SUMMARY
Targeted HIV cure strategies require definition of the mechanisms that maintain the virus. Here, we tracked HIV replication and the persistence of infected CD4 T cells in individuals with natural virologic control by sequencing viruses, T cell receptor genes, HIV integration sites and cellular transcriptomes. Our results revealed three mechanisms of HIV persistence operating within distinct anatomic and functional compartments. In lymph node, we detected viruses with genetic and transcriptional attributes of active replication in both T follicular helper (TFH) cells and non-TFH memory cells. In blood, we detected inducible proviruses of archival origin among highly differentiated, clonally expanded cells. Linking the lymph node and blood was a small population of circulating cells harboring inducible proviruses of recent origin. Thus, HIV replication in lymphoid tissue, clonal expansion of infected cells, and recirculation of recently infected cells act together to maintain the virus in HIV controllers despite effective antiviral immunity.
Graphical Abstract
eTOC BLURB
HIV persistence in people who can spontaneously control the infection involves different mechanisms within distinct anatomic and functional compartments.
INTRODUCTION
During chronic HIV infection, multiple mechanisms combine to ensure the persistence of virus-infected CD4 T cells despite innate and adaptive antiviral responses. Foremost among these is ongoing virus replication, which by itself can maintain an infected CD4 T cell pool in the absence of antiretroviral therapy (ART) (Ho et al. 1995). Even under ART, however, HIV-infected CD4 T cells remain detectable in blood and lymphoid tissue. This may partly reflect the persistence of memory cells that harbor replication-competent proviruses for long periods without expressing them (Chun, Carruth, et al. 1997; Chun, Stuyver, et al. 1997; Finzi et al. 1999; Finzi et al. 1997; Hermankova et al. 2003; Wong et al. 1997). That such cells can show a resting memory phenotype has led to their identification as a latent reservoir, and has spurred development of “shock and kill” HIV cure strategies (Archin et al. 2012; Rasmussen et al. 2014; Routy et al. 2012; Sogaard et al. 2015; Spivak et al. 2014). Nevertheless, recent studies have also demonstrated clonal expansion of HIV-infected CD4 T cells under ART (Cohn et al. 2015; Maldarelli et al. 2014; Simonetti et al. 2016; Wagner et al. 2014), raising questions about the intrinsic properties of infected cells in this setting (Kim and Siliciano 2016). The further characterization of mechanisms by which HIV-infected CD4 T cells persist under different conditions in vivo has thus emerged as a key research goal.
Here we investigated the mechanisms that maintain HIV in vivo through a detailed genetic analysis of virus sequences from CD4 T cell subsets in blood and lymphoid tissue. We chose people with natural control of the virus for this study. These individuals, termed HIV controllers, represent a rare group whose HIV-specific immune responses enable them to control the virus without ART (Migueles and Connors 2015; Walker and Yu 2013). Despite evidence of ongoing virus replication in HIV controllers not receiving ART (Boufassa et al. 2014; Chun et al. 2013; Fukazawa et al. 2015; Hatano et al. 2013; Mens et al. 2010; OConnell et al. 2010; Salgado et al. 2010), prior work has shown fewer CD4 T cells containing HIV DNA (Julg et al. 2010) and replication-competent HIV (Blankson et al. 2007) in HIV controllers than in non-controllers. We reasoned that this would allow us to sample more of the total virus population in these individuals and therefore obtain a comprehensive view of the infected CD4 T cell pool. Thus, we used sequencing not only to help infer mechanisms of HIV persistence during natural virologic control, but also to elucidate cellular processes that may maintain the virus both in HIV controllers and in non-controllers.
RESULTS
Distribution of HIV among blood CD4 T cell subsets in HIV controllers
We enrolled 14 HIV controllers, defined by plasma HIV RNA levels <1,000 copies/mL during chronic infection without ART, as well as 6 non-controllers with plasma HIV RNA levels >10,000 copies/mL off ART (Table S1). Participants had been documented HIV seropositive for a median of 15.5 years, with a median of 18 years in the controller group (range 4–30) and 6 years in the non-controller group (range 2–29; Mann-Whitney P = 0.1040 for controllers vs. non-controllers). Seven of 14 controllers and 2 of 6 non-controllers carried protective class I MHC alleles including multiple HLA-B57 subtypes and HLA-B2703. Blood CD4 T cell counts were higher in the controllers than in the non-controllers (Mann-Whitney P = 0.0064).
We first characterized naïve (TN), central memory (TCM), transitional memory (TTM), and effector memory (TEM) CD4 T cells in blood as hosts for the virus in these individuals by quantifying HIV nucleic acids in FACS-sorted cell subsets (Figure S1). In HIV controllers, fluorescence-assisted clonal amplification (FCA; Figure S2) revealed that TEM and TTM cells were more likely to contain HIV DNA ex vivo than were TCM cells (Figure 1A). By adjusting for absolute cell numbers and assuming one copy of HIV DNA per infected cell, we also found that the TEM and TTM subsets accounted for most of the infected CD4 T cells in blood in HIV controllers (Figure 1B), with a median 72.6% of total HIV DNA copies in TEM cells and a median 21.2% in TTM cells. Although we hypothesized that this characteristic distribution in HIV controllers reflected greater HIV expression and replication within TEM- and TTM-like cells in vivo, quantitative RTPCR revealed low or undetectable levels of unspliced and spliced HIV RNA in all CD4 T cell subsets from these individuals (Figure 1C). We also found no correlation across individual HIV controllers between the level of HIV genomic DNA in each subset and the absolute count of each subset in blood (TCM, Spearman r = −0.200, P = 0.492; TTM, r = −0.477, P = 0.087; TEM, r = 0.033, P = 0.916), as might have been expected with ongoing, active replication and resulting cell depletion.
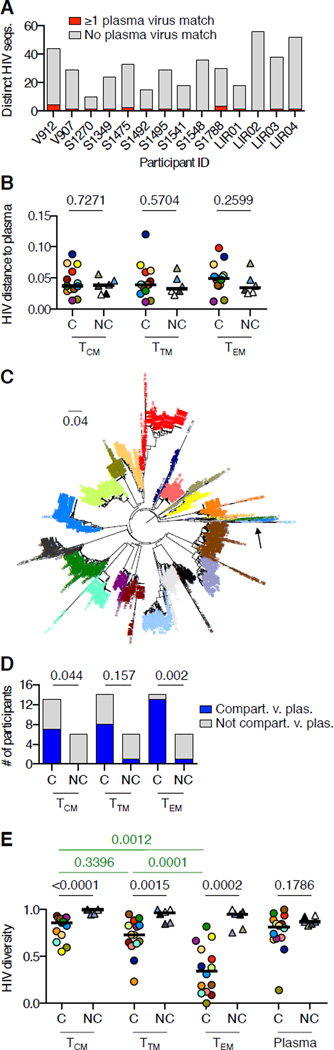
Figure 1.
HIV DNA and RNA levels in circulating CD4 T cell subsets from HIV controllers. (A) HIV DNA copies detected by FCA per 106 TN, TCM, TTM, or TEM cell equivalents. (B) Numbers of HIV-infected TN, TCM, TTM, and TEM cells per milliliter of blood, calculated by adjusting values in (A) for CD4 counts and proportions of CD4 T cells in each subset. The participant color code at right applies to all figures. Horizontal bars indicate median values in all figures. (C) Copies of unspliced (circles) and spliced (diamonds) HIV RNAs in TCM, TTM, and TEM cells from HIV controllers, measured by qRTPCR and normalized to values in (A). Undetectable values are plotted at the assay’s limit of detection (LOD) with open symbols. Wilcoxon signed rank test P values are shown. In panels (A) and (B), all Wilcoxon signed rank test P values for comparisons between TN and memory subsets in HIV controllers are <0.0001, and Mann-Whitney P values for comparisons between HIV controllers and non-controllers are <0.001 for all cell subsets. In panel (C), all Wilcoxon signed rank test P values for comparisons of unspliced or spliced RNA between subsets are >0.05. See also Figures S1–2.
HIV DNA sequence analysis in blood CD4 T cell subsets in HIV controllers
To clarify the roles of circulating CD4 TCM, TTM, and TEM cells as hosts for the virus in HIV controllers, we sequenced the single-template PCR products derived by FCA from these cell subsets and plasma virions. Compared to 608 HIV sequences from the non-controllers, the 1279 sequences from the HIV controllers showed two striking patterns (compare Figures S3 and S4). First, blood CD4 T cell-associated HIV sequences in each HIV controller differed markedly from that individual’s plasma virus sequences. HIV sequences in blood CD4 T cells from HIV controllers rarely matched plasma viruses (Figure 2A). Although genetic distances among HIV sequences might be expected to be greater in non-controllers than in controllers due to higher levels of virus replication in non-controllers, the genetic distances between cell-associated virus DNA sequences and plasma viruses in controllers were as great as those in non-controllers (Figure 2B). The large genetic distances observed in controllers were not due to dual infections because sequences from all study participants were genetically clustered by participant (Figure 2C). Moreover, whereas HIV DNA sequences in blood cells from non-controllers were genetically intermingled with plasma viruses, HIV DNA sequences in blood cells from controllers were frequently compartmentalized from plasma viruses (Figure 2D). This was especially true in the TEM subset, where compartmentalization from plasma viruses was seen in 13/14 controllers but only in 1/6 non-controllers.
Figure 2.
HIV DNA sequence analysis in circulating CD4 T cell subsets from HIV controllers. (A) Number of distinct HIV DNA sequences detected in blood CD4 T cells from each HIV controller, with number of sequences matching ≥1 plasma virus in red. (B) Average genetic distances between plasma HIV RNA sequences and HIV DNA sequences in TCM, TTM, or TEM cells in HIV controllers and non-controllers. Mann-Whitney P values are shown. (C) Phylogenetic analysis of all sequences in the study, with labels colored by participant. The arrow shows one clade in which G-to-A hypermutated sequences from multiple participants are intermingled. (D) Genetic compartmentalization between HIV DNA sequences in TCM, TTM, or TEM cells and plasma viruses in HIV controllers and non-controllers, as determined by Slatkin-Maddison testing with Bonferroni correction. Only 13 participants are shown for TCM cells because participant S1270 had no detectable HIV DNA in TCM cells. Fisher’s exact test P values for comparisons between controllers and non-controllers are shown. (E) Normalized Shannon diversities of plasma viruses and HIV DNA sequences in TCM, TTM, and TEM cells from HIV controllers and non-controllers. Mann-Whitney P values for comparisons between controllers and non-controllers are in black. Wilcoxon signed rank test P values for comparisons between subsets in HIV controllers are in green. All Wilcoxon signed rank test P values for comparisons between subsets in non-controllers are >0.05. See also Figures S3–4.
In addition to genetic divergence from plasma viruses, HIV DNA sequences in blood cells from controllers showed large clusters of identical sequences not commonly seen in non-controllers. As measured by normalized Shannon diversity, which increases with the proportion of unique sequences in a population, HIV DNA sequences were less diverse in controllers than in non-controllers within each CD4 T cell subset (Figure 2E). This was true despite similar levels of diversity in plasma viruses from the two groups (Figure 2E). Clusters of identical sequences were particularly prominent in TEM cells from controllers, with lower HIV diversity in TEM cells than in TTM or TCM cells (Figure 2E). Diversity of HIV DNA sequences in non-controllers was similar among TCM, TTM, and TEM subsets (Figure 2E). Thus, among memory CD4 T cell subsets in HIV controllers, the TEM subset accounted for most of the infected cells in blood, but generally harbored proviruses that were transcriptionally quiescent, markedly distinct from plasma viruses, and rich in clusters of identical sequences.
HIV clonality in blood CD4 T cell subsets from HIV controllers
Based on these results, we hypothesized that each cluster of identical HIV DNA sequences represented one CD4 T cell clone harboring a single HIV provirus amplified in vivo through cell proliferation. We also hypothesized that the TEM subset might be enriched for recurrent HIV sequences because memory CD4 T cells undergoing clonal expansion in vivo tend to differentiate into TEM cells. To test this, we first compared the clonotypic diversity of the three memory CD4 T cell subsets using T cell receptor beta chain (TCRB) deep sequencing. We found that TCRB diversity decreased progressively along the putative pathway of differentiation from TCM to TTM to TEM (Figure 3A), consistent with the hypothesis.
Figure 3.
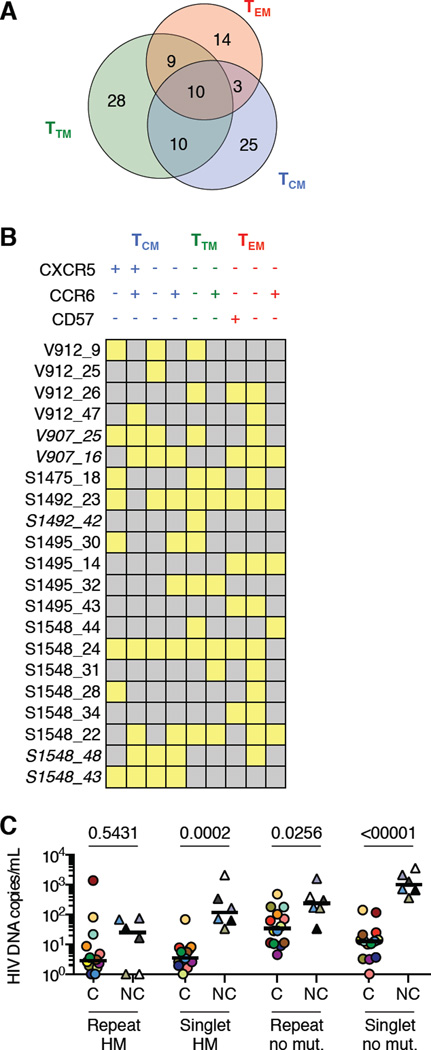
Clonality of cells and HIV DNA sequences in circulating CD4 T cell subsets from HIV controllers. (A) Normalized Shannon diversities of T cell receptor beta (TCRB) sequences from TCM, TTM, and TEM cells in HIV controllers. Wilcoxon signed rank test P values are shown. (B) Average genetic distances of plasma virus and TCM, TTM, and TEM cell-associated HIV DNA sequences from most recent common ancestral (MRCA) sequences in HIV controllers and non-controllers. Wilcoxon signed rank test P values are shown. (C) Number of recurrent HIV DNA sequences detected in circulating CD4 T cells of each HIV controller, with number of distinct sequences matching ≥1 plasma virus sequence in red. (D) Correlation between the abundance in blood of each recurrent HIV DNA sequence in circulating CD4 T cells from HIV controllers (x-axis) and the average genetic distance of that sequence to plasma viruses (y-axis). Each symbol represents one distinct sequence and is colored by participant. Spearman r and P values are shown. (E–F) HIV DNA sequences associated with expanded cellular clones in HIV controllers V907 (E) and S1349 (F), illustrated using dashed borders within phylogenetic trees. Gene locus names corresponding to the HIV integration sites in these expanded clones are shown. Each number in parentheses represents the percentage of all copies of HIV DNA in blood CD4 T cells from the individual deriving from the indicated HIV integrant. Sequences from participant V907 showing G-to-A hypermutation and associated with expanded cellular clones are shown as detached branches; other hypermutated sequences are omitted for clarity. The large sequence cluster in participant S1349 is shown separated from the tree for ease of viewing; an arrowhead shows the position of this sequence on the tree. See also Table S2.
We evaluated the alternative explanation for clusters of identical HIV DNA sequences in blood cells, whereby sequences closely related to the actively replicating virus pool and matching over the region we sequenced might accumulate through bursts of virus replication. Using plasma viruses as a genetic surrogate for actively replicating viruses, we observed the opposite pattern. In general, HIV sequences from blood TCM, TTM, and TEM cells in HIV controllers were ancestral to plasma viruses (Figure 3B). Sequences occurring at least twice rarely matched any plasma virus sequence (Figure 3C). Moreover, recurrent sequences that accounted for higher copy numbers in blood (i.e., occurred in more cells) were genetically more distant from plasma viruses (Figure 3D), with average genetic distances to plasma viruses frequently >5%. This suggested greater expansion among older HIV-infected CD4 T cell clones in HIV controllers, which was not apparent in sequences from non-controllers (Spearman r = −0.2706, P = 0.2117; not shown). Therefore, recurrent HIV DNA sequences in blood CD4 T cells from HIV controllers had genetic attributes of expanded cellular clones rather than virus replicative bursts.
We used HIV integration site analysis to prove the presence of clonally expanded, HIV-infected cells in blood from HIV controllers. Although too few infected cells to permit this analysis were available from some participants, we identified two HIV integration sites in clonally expanded cells for participant V907 and one such site for participant S1349. We then linked recurrent env sequences with their integration sites by PCR from the forward env primer to the integration site. Sequencing the env portions of these amplicons confirmed that clusters of identical sequences detected by FCA arose from expanded CD4 T cell clones (Figure 3E–F). These clones accounted for 91.3% and 95.5% of all amplifiable env DNA copies in blood CD4 T cells from these participants. Therefore, integration site analysis in HIV controllers confirmed both the presence of clonally expanded, HIV-infected cells and the predominance of these cells within the circulating, HIV-infected CD4 T cell pool.
We next characterized further the recurrent HIV DNA sequences detected in blood cells within the full HIV controller cohort. Excluding the 6/105 such sequences with <1% genetic distance from plasma viruses as possible replicative bursts, we analyzed the remaining 99 as coming from presumptive expanded clones (Table S2). These sequences were relatively abundant in blood, together accounting for between 32.7% and 96.8% of HIV DNA sequences in circulating CD4 T cells from each individual, at levels ranging between 0.32 and 1368.61 copies/mL for each sequence. Twenty-two of these sequences (22.2%) contained ≥1 stop codon associated with G-to-A hypermutation, and an additional sequence contained a frameshift in gp120. The remaining 76 sequences (76.8%) contained neither stop codons nor frameshifts over the region of env sequenced (HXB2 bases 7011–7502). We found 32 (32.3%) of these sequences in more than one memory CD4 T cell subset (Figure 4A) and many of these in multiple subsets within TCM, TTM, and TEM defined by the markers CCR6, CXCR5, and CD57 (Figure 4B). Therefore, confirmed and presumptive HIV-infected CD4 T cell clones in blood from HIV controllers showed evidence of in vivo maturation and functional differentiation. Finally, although repeated and hypermutated sequences accounted for a strikingly high proportion of all HIV DNA copies in blood CD4 T cells from HIV controllers, equal or higher absolute numbers of these sequences were detected in blood CD4 T cells from non-controllers (Figure 4C).
Figure 4.
Subset distribution and genetic attributes of HIV DNA sequences in circulating CD4 T cells. (A) Subset distribution of HIV sequences from presumptive expanded CD4 T cell clones in HIV controllers. (B) Distribution of HIV DNA sequences from presumptive expanded clones across subsets within TCM, TTM, and TEM populations defined by CXCR5, CCR6, and CD57. Each row represents one sequence, labeled by participant and a unique number. Yellow indicates that the sequence was detected in the given subset. Italics indicate G-to-A hypermutated sequences. (C) Levels of HIV DNA sequences in circulating CD4 T cells from HIV controllers and non-controllers categorized according to the number of occurrences (Repeat, >1 occurrence; Singlet, one occurrence) and the presence or absence of G-to-A hypermutation (HM, hypermutation detected; no mut., no lethal genetic defect detected). To allow display of these wide-ranging values – including several values of zero – on a logarithmic scale, each plotted value represents the measured value + 1. Sequences with lethal genetic defects other than hypermutation were rarely detected. See also Table S2.
Inducible proviruses in blood CD4 T cells from HIV controllers
Because some expanded HIV DNA sequences may have been lethally mutated outside the region we sequenced, we tested whether CD4 T cells harboring these sequences could produce virions by stimulating them through the T cell receptor and then sequencing RNA from virions released into the culture supernatant. Although these experiments revealed no virion production from most presumptive expanded clones (Figure S5, open bars), we recovered virion RNA matching two recurrent DNA sequences from TEM cells in participant S1270 (Figure S5, filled bars; Figure 5A, x-y plots, filled circles). One of these DNA sequences was highly expanded, divergent from plasma viruses, and close to the MRCA, consistent with an expanded clone carrying an ancestral provirus. The other appeared to be more recent (arrowhead), but contained a frameshift in gp120. Importantly, while abundant virion production was again detected from cells harboring the latter sequence in a repeat experiment, this was not the case for cells harboring the former sequence, even though this sequence occurred within many more cells in the culture (Figure 5B). Therefore, although virion production was occasionally inducible from proviruses bearing genetic signatures of expanded cellular clones, the inducibility of a given provirus was not always uniform among the cells that harbored it.
Figure 5.
RNA sequences of virions induced from circulating TCM, TTM, and TEM cells in HIV controllers. (A, x-y plots) Proximity of each HIV DNA sequence from circulating CD4 T cells or virions induced from TCM, TTM, and TEM cells to the participant’s MRCA (x-axis) and nearest genetic neighbor (NN) from plasma virus (y-axis). HIV DNA sequences detected once are shown as gray dots; recurrent HIV DNA sequences are shown as black circles scaled by the abundance of the sequence; and sequences detected in induced virion RNA but not in DNA from a second aliquot of cells (i.e., “unique induced” viruses) are shown as stars. Where an induced virion RNA sequence matched a recurrent HIV DNA sequence from blood cells, the circle corresponding to that DNA sequence is filled. The arrow shows one sequence from participant S1270 containing a lethal deletion within gp120. For alignment production, this deletion was filled with the participant’s consensus sequence; the measured genetic distance of this sequence to the plasma virus NN is therefore an underestimate. (A, hemispheres) Quantities of unique induced proviruses and all other HIV DNA sequences in circulating CD4 T cells from HIV controllers. Plots are scaled to show relative levels of HIV DNA in circulating CD4 T cells from the five participants. For participants with undetectable unique induced viruses from blood cells, the LOD of this measurement is shown. (B) Sequences in virions induced from blood cells in participant S1270. Cyan indicates sequences from an initial experiment; magenta indicates sequences from a repeat experiment. Sequences in which relative insertions were excised or deletions filled in alignment production are shown with gray arrows. See also Figure S5.
Importantly, we also detected a second class of inducible viruses with a distinct genetic signature in these experiments. In participants S1495, V912, and S1349, we found multiple induced viruses that had not been detected as recurrent DNA sequences in separate aliquots of the same cell populations (stars in Figure 5A, x-y plots). All but one of these “unique induced” viruses were closely related to plasma viruses and divergent from ancestral sequences, suggesting recent in vivo infection of the cells producing them. Assuming that recurrent virion RNA copies of unique induced sequences represented progeny virions from one cell, we calculated that cells harboring these proviruses accounted for 0.19–1.48% of all HIV-infected, circulating memory CD4 T cells from the individuals in whom they were detected (Figure 5A, hemispheres). Of note, unique induced viruses were detected in all circulating memory CD4 T cell subsets, and were found mainly in the two HIV controllers with the highest plasma viremia. Overall, therefore, inducible proviruses in circulating memory CD4 T cells from HIV controllers fell into two distinct categories: expanded, ancestral proviruses in TEM cells, and proviruses of recent origin in rare TCM, TTM, and TEM cells.
Lymphoid tissue viruses in HIV controllers
To help determine the origin of circulating CD4 T cells harboring unique induced viruses in HIV controllers, we characterized HIV in lymph node (LN) from four study participants (Table S3). Three of these four had detectable plasma viremia and showed high levels of HIV DNA in LN CD4 T cells. In these viremic HIV controllers, the highest levels of HIV DNA were observed in germinal center (GC) and non-germinal center (non-GC) TFH cells (Figure 6A). These TFH subsets accounted for 73.1–86.3% of the infected CD4 T cells in LN (Figure 6D). However, HIV-infected cells were also detected in LN TCM, CD57+, and TEM subsets, at levels that were higher than in TCM, TTM, and TEM cells from blood (Figure 6A). HIV DNA in both follicular and non-follicular LN memory CD4 T cells was also associated with higher HIV RNA levels than was HIV DNA in blood cells (Figure 6B–C). Detection of cell-associated HIV RNA by in situ hybridization using intact LN tissue samples confirmed that HIV RNA+ LN cells were present both inside and outside follicles in these individuals (Figure 6E).
Figure 6.
HIV DNA and RNA levels in LN CD4 T cell subsets. (A) Levels of HIV DNA measured by FCA in LN non-GC TFH and GC TFH; non-follicular LN subsets (CD57+ subset collected only for participant LIR02); and blood TCM, TTM, and TEM subsets. (B and C) Copies of unspliced (B) and spliced (C) HIV RNAs in LN memory CD4 T cell subsets, measured by qRTPCR and normalized to values in (A). Each cell subset is shown with a unique shape and colored by participant. Undetectable values are plotted at the assay LOD with open symbols. Mann-Whitney P values are shown. (A–C) include results from blood CD4 TCM, TTM, and TEM subsets that are also shown in Figure 1. (D) The percentage of all HIV DNA copies in LN memory CD4 T cells from each participant detected in each subset. (E) HIV RNA+ LN cells detected by in situ hybridization using 35S-labeled riboprobes in two study participants. White arrows indicate examples of HIV RNA+ cells. Some such cells were associated with areas of diffusely increased signal corresponding to the follicular dendritic cell network (follicular); others were outside such areas (extrafollicular). See also Figures S6–7 and Table S3.
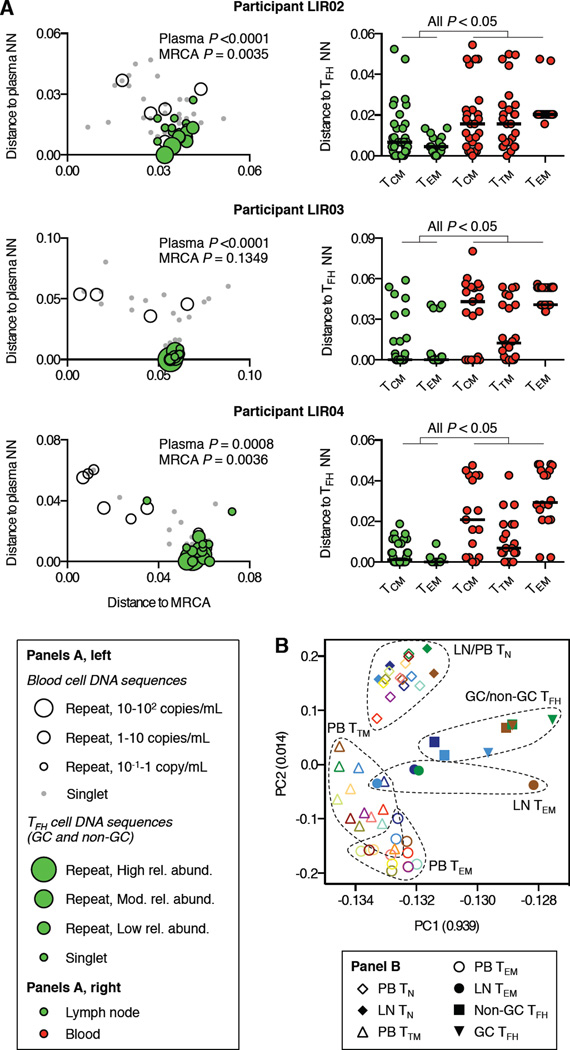
To clarify the relationship of HIV in TFH and non-TFH LN cells to viruses in blood cells and plasma, we analyzed HIV DNA sequences in LN cell subsets. We found that sequences from GC and non-GC TFH cells in the three viremic controllers (n = 113 sequences) were more closely related to plasma viruses than were HIV DNA sequences from blood cells (Figure 7A, left panels). In two participants, sequences in TFH cells were also significantly further from the MRCA than were blood cell-associated sequences. Although recurrent HIV DNA sequences were observed in TFH cells, these sequences too were closely related to plasma viruses and divergent from ancestral sequences, thus distinguishing them from recurrent HIV DNA sequences in blood cells. Importantly, analysis of 145 additional sequences from non-TFH TCM and TEM LN cells in these individuals showed a close relationship with viruses in TFH cells (Figure 7A, right panels, green symbols). By contrast, the sequences from circulating TCM, TTM, and TEM cells were significantly more distant from viruses in TFH cells (Figure 7A, right panels, red symbols). Thus, among CD4 T cells of a given maturation phenotype, we identified sharp distinctions in the genetic characteristics and transcriptional activity of cell-associated HIV populations detected in different anatomic sites. Whole transcriptome sequencing and principal component analysis (PCA) of cell subsets from these sites revealed that LN TEM cells had a transcriptional profile that was related to that of TFH cells and distinct from that of circulating CD4 TEM cells (Figure 7B).
Figure 7.
Analysis of HIV DNA sequences and host gene expression in LN CD4 T cell subsets from HIV controllers. (A, left), Proximity of each HIV DNA sequence from LN GC and non-GC TFH cells to each participant’s MRCA (x-axis) and plasma virus NN sequence (y-axis). HIV DNA sequences from blood cells are shown as in Figure 5; HIV DNA sequences from GC and non-GC TFH cells are shown as green-filled circles scaled by their relative abundance in LN. Sequences from GC and non-GC TFH cells were compared to those from blood cells for genetic distance to plasma virus NN and MRCA sequences. Mann-Whitney P values for these comparisons are shown. (A, right) Proximity of each HIV DNA sequence from non-TFH TCM and TEM LN cells and TCM, TTM, and TEM blood cells to the NN sequence from GC and non-GC TFH cells. All Mann-Whitney P values for comparisons between LN cell subsets and blood cell subsets are <0.05. (B) PCA of transcriptomes from blood (PB) and LN CD4 T cell subsets. Clusters of symbols representing samples of the same cell subset from multiple study participants are demarcated with dashed boundaries. See also Figures S6–7 and Table S3.
Finally, in one HIV controller with undetectable plasma viremia, levels of LN cell-associated HIV DNA were nearly undetectable (Table S3). In this individual, only a single, G-to-A-hypermutated copy of HIV was detected among all LN CD4 T cells studied, with no HIV DNA found in >2 × 104 TFH and GC TFH (Figure S6I). Flow cytometry confirmed the expected distribution of LN CD4 T cells among TN, TFH, and other memory subsets in this individual (Figure S6A–H). Furthermore, plasma virus sequences from this individual showed a relatively high average intragroup pairwise genetic distance (Figure S6J) and an average divergence from MRCA that was the highest out of all 20 participants (Figure S6K). Therefore, HIV-infected CD4 T cells were very rare in LN from one HIV controller even though plasma viruses showed genetic markers of diversification and ongoing evolution. This individual also showed TCM cell-associated HIV DNA sequences in blood that were more genetically distant from plasma viruses than were TEM-associated sequences. Among all 14 HIV controllers studied, the relative genetic proximity of TCM-associated HIV DNA sequences to plasma viruses was directly associated with plasma viremia (Figure S7).
DISCUSSION
In this study we investigated the mechanisms of virus persistence in HIV controllers using a combination of genetic analyses to track HIV replication and the persistence of HIV-infected cells in vivo. We found that the populations of HIV-infected CD4 T cells in blood and lymphoid tissue from these individuals differed markedly from one another. The infected cell pool in blood largely comprised archival proviruses within highly differentiated cells that appeared to be clonally expanded and that occasionally expressed virions when stimulated. In sharp contrast, the lymphoid tissue was often rich in TFH and other memory CD4 T cells bearing HIV genetic markers of recent infection and containing abundant virus transcripts. Despite these differences, however, we also detected rare circulating CD4 T cells that inducibly expressed HIV proviruses with the genetic signature of an actively replicating virus pool. These cells link the infected populations from blood and lymphoid tissue and may thus reflect the hematogenous dissemination of newly infected cells. Therefore, our findings suggest a single model in which HIV persists despite natural virologic control by three interrelated mechanisms: (1) ongoing infection of cells in lymphoid tissue, (2) survival and recirculation of some of these cells, and (3) long-term persistence of proviruses in clonally expanded cells.
Our results support this model through key advances in several areas. Consistent with previous studies of SIV-infected macaques and HIV-infected long-term nonprogressors (Fukazawa et al. 2015; Perreau et al. 2013), we detected viruses with genetic and transcriptional markers of active replication most abundantly within PD1high, TFH-enriched cell populations in hosts who control the virus spontaneously. However, by using single-copy sequence analysis to identify this virus population, we were also able to detect its dissemination to extrafollicular LN cells. This implies either HIV transmission across follicle boundaries or differentiation of infected TFH cells into non-TFH cells. Furthermore, the genetic similarity among plasma viruses, viruses in LN cells, and unique induced viruses from blood cells suggests that some infected LN cells survive their initial encounter with infectious virus long enough to recirculate. We also identified blood TEM cells that appeared to be clonally expanded as a source of archival virus in HIV controllers. Our finding that inducible proviruses of recent origin were present but rare amid the excess of archival proviruses in highly differentiated, clonally expanded blood cells explains the large genetic distance between HIV sequences in blood cells and plasma observed previously in controllers (Bailey et al. 2006; OConnell et al. 2010). Finally, although prior studies have found expanded clones of HIV-infected cells ex vivo (Cohn et al. 2015; Maldarelli et al. 2014; Wagner et al. 2014), virus production has been shown previously from only one clone in one individual (Maldarelli et al. 2014; Simonetti et al. 2016), and never in an HIV controller. Although CD4 T cell clonal proliferation may involve transcriptional processes that elicit HIV expression and thus select for lethal mutations among expanded proviruses, the dissociation of virion production from cellular stimulation supported by our findings provides one mechanism for proliferative self-renewal of HIV-producing CD4 T cells in vivo.
The heterogeneity that we observed among HIV-infected TEM cells from HIV controllers was also noteworthy. Most HIV DNA sequences in circulating TEM cells appeared to be associated with expanded cellular clones. We propose that this reflects the basic biology of TEM cells, which may also harbor infected cells that have clonally expanded in ART-treated individuals (von Stockenstrom et al. 2015). The relatively high levels of HIV DNA we found in blood TEM cells distinguish HIV controllers from ART-treated individuals (Chomont et al. 2009), and may stem from higher levels of virus replication and immune activation (Hunt et al. 2008; Hunt et al. 2011; Krishnan et al. 2014) stimulating HIV-infected cells to proliferate and differentiate in HIV controllers. At the same time, by sampling lymphoid tissue or virions induced from blood cells instead of blood cell DNA, we uncovered viruses of recent origin in rare TEM cells. Therefore, it appears that both archival and actively replicating virus populations persist within TEM cells in HIV controllers, but by distinct mechanisms. Importantly, lack of CD27 expression represents an inclusive definition for the TEM subset, and finer distinctions among subsets of CD27− cells may yield additional insights in future studies. Furthermore, the lifespan of circulating CD27− cells carrying inducible, recently acquired proviruses remains to be determined. Nonetheless, these considerations make clear that targeting less differentiated TCM and TTM cells is unlikely to eliminate all potentially infectious proviruses in HIV controllers.
Despite these diverse mechanisms of persistence, we detected profound imprints of effective antiviral defenses among HIV DNA sequences from HIV controllers. In peripheral blood, restricted replication of the virus was associated with a predominance of recurrent sequences within clonally expanded cells, a scarcity of cell-associated virus transcripts, and a small number of cells harboring unique induced viruses. While expanded G-to-A hypermutant viruses were abundant in circulating infected cells from some HIV controllers, this most likely reflects a predominance of non-replicating proviruses in these individuals, rather than excess activity of APOBEC3G (Abdel-Mohsen et al. 2013). In lymphoid tissue, the enrichment for recently infected cells containing abundant HIV transcripts within TFH populations is consistent with a key role for virus-specific CD8+ T cells in suppressing HIV replication outside follicles (Fukazawa et al. 2015). In fact, we documented this effect in individuals lacking major protective HLA-B alleles, suggesting that the suppression of extrafollicular virus replication may occur widely among HIV controllers. Finally, we propose that the relationship between plasma viremia and the genetic proximity of blood TCM-associated HIV DNA sequences to plasma viruses in HIV controllers reflects reduced dissemination of recently infected CD4 T cells in individuals with greater virologic control. When ongoing virus replication is very limited, rare recently infected cells — more readily detected by sequencing in the TCM subset than in the more heavily expanded TTM and TEM subsets — may occur too rarely for detection in a single blood sample. Taken to an extreme, the limited dissemination of recently infected CD4 T cells in blood may prevent the virus from spreading throughout the lymphoid tissue compartment, as for one participant in our study. Thus, in rare cases, natural antiviral responses may restrict the regional anatomic distribution of the virus.
Our findings reflect in vivo properties of HIV-infected CD4 T cell populations that may be generalizable to all infected individuals. Of particular interest is the uncertain significance of expanded CD4 T cell clones as barriers to cure. On the one hand, the rarity of virion production from expanded proviruses in our study mirrors results in ART-treated individuals, calling into question whether this mechanism alone could account for the near certainty of rebound viremia after ART. On the other hand, the recent temporal clustering of unique induced proviruses in our study suggests that newly infected cells recirculating from lymphoid tissue may disappear quickly if not maintained by clonal expansion. Complicating attempts to resolve these findings, however, is the evident difficulty of eliciting virus expression from some inducible proviruses in HIV controllers, as shown previously in ART-treated non-controllers (Ho et al. 2013). In addition to preventing full characterization of inducible proviruses ex vivo, this difficulty may reflect natural heterogeneity in the cellular response to stimulation that serves to protect some proviruses from antiviral defenses in vivo. Therefore, while our findings support the role of the B cell follicle in ongoing HIV replication in HIV controllers, we also demonstrate extrafollicular mechanisms that could maintain and disseminate the virus even if its replication inside follicles were disrupted. Were any approach to a functional or sterilizing cure to succeed either in HIV controllers or in non-controllers, it would need to target these multiple distinct cellular processes.
EXPERIMENTAL PROCEDURES
PBMC and LN samples
Participants were recruited from UCSF and NIAID, and gave informed consent for all procedures. Studies were approved by the UCSF and NIAID institutional review boards. PBMC were isolated from blood or leukapheresis by density gradient centrifugation. LN excisional biopsy samples were sectioned and filtered to generate single-cell suspensions.
Fluorescence-activated cell sorting (FACS)
Viable PBMC were stained and sorted on a FACSAria (BD). Subset definitions were as shown in Figure S1 for PBMC and in Figure S6 for LN cells.
Nucleic acid extraction
RNA and DNA were extracted in separate fractions from sorted cells lysed in RNAzol RT (MRC), according to the manufacturer’s instructions.
Recovery of HIV virion RNA
Virion RNA was extracted from whole plasma or pelleted virions using the QiaAmp vRNA mini kit (Qiagen), according to the manufacturer’s instructions.
Fluorescence-assisted clonal amplification (FCA) for single-copy HIV quantification and sequencing
Cell DNA or reverse-transcribed HIV virion RNA was amplified at limiting dilution in replicate PCR wells using primers targeting HXB2 positions 6908–7517, with SYBR green I as marker of dsDNA. Amplification was detected by real-time fluorescence and confirmed by melt curve (Figure S2). Single copies of HIV DNA were enumerated for each sample by limiting dilution calculations. Amplified env products were reamplified by nested PCR and Sanger sequenced. Because all primers in these PCRs targeted regions within the HIV genome, the resulting sequence data were considered to reflect all HIV DNA copies in samples from cells, including both integrated and unintegrated forms.
Quantification of HIV RNAs
Cellular total RNA samples were tested for HIV RNA by qRTPCR for unspliced (gag) RNA or transcripts spliced between the SD1 and SA4 sites (Purcell and Martin 1993). Copies of RNA were enumerated using standard curves generated from dilutions of synthetic RNAs.
HIV sequence analysis
Sanger sequence reads defining single HIV sequences spanning HXB2 base positions 7011–7502 were edited, analyzed for diversity by subset within each participant, aligned by participant, and subjected to phylogenetic analysis after identification and removal of G-to-A hypermutated sequences.
T cell receptor beta (TCRB) gene deep sequencing
Sequencing and annotation of expressed TCRB genes were performed as described (Gros et al. 2014). The normalized Shannon diversity was calculated for the set of TCRB sequences from each CD4 T cell subset by the same method used for populations of HIV sequences (see Supplemental Experimental Procedures), determining the maximum Shannon diversity for each sample based on the number of cells used in library preparation.
HIV integration site deep sequencing
Integration site analysis was performed as described (Maldarelli et al. 2014).
Sequencing of viruses induced in vitro
Sorted TCM, TTM, and TEM cells were stimulated in culture through the T cell receptor in the presence of antiretrovirals, and virions were pelleted from culture supernatants for RNA extraction after 4 and 8 days of culture. Virion RNA was reverse transcribed and amplified for single-copy sequencing by FCA.
In situ hybridization
Hybridization of 35S-labeled riboprobes for the detection of HIV RNA+ cells in LN tissue slices available from two study participants was performed as described (Rothenberger et al. 2015).
Whole transcriptome sequencing
Messenger RNA libraries were constructed as described (Sandler et al. 2014) and sequenced in 2x75-base paired-end runs on an Illumina HiSeq. High-quality reads were mapped to the Hg19 reference using Tophat (v2.0.8) with a reference annotation (Ensembl “Homo_sapiens.GRCh37.74.gtf”). Samples with low map rates were discarded. Transcript abundance was determined with Cufflinks (v2.1.1). PCA was performed using “princomp” in the R “stats” package. The abundance of each transcript was log transformed before PCA. To limit the influence of low transcript abundance levels, the level of each transcript was set to 1 if the measured level was <1.
Sequence data
Sequence data from this study can be retrieved from GenBank by searching the publication title. Integration site data can be retrieved at https://rid.ncifcrf.gov/index.php.
Supplementary Material
HIGHLIGHTS.
In HIV controllers, both TFH and non-TFH lymph node CD4 T cells contain HIV
Lymph node viruses in both TFH and non-TFH have attributes of active replication
Rare, recently infected cells that produce virus upon stimulation circulate in blood
Archival proviruses predominant in clonally expanded blood cells can be inducible
Acknowledgments
We thank the study participants, S. Kosakovsky-Pond for help with phylogenetic analysis, and C. Petrovas for helpful discussions. DD and EB are funded by the NIH Intramural Research Program. DD is also funded by the NIAID Division of AIDS and the NIH Office of AIDS Research. Additional funding came from AIDS Vaccine Discovery grant OPP1032325 from the Bill and Melinda Gates Foundation (RK), the Delaney AIDS Research Enterprise (AI096109; SDeeks), NIAID K24 (AI069994; SDeeks), the UCSF/Gladstone Institute of Virology & Immunology CFAR (P30 AI027763; SDeeks), and federal funds from the NCI (FM, SH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceptualization, EB and DD; Methodology, EB, XW, JC, TS, and DD; Investigation, EB, SDarko, LS, GW, DW, XW, AR, FL, DA, AV, DB, and KN; Formal Analysis, SDarko and JH; Resources, MH, RH, SMigueles, MC, SMoir, and SDeeks; Writing – Original Draft, EB and DD; Writing – Review and Editing, EB, SH, and DD; Supervision, RK, FM, SH, SDeeks, TS, and DD.
REFERENCES
- Abdel-Mohsen M, Raposo RA, Deng X, Li M, Liegler T, Sinclair E, Salama MS, Ghanem Hel D, Hoh R, Wong JK, et al. Expression profile of host restriction factors in HIV-1 elite controllers. Retrovirology. 2013;10:106. doi: 10.1186/1742-4690-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JR, Williams TM, Siliciano RF, Blankson JN. Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations. J Exp Med. 2006;203:1357–1369. doi: 10.1084/jem.20052319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankson JN, Bailey JR, Thayil S, Yang HC, Lassen K, Lai J, Gandhi SK, Siliciano JD, Williams TM, Siliciano RF. Isolation and characterization of replication-competent human immunodeficiency virus type 1 from a subset of elite suppressors. J Virol. 2007;81:2508–2518. doi: 10.1128/JVI.02165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boufassa F, Lechenadec J, Meyer L, Costagliola D, Hunt PW, Pereyra F, Deeks S, Pancino G, Taulera O, Lichterfeld M, et al. Blunted response to combination antiretroviral therapy in HIV elite controllers: an international HIV controller collaboration. PLoS One. 2014;9:e85516. doi: 10.1371/journal.pone.0085516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- Chun TW, Shawn Justement J, Murray D, Kim CJ, Blazkova J, Hallahan CW, Benko E, Costiniuk CT, Kandel G, Ostrowski M, et al. Effect of antiretroviral therapy on HIV reservoirs in elite controllers. J Infect Dis. 2013;208:1443–1447. doi: 10.1093/infdis/jit306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn LB, Silva IT, Oliveira TY, Rosales RA, Parrish EH, Learn GH, Hahn BH, Czartoski JL, McElrath MJ, Lehmann C, et al. HIV-1 integration landscape during latent and active infection. Cell. 2015;160:420–432. doi: 10.1016/j.cell.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- Fukazawa Y, Lum R, Okoye AA, Park H, Matsuda K, Bae JY, Hagen SI, Shoemaker R, Deleage C, Lucero C, et al. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat Med. 2015;21:132–139. doi: 10.1038/nm.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros A, Robbins PF, Yao X, Li YF, Turcotte S, Tran E, Wunderlich JR, Mixon A, Farid S, Dudley ME, et al. PD-1 identifies the patient-specific CD8(+) tumor-reactive repertoire infiltrating human tumors. J Clin Invest. 2014;124:2246–2259. doi: 10.1172/JCI73639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano H, Yukl SA, Ferre AL, Graf EH, Somsouk M, Sinclair E, Abdel-Mohsen M, Liegler T, Harvill K, Hoh R, et al. Prospective antiretroviral treatment of asymptomatic, HIV-1 infected controllers. PLoS Pathog. 2013;9:e1003691. doi: 10.1371/journal.ppat.1003691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermankova M, Siliciano JD, Zhou Y, Monie D, Chadwick K, Margolick JB, Quinn TC, Siliciano RF. Analysis of human immunodeficiency virus type 1 gene expression in latently infected resting CD4+ T lymphocytes in vivo. J Virol. 2003;77:7383–7392. doi: 10.1128/JVI.77.13.7383-7392.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, Lai J, Blankson JN, Siliciano JD, Siliciano RF. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt PW, Brenchley J, Sinclair E, McCune JM, Roland M, Page-Shafer K, Hsue P, Emu B, Krone M, Lampiris H, et al. Relationship between T cell activation and CD4+ T cell count in HIV-seropositive individuals with undetectable plasma HIV RNA levels in the absence of therapy. J Infect Dis. 2008;197:126–133. doi: 10.1086/524143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt PW, Landay AL, Sinclair E, Martinson JA, Hatano H, Emu B, Norris PJ, Busch MP, Martin JN, Brooks C, et al. A low T regulatory cell response may contribute to both viral control and generalized immune activation in HIV controllers. PLoS One. 2011;6:e15924. doi: 10.1371/journal.pone.0015924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julg B, Pereyra F, Buzon MJ, Piechocka-Trocha A, Clark MJ, Baker BM, Lian J, Miura T, Martinez-Picado J, Addo MM, et al. Infrequent recovery of HIV from but robust exogenous infection of activated CD4(+) T cells in HIV elite controllers. Clin Infect Dis. 2010;51:233–238. doi: 10.1086/653677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Siliciano RF. Reservoir expansion by T-cell proliferation may be another barrier to curing HIV infection. Proc Natl Acad Sci U S A. 2016 doi: 10.1073/pnas.1600097113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan S, Wilson EM, Sheikh V, Rupert A, Mendoza D, Yang J, Lempicki R, Migueles SA, Sereti I. Evidence for innate immune system activation in HIV type 1-infected elite controllers. J Infect Dis. 2014;209:931–939. doi: 10.1093/infdis/jit581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345:179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mens H, Kearney M, Wiegand A, Shao W, Schonning K, Gerstoft J, Obel N, Maldarelli F, Mellors JW, Benfield T, et al. HIV-1 continues to replicate and evolve in patients with natural control of HIV infection. J Virol. 2010;84:12971–12981. doi: 10.1128/JVI.00387-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migueles SA, Connors M. Success and failure of the cellular immune response against HIV-1. Nat Immunol. 2015;16:563–570. doi: 10.1038/ni.3161. [DOI] [PubMed] [Google Scholar]
- OConnell KA, Brennan TP, Bailey JR, Ray SC, Siliciano RF, Blankson JN. Control of HIV-1 in elite suppressors despite ongoing replication and evolution in plasma virus. J Virol. 2010;84:7018–7028. doi: 10.1128/JVI.00548-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med. 2013;210:143–156. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell DF, Martin MA. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol. 1993;67:6365–6378. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen TA, Tolstrup M, Brinkmann CR, Olesen R, Erikstrup C, Solomon A, Winckelmann A, Palmer S, Dinarello C, Buzon M, et al. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV. 2014;1:e13–e21. doi: 10.1016/S2352-3018(14)70014-1. [DOI] [PubMed] [Google Scholar]
- Rothenberger MK, Keele BF, Wietgrefe SW, Fletcher CV, Beilman GJ, Chipman JG, Khoruts A, Estes JD, Anderson J, Callisto SP, et al. Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proc Natl Acad Sci U S A. 2015;112:E1126–E1134. doi: 10.1073/pnas.1414926112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routy JP, Tremblay CL, Angel JB, Trottier B, Rouleau D, Baril JG, Harris M, Trottier S, Singer J, Chomont N, et al. Valproic acid in association with highly active antiretroviral therapy for reducing systemic HIV-1 reservoirs: results from a multicentre randomized clinical study. HIV Med. 2012;13:291–296. doi: 10.1111/j.1468-1293.2011.00975.x. [DOI] [PubMed] [Google Scholar]
- Salgado M, Brennan TP, OConnell KA, Bailey JR, Ray SC, Siliciano RF, Blankson JN. Evolution of the HIV-1 nef gene in HLA-B*57 positive elite suppressors. Retrovirology. 2010;7:94. doi: 10.1186/1742-4690-7-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler NG, Bosinger SE, Estes JD, Zhu RT, Tharp GK, Boritz E, Levin D, Wijeyesinghe S, Makamdop KN, del Prete GQ, et al. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature. 2014;511:601–605. doi: 10.1038/nature13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti FR, Sobolewski MD, Fyne E, Shao W, Spindler J, Hattori J, Anderson EM, Watters SA, Hill S, Wu X, et al. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci U S A. 2016 doi: 10.1073/pnas.1522675113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogaard OS, Graversen ME, Leth S, Olesen R, Brinkmann CR, Nissen SK, Kjaer AS, Schleimann MH, Denton PW, Hey-Cunningham WJ, et al. The Depsipeptide Romidepsin Reverses HIV-1 Latency In Vivo. PLoS Pathog. 2015;11:e1005142. doi: 10.1371/journal.ppat.1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivak AM, Andrade A, Eisele E, Hoh R, Bacchetti P, Bumpus NN, Emad F, Buckheit R, 3rd, McCance-Katz EF, Lai J, et al. A pilot study assessing the safety and latency-reversing activity of disulfiram in HIV-1-infected adults on antiretroviral therapy. Clin Infect Dis. 2014;58:883–890. doi: 10.1093/cid/cit813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Stockenstrom S, Odevall L, Lee E, Sinclair E, Bacchetti P, Killian M, Epling L, Shao W, Hoh R, Ho T, et al. Longitudinal Genetic Characterization Reveals That Cell Proliferation Maintains a Persistent HIV Type 1 DNA Pool During Effective HIV Therapy. J Infect Dis. 2015;212:596–607. doi: 10.1093/infdis/jiv092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner TA, McLaughlin S, Garg K, Cheung CY, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, Frenkel LM. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345:570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BD, Yu XG. Unravelling the mechanisms of durable control of HIV-1. Nat Rev Immunol. 2013;13:487–498. doi: 10.1038/nri3478. [DOI] [PubMed] [Google Scholar]
- Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, Richman DD. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.