote the expansion of Th17 cells, an effector T cell population implicated in systemic autoimmunity5,6.
Figure 4. Patients with mutant COPA demonstrate an increase in Th17 priming cytokines in BLCLs and an increase in Th17 cells.
(a) Cytokine expression was measured in BLCLs from CO1, CO4 (C.III.10), PA2 (C.V.1) and PA3 (C.IV.2) either untreated (white bars; −) or treated (grey, black bars; +) with TG. Samples were analyzed via qPCR for mRNA levels of IL23p19, IL12p40, IL12p35, IL4, IL1β, and IL6. Shown are normalized mRNA levels against the average value for healthy control 1 (CO1) in the absence of stimulation. Errors bars indicate the standard error for triplicates in the assay. Results are representative of 3 independent experiments. Nothing indicated P>0.05; *P<0.05; ***P<0.001. (b,c) CD4+ T cells from three COPA patients with stable disease (C.IV.1 and C.IV.2 were on mycophenolate; C.IV.15 was on no treatment) and three age-matched controls were analyzed for intracellular cytokines IL-17A, IFN-, and IL-13 after stimulation with PMA and Ionomycin. (b) Representative FACS plots for IL-17A, IFN-, and IL-13-producing CD4+ T cells are shown. Plots are gated on living CD3+/CD4+ T cells. (c) Graphs demonstrating the frequency of Th1, Th2 and Th17 cells in patients and controls. The frequency of IL-17A-secreting Th17 cells is significantly increased in patients compared to controls (patients mean 39% vs. control mean 19% **P<0.01; N=3) whereas patients show a significant decrease in IFN--producing Th1 cells (patient mean 4% vs. control mean 13% *P<0.05; N=3). No significant difference was found in the frequency of IL-13-secreting Th2 cells. Student’s unpaired t test was used for statistical evaluation.
Through WES analysis and genetic studies in five unrelated families, we provide evidence that point mutations located in a 14 amino acid region of the WD40 domain of COPA cause a systemic autoimmune syndrome manifested by lung and joint disease. Functional analyses demonstrate that mutant COPA is unable to bind proteins with dilysine-based motifs and provide evidence of defective intracellular trafficking. Specifically, we show that expression of mutant COPA in cells induces higher levels of ER stress, which in patients may be further exacerbated by impaired autophagy25. Finally, we establish a putative immunological mechanism for COPA mutations in disease by showing that mutant COPA via ER stress leads to a cytokine milieu that promotes the generation of Th17 cells, a cell population shown to mediate systemic autoimmunity5,6.
At first glance it may seem unusual that a ubiquitously expressed protein that is part of an essential trafficking pathway leads to an autoimmune syndrome restricted primarily to the lung and joints. However, there are several examples of human disorders in which widely expressed proteins lead to distinct clinical phenotypes34,35. For instance, four Mendelian syndromes characterized by disparate clinical phenotypes have been described that involve mutations in the COPII trafficking machinery36. It remains unclear in these disorders and in patients with COPA mutations why only certain organ systems are affected. At this early stage we can only speculate that in patients with COPA mutations, there are tissue-specific cells central to disease pathogenesis that are particularly sensitive to alterations COPA function. In addition, given the reduced penetrance of disease, there may be gender differences or other genetic and environmental factors (e.g. viral respiratory infection) that dictate whether a mutant COPA carrier develops clinical disease.
Future studies may elucidate how defects in COPA lead to ER stress and also autoimmunity. ER stress and the UPR have been strongly associated with the pathogenesis of several inflammatory and autoimmune disorders, in part through the intersection of several key pathways that lead to activation of NF-κB, a central pro-inflammatory regulator, as well as through alterations in antigen presentation10,18. Furthermore, because the COPA mutations identified are predicted to impair retrograde transport, proteins destined for return to the ER may instead escape the cell or be presented in tissue sites to prime autoreactive immune responses37. Interestingly, both ER stress and Th17 cells have been previously implicated in patients with ILD19,38,39 and inflammatory arthritis10,40,41, including rheumatoid arthritis5. Given our data, it is intriguing to speculate that Th17 cells in COPA patients are pathogenic, though further investigation is required to determine whether they mediate disease or arise as a result of systemic inflammation. Additional studies may also focus on regulatory T cells and examine whether their function is impaired. In conclusion, the discovery of a molecular link between a defect in the COPI vesicular trafficking machinery to a novel syndrome of autoimmunity defines a new starting point for understanding the role of intracellular transport in the induction of human autoimmunity and as a potential therapeutic target in disease.
METHODS
Study Subjects
Subjects were selected based upon their unusual Mendelian phenotype and with their written consent, studied via protocols approved by the Research Ethics Board of The Hospital of Sick Children and the Institutional Review Boards for the protection of human subjects of Baylor College of Medicine or the University of California San Francisco.
Genomic DNA preparation
DNA was isolated from clotted whole blood by using the Clotspin Baskets and the Gentra Puregene Blood Kit (Qiagen) according to the manufacturer’s instructions.
Exome sequencing
Library Preparation
DNA samples were constructed into Illumina paired-end pre-capture libraries according to the manufacturer’s protocol (Illumina Multiplexing_SamplePrep_Guide_1005361_D) with modifications as described in the BCM-HGSC protocol (https://hgsc.bcm.edu/sites/default/files/documents/Illumina_Barcoded_Paired-End_Capture_Library_Preparation.pdf). Libraries were prepared using Beckman robotic workstations (Biomek NXp and FXp models). Briefly, 1 ug of DNA was sheared into fragments of approximately 300–400 base pairs with the Covaris E210 system followed end-repair, A-tailing, and ligation of the Illumina multiplexing PE adaptors. Pre-capture ligation-mediated PCR (LM-PCR) was performed for 6–8 cycles of amplification using the 2X SOLiD Library High Fidelity Amplification Mix (a custom product manufactured by Invitrogen). Purification was performed with Agencourt AMPure XP beads after enzymatic reactions, and following the final purification, quantification and size distribution of the pre-capture LM-PCR product was determined using the LabChip GX electrophoresis system (PerkinElmer).
Capture Enrichment
The pre-capture libraries were pooled as a 4-plex (approximately 500 ng/sample, 1 ug per pool) and hybridized in solution to the HGSC CORE design42 (52Mb, NimbleGen) according to the manufacturer’s protocol (NimbleGen SeqCap EZ Exome Library SR User’s Guide) with minor revisions. Human COT1 DNA and full-length Illumina adaptor-specific blocking oligonucleotides were added into the hybridization to block repetitive genomic sequences and the adaptor sequences, followed by post-capture LM-PCR amplification using the 2X SOLiD Library High Fidelity Amplification Mix with 14 cycles of amplification. After the final SPRI bead purification, quantity and size of the capture library was analyzed using the Agilent Bioanalyzer 2100 DNA Chip 7500. The efficiency of the capture was evaluated by performing a qPCR-based quality assay on the four standard NimbleGen internal controls. Successful enrichment of the capture libraries was estimated to range from a 6 to 9 of ΔCt value over the non-enriched samples.
Sequencing
Library templates were prepared for sequencing using Illumina’s cBot cluster generation system with TruSeq PE Cluster Generation Kits. Briefly, these libraries were denatured and diluted in hybridization buffer in order to achieve a load density of ~800K clusters/mm42. The library pool was loaded in a single lane of a HiSeq flow cell, with 2% phiX control library spiked into the lane for run quality control. The sample libraries then underwent bridge amplification to form clonal clusters, followed by hybridization with the sequencing primer. The sequencing run was performed in paired-end mode using the Illumina HiSeq 2000 platform. Using the TruSeq SBS Kits, sequencing-by-synthesis reactions were extended for 101 cycles from each end, with an additional 7 cycles for the index read. With the sequencing run yielding an average of ~8.4 Gb per sample, samples achieved an average of 89% of the targeted exome bases covered to a depth of 20X or greater.
Primary Data Analysis Families A and B
Initial sequence analysis was performed using the HGSC Mercury analysis pipeline43 (https://www.hgsc.bcm.edu/content/mercury). In summary, the primary analysis software on the instrument produces. bcl files that are transferred off-instrument into the HGSC analysis infrastructure by the HiSeq Real-time Analysis module. Once the run is complete and all .bcl files are transferred, Mercury runs the vendor’s primary analysis software (CASAVA), which demultiplexes pooled samples and generates sequence reads and base-call confidence values (qualities). The next step is the mapping of reads to the GRCh37 Human reference genome (http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/) using the Burrows-Wheeler aligner (BWA43, http://bio-bwa.sourceforge.net/) and producing a BAM44 (binary alignment/map) file. The third step involves quality recalibration (using GATK, http://www.broadinstitute.org/gatk/), and where necessary the merging of separate sequence-event BAMs into a single sample-level BAM. BAM sorting, duplicate read marking, and realignment to improve in/del discovery all occur at this step. Confirmation of sequencing results: The candidate disease associated mutations identified by WES were independently confirmed by Sanger sequencing, and analyzed for familial segregation.
Primary Data Analysis Family C - D
Nine samples in Family C (C.III.2, C.III.10, C.IV.1, C.IV.2, C.IV.15, C.IV.16, C.V.1, C.V.8, C.V.9) and eleven samples in family D (D.I.2, D.II.1, D.II.3–5, D.III.1–6) were sent to the UCSF Genomics Core for whole exome sequencing. The library preparation, capture enrichment, and sequencing were performed similar to Families A and B. Sequencing reads were aligned to the hg19 reference genome using the Burrows-Wheeler Alignment tool (BWA v0.5.9) using the default parameter that allows for two mismatches. Then indexing, realignment, and duplicate removal were performed using Picard (v1.56) and Samtools (0.1.18). Variants were called and recalibrated using the Genome Analysis Toolkit (GATK)44,45, version 1.4-25-g23e7f1b and hard-filtered with the recommendations listed in the Best Practice Variant Detection in GATK v3 with 'QD < 2.0', 'MQ < 40.0', 'FS > 60.0', 'HaplotypeScore 13.0', 'MQRankSum < 12.5', 'ReadPosRankSum < −8.0' for SNPs. The filtered list was then annotated with snpEff v2.0.5 in GRCh37.64 with information from public databases to determine the significance of novel sequence alterations, including the National Heart Lung Blood Institute Exome Sequencing Server, 1000 Genomes and the Database of Single Nucleotide Polymorphisms (dbsnp132). A filtering analysis of the exome variants in the 9 subjects in family C was performed in Varsifter46. The filtering strategy was designed to find variants shared among 6 carriers that were not seen in 3 related controls or in 20 other whole exome sequences performed on unrelated control subjects. We also excluded variants with a minor allele frequency (MAF) > 0.001. This filtering analysis gave us a single rare, variant in COPA (cG721A, NM_004371.3) that was shared among affected subjects and not present in any control. The filtering strategy in family D was performed similar to that employed for family C to identify (c.G690T, NM_004371.3) that co-segregated with the disease phenotype.
Plasmids and stable cell lines
The pCMV6-Entry vector encoding Myc-DDK-tagged, or untagged human COPA (Origene) were used as the parental vectors for mutant plasmid generation. Mutant plasmids were created via site directed mutagenesis using the QuikChange Lightning Kit (Agilent Technologies) according to manufacturer’s instructions or a third party service (Epoch Life Sciences). CD8α sequence spanning residues 1 through 207 was subcloned from human cDNA and into the pCMV6-Entry vector. mTagGFP was added to the N-terminus of the CD8α sequence, and a sequence encoding residues KYKSRRSFIDEKKTN including the di-lysine motif KKTN was added to the C-terminus. Cell lines stably expressing the wild-type or E241K mutant COPA-FLAG fusion protein in the presence of doxycycline were generated with the Flp-In T-REx system (Invitrogen). Full length sequences were amplified from patient cDNA and subcloned into pcDNA5/FRT/TO with a C-terminal FLAG tag. Complete vectors were then transfected into Flp-In T-REx 293 cells (Invitrogen) and selected per manufacturer’s protocol.
Cell transfection
For gene expression studies HEK293 cells were transiently transfected with 5µg of plasmid DNA by using the TransIT®-293 Transfection Reagent (Mirus) according to the manufacturer’s instructions. For COPA knockdown experiments HEK293 cells were transfected with either COPA targeting siRNA or control scrambled siRNA (Thermo Scientific Dharmacon) by using Lipofectamine RNAiMAX transfection reagent (Life Technologies) following the manufacturer’s instructions. 48 hours later cells were harvested and processed for RNA isolation.
For microscopic analysis HEK293T cells stably expressing GFP-LC3 were electroporated using the Amaxa 4D nucleofector system according to the manufacturer’s protocol. Briefly 293T cells were grown to 80% confluence. 1×106 cells were then nucleofected with 2µg plasmid DNA encoding COPA mutants. 24 hours later cells were harvested and imaged as described in the microscopy method section.
Generation and stimulation of B lymphoblastoid cell lines (BLCL)
Exponentially growing B95-8 cells were seeded at 1×106 cells/mL and incubated for 3d at 37°C, 5% CO2. B95-8 supernatant was harvested by centrifugation at 300×g and 4°C and passed through a 0.45µm filter. PBMCs were suspended at 1–5×106 cells/ml in RPMI-1640 media with 20% FBS followed by an addition of 0.5–1 volumes of B95-8 supernatant with or without 10µg of a human TLR9 ligand (ODN 2006, Class B CpG oligonucleotide, Invivogen). Cells were incubated overnight at 37°C in the presence of 5% CO2 and 0.5µg/ml Cyclosporine A, and an equal volume of media was added the next day. Cells were cultured until they formed macroscopically visible clusters and then maintained at concentrations of 0.5–1×106 cells/ml. For ER stress induction, EBV-transformed cells were seeded at 1×106 cells/ml and incubated with 100nM thapsigargin (Sigma) for six hours. Cells were harvested, washed in PBS and processed for RNA isolation as described below.
RNA isolation and quantitative PCR analysis
RNA was isolated with the RNeasy Mini kit (Qiagen). Superscript-III Reverse Transcriptase (Invitrogen) was used to synthesize cDNA. Quantitative PCR was performed with TaqMan Gene Expression Assays from Life Technologies (COPA: Hs00189232_m1; Bip: Hs00607129_gH; GAPDH: Hs02758991_g1; IL23p19: Hs00900828_g1; IL12p35: Hs01073447_m1; IL12p40: Hs01011519_m1; IL6: Hs00985639_m1; IL1B: Hs01555410_m1; IL4: Hs00174122_m1) or by using Qiagen SYBR green with the following primers: β-actin fwd CAGCCATGTACGTTGCTATCCAGG; β-actin rev AGGTCCAGACGCAGGATGGCATG; CHOP fwd CAGAACCAGCAGAGGTCACA; CHOP rev AGCTGTGCCACTTTCCTTTC; ATF4 fwd GGGACAGATTGGATGTTGGAGA; ATF4 rev ACCCAACAGGGCATCCAAGT.
Routine and Immunohistochemical Staining of Lung Biopsy Tissues
Lung biopsies from the patients were fixed in 10% buffered formalin and routinely processed for paraffin embedding. The resulting formalin-fixed, paraffin-embedded lung biopsy tissues were sectioned, resulting in glass slides with 5µm tissue sections that underwent routine hematoxylin- eosin (H&E) staining and immunohistochemical staining. Tissue section staining was performed by the UCSF and/or Texas Children’s Hospital Pathology Laboratories certified by the College of American Pathologists (Northfield IL USA), employing procedures certified by the College of American Pathologists, with appropriate positive and negative controls, and using automated H&E (Leica Microsystems Inc., Buffalo IL USA), and immunohistochemical (Leica Bond III Automated Immunohistochemical and In Situ Hybridization Biosystem, Leica Microsystems Inc, Buffalo IL USA) staining systems. Epitope antigen retrieval was performed using a proprietary automated system employing citrate-based buffer and surfactant proprietary kits (Novocastra Bond Epitope Retrieval Solutions 1, Leica Microsystems Inc., Buffalo IL USA). Proprietary antibodies kits (Leica Microsystems Inc., Buffalo IL USA) for CD20, CD4 and CD8 were used with appropriate positive and negative control tissues. Immune infiltrates of organs were confirmed by an independent reading of the slides with a blinded observer.
For BiP staining, microwave antigen retrieval was performed on deparaffinized and rehydrated lung sections with sodium citrate (pH 5.5). After peroxidase block (3% H2O2) and blocking with 1% BSA and 3% goat serum, sections were stained with anti-BiP antibody (Abcam) over night. BiP staining was visualized with biotinylated goat anti-rabbit antibody (Jackson ImmunoResearch Laboratories) and Elite ABC reagent followed by DAB staining (Vector Laboratories). Slides were counter stained with Mayer’s Hematoxylin. Images were obtained using a standard histology microscope and captured with an AxioCam using AxioVision software (Carl Zeiss MicroImaging).
Binding assay and Immunoblot
Flp-In T-REx 293 cells described above were cultured for 2 days in the presence of doxycycline to induce wild-type or mutant COPA-FLAG overexpression. Cells were then lysed in 500 µL of lysis buffer (25mM Tris-HCl pH 7.4, 150mM NaCl, 1mM EDTA, 1% NP-40 and 5% glycerol). A synthetic peptide (Genscript) representing the cytoplasmic tail of yeast Wbp1 (CKKLETFKKTN) was covalently linked to SulfoLink resin (Pierce) per manufacturer’s protocol. Cell lysate containing 80 µg of protein was added to 20 µL of resin in 0.5 mL of binding buffer (200 mM NaCl, 20 mM Tris-HCl pH 7.4, 7.5 mM MgCl2, 5% glycerol, and 0.2% Triton X-100) and gently rotated at room temperature for 5 minutes. Resin was subsequently washed by inverting 8 times in 1 mL of binding buffer and lastly boiled in SDS-PAGE sample buffer to release bound COPA. For immunoprecipitation, Flp-In T-REx 293 cells cultured in the presence of doxycycline were transfected with the GFP-CD8-KKTN construct for 36h. Cells lysates were pre-cleared with sepharose beads and then incubated with anti-CD8 antibody covalently linked to sepharose beads. Protein-bead complexes were washed and boiled in SDS-PAGE sample buffer. Proteins were subjected to SDS-PAGE, transferred to PVDF, blocked, and probed with one of the following antibodies: a horseradish peroxidase(HRP)-conjugated monoclonal antibody against FLAG (Sigma), a mouse monoclonal anti-mGFP antibody (Origene), a rabbit anti-COPA antibody (Sigma), or a mouse anti-GAPDH antibody (Santa Cruz). Bands were detected by incubation with secondary HRP-coupled antibodies and SuperSignal West Femto (Pierce) chemiluminescence.
Flow Cytometry
PBMCs were suspended in FACS buffer (PBS, 2% FCS, 1% sodium azide) at 1×106 per well of 96 well culture plates and incubated for 4h in the presence of brefeldin A (Sigma) with or without PMA (70ng/ml) and ionomycin (700ng/ml). For evaluation of surface receptors, cells were stained with the viability Ghost Dye violet 510 (Tonbo) and monoclonal antibodies including anti-CD3 clone SKY7 (Biolegend) and anti-CD4 clone OKT4 (Biolegend). Cells were incubated with antibodies at room temperature for 20 min and then washed with FACS buffer. For intracellular staining cells were permeabilized with Cytofix/Cytoperm reagent (BD) and incubated for 30 min at room temperature after incubation with anti-IFN-γ clone 4S.B3 (eBioscience), anti-IL17A clone eBio64CAP17 (eBioscience) or anti-IL-13 clone PVM13-1 (eBioscience). Samples were acquired using a Fortessa flow cytometer (BD Bioscience) and analyzed using FlowJo software
Statistical Analysis
Statistical analyses were performed using Prism 6.0 (GraphPad Software). Statistical Comparison was made using either Mann-Whitney U-test or the Student’s t test as indicated in the figure legends. Welch’s t-test was applied when variances were unequal. Comparison between more than two experimental groups was made using one-way analysis of variance (ANOVA), followed by Dunnette’s post hoc test. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
Funding: NIH R01AI-067946 (JSO), The Jeffrey Modell Foundation (JSO), NIH K08HL095659 (AKS), The Foundation of the American Thoracic Society (AKS), The Pulmonary Fibrosis Foundation (AKS), The Nina Ireland Lung Disease Program (AKS), NIH (NHGRI/NHBLI) U54HG006542 (JRL), 2R01NS058529 (JRL), NIH K23NS078056 (WW), NIH AI053831 (LBW).
REFERENCES
- 1.Zenewicz LA, Abraham C, Flavell RA, Cho JH. Unraveling the genetics of autoimmunity. Cell. 2010;140:791–797. doi: 10.1016/j.cell.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Letourneur F, et al. Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell. 1994;79:1199–1207. doi: 10.1016/0092-8674(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 3.Eugster A, Frigerio G, Dale M, Duden R. COP I domains required for coatomer integrity, and novel interactions with ARF and ARF-GAP. EMBO J. 2000;19:3905–3917. doi: 10.1093/emboj/19.15.3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Popoff V, Adolf F, Brügger B, Wieland F. COPI budding within the Golgi stack. Cold Spring Harbor Perspectives in Biology. 2011;3:a005231. doi: 10.1101/cshperspect.a005231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leipe J, et al. Role of Th17 cells in human autoimmune arthritis. Arthritis Rheum. 2010;62:2876–2885. doi: 10.1002/art.27622. [DOI] [PubMed] [Google Scholar]
- 6.Miossec P, Korn T, Kuchroo VK. Interleukin-17 and type 17 helper T cells. N Engl J Med. 2009;361:888–898. doi: 10.1056/NEJMra0707449. [DOI] [PubMed] [Google Scholar]
- 7.Cheng MH, Anderson MS. Monogenic Autoimmunity. Annu Rev Immunol. 2012;30:393–427. doi: 10.1146/annurev-immunol-020711-074953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rieux-Laucat F, Casanova JL. Autoimmunity by haploinsufficiency. Science. 2014 doi: 10.1126/science.1260791. [DOI] [PubMed] [Google Scholar]
- 9.Garg S, et al. Lysosomal trafficking, antigen presentation, and microbial killing are controlled by the Arf-like GTPase Arl8b. Immunity. 2011;35:182–193. doi: 10.1016/j.immuni.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Todd DJ, Lee A-H, Glimcher LH. The endoplasmic reticulum stress response in immunity and autoimmunity. Nature Reviews Immunology. 2008;8:663–674. doi: 10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- 11.Strange C, Highland KB. Interstitial lung disease in the patient who has connective tissue disease. Clinics in Chest Medicine. 2004;25:549–559. vii. doi: 10.1016/j.ccm.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 12.Bamshad MJ, et al. The Centers for Mendelian Genomics: a new large-scale initiative to identify the genes underlying rare Mendelian conditions. Am. J. Med. Genet. A. 2012;158A:1523–1525. doi: 10.1002/ajmg.a.35470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fishelson M, Geiger D. Exact genetic linkage computations for general pedigrees. Bioinformatics. 2002;18(Suppl 1):S189–S198. doi: 10.1093/bioinformatics/18.suppl_1.s189. [DOI] [PubMed] [Google Scholar]
- 14.Brandizzi F, Barlowe C. Organization of the ER-Golgi interface for membrane traffic control. Nature Reviews Molecular Cell Biology. 2013;14:382–392. doi: 10.1038/nrm3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schröder-Köhne S, Letourneur F, Riezman H. Alpha-COP can discriminate between distinct, functional di-lysine signals in vitro and regulates access into retrograde transport. Journal of Cell Science. 1998;111(Pt 23):3459–3470. doi: 10.1242/jcs.111.23.3459. [DOI] [PubMed] [Google Scholar]
- 16.Goldberg J. Decoding of sorting signals by coatomer through a GTPase switch in the COPI coat complex. Cell. 2000;100:671–679. doi: 10.1016/s0092-8674(00)80703-5. [DOI] [PubMed] [Google Scholar]
- 17.Claerhout S, et al. Abortive autophagy induces endoplasmic reticulum stress and cell death in cancer cells. PLoS ONE. 2012;7:e39400. doi: 10.1371/journal.pone.0039400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasnain SZ, Lourie R, Das I, Chen AC-H, McGuckin MA. The interplay between endoplasmic reticulum stress and inflammation. Immunol. Cell Biol. 2012;90:260–270. doi: 10.1038/icb.2011.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanjore H, Blackwell TS, Lawson WE. Emerging evidence for endoplasmic reticulum stress in the pathogenesis of idiopathic pulmonary fibrosis. Am. J. Physiol. Lung Cell Mol. Physiol. 2012;302:L721–L729. doi: 10.1152/ajplung.00410.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samali A, Fitzgerald U, Deegan S, Gupta S. Methods for monitoring endoplasmic reticulum stress and the unfolded protein response. Int J Cell Biol. 2010;2010:830307. doi: 10.1155/2010/830307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogata M, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell. Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nature Reviews Immunology. 2013;13:722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abhisek Bhattacharya NTE. Autophagy and Autoimmunity Crosstalks. Front Immunol. 2013;4 doi: 10.3389/fimmu.2013.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Razi M, Chan EYW, Tooze SA. Early endosomes and endosomal coatomer are required for autophagy. The Journal of Cell Biology. 2009;185:305–321. doi: 10.1083/jcb.200810098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adolph TE, et al. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503:272–276. doi: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmer MT, Weaver CT. Autoimmunity: increasing suspects in the CD4+ T cell lineup. Nature Immunology. 2010;11:36–40. doi: 10.1038/ni.1802. [DOI] [PubMed] [Google Scholar]
- 27.Wheeler MC, et al. KDEL-retained antigen in B lymphocytes induces a proinflammatory response: a possible role for endoplasmic reticulum stress in adaptive T cell immunity. The Journal of Immunology. 2008;181:256–264. doi: 10.4049/jimmunol.181.1.256. [DOI] [PubMed] [Google Scholar]
- 28.Glatigny S, et al. Proinflammatory Th17 cells are expanded and induced by dendritic cells in spondylarthritis-prone HLA-B27-transgenic rats. Arthritis Rheum. 2012;64:110–120. doi: 10.1002/art.33321. [DOI] [PubMed] [Google Scholar]
- 29.DeLay ML, et al. HLA-B27 misfolding and the unfolded protein response augment interleukin-23 production and are associated with Th17 activation in transgenic rats. Arthritis Rheum. 2009;60:2633–2643. doi: 10.1002/art.24763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodall JC, et al. Endoplasmic reticulum stress-induced transcription factor, CHOP, is crucial for dendritic cell IL-23 expression. Proceedings of the national academy of Sciences. 2010;107:17698–17703. doi: 10.1073/pnas.1011736107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaffen SL, Jain R, Garg AV, Cua DJ. The IL-23–IL-17 immune axis: from mechanisms to therapeutic testing. Nature Reviews Immunology. 2014;14:585–600. doi: 10.1038/nri3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Amelsfort JMR, Jacobs KMG, Bijlsma JWJ, Lafeber FPJG, Taams LS. CD4(+)CD25(+) regulatory T cells in rheumatoid arthritis: differences in the presence, phenotype, and function between peripheral blood and synovial fluid. Arthritis Rheum. 2004;50:2775–2785. doi: 10.1002/art.20499. [DOI] [PubMed] [Google Scholar]
- 33.Brusko TM, Putnam AL, Bluestone JA. Human regulatory T cells: role in autoimmune disease and therapeutic opportunities. Immunological reviews. 2008;223:371–390. doi: 10.1111/j.1600-065X.2008.00637.x. [DOI] [PubMed] [Google Scholar]
- 34.Bolze A, et al. Ribosomal protein SA haploinsufficiency in humans with isolated congenital asplenia. Science. 2013;340:976–978. doi: 10.1126/science.1234864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gissen P, Maher ER. Cargos and genes: insights into vesicular transport from inherited human disease. J. Med. Genet. 2007;44:545–555. doi: 10.1136/jmg.2007.050294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russo R, Esposito MR, Iolascon A. Inherited hematological disorders due to defects in coat protein (COP)II complex. Am. J. Hematol. 2013;88:135–140. doi: 10.1002/ajh.23292. [DOI] [PubMed] [Google Scholar]
- 37.Weber CK, et al. Antibodies to the endoplasmic reticulum-resident chaperones calnexin, BiP and Grp94 in patients with rheumatoid arthritis and systemic lupus erythematosus. Rheumatology (Oxford) 2010;49:2255–2263. doi: 10.1093/rheumatology/keq272. [DOI] [PubMed] [Google Scholar]
- 38.Olewicz-Gawlik A, et al. Interleukin-17 and interleukin-23: importance in the pathogenesis of lung impairment in patients with systemic sclerosis. Int J Rheum Dis n/a–n/a. 2014 doi: 10.1111/1756-185X.12290. [DOI] [PubMed] [Google Scholar]
- 39.Weaver CT, Elson CO, Fouser LA, Kolls JK. The Th17 pathway and inflammatory diseases of the intestines, lungs, and skin. Annu Rev Pathol. 2013;8:477–512. doi: 10.1146/annurev-pathol-011110-130318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nistala K, et al. Interleukin-17-producing T cells are enriched in the joints of children with arthritis, but have a reciprocal relationship to regulatory T cell numbers. Arthritis Rheum. 2008;58:875–887. doi: 10.1002/art.23291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benham H, et al. Th17 and Th22 cells in psoriatic arthritis and psoriasis. Arthritis Research & Therapy. 2013;15:R136. doi: 10.1186/ar4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reid JG, et al. Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics. 2014;15:30. doi: 10.1186/1471-2105-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Depristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teer J. Variant Annotation and Viewing Exome Sequencing Data. 2011:1–23. in. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.