Summary
Cancer arises from genetic alterations that invariably lead to dysregulated transcriptional programs. These dysregulated programs can cause cancer cells to become highly dependent on certain regulators of gene expression. Here we discuss how transcriptional control is disrupted by genetic alterations in cancer cells, why transcriptional dependencies can develop as a consequence of dysregulated programs, and how these dependencies provide opportunities for novel therapeutic interventions in cancer.
Introduction
Gene dysregulation is a hallmark of cancer. Recent progress in our understanding of transcription and its role in cancer pathogenesis suggest that many new insights will soon be leveraged for patient benefit. Thus, the bulk of the phenotypes, including those affecting their clinical progression and therapeutic responsiveness, are likely to be strongly regulated by the dysregulated versions of transcriptional programs operating within cancer cells. Increasingly, the molecular regulators of these programs, notably proteins involved in transcriptional control, are coming into view as attractive targets of a new generation of drugs that perturb their functions and thus the transcriptional programs that they govern.
By now, extensive cancer genome sequencing studies have revealed recurrent somatic mutations in tumor cells that affect nearly every DNA, RNA and protein component of normal transcriptional control. These findings provide insights into the genes whose alterations influence the cancer state and identify potential therapeutic targets. A number of excellent reviews describe these alterations that affect cell signaling, transcription factors, enhancer elements, chromatin regulators and chromosome structure (Garraway and Lander, 2013; Kandoth et al., 2013; Lawrence et al., 2014; Stratton et al., 2009; Sur and Taipale, 2016; Vogelstein et al., 2013; Watson et al., 2013).
An alternative approach to understanding cancer and identifying therapeutic targets is to discover the key components on which the dysregulated transcriptional programs depend in cancer cells (Figure 1). Such transcriptional dependencies are not typically identified by cancer genome sequencing, but arise through focused mechanistic studies of gene control programs operating in both normal and neoplastic cells. We describe our current views of the transcriptional programs operating in normal cells, explain how these programs are altered in tumor cells, and discuss recent insights into components of transcriptional control on which certain cancer cells become dependent.
Figure 1. Genetic changes and dysregulated gene expression programs lead to cancer cell state.
The path to a cancer cell involves genetic alterations that lead to changes in the gene expression program. The dysregulated program can create dependencies on transcriptional regulators that make the tumor cells more sensitive to inhibition of these regulators than normal cells.
Transcriptional Programs in Normal Cells
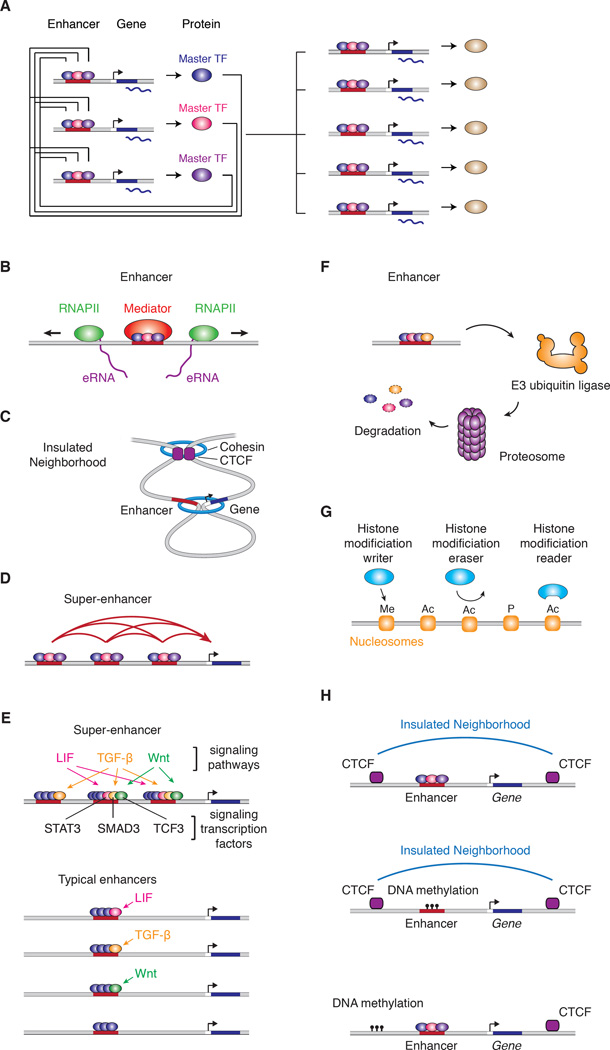
Cell identity – more specifically the identity of one or another differentiated cell type – is controlled in large part by the action of transcription factors (TFs) that recognize and bind specific sequences in the genome and thereby regulate gene expression. While nearly half of all the transcription factors encoded in the human genome are expressed in any one cell type (Vaquerizas et al., 2009), a small number of master TFs, sometimes called lineage regulators, are sufficient to establish control of the gene expression programs that define cell identity (Buganim et al., 2013; Graf and Enver, 2009; Lee and Young, 2013; Morris and Daley, 2013; Sancho-Martinez et al., 2012; Vierbuchen and Wernig, 2012; Yamanaka, 2012). Thus, the control of transcriptional programs that characterize normal differentiated cell states is dominated by these master TFs, which are expressed at high levels in selected cell types, tend to co-occupy most enhancers together with other master TFs, and typically regulate their own genes through an autoregulatory loop that forms the core transcriptional regulatory circuitry of a cell (Figure 2A) (Lee and Young, 2013). The master TFs of any one cell type can be found at the enhancers of a majority of the active cell-type specific genes, and may thus account for much of the organization of cell-type specific gene expression programs.
Figure 2. Key features of transcriptional regulation of gene expression programs.
A) In this model of cell-type specific core regulatory circuitry, master TFs co-regulate their own genes, forming an interconnected autoregulatory loop (left), as well as those of those of many other cell-type specific genes (right).
B) Enhancers are DNA elements bound by multiple TFs that recruit coactivators (such as Mediator) and RNA polymerase II, which can initiate transcription within enhancer sequences to produce enhancer RNAs (eRNAs).
C) SE constituents are physically connected. Interactions among SE constituent enhancers, and between SEs and a target gene are indicated by red arcs.
D) Human genome contains over 10,000 insulated neighborhoods, which are produced by multimerization of CTCF bound to two sites and reinforced with cohesin; enhancer-gene interactions occur predominantly within these neighborhoods.
E) SEs are enriched for signaling transcription factors, and their associated genes, which tend to play prominent roles in cell identity, are thus especially responsive to signaling.
F) Signaling TFs and other TFs that play important regulatory roles are themselves regulated by ubiquitin and proteasome-mediated destruction.
G) Chromatin regulators that act through “writing”, “erasing” or “reading” histone modifications.
H) DNA methylation can contribute to gene control by causing loss of TF binding at enhancers and loss of CTCF at insulated neighborhood loop anchors.
The master TFs bind cooperatively to enhancer DNA elements and recruit coactivators and the transcription apparatus (Bulger and Groudine, 2011; Levine et al., 2014; Long et al., 2016; Malik and Roeder, 2010; Ong and Corces, 2011; Spitz and Furlong, 2012). These TFs often activate transcription from the enhancer elements themselves (Figure 2B), producing enhancer RNAs (eRNAs) that bind certain TFs and cofactors and contribute to enhancer maintenance and dynamics (Lai et al., 2013; Li et al., 2016). Enhancers, which tend to be cell-type specific because they are generally established by cell-type specific master TFs, have been mapped in a broad spectrum of human tissue types by using epigenetic marks associated with enhancer activity (ENCODE Project Consortium et al., 2012; Roadmap Epigenomics et al., 2015).
Bound by master TFs, clusters of enhancers known as super-enhancers (SEs) regulate genes that play prominent roles in cell identity or specialized cellular function (Chapuy et al., 2013; Hnisz et al., 2013; Hnisz et al., 2015; Whyte et al., 2013). Enhancer-associated proteins and RNAs, including TFs, co-factors such as Mediator, chromatin regulators, signaling factors, RNA polymerase II (RNAPII), enhancer-associated chromatin marks (H3K27Ac) and eRNAs, are all found at especially high density at the constituent enhancers of SEs. The constituent enhancers of SEs physically associate with one another (Figure 2C) (Dowen et al., 2014; Ji et al., 2016; Kieffer-Kwon et al., 2013) and can function as independent or interdependent components of these large transcription-regulating complexes to drive high-level expression of their associated genes (Hah et al., 2015; Hay et al., 2016; Hnisz et al., 2015; Jiang et al., 2016; Shin et al., 2016).
Enhancers, and super-enhancers, become physically juxtaposed to target gene promoters by looping of the chromatin and, having become so, stimulate transcription from these promoters. Although enhancers can activate any gene, they are physically and functionally constrained to act within insulated neighborhoods (Figure 2D) (Hnisz et al., 2016a). Insulated neighborhoods are chromosomal loop structures formed by the interaction of two DNA sites bound by the CTCF protein and occupied by the cohesin complex. These chromosomal neighborhoods engender specific enhancer-gene interactions and are essential for normal gene activation and repression. The CTCF-CTCF loops that form insulated neighborhoods are the mechanistic basis of higher-order chromosome structures, such as topologically associating domains (TADs), and form a chromosome scaffold that is largely preserved throughout development (Gibcus and Dekker, 2013; Gorkin et al., 2014; Phillips-Cremins and Corces, 2013).
Normal cell states depend on signals received from the tissue microenvironments. Much of this contextual information is delivered by signaling pathways to SEs and, to a lesser extent, to typical enhancers (Figure 2E). SEs have been shown to integrate input from Wnt, TGFβ and LIF signaling pathways operating within embryonic stem cells (ESCs) (Hnisz et al., 2015) and BDNF and KCl signaling at c-Fos in neurons (Joo et al., 2016). This signal integration is thought to be a consequence of the ability of master TFs to recruit signal-activated TFs to enhancer sites previously established by the master TFs (Mullen et al., 2011; Trompouki et al., 2011). The extent to which extracellular and intracellular information is delivered to enhancers is underappreciated, due in part to the paucity of literature on nuclear signaling relative to cytoplasmic sequencing.
Enhancers that are responsive to various types of afferent signaling are thought to be highly dynamic because the activation activity of signaling TFs is linked to their destruction (Figure 2F). For example, activated TGFβ and BMP receptor kinases phosphorylate Smad transcription factors, which become fully functional transcriptional activators after being further phosphorylated by the transcriptional cyclin-dependent kinases CDK8 and CDK9. However, following their initial activation, these phosphorylated Smads are recognized by specific ubiquitin ligases operating in the nucleus, leading to their proteasome-mediated destruction (Alarcon et al., 2009). Remarkably, ubiquitylation and degradation of many transcriptional activators occurs at enhancer/promoter sites and can be required for efficient transcription (Geng et al., 2012; Thomas and Tyers, 2000).
Multiple cyclin-dependent kinases (CDKs), including CDK7, 8, 9, 12 and 13, are dynamic effectors of gene control and function, in part, by phosphorylating serine residues in the C-terminal domain of RNA polymerase II (Eick and Geyer, 2013). For example, CDK7 and CDK9 contribute to control of transcription initiation and elongation, respectively (Jonkers and Lis, 2015). Transcription factors may thus control initiation and/or elongation by their interactions with these CDKs.
DNA is packaged into nucleosomes, which consist of histones that are substrates for chromatin regulators that can modify various amino acid residues or bind in a modification-dependent manner to these histones (Figure 2G). The roles of these diverse chromatin regulators in gene control have been reviewed extensively elsewhere (Campos and Reinberg, 2009; Kouzarides, 2007; Piunti and Shilatifard, 2016; Soshnev et al., 2016; Tessarz and Kouzarides, 2014). The positioning of nucleosomes on DNA can also influence gene control, for example by limiting access of TFs to regulatory sequences, and ATP-dependent remodeling complexes influence transcriptional states by mobilizing nucleosomes (Kadoch and Crabtree, 2015). The common functional theme of these regulators is that they facilitate maintenance of positive or negative gene expression states.
DNA methylation contributes to gene control at three levels. Methylation of enhancer and promoter sites can prevent TF binding and thus silences genes (Figure 2H) (Ziller et al., 2013). Methylation of CTCF loop anchor sites prevents CTCF binding and can thus alter insulated neighborhood structure (Ghirlando and Felsenfeld, 2016; Liu et al., 2016b). Cytosine methylation and hydroxymethylation present spatially positioned chemical motifs that can be recognized by chromatin-associated proteins (e.g., meCP2), thereby influencing transcriptional regulation (Baubec et al., 2013; Liu et al., 2016a; Mellen et al., 2012).
In summary, normal cells have transcriptional programs that are established and maintained by master transcription factors that regulate genes by binding specific enhancer elements, which in turn interact with genes within insulated neighborhoods. The maintenance of normal cell states depends on the tissue environment, and such information is delivered by signaling pathways ultimately to enhancers. Maintenance of cell identity and dynamics of cell states also depend on a large number of histone readers, writers and erasers, as well as regulators of DNA methylation, that together ensure chromatin states are appropriate for positive and negative gene regulation.
Transcriptional dysregulation in cancer
The complements of genetic alterations that collaborate to transform normal cells into neoplastic derivatives exhibit a high degree of tissue specificity. Tissuespecific enhancers are structurally altered to drive expression of oncogenes (e.g. TMPRSS2-ERG in prostate cancer, IgH-locus alterations in B-cell malignancies, TCR-locus alterations in T-cell malignancies) (Sur and Taipale, 2016). In addition, many oncogenic signaling pathways require cell-specific chromatin contexts (e.g., NOTCH1 activation in T-cell but not B-cell leukemia, and EZH2 activation in B-cell but not T-cell lymphoma). Because many tissue-specific differentiation programs (specifically those that define cell identity) persist in cancer cells, it is clear that cancer arises from the collaborative interplay of oncogenic events acquired during multi-step tumor formation with the tissue-specifying gene expression programs that survive neoplastic progression and continue to influence cancer cell behavior.
This transcriptional dysregulation arises in cancer from disease-defining genetic alterations either indirectly, via mutation of signaling factors converging on transcriptional control, or directly, via genetic alterations in gene control factors themselves. Cancer-associated genetic alterations can affect proteins participating in nearly all levels of transcriptional control, including trans-factors (transcription factors, signaling proteins, cofactors, chromatin regulators and chromosome structuring proteins) and cis-elements (enhancers, promoters and insulators) (Figure 3). Many excellent reviews have described an ever-expanding catalogue of these alterations (Bywater et al., 2013; Garraway and Lander, 2013; Kandoth et al., 2013; Lawrence et al., 2014; Stratton et al., 2009; Sur and Taipale, 2016; Vogelstein et al., 2013; Watson et al., 2013). Here we discuss a subset of these alterations, specifically those that lead to the most profound changes in the gene expression program, thereby driving the malignant cell state. We focus on these programs because they may direct the discovery and development of new classes of cancer therapeutics designed to target vulnerabilities of cancer cells, in particular their addiction to certain transcriptional programs.
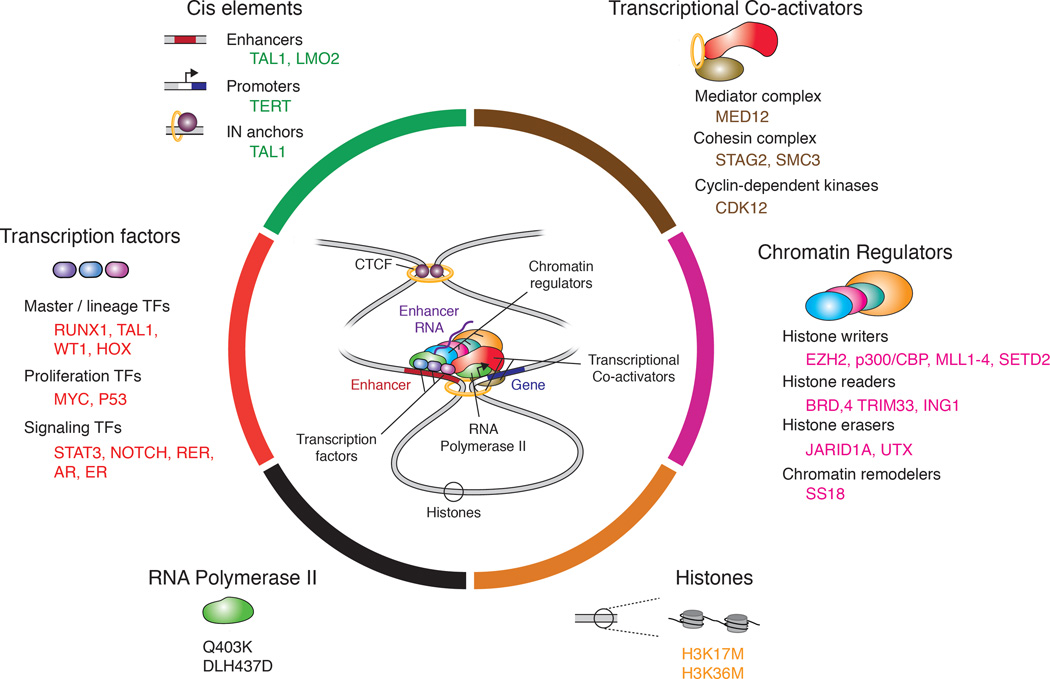
Figure 3. Components of gene control altered in cancer.
Examples of three types of cis-factors (enhancers, promoters and insulators) and various trans-factors (transcription factors, cofactors, chromatin regulators, RNA polymerase II and histones) that acquire recurrent somatic mutations in cancer cells.
Trans-factors
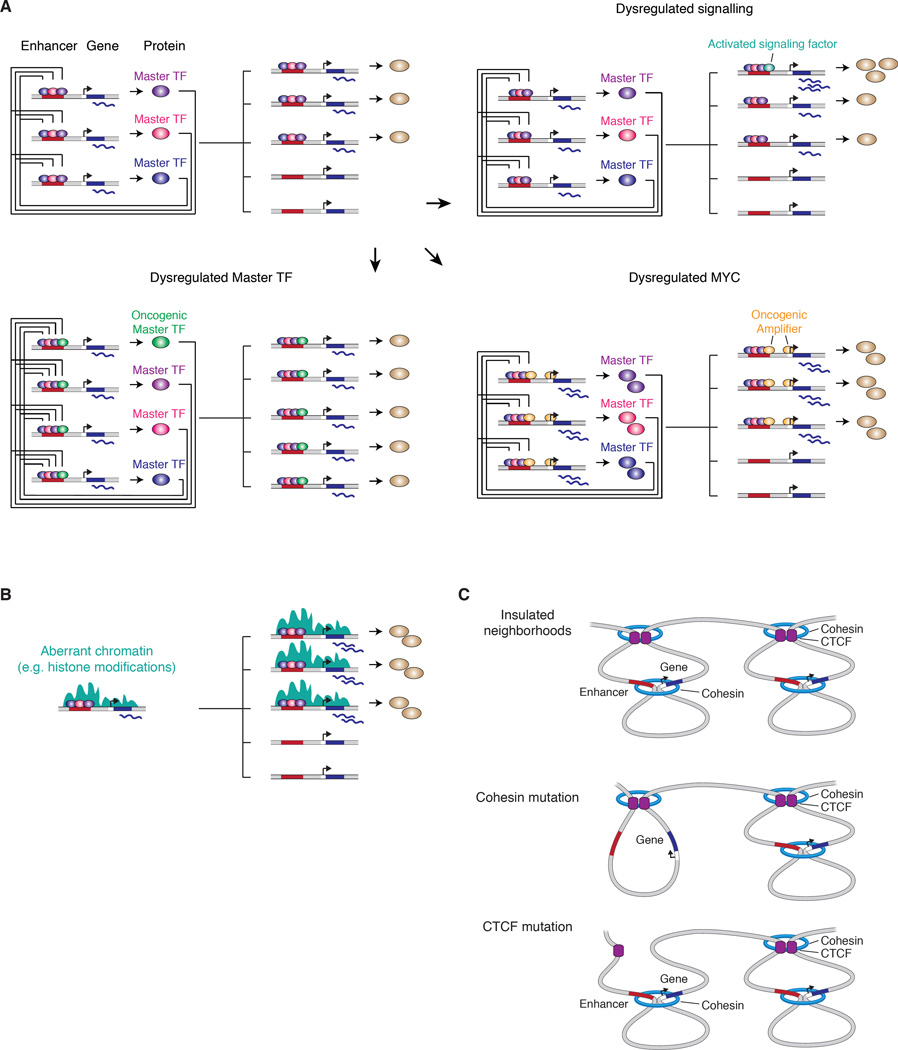
The TFs that are deregulated in cancer cells and have the potential to produce profound changes in gene expression programs can be considered to fall into three classes: master TFs involved in organizing cell identity, proliferation control TFs that amplify transcriptional output, and signaling TFs involved in dynamic changes in the control machinery occurring in response to extracellular signals. The activation of a master TF that is normally expressed in early development, such as the pluripotency TF OCT4, or a master TF that is normally expressed early in a specific lineage, such as TAL1 in T cells, can alter core regulatory circuitry and activate additional genes that are normally expressed in more embryonic states (Figure 4A) (Sanda et al., 2012). Genes encoding the MYC and TP53 proliferation control TFs, a classic oncogene and tumor suppressor gene, are among the most frequently mutated genes in cancer. MYC can have profound effects because it can function to amplify the entire gene expression program (Figure 4A) (Lin et al., 2012; Nie et al., 2012), and P53 is a powerful tumor suppressor because of its ability to arrest progress through the cell cycle or induce apoptosis (Lane and Levine, 2010). Dysregulation of signaling pathways is a common feature of cancer cells; a dysregulated signaling TF can profoundly change the gene expression program through its binding to enhancers occupied by master TFs (Figure 4A) (Mullen et al., 2011; Trompouki et al., 2011), and dysregulated signaling can even stimulate super-enhancer formation (Brown et al., 2014; Hnisz et al., 2015).
Figure 4. Common mechanisms of dysregulation of gene expression programs in cancer involving trans-factors.
A) Model of a transcriptional regulatory circuit controlling the gene expression program in normal cells is shown top left. Dysregulation of gene expression programs in cancer cells can occur through dysregulation an oncogenic master TFs (bottom left), dysregulation of a transcriptional amplifier (e.g. MYC) (bottom right), and dysregulated signaling (top right).
B) Model of aberrant chromatin modification affecting gene expression programs in cancer cells.
C) Model of effect of cohesin mutations and CTCF mutations on enhancer-promoter interactions and insulated neighborhoods in cancer cells.
These TFs signal to RNAPII through transcriptional cofactors, defined here as regulatory components that do not bind directly to DNA in a sequence-specific manner. Exemplifying this class of transcriptional signaling proteins are the components of the Mediator complex, which is recruited to enhancer-promoter regions by TFs in the context of transcription activation (Allen and Taatjes, 2015; Kagey et al., 2010). Genetic alterations of Mediator complex-encoding genes are observed frequently in prostate cancer and in many uterine myomas (Allen and Taatjes, 2015; Barbieri et al., 2012; Makinen et al., 2011). Beyond these discrete diseases, the genome-wide activities of Mediator in gene control would be expected to function broadly in all cancer-associated transcription. Interestingly, few cancer-associated alterations are identified in the core RNAPII complex itself (Clark et al., 2016), suggesting that coordinated transcriptional signaling upstream from polymerase favors the neoplastic cell state more than alterations of this core complex.
Efficient transcriptional signaling from enhancers to promoters is often chromatin-dependent, being mediated by specialized transcriptional cofactors that physically associate with and/or biochemically modify the genome to reinforce gene activation or repression. Chromatin regulators function globally, so their dysregulation can also have profound effects on the gene expression program of cells (Jones et al., 2016). In some tumors, chromatin regulators have become fused to transcriptional cofactors, producing gene-specific effects, such as those observed in acute lymphoblastic leukemia cells with MLL-AF4 fusions (Figure 4B) (Guenther et al., 2008; Krivtsov et al., 2008).
Insulated neighborhoods contribute to proper positive and negative gene control, so alterations in chromosome-structuring proteins that establish and maintain insulated neighborhood boundaries would be expected to have profound effects on a cell’s overall gene expression program. Cancer genome sequencing has revealed that somatic mutations occur in CTCF and cohesin coding sequences in various solid tumors and leukemias (Lawrence et al., 2014), and it seems likely that these mutations contribute to oncogenesis by altering insulated neighborhoods throughout the genome, perhaps rendering chromatin generally more permissive to oncogenic transcriptional signaling (Figure 4C) (Viny et al., 2015).
Cis-elements
Cis-regulatory elements within the genome that contribute to cancer pathogenesis were first recognized in pioneering studies of cancer-associated chromosomal translocation [e.g. IgH-MYC in Burkitt’s lymphoma (Taub et al., 1982)]. Following early efforts in cancer genome sequencing, which concentrated on coding regions of the human genome, recent focused and genome-wide sequencing efforts have revealed frequent alteration of cis-elements in both solid and hematological tumors. Two types of cis-elements that play prominent roles in cancer biology - SEs and insulators – are discussed here.
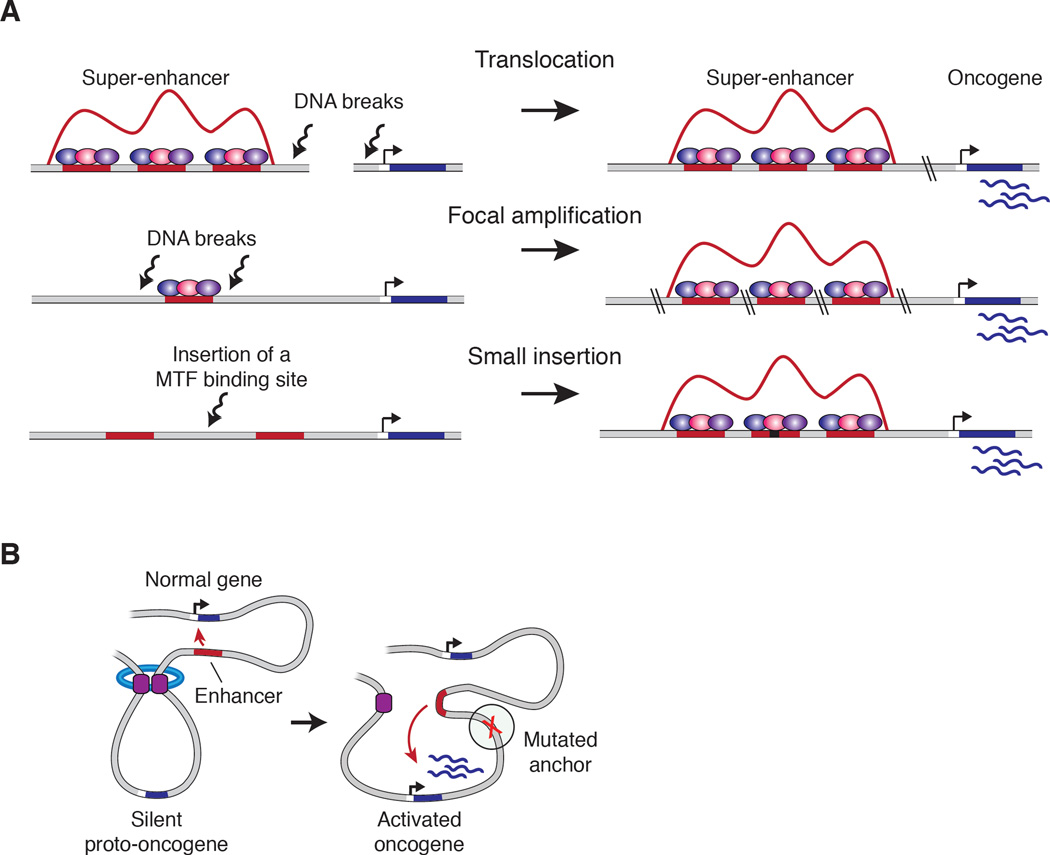
In normal cells, SEs – which, as we noted above are large clusters of enhancers that bind high densities of transcriptional components - control genes that play prominent roles in specific cell identities. Tumor cells acquire SEs at oncogenic driver genes and they do so through many different mechanisms (Figure 5A) (Chapuy et al., 2013; Drier et al., 2016; Hnisz et al., 2013; Kennedy et al., 2015; Loven et al., 2013; Tomazou et al., 2015; Wang et al., 2015; Yang et al., 2015). The genetic mechanisms that lead to SE acquisition in cancer include DNA translocation (Affer et al., 2014; Drier et al., 2016; Groschel et al., 2014; Northcott et al., 2014; Walker et al., 2014), focal amplification (Hnisz et al., 2013; Shi et al., 2013; Zhang et al., 2016b), and nucleation by small INDELs that create master TF binding sites (Mansour et al., 2014). Additional, epigenomic mechanisms also lead to SE formation in cancer, such as those associated with oncogenic TF overexpression (Hnisz et al., 2013), the global function of oncogenic TF fusions (e.g. EWS-FLI) (Kennedy et al., 2015; Tomazou et al., 2015), and the consequences of upstream oncogenic signaling (e.g., RAS-dependent signaling to chromatin) (Nabet et al., 2015).
Figure 5. Common mechanisms of dysregulation of gene expression programs in cancer involving cis-factors.
A) Mechanisms leading to the acquisition of super-enhancers to drive oncogenes in cancer cells: translocation of an existing super-enhancer, focal amplification of an enhancer element, and nucleation of super-enhancer through somatic insertion of transcription factor binding sites.
B) Activation of silent proto-oncogenes by somatic mutations that disrupt insulated neighborhood anchor sites.
Mutations that alter insulator sequences of oncogene-containing insulated neighborhoods appear to make important contributions to the dysregulation of gene expression observed in some cancers (Figure 5B) (Flavahan et al., 2016; Hnisz et al., 2016b; Katainen et al., 2015). Somatic mutations occur frequently and recurrently in loop anchors of oncogene-containing insulated neighborhoods in a broad spectrum of cancer cells. Thus, the CTCF DNA-binding motif in loop anchor regions is among the most altered human transcription factor-binding sequences in cancer cells (Hnisz et al., 2016b; Ji et al., 2016; Katainen et al., 2015). DNA hypermethylation occurs in some cancer cells, and tumor-specific DNA methylation has been implicated in the disruption of CTCF binding, alterations of chromosome structure and dysregulation of oncogene expression in a subset of gliomas (Flavahan et al., 2016). Moreover, chromosomal rearrangements that disrupt insulated neighborhoods can activate oncogenes without altering the sequences of the oncogenes themselves (Groschel et al., 2014; Hnisz et al., 2016b).
Transcriptional addiction and cancer therapeutics
The concept of oncogene addiction (Weinstein and Joe, 2006) refers to the behavior of cancer cells that exhibit an absolute dependence on oncogenes that were initially acquired during multi-step tumorigenesis and remain critical to the ongoing proliferation and viability of these cells long after they have progressed to a fully neoplastic state. As of late, this concept has been extended to include other changes acquired during tumor progression that fostered the early development of a tumor and continue to be absolutely essential to its continued growth. Included among these are the dysregulated transcriptional programs operating in certain tumor cells, yielding the concept of transcriptional addiction.
Various types of transcriptional addiction appear to operate in specific subsets of cancer. Thus, the majority of human cancers exhibit genetic amplification or transcriptional dysregulation of MYC, which is accompanied by an anabolic transcriptional response driving proliferation and metabolic adaptation (Beroukhim et al., 2010). Research in model systems of MYC addiction and withdrawal have validated addiction to this master regulatory transcription factor in solid and hematologic malignancies, both with and without structural changes to the MYC locus itself (Felsher and Bishop, 1999; Jain et al., 2002; Soucek et al., 2008; Soucek et al., 2013). As for many TFs, direct pharmacologic inhibition of MYC remains an elusive challenge in drug discovery. We discuss below the strategy of leveraging mechanistic insights into transcriptional dysregulation toward a more immediate therapeutic benefit in cancer.
Direct inhibition of oncogenic TFs and cofactors
Gene-targeted therapy has emerged as a paradigm of cancer medicine. Where available and where actionable, somatic alterations of driver oncogenes has provided strong guidance to the discovery and focused development of cancer therapeutics (Darnell, 2002; Pagliarini et al., 2015; Stuart and Sellers, 2009). Among the large number of somatically altered oncogenic transcription factors, only very few have been successfully approached by coordinated efforts in drug discovery. Most compounds of this class are, at best, experimental tools for the study of TFs in cell biology, including our own work to inhibit the NOTCH1 transactivation complex with constrained alpha helical peptides (Moellering et al., 2009). Nonetheless, in the longer term, small molecule agents directed at oncogenic TFs have the promise to confer significant clinical responses for patients bearing certain types of genetically defined cancers.
Among the most impactful examples of direct inhibition of an oncogenic TF is the development of all-trans retinoic acid (ATRA) as therapy for acute promyelocytic leukemia (APML). ATRA was initially investigated owing to phenotypic evidence of differentiation in cultivated APML cells, an observation made without the guidance of cancer genetics. In 1985, a 5 year-old girl with anthracycline-refractory APML was administered ATRA at Shanghai Children’s Hospital, achieving a complete remission (CR) and ultimately long-term remission from ATRA with chemotherapy (Wang and Chen, 2008). A first case series reported by Zhen-Yi Wang three years later would establish ATRA as a highly effective therapy for APML, conferring CR in 23 of 24 patients as a single agent (Huang et al., 1988). Eleven years after the identification of the t(15;17) translocation by Rowley (Rowley et al., 1977), and seven years after the development of ATRA, Chen and colleagues reported that the reciprocal t(15;17) translocation encodes a novel oncogene comprising the gene encoding the retinoic acid receptor alpha (RARA) fused to a second gene (PML) specifying a novel Krüppel-like zinc finger protein (Chen et al., 1993). The ensuing decade of laboratory research would firmly establish ATRA as targeted therapy for APML via modulation and destabilization of the encoded chimeric PML-RARA oncogenic transcription factor (Wang and Chen, 2008). Notably, innovation in clinical investigation would ultimately pair ATRA with arsenic trioxide (also associated with PML-RARA degradation) in low- to intermediate-risk APML as effective first-line treatment, equivalent in efficacy to chemotherapy in this disease (Lo-Coco et al., 2013). APML exemplifies biology of transcriptional addiction, revealed through the effective development of transcriptional therapy with curative intent.
An emerging example of direct inhibition of an oncogenic transcriptional cofactor is bromodomain inhibition in carcinomas harboring fusions of BET bromodomain co-activators. Among the most aggressive subtypes of lung and head and neck cancer are tumors expressing the chimeric, oncogenic transcription factors BRD4-NUT or BRD3-NUT (so-called NUT midline carcinoma or NMC). NMC is a poorly differentiated, chemoresistant and aggressive malignancy that lacks effective, FDA-approved therapy. In 2010, we reported the first effective inhibitors of human bromodomains targeting the BET family (BRD2, BRD3 and BRD4), exemplified by the chemical probe JQ1 (Filippakopoulos et al., 2010). Using preclinical models of NMC, JQ1 was shown to displace BRD4 from chromatin, resulting in potent and irreversible squamous differentiation. In murine models harboring primary human NMC xenografts, JQ1 prompted a robust pro-differentiation and anti-proliferative response associated with durable responses by PET-CT imaging. Based on this rationale, drug-like derivatives of JQ1 have been transitioned to clinical investigation by our group and others. Early reports of index trials confirm unambiguous antitumor activity in advanced disease (Stathis et al., 2016).
Targeting tissue identity and homeostasis
Oncogenic events occur in the context of cell identity, which is established and maintained by TF-defined core regulatory circuits and signals from the tissue environment (Lee and Young, 2013). Targeting transcriptional identity has thus emerged as an important therapeutic strategy in cancer. Initial efforts have attempted to deplete the tissue type in which the tumor arose, or interfere with the signaling pathways that contribute to cell identity by connecting to cell-type-specific core regulatory circuits.
Tissue depletion may be accomplished surgically, as with excision of neoplastic and surrounding unaffected tissue in mastectomy or radical prostatectomy. Medical approaches also exist, as with anti-CD20 monoclonal antibody therapy or CD19-directed chimeric antigen receptor T-cell (CART) therapy to eradicate B-cell acute lymphoblastic leukemia (B-ALL). A consequence of B-cell depletion therapy is the temporary or permanent depletion of systemic, normal B-cells as a considerable side effect.
In contrast to tissue depletion, direct targeting of cell identity undermines the requisite interaction of oncogenic and cellular circuitry. The best-studied example of this strategy led to the development of nuclear hormone receptor antagonists. Nuclear hormone receptors facilitate differentiation and growth of discrete tissues during development. In post-pubertal adults, the estrogen receptor (ER) and androgen receptor (AR) TFs maintain tissue homeostasis and sex hormone responsive function of breast and prostate tissues, respectively, among other tissues. In low-grade breast and prostate cancer expressing ER and AR, respectively, these TFs are not oncogenes per se, and are genetically unaltered in the majority of patients. Indeed, these tumors are characterized each by diverse genetic alterations to oncogenes and tumor suppressors, the majority of which lack direct-acting agents (Kandoth et al., 2013; Lawrence et al., 2014; Vogelstein et al., 2013). The impactful and important contribution of anti-estrogen and anti-androgen therapy in these diseases, then, defines the opportunity of disrupting transcriptional pathways of cellular identity alone and in combination with oncogene-targeted therapy as available. Further, the broad use of glucocorticoids in lymphoid malignancies exemplifies the same conceptual strategy (Inaba and Pui, 2010), and establishes the feasibility of agonizing a core transcriptional pathway for therapeutic benefit.
Context-specific transcriptional dependencies
As discussed earlier, eukaryotic transcription is a dynamic network with multiprotein complexes collaborating as nodes of activating, repressing, remodeling and insulating function. More than a thousand human proteins contribute to gene control at the level of nuclear chromatin, spatially distributed to tens of thousands of sites in the human genome (ENCODE Project Consortium et al., 2012; Levine et al., 2014; Roadmap Epigenomics et al., 2015). Despite this complexity, specific oncogenic impulses can engender exceptional reliance on discrete protein complexes and even individual factors. The identification, validation and prosecution of these targets can yield important mechanistic insights and therapeutic opportunities (Table 1).
Table 1.
Examples of transcriptional dependencies that occur as a consequence of transcriptional dysregulation
| Cancer type | Component of transcriptional control perturbed by genetic change |
Genetic change | Transcriptional dependency |
|---|---|---|---|
| T cell leukemia | Master transcription factor |
TAL1 overexpression |
CDK7 |
| Multiple myeloma | Enhancer/ Super-enhancer |
IgH-MYC translocation |
BRD4 |
| Glioma | Insulated neighborhood anchor |
IDH1 mutation |
PDGFRA |
| Mixed lineage leukemia | Chromatin regulator |
MLL-AF9 fusion |
DOT1L BRD4 |
| T cell leukemia | Signaling factor |
NOTCH1 mutation |
BRD4 |
Indeed, our research on BET bromodomains began with a mechanistic hypothesis, specifically that BRD4 might mediate transcriptional addiction to MYC. Because MYC had been shown to play a key role in the transcriptional elongation operating at many genes, we hypothesized that BRD4 acted by mediating chromatin-dependent transcription elongation signaling to RNAP II. BRD4 possesses twin acetyl-lysine recognition domains (bromodomains), suggesting that BRD4 might localize to hyperacetylated regions of euchromatin occupied in cancer by MYC and associated lysine acetyltransferases. BRD4 binds the pTEFb elongation factor, establishing the possibility that BRD4 may function to facilitate elongation downstream of MYC function in cancer (Bisgrove et al., 2007; Rahl et al., 2010). Indeed, in models of MYC-addicted hematologic cancers, BRD4 localizes with MYC throughout the active genome, and contributes to the MYC-mediated proximal promoter pause release that enables elevated transcriptional elongation (Chapuy et al., 2013; Loven et al., 2013). These mechanistic studies and accompanying translational research establish BRD4 inhibition as a therapeutic strategy to inhibit MYC-dependent transcriptional signaling in multiple myeloma, diffuse large B-cell lymphoma and mixed lineage leukemia (MLL) (Chapuy et al., 2013; Dawson et al., 2011; Delmore et al., 2011; Zuber et al., 2011).
Of relevance to the present subject of targeting transcriptional addiction in cancer, these studies established that pharmacologic inhibition of a transcription factor or co-factor that is presumed to act widely on countless genes throughout the genome can nevertheless exert highly selective effects on gene control. Thus, the mechanistic investigation of this phenomenon identified regions of disproportionately high levels BRD4 occupancy in cis-regulatory regions of many genes that were associated with massive Mediator enrichment, i.e., super-enhancers (Loven et al., 2013). Moreover, BRD4 function has emerged as a transcriptional addiction in MYCN-amplified neuroblastomas (Puissant et al., 2013), a positive regulator of anti-apoptotic gene expression in AMLs (Dawson et al., 2011), and as a mediator of resistance to Notch pathway inhibition in T-ALLs (Yashiro-Ohtani et al., 2014).
Several features of TF gene regulation are thought to contribute to the exceptional sensitivity of large SEs to BRD4 inhibition in tumor cells. TFs and their mRNAs generally have short half-lives, so the genes that encode key TFs may evolve SEs in order to maintain a high transcriptional output of these short-lived proteins (Figure 6A). Many TFs bind and auto-regulate the genes that encode them, so that disruption of TF levels may have an especially pronounced effect on auto-regulated SE control of these genes (Figure 6A). The cooperative features of enhancer components may make SEs especially vulnerable to inhibition of enhancer factors (Figure 6B). Genes encoding tumor cell master TFs acquire especially large SEs, with exceptionally high densities of enhancer factors, and this may further contribute to the exceptional vulnerability of SE-driven oncogenic TFs seen with inhibition of BRD4 (Bhagwat et al., 2016; Chapuy et al., 2013; Loven et al., 2013; Shi and Vakoc, 2014). SE-driven transcription may also be especially sensitive to disruptions in cooperative interactions that contribute to the 3D chromatin architecture at SEs (Dowen et al., 2014; Ji et al., 2016; Kieffer-Kwon et al., 2013).
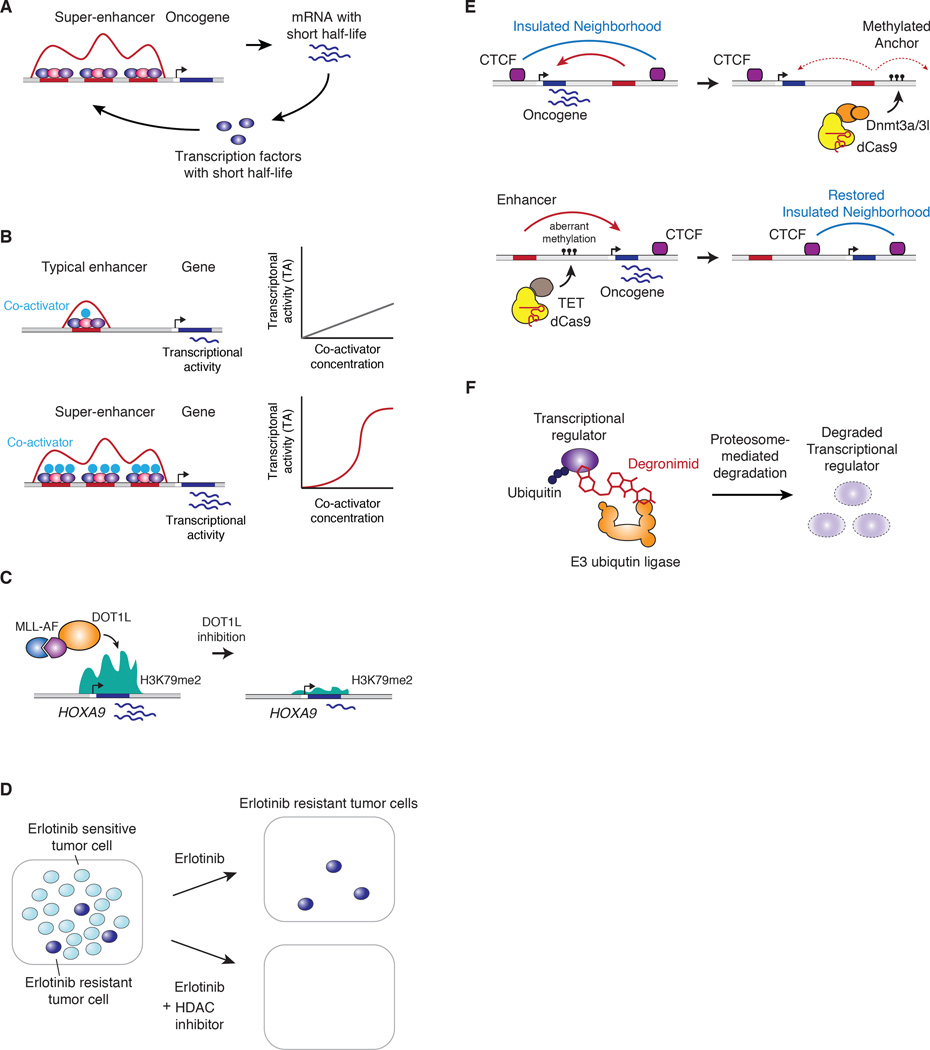
Figure 6. Drugging transcriptional dependencies.
A) Model of the features of super-enhancer –associated oncogene control that contribute to transcriptional dependencies.
B) Model of the mechanistic basis of higher transcriptional activity and vulnerability of super-enhancers: co-operative interactions between the co-activators recruited to these sites.
C) Therapeutic strategy to inhibit transcriptional dependencies due to aberrant recruitment of DOT1L to driver genes in MLL-AF fusion leukemias.
D) Therapeutic strategy to attack small populations of drug-tolerant tumor cells with histone deacetylase inhibitors.
E) Therapeutic strategy to downregulate oncogenes through manipulation and repair of insulated neighborhoods.
F) Targeted degradation of transcriptional regulators using degronimids that recruit E3-ubiquitin ligase for proteasome-mediated degradation.
The transcriptional CDKs have likewise emerged as compelling targets for transcriptional therapy in cancer. CDK7 is a key component of the general transcription initiation apparatus, which governs RNAPII activity by phosphorylating residues on its C-terminal domain. ChIP-seq data indicates that CDK7 densely occupies the SEs that drive expression of oncogenes in a wide variety of cancers, including T-ALL (Kwiatkowski et al., 2014), NSCLC (Christensen et al., 2014), neuroblastoma (Chipumuro et al., 2014), and triple-negative breast cancer (Wang et al., 2015). Small molecule inhibition of CDK7 by THZ1 leads to rapid loss of transcripts for TFs that contribute to oncogenesis and whose expression is driven by SEs in these cells, consistent with the idea that these SE-driven TF genes are especially vulnerable to inhibition of CDK7. The preferential loss of SE-driven TF gene expression, along with expression of genes involved in the DNA damage response, also occurs when tumor cells are treated with inhibitors of CDK12 and CDK13 (Zhang et al., 2016a). Interestingly, inhibition of the CDK8 and CDK19, components of Mediator that contribute to transcriptional repression, leads to hyperactivation of SE-driven genes in AML, and this is as deleterious to the leukemia cells as BRD4 inhibition, which has the opposite effect on these SE-driven genes (Pelish et al., 2015). These and other results show that the leukemia cells are addicted to a specific level of SE-associated gene transcription (Pelish et al., 2015).
The tumor-promoting effect of fusion oncogenes can produce actionable transcriptional dependencies. For example, many AML and MLL leukemias contain oncogenic fusions between MLL and AF genes; the AF part of the fusion product binds the DOT1L histone methyltransferase, which plays important roles in the control of transcriptional elongation by dimetylating histone H3 on lysine 79 (H3K79) (Cai et al., 2015; Deshpande et al., 2013). The MLL-AF fusions thus recruit DOT1L to MLL-target genes, including the TF genes HOXA9 and MEIS1, and the hypermethylation of histones leads to aberrant expression of those genes, which in turn drive leukemogenesis (Figure 6C) (Chen and Armstrong, 2015). These leukemias are especially vulnerable to genetic and chemical perturbation of DOT1L, and DOT1L inhibitors are now in clinical trials for these malignancies (Daigle et al., 2013).
Evidence that tumor cells can develop additional dependencies on specific transcriptional and chromatin regulators, rendering them susceptible to transcriptional drugs, has emerged from studies of drug resistance. Treatment of tumor cells with various anti-cancer agents can select for populations of drug-tolerant cells that have become dependent on specific chromatin regulators such as histone demethylases (KDM5A) and deacetylases (HDACs) (Figure 6D) (Sharma et al., 2010). The drug-tolerant tumor cells can in turn be ablated with histone deacetylase inhibitors, establishing a paradigm of combination therapy using inhibitors of chromatin regulators against drug resistance (Sharma et al., 2010).
Targeting chromosomal neighborhoods and associated addictions
As described above, insulated neighborhoods, whose ends are established by CTCF anchor sites, are frequently perturbed in cancer genomes by somatic mutations or aberrant DNA methylation; these perturbations have been implicated in the activation of cellular proto-oncogenes (Flavahan et al., 2016; Hnisz et al., 2016b; Katainen et al., 2015). Cancer cells containing aberrantly methylated neighborhood anchors can develop transcriptional dependencies that are amenable to small molecule therapeutics (Flavahan et al., 2016). The recent development of genetic and epigenetic editing technologies that can manipulate or repair chromosomal neighborhoods suggests novel therapeutic strategies for such modified cancer genomes (Amabile et al., 2016; Doudna and Charpentier, 2014; Liu et al., 2016b).
As an example, the expression of active genes can be reduced by disrupting the boundaries of its insulated neighborhood, which can, in turn, cause enhancers within a neighborhood to loop to other gene targets outside the neighborhood (Dowen et al., 2014). Targeted methylation of a neighborhood anchor site with a dCas9-DNA-methyltransferase-3 fusion protein has been shown to disrupt the neighborhood through the eviction of CTCF (Liu et al., 2016b). Thus, targeted methylation of an oncogene enhancer and/or the anchor sites of oncogene-containing neighborhoods might lead to selective downregulation of oncogene expression (Figure 6E).
In glioma cells that harbor mutations in the IDH1 gene, hypermethylation of insulated neighborhood anchor sites occurs, which leads to loss of CTCF binding at those anchors and activation of oncogenes that were previously silent in the intact neighborhoods (Flavahan et al., 2016). This suggests that restoration of the perturbed neighborhood boundaries might lead to silencing of the oncogene. Targeted DNA 5-hydroxy-methylation with a dCas9-TET fusion protein has recently been demonstrated (Amabile et al., 2016; Liu et al., 2016b), and this strategy could be used to restore an insulated neighborhood whose anchor site has been disrupted by aberrant DNA methylation (Figure 6E).
The identification of oncogenes activated by perturbations of insulated neighborhood CTCF anchor sites by aberrant methylation has revealed transcriptional dependencies of these tumor cells. For example, the IDH1-mutant glioma cells described above overexpress the PDGFRA gene, due to hypermethylation of the CTCF anchor site of the insulated neighborhood containing PDGFRA, which disrupts a boundary of the neighborhood and allows enhancers located elsewhere to activate PDGFRA. The growth of these cells is inhibited by small molecules that block PDGFRA, while gliomas containing a wild type IDH1 gene and an intact PDGFR neighborhood are unaffected by such inhibition (Flavahan et al., 2016). In this case, the dependence of the cancer cells in these tumors on PDGFRA was not identified by cancer genome sequencing (which revealed the IDH1 mutation), but by hypothesis-driven study of the effect of IDH1-associated hypermethylation on transcriptional control in these tumor cells.
Inhibition of DNA methyltransferases (DNMTs) by 5-azanucleoside drugs such as 5-azacitidine have broad impacts on gene expression. Though toxic in high doses, these compounds have shown therapeutic benefit at optimized low doses, and are approved for the treatment of myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) (Jones et al., 2016). These drugs were notable for their ability to alter the identities of cultured mammalian cells (Taylor and Jones, 1979) and are thought to activate tumor suppressor genes that have been silenced by DNA methylation and to reverse the tumor promoting effects of cancer-specific genetic alterations (Jones et al., 2016). With our new understanding of the roles of insulated neighborhoods and DNA methylation in gene control, it is possible that a major effect of these drugs is to alter neighborhood structures and thus the gene expression programs to which the tumor cells are addicted.
Future challenges
The fields of cancer biology and transcriptional biology are rapidly maturing and converging, establishing a compelling role for transcriptional dysregulation in cancer. We think it is imperative to commit to more aggressive study of the mechanisms that produce transcriptional addiction and learn how to exploit these for new therapies. Despite tremendous progress, there exist significant challenges ahead before these nascent insights can be broadly leveraged for patient benefit.
Where feasible, targeting oncogenic TFs in cancer (e.g., PML-RARA) has demonstrated profound clinical benefit. However, therapeutics capable of disrupting oncogenic TFs are lacking. We need a new science of transcriptional therapeutics and a generational commitment to the pursuit of the high-hanging fruit. Oncogenic MYC remains a holy grail of cancer therapy, yet there are as yet no compounds that directly target this TF. One solution may be new platforms of discovery chemistry capable of identifying and optimizing TF-directed compounds, delivery solutions to bring biomolecules nimbly across cell membranes and, quite plausibly, entirely new classes of agents. As an example, the general chemical strategy for drug-induced degradation of targeted proteins provides one path to this goal (Figure 6F) (Winter et al., 2015).
Where known, targeting tissue-specifying master TFs in cancer (ER/PR, AR) has profound clinical benefit. However, our knowledge of tissue-specifying TFs and dependencies remains limited. Here there is a need for more epigenome-based insights into cancer core regulatory circuitry, elaborated from primary human tumors. Some important insights into specific TF dependencies have recently emerged using such approaches [e.g., OCA-B in DLBCL (Chapuy et al., 2013); LMX1A in a medulloblastoma subtype (Lin et al., 2016)].
The development of clinically useful inhibitors of various components of the transcriptional apparatus will require further mechanistic understanding of the role of transcriptional addiction in cancer pathogenesis. As an example, CDK9 inhibition is highly active in CLL; at present, however, the drugs that have been developed exhibit only a narrow therapeutic index and no biomarkers of pharmacologic efficacy exist. Clarity on the mechanisms that create such dependencies may provide solutions such as therapeutic synergies.
A ubiquitous challenge of targeted cancer therapy is the emergence of tumor cells resistant to the therapeutic agent. While emerging evidence suggests that transcriptional inhibitors can suppress the emergence of drug resister cells when combined with other therapeutic agents (Sharma et al., 2010; Yashiro-Ohtani et al., 2014), recent studies indicate that tumor cells can develop resistance against the transcriptional inhibitors (Fong et al., 2015; Rathert et al., 2015; Shu et al., 2016). Future investigations will need to evaluate the effects of combination therapies that include transcriptional inhibitors.
While genome structural alterations have long been understood to have roles in cancer, the science of genome structure and function is still in its infancy. More complete mechanistic understanding of insulators, CTCF, and regional roles of chromatin-associated complexes is needed in cancer. Careful dissection of genome structure and its regulators in experimental and translational model systems will surely reveal new targets.
Improved understanding of dysregulated transcription in the context of the cancer cell heterogeneity present in individual tumors will be important for the development of effective therapies. However, precise measures of this heterogeneity at the level of the epigenome is currently limited by the fact that the generation of genome-wide maps of DNA-associated proteins often requires samples of >106 cells. As a consequence, current analyses obscure the heterogeneity created by intermingled clonal subpopulations within individual tumors that may differ from one another in important ways. The need to understand transcriptional control in, for example, tumor-initiating cells, AKT-low dormant cells, and metastatic carcinoma cells generated by epithelial-mesenchymal transitions, will require single-cell integrated epigenomic analyses. Recent insights into intratumoral heterogeneity in primary glioblastoma, obtained using single-cell RNA-seq, highlight this need (Patel et al., 2014).
Finally, we need to study the dynamics of transcriptional control in the most relevant model system of cancer – patients. We do not presently have the capability to study the dynamic effects of treatment in order to elucidate mechanisms of response and resistance. Indeed, future technologies that reveal epigenetic responses to targeted therapy are likely to provide novel, critically important mechanistic insights that will greatly benefit patients.
Acknowledgments
We are grateful to Brian Abraham, Brad Bernstein, Daniel Day, Matthew Fearer, Isaac Klein, Nick Kwiatkowski, Charles Lin, Marc Mansour, Peter Rahl, Jussi Taipale, William Tansey, Marc Timmers, Chris Vakoc and Robert Weinberg for critical comments. Supported by an NIH Grant HG002668 (R.A.Y.), an Erwin Schrödinger Fellowship (J3490) from the Austrian Science Fund (D.H.) and a Margaret and Herman Sokol Postdoctoral Award (D.H.). R.A.Y. and J.E.B are a founders and shareholders of Syros Pharmaceuticals. R.A.Y. is a founder and shareholder of Marauder Therapeutics. J.E.B. is a shareholder and executive of Novartis Pharmaceuticals, and a shareholder of Acetylon Pharmaceuticals. Syros, Novartis and Acetylon are discovering and developing therapeutics directed at transcriptional pathways in cancer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Affer M, Chesi M, Chen WD, Keats JJ, Demchenko YN, Tamizhmani K, Garbitt VM, Riggs DL, Brents LA, Roschke AV, et al. Promiscuous MYC locus rearrangements hijack enhancers but mostly super-enhancers to dysregulate MYC expression in multiple myeloma. Leukemia. 2014;28:1725–1735. doi: 10.1038/leu.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen BL, Taatjes DJ. The Mediator complex: a central integrator of transcription. Nature reviews Molecular cell biology. 2015;16:155–166. doi: 10.1038/nrm3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amabile A, Migliara A, Capasso P, Biffi M, Cittaro D, Naldini L, Lombardo A. Inheritable Silencing of Endogenous Genes by Hit-and-Run Targeted Epigenetic Editing. Cell. 2016;167:219–232. e214. doi: 10.1016/j.cell.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nature genetics. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baubec T, Ivanek R, Lienert F, Schubeler D. Methylation-dependent and - independent genomic targeting principles of the MBD protein family. Cell. 2013;153:480–492. doi: 10.1016/j.cell.2013.03.011. [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat AS, Roe JS, Mok BY, Hohmann AF, Shi J, Vakoc CR. BET Bromodomain Inhibition Releases the Mediator Complex from Select cis-Regulatory Elements. Cell reports. 2016;15:519–530. doi: 10.1016/j.celrep.2016.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove DA, Mahmoudi T, Henklein P, Verdin E. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13690–13695. doi: 10.1073/pnas.0705053104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JD, Lin CY, Duan Q, Griffin G, Federation AJ, Paranal RM, Bair S, Newton G, Lichtman AH, Kung AL, et al. NF-kappaB Directs Dynamic Super Enhancer Formation in Inflammation and Atherogenesis. Molecular cell. 2014 doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nature reviews Genetics. 2013;14:427–439. doi: 10.1038/nrg3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bywater MJ, Pearson RB, McArthur GA, Hannan RD. Dysregulation of the basal RNA polymerase transcription apparatus in cancer. Nat Rev Cancer. 2013;13:299–314. doi: 10.1038/nrc3496. [DOI] [PubMed] [Google Scholar]
- Cai SF, Chen CW, Armstrong SA. Drugging Chromatin in Cancer: Recent Advances and Novel Approaches. Molecular cell. 2015;60:561–570. doi: 10.1016/j.molcel.2015.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: annotating chromatin. Annual review of genetics. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, Rahl PB, Sun HH, Yeda KT, Doench JG, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CW, Armstrong SA. Targeting DOT1L and HOX gene expression in MLL-rearranged leukemia and beyond. Exp Hematol. 2015;43:673–684. doi: 10.1016/j.exphem.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Brand NJ, Chen A, Chen SJ, Tong JH, Wang ZY, Waxman S, Zelent A. Fusion between a novel Kruppel-like zinc finger gene and the retinoic acid receptor-alpha locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. The EMBO journal. 1993;12:1161–1167. doi: 10.1002/j.1460-2075.1993.tb05757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipumuro E, Marco E, Christensen CL, Kwiatkowski N, Zhang T, Hatheway CM, Abraham BJ, Sharma B, Yeung C, Altabef A, et al. CDK7 Inhibition Suppresses Super-Enhancer-Linked Oncogenic Transcription in MYCN-Driven Cancer. Cell. 2014;159:1126–1139. doi: 10.1016/j.cell.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen CL, Kwiatkowski N, Abraham BJ, Carretero J, Al-Shahrour F, Zhang T, Chipumuro E, Herter-Sprie GS, Akbay EA, Altabef A, et al. Targeting Transcriptional Addictions in Small Cell Lung Cancer with a Covalent CDK7 Inhibitor. Cancer cell. 2014;26:909–922. doi: 10.1016/j.ccell.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark VE, Harmanci AS, Bai H, Youngblood MW, Lee TI, Baranoski JF, Ercan-Sencicek AG, Abraham BJ, Weintraub AS, Hnisz D, et al. Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nature genetics. 2016;48:1253–1259. doi: 10.1038/ng.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle SR, Olhava EJ, Therkelsen CA, Basavapathruni A, Jin L, Boriack-Sjodin PA, Allain CJ, Klaus CR, Raimondi A, Scott MP, et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood. 2013;122:1017–1025. doi: 10.1182/blood-2013-04-497644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE., Jr Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande AJ, Chen L, Fazio M, Sinha AU, Bernt KM, Banka D, Dias S, Chang J, Olhava EJ, Daigle SR, et al. Leukemic transformation by the MLL-AF6 fusion oncogene requires the H3K79 methyltransferase Dot1l. Blood. 2013;121:2533–2541. doi: 10.1182/blood-2012-11-465120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, et al. Control of cell identity genes occurs in insulated neighborhoods in Mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drier Y, Cotton MJ, Williamson KE, Gillespie SM, Ryan RJ, Kluk MJ, Carey CD, Rodig SJ, Sholl LM, Afrogheh AH, et al. An oncogenic MYB feedback loop drives alternate cell fates in adenoid cystic carcinoma. Nature genetics. 2016 doi: 10.1038/ng.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick D, Geyer M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem Rev. 2013;113:8456–8490. doi: 10.1021/cr400071f. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Molecular cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suva ML, Bernstein BE. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong CY, Gilan O, Lam EY, Rubin AF, Ftouni S, Tyler D, Stanley K, Sinha D, Yeh P, Morison J, et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature. 2015;525:538–542. doi: 10.1038/nature14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Geng F, Wenzel S, Tansey WP. Ubiquitin and proteasomes in transcription. Annu Rev Biochem. 2012;81:177–201. doi: 10.1146/annurev-biochem-052110-120012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirlando R, Felsenfeld G. CTCF: making the right connections. Genes & development. 2016;30:881–891. doi: 10.1101/gad.277863.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Dekker J. The hierarchy of the 3D genome. Molecular cell. 2013;49:773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorkin DU, Leung D, Ren B. The 3D genome in transcriptional regulation and pluripotency. Cell stem cell. 2014;14:762–775. doi: 10.1016/j.stem.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- Groschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman BA, Erpelinck C, van der Velden VH, Havermans M, Avellino R, van Lom K, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157:369–381. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Lawton LN, Rozovskaia T, Frampton GM, Levine SS, Volkert TL, Croce CM, Nakamura T, Canaani E, Young RA. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes & development. 2008;22:3403–3408. doi: 10.1101/gad.1741408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Benner C, Chong LW, Yu RT, Downes M, Evans RM. Inflammation-sensitive super enhancers form domains of coordinately regulated enhancer RNAs. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E297–E302. doi: 10.1073/pnas.1424028112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay D, Hughes JR, Babbs C, Davies JO, Graham BJ, Hanssen LL, Kassouf MT, Oudelaar AM, Sharpe JA, Suciu MC, et al. Genetic dissection of the alpha-globin super-enhancer in vivo. Nature genetics. 2016;48:895–903. doi: 10.1038/ng.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Day DS, Young RA. Insulated Neighborhoods: Structural and Functional Units of Mammalian Gene Control. Cell. 2016a;167:1188–1200. doi: 10.1016/j.cell.2016.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Schuijers J, Lin CY, Weintraub AS, Abraham BJ, Lee TI, Bradner JE, Young RA. Convergence of Developmental and Oncogenic Signaling Pathways at Transcriptional Super-Enhancers. Molecular cell. 2015 doi: 10.1016/j.molcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016b;351:1454–1458. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ME, Ye YC, Chen SR, Chai JR, Lu JX, Zhoa L, Gu LJ, Wang ZY. Use of all-trans retinoic acid in the treatment of acute promyelocytic leukemia. Blood. 1988;72:567–572. [PubMed] [Google Scholar]
- Inaba H, Pui CH. Glucocorticoid use in acute lymphoblastic leukaemia. Lancet Oncol. 2010;11:1096–1106. doi: 10.1016/S1470-2045(10)70114-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, Sundberg CD, Bishop JM, Felsher DW. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science. 2002;297:102–104. doi: 10.1126/science.1071489. [DOI] [PubMed] [Google Scholar]
- Ji X, Dadon DB, Powell BE, Fan ZP, Borges-Rivera D, Shachar S, Weintraub AS, Hnisz D, Pegoraro G, Lee TI, et al. 3D Chromosome Regulatory Landscape of Human Pluripotent Cells. Cell stem cell. 2016;18:262–275. doi: 10.1016/j.stem.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T, Raviram R, Snetkova V, Rocha PP, Proudhon C, Badri S, Bonneau R, Skok JA, Kluger Y. Identification of multi-loci hubs from 4C-seq demonstrates the functional importance of simultaneous interactions. Nucleic acids research. 2016 doi: 10.1093/nar/gkw568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nature reviews Genetics. 2016;17:630–641. doi: 10.1038/nrg.2016.93. [DOI] [PubMed] [Google Scholar]
- Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nature reviews Molecular cell biology. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo JY, Schaukowitch K, Farbiak L, Kilaru G, Kim TK. Stimulus-specific combinatorial functionality of neuronal c-fos enhancers. Nature neuroscience. 2016;19:75–83. doi: 10.1038/nn.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci Adv. 2015;1:e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katainen R, Dave K, Pitkanen E, Palin K, Kivioja T, Valimaki N, Gylfe AE, Ristolainen H, Hanninen UA, Cajuso T, et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nature genetics. 2015;47:818–821. doi: 10.1038/ng.3335. [DOI] [PubMed] [Google Scholar]
- Kennedy AL, Vallurupalli M, Chen L, Crompton B, Cowley G, Vazquez F, Weir BA, Tsherniak A, Parasuraman S, Kim S, et al. Functional, chemical genomic, and super-enhancer screening identify sensitivity to cyclin D1/CDK4 pathway inhibition in Ewing sarcoma. Oncotarget. 2015;6:30178–30193. doi: 10.18632/oncotarget.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer-Kwon KR, Tang Z, Mathe E, Qian J, Sung MH, Li G, Resch W, Baek S, Pruett N, Grontved L, et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155:1507–1520. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer cell. 2008;14:355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski N, Zhang T, Rahl PB, Abraham BJ, Reddy J, Ficarro SB, Dastur A, Amzallag A, Ramaswamy S, Tesar B, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature. 2014;511:616–620. doi: 10.1038/nature13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D, Levine A. p53 Research: the past thirty years and the next thirty years. Cold Spring Harbor perspectives in biology. 2010;2:a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, Cattoglio C, Tjian R. Looping back to leap forward: transcription enters a new era. Cell. 2014;157:13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nature reviews Genetics. 2016;17:207–223. doi: 10.1038/nrg.2016.4. [DOI] [PubMed] [Google Scholar]
- Lin CY, Erkek S, Tong Y, Yin L, Federation AJ, Zapatka M, Haldipur P, Kawauchi D, Risch T, Warnatz HJ, et al. Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature. 2016;530:57–62. doi: 10.1038/nature16546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MY, DeNizio JE, Schutsky EK, Kohli RM. The expanding scope and impact of epigenetic cytosine modifications. Curr Opin Chem Biol. 2016a;33:67–73. doi: 10.1016/j.cbpa.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R. Editing DNA Methylation in the Mammalian Genome. Cell. 2016b;167:233–247. e217. doi: 10.1016/j.cell.2016.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo-Coco F, Avvisati G, Vignetti M, Thiede C, Orlando SM, Iacobelli S, Ferrara F, Fazi P, Cicconi L, Di Bona E, et al. Retinoic acid and arsenic trioxide for acute promyelocytic leukemia. The New England journal of medicine. 2013;369:111–121. doi: 10.1056/NEJMoa1300874. [DOI] [PubMed] [Google Scholar]
- Long HK, Prescott SL, Wysocka J. Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell. 2016;167:1170–1187. doi: 10.1016/j.cell.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinen N, Mehine M, Tolvanen J, Kaasinen E, Li Y, Lehtonen HJ, Gentile M, Yan J, Enge M, Taipale M, et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science. 2011;334:252–255. doi: 10.1126/science.1208930. [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nature reviews Genetics. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, Etchin J, Lawton L, Sallan SE, Silverman LB, et al. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014 doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, Kung AL, Gilliland DG, Verdine GL, Bradner JE. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462:182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA, Daley GQ. A blueprint for engineering cell fate: current technologies to reprogram cell identity. Cell research. 2013;23:33–48. doi: 10.1038/cr.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabet B, P OB, Reyes JM, Shieh K, Lin CY, Will CM, Popovic R, Ezponda T, Bradner JE, Golden AA, et al. Deregulation of the Ras-Erk Signaling Axis Modulates the Enhancer Landscape. Cell reports. 2015;12:1300–1313. doi: 10.1016/j.celrep.2015.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Lee C, Zichner T, Stutz AM, Erkek S, Kawauchi D, Shih DJ, Hovestadt V, Zapatka M, Sturm D, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511:428–434. doi: 10.1038/nature13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nature reviews Genetics. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini R, Shao W, Sellers WR. Oncogene addiction: pathways of therapeutic response, resistance, and road maps toward a cure. EMBO Rep. 2015;16:280–296. doi: 10.15252/embr.201439949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelish HE, Liau BB, Nitulescu II, Tangpeerachaikul A, Poss ZC, Da Silva DH, Caruso BT, Arefolov A, Fadeyi O, Christie AL, et al. Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature. 2015;526:273–276. doi: 10.1038/nature14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Corces VG. Chromatin insulators: linking genome organization to cellular function. Molecular cell. 2013;50:461–474. doi: 10.1016/j.molcel.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piunti A, Shilatifard A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science. 2016;352:aad9780. doi: 10.1126/science.aad9780. [DOI] [PubMed] [Google Scholar]
- Puissant A, Frumm SM, Alexe G, Bassil CF, Qi J, Chanthery YH, Nekritz EA, Zeid R, Gustafson WC, Greninger P, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer discovery. 2013;3:308–323. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathert P, Roth M, Neumann T, Muerdter F, Roe JS, Muhar M, Deswal S, Cerny-Reiterer S, Peter B, Jude J, et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature. 2015;525:543–547. doi: 10.1038/nature14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley JD, Golomb HM, Dougherty C. 15/17 translocation, a consistent chromosomal change in acute promyelocytic leukaemia. Lancet. 1977;1:549–550. doi: 10.1016/s0140-6736(77)91415-5. [DOI] [PubMed] [Google Scholar]
- Sancho-Martinez I, Baek SH, Izpisua Belmonte JC. Lineage conversion methodologies meet the reprogramming toolbox. Nature cell biology. 2012;14:892–899. doi: 10.1038/ncb2567. [DOI] [PubMed] [Google Scholar]
- Sanda T, Lawton LN, Barrasa MI, Fan ZP, Kohlhammer H, Gutierrez A, Ma W, Tatarek J, Ahn Y, Kelliher MA, et al. Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer cell. 2012;22:209–221. doi: 10.1016/j.ccr.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Molecular cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Whyte WA, Zepeda-Mendoza CJ, Milazzo JP, Shen C, Roe JS, Minder JL, Mercan F, Wang E, Eckersley-Maslin MA, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes & development. 2013;27:2648–2662. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin HY, Willi M, Yoo KH, Zeng X, Wang C, Metser G, Hennighausen L. Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nature genetics. 2016;48:904–911. doi: 10.1038/ng.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM, Janiszewska M, Huh SJ, Liang Y, Ryan J, et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature. 2016;529:413–417. doi: 10.1038/nature16508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnev AA, Josefowicz SZ, Allis CD. Greater Than the Sum of Parts: Complexity of the Dynamic Epigenome. Molecular cell. 2016;62:681–694. doi: 10.1016/j.molcel.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Whitfield JR, Sodir NM, Masso-Valles D, Serrano E, Karnezis AN, Swigart LB, Evan GI. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes & development. 2013;27:504–513. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nature reviews Genetics. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- Stathis A, Zucca E, Bekradda M, Gomez-Roca C, Delord JP, de La Motte Rouge T, Uro-Coste E, de Braud F, Pelosi G, French CA. Clinical Response of Carcinomas Harboring the BRD4-NUT Oncoprotein to the Targeted Bromodomain Inhibitor OTX015/MK-8628. Cancer discovery. 2016;6:492–500. doi: 10.1158/2159-8290.CD-15-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D, Sellers WR. Linking somatic genetic alterations in cancer to therapeutics. Current opinion in cell biology. 2009;21:304–310. doi: 10.1016/j.ceb.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Sur I, Taipale J. The role of enhancers in cancer. Nat Rev Cancer. 2016;16:483–493. doi: 10.1038/nrc.2016.62. [DOI] [PubMed] [Google Scholar]
- Taub R, Kirsch I, Morton C, Lenoir G, Swan D, Tronick S, Aaronson S, Leder P. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:7837–7841. doi: 10.1073/pnas.79.24.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SM, Jones PA. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell. 1979;17:771–779. doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nature reviews Molecular cell biology. 2014;15:703–708. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- Thomas D, Tyers M. Transcriptional regulation: Kamikaze activators. Current biology : CB. 2000;10:R341–R343. doi: 10.1016/s0960-9822(00)00462-0. [DOI] [PubMed] [Google Scholar]
- Tomazou EM, Sheffield NC, Schmidl C, Schuster M, Schonegger A, Datlinger P, Kubicek S, Bock C, Kovar H. Epigenome mapping reveals distinct modes of gene regulation and widespread enhancer reprogramming by the oncogenic fusion protein EWS-FLI1. Cell reports. 2015;10:1082–1095. doi: 10.1016/j.celrep.2015.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, DiBiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147:577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nature reviews Genetics. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Wernig M. Molecular roadblocks for cellular reprogramming. Molecular cell. 2012;47:827–838. doi: 10.1016/j.molcel.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viny AD, Ott CJ, Spitzer B, Rivas M, Meydan C, Papalexi E, Yelin D, Shank K, Reyes J, Chiu A, et al. Dose-dependent role of the cohesin complex in normal and malignant hematopoiesis. The Journal of experimental medicine. 2015;212:1819–1832. doi: 10.1084/jem.20151317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BA, Wardell CP, Brioli A, Boyle E, Kaiser MF, Begum DB, Dahir NB, Johnson DC, Ross FM, Davies FE, et al. Translocations at 8q24 juxtapose MYC with genes that harbor superenhancers resulting in overexpression and poor prognosis in myeloma patients. Blood cancer journal. 2014;4:e191. doi: 10.1038/bcj.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang T, Kwiatkowski N, Abraham BJ, Lee TI, Xie S, Yuzugullu H, Von T, Li H, Lin Z, et al. CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell. 2015;163:174–186. doi: 10.1016/j.cell.2015.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Chen Z. Acute promyelocytic leukemia: from highly fatal to highly curable. Blood. 2008;111:2505–2515. doi: 10.1182/blood-2007-07-102798. [DOI] [PubMed] [Google Scholar]
- Watson IR, Takahashi K, Futreal PA, Chin L. Emerging patterns of somatic mutations in cancer. Nature reviews Genetics. 2013;14:703–718. doi: 10.1038/nrg3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein IB, Joe AK. Mechanisms of disease: Oncogene addiction--a rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol. 2006;3:448–457. doi: 10.1038/ncponc0558. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe-Paganon S, Bradner JE. DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348:1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell stem cell. 2012;10:678–684. doi: 10.1016/j.stem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Yang H, Schramek D, Adam RC, Keyes BE, Wang P, Zheng D, Fuchs E. ETS family transcriptional regulators drive chromatin dynamics and malignancy in squamous cell carcinomas. eLife. 2015;4 doi: 10.7554/eLife.10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiro-Ohtani Y, Wang H, Zang C, Arnett KL, Bailis W, Ho Y, Knoechel B, Lanauze C, Louis L, Forsyth KS, et al. Long-range enhancer activity determines Myc sensitivity to Notch inhibitors in T cell leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E4946–E4953. doi: 10.1073/pnas.1407079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Kwiatkowski N, Olson CM, Dixon-Clarke SE, Abraham BJ, Greifenberg AK, Ficarro SB, Elkins JM, Liang Y, Hannett NM, et al. Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors. Nat Chem Biol. 2016a;12:876–884. doi: 10.1038/nchembio.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Choi PS, Francis JM, Imielinski M, Watanabe H, Cherniack AD, Meyerson M. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nature genetics. 2016b;48:176–182. doi: 10.1038/ng.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]