Abstract
Filamentous fungi are renowned for the production of a diverse array of secondary metabolites (SMs) where the genetic material required for synthesis of a SM is typically arrayed in a biosynthetic gene cluster (BGC). These natural products are valued for their bioactive properties stemming from their functions in fungal biology, key among those protection from abiotic and biotic stress and establishment of a secure niche. The producing fungus must not only avoid self-harm from endogenous SMs but also deliver specific SMs at the right time to the right tissue requiring biochemical aid. This review highlights functions of BGCs beyond the enzymatic assembly of SMs, considering the timing and location of SM production and other proteins in the clusters that control SM activity. Specifically, self-protection is provided by both BGC-encoded mechanisms and non-BGC subcellular containment of toxic SM precursors; delivery and timing is orchestrated through cellular trafficking patterns and stress- and developmental-responsive transcriptional programs.
Secondary metabolites, or natural products, are produced by a wealth of microorganisms and plants but are particularly abundant in fllamentous fungi primarily belonging to the ascomycete taxon Pezizomycotina. Typically the genetic material required to synthesize any particular SM is arranged in a multigene biosynthetic gene cluster (BGC) reminiscent of bacterial operons. Upon synthesis, these metabolites confer a variety of survival functions on the producing organism. These range from protection from abiotic and biotic stresses, such as UV radiation, desiccation, predation from insects and competition with other microbes, as well as participation in metal homeostasis. These small molecules even serve as signals required for differentiation or to direct symbioses or parasitism with other organisms1–3. The very properties that confer survival attributes to the fungus have lent themselves to profitable pharmacological applications and thus ensure continued interest in these fascinating molecules.
Fungal SMs are perhaps best known as potent antimicrobial agents, playing important parts in defense and niche securement for the producing fungus, and these activities have been directly co-opted for human medicine. For example, penicillin, anidulafungin and griseofulvin are well-known fungal metabolites that have been put into service as antimicrobials. Gliotoxin is produced by Aspergillus fumigatus and serves as a virulence factor in invasive aspergillosis4. It was first explored in the 1940s as antimicrobial5,6, and its antifungal properties allow its application as an antibiotic marker in fungal transformation7. However, gliotoxin and other antimicrobial SMs have also been explored for their potential relevance to other clinical goals. In the case of gliotoxin, its toxic properties have largely precluded use as an antitumor agent, although it is still under consideration for treatment of some cancers8,9. The statins (for example, lovastatin) are efficacious antiparasitic and antifungal agents10,11, yet are routinely used as cholesterol-lowering drugs. Fumagillin and its analogs are used as antifungals in treatment of microsporidial diseases12, including honey bee colony collapse13, but have also shown promise in clinical trials for obesity14 and cancer15. Cyclosporin A (also called cyclosporine), although touted for its immunosuppressive properties, is also a potent antifungal; both functions are achieved through targeting the same molecule, cyclophilin, in humans and fungi16. Finally, the ergot alkaloids are known for their treatment of migraines, beauvericin is employed as an insecticide, and the gibberellins are excellent growth hormones.
To achieve these functional outcomes, fungal SMs act as weapons against a plethora of cellular proteins, many of them present in the SM-producing fungi. As a result, fungi must develop mechanisms to avoid injuring themselves by targeting these same proteins. Alternatively, for SMs that serve as protective armor, fungi must accurately place these molecules in the tissues requiring biochemical shielding. Filamentous fungi are multicellular eukaryotes and, like plants and animals but unlike yeast, differentiate into elaborate and distinct morphologies serving different survival roles. Metabolites providing UV protection of windblown dispersal spores would probably therefore differ from metabolites that have defensive roles, such as protecting against insect predators of sessile fruiting bodies. Elucidation of mechanisms of protection from self-harm and environmental stressors not only unmasks basic cellular processes in the producing fungi but may serve to predict possible avenues of resistance in applied therapeutics (for example, development of antimicrobial or cancer cell resistance). This article addresses our emerging understanding of how fungi self-protect from their own metabolites in an orchestrated manner, involving encoded BGC protective devices coupled with cellular trafficking patterns and timed delivery to specific tissue destinations.
Self-protection encoded in the BGC
For fungal SMs that are antifungal agents, the producing fungi must be resistant to their own metabolites or face self-destruction. How do fungi control their own chemicals? The answer can lie in the gene content of the BGC. Genes encoding fungal SMs are clustered into a set of contiguous genes specific for a SM and usually contain all of the activities required for product formation. Although BGCs are often discussed in terms of the biosynthetic modules and tailoring enzymes that determine the chemical structure of the natural product, the clusters also contain gene(s) encoding transcription factors specific for the cluster and ‘other’ gene(s). These ‘other’ genes often encode proteins whose activities appear to be incongruous with or unnecessary for formation of the SM and thus are often given limited attention in descriptions of new biosynthetic gene clusters. However, renewed interest has demonstrated they have important roles in self-protection mechanisms. There are at least three potential in-cluster self-protection strategies—duplication of the SM target (sometimes including SM-resistant forms of the target), detoxification of the SM and efflux of the SM—with many clusters displaying two or more of these properties (Fig. 1).
Figure 1. Fungal BGCs can contain genes encoding one or more self-protective devices.
Duplication (1) of the target of the BGC product can occur as a resistant form of the target that sustains activity even when the target is inhibited or in a form that is still sensitive but increases the target pool, allowing for some escape and, hence, activity. Other devices include enzymes that chemically modify the BGC product to fully or partially detoxify it (2) and transporters that export the BGC product outside of the cell (3).
Duplicated or resistant target
As indicated above, many fungal SMs are valued for specific activities; this specificity of action is linked to the specific biomolecule targeted by the SM. For example, lovastatin blocks cholesterol synthesis by inhibiting HMG-CoA reductase, a crucial step in the cholesterol biosynthetic pathway. This enzyme is also required for synthesis of the fungal sterol ergosterol, explaining lovastatin’s antifungal activity17. To prevent toxicity in the host strain, the Aspergillus terreus lovastatin gene cluster encodes a second HMG-CoA reductase proposed to be resistant to lovastatin18. This motif of an in-cluster HMG-CoA reductase is maintained in related statin SM clusters19 and can be readily identified in bioinformatics searches.
Fumagillin targets the enzyme methionine aminopeptidase-2, which is required for hydrolytic removal of N-terminal methio-nine residues from nascent proteins. As with the lovastatin cluster, the fumagillin gene cluster in A. fumigatus contains an additional methionine aminopeptidase-2–encoding gene20. Analogous clusters in other genera maintain an incluster methionine aminopeptidase-2–encoding gene. It is unknown whether the duplicated enzyme is resistant to fumagillin or simply serves to increase the pool of methionine aminopeptidase-2 such that fumagillin is unable to inhibit all of the enzymes. Finally, cyclosporin A binds to cyclophilin, leading to inhibition of the calcium-calmodulin phosphatase calcineurin. Interestingly, the cyclosporin A BGC encodes cyclophilin21, but this gene does not appear to be duplicated in the available Tolypocladium genome, suggesting the sequence encoded in the BGC may somehow facilitate resistance to cyclosporin.
These data support a tactic where identification of nonbiosynthetic genes in BGCs might reveal natural product targets and facilitate the search for useful clusters to characterize. Indeed, by searching for a cluster containing the gene encoding mycophenolic acid target IMP dehydrogenase, the mycophenolic acid BGC was identified in Penicillium brevicompactum22. A systematic assessment of these target proteins to determine whether and how they are resistant to the BGC SM, as opposed to merely flooding the system with excess target molecules, could aid in our understanding of how the SM disables target activity.
Detoxification
In some clusters, additional enzymes are present that limit the toxicity of SMs by altering their chemical structure (thus changing the ability of the molecule to bind its target) or by eliminating or blocking functional groups important for binding the target. For example, GliT, a gliotoxin oxidoreductase encoded in the A. fumigatus gliotoxin BGC (gli) modifies the gliotoxin structure to generate a less-toxic molecule23,24.
Although most SM clusters appear to contain all the genes necessary to make, regulate and protect the fungi from the active molecule, there are examples where potential protective activities lie outside of the BGC. GtmA, a gliotoxin bis-thiomethyltransferase, lies outside of the cluster and acts to negatively regulate gliotoxin synthesis through production of bisdethiobis(methylthio)gliotoxin, which acts as a gli BGC off switch24. Trichothecene mycotoxins are protein synthesis inhibitors produced by many Fusarium spp. The acetyltransferase Tri101 acetylates the C3 hydroxyl group of trichothecenes and their precursors making them less toxic to other fungi25, and transgenic plants expressing Tri101 activity show enhanced resistance to the phytotoxic properties of tri-chothecenes25,26. However, deletion of Tri101 does not inhibit growth of Fusarium sporotrichioides on trichothecene-containing medium27.
There are few examples of BCGs encoding detoxifying enzymes at present. This could reflect the tendency to disregard enzymes that do not obviously fit into a biosynthetic scheme. A reinvestigation of such genes and their enzyme activities, for example, stcT, encoding a putative glutathione-S-transferase in the sterigmatocystin gene cluster28, might reveal some protective or detoxifying function.
Transporters
These membrane proteins are perhaps the best-known way that fungi and other species rid themselves of toxic materials, and thus it is no surprise that BGCs make use of this strategy. Genes encoding transporters (primarily belonging to the ABC superfamily and major facilitator superfamily (MFS)) are commonly found in fungal BGCs. For example, of the 35 BGCs in A. fumigatus, 24 contain one or more putative transporters. The transporter gene in the gliotoxin gene cluster, gliA, encodes an effective efflux pump important in resistance to gliotoxin24. This mechanism is common for the epipolythiodioxopiperazine (ETP) toxins, including gliotoxin, as other ETPs, such as sirodesmin, also encode self-protective transporters in the cluster29. Tri12 belonging to the MSF-type transporter family affords self-protection to trichothecenes27,30. Transporters can also be coupled to the transcriptional regulation of the cluster as a secondary protection mechanism. Deletion of the gene encoding MSF-type transporter Bik6 in the bikaverin gene cluster results in significant reduction of bikaverin production, mediated in part through a negative transcriptional feedback loop regulating expression of the enzymatic genes31.
Some BCG transporters appear to have no impact on SM production or fungal growth32. However, this does not necessarily mean that export and efflux of the SM are unimportant. For example, the gene cluster for the photoactivated perylenequinone toxin cercosporin contains a transporter, yet endogenous resistance to the toxin is provided by two noncluster transporters33. The roles of BGC-encoded transporters that appear to have no phenotype on the fungus when deleted remain unclear. One possible explanation lies in a redundancy of function.
There is also evidence that transporters can protect fungi from SMs originating from BCGs that do not contain pumps. The zea-ralenone cluster does not contain a transporter, yet its synthesis in Gibberella zeae is dependent on the noncluster ABC transporter ZRA1 (ref. 34). Interestingly, zra1 expression is positively regulated by the zearalenone in-cluster transcription factor ZEB2, providing a link between the SM and its protection mechanism even though they are not physically clustered. As ABC transporters are known to display some promiscuity in their substrates, it is likely that many transporters protect from SMs produced by other fungi35. This can be illustrated in Neurospora crassa, a fungus sensitive to cer-cosporin that possesses a transporter conferring some resistance to the toxin36, or in the biocontrol fungus Clonostachys rosea, which contains an ABC transporter protective against the Fusarium mycotoxin zearalenone as well as several fungicides37.
Although the general supposition is that transporters act to export the final BGC product out of the cell, this view misses other potential roles of BGC transporters, including transport of SM precursors into the cell or SM enzymes or pathway intermediates throughout subcellular destinations, as discussed below.
Cellular trafficking
Fungi have not only evolved an impressive set of in-cluster mechanisms to self-protect but also created a complex cellular trafficking highway that compartmentalizes biosynthesis steps along sub-cellular routes, ultimately leading to secretion at the cell surface. BGCs encode many enzymatic steps generating several precursors, some of which exhibit substantial toxicities. Where each enzyme is located and how cargo passes from one biosynthetic step to another presents an engineering challenge for the fungus, and it is becoming increasingly apparent that subcellular compartmentalization is the solution. This strategy appears to provide several aspects crucial for successful SM synthesis, such as access to SM precursor pools, protection from toxic intermediates and trafficking to the secretion machinery.
Cellular detection of secondary metabolite enzymes—chiefly by transmission electron microscopy, vacuole-vesicle fractionation or fluorescence-tagged proteins—has provided a complex pattern of enzyme trafficking through various subcellular compartments38,39. Pathway enzymes can be located in vacuoles, peroxisomes, endosomes, the endoplasmic reticulum (ER) and Golgi apparatus, or the cytosol. Figure 2 illustrates the current understanding of cellular trafficking patterns for three well-characterized SMs: aflatoxin, penicillin and trichothecenes.
Figure 2. Subcellular trafficking models for biosynthesis of aflatoxin, penicillin and trichothecene.
(a) Aflatoxin synthesis is proposed to originate in the peroxisome (1); synthesis then proceeds from this organelle through multiple fusion events incorporating materials from the ER and ribosomally derived vesicles (2) to yield endosomes that fuse, eventually leading to large bodies containing substantial concentrations of aflatoxin (3). Contents of the aflatoxisomes are released to the environment in a yet unidentified manner. (b) Penicillin synthesis originates at the interface of the vacuole and cytosol (1). The first intermediate is released in the cytosol and processed by a second cytosolic enzyme (2), and the product from the second enzyme is transported to the peroxisome, where synthesis is completed (3). Two transporters, one on the vacuole and one on the peroxisome, are required for synthesis. The final product is released to the environment in a yet unidentified manner. (c) Trichothecene synthesis probably originates in a vacuole (1), with multiple steps associated with vesicle fusion events (2), with sufficient fusions leading to a toxisome (3). An in-cluster transporter is proposed to release the end product from the toxisome and outside of the cell but may also act to transport materials into the cell and between vesicles38,39.
Peroxisomes
Peroxisomes, also known as microbodies, are implicated in many cellular processes, including β-oxidation and biogenesis of Woronin bodies (dense organelles plugging septal pores in ascomycete fungi to prevent cytoplasmic bleeding). The first enzymatic steps of aflatoxin and sterigmatocystin synthesis appear to occur in the peroxisome, as do various steps of many other SM pathways, including those responsible for making paxilline, AK-toxin, penicillin, cephalosporin and some siderophores38. Although it has not been demonstrated per se, the assumption is that acetyl-CoA generated by β-oxidation may be incorporated into the synthesis of metabolites using acyl-CoA pools as starter or extender units.
Many, but not all, of the SM enzymes localized to the peroxi-some contain peroxisomal localization motifs. Transporters may also be localized to organelles. The non-BGC transporter PaaT, which possesses a peroxisomal binding motif and is involved in penicillin synthesis—presumably through translocation of the side-chain precursors phenylacetic acid and phenoxyacetic acid into the peroxisome—has been characterized in Penicillium chrysogenum40.
Cytosol
Many SM enzymes localize to the cytosol, a milieu for accruing and trafficking enzymes, substrates and co-factors between subcellular compartments and housing cytoskeleton machinery potentially involved in vesicle movement. Detailed studies of the aflatoxin biosynthetic pathway suggest a transient localization of pathway enzymes in the cytosol during movement of precursors from one subcellular compartment to another (Fig. 2). Additional SMs with at least one step localized to the cytosol include gibberellins, trichothecenes, fumiquinazolines, penicillin and cephalosporin39.
Whether or not these enzymatic reactions specifically require cytosolic conditions or are just in transit to other sites in the cell is largely unknown. Some insight into this question comes from a recent study to improve penicillin yield in Aspergillus nidulans. The A. nidulans penicillin BGC consists of three genes, encoding the nonribosomal peptide AcvA (located on the cytosolic side of vacuoles), the cyclase IpnA (located in the cytosol) and the final ligase-acyltransferase step by AatA in the peroxisome. Re-engineering the location of IpnA to the peroxisomes results in a complete loss of penicillin synthesis, whereas redirecting the AcvA to the peroxi-some increases penicillin production three-fold41. Thus, in the case of IpnA, the cytosolic location appears to be critical for function.
Vesicles, vacuoles, endosomes and toxisomes
The intracellular endomembrane network is composed of vesicles and vacuoles that bud from various membranous subcellular compartments. Vesicles are double membrane–bound organelles that can bud from the ER or Golgi apparatus, peroxisome, mitochondria, nucleus and even the cytoplasmic membrane. Endosomes are noted for their ability to receive extracellular materials, recycle molecules and mobilize cargo along the microtubule42. Vacuoles, important sites for recycling and storage, are single membrane–bound organelles that can form from fusion of vesicles and endosomes. The discovery of PenV, a vacuolar transporter important in penicillin synthesis, putatively through supplying amino acids to the vacuole-anchored AcvA43, presents a specific example of how materials may be transported from the vacuole to the cytosol (Fig. 2).
Fusion, exchange of material and movement of these organelles allow fungi to direct SM intermediates and enzymes along a cellular pathway to achieve deposition and secretion at cell membranes. Toxisomes, first called aflatoxisomes in aflatoxin synthesis44, are products of various vesicle-vacuole fusions that house several biosynthetic steps of SM pathways and provide self-protection from toxic intermediates (Fig. 2). Final export from toxisomes (or any cellular compartment) out of the cell is not clear but hypothesized to involve exocytosis and/or active transport.
Regardless of how BGC materials are trafficked through the fungus, the impact of high production of a SM on hyphal morphology can be substantial. A case in point is illustrated by a study showing massive alterations in Aspergillus parasiticus hyphal morphology during aflatoxin-inducing conditions where a high proportion of subcellular materials are tied up in aflatoxi-some biogenesis45.
Expression pathways and delivery to specific tissues
SMs are, by definition, molecules that are not needed for routine cellular function; thus, fungi do not and cannot sustain continuous flux to SM pathways without cost to growth and development. Synthesis should be optimized and occur only during conditions requiring their use. Furthermore, SMs may actively and preferentially accumulate in specific tissues. Filamentous fungi differentiate into several morphologies in response to the environmental milieu and, as a result, SM localization can be specific to asexual or sexual spores, hyphae or resistant survival structures (for example, sclerotia, which are large masses of hardened, tightly adhered mycelium able to survive environmental extremes and insect predation). How are these aspects of secondary metabolism achieved? Here transcriptional cascades, which can include the transcription factors encoded in many BGCs, become of central importance.
Expression of BGCs in response to stress
Numerous reports have demonstrated that fungi synthesize SMs in response to environmental stress. For example, BGC activation occurs during fungal encounters with other microbes and insects, and a particular coupling exists between oxidative stress pathways and transcriptional activation of BGCs46,47.
Basic leucine zipper (bZIP) proteins are the primary conduits linking secondary metabolism with oxidative stress. bZIP proteins, well characterized in Saccharomyces cerevisiae as yeast activator protein (Yap) transcription factors48, are crucial for fungal responses to several types of abiotic stress, most notably oxidative (ROS) and osmotic stress. These proteins, including the two canonical ROS response bZIP proteins AtfA/Atf1 and NapA/Yap1 are also critical mediators of the SM response in filamentous fungi (Table 1). Some BGCs are regulated positively and some negatively by bZIP proteins, and this regulation is dependent on environmental conditions. The reasoning behind the linkage of stress responses with altered secondary metabolism is several-fold and involves the hypotheses that some SMs (for example, aflatoxin) protect from ROS damage, that coupling several modes of protection (for example, antioxidant enzyme and toxin synthesis) under one bZIP regulon is eficient, and that, in the case of SMs negatively regulated by bZIPs, suppressing their synthesis allows for valuable energy to be shunted to other critical responses specific for the particular stress at hand.
Table 1.
SMs regulated by bZIP proteins
| Fungus | SM | bZIP | Ref. |
|---|---|---|---|
| Aspergillus parasiticus | Aflatoxin | AtfB | 75 |
| Aflatoxin | Apyap1 | 76 | |
| Aspergillus nidulans | Sterigmatocystin | RsmA | 52 |
| Asperthecin | RsmA | 52 | |
| Sterigmatocystin | NapA | 77 | |
| Emericellin | NapA | 77 | |
| Asperthecin | NapA | 77 | |
| Shamixanthone | NapA | 77 | |
| Epishamixanthone | NapA | 77 | |
| Various | AtfA | 78 | |
| Pestalotiopsis fici | Isosulochrin | PfzipA | 79 |
| Ficipyronea | PfzipA | 79 | |
| Iso-A82775C | PfzipA | 79 | |
| Pestaloficiol M | PfzipA | 79 | |
| Res1214-1 | PfzipA | 79 | |
| Aspergillus fumigatus | Pseurotin | HapX | 54 |
| Fumagillin | HapX | 54 | |
| Fumitremorgin | HapX | 54 | |
| Hexadehydroastechrome | HapX | 54 | |
| Gliotoxin | RsmA | 80 | |
| Gliotoxin | FlbB | 81 | |
| Fusarium graminearum | Trichothecenes | Fgap1 | 82 |
| Deoxynivalenol | Atf1 | 83 | |
| Fusarium fujikuroi | Bikaverine | MeaB | 84 |
| Aspergillus flavus | Aflatoxin | MeaB | 85 |
| Botrytis cinerea | Botrydial | Bcatf1 | 86 |
| Botcinic acid | Bcatf1 | 86 | |
| Abscisic acid | Bcatf1 | 86 | |
| Aspergillus ochraceus | Ochratoxin | AoYap1 | 87 |
As mentioned above, studies have also shown that encounters with other microbes or insects can induce BGC expression and metabolite production in fungi49,50. These SMs can function as antimicrobials or as feeding deterrents and toxins toward insects. There is evidence that some of the bZIPs regulating secondary metabolism in fungi are also important in the response toward other organisms. Aspergillus napA—involved in negatively regulating a subset of A. nidulans SMs (Table 1)—is induced by Pseudomonas aeruginosa redox SMs and required for a defensive sporulation response in this genus51. Aspergillus RsmA, a positive regulator of the feeding deterrent sterigmatocystin, confers protection from the fungivore Folsomia candida, presumably through providing a chemical shield52. RsmA directly regulates the sterig-matocystin BGC transcription factor AflR by binding to a Yap site in the aflR promoter; mutations of this site result in strains with greatly reduced aflR expression and sterigmatocystin synthesis.
Our knowledge of what ligands trigger BGC responses to abiotic and biotic stressors and the transcriptional pathways transmitting the triggers is far from complete. LaeA and VeA, members of the conserved fungal velvet complex linking morphological development with light, play a role in many of these cascades as do nitrogen53 and metal54 imbalances and epigenetic modifications of the chromatin code55. Contrasting and comparison of the transcriptional profiles of fungi under different environmental milieus and organism encounters may allow the identification of other cascades with specificity toward BGC expression.
Developmental delivery of SMs to spores
Most fungi disperse by airborne asexual spores that can travel long distances at considerable altitudes, where exposure to UV radiation is significant. Aromatic and pigmented SMs—often called melanins—have been shown by several studies to protect spores from UV damage56–58. In radiotrophic fungi, some evidence suggests that melanins function to enhance fungal growth via capture of ionizing radiation for energy conversion59. Spore pigments can also be critical in the development of normal asexual and sexual spore structures60,61. Deletion of sporulation-specific BGC genes can result in absolute loss of sexual development in fungi, as seen in the requirement for a polyketide for fertile structures in Sordaria macrospora62 or the contribution of numerous SMs to development of sclerotia (sites of sexual sporulation and resistance structures to insect feeding) in Aspergillus flavus63–65.
Although the linkage with SM synthesis and sporulation has been long noted66, it is only recently that a process for this journey has been elucidated. Asexual spore development in Aspergillus and Penicillium species is mediated by the transcription factor BrlA67. Deletion of brlA yields a fungus unable to produce asexual spores, although hyphal and sexual spore production remains intact. Coupled with the loss of asexual spores is the loss or diminishment of a subset of SMs in BrlA-null mutants68. A compilation of studies using brlA mutant strains and/or spore extraction techniques have led to the identification of several A. fumigatus SMs specific to the asexual spore, including dihydroxynaphtalene (DHN) melanin, endocrocin, trypacidin, fumigaclavine and fumiquinazoline C69–73. Interestingly, with the exception of trypacidin, these spore clusters lack a BGC transcription factor, which may facilitate eficient regulation by BrlA.
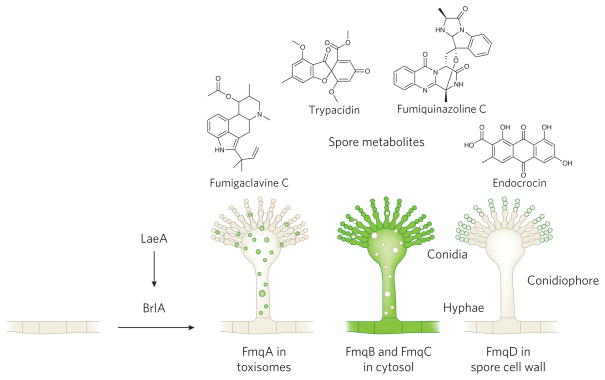
An understanding of the transcriptional machinery required for production of SMs specifically in the spore first emerged from the finding that all five of the characterized A. fumigatus spore BGCs and brlA were transcriptionally regulated by LaeA, a global regulator of fungal secondary metabolism74. This allowed for a model where LaeA regulation of spore metabolites was indirect and mediated through BrlA (Fig. 3). This hypothesis was examined by dissecting fumiquinazoline C synthesis in A. fumigatus. A complete vision of fumiquinazoline C placement to the asexual spore cell wall was achieved by coupling fluorescence microscopy with metabolic profiling of fmq (fumiquinazoline) cluster and brlA mutants over a timeline of fungal development73. The journey requires BrlA regulation of gene expression and conidiophore formation, trafficking of fmq enzyme or precursor machineries from a toxisome-like origin through the cytosol to the ER or Golgi apparatus and ends with localization of FmqD, the terminal enzyme of the fumiquinazoline biosynthetic pathway, to the spore wall (Fig. 3).
Figure 3. A transcriptional conduit from LaeA to BrlA regulates production of spore secondary metabolites.
BrlA induction results in morphological development of the asexual sporulation structure (the conidiophore) and expression of spore-specific BGCs, including the fmq BGC genes responsible for making fumiquinazoline C. fmq genes are expressed only when BrlA is active, with FmqA localizing to a toxisome-like structure, FmqB and FmqC in the cytosol, and FmqD, the terminal enzyme, localizing to the spore cell wall73.
The joint regulation of other spore SMs by LaeA and BrlA defines the transcriptional regulatory cascades leading to production of these metabolites in the spore but does not reveal the cellular trafficking patterns leading to secretion from the cell wall (or in the case of DHN melanin, incorporation into the cell wall). These pathways are likely to be as varied as the trafficking patterns illustrated for aflatoxin, trichothecenes and penicillin (Fig. 2) and involve shuttling to various subcellular compartments. Certainly cell biological studies will help clarify the patterns and mechanisms of placement of specific SMs to specific tissues—asexual spores as well as sexual tissues and resistant morphologies.
Conclusion
Canonical fungal BGCs often contain not only the genetic material to synthesize SMs but also mechanisms to self-protect from injurious bioactive properties of these metabolites. These mechanisms reveal clues of SM activity targets and can be predictive of modes of resistance in future applications of these compounds. Self-protection may also underlie the emerging cell biological studies illustrating variation in subcellular compartmentalization and trafficking of SM pathway precursors and enzymes. These studies are in their infancy, with promise to expand into the role of molecular motors in intracellular transport of SM machinery through the fungal thallus. However, cellular trafficking alone cannot ensure that SMs are produced at the right time and in the correct tissues; transcriptional programs that dictate when and where BGCs should be expressed are also coupled to these activities.
Advances in genetics, chemistry and cell biology have allowed for a clearer understanding of the process of specialized metabolism in filamentous fungi and also some insight into why the yeast genome, a fungus exhibiting little deviation in morphologies, is devoid of BGCs. Future studies are likely to continue to fill in holes in the burgeoning field of fungal secondary metabolism. By taking advantage of the modes of BGC self-protection described here, bioinformatic examination of the vast number of sequenced ascomycete genomes will most assuredly provide new information on SM targets and potential mechanisms toward developing resistance to SMs of pharmacological interest. Subcellular compartmentalization, which has a critical role in successful synthesis of SMs, needs further study and may present roadblocks in heterologous expression of fungal BGCs in bacteria or yeast, where cellular compartments may be lacking or inadequate. Finally, the integration of transcriptional cascades governing fungal differentiation processes and responses to environmental signals promise to tie into BGC activation in a logical and reasoned manner that should help address the challenge of activation of cryptic gene clusters.
Acknowledgments
This work was supported by US National Institutes of Health R01 Al065728-09 and R01GM112739-01 to N.P.K.
Footnotes
Competing financial interests
The author declares no competing financial interests.
References
- 1.Demain AL, Fang A. The natural functions of secondary metabolites. Adv Biochem Eng Biotechnol. 2000;69:1–39. doi: 10.1007/3-540-44964-7_1. [DOI] [PubMed] [Google Scholar]
- 2.Rohlfs M, Churchill AC. Fungal secondary metabolites as modulators of interactions with insects and other arthropods. Fungal Genet Biol. 2011;48:23–34. doi: 10.1016/j.fgb.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 3.Stergiopoulos I, Collemare J, Mehrabi R, De Wit PJ. Phytotoxic secondary metabolites and peptides produced by plant pathogenic Dothideomycete fungi. FEMS Microbiol Rev. 2013;37:67–93. doi: 10.1111/j.1574-6976.2012.00349.x. [DOI] [PubMed] [Google Scholar]
- 4.Dagenais TR, Keller NP. Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin Microbiol Rev. 2009;22:447–465. doi: 10.1128/CMR.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson JR, Bruce WF, Dutcher JD. Gliotoxin, the antibiotic principle of Gliocladium fimbriatum I Production, physical and biological properties. J Am Chem Soc. 1943;65:2005–2009. [Google Scholar]
- 6.Brian PW, Hemming HG. Gliotoxin, a fungistatic metabolic product of Trichoderma viride. Ann Appl Biol. 1945;32:214–220. doi: 10.1111/j.1744-7348.1945.tb06238.x. [DOI] [PubMed] [Google Scholar]
- 7.Carberry S, et al. Gliotoxin effects on fungal growth: mechanisms and exploitation. Fungal Genet Biol. 2012;49:302–312. doi: 10.1016/j.fgb.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Vigushin DM, et al. Gliotoxin is a dual inhibitor of farnesyltransferase and geranylgeranyltransferase I with antitumor activity against breast cancer in vivo. Med Oncol. 2004;21:21–30. doi: 10.1385/MO:21:1:21. [DOI] [PubMed] [Google Scholar]
- 9.Reece KM, et al. Epidithiodiketopiperazines (ETPs) exhibit in vitro antiangiogenic and in vivo antitumor activity by disrupting the HIF-1α/p300 complex in a preclinical model of prostate cancer. Mol Cancer. 2014;13:91. doi: 10.1186/1476-4598-13-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nyilasi I, et al. Susceptibility of clinically important dermatophytes against statins and different statin-antifungal combinations. Med Mycol. 2014;52:140–148. doi: 10.3109/13693786.2013.828160. [DOI] [PubMed] [Google Scholar]
- 11.Haughan PA, Chance ML, Goad LJ. Synergism in vitro of lovastatin and miconazole as anti-leishmanial agents. Biochem Pharmacol. 1992;44:2199–2206. doi: 10.1016/0006-2952(92)90347-l. [DOI] [PubMed] [Google Scholar]
- 12.Desoubeaux G, et al. Successful treatment with fumagillin of the first pediatric case of digestive microsporidiosis in a liver-kidney transplant. Transpl Infect Dis. 2013;15:E250–E259. doi: 10.1111/tid.12158. [DOI] [PubMed] [Google Scholar]
- 13.Maggi M, et al. Effects of the organic acids produced by a lactic acid bacterium in Apis mellifera colony development, Nosema ceranae control and fumagillin efficiency. Vet Microbiol. 2013;167:474–483. doi: 10.1016/j.vetmic.2013.07.030. [DOI] [PubMed] [Google Scholar]
- 14.Howland RH. Aspergillus, angiogenesis, and obesity: the story behind beloranib. J Psychosoc Nurs Ment Health Serv. 2015;53:13–16. doi: 10.3928/02793695-20150219-01. [DOI] [PubMed] [Google Scholar]
- 15.Kornienko A, et al. Toward a cancer drug of fungal origin. Med Res Rev. 2015 Apr 8; doi: 10.1002/med.21348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viaud MC, Balhadere PV, Talbot NJ. A Magnaporthe grisea cyclophilin acts as a virulence determinant during plant infection. Plant Cell. 2002;14:917–930. doi: 10.1105/tpc.010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chamilos G, Lewis RE, Kontoyiannis DP. Lovastatin has significant activity against zygomycetes and interacts synergistically with voriconazole. Antimicrob Agents Chemother. 2006;50:96–103. doi: 10.1128/AAC.50.1.96-103.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nyilasi I, et al. Susceptibility of clinically important dermatophytes against statins and different statin-antifungal combinations. Med Mycol. 2014;52:140–148. doi: 10.3109/13693786.2013.828160. [DOI] [PubMed] [Google Scholar]
- 19.Abe Y, et al. Effect of increased dosage of the ML-236B (compactin) biosynthetic gene cluster on ML-236B production in Penicillium citrinum. Mol Genet Genomics. 2002;268:130–137. doi: 10.1007/s00438-002-0736-8. [DOI] [PubMed] [Google Scholar]
- 20.Wiemann P, et al. Prototype of an intertwined secondary-metabolite supercluster. Proc Natl Acad Sci USA. 2013;110:17065–17070. doi: 10.1073/pnas.1313258110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bushley KE, et al. The genome of Tolypocladium inflatum: evolution, organization, and expression of the cyclosporin biosynthetic gene cluster. PLoS Genet. 2013;9:e1003496. doi: 10.1371/journal.pgen.1003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Regueira TB, et al. Molecular basis for mycophenolic acid biosynthesis in Penicillium brevicompactum. Appl Environ Microbiol. 2011;77:3035–3043. doi: 10.1128/AEM.03015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scharf DH, et al. Transannular disulfide formation in gliotoxin biosynthesis and its role in self-resistance of the human pathogen Aspergillus fumigatus. J Am Chem Soc. 2010;132:10136–10141. doi: 10.1021/ja103262m. [DOI] [PubMed] [Google Scholar]
- 24.Dolan SK, O’Keeffe G, Jones GW, Doyle S. Resistance is not futile: gliotoxin biosynthesis, functionality and utility. Trends Microbiol. 2015;23:419–428. doi: 10.1016/j.tim.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Kimura M, et al. Trichothecene 3-O-acetyltransferase protects both the producing organism and transformed yeast from related mycotoxins. Cloning and characterization of Tri101. J Biol Chem. 1998;273:1654–1661. doi: 10.1074/jbc.273.3.1654. [DOI] [PubMed] [Google Scholar]
- 26.Ohsato S, et al. Transgenic rice plants expressing trichothecene 3-O-acetyltransferase show resistance to the Fusarium phytotoxin deoxynivalenol. Plant Cell Rep. 2007;26:531–538. doi: 10.1007/s00299-006-0251-1. [DOI] [PubMed] [Google Scholar]
- 27.Alexander NJ, McCormick SP, Hohn TM. TRI12, a trichothecene efflux pump from Fusarium sporotrichioides: gene isolation and expression in yeast. Mol Gen Genet. 1999;261:977–984. doi: 10.1007/s004380051046. [DOI] [PubMed] [Google Scholar]
- 28.Andersen MR, et al. Accurate prediction of secondary metabolite gene clusters in filamentous fungi. Proc Natl Acad Sci USA. 2013;110:E99–E107. doi: 10.1073/pnas.1205532110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardiner DM, Jarvis RS, Howlett BJ. The ABC transporter gene in the sirodesmin biosynthetic gene cluster of Leptosphaeria maculans is not essential for sirodesmin production but facilitates self-protection. Fungal Genet Biol. 2005;42:257–263. doi: 10.1016/j.fgb.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Menke J, Dong Y, Kistler HC. Fusarium graminearum Tri12p influences virulence to wheat and trichothecene accumulation. Mol Plant Microbe Interact. 2012;25:1408–1418. doi: 10.1094/MPMI-04-12-0081-R. [DOI] [PubMed] [Google Scholar]
- 31.Wiemann P, et al. Biosynthesis of the red pigment bikaverin in Fusarium fujikuroi: genes, their function and regulation. Mol Microbiol. 2009;72:931–946. doi: 10.1111/j.1365-2958.2009.06695.x. [DOI] [PubMed] [Google Scholar]
- 32.Chang PK, Yu J, Yu JH. aflT, a MFS transporter-encoding gene located in the aflatoxin gene cluster, does not have a significant role in aflatoxin secretion. Fungal Genet Biol. 2004;41:911–920. doi: 10.1016/j.fgb.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 33.Amnuaykanjanasin A, Daub ME. The ABC transporter ATR1 is necessary for efflux of the toxin cercosporin in the fungus Cercospora nicotianae. Fungal Genet Biol. 2009;46:146–158. doi: 10.1016/j.fgb.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 34.Lee S, Son H, Lee J, Lee YR, Lee YW. A putative ABC transporter gene, ZRA1, is required for zearalenone production in Gibberella zeae. Curr Genet. 2011;57:343–351. doi: 10.1007/s00294-011-0352-4. [DOI] [PubMed] [Google Scholar]
- 35.Perlin MH, Andrews J, Toh SS. Essential letters in the fungal alphabet: ABC and MFS transporters and their roles in survival and pathogenicity. Adv Genet. 2014;85:201–253. doi: 10.1016/B978-0-12-800271-1.00004-4. [DOI] [PubMed] [Google Scholar]
- 36.Beseli A, Amnuaykanjanasin A, Herrero S, Thomas E, Daub ME. Membrane transporters in self resistance of Cercospora nicotianae to the photoactivated toxin cercosporin. Curr Genet. 2015 Apr 11; doi: 10.1007/s00294-015-0486-x. http://dx.doi.org/10.1007/s00294-015-0486-x. [DOI] [PubMed]
- 37.Dubey MK, Jensen DF, Karlsson M. An ATP-binding cassette pleiotropic drug transporter protein is required for xenobiotic tolerance and antagonism in the fungal biocontrol agent Clonostachys rosea. Mol Plant Microbe Interact. 2014;27:725–732. doi: 10.1094/MPMI-12-13-0365-R. [DOI] [PubMed] [Google Scholar]
- 38.Kistler HC, Broz K. Cellular compartmentalization of secondary metabolism. Front Microbiol. 2015;6:68. doi: 10.3389/fmicb.2015.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim FY, Keller NP. Spatial and temporal control of fungal natural product synthesis. Nat Prod Rep. 2014;31:1277–1286. doi: 10.1039/c4np00083h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fernández-Aguado M, Ullan RV, Teijeira F, Rodriguez-Castro R, Martin JF. The transport of phenylacetic acid across the peroxisomal membrane is mediated by the PaaT protein in Penicillium chrysogenum. Appl Microbiol Biotechnol. 2013;97:3073–3084. doi: 10.1007/s00253-012-4425-1. [DOI] [PubMed] [Google Scholar]
- 41.Herr A, Fischer R. Improvement of Aspergillus nidulans penicillin production by targeting AcvA to peroxisomes. Metab Eng. 2014;25:131–139. doi: 10.1016/j.ymben.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 42.Steinberg G. Endocytosis and early endosome motility in filamentous fungi. Curr Opin Microbiol. 2014;20:10–18. doi: 10.1016/j.mib.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernández-Aguado M, Teijeira F, Martin JF, Ullan RV. A vacuolar membrane protein affects drastically the biosynthesis of the ACV tripeptide and the β-lactam pathway of Penicillium chrysogenum. Appl Microbiol Biotechnol. 2013;97:795–808. doi: 10.1007/s00253-012-4256-0. [DOI] [PubMed] [Google Scholar]
- 44.Chanda A, et al. A key role for vesicles in fungal secondary metabolism. Proc Natl Acad Sci USA. 2009;106:19533–19538. doi: 10.1073/pnas.0907416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banerjee S, et al. Quantitative acoustic contrast tomography reveals unique multiscale physical fluctuations during aflatoxin synthesis in Aspergillus parasiticus. Fungal Genet Biol. 2014;73:61–68. doi: 10.1016/j.fgb.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 46.Hong SY, Roze LV, Linz JE. Oxidative stress-related transcription factors in the regulation of secondary metabolism. Toxins (Basel) 2013;5:683–702. doi: 10.3390/toxins5040683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Montibus M, Pinson-Gadais L, Richard-Forget F, Barreau C, Ponts N. Coupling of transcriptional response to oxidative stress and secondary metabolism regulation in filamentous fungi. Crit Rev Microbiol. 2015;41:295–308. doi: 10.3109/1040841X.2013.829416. [DOI] [PubMed] [Google Scholar]
- 48.Rodrigues-Pousada C, Menezes RA, Pimentel C. The Yap family and its role in stress response. Yeast. 2010;27:245–258. doi: 10.1002/yea.1752. [DOI] [PubMed] [Google Scholar]
- 49.Netzker T, et al. Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front Microbiol. 2015;6:299. doi: 10.3389/fmicb.2015.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rohlfs M. Fungal secondary metabolite dynamics in fungus-grazer interactions: novel insights and unanswered questions. Front Microbiol. 2014;5:788. doi: 10.3389/fmicb.2014.00788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng H, et al. Redox metabolites signal polymicrobial biofilm development via the NapA oxidative stress cascade in Aspergillus. Curr Biol. 2015;25:29–37. doi: 10.1016/j.cub.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yin WB, et al. An Aspergillus nidulans bZIP response pathway hardwired for defensive secondary metabolism operates through aflR. Mol Microbiol. 2012;83:1024–1034. doi: 10.1111/j.1365-2958.2012.07986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tudzynski B. Nitrogen regulation of fungal secondary metabolism in fungi. Front Microbiol. 2014;5:656. doi: 10.3389/fmicb.2014.00656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiemann P, et al. Perturbations in small molecule synthesis uncovers an iron-responsive secondary metabolite network in Aspergillus fumigatus. Front Microbiol. 2014;5:530. doi: 10.3389/fmicb.2014.00530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brakhage AA. Regulation of fungal secondary metabolism. Nat Rev Microbiol. 2013;11:21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- 56.Akamatsu HO, Chilvers MI, Stewart JE, Peever TL. Identification and function of a polyketide synthase gene responsible for 1,8-dihydroxynaphthalene-melanin pigment biosynthesis in Ascochyta rabiei. Curr Genet. 2010;56:349–360. doi: 10.1007/s00294-010-0306-2. [DOI] [PubMed] [Google Scholar]
- 57.Esbelin J, Mallea S, Ram AF, Carlin F. Role of pigmentation in protecting Aspergillus niger conidiospores against pulsed light radiation. Photochem Photobiol. 2013;89:758–761. doi: 10.1111/php.12037. [DOI] [PubMed] [Google Scholar]
- 58.Imshenetsky AA, Lysenko SV, Lach SP. Microorganisms of the upper layer of the atmosphere and the protective role of their cell pigments. Life Sci Space Res. 1979;17:105–110. doi: 10.1016/b978-0-08-023416-8.50017-9. [DOI] [PubMed] [Google Scholar]
- 59.Dadachova E, et al. Ionizing radiation changes the electronic properties of melanin and enhances the growth of melanized fungi. PLoS ONE. 2007;2:e457. doi: 10.1371/journal.pone.0000457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu Y, et al. A PKS gene, pks-1, is involved in chaetoglobosin biosynthesis, pigmentation and sporulation in Chaetomium globosum. Sci China Life Sci. 2012;55:1100–1108. doi: 10.1007/s11427-012-4409-5. [DOI] [PubMed] [Google Scholar]
- 61.Islamovic E, et al. Transcriptome analysis of a Ustilago maydis ust1 deletion mutant uncovers involvement of laccase and polyketide synthase genes in spore development. Mol Plant Microbe Interact. 2015;28:42–54. doi: 10.1094/MPMI-05-14-0133-R. [DOI] [PubMed] [Google Scholar]
- 62.Schindler D, Nowrousian M. The polyketide synthase gene pks4 is essential for sexual development and regulates fruiting body morphology in Sordaria macrospora. Fungal Genet Biol. 2014;68:48–59. doi: 10.1016/j.fgb.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 63.Cary JW, et al. An Aspergillus flavus secondary metabolic gene cluster containing a hybrid PKS-NRPS is necessary for synthesis of the 2-pyridones, leporins. Fungal Genet Biol. 2015;81:88–97. doi: 10.1016/j.fgb.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 64.Cary JW, et al. Functional characterization of a veA-dependent polyketide synthase gene in Aspergillus flavus necessary for the synthesis of asparasone, a sclerotium-specific pigment. Fungal Genet Biol. 2014;64:25–35. doi: 10.1016/j.fgb.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 65.Forseth RR, et al. Homologous NRPS-like gene clusters mediate redundant small-molecule biosynthesis in Aspergillus flavus. Angew Chem Int Edn Engl. 2013;52:1590–1594. doi: 10.1002/anie.201207456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Calvo AM, Wilson RA, Bok JW, Keller NP. Relationship between secondary metabolism and fungal development. Microbiol Mol Biol Rev. 2002;66:447–459. doi: 10.1128/MMBR.66.3.447-459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Park HS, Yu JH. Genetic control of asexual sporulation in filamentous fungi. Curr Opin Microbiol. 2012;15:669–677. doi: 10.1016/j.mib.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 68.Qin Y, et al. Penicillium decumbens BrlA extensively regulates secondary metabolism and functionally associates with the expression of cellulase genes. Appl Microbiol Biotechnol. 2013;97:10453–10467. doi: 10.1007/s00253-013-5273-3. [DOI] [PubMed] [Google Scholar]
- 69.Mulinti P, et al. Accumulation of ergot alkaloids during conidiophore development in Aspergillus fumigatus. Curr Microbiol. 2014;68:1–5. doi: 10.1007/s00284-013-0434-2. [DOI] [PubMed] [Google Scholar]
- 70.Tsai HF, Wheeler MH, Chang YC, Kwon-Chung KJ. A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J Bacteriol. 1999;181:6469–6477. doi: 10.1128/jb.181.20.6469-6477.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berthier E, et al. Low-volume toolbox for the discovery of immunosuppressive fungal secondary metabolites. PLoS Pathog. 2013;9:e1003289–e1003289. doi: 10.1371/journal.ppat.1003289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gauthier T, et al. Trypacidin, a spore-borne toxin from Aspergillus fumigatus, is cytotoxic to lung cells. PLoS ONE. 2012;7:e29906. doi: 10.1371/journal.pone.0029906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lim FY, Ames B, Walsh CT, Keller NP. Co-ordination between BrlA regulation and secretion of the oxidoreductase FmqD directs selective accumulation of fumiquinazoline C to conidial tissues in Aspergillus fumigatus. Cell Microbiol. 2014;16:1267–1283. doi: 10.1111/cmi.12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perrin RM, et al. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 2007;3:e50. doi: 10.1371/journal.ppat.0030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roze LV, Chanda A, Wee J, Awad D, Linz JE. Stress-related transcription factor AtfB integrates secondary metabolism with oxidative stress response in aspergilli. J Biol Chem. 2011;286:35137–35148. doi: 10.1074/jbc.M111.253468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reverberi M, et al. Apyap1 affects aflatoxin biosynthesis during Aspergillus parasiticus growth in maize seeds. Food Addit Contam. 2007;24:1070–1075. doi: 10.1080/02652030701553244. [DOI] [PubMed] [Google Scholar]
- 77.Yin WB, et al. bZIP transcription factors affecting secondary metabolism, sexual development and stress responses in Aspergillus nidulans. Microbiology. 2013;159:77–88. doi: 10.1099/mic.0.063370-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Emri T, et al. Core oxidative stress response in Aspergillus nidulans. BMC Genomics. 2015;16:478. doi: 10.1186/s12864-015-1705-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang X, et al. The bZIP transcription factor PfZipA regulates secondary metabolism and oxidative stress response in the plant endophytic fungus Pestalotiopsis fici. Fungal Genet Biol. 2015;81:221–228. doi: 10.1016/j.fgb.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 80.Sekonyela R, et al. RsmA regulates Aspergillus fumigatus gliotoxin cluster metabolites including Cyclo(L-Phe-L-Ser), a potential new diagnostic marker for invasive aspergillosis. PLoS ONE. 2013;8:e62591. doi: 10.1371/journal.pone.0062591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xiao P, Shin KS, Wang T, Yu JH. Aspergillus fumigatus fibB encodes two basic leucine zipper domain (bZIP) proteins required for proper asexual development and gliotoxin production. Eukaryot Cell. 2010;9:1711–1723. doi: 10.1128/EC.00198-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Montibus M, et al. The bZIP transcription factor Fgap1 mediates oxidative stress response and trichothecene biosynthesis but not virulence in Fusarium graminearum. PLoS ONE. 2013;8:e83377. doi: 10.1371/journal.pone.0083377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Van Nguyen T, Kroger C, Bonnighausen J, Schafer W, Bormann J. The ATF/CREB transcription factor Atf1 is essential for full virulence, deoxynivalenol production, and stress tolerance in the cereal pathogen Fusarium graminearum. Mol Plant Microbe Interact. 2013;26:1378–1394. doi: 10.1094/MPMI-04-13-0125-R. [DOI] [PubMed] [Google Scholar]
- 84.Wagner D, et al. The bZIP transcription factor MeaB mediates nitrogen metabolite repression at specific loci. Eukaryot Cell. 2010;9:1588–1601. doi: 10.1128/EC.00146-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Amaike S, et al. The bZIP protein MeaB mediates virulence attributes in Aspergillus flavus. PLoS ONE. 2013;8:e74030. doi: 10.1371/journal.pone.0074030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Temme N, et al. BcAtf1, a global regulator, controls various differentiation processes and phytotoxin production in Botrytis cinerea. Mol Plant Pathol. 2012;13:704–718. doi: 10.1111/j.1364-3703.2011.00778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Reverberi M, et al. Aoyap1 regulates OTA synthesis by controlling cell redox balance in Aspergillus ochraceus. Appl Microbiol Biotechnol. 2012;95:1293–1304. doi: 10.1007/s00253-012-3985-4. [DOI] [PubMed] [Google Scholar]