Background: Caveolin-1 (Cav1) requires the caveolin scaffolding domain for caveola formation.
Results: The Cav1 scaffolding domain and oligomerization domain are tightly juxtaposed to the membrane in caveolae.
Conclusion: Concerted membrane association of the oligomerization, scaffolding, and intramembrane domains are critical for caveola biogenesis and membrane deformation.
Significance: Understanding the membrane association of Cav1 is critical for dissecting how the protein regulates caveola formation and achieves regulation over cellular signaling.
Keywords: caveolae, caveolin, electron microscopy (EM), electron tomography, protein domain, biochemistry, membrane deformation
Abstract
The generation of caveolae involves insertion of the cholesterol-binding integral membrane protein caveolin-1 (Cav1) into the membrane, however, the precise molecular mechanisms are as yet unknown. We have speculated that insertion of the caveolin scaffolding domain (CSD), a conserved amphipathic region implicated in interactions with signaling proteins, is crucial for caveola formation. We now define the core membrane-juxtaposed region of Cav1 and show that the oligomerization domain and CSD are protected by tight association with the membrane in both mature mammalian caveolae and a model prokaryotic system for caveola biogenesis. Cryoelectron tomography reveals the core membrane-juxtaposed domain to be sufficient to maintain oligomerization as defined by polyhedral distortion of the caveolar membrane. Through mutagenesis we demonstrate the importance of the membrane association of the oligomerization domain/CSD for defined caveola biogenesis and furthermore, highlight the functional significance of the intramembrane domain and the CSD for defined caveolin-induced membrane deformation. Finally, we define the core structural domain of Cav1, constituting only 66 amino acids and of great potential to nanoengineering applications, which is required for caveolin-induced vesicle formation in a bacterial system. These results have significant implications for understanding the role of Cav1 in caveola formation and in regulating cellular signaling events.
Introduction
Caveolae are small (50–80 nm in diameter) bulb-shaped invaginations of the plasma membrane (PM)4 that function in a multitude of fundamental cellular processes, including the mechanosensation response, cholesterol homeostasis, and the regulation of cellular signaling (1–6). An essential structural protein of caveolae is Caveolin-1 (Cav1) (7), a 22-kDa cholesterol-binding membrane protein that has been hypothesized to adopt a hairpin-like structure in the membrane, with both the N and C termini facing the cellular cytoplasm (8).
The C terminus of Cav1 is composed of two separate domains, the intramembrane domain (IMD, amino acids 102–135) and the remaining residues (amino acids 136–178). It has been hypothesized that the C terminus functions in lipid association as truncations consisting of this region in Cav1 and Caveolin-3 (Cav3) were constitutively associated with intracellular lipid droplets (9), and the C terminus is triply palmitoylated during exportation from the Golgi. These post-translational modifications are not important for caveola formation but have been shown to exert local effects on the insertion of Cav3 within the membrane (10). Additionally, the C terminus has been implicated in Cav1 homo-oligomerization as this region can co-immunoprecipitate both the N and C termini of the protein (11, 12). However, expression of a protein lacking the final 31 amino acids of Cav1 (Cav1-(1–147)) resulted in normal caveolar formation when expressed in Cav1−/− mouse embryonic fibroblasts (13). The IMD is a highly hydrophobic domain that is critical for caveolar formation (13, 14). NMR spectroscopy and chemical shift indexing demonstrated residues 97–136 adopt a “horseshoe” structure within a micelle membrane (15); residues 97–107 comprising α-helix 1, separated from α-helix 2 (residues 111–129) by an unstructured region (amino acids 108–110), and flanked by a second unstructured region (residues 130–136). This horseshoe conformation has been postulated to form a “wedge” between the inner and outer leaflet of the membrane allowing for membrane deformation, and is potentially responsible for the characteristic caveolar morphology (15–17).
Structural analysis of the N terminus of Cav1 has remained elusive, with conflicting reports suggesting both α-helical (16, 18–21) and β-sheet (22, 23) organizations. The N terminus encompasses amino acids 1–101 and is commonly represented with three separable domains comprising the membrane-associated caveolin scaffolding domain (CSD, residues 81–101) and the oligomerization domain (OD, amino acids 61–101) that partially overlaps with the proposed N-terminal soluble domain (residues 1–80) (24). In cell systems, de novo expression of Cav1 truncations demonstrated the deletion of the first 48 amino acids of the N terminus does not affect caveolar morphology but further deletion of the next 11 amino acids abrogates caveola formation (13). The CSD is a critical domain for caveolin oligomerization and caveola formation (13) but has also been proposed to regulate cellular signaling events. The interaction between the scaffolding domain and a consensus sequence termed the caveolin binding motif (CBM, proposed to have the sequence ΩXΩXXXXΩ, ΩXXXXΩXXΩ, or ΩXΩXXXXΩXXΩ where Ω is an aromatic residue and X is any amino acid) has been hypothesized to regulate signaling via direct protein-protein-mediated sequestration of interacting proteins in an inactive state within caveolae (2, 25–27). However, a number of observations have questioned the likelihood of Cav1 regulating signaling in this manner. Most critically, many of the proposed CBMs appear unavailable for interaction with caveolin, often comprising the highly hydrophobic core structural backbone of the proposed binding protein (28, 29). Putative CBMs within proteins for which structural information is available do not adopt specific structures and no particular enrichment of CBMs has been observed in organisms with caveolae compared with those without (28, 29). Finally, the constitutive and irreversible association of signaling proteins with the caveolin CSD does not explain how reversible interactions with signaling proteins can occur. The availability of the CSD for interaction with CBMs has received less attention yet remains another crucial aspect of the caveolin signaling hypothesis.
Several studies have sought to ascribe both structural and biochemical descriptions to homo-oligomerized Cav1, observing different sized oligomers of 200, 400, and 600 kDa (30) up to 3.3 MDa (31) and different organizations, from filamentous and ring-shaped (19) to ovoid complexes (30). Recently, it has been demonstrated that the heterologous expression of Cav1 in Escherichia coli resulted in the generation of caveola-like structures (32). It was demonstrated that heterologous caveolae (h-caveolae) could be formed efficiently to high concentrations, easily purified, and did not suffer from many of the drawbacks of studying caveolin proteins in mammalian cell systems, mutational analysis regularly results in exportation problems of the Golgi complex (13, 33). h-Caveolae closely mimic the size of mammalian caveolae, and possess similar numbers of Cav1 proteins and mutations in Cav1 that do not give rise to caveolae in mammalian cells systems, similarly, did not form h-caveolae (32). The intrinsic propensity for Cav1 to generate vesicles in a heterologous system underlies the basic function of the protein in the general cellular context. Cav1, like many other proteins in the mammalian cell, is involved in sculpting cellular membranes although how this deformation is achieved remains poorly understood (34).
In the current study we utilize this heterologous expression system and purified mammalian caveolae to analyze the insertion of Cav1 in the membrane, map the exact amino acids tightly associated with the membrane, and analyze the effects of deletions on h-caveola formation and structural regulation. We define the minimal domain of Cav1 required for h-caveola formation in bacteria and show that, remarkably, uniform vesicle formation can occur upon the expression of less than 100 amino acids.
Experimental Procedures
Expression, Purification, and Cleavage of h-Caveolae
E. coli (Rosetta pLysS DE3) were grown at 37 °C to an A600 nm of 1 in Terrific Broth. Caveolin fusion protein constructs, under the T7 RNA polymerase promoter, were induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside (Astral Scientific) to express for either 3 h (for EM) or overnight at 30 °C (for purification). E. coli were lysed using a Cell Disruptor (Constant Systems) and h-caveolae were purified using affinity chromatography with amylose resin (New England Biolabs), eluted with maltose (100 mm) in phosphate-buffered saline (150 mm NaCl, 15 mm K2HPO4/KH2PO4, pH 7.4), and concentrated with 100,000 MWCO Amicon Ultracel Centifugal Filter (Millipore). The MBP-CAV1-His6 fusion protein was engineered with two tobacco etch virus protease cleavage sites between the maltose-binding protein (MBP) and CAV1. Purified h-caveolae were incubated with AcTEV (Invitrogen) at 4 °C overnight to remove the MBP affinity tag.
Coomassie Staining and Western Blot Analysis
Purified h-caveolae were boiled in Laemmli sample buffer (63 mm Tris-HCl, pH 6.8, 0.2 m DTT, 2.5% SDS, and 1% bromphenol blue) and proteins were separated using SDS-PAGE (15% acrylamide). Gel staining was performed with Coomassie stain (0.25% Coomassie Brilliant Blue R, 40% methanol, and 10% acetic acid in water). Western blots were blocked in 5% milk powder dissolved in Tris-buffered saline with Tween (TBST, pH 7.4, 10 mm Tris-HCl, 150 mm NaCl, and 0.1% (v/v) Tween 20) and incubated with various antibodies (MBP, New England Biolabs; polyclonal caveolin, BD Transduction Labs, VIPN (30) and Concav (45)) and visualized by chemiluminescence (Pierce).
Proteinase K Digestion
Purified h-caveolae were incubated with ProK (∼20 μg of ProK/mg of h-caveolae, New England Biolabs) at 37 °C for 30 min.
Cloning
Site-directed mutagenesis was performed to generate the ΔN, ΔC, and ΔNC truncation mutants and the S80E, F81E, and W85E point mutations. Primer sequences were as follows: ΔN-forward: GCTGATATCGGATCCATCGACCTGGTCAAC, ΔN-reverse: GTTGACCAGGTCGATGGATCCGATATCAGC; ΔC-forward, GTGCATCAGCCGTGTCATCGAAGGTCGTCATC, ΔC-reverse: GATGACGCACTTCGATGACACGGCTGATGCAC; S80E-forward: GAACCAGAA GGAACACACGAGTTTGATGGCATCTGGAAG, S80E-reverse: CTTCCAGATGCCATCAAACTCGTGTGTTCCTTCTGGTTC, F81E-forward: CAGAAGGAACACACAGTGAAGATGGCATCTGGAAGG, F81E-reverse: CCTTCCAGATGCCATCTTCACTGTGTGTTCCTTCTG, W85E-forward: CAGTTTTGATGGCATCGAAAAGGCCAGCTTCACCAC, W85E-reverse: GTGGTGAAGCTGGCCTTTTCGATGCCATCAAAACTG, His6 removal-forward: GCAGAAAGAAACAATCTAAGGTCGTCATCATC, His6 removal-reverse: GATGATGACGACCTTAGATTGTTTCTTTCTGC.
Polymerase chain reaction was performed using HF Phusion (New England Biolabs) DNA polymerase. The library of Cav1 truncations was designed in MacVector and sequences were synthesized as gBlocks by Integrated DNA Technologies (IDT) and inserted into a pNMTMA vector (as described previously in Ref. 32) by cohesive-end cloning as per the manufacturer's instructions.
Purification of Caveolae
Madin-Darby canine kidney (MDCK) cells stably expressing GFP or GFP-Cavin-1 were scraped at 4 °C in TNE buffer (50 mm Tris-HCl, pH 7.4, 100 mm NaCl, and 0.1 mm EDTA), syringe lysed, and spun at high speed (100,000 × g) in an ultracentifuge. The supernatant was discarded and the pellet was resuspended in TNE buffer and incubated with MBP-tagged GFP-trap. GFP/GFP-Cavin1·GFP-trap complex was bound to amylose resin (New England Biolabs) by the MBP tag. The MBP-bound protein complex was washed repeatedly with high salt PBS and GFP/GFP-Cavin-1 protein was eluted with maltose and concentrated in 100,000 MWCO Amicon Ultracel Centrifugal Filters (Millipore). ProK treatment was performed as described previously, however, 0.5% SDS for 30 min at 37 °C was utilized for detergent treatment as caveolae are resistant to disruption by Triton X-100.
Preparation of Liposomes
E. coli extracted lipids (Avanti) in chloroform (Sigma) were deposited on the sides of a round-bottom flask by evaporating the organic solvent under a nitrogen atmosphere. To ensure the complete removal of chloroform, a formed lipid layer was stored overnight under vacuum. Dried lipids were then hydrated with an aqueous buffer (20 mm HEPES-KOH, pH 7.5, 150 mm NaCl) at a concentration 1 mg/ml. To prepare 50-nm liposomes, a mini-extruder block (Avanti) was heated to 65 °C. Liposomes were extruded 21 times through 50-nm polycarbonate filters (Avanti) and plunge frozen immediately after preparation.
Mass Spectrometry
Protein bands were excised and destained in 50% acetonitrile, 25 mm NH4HCO3, reduced with 20 mm dithiothreitol (DTT), alkylated with 50 mm iodoacetamide, adjusted to pH 8 with 50 mm NH4HCO3, dehydrated with 100% acetonitrile, and dried with a SpeedVac. The dried gel pieces were rehydrated and incubated overnight with 0.2 μg of trypsin per gel band (in 10% acetonitrile, 50 mm NH4HCO3). The digest was terminated with 5% formic acid and peptides were extracted with a solution of 60% acetonitrile, 1% formic acid. The extracted peptides were dried and resuspended in 5% formic acid for mass spectrometric analysis.
Tryptic peptides were subjected to liquid chromatography-tandem mass spectrometry using an Agilent 6520 QTOF coupled with a Chip CUBE and 1200 HPLC. The nano pump flow was set at 0.3 μl/min and the capillary pump flow at 4 μl/min. The HPLC-chip contained a 160-nl C18 trapping column, and a 150-mm C18 resolving column (Agilent G4240–62010). Buffer A was 0.1% formic acid and buffer B was 0.1% formic acid in 90% acetonitrile. The samples were analyzed with a 50-min LC gradient, which increased from 5 to 50% Buffer B in 45 min, increased up to 95% Buffer B in 1 min, was held at 95% Buffer B for 3 min, and then was returned to 10% buffer B in 1 min. The mass spectrometer was programmed to acquire 8 precursor MS1 spectra and 4 MS/MS spectra per second. Dynamic exclusion was applied after 2 spectra and released within 0.25 min. LC-MS/MS raw data generated was processed with Agilent's Spectrum Mill. Data were extracted with carbamidomethylated cysteine as fixed modifications. The data were searched against the species Canis familiaris of the SwissProt database, with carbamidomethylated cysteine as fixed modification, a maximum missed cleavage of 2, precursor mass tolerance of ±20, and product mass tolerance of ±50. Results were filtered by protein score of >11.0, peptide score of >10, and % Scored Peak Intensity of >60.
Electron Microscopy
Negative staining and immunolabeling of purified h-caveolae, caveolae, and bacteria expressing different caveolin truncations were performed as previously described (32). Imaging was performed on a JEOL 1011 electron microscope at 80 kV fitted with a Morada 4K × 4K soft imaging camera (Olympus) with 2-fold binning.
Cryo-electron tomography (ET) was performed as described previously (32). Images were recorded using LC-1100 Direct Electron 4K × 4K camera (at a binning of 2) under the control of SerialEM (35). Tilt-series were reconstructed using 10-nm fiducial markers and weighted back-projection in IMOD (36). Cryo-tomograms were subjected to a mild de-noising, non-linear anisotropic diffusion filter, to improve the signal-to-noise ratio (37).
Electron tomography was performed on E. coli preserved by high-pressure freezing (EMPact2, Leica) and freeze substitution (AFS2, Leica) in 1% osmium tetroxide OsO4 and 0.1% UA then subjected to temperature ramping and cells were embedded in epon. Thick sections (300 nm) were cut on an ultramicrotome (UC6, Leica). Dual-axis tilt series were acquired on a TECNAI F30 (FEI) electron microscope encompassing a tilt range of −60 to +60° at 2° increments. Image acquisition was performed with a Direct Electron LC-1100 4K × 4K camera with 2-fold binning under the control of SerialEM (35). Reconstructions were performed using weighted back-projection in IMOD. Semi-automated segmentation was employed (38, 39) to generate a three-dimensional representation of h-caveolae. Morphometric analysis of h-caveola diameters was performed as follows; individual h-caveola were scanned through the “z” dimension to determine the central region and measured, outer-membrane to outer-membrane, such that the bilayer was spanned twice (excluding the protein coat). Two-tailed Student's t-test were performed to determine significance.
Results
Mapping the Membrane-protected Domains of Cav1
The C-terminal domain (residues 102–178) and the CSD (residues 81–101) of Cav1 have been shown to possess membrane-binding affinity (11, 15, 40, 41) and have been predicted by multiple different studies to adopt a predominantly α-helical structure (14–16, 22, 42, 43). Despite these observations, the exact residues tightly associated with the membrane have remained unclear. A summary of predicted and experimental structural data including membrane-associated domains is represented in Fig. 1. Using our model heterologous system and biochemical approaches coupled with enzymatic degradation of the soluble domains of Cav1, we investigated the exact residues tightly associated with the membrane. For all subsequent experiments, unless otherwise stated, MBP-Cav1-His6 h-caveolae (∼72 kDa, the standard caveogenic fusion protein characterized previously (32)) were purified using the MBP tag then treated with a tobacco etch virus protease to specifically cleave the MBP tag from the membrane-associated Cav1-His6 (∼22–24 kDa). Purified Cav1-His6 h-caveolae were treated with a nonspecific protease, proteinase K (ProK), to degrade accessible Cav1 epitopes.
FIGURE 1.

Alignments of observed and predicted structural organizations for full-length caveolin 1 and peptides of Cav1. Red = α-helix, light blue = β-sheet, and gray = predicted transmembrane domain.
After ProK treatment several small molecular mass bands, of ∼16–17 and 11–14 kDa, can be observed by Coomassie staining (Fig. 2A). To determine whether these remaining small molecular weight bands were Cav1 domains, liquid chromatography tandem mass spectrometry (LC MS/MS) was performed on the largest and most abundant fragment after ProK treatment. Control bands (undigested) showed a high abundance of tryptic peptides of Cav1 (Table 1) comprising amino acids 5 to 172, although the IMD (residues 102–136) was undetected. This highly hydrophobic domain does not possess tryptic cleavage sites and therefore is likely to result in a peptide too large for the detection by LC MS/MS analysis employed here. The undigested bands after ProK treatment were Cav1 peptides, but only constituted 41.6% of the protein (Table 1). Mapping of these domains revealed Cav1 lacked the immediate N-terminal domain but highly abundant peptides comprising a complete sequence from amino acids 48–102, as well as 135–148, were observed (Table 1). Interestingly, residues from the C terminus of Cav1, amino acids 165–172, were also highly abundant (Table 1), suggesting that this peptide may be involved in lipid interactions.
FIGURE 2.
Cav1 is protected from ProK digestion by tight association with the bilayer. A, Coomassie stain of purified MBP-Cav1-His6 h-caveolae ± ProK. Small molecular mass bands of approximately 16–17 and 11–14 kDa remained. B, Western blot analysis of Cav1-His6 (after tobacco etch virus cleavage) using the α-MBP, VIP-N, polyclonal and Concav α-caveolin antibodies. VIP-N recognizes amino acids 1–20, the polyclonal α-caveolin has multiple recognition domains between amino acids 1 and 97, and the Concav antibody binds residues 67–81. C, Coomassie stain of purified Cav1-His6 h-caveolae (tobacco etch virus cleaved) ± 0.5% Triton X-100 for 1 h prior to ProK digestion. The small molecular weight fragments of Cav1 are lost when h-caveolae are disrupted prior to ProK treatment. D, negative staining (1% uranyl acetate) of purified h-caveolae show ProK digestion does not disrupt h-caveola morphology, however, a complete loss of morphology is evident in the Triton X-100-treated conditions. Scale bar, 100 nm. E, refined model of Cav1 association with the membrane.
TABLE 1.
Summary of detected peptides after ProK treatment

To confirm our proteomic analysis, the small molecular weight bands were screened with a panel of antibodies directed against different domains of Cav1. An α-MBP antibody was utilized as a control to assess the effectiveness of ProK treatment, after which almost all reactivity was lost (Fig. 2B). The protected fragment after ProK digestion lacked reactivity to the VIP-N antibody (recognizes the first 20 amino acids of Cav1) after ProK digestion suggesting the first 20 residues of Cav1 are not tightly associated with the membrane and confirming our proteomic analysis. However, reactivity was observed with both the polyclonal α-caveolin antibody (directed against multiple recognition sites between amino acids 1–97) and the Concav α-caveolin antibody that recognizes a region within the OD immediately adjacent to the CSD (amino acids 67–81) (Fig. 2B). To determine whether these domains are protected from ProK-mediated digestion by their membrane association, we performed ProK treatment before and after membrane disruption with the non-ionic detergent Triton X-100. A complete degradation of the protected small molecular weight fragments of Cav1 was observed when ProK treatment was performed after Triton X-100 treatment (Fig. 2C), which corresponded with a complete disruption of h-caveola structure (Fig. 2D). Furthermore, immunoelectron microscopy (immuno-EM) of purified h-caveolae demonstrated that the Concav epitope was unavailable for binding in fully formed h-caveolae unless pre-treated with Triton X-100 (data not shown). These data suggest that tight association with the membrane results in protection from ProK-mediated digestion. The protected residues of Cav1 are schematically represented in Fig. 2E, this analysis closely mimics previous observations that demonstrated the minimal caveogenic domain of Cav1 to encompass amino acids 49–147 (13).
The OD of Cav1 Is Protected from ProK Digestion in Mature Mammalian Caveolae
To examine whether this model of membrane insertion/association is consistent between h-caveolae and mature mammalian caveolae, caveolae were purified from MDCK cells using a novel pull-down method (schematic in Fig. 3A) exploiting stable expression of GFP-tagged Cavin-1, a specific marker of PM caveolae (44). Previous observations have demonstrated Cav1 is present in multiple different subcellular domains and undergoes different conformational changes during its maturation/export to the PM (8, 45). Cav1 is also present in non-caveolar pools at the PM suggesting Cav1 itself is an unsuitable candidate for the immunoprecipitation of mature PM caveolae (46). Cavin1, however, is exclusively localized to caveolae when present at the membrane and therefore is ideal for the isolation of mature PM caveolae (44). An 8-fold enrichment of purified caveolin-positive vesicles was observed comparing MDCK cells expressing Cavin1-GFP with cells expressing GFP alone (Fig. 3, B–D). Purified mammalian caveolae were subjected to ProK treatment, after which, the remaining fragment of Cav1 demonstrated a shift in molecular mass from ∼23 to ∼16–17 kDa, and demonstrated strong reactivity to both the Concav and polyclonal α-caveolin antibodies. Reactivity to both the VIP-N epitope and the GFP tag from Cavin1 was completely lost after ProK treatment, indicating degradation was effective and confirming the first 20 amino acids of Cav1 are exposed for degradation (Fig. 3E). No reactivity to Cav1 was observed in purified caveolae pre-treated with 0.5% SDS for 30 min prior to ProK digestion (SDS was used as mammalian caveolae are resistant Triton X-100 disruption). Previous work has demonstrated the Concav α-caveolin antibody is only reactive against the intracellular pool of Cav1 in mammalian cells and not against Cav1 at the PM unless pre-treated with cholesterol perturbing agents (47). Together with our data this indicates that the OD adjacent to CSD is tightly associated with the membrane in both mature mammalian caveolae and h-caveolae.
FIGURE 3.
The OD is protected by the PM in mature caveolae. A, schematic of the method used for mammalian PM caveolae purification, as described in detail under “Experimental Procedures.” B, immunolabeling of GFP expressing MDCK cells shows very few caveolin positive vesicles (polyclonal α-caveolin primary, 10-nm Protein A Gold secondary). C, immunolabeling of immunoprecipitated vesicles from MDCK cells expressing Cavin1-GFP demonstrated abundant and specific labeling of vesicles within the size range of caveolae. D, quantification of Cav1-positive vesicles. E, purified caveolae from MDCK cells expressing GFP-Cavin1 digested with ProK alone or treated with 0.5% SDS for 30 min at 37 °C prior to ProK digestion. Scale bars, 100 nm.
Membrane-associated Domains of Cav1 Are Sufficient for Membrane Deformation
We have previously shown that formation of h-caveolae in bacteria depends on caveolin oligomerization and gives rise to the polyhedral tertiary structure of h-caveolae (32). High-resolution cryo-ET revealed distinct membrane organization, defined as clear angles in the membrane through the medial plane of each h-caveola, and indicate that caveolin oligomers (directly or indirectly) cause distortion of the h-caveolar membrane (Fig. 4A). This is particularly evident in comparison to liposomes of similar bacterial lipid composition and size (Fig. 4C) that show a completely circular profile through the medial plane. To test whether membrane distortion was a feature of the membrane-associated domains of Cav1 or were imposed on the membrane by interactions on the exposed cytoplasmic face of h-caveola, h-caveolae treated with ProK were examined by cryo-ET. ProK-treated h-caveolae showed similar membrane distortion when compared with the control samples (Fig. 4B). Proteinase treatment resulted in a reduction in the density of the h-caveola coat, likely the MBP affinity tag, when compared with the control. These results indicate that the proteolytically protected fragments of Cav1 are sufficient to maintain three-dimensional structural integrity of h-caveolae.
FIGURE 4.
Membrane-associated domains of Cav1 are sufficient for membrane-deformation. A, optical slices through the medial plane of MBP-Cav1-His6 h-caveolae, abundant protein coat is observed around each h-caveola. Scale bar = 5 nm. B, optical slices through the medial plane of MBP-Cav1-His6 h-caveolae + ProK − no protein coat was observed to each h-caveola. Scale bar = 5 nm. The medial planes of MBP-Cav1-His6 h-caveolae ± ProK possess definitive polyhedral tertiary structure as defined by the clear angled profile to the membrane. C, comparative medial plane slices through liposomes demonstrating circular profiles. Scale bars = 5 nm. D, diagrammatic representation of Cav1 truncations generated for biochemical and structural analysis. E, Western blot time course of induction (α-MBP) of the Cav1 truncations. F, MBP-Cav1-His6 construct generated h-caveola consistent with previous observations(32). G, ΔN generated h-caveolae of regular size efficiently. H, ΔC was less efficient in generating regular size and structurally uniform h-caveolae. I, ΔNC generated h-caveolae of regular size efficiently. Labeling for F–I, α-MBP antibody primary (1:100 dilution) and 10-nm PAG secondary (1:70 dilution). Scale bars, 100 nm. J, Western blot analysis with the Concav antibody demonstrates ProK treatment of ΔNC does not reduce the molecular weight.
The data obtained from our proteomic, biochemical, and cryo-ET analyses indicated the importance of the central domain of Cav1 for maintaining the three-dimensional structure of h-caveolae. Therefore, we generated a series of truncation mutants of the caveolin fusion protein: an N-terminal truncation, ΔN, (Cav1-(49–178)), a C-terminal truncation, ΔC (Cav1-(1–147)), and an N and C termini truncation, ΔNC (Cav1-(49–147)) (Fig. 4D) to analyze the function of these domains in regulating the formation of h-caveolae. A time course of the expression of the Cav1 truncations demonstrated efficient and comparable protein expression after 3 h of induction (Fig. 4E). To further understand the role these domains have in the oligomeric arrangement of Cav1, the ability of these constructs to form h-caveolae was assessed using immuno-EM of cryosections. The full-length MBP-Cav1-His6 construct generated h-caveolae (Fig. 4F) as previously described (32). Both the ΔN and ΔNC constructs efficiently generated h-caveolae of consistent size and shape (Fig. 4, G and I). The ΔC truncation, however, generated more pleomorphic structures with larger diameters and with reduced efficiency of formation (Fig. 4H; see also Fig. 5, C and E). It was confirmed by ProK-mediated digestion that the ΔNC truncation represents the core membrane-associated domain by Western blot analysis, as ProK degradation did not result in a reduction in the size of the molecular weight of Cav1 (Fig. 4J).
FIGURE 5.
Three-dimensional characterization of h-caveolae generated by Cav1 truncations. A, an optical slice from a reconstructed tomogram of an E. coli expressing full-length MBP-Cav1-His6 h-caveolae. B–D, optical slices of E. coli expressing ΔN, ΔC, and ΔNC. Red asterisks highlight small h-caveolae. E, distribution of the diameters of h-caveolae revealed significant changes in h-caveolae structure and size compared with the full-length protein. Significant differences between the means of populations are indicated by asterisks on the right comparing two populations and illustrating differences with p < 0.01 (**) and p < 0.001 (***), respectively. Diameters were measured from the outer leaflet to the corresponding outer leaflet through the medial plane of each h-caveola. IMODinfo was used to export diameter measurements from IMOD for two-tailed Student's t-test statistical analyses. Scale bar, 50 nm.
ET was utilized on high-pressure frozen freeze-substituted E. coli expressing the various truncations after 3 h of induction to dissect changes in the three-dimensional architecture of h-caveolae in response to the N- and C-terminal truncations. Consistent with our previous observations, ΔC Cav1 h-caveolae were significantly larger (average diameter of full-length = 39.3 nm (n = 559) and ΔC = 43.3 nm (n = 300)) and also demonstrated greater variation in size (standard deviation of full-length = 11.4 nm compared with ΔC truncation = 13.0 nm) than full-length Cav1 h-caveolae (Fig. 5, A, C, and E). The N termini truncation mutants, however, demonstrated a significant reduction in both the diameter, Δn = 29.0 nm (n = 545) and ΔNC = 31.0 nm (n = 356), and the degree of variation in diameter (standard deviation of ΔN h-caveolae = 5.8 nm and ΔNC h-caveolae = 7.1 nm) (Fig. 5, B, D, and E). Through high-resolution electron tomography-based approaches we have determined that the deletion of the N terminus, whereas not essential for h-caveola formation, results in a more monodisperse population of h-caveola diameters.
Dissecting the Membrane Association of the OD and CSD
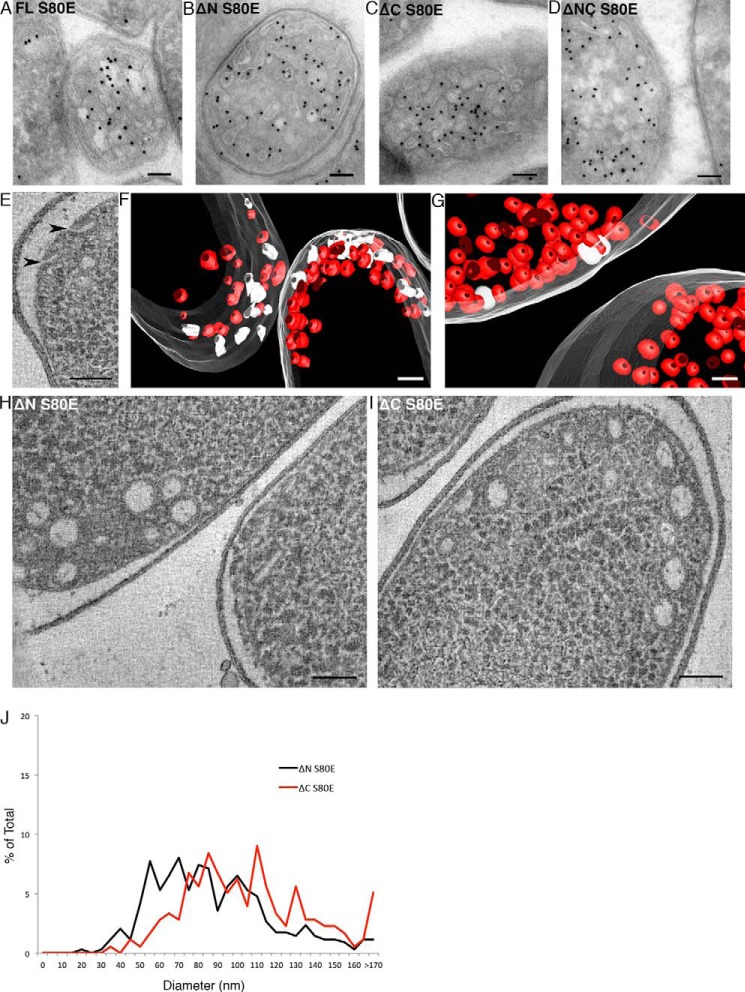
Our data suggest that the OD and CSD are protected from ProK enzymatic digestion by their tight association with the membrane. To further examine the legitimacy of this claim we mutated a single amino acid, Ser80, to a negatively charged glutamate residue. Ser80 is located in the middle of the OD and adjacent to the CSD, which is critical for lipid binding and caveola and h-caveola structural integrity (13, 32, 48). As previously demonstrated, the introduction of this S80E point mutation disrupted h-caveola formation with the full-length MBP-Cav1-His6 fusion protein (Fig. 6A) (32). We confirmed this result with the ΔN, ΔC, and ΔNC truncations by immuno-EM of cryosections; the S80E point mutant resulted in complete loss of defined h-caveola formation and dramatic tubulation of the cytoplasmic membrane (Fig. 6, B–D, respectively). Additionally, electron tomography and segmentation analysis demonstrated an increased number of connections maintained with the cytoplasmic membrane (Fig. 6, E and F) when compared with a non-mutated control (Fig. 6G) suggesting the introduction of the S80E point mutation results in a reduction in the potential to perform fission from the bulk membrane. Furthermore, ET revealed a complete loss of defined h-caveola formation (average diameter ΔN S80E = 89.9 nm, standard deviation = 30.6 nm: average diameter ΔC S80E = 110.1 nm and standard deviation = 41.2 nm) (Fig. 6, H–J). These data are consistent with a model whereby normal association of the OD and CSD with the membrane is critical for defined h-caveola formation and the addition of a single negatively charged residue at Ser80 results in the forced separation between the CSD/OD and membrane.
FIGURE 6.
Mutating Ser80 abrogates defined h-caveolae formation. A–D, immunolabeled frozen sections of E. coli expressing Cav1S80E, ΔN S80E, ΔC S80E, and ΔNC S80E, respectively. Sections were labeled with an α-MBP antibody (1:100) and 10-nm Protein A gold (1:70). A loss of defined h-caveolae formation and a marked increase in the number of tubules were observed. Scale bar, 100 nm. E, optical slice of reconstructed electron tomogram employed to analyze the structure of h-caveolae generated by S80E point mutation in three dimensions. An increase in the number of cytoplasmic membrane connected h-caveolae was observed. Black arrowheads indicate membrane-connected h-caveolae. Scale bar, 100 nm. F, segmentation of ΔC S80E, abundant surface connected h-caveolae (solid white) were observed when compared with h-caveolae where no connections were resolved (red). Cytoplasmic membrane, translucent white. Scale bar, 100 nm. G, segmentation of ΔC shows the reduced number of surface-connected membrane invaginations (surface connected h-caveolae, solid white; disconnected h-caveolae, red, cytoplasmic membrane, translucent white). Scale bar, 100 nm. H and I, the diameter of ΔN S80E (n = 337) and ΔC S80E (n = 178) h-caveolae were analyzed, respectively, at their mid-section. Scale bar, 100 nm. J, quantification revealed a significant loss of defined h-caveolae formation. Statistical significance was determined by two-tailed t-tests by comparison to the distribution of diameters of ΔN (p < 0.01) and ΔC (p < 0.01), respectively.
To confirm the importance of the membrane insertion of this domain in defined h-caveola structure formation we generated additional mutants; a ΔN construct lacking the polyhistidine tag and two additional point mutants in aromatic side chains of the OD/CSD predicted to insert into the lipid bilayer, ΔN Phe81 to glutamate (F81E) and ΔN Trp85 to glutamate (W85E). The ΔN construct that lacked the polyhistidine tag generated morphologically typical h-caveolae (Fig. 7A), confirming the potentially charged residues that comprise the His-tag do not affect formation. The two point mutants, however, resulted in a loss of defined vesicular h-caveola structure (Fig. 7, B and C) resembling the tubular invaginations generated by the ΔN S80E mutant (Fig. 6B).
FIGURE 7.
The Ser80 to Trp85 region is critical for efficient association of Cav1 with the bilayer. A, immuno-EM of MBP-ΔN-Cav1 without the polyhistidine tag to confirm the potentially charged residues that comprise the C-terminal tag do not effect h-caveola formation, h-caveolae were morphological identical to h-caveolae from the MBP-ΔN-Cav1-His6 construct. Scale bar = 100 nm. B, ΔN F81E results in heterogeneous h-caveola formation. Scale bar = 100 nm. C, ΔN W85E point mutation dramatically effects h-caveolae morphology with extensive tubulation of the membrane. Scale bar = 100 nm. D, Western blot of Concav α-caveolin antibody demonstrated reduced abundance of recognition domain in ΔN S80E, ΔN F81E, and ΔN W85E h-caveolae treated with ProK compared with ΔN and confirms the accessibility of this domain for digestion after the introduction of a glutamate residue. E, quantification of band intensities comparing ± ProK. Band intensities were not significantly reduced with ΔN ± ProK, however, ΔN S80E ± ProK resulted in a significant reduction in the abundance of the epitope (p = 0.0003; n = 3). The recognition of this epitope was also significantly reduced in the ΔN F81E and ΔN W85E point mutants (p = 0.0007, p = 0.018 respectively; n = 2). Statistical significance was determined by two-tailed t-tests. F, schematic of Cav1 membrane association after introduction of the negatively charged residue.
To further test this hypothesis, we analyzed the effect and extent of ProK degradation on the ΔN construct in comparison to ΔN S80E, ΔN F81E, and ΔN W85E point mutants (the ΔN and ΔN point mutants were used because these constructs expressed equivalently). In agreement with our hypothesis, the substitution of a glutamate residue at Ser80 resulted in almost complete proteolytic degradation of Cav1 by Western blot analysis of the OD/CSD with the Concav antibody (Fig. 7D) and a significant reduction in band intensity was observed comparing the fold-changes of ΔN S80E ± ProK but not ΔN ± ProK (Fig. 7E). The extrusion of ΔN F81E and ΔN W85E OD/CSD domains were confirmed by Western blot as these domains were largely digested by protease treatment (Fig. 7, D and E). These results suggest that the disruption of the association/insertion of the OD/CSD with the membrane by the addition of the S80E, F81E, or W85E point mutations is causal in the abrogating defined h-caveola structure (a schematic is represented in Fig. 7F).
The IMD and CSD Are Critical for h-Caveola Formation
Next we investigated the minimal required structural domains for h-caveola formation. We generated an extensive library of Cav1 truncations and analyzed the ability of these different truncations to generate h-caveolae by immuno-EM of frozen sections. For this assay we have defined the formation of morphologically typical h-caveolae by the appearance of gold-labeled circular membranous profiles within the cytoplasm of expressing E. coli, as shown in Fig. 4, F–I. This is indicative of formation of the uniformly shaped spherical free vesicles typical of h-caveolae as shown by tomography (Fig. 5 and Ref. 32). As both the IMD and CSD have previously been demonstrated to be essential domains for caveolar formation (13, 14, 33) we first interrogated the structural outcomes of deleting these domains. The removal of the IMD (Cav1-(49–97,134–178)) completely abrogated h-caveola formation and membrane association (Fig. 8A), whereas the removal of the CSD (Cav1-(49–81,97–178)) resulted in the accumulation of bacterial membranes in the cytoplasm of expressing cells but did not result in a defined structure (Fig. 8B).
FIGURE 8.
A library of Cav1 mutants reveal the critical domains for h-caveola formation. Immuno-EM of frozen sections of E. coli expressing various Cav1 truncations. A schematic of each truncation is depicted above a representative image of each mutant. Dotted lines represent deleted domains. Red lines are representative traces of h-caveola morphologies generated by each truncation. A, Cav1-(49–97,134–178); B, Cav1-(49–81,97–178); C, Cav1-(49–134); D, Cav1-(49–101); E, Cav1-(60–101); F, Cav1-(81–97); G, Cav1-(60–147); H, Cav1-(81–147); I, Cav1-(81–134); J, Cav1-(97–147); K, Cav1-(97–134). Scale bars = 100 nm.
Maintaining the core structural domains for h-caveola formation, we then performed sequential truncation of the C terminus and defined two different phenotypes; the removal of 13 further residues from the core membrane-associated domain (Cav1-(49–134)) caused a loss of defined h-caveolae structure and extensive tubulation (Fig. 8C). However, the additional removal of the IMD resulted in a completely soluble protein (Cav1-(49–101), Cav1-(60–101), and Cav1-(81–97); Fig. 8, D–F). Intriguingly, the further removal of the N terminus did not significantly affect h-caveola formation as both Cav1-(60–147) and Cav1-(81–147) retained the ability to generate morphologically typical h-caveolae (Fig. 8, G and H), suggesting, in our model prokaryotic system, the first 20 residues of the OD are not critical for h-caveola formation. Finally, the truncation of the remaining domains completely abrogated h-caveola structure; the CSD and IMD (Cav1-(81–134)) alone were unable to localize to membranes (Fig. 7I), whereas constructs consisting of the IMD alone (Cav1-(97–147) and Cav1-(97–134)) were localized to the cytoplasmic membrane but did not bend the membrane (Fig. 8, J and K). These data suggest that both the IMD and the CSD are essential for h-caveola structure and for the first time we define a minimal h-caveogenic domain, a short sequence of only 66 amino acids (Cav1-(81–147)) that can generate vesicles when expressed in E. coli.
Discussion
This study of caveolae produced in a model prokaryotic system has facilitated novel insights into Cav1, how it associates with the membrane, and the role different domains have in regulating h-caveola formation and structure. Using a variety of techniques, including negative staining electron microscopy, electron tomography, biochemical approaches, mass spectrometry, cryosectioning, and immunolabeling and cryo-electron tomography, we have identified Cav1 peptides that are protected from protease degradation and determined the core structural domain required defined h-caveola formation. These results have fundamental implications for understanding the topology of Cav1 in the membrane and how Cav1 could regulate signaling events in the cell.
Cav1 has, to date, been represented as a protein with two separable domains, the largely soluble N-terminal domain (11, 41) and the hydrophobic C terminus (12, 41). The results presented here indicate that a reassessment of these boundaries is required. We have shown that the core membrane-associated domain of Cav1 constitutes residues 49–147; close to the minimal domain for caveola formation in mammalian cells (13).
To investigate the importance of the core membrane-associated domain in h-caveola structure formation we generated a library of truncations and assayed h-caveola formation concurrently with the membrane attachment by immuno-EM. As Cav1 is an integral membrane protein, the conservation of membrane association is critical for elucidating domain function but is often overlooked in biochemical assays of caveolin oligomerization (11, 12). Our results demonstrated that both the mutation of the CSD and the deletion of this domain abrogated the defined vesicular structure, reiterating the importance of the CSD for caveola formation (13). Additionally, we demonstrated the IMD is critical for h-caveola formation. In our assays the IMD, in concert with amino acids 135–147, mediated the retention of the protein within the bacterial membrane; the removal of these domains resulted in the association of Cav1 to the cytoplasm of expressing cells. It is known that mutation of this region abrogates caveola formation in mammalian cells and inhibits the export of the protein from the Golgi complex to the PM (13, 14, 33). Intriguingly, large regions of the N terminus could be removed without affecting h-caveola formation, in some cases resulting in a more regular structure. It is possible that the removal of the non-membrane associated domains (amino acids 1–48) may result in the tighter packing of caveolin molecules in the membrane, as fluorescence-based techniques do not show significant differences in the number of caveolins per vesicle comparing the full-length and the ΔN construct (data not shown).
Understanding how Cav1 associates with the membrane is critical for understanding the function of the protein in the general cellular context. The results presented here demonstrate a tight association between the OD/CSD and the lipid bilayer in both mammalian caveolae and our bacterial system. Our observation that the membrane association of this domain, and caveola formation, can be disrupted by the insertion of charged residues at Ser80, Phe81, or Trp85 strongly links caveola formation to the stable insertion of this domain of caveolin within the membrane, consistent with a wedge mechanism for cytoplasmic leaflet expansion and curvature generation (34). The disruption of OD/CSD membrane association by the phosphomimetic S80E construct is suggestive of a potential regulation mechanism for the disassembly of caveolae. Although no in vivo data suggests Ser80 is directly phosphorylated (for example, see Ref. 49), Cav1 can be phosphorylated at multiple different residues (50) and several studies have proposed Ser80 as a potential phosphorylation site (16, 48). As these mutations increase the availability of the CSD for protease-mediated degradation and concurrently disrupt caveola formation, it is possible that the extrusion of this site from the membrane is a critical step in destabilizing the caveola unit in response to a mechanical stress (6). This hypothesis is strengthened by the observation that the S80E point mutant cannot form caveolae (13) and this region is masked by the membrane in mature PM-associated caveolae but exposed after the disruption of caveola structure by cholesterol depletion (47). This region is also exposed in Golgi-localized caveolin (45).
Although the introduction of charged residues within the crucial Ser80 to Trp85 region abrogates the formation of spherical vesicles of regular size and shape (characteristic of the wild-type caveolin), the mutated caveolin molecule still induces the formation of tubular elements that show limited fission from the bacterial cytoplasmic membrane. We have previously proposed a model in which a polyhedral caveolin cage is generated through a two-step process, linear extension and subsequent trimerization of preassembled linear oligomers (32). This can arise through distinct oligomeric signals within the membrane-associated region that we now pinpoint to residues 81–147. An analogy can be made between the combined activity of epsin (helix insertion) and clathrin (cage formation) to the activity of caveolin, which we propose acts as a hybrid protein with combined membrane insertion and cage assembly properties. We hypothesize in wild-type Cav1 the membrane association of the amphipathic helices of oligomerized CSDs function to promote vesiculation through destabilization of the membrane, in a manner similar to the coordinated function of epsin and clathrin in the internalization of clathrin-coated pits (51). The mutation of the CSD in the critical Ser80 to Trp85 region compromises the insertion of these helices, which is reflected by the protease sensitivity of these mutants. As such, the extrusion of the CSD from the membrane alters the membrane remodeling properties of Cav1 resulting in a BAR domain-like phenotype, greater tubulation, and reduced membrane fission (51).
These findings have wide reaching implications for the control of cellular signaling events as the CSD is often presented as the key binding domain of Cav1. There is a large body of literature discussing the scaffolding domain and how it potentially participates in signal regulation via the “caveolin signaling hypothesis” (2–4, 52–54). This proposes Cav1 mediates signal regulation through a series of direct protein-protein interactions between binding motifs on signaling proteins and the CSD, which, in turn facilitates the sequestration of these proteins in inactive forms within caveolae. Our findings argue against this model and suggest that the CSD is likely to be inaccessible for interaction with cytoplasmic proteins as this region is inaccessible to proteases and antibodies unless caveola membranes are disrupted with detergents. Although detergent treatment could also potentially affect Cav1-Cav1 interactions that are dependent on membrane integrity (and thus make this domain accessible to proteases indirectly) the increased protease digestion observed upon replacement of Ser80, Phe81, and Trp85 with glutamate residues argues for the membrane insertion model of this region in the wild-type protein and thus against direct interactions between the CSD and a caveolin binding motif on a cytoplasmic protein. Finally, we have characterized the core structural domain for h-caveola formation and, strikingly, this comprises only 66 amino acids (residues 81–147). The need for tissue-specific delivery of drugs, such as anti-cancer therapies, is critical to improve the efficacy of treatment and reduce nonspecific deleterious side effects of these cytotoxic compounds (55). As a tool for biotechnology our prokaryotic h-caveolae system is a significant advance on currently available methods. It is cost effective, easily manufactured, and purified in a single-step but has the added advantages of being a genetically encoded nanovesicle that is easily modified for directed targeting (32). This study has demonstrated significant reduction in the structural heterogeneity to h-caveolae by the removal of the soluble N-terminal domain of Cav1, this system now generates more monodisperse nanovesicles. Further understanding at the atomic level of how caveolin-lipid interactions allow a 66-amino acid membrane-inserted polypeptide to generate a uniform nanovesicle should provide insights into both the fundamental properties of caveolin in forming caveolae and elucidate how further modifications can be made to optimize the properties of genetically encoded nanovesicle.
Author Contributions
N. A. and R. G. P. designed experiments, interpreted data, and edited the manuscript. N. A. performed all experiments and wrote the manuscript. J. R. provided technical assistance for Figs. 7 and 8, S. O. for Fig. 3, N. L. for Fig. 4, and C. F. with EM. D. L. and M. M. H. provided proteomic expertise. P. W. and B. M. C. edited the manuscript. B. M. C. performed the alignments/predictions in Fig. 1. R. G. P. supervised the work. All authors reviewed the results and approved the final manuscript.
Acknowledgments
We acknowledge the use of the Australian Microscopy and Microanalysis Research Facility at the Center for Microscopy and Microanalysis at The University of Queensland. We also acknowledge Professor Ulf Skoglund for generously donating his time to peruse the manuscript. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, CA, supported by National Institutes of Health Grant P41 RR001081.
This work was supported in part by a fellowship and National Health and Medical Research Council (NHMRC) of Australia Grants APP1037320, APP1058565, and APP569542 (to R. G. P.), and APP1045092 (to R. G. P. and N. A.). The authors declare that they have no conflicts of interest with the contents of this article.
- PM
- plasma membrane
- CSD
- caveolin scaffolding domain
- OD
- oligomerization domain
- IMD
- intramembrane domain
- h-caveolae
- heterologous caveolae
- MDCK
- Madin-Darby canine kidney
- MBP
- maltose-binding protein
- ProK
- proteinase K
- ET
- electron tomography
- CBM
- caveolin binding motif.
References
- 1. Ariotti N., Fernández-Rojo M. A., Zhou Y., Hill M. M., Rodkey T. L., Inder K. L., Tanner L. B., Wenk M. R., Hancock J. F., Parton R. G. (2014) Caveolae regulate the nanoscale organization of the plasma membrane to remotely control Ras signaling. J. Cell Biol. 204, 777–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Couet J., Sargiacomo M., Lisanti M. P. (1997) Interaction of a receptor tyrosine kinase, EGF-R, with caveolins: caveolin binding negatively regulates tyrosine and serine/threonine kinase activities. J. Biol. Chem. 272, 30429–30438 [DOI] [PubMed] [Google Scholar]
- 3. Engelman J. A., Chu C., Lin A., Jo H., Ikezu T., Okamoto T., Kohtz D. S., Lisanti M. P. (1998) Caveolin-mediated regulation of signaling along the p42/44 MAP kinase cascade in vivo: a role for the caveolin-scaffolding domain. FEBS Lett. 428, 205–211 [DOI] [PubMed] [Google Scholar]
- 4. Galbiati F., Volonte D., Engelman J. A., Watanabe G., Burk R., Pestell R. G., Lisanti M. P. (1998) Targeted downregulation of caveolin-1 is sufficient to drive cell transformation and hyperactivate the p42/44 MAP kinase cascade. EMBO J. 17, 6633–6648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Murata M., Peränen J., Schreiner R., Wieland F., Kurzchalia T. V., Simons K. (1995) VIP21/caveolin is a cholesterol-binding protein. Proc. Natl. Acad. Sci. U.S.A. 92, 10339–10343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sinha B., Köster D., Ruez R., Gonnord P., Bastiani M., Abankwa D., Stan R. V., Butler-Browne G., Vedie B., Johannes L., Morone N., Parton R. G., Raposo G., Sens P., Lamaze C., Nassoy P. (2011) Cells respond to mechanical stress by rapid disassembly of caveolae. Cell 144, 402–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Park D. S., Woodman S. E., Schubert W., Cohen A. W., Frank P. G., Chandra M., Shirani J., Razani B., Tang B., Jelicks L. A., Factor S. M., Weiss L. M., Tanowitz H. B., Lisanti M. P. (2002) Caveolin-1/3 double-knockout mice are viable, but lack both muscle and non-muscle caveolae, and develop a severe cardiomyopathic phenotype. Am. J. Pathol. 160, 2207–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dupree P., Parton R. G., Raposo G., Kurzchalia T. V., Simons K. (1993) Caveolae and sorting in the trans-Golgi network of epithelial cells. EMBO J. 12, 1597–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pol A., Luetterforst R., Lindsay M., Heino S., Ikonen E., Parton R. G. (2001) A caveolin dominant negative mutant associates with lipid bodies and induces intracellular cholesterol imbalance. J. Cell Biol. 152, 1057–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim J. H., Peng D., Schlebach J. P., Hadziselimovic A., Sanders C. R. (2014) Modest effects of lipid modifications on the structure of caveolin-3. Biochemistry 53, 4320–4322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schlegel A., Lisanti M. P. (2000) A molecular dissection of caveolin-1 membrane attachment and oligomerization: two separate regions of the caveolin-1 C-terminal domain mediate membrane binding and oligomer/oligomer interactions in vivo. J. Biol. Chem. 275, 21605–21617 [DOI] [PubMed] [Google Scholar]
- 12. Song K. S., Tang Z., Li S., Lisanti M. P. (1997) Mutational analysis of the properties of caveolin-1: a novel role for the C-terminal domain in mediating homo-typic caveolin-caveolin interactions. J. Biol. Chem. 272, 4398–4403 [DOI] [PubMed] [Google Scholar]
- 13. Kirkham M., Nixon S. J., Howes M. T., Abi-Rached L., Wakeham D. E., Hanzal-Bayer M., Ferguson C., Hill M. M., Fernandez-Rojo M., Brown D. A., Hancock J. F., Brodsky F. M., Parton R. G. (2008) Evolutionary analysis and molecular dissection of caveola biogenesis. J. Cell Sci. 121, 2075–2086 [DOI] [PubMed] [Google Scholar]
- 14. Rieth M. D., Lee J., Glover K. J. (2012) Probing the caveolin-1 P132L mutant: critical insights into its oligomeric behavior and structure. Biochemistry 51, 3911–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee J., Glover K. J. (2012) The transmembrane domain of caveolin-1 exhibits a helix-break-helix structure. Biochim. Biophys. Acta 1818, 1158–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Parton R. G., Hanzal-Bayer M., Hancock J. F. (2006) Biogenesis of caveolae: a structural model for caveolin-induced domain formation. J. Cell Sci. 119, 787–796 [DOI] [PubMed] [Google Scholar]
- 17. Yang G., Xu H., Li Z., Li F. (2014) Interactions of caveolin-1 scaffolding and intramembrane regions containing a CRAC motif with cholesterol in lipid bilayers. Biochim. Biophys. Acta 1838, 2588–2599 [DOI] [PubMed] [Google Scholar]
- 18. Epand R. M., Sayer B. G., Epand R. F. (2005) Caveolin scaffolding region and cholesterol-rich domains in membranes. J. Mol. Biol. 345, 339–350 [DOI] [PubMed] [Google Scholar]
- 19. Fernandez I., Ying Y., Albanesi J., Anderson R. G. (2002) Mechanism of caveolin filament assembly. Proc. Natl. Acad. Sci. U.S.A. 99, 11193–11198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Le Lan C., Gallay J., Vincent M., Neumann J. M., de Foresta B., Jamin N. (2010) Structural and dynamic properties of juxta-membrane segments of caveolin-1 and caveolin-2 at the membrane interface. Eur. Biophys. J. 39, 307–325 [DOI] [PubMed] [Google Scholar]
- 21. Le Lan C., Neumann J. M., Jamin N. (2006) Role of the membrane interface on the conformation of the caveolin scaffolding domain: a CD and NMR study. FEBS Lett. 580, 5301–5305 [DOI] [PubMed] [Google Scholar]
- 22. Hoop C. L., Sivanandam V. N., Kodali R., Srnec M. N., van der Wel P. C. (2012) Structural characterization of the caveolin scaffolding domain in association with cholesterol-rich membranes. Biochemistry 51, 90–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Spisni E., Tomasi V., Cestaro A., Tosatto S. C. (2005) Structural insights into the function of human caveolin 1. Biochem. Biophys. Res. Commun. 338, 1383–1390 [DOI] [PubMed] [Google Scholar]
- 24. Sargiacomo M., Scherer P. E., Tang Z., Kübler E., Song K. S., Sanders M. C., Lisanti M. P. (1995) Oligomeric structure of caveolin: implications for caveolae membrane organization. Proc. Natl. Acad. Sci. U.S.A. 92, 9407–9411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Benferhat R., Krust B., Rey-Cuillé M. A., Hovanessian A. G. (2009) The caveolin-1 binding domain of HIV-1 glycoprotein gp41 (CBD1) contains several overlapping neutralizing epitopes. Vaccine 27, 3620–3630 [DOI] [PubMed] [Google Scholar]
- 26. Benferhat R., Martinon F., Krust B., Le Grand R., Hovanessian A. G. (2009) The CBD1 peptide corresponding to the caveolin-1 binding domain of HIV-1 glycoprotein gp41 elicits neutralizing antibodies in cynomolgus macaques when administered with the tetanus T helper epitope. Mol. Immunol. 46, 705–712 [DOI] [PubMed] [Google Scholar]
- 27. Li S., Couet J., Lisanti M. P. (1996) Src tyrosine kinases, Gα subunits, and H-Ras share a common membrane-anchored scaffolding protein, caveolin: caveolin binding negatively regulates the auto-activation of Src tyrosine kinases. J. Biol. Chem. 271, 29182–29190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Byrne D. P., Dart C., Rigden D. J. (2012) Evaluating caveolin interactions: do proteins interact with the caveolin scaffolding domain through a widespread aromatic residue-rich motif? PloS One 7, e44879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Collins B. M., Davis M. J., Hancock J. F., Parton R. G. (2012) Structure-based reassessment of the caveolin signaling model: do caveolae regulate signaling through caveolin-protein interactions? Dev. Cell 23, 11–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Monier S., Parton R. G., Vogel F., Behlke J., Henske A., Kurzchalia T. V. (1995) VIP21-caveolin, a membrane protein constituent of the caveolar coat, oligomerizes in vivo and in vitro. Mol. Biol. Cell 6, 911–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hayer A., Stoeber M., Bissig C., Helenius A. (2010) Biogenesis of caveolae: stepwise assembly of large caveolin and cavin complexes. Traffic 11, 361–382 [DOI] [PubMed] [Google Scholar]
- 32. Walser P. J., Ariotti N., Howes M., Ferguson C., Webb R., Schwudke D., Leneva N., Cho K. J., Cooper L., Rae J., Floetenmeyer M., Oorschot V. M., Skoglund U., Simons K., Hancock J. F., Parton R. G. (2012) Constitutive formation of caveolae in a bacterium. Cell 150, 752–763 [DOI] [PubMed] [Google Scholar]
- 33. Lee H., Park D. S., Razani B., Russell R. G., Pestell R. G., Lisanti M. P. (2002) Caveolin-1 mutations (P132L and null) and the pathogenesis of breast cancer: caveolin-1 (P132L) behaves in a dominant-negative manner and caveolin-1 (−/−) null mice show mammary epithelial cell hyperplasia. Am. J. Pathol. 161, 1357–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stachowiak J. C., Brodsky F. M., Miller E. A. (2013) A cost-benefit analysis of the physical mechanisms of membrane curvature. Nat. Cell Biol. 15, 1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mastronarde D. N. (2005) Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 [DOI] [PubMed] [Google Scholar]
- 36. Kremer J. R., Mastronarde D. N., McIntosh J. R. (1996) Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 [DOI] [PubMed] [Google Scholar]
- 37. Frangakis A. S., Hegerl R. (2001) Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J. Struct. Biol. 135, 239–250 [DOI] [PubMed] [Google Scholar]
- 38. Noske A. B., Costin A. J., Morgan G. P., Marsh B. J. (2008) Expedited approaches to whole cell electron tomography and organelle mark-up in situ in high-pressure frozen pancreatic islets. J. Struct. Biol. 161, 298–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Richter T., Floetenmeyer M., Ferguson C., Galea J., Goh J., Lindsay M. R., Morgan G. P., Marsh B. J., Parton R. G. (2008) High-resolution 3D quantitative analysis of caveolar ultrastructure and caveola-cytoskeleton interactions. Traffic 9, 893–909 [DOI] [PubMed] [Google Scholar]
- 40. Ingelmo-Torres M., González-Moreno E., Kassan A., Hanzal-Bayer M., Tebar F., Herms A., Grewal T., Hancock J. F., Enrich C., Bosch M., Gross S. P., Parton R. G., Pol A. (2009) Hydrophobic and basic domains target proteins to lipid droplets. Traffic 10, 1785–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schlegel A., Schwab R. B., Scherer P. E., Lisanti M. P. (1999) A role for the caveolin scaffolding domain in mediating the membrane attachment of caveolin-1: the caveolin scaffolding domain is both necessary and sufficient for membrane binding in vitro. J. Biol. Chem. 274, 22660–22667 [DOI] [PubMed] [Google Scholar]
- 42. Rui H., Root K. T., Lee J., Glover K. J., Im W. (2014) Probing the U-shaped conformation of caveolin-1 in a bilayer. Biophys. J. 106, 1371–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang G., Dong Z., Xu H., Wang C., Li H., Li Z., Li F. (2015) Structural study of caveolin-1 intramembrane domain by CD and NMR. Biopolymers 104, 11–20 [DOI] [PubMed] [Google Scholar]
- 44. Hill M. M., Bastiani M., Luetterforst R., Kirkham M., Kirkham A., Nixon S. J., Walser P., Abankwa D., Oorschot V. M., Martin S., Hancock J. F., Parton R. G. (2008) PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell 132, 113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Luetterforst R., Stang E., Zorzi N., Carozzi A., Way M., Parton R. G. (1999) Molecular characterization of caveolin association with the Golgi complex: identification of a cis-Golgi targeting domain in the caveolin molecule. J. Cell Biol. 145, 1443–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Head B. P., Insel P. A. (2007) Do caveolins regulate cells by actions outside of caveolae? Trends Cell Biol. 17, 51–57 [DOI] [PubMed] [Google Scholar]
- 47. Pol A., Martin S., Fernández M. A., Ingelmo-Torres M., Ferguson C., Enrich C., Parton R. G. (2005) Cholesterol and fatty acids regulate dynamic caveolin trafficking through the Golgi complex and between the cell surface and lipid bodies. Mol. Biol. Cell 16, 2091–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fielding P. E., Chau P., Liu D., Spencer T. A., Fielding C. J. (2004) Mechanism of platelet-derived growth factor-dependent caveolin-1 phosphorylation: relationship to sterol binding and the role of serine-80. Biochemistry 43, 2578–2586 [DOI] [PubMed] [Google Scholar]
- 49. Humphrey S. J., Yang G., Yang P., Fazakerley D. J., Stöckli J., Yang J. Y., James D. E. (2013) Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab. 17, 1009–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nomura R., Fujimoto T. (1999) Tyrosine-phosphorylated caveolin-1: immunolocalization and molecular characterization. Mol. Biol. Cell 10, 975–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Boucrot E., Pick A., Çamdere G., Liska N., Evergren E., McMahon H. T., Kozlov M. M. (2012) Membrane fission is promoted by insertion of amphipathic helices and is restricted by crescent BAR domains. Cell 149, 124–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Foti M., Porcheron G., Fournier M., Maeder C., Carpentier J. L. (2007) The neck of caveolae is a distinct plasma membrane subdomain that concentrates insulin receptors in 3T3-L1 adipocytes. Proc. Natl. Acad. Sci. U.S.A. 104, 1242–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. García-Cardeña G., Martasek P., Masters B. S., Skidd P. M., Couet J., Li S., Lisanti M. P., Sessa W. C. (1997) Dissecting the interaction between nitric oxide synthase (NOS) and caveolin: functional significance of the nos caveolin binding domain in vivo. J. Biol. Chem. 272, 25437–25440 [DOI] [PubMed] [Google Scholar]
- 54. Lisanti M. P., Tang Z., Scherer P. E., Kübler E., Koleske A. J., Sargiacomo M. (1995) Caveolae, transmembrane signalling and cellular transformation. Mol. Membr. Biol. 12, 121–124 [DOI] [PubMed] [Google Scholar]
- 55. Couvreur P., Vauthier C. (2006) Nanotechnology: intelligent design to treat complex disease. Pharm. Res. 23, 1417–1450 [DOI] [PubMed] [Google Scholar]