Abstract
Changes in plasma concentration of small organic metabolites could be due to their altered production or urinary excretion and changes in their urine concentration may be due to the changes in their filtered load, tubular reabsorption, and/or altered urine volume. Therefore, these factors should be considered in interpretation of the changes observed in plasma or urine of the target metabolite(s). Fasting plasma and urine samples from 180 CKD patients and 120 age-matched healthy controls were determined by UPLC-HDMS-metabolomics and quantitative real-time RT-PCR techniques. Compared with healthy controls, patients with CKD showed activation of NF-κB and up-regulation of pro-inflammatory and pro-oxidant mRNA and protein expression as well as down-regulation of Nrf2-associated anti-oxidant gene mRNA and protein expression, accompanied by activated canonical Wnt/β-catenin signaling. 124 plasma and 128 urine metabolites were identified and 40 metabolites were significantly altered in both plasma and urine. Plasma concentration and urine excretion of 25 metabolites were distinctly different between CKD and controls. They were related to amino acid, methylamine, purine and lipid metabolisms. Logistic regression identified four plasma and five urine metabolites. Parts of them were good correlated with eGFR or serum creatinine. 5-Methoxytryptophan and homocystine and citrulline were good correlated with both eGFR and creatinine. Clinical factors were incorporated to establish predictive models. The enhanced metabolite model showed 5-methoxytryptophan, homocystine and citrulline have satisfactory accuracy, sensitivity and specificity for predictive CKD. The dysregulation of CKD was related to amino acid, methylamine, purine and lipid metabolisms. 5-methoxytryptophan, homocystine and citrulline could be considered as additional GFR-associated biomarker candidates and for indicating advanced renal injury. CKD caused dysregulation of the plasma and urine metabolome, activation of inflammatory/oxidative pathway and Wnt/β-catenin signaling and suppression of antioxidant pathway.
Keywords: Chronic kidney disease, Oxidative stress, Inflammation, Wnt/beta-catenin signaling, Metabolomics, Clinical factors
Graphical abstract

Highlights
-
•
Chronic kidney disease (CKD) is a serious public health problem.
-
•
Redox signaling and wnt/β-catenin pathway were studied in CKD patients.
-
•
Clinical phenotypes and metabolites in CKD patients were studied by metabolomics.
-
•
Plasma and urine metabolites related to amino acid, purine and lipid metabolisms.
-
•
Four plasma and five urine metabolites were good correlated with GFR or creatinine.
1. Introduction
The body's metabolic processes result in formation numerous systemic or tissue specific small molecular size organic metabolites released in the circulation. Via glomerular filtration, tubular reabsorption and secretion, kidney plays a key role in disposition of these metabolites. In addition kidney generates numerous metabolites including amino acids. Therefore changes in kidney function can affect the blood level and urinary excretion of metabolites in humans and animals. Moreover, the underlying causes of chronic kidney disease (CKD) such as diabetes and autoimmune disorders independently impact generation of metabolites and compound the effects of kidney disease. Technological advances which allow comprehensive determination of small molecular weight metabolites have provided an important tool to measure the full spectrum of endogenous metabolites in biological samples [1], [2], [3]. Application of these sophisticated systems has provided a valuable tool for early diagnosis, identification of reliable biomarkers, elucidation of the mechanism of various diseases, and monitoring of their progression and response to therapeutic interventions. Several studies have demonstrated marked changes in the plasma or urine concentrations of numerous metabolites in patients [4], [5] and animals with CKD [6], [7], [8], [9]. Although altered metabolic changes have been identified in CKD, in many cases the underlying mechanisms of the observed changes remain unclear. To our knowledge, simultaneous determination of plasma level and the rate of urinary excretion of metabolites have not been reported in patients with CKD and healthy controls. Simultaneous determination of plasma level and the rate of urinary excretion of metabolites are critical because the rise in plasma concentration could be due to either increased production and or reduced urinary excretion of the given metabolite(s). Likewise the rise in urine level may be due either to increased abundance or reduced tubular reabsorption of the target metabolite(s) whereas the decline in the urine concentration may be due to the impaired urinary concentrating capacity which is invariably present in CKD or polyuric states e.g. diuretic therapy or osmotic diuresis as seen with glucosuria in diabetes. Therefore, these variables should be carefully considered in interpretation of the mechanism of the observed changes in plasma and urine levels of the identified metabolite(s) and selection of biomarker metabolite(s) for CKD. In order to eliminate the error caused by differences in urinary concentrating capacity in the present study the urine metabolite data were presented as the ratios with the corresponding urine creatinine concentrations. In addition to determine the difference in the rate of production versus decreased glomerular filtration or tubular reabsorption of the target metabolites, changes in plasma levels were viewed in the context of their urinary excretion.
Oxidative stress and inflammation played a central role in the development and progression of pathogenesis of CKD [10], [11]. Under physiological conditions, oxidative stress provokes the anti-oxidant and cytoprotective proteins upregulation to prevent dysfunction and kidney damage. The process was mediated by activated nuclear factor-erythroid-2-related factor 2 (Nrf2) which adjust the basal activity and coordinated induction of many genes that encode anti-oxidant and phase 2 detoxifying enzymes and associated with proteins [11]. Oxidative stress and inflammation are closely interacted as they caused a vicious cycle in which oxidative stress triggers inflammation by several mechanisms including transcription factor kappa B (NF-κB) activation which caused the activation and recruitment of immune cells. Inflammation, in turn, triggered oxidative stress via production of reactive oxygen and nitrogen species by the activation of leukocytes and resident cells. Taken together, these events promote kidney injury by inflicting necrosis and fibrosis. Canonical Wnt/β-catenin signaling pathway was an evolutionarily conserved developmental signaling cascade that played a vital role in the regulation of organ development and tissue homeostasis [12]. Despite being relatively silent in normal adult kidneys, Wnt/β-catenin signaling is activated in the different CKD, such as obstructive nephropathy, adriamycin nephropathy, chronic allograft nephropathy and diabetic nephropathy [13], [14], [15], [16], which was accompanied by oxidative stress and inflammation. It was reported that activated oxidative stress and inflammation were closed associated with dysregulation of amino acid, uremic toxin (indoxyl sulfate, uric acid), bile acid, fatty acid, triglyceride, glycerophospholipid metabolisms in rats with chronic kidney disease [17], [18].
Application of UPLC-QTOF/HDMS is highly suitable for large-scale metabolomic evaluation [19]. In this study, a non-targeted and targeted metabolomics approach was performed to simultaneously determine the plasma and urine metabolites from CKD patients and controls (Fig. 1). The study uncovers the relationship between identified metabolites and gene expression of inflammation/Wnt/β-catenin signaling cascade to illuminate the molecular pathogenesis of patients with advanced CKD.
Fig. 1.
Flow diagram of metabolomic analysis was used to show the overview of study design.
2. Materials and methods
2.1. Chemicals and reagents
Creatinine, hippuric acid, uric acid, hypoxanthine, xanthine, myoinositol, cis-aconitic acid, indoxyl sulfate and aldosterone were obtained from the National Institutes for Food and Drug Control (Beijing, China). Amino acids including l-methionine, l-lysine, l-phenylalanine, l-homoserine, homocysteine, l-tyrosine, l-glutamine, citrulline, l-arginine, l-cysteine, l-glutamic acid, l-alanine and L-aspartic acid were purchased from Amresco Company. Kynurenine, kynurenic acid, dopamine, 1-methyladenosine, l-xanthosine, xanthurenic acid, indole, p-cresol sulfate, uracil, p-cresol, guanosine, succinic acid, deoxyuridine, guanine, taurine and thymine were purchased from Sigma Company or Aladdin Company. Antibodies against nuclear factor kappa B p65, Nrf2, cyclooxygenase-2 (COX-2), 12-lipoxygenase (12-LP), etc. were purchased from Santa Cruz Biotechnology or Abcam Company.
2.2. Participants
The participants included 120 adult CKD patients and 80 healthy controls recruited for the discovery phase and additional 60 CKD patients and 40 healthy controls recruited for the validation phase between February 2013 and November 2014 at the Traditional Chinese Medicine Hospital and the Fourth Hospital of Xi’an. Patients with stage 4–5 CKD based on estimated glomerular filtration rate (eGFR) for at least 3 months were enrolled in the study. Patients with acute kidney injury, liver disease, gastrointestinal pathology, active vasculitis, or cancer, and those who required dialysis, immunosuppressive or chemotherapy therapy or had received kidney transplant were excluded. Baseline demographic and clinical data and list of medications were collected from patients' medical records. 120 age-matched healthy controls with no history of kidney disease were enrolled. The study was approved by the Ethical Committee and all patients provided informed consent prior to entering the study.
Blood and urine samples were obtained after an overnight fasting and plasma was separated and stored at −80 °C. BMI was calculated from body weight (kg) divided by height squared (m2) based on World Health Organization (WHO) categories. Systolic (SBP) and diastolic blood pressure (DBP) was measured after a five minutes rest in the sitting position. Estimated glomerular filtration rate (eGFR) was calculated using the Modification of Diet of Renal Disease equation. Serum concentrations of urea nitrogen (BUN), creatinine, albumin, triglycerides, total cholesterol (TC), high-density lipoprotein-cholesterol (HDL-C), low-density lipoprotein-cholesterol (LDL-C), uric acid, intact parathyroid hormone (iPTH), potassium, sodium, calcium and phosphate were determined using an Olympus AU640 automatic analyzer. Urine total protein was measured by colorimetric assay. White blood cell count (WBC), red blood cell count (RBC) and haemoglobin were determined by HF-3800 analyzer.
2.3. Gene expression study by quantitative real-time polymerase chain reaction (q-RT-PCR)
Red Blood Cell Lysis Buffer (Tiangen, China) was added into fresh patient's plasma. After dissociating red blood cells, biological samples were centrifuged at 450g for 10 min and save the pellet. The pellet was suspended by a High Pure RNA Isolation Kit (RNAiso Plus, Takara Bio, Japan) according to the manufacturer's guide, including DNase-free treatment. Primers applied for quantitative real-time RT-PCR (q-RT-PCR) are listed in Table 1. The quantity and purity of the RNA preparations were determined by measuring the optical densities at 260 and 280 nm. Quantitative real-time RT-PCR (qRT-PCR) was carried out by the Bio-Rad PCR system. Total RNA was reverse transcribed by a Transcriptor First Strand cDNA Synthesis Kit according to the instructions of the manufacturer (Roche, Germany) using Bio-Rad T100™ system. Quantitative real-time RT-PCR (q-RT-PCR) was performed on Bio-Rad CFX 96 Touch ™ system (Bio-Rad, USA). The PCR reaction mixture in a 20-µL volume contained 10 µL of SYBR® Premix Ex Taq™ II (Takara Bio, Japan), 1.0 µL of reverse transcription product (1 μg cDNA), 0.4 µL of sense and antisense primer sets and 8.2 µL of double distilled water. The mRNA levels of genes were calculated by normalizing with β-actin. Initial activation at 95 °C for 30 s, followed by 40 cycles with denaturation at 94 °C for 5 s, annealing at 60 °C for 30 s, and extension was at 65 °C for 5 s. Reactions were replicated three times per experiment and experiments were replicated three times to verify the positive results.
Table 1.
Reference genes evaluated in this study: species, primer sequences and product sizes.
| Gene | Species | Forward | Reverse | Product Size (bp) |
|---|---|---|---|---|
| IκBα | Homo sapiens | CTCCGAGACTTTCGAGGAAATAC | CCATTGTAGTTGGTAGCCTTCA | 134 |
| NF-κB p65 | Homo sapiens | CTGTCCTTTCTCATCCCATCTT | TCCTCTTTCTGCACCTTGTC | 139 |
| COX−2 | Homo sapiens | GTTCCAGACAAGCAGGCTAATA | CCACTCAAGTGTTGCACATAATC | 78 |
| MCP−1 | Homo sapiens | CCCAGTCACCTGCTGTTAT | ACAGCTTCTTTGGGACACTT | 102 |
| iNOS | Homo sapiens | TGCCAAGCTGAAATTGAATGAG | CTTCGCCTCGTAAGGAAATACA | 112 |
| p47phox | Homo sapiens | GGTTCTGTCAGATGAAAGCAAAG | GTATGGCTCACCTGCATAGTT | 113 |
| 12-LO | Homo sapiens | TCCGCTACACCATGGAAATC | CTGTGCTCACTGCCTTATCA | 84 |
| p67phox | Homo sapiens | GTGGAGGCACTCTTCAGTTATG | CAGCCATTCTTCATTCACCTTTG | 102 |
| Rac1 | Homo sapiens | GAATCTGGGCTTATGGGATACA | ATCTGTTTGCGGATAGGATAGG | 76 |
| gp91phox | Homo sapiens | ACCCTCCTATGACTTGGAAATG | TGATGACCACCTTCTGTTGAG | 99 |
| keap1 | Homo sapiens | CACAACAGTGTGGAGAGGTATG | CGGCATAAAGGAGACGATTGA | 115 |
| Nrf2 | Homo sapiens | GTTGCCCACATTCCCAAATC | CGTAGCCGAAGAAACCTCAT | 105 |
| HO−1 | Homo sapiens | CCAGCAACAAAGTGCAAGATTC | CCACCAGAAAGCTGAGTGTAAG | 117 |
| GPX | Homo sapiens | GGGCAAGGTACTACTTATCGAG | CTCGTTCATCTGGGTGTAGTC | 76 |
| Gclc | Homo sapiens | GACCCATGGAGGTGCAATTA | AACCTTTGACAGTGGAATGAGA | 119 |
| NQO−1 | Homo sapiens | AGAGGTACAGGATGAGGAGAAA | ATCTGGTTGTCAGTTGGGATG | 87 |
| Wnt1 | Homo sapiens | CGGCGTTTATCTTCGCTATCA | GTAGTCACACGTGCAGGATT | 95 |
| Wnt2 | Homo sapiens | CGGGAATCTGCCTTTGTTTATG | TTGGATCACAGGAACAGGATTT | 103 |
| Wnt2b | Homo sapiens | TGCAGTGACAACATCCACTAC | GCGACCACAGCGGTTATTA | 117 |
| Wnt3 | Homo sapiens | CTGACTTCGGCGTGTTAGT | CCTCGTTGTTGTGCTTGTTC | 93 |
| Wnt3a | Homo sapiens | CAAGATTGGCATCCAGGAGT | ATGAGCGTGTCACTGCAAAG | 173 |
| Wnt4 | Homo sapiens | ATGGAAGTACACCCTCTGG | CCTGGAAGGACCCACAGATA | 201 |
| Wnt5a | Homo sapiens | GGGTGGGAACCAAGAAAAAT | TGGAACCTACCCATCCCATA | 194 |
| Wnt6 | Homo sapiens | GAGAGTGCCAGTTCCAGTTC | GTCTCCCGAATGTCCTGTTG | 94 |
| Wnt7a | Homo sapiens | CGTGCTCAAGGACAAGTACA | GTACGACAGTGGCTTCTTGAT | 103 |
| Wnt7b | Homo sapiens | CCTGCTGAAGGAGAAGTACAA | GTCTCCATGGGCTTCTGATAG | 120 |
| Wnt8a | Homo sapiens | CAGAGGCGGAACTGATCTTT | TGTTGTGGCTGTTCTGTAGG | 111 |
| Wnt8b | Homo sapiens | CCAATCGGGAGACAGCATTT | AGTCATCACAGCCACAGTTATC | 108 |
| Wnt9a | Homo sapiens | GCGGAGACAACCTTAAGTACAG | ACGAGGTTGTTGTGGAAGTC | 103 |
| Wnt9b | Homo sapiens | TGTAAGTGCCATGGCGTATC | CCGAGTCATAGCGCAGTTT | 106 |
| Wnt10a | Homo sapiens | GTGCTCCTGTTCTTCCTACTG | CACACTGTGTTGGCATTGAG | 113 |
| Wnt10b | Homo sapiens | TCTGACAAGGGGACAGAACC | TCATTGGCTTAGACCCGACT | 240 |
| Wnt16 | Homo sapiens | CCAGTTCAGACACGAGAGATG | GCAGCCATCACAGCATAAATAA | 138 |
| β-catenin | Homo sapiens | ACAAGCCACAAGATTACAAG | ATCAGCAG TCTCATTCCAA | 92 |
| β-actin | Homo sapiens | AGGCATCCTCACCCTGAAGTA | CACACGCAGCTCATTGTAGA | 103 |
2.4. Protein expression study by Western blot analysis
Western blot for protein expression were done in plasma samples. The total protein concentration was determined using Pierce™ BCA Protein Assay Kit (23227, Thermo Scientific, USA) according to the manufacturer's instructions. Protein expression was performed by Western blot analyses as described previously [17], [18]. The ratio of each protein was calculated by densitometry using free ImageJ software (version 1.48 v, NIH, Bethesda, MD, USA) and Band densities were normalized by Histone H3 or GAPDH expression levels.
2.5. Sample preparation
The plasma samples were thawed at room temperature prior to analysis. 400 µL acetonitrile was added to 200 µL plasma and vortex-mixed vigorously for three min. The mixture was settled for ten min, and then centrifuged at 13,000 rpm for ten min at 4 °C. The 400 µL supernatant were pipetted out and lyophilized. Urine samples were thawed and then centrifuged at 13,000 rpm for ten min to remove solid materials. The supernatant was diluted at a ratio of 3:1 with distilled water, vortex mixed for UPLC-MS analysis.
2.6. UPLC-HDMS analysis for plasma samples
The plasma samples were analyzed by a 2.1 mm×100 mm ACQUITY 1.8 µm HSS T3 using a Waters Acquity™ UPLC system equipped with a Waters Xevo™ G2 QTof MS. A gradient of acetonitrile (A) and 0.1% formic acid in water (B) and used as follows: a linear gradient of 0–1.0 min, 0.0% A; 1.0–16.0 min, 0.0–100% A; 16.0–20.0 min, 100% A and 20.0–22.0 min, 100–0.0% A. The flow rate was 0.40 ml/min. The autosampler was maintained at 4 °C. The lyophilized plasma samples were dissolved in 100 µL of acetonitrile/water (4:1). Every 2 µL sample solution was injected for each run.
MS of the optimal conditions were as follows: capillary voltage: 2.5 kV, cone voltage: 30 V, desolvation gas temperature: 500 °C, source temperature: 120 °C, desolvation gas flow: 600 L/h, cone gas flow: 50 L/h, The scan range was from 50 to 1200 m/z. Leucine–enkephalin was used for accurate mass acquisition. Waters MassLynx v4.1 was used for all the acquisition and analysis of data in both positive ion mode and negative ion mode.
2.7. UPLC-HDMS analysis for urine samples
The urine samples were analyzed by a 2.1 mm×100 mm ACQUITY 1.8 µm HSS T3 using a Waters Acquity™ UPLC system equipped with a Waters Xevo™ G2 QTof MS. A gradient of acetonitrile (A) and 0.1% formic acid in water (B) and used as follows: a linear gradient of 0–1.0 min, 1.0% A; 1.0–8.0 min, 1.0–40% A; 8.0–9.0 min, 40.0–99% A; 9.0–12.0 min, 99% and 12.0–15.0 min, 99.0–1.0% A. The flow rate was 0.40 ml/min. The autosampler was maintained at 4 °C. The lyophilized plasma samples were dissolved in 100 µL of acetonitrile/water (4:1). Every 2 µL sample solution was injected for each run.
MS of the optimal conditions were as follows: capillary voltage: 3.0 kV, cone voltage: 40 V, desolvation gas temperature: 500 °C, source temperature: 110 °C, desolvation gas flow: 800 L/h, cone gas flow: 50 L/h, The scan range was from 50 to 1200 m/z. Leucine–enkephalin was used for accurate mass acquisition. Waters MassLynx v4.1 was used for all the acquisition and analysis of data in both positive ion mode and negative ion mode.
2.8. Data analysis, model development, biomarker selection and cross validation
The acquired raw data from UPLC-HDMS analysis in positive and negative ion modes were first pre-processed by Markerlynx XS and Progenesis QI (Waters, Manchester, U.K.). Orthogonal partial least square-discriminant analysis (OPLS-DA) and principal component analysis (PCA) was performed to discriminate between patients with CKD and healthy controls. The variables were selected based on variable importance in the projection (VIP >1.5) from the normalized peak intensity. We reduced the resulting matrix by removing any ion perks with zero value in the samples to obtain consistent differential variables. The urine data of patients with CKD and healthy controls were normalized by variable intensity/creatinine intensity. Variables or normalized variables were selected by one-way analysis of variance (ANOVA) with a threshold of P<0.05 in SPSS 19.0 (SSPS Inc., Chicago, USA). Based on reported literatures, significantly variables were identified and confirmed by comparing MS data, MS/MS fragments, molecular weights and elemental compositions with the available reference chemicals.
Identified metabolites were subjected to further statistical analysis by univariate and multivariate statistical methods. Fold change (FC) was calculated based on mean ratios for CKD/controls. Metabolites were also selected by Mann-Whitney U test with a threshold of P<0.05. The resultant P values from ANOVA were further adjusted by a false discovery rate (FDR) based on the Hochberg-Benjamini method. Significantly altered variables were defined and further identified by a VIP >1.5, P<0.05 and FDR <0.05. Variables or metabolites are visualized using heat map and z-score plots analyses. The z-score of metabolites was calculated according to reference distribution of the control samples. Then each metabolite was centered by the control mean and scale by the control standard deviation. Analysis of Pearson correlation coefficient was performed to find the correlations between the potential biomarkers.
2.9. Binary logistic regression (BLR) and ROC curve analysis
Based on the binary outcome of CKD and control as dependent variables, we developed a BLR model to find the best combination of plasma and urine differential metabolites. PLS-DA-based ROC analysis was performed for the selection of candidate biomarkers.
3. Results
3.1. General data
The general clinical and demographic data are presented in Table 2. CKD patients had significantly higher SBP and DBP and significantly lower eGFR compared with healthy controls. Serum BUN, creatinine, uric acid, potassium and phosphate levels as well as urine proteins were significantly higher, while serum albumin and calcium were significantly lower in CKD patients.
Table 2.
Summary of clinical and demographic baseline characteristics of patients with CKD and health control subjects.
| Variable | Discover phase |
Validation phase |
||
|---|---|---|---|---|
| Control | Patient with CKD | Control | Patient with CKD | |
| Number | 80 | 120 | 40 | 60 |
| Age (years) | 53.7 (11.2) | 57.3 (14.5) | 54.1 (9.6) | 57.1 (15.8) |
| Men (%) | 56.3 | 58.3 | 57.5 | 58.3 |
| Body mass index (kg/m2) | 24.3 (3.5) | 25.4 (5.2) | 23.3 (2.8) | 25.9 (4.9) |
| Etiology of advanced CKD | NA | 70 hypertension | NA | 30 hypertension |
| 26 glomerular disease | 16 glomerular disease | |||
| 14 chronic nephritis | 8 chronic nephritis | |||
| 10 obstructive uropathy | 6 obstructive uropathy | |||
| CKD vintage (months) | NA | 38(32) | NA | 39 (34) |
| Lipid-lowering drug use (%) | NA | 24.5 | NA | 26.9 |
| Antihypertensive drug use (%) | NA | 87.9 | NA | 86.9 |
| SBP (mm Hg) | 121.7 (12.6) | 144.2 (15.2)b | 119.5 (13.3) | 141.2 (16.8)b |
| DBP (mm Hg) | 75.1 (11.3) | 82.4 (12.5)b | 74.6 (10.3) | 83.7 (10.8)b |
| eGFR (ml/min/1.73 m2) | 99.5 (15.1) | 14.4 (5.1)b | 97.4 (23.2) | 13.2 (4.2)b |
| BUN (mmol/L) | 5.11 (1.05) | 33.0 (16.2)b | 5.06 (0.87) | 34.6 (17.6)b |
| Serum creatinine (μmol/L) | 68.7 (14.1) | 516 (208)b | 73.8 (17.2) | 486 (216)b |
| Urine proteins (g/24 h) | NA | 1.86 (1.49)b | NA | 1.77 (1.65)b |
| Uric acid (μmol/L) | 303.3(87.6) | 460 (125)b | 289.4 (81.5) | 483 (129)b |
| Potassium (mmol/L) | 3.89 (1.23) | 4.63 (0.71)b | 3.94 (1.17) | 4.62 (0.79)b |
| Sodium (mmol/L) | 140.7 (3.2) | 141.4 (4.9) | 138.6 (3.8) | 139.2 (4.7) |
| Calcium (mmol/L) | 2.28 (0.42) | 2.14 (0.29)a | 2.19 (0.38) | 2.04 (0.41)a |
| Phosphate (mmol/L) | 1.08 (0.23) | 1.45 (0.34)b | 1.03 (0.21) | 1.51 (0.38)b |
| Albumin (g/L) | 48.5 (7.9) | 35.7 (6.1)b | 47.1 (8.4) | 36.6 (6.4)b |
| White blood cell (×109/L) | 6.63 (2.12) | 8.79 (2.75)b | 6.58 (1.97) | 9.29 (3.62)b |
| Red blood cell (×1012/L) | 4.51 (1.45) | 3.33 (0.78)b | 4.26 (1.51) | 3.13 (0.87)b |
| Haemoglobin (g/L) | 145.3 (12.8) | 99.6 (22.3)b | 142.4 (13.2) | 93.3 (25.6)b |
Results are summarized as mean (SD) and categorical variables are given as percentages.
N/A, not available.
P<0.05;
P<0.001.
3.2. Differential gene expression profile of IκBα/NF-κB, Keap1/Nrf2 and Wnt/β-catenin signaling
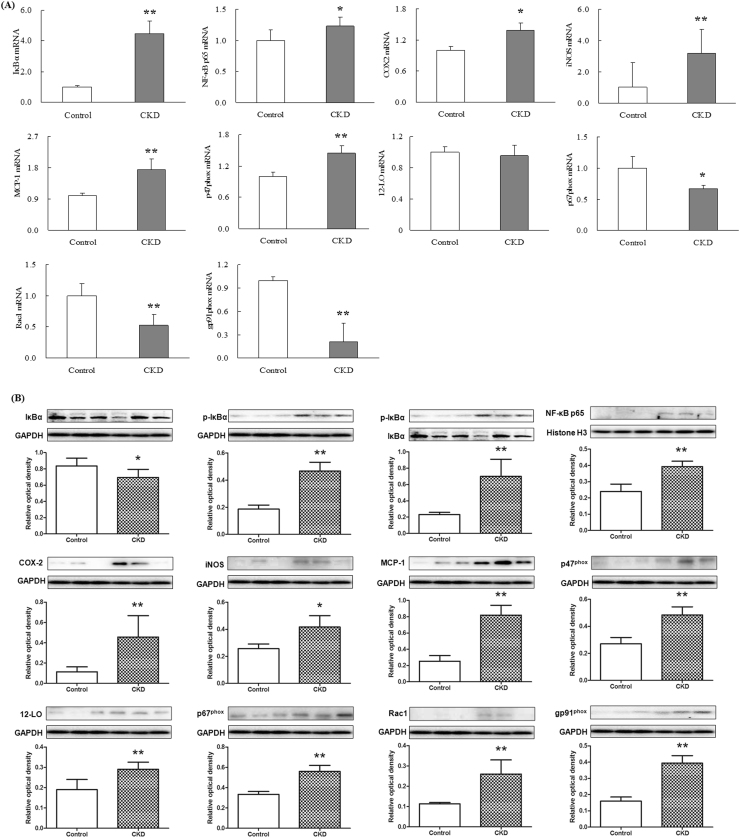
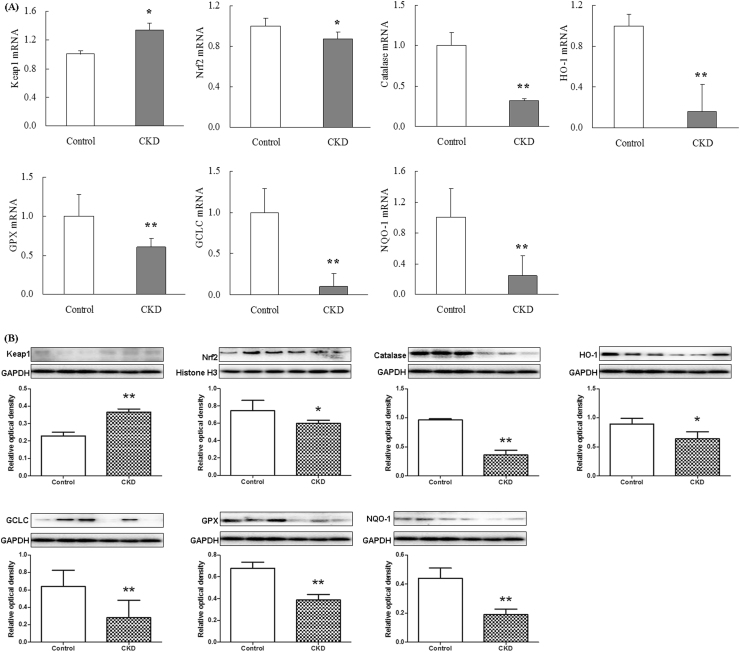
The plasma in patients with advanced CKD showed a marked up-regulation in nuclear translocation of p65 mRNA expression, indicating activation of NF-κB signaling. Activation of IκBα/NF-κB was accompanied by significant up-regulation of inflammatory genes including cyclooxygenase-2 (COX-2), inducible nitric oxide synthase (iNOS) and monocyte chemotactic protein-1 (MCP-1), and up-regulation of pro-oxidant gene p47phox and down-regulation of anti-oxidant system including Nrf2, catalase, heme oxygenase 1 (HO-1), glutathione peroxidase (GPX), glutamate-cysteine ligase catalytic subunit (GCLC) and NAD(P)H quinone dehydrogenase 1 (NQO1) (Figs. 2A and 3A). But p67Phox, Rac1 and gp91phox mRNA expression was significant downregulated in patients with advanced CKD. Western blot showed that activation of NF-κB p65 and significantly increased p-IκBα/IκBα ratio were accompanied by significant upregulation of inflammatory proteins including COX-2, iNOS, MCP-1, p47phox, 12-LO, p67phox, Rac1 and gp91phox and downregulation of anti-oxidant system including Nrf2, catalase, HO-1, GPX, GCLC and NQO1 (Figs. 2B and 3B). Activation of inflammatory and oxidative signaling pathways in patients with advanced CKD was accompanied by significant up-regulation of 15 Wnt genes including Wnt1, 2, 2b, 3, 4, 5a, 6, 7a, 7b, 8a, 8b, 9a, 9b, 10a and 16 and β-catenin genes in patients with advanced CKD. However, Wnt10b was significant downregulated and Wnt3a was not detected in patients with advanced CKD (Fig. 4A). Western blot showed that protein expressions of nuclear and cytoplasmic β-catenin and nuclear active β-catenin were dramatically upregulated in patients with CKD (Fig. 4B). Canonical Wnt/β-catenin signaling pathways was activated in patients with advanced CKD. Taken together, these findings point to activation of the pro-inflammatory, pro-oxidant, and down-regulation of Nrf2-mediated antioxidant and phase 2 detoxifying enzymes and related proteins accompanied by activated Wnt/β-catenin signaling pathways.
Fig. 2.
NF-κB target gene and protein expression in plasma from CKD patients. Quantitative real-time RT-PCR and Western blot depicting nuclear content of p65 active subunit of NF-κB and expression of COX-2, iNOS, MCP-1, P47Phox, p67Phox, Rac1 and gp91phox in the plasma of the healthy controls and patients with advanced CKD. *P<0.05, **P<0.01 compared with the healthy controls.
Fig. 3.
Anti-oxidative stress Nrf2 and Nrf2 target gene and protein expression in plasma from CKD patients. Quantitative real-time RT-PCR and Western blot depicting Nrf2, catalase, HO-1, GPX, GCLC and NQO1 in the plasma of the healthy controls and patients with advanced CKD. *P<0.05, **P<0.01 compared with the healthy controls.
Fig. 4.
Total 16 Wnt and β-catenin gene expression as well as β-catenin and active β-catenin protein expression in plasma from CKD patients. Quantitative real-time RT-PCR including Wnt1, 2, 2b, 3, 4, 5a, 6, 7a, 7b, 8a, 8b, 9a, 9b, 10a, 10b and 16 and β-catenin as well as nuclear and cytoplasmic β-catenin and active β-catenin protein expression in the plasma of the healthy controls and patients with advanced CKD. *P<0.05, **P<0.01 compared with the healthy controls.
3.3. Important differential plasma and urine metabolites
Initially, variables were selected according to the VIP values from S-plots, which reflect the influence of each variable in the two groups. 317 and 199 variables from plasma samples had VIP values greater than 1.5 in positive ion and negative ion modes, respectively. Subsequently, 212 and 198 variables from plasma samples had a P<0.05 based one-way ANOVA in positive ion and negative ion modes, respectively. After excluding xenobiotics and different fragment ions from the same metabolites 65 and 59 metabolites were identified in positive ion and negative ion modes, respectively (Table S1). Similarly, 353 and 198 variables from urine samples had a VIP >1.5 in positive ion and negative ion modes, respectively. Subsequently, 238 and 134 variables from urine samples had a P<0.05 based on one-way ANOVA in positive ion and negative ion modes, respectively. After excluding the xenobiotics and different fragment ions from the same metabolites 89 and 39 metabolites were identified in positive ion and negative ion modes, respectively (Table S2). PCA score plots and heatmap display showed that plasma or urine metabolites could separate patients with CKD from control (Fig. 5A, B and S1). Using a combination of the VIP values and one-way ANOVA analysis, 124 plasma metabolites and 128 urine metabolites were identified (Appendix A, Appendix A). The z-score showed that the metabolites in plasma and urine were significantly altered in CKD (Fig. 5C and D). For each of these 124 plasma metabolites and 128 urine metabolites, Fig. 6A and B shows the mean ratio of each plasma or urinary metabolite in CKD patients versus controls, plotted against their corresponding minus logarithm of P value (Tables S1 and S2). Further, 119 out of 125 plasma metabolites and 106 out of 128 urine metabolites had a P<0.05 based on the combination of Mann-Whitney U test and Bonferroni-adjusted FDR methods (Tables S1 and S2).
Fig. 5.
Metabolomic profiling of plasma and urine samples from two groups identifies metabolites that distinguish patients with CKD from controls. Outlier analysis of 124 identified metabolites in plasma (B) and 128 identified metabolites in urine (C). PCA of metabolites from 120 CKD samples and 80 control samples. Different principal components have a different contribution to separating CKD from controls in this study. Green crosses and red triangles represent controls and CKD, respectively. (D) Control-based z-score plot of metabolomic alterations from plasma and urine in control and patients with CKD. Each point represents an individual metabolite in one sample. Z-score plots for the data normalized to the mean of the control samples. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Fig. 6.
The geometric mean ratio of each metabolite for individuals in CKD patients versus controls. The y-axis shows minus logarithm of P value. The x-axis shows the logarithm of ratio of CKD/control of each plasma sample (A) or urine sample (B). The log2(CKD/Control) with a value >0 indicated a relatively higher intensity present in CKD patients, whereas a value <0 indicated a relatively lower intensity compared with the healthy subjects. Diagnostic performances of the 25 metabolites in both plasma (D) and urine (E) based on the PLS-DA model. The black dots and black circles with red squares are for the incorrectly predicted samples in patient with CKD and controls, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
40 metabolites were significantly altered in both plasma and urine (Table 3). Compared with the controls, 18 metabolites were significantly increased in CKD plasma while they were significantly decreased in CKD urine; 6 metabolites were significantly decreased in CKD plasma while they were significantly increased in CKD urine; 11 metabolites were significantly increased in both plasma and urine of CKD patients; and 5 metabolites were significantly decreased in both plasma and urine of CKD patients. Additionally, 79 out of 119 plasma metabolites were significantly altered in CKD patients and 66 out of 106 urine metabolites were significantly altered in CKD patients.
Table 3.
plasma and urine differential metabolites in CKD patients compared to controls.
| Plasma |
Urine |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Metabolites |
VIPa |
FCb |
Pc |
FDRd |
AUCe |
95%Cl |
Sensitivity (%) |
Specificity (%) |
VIPa |
FCb |
Pc |
FDRd |
AUC |
95%Cl |
Sensitivity (%) |
Specificity (%) |
| Increased 18 metabolites in plasma | Decreased 18 metabolites in urine | |||||||||||||||
| p-Cresol sulfate | 22.99 | 10.1 | 1.08E−13 | 2.28E−13 | 0.86 | 0.822–0.916 | 89.6 | 87.6 | 19.98 | 0.29 | 2.55E−27 | 2.72E−26 | 0.94 | 0.910–0.971 | 93.6 | 87.9 |
| Indoxyl sulfate | 18.75 | 5.10 | 8.59E−29 | 8.45E−28 | 0.96 | 0.936–0.980 | 95.6 | 89.7 | 2.66 | 0.85 | 4.61E−12 | 1.20E−11 | 0.88 | 0.826–0.924 | 89.6 | 88.7 |
| 1-Methylguanosine | 10.52 | 4.59 | 1.03E−46 | 6.08E−45 | 0.99 | 0.986–1.00 | 99.5 | 97.1 | 7.33 | 0.55 | 1.58E−22 | 1.35E−21 | 0.92 | 0.865–0.958 | 90.8 | 92.8 |
| Hippuric acid | 4.95 | 5.98 | 2.42E−11 | 3.96E−11 | 0.87 | 0.831–0.923 | 88.7 | 93.5 | 9.32 | 0.61 | 3.74E−19 | 1.91E−18 | 0.88 | 0.834–0.928 | 86.9 | 88.2 |
| Phenylalanine | 3.95 | 1.90 | 7.91E−22 | 3.89E−21 | 0.91 | 0.840–0.927 | 87.5 | 88.6 | 6.91 | 0.43 | 6.27E−22 | 5.02E−21 | 0.91 | 0.866–0.945 | 94.3 | 91.9 |
| p-Cresol | 3.90 | 8.30 | 1.19E−18 | 4.37E−18 | 0.95 | 0.919–0.972 | 92.5 | 96.1 | 4.23 | 0.44 | 1.30E−17 | 5.74E−17 | 0.91 | 0.869–0.945 | 90.5 | 87.2 |
| Octanoylcarnitine | 3.03 | 9.08 | 2.39E−11 | 2.98E−11 | 0.94 | 0.889–0.960 | 92.5 | 94.6 | 2.82 | 0.49 | 2.89E−05 | 4.46E−05 | 0.87 | 0.816–0.917 | 89.5 | 85.6 |
| Myoinositol | 2.97 | 6.83 | 7.51E−15 | 1.70E−14 | 0.90 | 0.837–0.928 | 89.6 | 87.2 | 3.82 | 0.67 | 1.10E−34 | 1.76E−33 | 0.88 | 0.833–0.937 | 85.7 | 89.6 |
| 11-Oxohexadecanoic acid | 2.70 | 3.39 | 1.72E−06 | 2.03E−06 | 0.99 | 0.991–1.00 | 98.6 | 99.7 | 2.91 | 0.36 | 3.38E−16 | 1.27E−15 | 0.89 | 0.840–0.932 | 87.5 | 89.1 |
| Dopamine | 2.39 | 8.63 | 2.44E−17 | 8.46E−17 | 0.88 | 0.862–0.943 | 88.8 | 96.4 | 3.22 | 0.21 | 3.39E−63 | 1.45E−61 | 0.89 | 0.843–0.942 | 89.6 | 95.4 |
| 4-Aminohippuric acid | 2.26 | 3.71 | 1.77E−18 | 3.49E−18 | 0.87 | 0.817–0.918 | 89.6 | 93.2 | 7.35 | 0.10 | 1.40E−35 | 2.99E−34 | 0.95 | 0.916–0.978 | 98.8 | 93.4 |
| Indole−3-carboxylic acid | 2.09 | 3.54 | 1.99E−11 | 3.35E−11 | 0.85 | 0.791–0.902 | 86.5 | 89.6 | 2.56 | 0.47 | 2.60E−55 | 8.32E−54 | 0.88 | 0.822–0.926 | 89.6 | 85.6 |
| Xanthine | 1.94 | 4.19 | 2.06E−13 | 4.06E−13 | 0.87 | 0.779–0.917 | 91.2 | 87.6 | 4.20 | 0.47 | 1.11E−04 | 1.60E−04 | 0.87 | 0.823–0.914 | 87.5 | 86.2 |
| Hypoxanthine | 1.87 | 5.38 | 9.67E−16 | 2.28E−15 | 0.94 | 0.934–0.986 | 93.5 | 99.7 | 8.61 | 0.19 | 6.38E−22 | 4.80E−21 | 0.96 | 0.928–0.983 | 97.5 | 95.2 |
| Succinic acid | 1.86 | 5.02 | 1.46E−16 | 4.52E−16 | 0.91 | 0.865–0.953 | 92.5 | 98.7 | 2.44 | 0.14 | 1.17E−10 | 2.67E−10 | 0.86 | 0.809–0.908 | 87.7 | 86.9 |
| Kynurenine | 1.73 | 3.60 | 6.35E−16 | 1.56E−15 | 0.85 | 0.805–0.904 | 78.1 | 83.6 | 6.04 | 0.24 | 2.24E−21 | 1.59E−20 | 0.95 | 0.913–0.972 | 99.5 | 97.6 |
| Glutamine | 4.32 | 2.14 | 1.84E−06 | 2.36E−06 | 0.76 | 0.689–0.825 | 74.3 | 78.6 | 3.53 | 0.42 | 8.53E−18 | 3.90E−17 | 0.87 | 0.825–0.918 | 87.6 | 86.9 |
| Targinine | 3.94 | 2.14 | 9.05E−03 | 9.54E−03 | 0.62 | 0.537–0.686 | 68.7 | 72.6 | 2.67 | 0.70 | 8.32E−04 | 1.15E−03 | 0.82 | 0.754–0.880 | 78.9 | 92.5 |
| Increased 11 metabolites in plasma | Increased 11 metabolites in urine | |||||||||||||||
| Trimethylamine | 10.65 | 1.50 | 6.97E−20 | 1.46E−19 | 0.89 | 0.856–0.964 | 83.8 | 92.5 | 2.45 | 1.56 | 4.31E−17 | 5.44E−16 | 0.84 | 0.774–0.901 | 87.5 | 85.6 |
| Uridine | 2.26 | 4.70 | 9.43E−22 | 4.28E−21 | 0.92 | 0.874–0.960 | 92.6 | 99.5 | 3.56 | 3.65 | 7.55E−06 | 1.26E−05 | 0.87 | 0.817–0.911 | 78.9 | 84.6 |
| Trimethylamine-N-oxide | 3.71 | 3.01 | 8.98E−20 | 5.37E−20 | 0.89 | 0.851–0.934 | 87.8 | 92.5 | 2.49 | 1.24 | 1.56E−03 | 1.74E−03 | 0.87 | 0.814–0.918 | 88.8 | 89.3 |
| Deoxyadenosine | 3.05 | 5.28 | 5.24E−36 | 4.26E−35 | 0.97 | 0.934–0.996 | 99.6 | 94.3 | 4.15 | 1.69 | 1.53E−69 | 9.79E−68 | 0.89 | 0.838–0.933 | 87.5 | 97.5 |
| Oleamide | 5.28 | 1.71 | 6.84E−06 | 7.93E−06 | 0.86 | 0.816–0.913 | 82.6 | 86.7 | 10.03 | 1.47 | 6.31E−16 | 2.13E−15 | 0.88 | 0.829–0.917 | 84.2 | 87.5 |
| Aldosterone | 4.47 | 5.72 | 7.23E−35 | 5.22E−34 | 0.96 | 0.915–0.972 | 95.8 | 94.3 | 3.41 | 5.32 | 3.07E−08 | 5.95E−08 | 0.97 | 0.948–0.991 | 98.5 | 99.6 |
| Uric acid | 4.37 | 2.15 | 9.08E−24 | 2.46E−23 | 0.90 | 0.871–0.956 | 87.5 | 92.3 | 9.16 | 1.53 | 7.22E−12 | 1.81E−11 | 0.89 | 0.835–0.929 | 87.5 | 86.6 |
| cis-Aconitic acid | 2.33 | 2.94 | 2.43E−09 | 3.59E−09 | 0.82 | 0.752–0.883 | 86.5 | 76.3 | 3.2 | 2.75 | 5.00E−43 | 1.28E−41 | 0.90 | 0.850–0.942 | 91.8 | 86.9 |
| Creatinine | 3.84 | 10.8 | 2.25E−18 | 4.31E−18 | 0.97 | 0.938–0.982 | 94.6 | 92.5 | 4.97 | 1.02 | 4.09E−02 | 7.20E−01 | 0.50 | 0.423–0.575 | 54.2 | 68.7 |
| Homoserine | 2.28 | 1.19 | 4.45E−03 | 4.45E−03 | 0.60 | 0.503–0.666 | 65.5 | 59.9 | 4.09 | 1.23 | 1.88E−17 | 8.02E−17 | 0.87 | 0.813–0.915 | 82.1 | 91.5 |
| Phenol sulfate | 11.87 | 2.05 | 1.03E−03 | 1.15E−03 | 0.60 | 0.529–0.680 | 65.8 | 82.2 | 8.56 | 4.18 | 8.07E−03 | 9.33E−03 | 0.90 | 0.846–0.947 | 92.5 | 95.5 |
| Decreased 6 metabolites in plasma | Increased 6 metabolites in urine | |||||||||||||||
| Taurine | 13.46 | 0.64 | 1.22E−36 | 1.13E−35 | 0.97 | 0.925–0.992 | 93.2 | 97.6 | 1.82 | 2.40 | 1.66E−14 | 5.06E−14 | 0.89 | 0.843–0.937 | 86.5 | 98.9 |
| Tryptophan | 9.26 | 0.52 | 2.22E−26 | 7.22E−26 | 0.86 | 0.812–0.922 | 85.6 | 89.2 | 3.55 | 3.44 | 2.92E−20 | 1.78E−19 | 0.92 | 0.899–0.954 | 88.5 | 93.3 |
| Cysteine | 2.95 | 0.20 | 1.88E−37 | 2.04E−36 | 0.91 | 0.866–0.952 | 87.6 | 93.5 | 1.71 | 2.24 | 1.48E−19 | 8.61E−19 | 0.91 | 0.870–0.945 | 92.1 | 88.9 |
| Deoxyuridine | 6.60 | 0.31 | 7.56E−30 | 8.92E−29 | 0.90 | 0.844–0.944 | 91.2 | 95.4 | 2.42 | 8.96 | 2.14E−08 | 4.35E−08 | 0.94 | 0.905–0.969 | 93.7 | 90.8 |
| Kynurenic acid | 2.21 | 0.20 | 6.49E−19 | 2.55E−18 | 0.91 | 0.883–0.955 | 85.6 | 98.6 | 4.46 | 1.80 | 4.26E−35 | 7.79E−34 | 0.88 | 0.839–0.932 | 92.3 | 87.7 |
| Carnitine | 2.65 | 0.38 | 2.35E−03 | 2.43E−03 | 0.61 | 0.551–0.689 | 67.5 | 62.4 | 2.57 | 1.97 | 1.31E−03 | 1.78E−03 | 0.71 | 0.645–0.780 | 71.6 | 75.3 |
| Decreased 5 metabolites in plasma | Decreased 5 metabolites in urine | |||||||||||||||
| Inosine | 10.06 | 0.36 | 5.48E−24 | 4.04E−23 | 0.91 | 0.873–0.950 | 93.5 | 94.3 | 4.85 | 0.62 | 8.43E−05 | 1.23E−04 | 0.74 | 0.678–0.812 | 78.5 | 73.6 |
| 1-Methylxanthine | 4.63 | 0.61 | 3.36E−25 | 9.94E−25 | 0.89 | 0.882–0.954 | 92.5 | 87.6 | 1.77 | 0.09 | 2.07E−05 | 3.23E−05 | 0.86 | 0.813–0.910 | 84.5 | 76.9 |
| Lysine | 7.38 | 0.50 | 7.50E−21 | 3.16E−20 | 0.88 | 0.819–0.933 | 89.6 | 91.5 | 15.05 | 0.19 | 2.76E−88 | 3.53E−86 | 1.00 | 1.00–1.00 | 100 | 100 |
| Alanine | 3.53 | 0.93 | 1.02E−03 | 1.08E−03 | 0.64 | 0.581–0.821 | 81.5 | 56.1 | 4.57 | 0.25 | 3.30E−28 | 3.84E−27 | 0.87 | 0.818–0.922 | 87.5 | 83.7 |
| N-acetyltryptophan | 39.73 | 0.21 | 6.48E−17 | 2.12E−16 | 0.66 | 0.553–0.764 | 92.1 | 67.8 | 3.32 | 0.32 | 1.10E−06 | 1.93E−06 | 0.83 | 0.755–0.880 | 74.9 | 83.6 |
VIP was obtained from OPLS-DA model with a threshold of 1.5.
FC was obtained by comparing those metabolites in patients with CKD with controls; FC with a value >1 indicated a relatively higher intensity present in patients with CKD, whereas a value <1 indicated a relatively lower intensity compared with controls.
P values from one-way ANOVA.
FDR value was obtained from the adjusted P value of FDR correction by Benjamini-Hochberg method.
The AUC with 95%CI was obtained from PLS-DA-based ROC curves of the identified metabolites.
3.4. ROC curve and prediction model
To assess the potential value of the differential metabolites that can discriminate CKD from controls, PLS-DA-based ROC curve was prepared. 25 out of 40 significantly altered metabolites in both plasma and urine had AUC value greater than 0.85 and displayed sensitivities and specificities above 85% (Table 3). We further performed a combination ROC curves for each metabolite (data not show). The AUC value was larger than each metabolite in plasma or urine. Additionally, predicted class probabilities showed that 116 out of 120 plasma samples from CKD were correctly grouped (96.7% sensitivity). All 80 controls were located in control area (100% specificity) (Fig. 6C). 120 urine samples from CKD were correctly grouped (100% sensitivity) and 79 out of 80 controls were located in control area (98.7% specificity) (Fig. 6D).
Though 39 metabolites only in plasma had an AUC ≥0.85, only 27 metabolites displayed sensitivities and specificities of above 85% (data not show). PCA and heatmap of 27 plasma metabolites showed that CKD and control can be clearly separated (Fig. 7A and B). To understand the functional role of these altered plasma metabolites, the KEGG metabolic library was used by Metaboanalyst (Fig. S2A). The top five pathways in plasma by p value (top four) or impact (top one) were shown in Table S3. Altered these plasma pathways in CKD indicates that disturbed certain central metabolites have an important effect on multiple pathways that are interconnected. We further constructed BLR model to assess these metabolites for the discrimination between CKD and controls. Canavaninosuccinate, 5-methoxytryptophan (5-MTP), homocystine and leucine were identified by forward stepwise analysis. Compared with the controls, these metabolites were significantly altered in CKD (Fig. 7D). These metabolites have a high AUC, sensitivity and specificity and thus could be considered as suitable plasma biomarkers to best predict CKD status (Fig. 7C). To further validate four metabolites that could be useful for predicting CKD, we analyzed the correlation between each metabolites and eGFR or creatinine. Except for leucine, canavaninosuccinate, 5-MTP and homocystine were good positively or negatively correlated with eGFR (R>0.9318) (Fig. 7E). 5-MTP and homocystine were good correlated with creatinine (R>0.9507) (Fig. 7F).
Fig. 7.
Multivariate analyses and correlation analysis of four significantly altered plasma metabolites in CKD patients. (A) The PCA score scatter plot using 27 differential metabolites from plasma sample between the CKD patients and controls. (B) Heatmap of 27 differential metabolites from plasma sample between the CKD patients and control. Red and blue in heatmap indicates increased and decreased levels, respectively. Rows: sample; columns: metabolite. THCA: trihydroxycoprostanoic acid. (C) PLS-DA-based ROC curves for the predictive power of four plasma biomarkers and for distinguishing CKD from controls. (D) Bar graphs of significant changes of four plasma biomarkers between CKD and controls. Abundance is represented as the relative intensity (y axis) of different groups (x axis). The statistical significance of differences between the two groups was marked. ***P<0.001 compared to the controls. (E) Correlation between canavaninosuccinate, 5-methoxytryptophan, homocystine and leucine levels (peak intensity) measured by the UPLC-MS and eGFR by calculated formula. (F) Correlation between canavaninosuccinate, 5-methoxytryptophan, homocystine and leucine levels (peak intensity) measured by the UPLC-MS and serum creatinine (µmol/L) measured by the clinical laboratory. The x-axes show the eGFR value or serum creatinine concentration. The y-axis shows the peak intensity of each plasma sample. The correlation coefficient is shown in each graph. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Similarly, 21 urine metabolites had high sensitivity and specificity to clearly separate CKD from the control group (Fig. 8A and B). The top six pathways in plasma by p value (top four) or impact (top two) were shown in Table S4. Altered these plasma pathways in CKD indicates that disturbed certain central metabolites have an important effect on multiple pathways that are interconnected. Five metabolites including 1-methyladenosine, spermidine, xanthosine, xanthurenic acid and citrulline were identified by BLR. Compared with the controls, these metabolites were significantly altered in CKD patients (Fig. 8D) and thus could be considered as suitable urine biomarkers of advanced CKD (Fig. 8C). Xanthosine, xanthurenic acid and citrulline were good positively or negatively correlated with eGFR (R>0.9355) (Fig. 8E). Only xanthosine was good correlated with creatinine (R>0.9223) (Fig. 8F).
Fig. 8.
Multivariate analyses and correlation analysis of five significantly altered urinary metabolites in CKD patients. (A) The PCA score scatter plot using 21 differential metabolites from urine sample between the CKD patients and controls. (B) Heatmap of 27 differential metabolites from urine sample between the CKD patients and control. Red and blue in heatmap indicates increased and decreased levels, respectively. Rows: sample; columns: metabolite. THCA: trihydroxycoprostanoic acid. (C) PLS-DA-based ROC curves for the predictive power of five urine metabolites and for distinguishing CKD from controls. (D) Bar graphs of significant changes of five urine metabolites between CKD and controls. Abundance is represented as the relative intensity (y axis) of different groups (x axis). The statistical significance of differences between the two groups was marked. ***P<0.001 compared to the controls. (E) Correlation between 1-methyladenosine, spermidine, xanthosine, xanthurenic acid and citrulline levels (peak intensity) measured by the UPLC-MS and eGFR by calculated formula. (F) Correlation between 1-methyladenosine, spermidine, xanthosine, xanthurenic acid and citrulline levels (peak intensity) measured by the UPLC-MS and serum creatinine (µmol/L) measured by the clinical laboratory. The x-axes show the eGFR value or serum creatinine concentration. The y-axis shows the peak intensity of each urine sample. The correlation coefficient is shown in each graph. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
3.5. Differential metabolite validation
For validation of 25 urine and plasma metabolites, these metabolites were measured in the plasma and urine of an independent cohort. 19 metabolites had a P<0.05 and AUC ≥0.85 and a sensitivity and specificity above 85% (Table S5). Predicted class probability showed that plasma samples achieved a sensitivity of 88.3% and a specificity of 92.5%, respectively (Fig. S3A). Urine samples achieved a sensitivity of 91.7% and a specificity of 92.5%, respectively (Fig. S3B).
To evaluate the diagnostic power of the four potential plasma and five potential urine biomarkers for CKD, data from the above-mentioned independent cohort were analyzed using the previously described models. Four plasma biomarkers could separate CKD from controls (Fig. 9A) and had a high sensitivity and specificity (Fig. 9B). The sensitivity and specificity of the combination of four plasma biomarkers was 93.3% and 92.5%, respectively (Fig. 9C). Similarly, five urine biomarkers could separate CKD from controls (Fig. 9D) with high sensitivity and specificity (Fig. 9E). The sensitivity and specificity of the combination of four plasma biomarkers were 91.9% and 92.5%, respectively (Fig. 9F). Thus four plasma and five urine metabolites could be considered as suitable biomarkers of advanced CKD.
Fig. 9.
Biomarker validations from four plasma metabolites and five urine metabolites from additional 60 CKD patients and 40 age-matched healthy controls. The three-dimensional PCA score scatter plot using four metabolites (canavaninosuccinate, 5-methoxytryptophan, homocystine and leucine) in plasma (A) and five metabolites (1-methyladenosine, spermidine, xanthosine, xanthurenic acid and citrulline) in urine (D) between the patients with CKD and controls. PLS-DA-based ROC curves for the predictive power of four plasma biomarkers (B) and five urine biomarkers (E) for distinguishing CKD from controls. Diagnostic performance of the four plasma biomarkers (C) and five urine biomarkers (F) based on the PLS-DA model. The black dots or black circles with red squares are for the incorrectly predicted samples in patient with CKD and controls, respectively. (G) IPA with MetPA of 25 potential biomarkers combined plasma and urine. The size and color of each circle was based on pathway impact value and p-value, respectively. (H) Visualization of the remarkably disturbed metabolic pathways in plasma and urine by MetScape analysis. The identified metabolites in the current study were shown by red hexagons. Hexagons with green lines means that the significantly changes of the identified metabolite in CKD had statistical significance (P<0.05). The size of hexagons showed the fold change of the differential metabolite in CKD relative to control. In addition, pink hexagons showed metabolites participating in the metabolic pathway but not been identified in the current study. CKD were associated with purine metabolism, TCA cycle, aminoacyl-tRNA biosynthesis, nitrogen metabolism, taurine and hypotaurine metabolism, biotin metabolism, pantothenate and CoA biosynthesis, inositol phosphate metabolism, galactose metabolism, ascorbate and aldarate metabolism, primary bile acid biosynthesis, pyrimidine metabolism and steroid hormone biosynthesis. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
3.6. Metabolite and phenotype
Plasma metabolites 5-MTP and homocystine and urinary metabolite citrulline were good correlated with both eGFR and creatinine. Clinical factors, such as age, gender, CKD vintage and medication was usually incorporated to establish diagnostic models, and the variables were used to enhance metabolite biomarker models [20]]. To enhance 5-MTP, homocystine and citrulline models, three clinical factors (age, gender and CKD vintage) were selected to be each metabolite for inclusion in the predictive model. The enhanced metabolite model showed excellent AUC for differentiating patients with CKD from controls, with high sensitivity and specificity (Fig. 10).
Fig. 10.
ROC curves analysis of enhanced PLS−DA model for the predictive power of combined each metabolite (5-MTP, homocystine and citrulline) and three general clinical parameters (age, gender and CKD vintage) for differentiating patients with CKD from controls.
4. Discussion
Oxidative stress and inflammation plays a key role in the pathogenesis of various CKD and its adverse complications. The present gene expression and metabolomics data indicated the activated inflammation in patients with CKD. The patients with CKD showed a significant increase in nuclear translocation of p65, up-regulation of inflammatory and pro-oxidant gene expressions as well as down-regulation of anti-oxidant system gene expressions. Leal et al. determined the levels of NF-κB and Nrf2 mRNA in nondialysis and hemodialysis patients and healthy controls. NF-κB mRNA expression was increased and Nrf2 mRNA expression was decreased in nondialysis patients compared with healthy individuals, but did not arrive at statistical significance [21] This study also demonstrated that NF-κB mRNA expression was significantly increased and Nrf2 mRNA expression was significantly decreased in hemodialysis patients compared with nondialysis patients. Another study indicated that hemodialysis patients had significantly up-regulation of NF-κB mRNA expression and dramatically down-regulation of Nrf2 and NQO1 mRNA expressions compared with healthy controls. Plasma levels of TNF-α and malondialdehyde were significantly increased in hemodialysis patients compared with healthy controls [22] It has been reported that NF-kB p65 mRNA and NF-kB p65 protein expression levels were both significantly higher in type 2 diabetic nephropathy compared with healthy controls [23] Additionally, many studies have demonstrated significantly up-regulation of oxidative stress and inflammation proteins and down-regulation of anti-oxidant system proteins in patients with CKD [24], [25], [26], [27].
Wang et al. reported that an increase in mRNA expression of β-catenin and axin2 in kidney tissues of patients with lupus nephritis compared with control kidney tissues, accompanied by an increase of β-catenin protein expression [28] Their study also showed that β-catenin mRNA expression was significantly higher in lupus nephritis patients without renal interstitial fibrosis compared to those with renal interstitial fibrosis and increased β-catenin mRNA expression positively correlated with the creatinine clearance rate (Ccr). Another study showed that canonical Wnt/β-catenin and Wnt/Ca2+ signaling were both dramatically changed with the development of chronic kidney damage [15]. Wnt3, lymphoid enhancer-binding factor 1 and T-cell factor 1 showed differential regulation in canonical Wnt/β-catenin pathway. Target genes fibronectin 1, CD44, matrix metalloproteinase 7 and nitric oxide synthase 2 were up-regulated in the progression of renal damage. Altered Wnt/Ca2+ pathway was demonstrated by up-regulation expression of Wnt6, Wnt7a, protein kinase C, Cam Kinase II and nuclear factor of activated T-cell transcription factors and the target gene vimentin. Taken together, these findings point to activation of the pro-inflammatory, pro-oxidant, and down-regulation of Nrf2-mediated antioxidant system were accompanied by activated Wnt/β-catenin signaling pathways.
On the basis of our model construction, multiple step metabolite selection and cross validation, 124 plasma metabolites and 128 urine metabolites were identified and 40 metabolites were significantly altered in both plasma and urine. 25 differential metabolites from combined plasma and urine were successfully identified which robustly separated CKD patients from controls and they reflected dysregulation of 30 metabolic pathways (Fig. 9G and H). Additionally, 27 plasma metabolites and 21 urine metabolites were related to 11 and 19 metabolic pathways, respectively (Fig. S4). Four plasma metabolites (canavaninosuccinate, 5-MTP, homocystine, leucine) and five urinary metabolites (1-methyladenosine, spermidine, xanthosine, xanthurenic acid, and citrulline) were identified by binary logistic regression. Part of them was good correlated with eGFR or serum creatinine. These abnormal altered metabolites could be restored by treatment of enalapril and lisinopril. Plasma metabolites 5-MTP and homocystine and urinary metabolite citrulline were good correlated with both eGFR and creatinine. General clinical factors were incorporated to establish diagnostic models. The enhanced metabolite model showed 5-MTP, homocystine and citrulline have an excellent AUC with high sensitivity and specificity for predictive CKD. They could be considered as additional GFR-associated biomarker candidates and for indicating advanced renal injury.
Plasma levels of 4 and 6 AA as well as urine levels of 2 and 22 AA were significantly increased and decreased in CKD, respectively (Tables S1 and S2). Previous studies have shown significant changes in plasma and urine AA profiles and significant reduction of plasma concentrations of most AA in CKD patients [29], [33]. Increased phenylalanine and alanine as well as decreased tryptophan were observed in plasma of advanced CKD patients [33]. Other studies showed increased plasma phenylalanine and decreased tryptophan in CKD patients [30] and decreased urine phenylalanine in glomerulonephritides patients [31]. Another study demonstrated the association of altered plasma valine, alanine, glutamate and glycine with different stages of CKD [32]. Additionally, urine asparagine, leucine, proline and citrulline were significant increased in advanced CKD patients [3], [34]. The present study identified 5-HTP, homocystine and leucine as plasma biomarker candidates. 5-MTP is synthesized from tryptophan via tryptophan hydroxylase-1 and hydroxyindole O-methyltransferase [35]. It has been reported that fibroblasts produce and release 5-MTP which promotes COX-2 over-expression [36]. In fact CKD is associated with expansion of fibroblasts and upregulation of COX-2 expression in the kidney and cardiovascular tissues [37]. For this reason 5-MTP synthesis is a valuable lead for new therapeutic targets and anti-inflammatory drug development. 5-HTP was first identified as a significant metabolite in advanced CKD. Plasma homocystine has been demonstrated to increase in parallel with the decline of the GFR [38]. Several reports have shown that hemodialysis lowers plasma homocystine only transiently [39], [40] and elevated plasma homocystine can not be lowered efficiently by peritoneal dialysis [41]. Interestingly, decreased plasma leucine was reported in uremic patients, stages 4–5 CKD and diabetic nephropathy [42], [43]. In the present study citrulline was identified as urine biomarker. Several studies have shown increased urine citrulline in advanced CKD [33]. The CKD-induced changes in AA metabolism have been, in part, attributed to the systemic inflammation and metabolic acidosis which are common features of CKD [44]. In fact compared with the controls, CKD patients showed significant increase in plasma TNF-α and IL-6 levels reflecting the presence of systemic inflammation. Several key oxidant pathways can lead to the excessive generation of oxidative intermediates during disease progression. Amino acids were natural targets of these pathways and serve as markers of organ injury in addition to serving as a reactive oxygen and nitrogen species intermediate [45]. The protein phenylalanine residues subjected to hydroxyl radical damage form meta-tyrosine and ortho-tyrosine. Oxidation reactions of tyrosine residues include cross-linking (to form oo′-dityrosine; mediated by tyrosine radicals, reactive oxygen species and reactive nitrogen species), chlorination (to form 3-chlorotyrosine; catalyzed by myeloperoxidase) and nitration (to from 3-nitrotyrosine; mediated by reactive nitrogen species) [45].
Abnormal AA metabolites including indoxyl sulfate, indole-3-carboxylic acid, kynurenine, kynurenic acid, 4-aminohippuric acid, hippuric acid and xanthurenic acid (byproducts of tryptophan metabolism), p-cresol, p-cresol sulfate, dopamine (byproduct of tyrosine metabolism), taurine (byproduct of cystcine metabolism) and octanoylcarnitine were observed in plasma and urine of CKD patients. As noted above under normal condition filtered AA is fully reabsorbed in proximal tubules. Tubulointerstitial injury is a constant feature of all forms of CKD and the degree of tubulointerstitial injury correlates better than glomerular injury with the prognosis of the disease [46]. Nkuipou-Kenfack et al. found a strong association between plasma and urinary metabolites and peptides with kidney function and CKD progression [34]. They identified serum hydroxykynurenine as a biomarker [34]. The first and rate-limiting step in tryptophan conversion to kynurenine is catalyzed by indoleamine 2,3-dioxygenase, kynurenine is then metabolized to kynurenic acid. Inflammatory states have been shown to increase production of kynurenine by upregulation of indoleamine 2,3-dioxygenase in endothelial cells [47]. Zinellu et al. found a correlation between kynurenine/tryptophan ratio and oxidative stress indices malondialdehyde and allantoin/uric acid ratio [48], which also confirms previous study on the association between oxidative stress and inflammation in nephropathic pre-dialyzed patients [49], [50] Thus systemic inflammation which is invariably present in CKD patients must contribute to increased plasma kynurenine. It has been reported that anti-oxidant taurine can ameliorate oxidative stress and inflammation in CKD [48], [51]. Elevated plasma indoxyl sulfate and hippuric acid have been regarded as biomarkers for predicting the stages of CKD [52]. Additionally, other uremia solutes p-cresol and p-cresol sulfate are significantly altered in plasma and urine of CKD patients [5], [53]. Accumulated evidence indicated that oxidative stress and inflammation in patients with CKD were associated with marked alteration of amino acid concentrations in plasma and urine.
TMA and TMAO are byproducts of bacterial metabolism of phosphatidylcholine, choline, or L-carnitine. Plasma TMA and TMAO are elevated in CKD patients and contribute to the cardiovascular disease and overall mortality in this population. Both plasma and urine TMA and TMAO were significantly elevated in our CKD patients. Plasma TMAO concentration in CKD patients has been shown to directly correlate with plasma concentration of urea and creatinine, indicating close association of TMAO with the degree of renal insufficiency [54], [55]. Additionally, increased urine TMAO in CKD patients observed in our study is consistent with previous study [56]. The excellent review reported that intestine and its microbial bacteria were the main source of several well-known pro-inflammatory uremic toxins such as TMA, TMAO, indoxyl sulfate, p-cresol sulfate in patients with CKD [57]. Taken together, increased TMA and TMAO were closely associated with oxidative stress and inflammation in patients with CKD.
Urine spermidine was significantly reduced in our CKD patients. Several studies have demonstrated elevated plasma polyamine in uremia [58], [59]. Serum spermine has been shown to inversely correlate with hematocrit and directly correlate with the serum creatinine and BUN and significantly fall with hemodialysis in ESRD patients [59]. Elevation of plasma spermidine in CKD patients was associated with and largely due to its reduced urinary excretion [58]. It has been reported that dysregulation of the polyamine pathway leads to inflammation, renal failure and diabetes [60].
Plasma 1-methylguanosine level was increased, whereas its urinary excretion was decreased confirming the result of a previous study of uremic patients [61]. In contrast plasma deoxyuridine level was decreased whereas its urinary excretion was increased in our CKD patients. These findings are consistent with previous studies in CKD and diabetic nephropathy patients [5], [62]. Increased uridine and deoxyadenosine as well as decreased inosine in both plasma and urine were observed in our CKD patients confirming the results of previous studies [5], [61].
Plasma concentration of purine metabolites, hypoxanthine and xanthine were increased whereas their urinary excretion was decreased in our CKD. Additionally, increased plasma uric acid and decreased 1-methylxanthine in both plasma and urine was observed. The previous studies have shown increased plasma hypoxanthine, xanthine and uric acid in CKD patients [5], [63], which are consistent with the results of the present study. In confirmation of earlier studies [56], [64] increased plasma and decreased urine myoinositol were found in our CKD patients. Since myoinositol is a second messenger in inflammatory cells its increased plasma level in our CKD patients may reflect the associated inflammatory state. Besides the above-mentioned metabolites which were found in both plasma and urine, 1-methyladenosine and xanthosine were detected only in urine of the CKD patients. The urinary excretion of 1-methyladenosine was significantly decreased in our CKD group. This accounts for the previously reported accumulation of this modified ribonucleoside in the uremic serum [61]. In agreement with the previous study [61] urinary excretion of xanthosine was significantly increased in our CKD patients. Plasma concentration of 1-methyladenosine was significantly elevated in our CKD group. This was associated with reduced urinary excretion of this modified ribonucleoside.
Plasma succinic acid and cis-aconitic acid were elevated whereas plasma citric acid was reduced in CKD patients. This was associated with increased urine cis-aconitic acid and decreased urine oxoglutaric acid and succinic acid levels. These findings are consistent with reported results [65], [66]. The observed changes in plasma and urine TCA intermediates point to the systemic and renal tubular mitochondrial dysfunction in CKD patients. One study demonstrated that mitochondrial localization of heme oxygenase 1 with increased heme oxygenase activity in the mitochondrial fraction of Mito-HO-1 renal epithelial cells was observed and decreased tricarboxylic acid cycle intermediates following hypoxia was significantly mitigated in Mito-HO-1 renal epithelial cells [67].
5. Conclusion
Plasma concentration and urine excretion of 25 metabolites from 30 metabolic pathways were distinctly different between the CKD and control groups. The combined plasma and urinary metabolite were related to amino acid, methylamine, purine, and lipid metabolisms and TCA cycle. These were associated with activation of NF-κB, up-regulation of pro-inflammatory and pro-oxidant and down-regulation of Nrf2 activity and its down-stream antioxidant and cytoprotective proteins, accompanied by activated canonical Wnt/β-catenin signaling. Plasma metabolites 5-MTP and homocystine and urinary metabolite citrulline were good correlated with both eGFR and creatinine. General clinical factors were incorporated to establish diagnostic models. The enhanced metabolite model showed 5-MTP, homocystine and citrulline have high sensitivity and specificity for predictive CKD. Plasma 5-MTP in CKD was identified as novel biomarker candidates. They could be considered as additional GFR-associated biomarker candidates and for indicating advanced renal injury.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Nos. 81673578, 81603271), the Program for the New Century Excellent Talents in University from Ministry of Education of China (No. NCET-13-0954) and the project As a Major New Drug to Create a Major National Science and Technology Special (No. 2014ZX09304307-002).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2017.03.017.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Zhao Y. UPLC-based metabonomic applications for discovering biomarkers of diseases in clinical chemistry. Clin. Biochem. 2014;47(15):16–26. doi: 10.1016/j.clinbiochem.2014.07.019. [DOI] [PubMed] [Google Scholar]
- 2.Zhao Y. Intrarenal metabolomic investigation of chronic kidney disease and its TGF-β1 mechanism in induced-adenine rats using UPLC Q-TOF/HSMS/MSE. J. Proteome Res. 2013;12(2):692–703. doi: 10.1021/pr3007792. [DOI] [PubMed] [Google Scholar]
- 3.Zhao Y. Metabolomics analysis reveals the association between lipid abnormalities and oxidative stress, inflammation, fibrosis, and Nrf2 dysfunction in aristolochic acid-induced nephropathy. Sci. Rep. 2015;5:12936. doi: 10.1038/srep12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duranton Plasma and urinary amino acid metabolomic profiling in patients with different levels of kidney function. Clin. J. Am. Soc. Nephrol. 2014;9(1):37–45. doi: 10.2215/CJN.06000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H. Metabolomic signatures of chronic kidney disease of diverse etiologies in the rats and humans. J. Proteome Res. 2016;15(10):3802–3812. doi: 10.1021/acs.jproteome.6b00583. [DOI] [PubMed] [Google Scholar]
- 6.Zhang H. An integrated lipidomics and metabolomics reveal nephroprotective effect and biochemical mechanism of Rheum officinale in chronic renal failure. Sci. Rep. 2016;6:22151. doi: 10.1038/srep22151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H. Metabolomics insights into chronic kidney disease and modulatory effect of rhubarb against tubulointerstitial fibrosis. Sci. Rep. 2015;5:14472. doi: 10.1038/srep14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Y. Serum metabonomics study of adenine-induced chronic renal failure rat by ultra performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry. Biomarkers. 2012;17(1):48–55. doi: 10.3109/1354750X.2011.637180. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y. Urinary metabonomics study on biochemical changes in an experimental model of chronic renal failure by adenine based on UPLC Q-TOF/MS. Clin. Chim. Acta. 2012;413(5–6):642–649. doi: 10.1016/j.cca.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 10.Pergola E. BEAM Study Investigators. Bardoxolone methyl and kidney function in CKD with type 2 diabetes. N. Engl. J. Med. 2011;365(4):327–336. doi: 10.1056/NEJMoa1105351. [DOI] [PubMed] [Google Scholar]
- 11.Ruiz Targeting the transcription factor Nrf2 to ameliorate oxidative stress and inflammation in chronic kidney disease. Kidney Int. 2013;83(6):1029–1041. doi: 10.1038/ki.2012.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clevers R. Nusse. Wnt/β-catenin signaling and disease. Cell. 2012;149(6):1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 13.Surendran, Schiavi S., Hruska K.A. Wnt-dependent beta-catenin signaling is activated after unilateral ureteral obstruction, and recombinant secreted frizzled-related protein 4 alters the progression of renal fibrosis. J. Am. Soc. Nephrol. 2005;16(8):2373–2384. doi: 10.1681/ASN.2004110949. [DOI] [PubMed] [Google Scholar]
- 14.He Blockade of Wnt/b-catenin signaling by paricalcitol ameliorates proteinuria and kidney injury. J. Am. Soc. Nephrol. 2011;22(1):90–103. doi: 10.1681/ASN.2009121236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Toerne Wnt pathway regulation in chronic renal allograft damage, Am. J. Transpl. 2009;9(10):2223–2239. doi: 10.1111/j.1600-6143.2009.02762.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Implication of dysregulation of the canonical wingless-type MMTV integration site (WNT) pathway in diabetic nephropathy. Diabetologia. 2012;55(1):255–266. doi: 10.1007/s00125-011-2314-2. [DOI] [PubMed] [Google Scholar]
- 17.Chen Metabolomics insights into activated redox signaling and lipid metabolism dysfunction in chronic kidney disease progression. Redox Biol. 2016;10:168–178. doi: 10.1016/j.redox.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Q. The link between phenotype and fatty acid metabolism in advanced chronic kidney disease. Nephrol. Dial. Transpl. 2017 doi: 10.1093/ndt/gfw415. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y., Lin R.C. UPLC-MSE application in disease biomarker discovery: the discoveries in proteomics to metabolomics. Chem. Biol. Inter. 2014;215:7–16. doi: 10.1016/j.cbi.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 20.Rhee P. A genome-wide association study of the human metabolome in a community-based cohort. Cell Metab. 2013;18(1):130–143. doi: 10.1016/j.cmet.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leal O. NRF2 and NF‑κB mRNA expression in chronic kidney disease: a focus on nondialysis patients. Int. Urol. Nephrol. 2015;47(12):1985–1991. doi: 10.1007/s11255-015-1135-5. [DOI] [PubMed] [Google Scholar]
- 22.Pedruzzi M. Systemic inflammation and oxidative stress in hemodialysis patients are associated with down-regulation of Nrf2. J. Nephrol. 2015;28(4):495–501. doi: 10.1007/s40620-014-0162-0. [DOI] [PubMed] [Google Scholar]
- 23.Yi Nuclear NF-κB p65 in peripheral blood mononuclear cells correlates with urinary MCP-1, RANTES and the severity of type 2 diabetic nephropathy. PLoS One. 2014;9(6):e99633. doi: 10.1371/journal.pone.0099633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Locatelli Oxidative stress in end-stage renal disease: an emerging threat to patient outcome. Nephrol. Dial. Transplant. 2003;18(7):1272–1280. doi: 10.1093/ndt/gfg074. [DOI] [PubMed] [Google Scholar]
- 25.Pergola E. Effect of bardoxolone methyl on kidney function in patients with T2D and stage 3b-4 CKD. Am. J. Nephrol. 2011;33(5):469–476. doi: 10.1159/000327599. [DOI] [PubMed] [Google Scholar]
- 26.Suresh D.R., Delphine S., Agarwal R. Biochemical markers of oxidative stress in predialytic chronic renal failure patients. Hong Kong J. Nephrol. 2008;10(2):69–73. [Google Scholar]
- 27.Cardozo F., Stockler-Pinto M.B., Mafra D. Brazil nut consumption modulates Nrf2 expression in hemodialysis patients: a pilot study. Mol. Nutr. Food Res. 2016;60(7):1719–1724. doi: 10.1002/mnfr.201500658. [DOI] [PubMed] [Google Scholar]
- 28.Wang D. Aberrant activation of the WNT/β-catenin signaling pathway in lupus nephritis. PLoS One. 2014;9(1):e84852. doi: 10.1371/journal.pone.0084852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garibotto Amino acid and protein metabolism in the human kidney and in patients with chronic kidney disease. Clin. Nutr. 2010;29(4):424–433. doi: 10.1016/j.clnu.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Jia Serum metabonomics study of chronic renal failure by ultra performance liquid chromatography coupled with Q-TOF mass spectrometry. Metabolomics. 2008;4(2):183–189. [Google Scholar]
- 31.Psihogios G. Evaluation of tubulointerstitial lesions' severity in patients with glomerulonephritides: an NMR-based metabonomic study. J. Proteome Res. 2007;6(9):3760–3770. doi: 10.1021/pr070172w. [DOI] [PubMed] [Google Scholar]
- 32.Qi A pilot metabolic profiling study in serum of patients with chronic kidney disease based on 1H NMR-spectroscopy. Clin. Transl. Sci. 2012;5(5):379–385. doi: 10.1111/j.1752-8062.2012.00437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rhee P. A combined epidemiologic and metabolomic approach improves CKD prediction. J. Am. Soc. Nephrol. 2013;24(8):1330–1338. doi: 10.1681/ASN.2012101006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nkuipou-Kenfack Assessment of metabolomic and proteomic biomarkers in detection and prognosis of progression of renal function in chronic kidney disease. PLoS One. 2014;9(5):e96955. doi: 10.1371/journal.pone.0096955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu K., Cheng H.H., Chang T.C. 5-methoxyindole metabolites of L-tryptophan: control of COX-2 expression, inflammation and tumorigenesis. J. Biomed. Sci. 2014;21:17. doi: 10.1186/1423-0127-21-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng H. Control of cyclooxygenase-2 expression and tumorigenesis by endogenous 5-methoxytryptophan. Proc. Natl. Acad. Sci. USA. 2012;109(33):13231–13236. doi: 10.1073/pnas.1209919109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim J., Vaziri N.D. Contribution of impaired Nrf2-Keap1 pathway to oxidative stress and inflammation in chronic renal failure. Am. J. Physiol. Ren. Physiol. 2010;298(3):F662–F671. doi: 10.1152/ajprenal.00421.2009. [DOI] [PubMed] [Google Scholar]
- 38.Chauveau Hyperhomocysteinemia, a risk factor for atherosclerosis in chronic uremic patients. Kidney Int. 1993;43(Suppl 41):S72–S77. [PubMed] [Google Scholar]
- 39.Kang S. Plasma protein-bound homocyst(e)ine in patients requiring chronic haemodialysis. Clin. Sci. 1983;65(3):335–336. doi: 10.1042/cs0650335. [DOI] [PubMed] [Google Scholar]
- 40.Laidlaw A. Sulfur amino acids in maintenance hemodialysis patients. Kidney Int. 1987;32(Suppl 22):S191–S196. [PubMed] [Google Scholar]
- 41.Vychytil Peritoneal elimination of homocysteine moieties in continuous ambulatory peritoneal dialysis patients. Kidney Int. 1999;55(5):2054–2061. doi: 10.1046/j.1523-1755.1999.00437.x. [DOI] [PubMed] [Google Scholar]
- 42.Tao GC-MS with ethyl chloroformate derivatization for comprehensive analysis of metabolites in serum and its application to human uremia. Anal. Bioanal. Chem. 2008;391(8):2881–2889. doi: 10.1007/s00216-008-2220-8. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Metabonomics research of diabetic nephropathy and type 2 diabetes mellitus based on UPLC-oaTOF-MS system. Anal. Chim. Acta. 2009;650(1):16–22. doi: 10.1016/j.aca.2009.02.027. [DOI] [PubMed] [Google Scholar]
- 44.Suliman E. Inflammation contributes to low plasma amino acid concentrations in patients with chronic kidney disease. Am. J. Clin. Nutr. 2005;82(2):342–349. doi: 10.1093/ajcn.82.2.342. [DOI] [PubMed] [Google Scholar]
- 45.Okamura M., Pennathur S. The balance of powers: redox regulation of fibrogenic pathways in kidney injury. Redox Biol. 2015;6:495–504. doi: 10.1016/j.redox.2015.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nath A. Tubulointerstitial changes as a major determinant in the progression of renal damage. Am. J. Kidney Dis. 1992;20(1):1–17. doi: 10.1016/s0272-6386(12)80312-x. [DOI] [PubMed] [Google Scholar]
- 47.Wang Kynurenine is an endothelium-derived relaxing factor produced during inflammation. Nat. Med. 2010;16(3):279–285. doi: 10.1038/nm.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zinellu Impact of cholesterol lowering treatment on plasma kynurenine and tryptophan concentrations in chronic kidney disease: relationship with oxidative stress improvement. Nutr. Metab. Cardiovasc. Dis. 2015;25(2):153–159. doi: 10.1016/j.numecd.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 49.Cachofeiro Oxidative stress and inflammation, a link between chronic kidney disease and cardiovascular disease. Kidney Int. Suppl. 2008;111:S4–S9. doi: 10.1038/ki.2008.516. [DOI] [PubMed] [Google Scholar]
- 50.Goicoechea Effects of atorvastatin on inflammatory andfibrinolytic parameters in patients with chronic kidney disease. J. Am. Soc. Nephrol. 2006;17(12 Suppl 3):S231–S235. doi: 10.1681/ASN.2006080938. [DOI] [PubMed] [Google Scholar]
- 51.Latchoumycandane L.E. Nagy, McIntyre T.M. Chronic ethanol ingestion induces oxidative kidney injury through taurine-inhibitable inflammation. Free Radic. Biol. Med. 2014;69:403–416. doi: 10.1016/j.freeradbiomed.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kobayashi A metabolomics-based approach for predicting stages of chronic kidney disease. Biochem. Biophys. Res. Commun. 2014;445N(2):412–416. doi: 10.1016/j.bbrc.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 53.Niewczas A. Uremic solutes and risk of end-stage renal disease in type 2 diabetes metabolic study. Kidney Int. 2014;85(5):1214–1224. doi: 10.1038/ki.2013.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bell D. Nuclear magnetic resonance studies of blood plasma and urine from subjects with chronic renal failure: identification of trimethylamine-N-oxide. Biochim. Biophys. Acta. 1991;1096(2):101–107. doi: 10.1016/0925-4439(91)90046-c. [DOI] [PubMed] [Google Scholar]
- 55.Tang H. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ. Res. 2015;116(3):448–455. doi: 10.1161/CIRCRESAHA.116.305360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Posada-Ayala Identification of a urine metabolomic signature in patients with advanced-stage chronic kidney disease. Kidney Int. 2014;85(1):103–111. doi: 10.1038/ki.2013.328. [DOI] [PubMed] [Google Scholar]
- 57.Vaziri D., Zhao Y.Y., Pahl M.V. Altered intestinal microbial flora and impaired epithelial barrier structure and function in CKD: the nature, mechanisms, consequences and potential treatment. Nephrol. Dial. Transpl. 2016;31(5):737–746. doi: 10.1093/ndt/gfv095. [DOI] [PubMed] [Google Scholar]
- 58.Swendseid E., Panaqua M., Kopple J.D. Polyamine concentrations in red cells and urine of patients with chronic renal failure. Life Sci. 1980;26(7):533–539. doi: 10.1016/0024-3205(80)90316-1. [DOI] [PubMed] [Google Scholar]
- 59.Saito Serum levels of polyamines in patients with chronic renal failure. Kidney Int. 1983;16:S234–S237. [PubMed] [Google Scholar]
- 60.Park H., Igarashi K. Polyamines and their metabolites as diagnostic markers of human diseases. Biomol. Ther. 2013;21(1):1–9. doi: 10.4062/biomolther.2012.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Niwa N., Takeda H., Yoshizumi RNA metabolism in uremic patients: accumulation of modified ribonucleosides in uremic serum. Kidney Int. 1998;53(6):1801–1806. doi: 10.1046/j.1523-1755.1998.00944.x. [DOI] [PubMed] [Google Scholar]
- 62.Xia F. Ultraviolet and tandem mass spectrometry for simultaneous quantification of 21 pivotal metabolites in plasma from patients with diabetic nephropathy. J. Chromatogr. B. 2009;877(20–21):1930–1936. doi: 10.1016/j.jchromb.2009.05.047. [DOI] [PubMed] [Google Scholar]
- 63.Caussé Simultaneous determination of allantoin, hypoxanthine, xanthine, and uric acid in serum/plasma by CE. Electrophoresis. 2007;28(3):381–387. doi: 10.1002/elps.200600205. [DOI] [PubMed] [Google Scholar]
- 64.Mutsaers A. Optimized metabolomic approach to identify uremic solutes in plasma of stage 3–4 chronic kidney disease patients. PLoS One. 2013;8(8):e71199. doi: 10.1371/journal.pone.0071199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Toyohara Metabolomic profiling of uremic solutes in CKD patients. Hypertens. Res. 2010;33(9):944–952. doi: 10.1038/hr.2010.113. [DOI] [PubMed] [Google Scholar]
- 66.Kienana Elucidating time-dependent changes in the urinary metabolome of renal transplant patients by a combined 1H NMR and GC-MS approach. Mol. Biosyst. 2015;11(9):2493–2510. doi: 10.1039/c5mb00108k. [DOI] [PubMed] [Google Scholar]
- 67.Bolisetty Mitochondria-targeted heme oxygenase-1 decreases oxidative stress in renal epithelial cells. Am. J. Physiol. Ren. Physiol. 2013;305(3):F255–F264. doi: 10.1152/ajprenal.00160.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material