Abstract
Genome-wide analyses of changes in gene expression, transcription factor occupancy on DNA, histone modification patterns on chromatin, genomic copy number variation, and nucleosome positioning have become popular in many modern laboratories, yielding a wealth of information during health and disease states. However, most of these studies have overlooked an inherent normalization problem that must be corrected with spike-in controls. Here we describe the reason why spike-in controls are so important and explain how to appropriately design and use spike-in controls for normalization. We also suggest ways to retrospectively renormalize data sets that were wrongly interpreted due to omission of spike-in controls.
TEXT
All analyses that use microarrays or next-generation sequencing platforms to compare changes between two or more experimental conditions are based on an assumption that is often wrong. The assumption is that the overall yields of the sample to be analyzed, be it DNA or RNA, are identical per cell under different experimental conditions. Accordingly, researchers usually take the same amount of total RNA or DNA for analysis on their microarray or sequencing platform, and the resulting data are normalized to each other so that the total amounts of signals from each experimental condition (e.g., reads per million [RPM] normalization for sequencing and quantile normalization for microarray) are identical. However, this assumption is flawed when cells from different experimental conditions do not yield identical amounts of DNA or RNA. In addition, common pipelines for analysis of next-generation sequencing data normalize to the total number of sequence reads being the same for each sample. This happens only when the sum of increases over the genome is equal to the sum of decreases over the genome, which is rarely the case. For these reasons, experiments have been, and continue to be, wrongly interpreted using conventional normalization approaches. To prevent this, a spike-in control has to be added in an amount proportional to the number of cells for subsequent normalization of the data, in order to allow accurate interpretations of whether there are increases or decreases in signal at each region of the genome between samples.
WHEN DO YOU NEED A SPIKE-IN CONTROL?
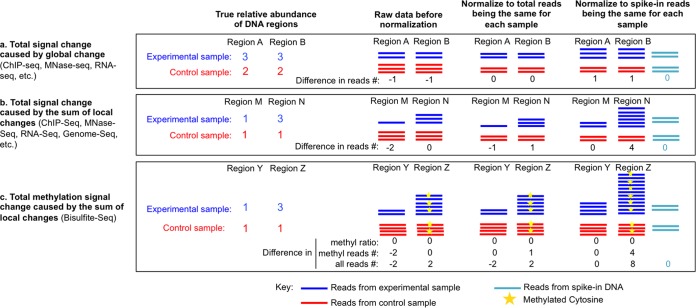
Spike-in controls are needed in all types of genome-wide profiling analyses by microarray or sequencing where changes in absolute amounts of the total signal are suspected to occur between different experimental conditions. This is the case whether the signal is RNA, DNA, nucleosome occupancy as detected by protection from micrococcal nuclease (MNase) digestion, or factor occupancy or histone modification patterns as detected by chromatin immunoprecipitation (ChIP). The striking importance of a spike-in control is most apparent when there is global signal change, which happens in similar trends at all genomic locations across the whole genome (Fig. 1a). However, because local signal changes at a subset of genomic locations may also lead to a change in total signal, spike-in controls are required for all genome-wide experiments to ensure accurate comparison between experimental conditions. For example, when there is significant increase of signal at some local regions but no signal decrease at any other regions, normalizing total signals to be the same introduces an artificial decrease in signal at the other regions (Fig. 1b). Similarly, in DNA copy number variation analyses, spike-in controls are important for analyzing repeat regions of the genome (Fig. 1b). Even for DNA methylation analyses, spike-in controls may also be required to detect changes in the total number of methylated CpGs, e.g., at regions bearing DNA copy number variations (Fig. 1c). The fundamental need for a spike-in control to facilitate accurate interpretation of virtually all genome-wide analyses cannot be overstated.
FIG 1.
Schematic to show why sequencing experiments require spike-in controls for accurate comparison between samples. Examples are shown for specific regions of the genome. (a) When the same degree of change happens everywhere on the genome, normalizing total sequencing reads to the same number hides the change, whereas normalizing spike-in reads to the same number reveals the global change of read density. (b) When signal increases happen at specific genomic regions, normalizing total sequencing reads between samples introduces artifactual reductions in the number of reads from other regions of the genome, which is falsely interpreted as being reduced under the specific experimental condition. Such artificial changes can be avoided by using spike-in controls as a reference for normalization. (c) Differences in copy numbers of methylated DNA, such as at repeat regions, can be detected accurately only with a spike-in reference, although the methylation ratio per se may be analyzed correctly without spike-in controls.
MNase-seq
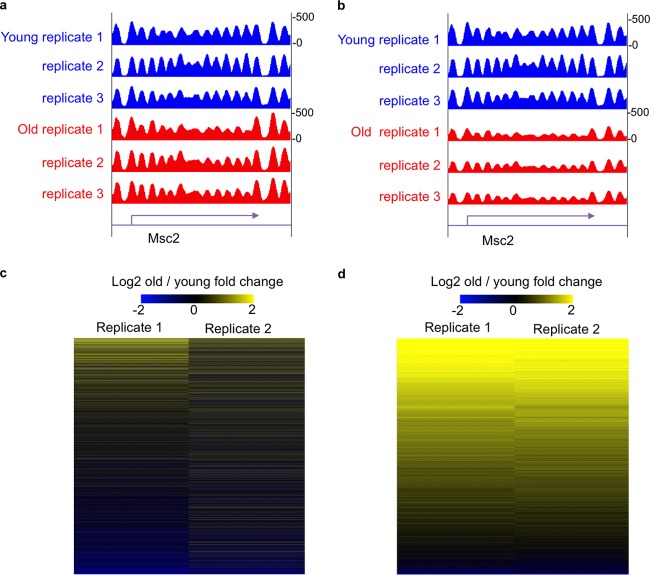
We recently discovered that aged yeast cells have half the normal amount of core histone proteins that constitute the protein-DNA structure called chromatin, and we showed that this loss of histones is a cause of aging (1). However, upon performing global nucleosome positioning analysis by MNase sequencing (MNase-seq) following standard protocols, we found that the occupancy of nucleosomes along the genome was unchanged during aging (Fig. 2a) (2). These contradictory results led us to question the standard practice of adding equal amounts of the DNA from different experimental conditions without normalization controls. Below, we describe the critical aspects of the design and use of spike-in normalization controls, but suffice it to say here that the addition of our spike-in control at equal amounts per cell prior to library construction, followed by normalization of our data to the spike-in control, revealed a clear 50% reduction of nucleosome occupancy over the entire genome in aged cells (Fig. 2b). This realization was critical for our subsequent properly normalized analysis of the transcriptional changes, histone modification changes, and genomic instability changes that occur as a consequence of global histone depletion during aging (2). In the future, it will be important to use spike-in control normalization to revisit many key questions. For example, is the transcriptional amplification and genomic instability that occurs in cancer also a consequence of potential global nucleosome depletion?
FIG 2.
The power of the spike-in control. (a) Snapshot of genome track showing nucleosome occupancy determined by MNase-seq in young and old cells without spike-in control. (b) Snapshot of the same region of the genome showing nucleosome occupancy determined by MNase-seq in young and old cells with spike-in normalization. (c) Heat map showing gene expression fold change determined by RNA-seq in young and old cells without spike-in control normalization. (d) Heat map showing gene expression fold change determined by RNA-seq in young and old cells with spike-in control normalization. Here, we used global-scaling normalization to a spike-in control, which is ideal for normalizing for global changes between experimental conditions.
RNA-seq
The enlightening experience with how normalization of our MNase-seq analyses completely changed the interpretation of the data led us to include spike-in controls in our subsequent high-throughput RNA sequencing (RNA-seq) analysis to identify transcription changes during replicative aging. We revisited the issue of transcriptional changes during aging that had been investigated in previously published literature by others, because we expected that global nucleosome depletion would lead to global transcriptional induction. As shown in Fig. 2c and d, the difference in the interpretation of RNA-seq data with and without spike-in normalization is striking. By appropriately normalizing our RNA-seq analysis, we discovered that all 6,000-plus genes in the yeast genome are transcriptionally induced during aging as a consequence of the global nucleosome depletion (2). This is in stark contrast to the interpretation made from similar analyses without normalization controls that led to the conclusion that most genes did not change during aging but that a few hundred were induced and a few hundred were repressed (3). This was also reminiscent of the previous interpretation of gene expression changes upon experimental histone depletion that led to the conclusion that most genes were not regulated by chromatin, as only a few hundred were induced and a few hundred were repressed (4). These kinds of erroneous interpretations of improperly controlled gene expression analyses not only are wrong but also can completely misdirect all subsequent analyses.
During the course of our analyses on gene expression changes during aging, Rick Young's group from the Massachusetts Institute of Technology (MIT) independently arrived at the same realization of the importance of spike-in controls for RNA-seq (5). Indeed, this allowed him to show that the cMyc oncogene, which was previously considered to be a gene-specific transcriptional activator, was in fact a genome-wide elongation factor that upregulates the transcription of virtually all genes in the genome when it is overexpressed (6). This provided a compelling example of how spike-in controls can totally change our understanding of biology. This is particularly the case when there are global changes in RNA transcription between different experimental conditions, as is the case during aging and upon overexpression of cMyc. We know that this is the case in other biological situations and encourage you all to include spike-in controls in your future genome-wide RNA-seq and microarray analyses and to revisit past analyses that were wrongly interpreted.
CHIP-seq
In addition, we strongly encourage the use of spike-in controls in chromatin immunoprecipitation sequencing (ChIP-seq) analyses of factor occupancy and histone modification patterns on the genome. We used spike-in controls during our global mapping of the DNA damage response marker of phosphorylation on serine 129 of histone H2A (called gamma H2A) during aging (2). This histone modification is about three times more abundant in old cells (2), and without a spike-in normalization control, our interpretation of where it, and hence DNA damage, increases and decreases on the genome would have been wrong. It is noteworthy that the changes in gamma H2A signal that we saw during aging were not global but instead were unchanged in most of the genome but were increased greatly in several repetitive regions of the genome such as the ribosomal DNA (rDNA) and mitochondrial DNA that was transferred to the nucleus during aging (2). If we had used normalization for total sequence reads, we would have concluded that there were decreases in gamma H2A signal over most of the genome and subtle increases at the rDNA and mitochondrial DNA in old cells. This interpretation would have been wrong and would have misled future analyses by others and us. Basically, the standard normalization protocol for ChIP-seq gives you the correct enrichment and depletion patterns for proteins or posttranslational modifications only if the total amounts of a particular protein or posttranslational modification on chromatin are identical under the different experimental conditions. When the total histone modification levels are not identical under the different experimental conditions, the histone modification is interpreted as depleted at genomic regions where it is unchanged or increased or vice versa. Conventional ChIP analyses are not subject to the need for a spike-in control, because these analyses should always already be normalized to the input DNA for each DNA sequence. However, unrelated IgG or low-depth input sequencing for ChIP-seq is an insufficient normalization control because the results cannot reflect global changes in the ChIP target, as was the case in our analysis of histone content during aging.
More recently, the exogenous epigenome was used as a spike-in control to define global changes of histone modifications over the human genome (7). To do this, the researchers added Drosophila melanogaster chromatin on a per-cell basis, to allow comparison between different ChIP-seq samples. As a proof of principle, they tested EPZ5676, an inhibitor of the enzyme DOT1L, a protein that catalyzes the dimethylation of histone H3 on lysine 79 (H3 K79me2). To test the effect of EPZ5676, human cells were treated with EPZ5676 and then compared with control cells. Using traditional RPM normalization, only small differences between the treated and control cells in locus-specific ChIP-seq profiles or metagene profiles for H3 K79me2 could be observed, despite clear evidence by Western blotting that the total level of H3 K79me2 was severely depleted by EPZ5676 treatment. After normalization to sequencing reads mapped to the Drosophila genome as a control, EPZ5676 was seen to strikingly reduce the global enrichment level of H3K79me2 in the human genome. Because they used combinations of known ratios of Drosophila cells to their experimental cells, in this case, human cells, prior to isolation of nuclei, this method also normalized for variations in many of the earlier steps involved in chromatin immunoprecipitation. However, this method does depend on the antibodies recognizing the histone modification of interest in both the experimental chromatin and Drosophila chromatin.
gDNA-seq
We even used spike-in controls during genomic DNA sequencing (gDNA-seq) of the young and old genomes in order to discover amplification of approximately one-third of chromosome XII in 15% of the population of cells during mitotic aging (2). Spike-in controls are even more important during gDNA-seq to accurately determine chromosome ploidy and whether regions of the genome are in fact amplified or depleted in cancer.
WHAT EXACTLY IS A SPIKE-IN CONTROL?
A spike-in control for all applications should constitute multiple different DNA sequences that are not from your organism of interest but have the GC content equivalent to the GC content of your organism of interest. For high-throughput sequencing platforms, the spike-in control DNA fragments should also be approximately the same length as your DNA fragments prior to library construction. For MNase-seq, the spike-in control DNA fragments that we used were around 150 bp in length, which is the length of DNA contained within a mononucleosome. This spike-in control can also be used for ChIP-seq and gDNA-seq after the experimental DNA samples are fragmented to around 150 bp in length. Our spike-in control for these applications was generated by PCR amplification of bacterial plasmid-derived sequences that show minimal homology and GC content similar to that of the genome of Saccharomyces cerevisiae. The DNA fragments could also be generated by artificial DNA synthesis. Following accurate quantitation, the fragments are then combined in a certain molecular ratio, for example, 1:2:4. Other ratios are acceptable, but the ratio has to be known to ensure that there is linear amplification during the data analysis stage.
For our RNA-seq analyses, we have been using a commercial Life Technologies external RNA control consortium (ERCC) spike-in control mix (8). The phase IV test set of ERCC spike-in control mixes comprises preformulated sets of 96 polyadenylated transcripts from the ERCC plasmid reference library. They also contain a poly(A)-positive [poly(A)+] tail mimic in the DNA template. Of the transcripts, 79 have GC content ranging from 31% to 51%. The other 17 transcripts have extremely low GC content of approximately 5%. The transcripts also have diverse lengths of 273 to 2,022 nucleotides. The concentrations of the transcripts in each spike-in mix span an approximately millionfold range following a Latin-square design. As such, they allow normalization of experimental RNA samples that have very different GC contents, lengths, and abundances. The ERCC spike-in control mix is suitable for use with next-generation sequencing platforms for almost all eukaryotes, from yeast to humans. ERCC RNAs show minimal sequence homology with endogenous transcripts from sequenced eukaryotes; e.g., only 0.5% and 0.01% of sequencing reads from these RNA were aligned to the Drosophila melanogaster and human genomes. The ERCC spike-in control mix was originally developed to enable the performance assessment of technology platforms with regard to the sensitivity and linearity of amplification, but it is also useful for the purposes of comparison of RNA levels from different experimental conditions. The efficiency of enrichment of the ERCC spike-in controls during library preparation has been shown to be dependent on the RNA purification protocols. For example, the ERCC controls are less efficiently enriched by poly(A) purification than by RiboZero enrichment of RNA (9). However, as long as the same RNA enrichment method is used for the samples being compared, this should not be a concern. High consistency of the RNA-seq data obtained using ERCC controls has been seen in comparisons of duplicate library preparations from the same RNA sample and in comparisons of different studies using the poly(A) selection protocol. In other laboratories (10), technical variations in spike-in control amplification during library preparation were observed, but the variations were removed using “Remove Unwanted Variation” (RUV) normalization (discussed in more detail below).
Another type of spike-in may be an exogenous genome or epigenome. As described above, a recent study successfully used the Drosophila epigenome as a normalization control for ChIP-seq analysis of H3K79me2 in human cells (7).
HOW TO USE A SPIKE-IN CONTROL
During your experiment, you must keep track of how many cells you isolated your samples from, for each different experimental condition, ideally performing the isolation from the same number of cells for each experimental condition. If counting the cells is not possible (for example, if the experimental samples are human tissues), one could normalize back to the total DNA content of your starting sample prior to RNA isolation, ChIP, or DNA isolation, but you would have to be sure that there were no changes in chromosome ploidy or genomic instability between your experimental conditions. The spike-in control mix is added at the same amount per cell for each sample from the different experimental conditions.
Random sampling, overall library complexity, and sequencing depth always limit RNA-seq detection. In one sequencing experiment with ERCC spike-ins, 5 of the 96 control fragments were not detected due to their low concentration, where the expected read numbers for each of those 5 spike-ins were between 0.6 and 2.5 in the total 10 million reads (8). Therefore, to detect all spike-in fragments, one might want to add spike-ins to a concentration that allows over 2.5 sequencing reads for each spike-in fragment in the final output. It is also important not to add too much spike-in control; otherwise, too many sequencing reads will be spike-in control derived. A reasonable ratio of your total sample to spike-in control can be from 1,000:1 to 50:1, such that 0.1% to 2% of the sequencing reads would be derived from the spike-in.
HOW TO NORMALIZE DATA TO THE SPIKE-IN CONTROL
Sequencing read counts show a strong linear correlation (Pearson's r > 0.96) with ERCC spike-in RNA concentrations over 6 orders of magnitude, demonstrating that read counts can be used as direct estimates of RNA abundance (8). The spike-in read counts from different libraries also show strong correlation (Pearson's r > 0.98), demonstrating that spike-in read counts are not influenced by the complexity of the endogenous RNAs. When customized spike-in controls need to be designed, e.g., for ChIP-seq, sequencing reads from spike-in controls should be analyzed to ensure that they are amplified according to the ratio in which they were combined. For example, if three spike-in fragments are combined in a ratio of 1:2:4, this ratio should be reflected in the read numbers from each experimental condition, e.g., 100:200:400 from one experimental condition and 300:600:1200 from another experimental condition.
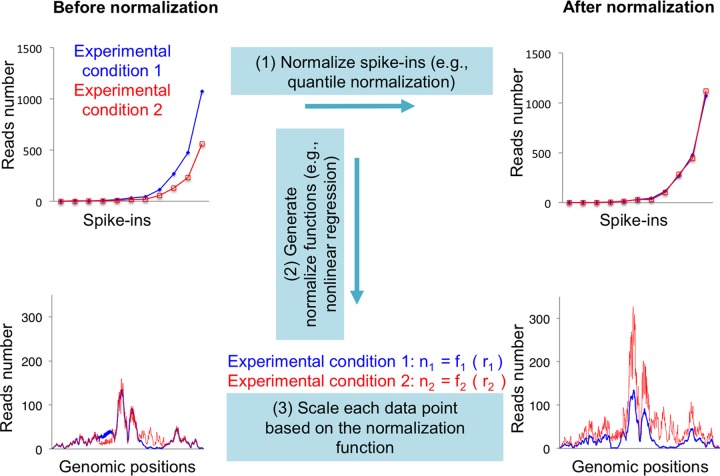
In order to analyze data based on the spike-in control, one needs to first normalize the spike-in read counts to the concentration values in the sample, e.g., 150, 300, and 600 copies per cell (Fig. 3, step 1). In a simple scenario, for example, when there is a linear correlation between read counts and sample concentration, the normalization can be performed based on a global linear scaling. By comparing the normalized read numbers to the raw read numbers of each spike-in, one is able to generate a normalization function for each experiment (Fig. 3, step 2), e.g., n = 1.5 × r for one experimental condition and n = 0.5 × r for another experimental condition, where n represents normalized read numbers and r is raw read numbers. Next, you use these functions to normalize number of reads mapped to each nucleotide across the whole genome (Fig. 3, step 3).
FIG 3.
Schematic of normalization of sequencing data with spike-in controls. At step 1, numbers of raw reads (top left, y axis) of each spike-in (top left and right, x axis) need to be normalized to be the same between experimental conditions. At step 2, by comparing numbers of normalized reads (top right, y axis) to numbers of raw reads, a normalize function can be generated specifically for each experimental condition (middle). These functions are used at step 3 to normalize read numbers at each genomic position (from bottom left to bottom right). This is an example of global-scaling normalization to a spike-in control, which is ideal for normalizing for global changes between experimental conditions.
In more-complicated situations, one may need dozens of spike-in fragments and more-sophisticated normalization algorithms in order to correct sequencing bias. For example, high-density fragments may appear to be more easily amplified than low-density fragments or the correlation between read counts and sample concentration may appear to be nonlinear. One solution is to conduct quantile normalization for spike-in read counts between samples and then use a nonlinear regression model to simulate the normalization function for each sample. Read counts for each gene or at each base pair in the genome can then be scaled based on the regression function. Another potential problem in spike-ins is library preparation effects. The proportions of reads mapping to the ERCC spike-ins have been observed to differ between libraries in some laboratories (10), although the proportion is stable between sequencing runs of the same library. This type of technical variation can be effectively eliminated using RUV normalization (10). This normalization method still uses a set of negative-control genes whose expression is assumed to not change, such as spike-in controls, but it also estimates unwanted factors for all genes. As such, the RUV method uses assumptions that are different from and more general than the assumptions of regression-based and global-scaling methods of normalization, which require unwanted technical effects to be roughly the same for the spike-in controls and for the rest of the genes (10). In this way, RUV can correct for technical errors in the amplification of the spike-in controls and is useful in situations where there are likely to be only gene-specific changes between samples. However, the RUV method is not appropriate for use when global changes between samples are suspected, because these would be normalized away by RUV. In such situations, it is useful to include spike-ins to ensure that the change in the total signal can be normalized. Therefore, we propose that spike-in normalization should be used in combination with other normalization methods such as the RUV in complicated situations.
CAN DATA SETS BE NORMALIZED RETROSPECTIVELY?
Genome-wide data have been generated for over a million different samples over the past several decades without the appropriate normalization controls. Are these data sets still useful, or should they be discarded and their interpretations ignored? We propose that these data sets can be salvaged and reinterpreted. For example, for gene expression analyses, this would require identification of marker genes (from spike-in controlled analyses) that are stably expressed across cell types, followed by renormalization of the data sets to the expression of the marker genes. Alternatively, quantitative reverse transcription-PCR (RT-PCR) analysis of specific marker gene transcripts from the relevant cell lines could be used to determine the difference in their expression levels, followed by renormalization of the data sets according to the differences in expression of these marker genes. Similarly, if regions of the genome that have quantifiable factor occupancy or histone modification patterns through all cell types/disease states can be identified, these could be used to renormalize the data sets for ChIP-seq analyses.
ACKNOWLEDGMENTS
This work was supported by NIH grants RO1GM64475 and RO1CA95641 and a CPRIT rising star award to J.K.T. and by CPRIT RP11047-C3 and NIH RO1HG007538 to W.L.
We declare that we have no conflicts of interest.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK. 2010. Elevated histone expression promotes life span extension. Mol Cell 39:724–735. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu Z, Chen K, Xia Z, Chavez M, Pal S, Seol JH, Chen CC, Li W, Tyler JK. 2014. Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev 28:396–408. doi: 10.1101/gad.233221.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lesur I, Campbell JL. 2004. The transcriptome of prematurely aging yeast cells is similar to that of telomerase-deficient cells. Mol Biol Cell 15:1297–1312. doi: 10.1091/mbc.E03-10-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wyrick JJ, Holstege FC, Jennings EG, Causton HC, Shore D, Grunstein M, Lander ES, Young RA. 1999. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- 5.Lovén J, Orlando DA, Sigova AA, Lin CY, Rahl PB, Burge CB, Levens DL, Lee TI, Young RA. 2012. Revisiting global gene expression analysis. Cell 151:476–482. doi: 10.1016/j.cell.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orlando DA, Chen MW, Brown VE, Solanki S, Choi YJ, Olson ER, Fritz CC, Bradner JE, Guenther MG. 2014. Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep 9:1163–1170. doi: 10.1016/j.celrep.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 8.Jiang L, Schlesinger F, Davis CA, Zhang Y, Li R, Salit M, Gingeras TR, Oliver B. 2011. Synthetic spike-in standards for RNA-seq experiments. Genome Res 21:1543-1551. doi: 10.1101/gr.121095.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qing T, Yu Y, Du T, Shi L. 2013. mRNA enrichment protocols determine the quantification characteristics of external RNA spike-in controls in RNA-Seq studies. Sci China Life Sci 56:134–142. doi: 10.1007/s11427-013-4437-9. [DOI] [PubMed] [Google Scholar]
- 10.Risso D, Ngai J, Speed TP, Dudoit S. 2014. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol 32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]