Introductory paragraph
Many anaerobic microbial parasites possess highly modified mitochondria known as mitochondrion-related organelles (MROs). The best-studied of these are the hydrogenosomes of Trichomonas vaginalis and Spironucleus salmonicida, which produce ATP anaerobically through substrate-level phosphorylation with concomitant hydrogen production; and the mitosomes of Giardia intestinalis, which are functionally reduced and lack any role in ATP production. However, in order to understand the metabolic specialisations that these MROs underwent in adaptation to parasitism, data from their free-living relatives are needed. Here, we present a large-scale comparative transcriptomic study of MROs across a major eukaryotic group, Metamonada, examining lineage-specific gain and loss of metabolic functions in the MROs of Trichomonas, Giardia, Spironucleus, and their free-living relatives. Our analyses uncover a complex history of ATP production machinery in diplomonads such as Giardia, and their closest relative, Dysnectes; and a correlation between the glycine cleavage machinery and lifestyles. Our data further suggest the existence of a previously undescribed biochemical class of MRO that generates hydrogen but is incapable of ATP synthesis.
Mitochondrion-related organelles (MROs) are highly modified mitochondria found in diverse anaerobic and microaerophilic eukaryotes; they vary widely in the ancestral mitochondrial pathways that they have retained, and in their role in anaerobic ATP production (reviewed in ref. 1). The greatest functional diversity of MROs is found in the excavate group Metamonada. This diversity encompasses the hydrogenosomes of Trichomonas vaginalis and Spironucleus salmonicida, which produce ATP anaerobically through substrate-level phosphorylation with concomitant hydrogen production1–4; the mitosomes of Giardia intestinalis, which are highly functionally reduced and lack any role in ATP production1,5; and Monocercomonoides sp., the first known eukaryote to have lost its MROs entirely6. However, little is known of the metabolic properties of MROs in free-living metamonads, nor how the metabolically specialised MROs of parasitic metamonads evolved from those of their free-living ancestors. The free-living, bacterivorous Carpediemonas-like organisms (CLOs7,8) are the closest relatives of these parasitic metamonads, and are key to resolving this question. Here, we present the first large-scale comparative transcriptomic study of MROs across Metamonada. By sequencing the transcriptomes of 8 free-living or commensal metamonads, and comparing their predicted MRO proteomes with publicly available proteome and transcriptome data, we examine lineage-specific gain and loss of metabolic functions in MROs in 18 metamonads.
Results
A robust tree of the Metamonada
To elucidate the relationships between T. vaginalis, G. intestinalis, and their free-living close relatives, we mined 18 metamonad transcriptomes or genomes for orthologues of 159 conserved proteins to add to a phylogenomic super-matrix representing the full breadth of eukaryote diversity. Phylogenetic analyses of this dataset (Fig. 1A) show that Metamonada form a highly supported clade with three major subgroups, Preaxostyla, Parabasalia and Fornicata; Preaxostyla is the earliest diverging lineage. All CLOs branched within Fornicata, forming a long series of nested clades, with Dysnectes brevis being the closest relative of diplomonads7, and Carpediemonas membranifera forming the deepest branch. Branching order and position of all CLOs received maximum bootstrap and posterior probability support in most analyses, except in two cases where bootstrap support nevertheless remained strong (i.e. >85%, see Supplementary Figs. 1–3).
Fig. 1.
Phylogeny of Metamonada and distribution of MRO-localising proteins. a. PhyloBayes tree of eukaryotes inferred from the phylogenomic dataset of 159 proteins (39,089 sites) and 94 taxa. The taxa other than Metamonada are not shown in detail. The topology shown was recovered by 3/4 PhyloBayes chains, and in all ML analyses. Parasitic species and lineages are highlighted in red. The bar graph indicates the percent coverage of the amino acid sites used in the phylogenomic analysis present in each transcriptome. See Supplementary Figs. 1, 2 and 3 for more detail and support values. b. Distribution of MRO-targeted proteins in Metamonada. L, H, P, and T: L, H, P, and T proteins of the Glycine cleavage system, SHMT: Serine hydroxymethyltransferase, NuoE: NADH:ubiquinone oxidoreductase 24 kD subunit, NuoF: NADH:ubiquinone oxidoreductase, NADH-binding 51 kD subunit, Fdx: ferredoxin, HydA: [FeFe]-hydrogenase, HydE-G: [FeFe]-hydrogenase maturation proteins E-G, SCSa: Succinyl-CoA synthase alpha subunit, SCSb: Succinyl-CoA synthase beta subunit, ASCT: Acetate:succinyl-CoA transferase, ACS1 and ACS2: acetyl-CoA synthase 1 and acetyl-CoA synthase 2. Note that Monocercomonoides sp. has undergone a complete secondary loss of MROs6.
Mitochondrial targeting sequences and recognition proteins are reduced in CLOs
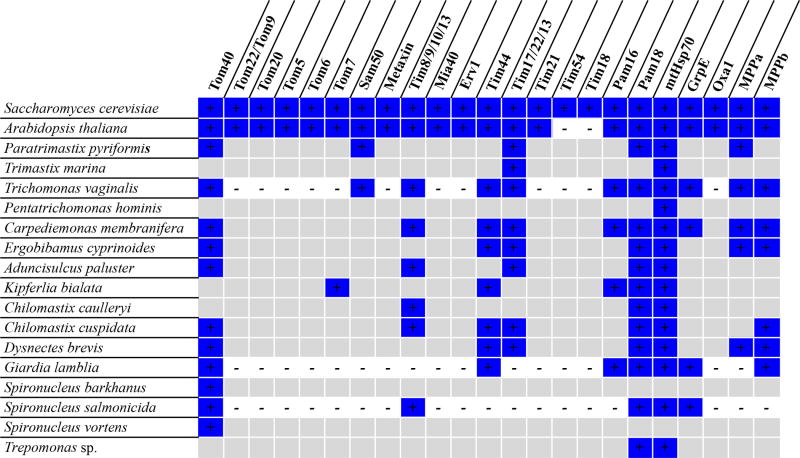
Using hidden Markov model searches, we identified the core conserved mitochondrial protein transport components Tom40, Tim44, Sam50, Tim17/22/23, Pam16, Pam18, mtHsp70 and GrpE, as well as mitochondrial processing peptidase subunits, from one or more CLOs, although many accessory subunits were not detected (Fig. 2). This complement of mitochondrial protein import proteins is comparable to that of T. vaginalis hydrogenosomes9. A poorly conserved homologue of Tim44 has long remained unidentified, but was recently described in G. intestinalis10. It is therefore possible that additional protein import components are in fact present in CLOs, but could not be detected by our methods.
Fig. 2.
Predicted mitochondrial protein import proteins in metamonads. Blue +: present; white −: absent from genome; grey, no symbols: absent from transcriptome. Data from Saccharomyces cerevisiae9 and Arabidopsis thaliana31 are provided for comparison.
In most eukaryotes, the majority of mitochondrial proteins are targeted to the mitochondria by N-terminal mitochondrial targeting sequences (MTS)11, which are cleaved by the heterodimeric mitochondrial processing peptidase (MPP) in the matrix12. However, some proteins that are known to localise to the Giardia, S. salmonicida and Trichomonas MROs have no predicted MTS, and lack any N-terminal extensions relative to prokaryotic homologues4,13,14. Likewise, we did not find MTS predictions for many proteins of clear mitochondrial origin identified in the transcriptome data from CLOs (e.g. mitochondrial chaperone Cpn60 and mitochondrial iron-sulfur cluster assembly proteins IscS and IscU; Supplementary Data 1). This reduction in the prevalence of MTS was likely a feature of the last common ancestor of Metamonada, and suggests the existence of unknown MRO-targeting signals in addition to canonical MTSs (Supplementary Data 1).
In Trichomonas and Giardia, the decreased number of canonical MTS is accompanied by reductive and/or divergent evolution of the MPP13, and hydrogenosome/mitosome protein import machineries are dependent on only a low membrane potential (i.e., Δψ) following the loss of electron transport14. Giardia possesses only the MPP-beta subunit, which appears to function as a homodimer13. The Trichomonas genome encodes divergent homologues of the alpha- and beta-subunits, which appear to function as a canonical alpha/beta-subunit heterodimer but recognise only short mitochondrial targeting sequences13. We recovered both subunits of MPP in the transcriptome data from Ca. membranifera, Ergobibamus cyprinoides, and D. brevis, but identified only the beta-subunit in Chilomastix cuspidata (Fig. 2).
Loss of the glycine cleavage system correlates with a parasitic lifestyle
The glycine cleavage system (GCS) consists of four proteins: GCSH, GCSL, GCSP, GCST. The GCS catalyzes the reversible conversion of glycine to ammonium and carbon dioxide, with the associated reduction of NAD+ to ultimately produce 5,10-methylene-tetrahydrofolate (5,10-CH2-THF, which is essential for serine production in mitochondria and MROs15), and NADH. Previous studies showed a patchy distribution of these four enzymes across Metamonada (Fig. 1B; see also ref.s 1,16–18), and as a result the evolutionary history of the GCS remained unclear16. We identified all components of the GCS encoded in D. brevis, Aduncisulcus paluster, Ca. membranifera, E. cyprinoides, Kipferlia bialata, and Ch. cuspidata. The 5,10-CH2-THF generated by the GCS is used by serine hydroxymethyl transferase (SHMT)15. We identified SHMT proteins encoded in the transcriptomes of some CLO species, usually with a clear predicted MTS (Supplementary Data 1).
The GCSP gene had an unexpectedly complex phylogenetic history. The eukaryotic-type GCSP contains two distinct N-terminal and C-terminal domains. These domains are consistently recovered as two separate contigs from the metamonad transcriptomes, an arrangement previously reported only in Paratrimastix pyriformis19. This split arrangement is also found in some alphaproteobacteria; Metamonada form a clade with these alphaproteobacteria, separate from other eukaryotes (Supplementary Figs. 4 and 5, Supplementary Data 2). For all other components of the GCS, CLOs branched with other eukaryotes (Supplementary Figs. 6–8). Whether metamonad GCSP proteins are truly present in the split form remains to be confirmed, and the possibility of trans-splicing to create a transcript encoding a single protein also cannot be excluded. However, the consistent recovery of two separate contigs for this protein, and the phylogenetic affinity to the split alphaproteobacterial GCSP proteins, suggest that GCSP is truly encoded as two single-domain proteins in metamonads.
An interesting evolutionary pattern emerges from the distribution of GCS across Metamonada: parasitic and secondarily free-living lineages always lack a complete GCS while ancestrally free-living lineages possess the complete system (Figs. 1A and 1B). A similar correlation between GCS distribution and lifestyle emerges when considering metabolically reduced MROs of other eukaryotic lineages (Supplementary Table 1, ref. 1). An apparent exception is the presence of only GCSH and GCSL in Trichomonas; however, the retention of these particular proteins by some parasites can be explained by the fact that they function in other pathways, such as peroxide detoxification20. GCS proteins are also absent from the transcriptome data in the free-living diplomonad Trepomonas. Phylogenies of diplomonads indicate that Trepomonas is secondarily free-living (Fig. 1, ref. 21), suggesting that the GCS was lost in a parasitic ancestor. Although parasitic/commensal taxa may be able to rely on the host for some metabolites, it is not known whether Trepomonas has an alternative way to cleave glycine into carbon dioxide and ammonium. We examined the metabolic pathways of Trepomonas predicted by the KEGG Automatic Annotation pipeline, in order to identify candidate proteins that might allow this organism to metabolise glycine. However, we did not identify any such candidates. Further experimental studies will be required to understand how Trepomonas can function without a canonical GCS.
MROs in all CLO lineages are likely capable of H2 production
The GCS is localised exclusively in mitochondria and MROs, and its activity is sensitive to the redox balance of NAD+/NADH22, resulting in a need for NADH reoxidation in these compartments. From the transcriptomes of Ca. membranifera, Ch. cuspidata, D. brevis, E. cyprinoides, K. bialata, and trichomonads, we identified two subunits of NADH dehydrogenase, NuoE and NuoF (Fig. 1; Supplementary Data 1), and no other electron transport chain components. These homologues of mitochondrial electron transport chain Complex I subunits are found exclusively in mitochondria and MROs, but have not previously been identified in preaxostylans or diplomonads (Fig. 1B; see also ref.s 4,5,19). In Trichomonas hydrogenosomes, [FeFe]-hydrogenase generates H2 from protons via the oxidation of ferredoxin (Fdx−) coupled with the oxidation of NADH by NuoE and NuoF2,3 (NADH is regenerated by the GCS, see above). We identified an MRO-type 2Fe-2S Fdx homologous to that of Trichomonas in all of the CLOs (Supplementary Data 1). The [FeFe]-hydrogenase HydA and its associated maturases, HydE, HydF, and HydG, were identified in almost all metamonad lineages (Fig. 1B and Supplementary Data 1); each of the maturases showed phylogenetic affinity with homologues from other anaerobic eukaryotes (Supplementary Figs. 9–11). Conversely, HydA in Metamonada may have multiple origins based on the phylogeny of this protein family (Supplementary Fig. 12). However, as with the maturases, we are unable to draw unambiguous conclusions as to the origins of these proteins because of the lack of bootstrap support across many parts of the tree. MTSs were identified for some of these proteins, such as HydE and HydF from D. brevis and HydA and HydG from Ca. membranifera (Supplementary Data 1), further supporting their location within the MROs. Thus, it is likely that the MROs of all CLOs, like the hydrogenosomes of T. vaginalis, are capable of H2 production, as well as NADH production (GCS), NADH reoxidation (NuoE/NuoF), and electron transfer (Fdx) (Fig. 3).
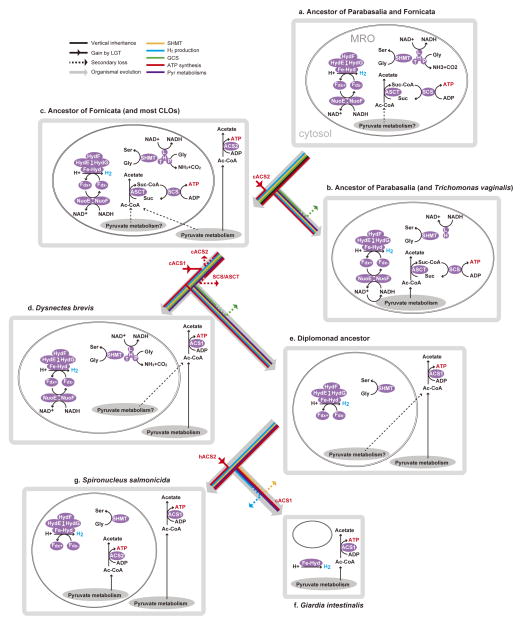
Fig. 3.
Evolutionary transitions of metabolic pathways of MROs from the common ancestor of Parabasalia and Fornicata to representative extant species. a. Ancestor of Parabasalia and Fornicata. b. Ancestor of Parabasalia. c. Ancestor of Fornicata. d. Dysnectes brevis. e. Ancestor of diplomonads. f. Giardia intestinalis. g. Spironucleus salmonicida. In this figure, only glycine cleavage (via the glycine cleavage system), hydrogen production, and ATP production are shown (we cannot confidently assign cytosolic or MRO localization of proteins involved in pyruvate metabolism, other than in Giardia, Trichomonas and Spironucleus). cACS1/cACS2, cytosolic acetyl-CoA synthase 1/cytosolic acetyl-CoA synthase 2; hACS2, hydrogenosomal acetyl-CoA synthase 2.
MROs in D. brevis are incapable of ATP production
In the hydrogenosomes in Trichomonas spp., ATP is generated through substrate-level phosphorylation by succinyl-CoA synthase (SCS), with the associated conversion of acetyl-CoA to succinyl-CoA by acetate:succinyl-CoA transferase (ASCT)3,23. These enzymes are found exclusively in mitochondria and MROs24,25. In contrast, ACS (acetyl-CoA synthase) generates ATP directly from the conversion of acetyl-CoA to acetate in both the MROs and cytosol of S. salmonicida4, and in the cytosol of Giardia spp.25,26. We identified the alpha- and beta-subunits of a eukaryotic-type SCS in Ca. membranifera, E. cyprinoides, A. paluster, and K. bialata (as well as in parabasalid species). These subunits showed phylogenetic affinity to mitochondrion-targeted homologues of other eukaryotes (Supplementary Figs. 13 and 14). ASCT homologues were also found in three of the four CLOs that have SCS (Ca. membranifera, A. paluster and K. bialata; Fig. 1B). ASCT is divided into three classes, known as subtypes 1A, 1B and 1C; T. vaginalis is known to possess ASCT1C25. ASCT1B was identified in three CLOs; unlike in trichomonads, no CLO transcriptome encoded ASCT1C alone. Interestingly, K. bialata has both subtype 1B and subtype 1C. Thus, although the separate phylogenetic analyses of 1B and 1C do not show strong evidence for either vertical inheritance or LGT into Metamonada, it is possible that possession of both of these enzyme subtypes represents the ancestral state for metamonads (Supplementary Figs. 15 and 16). The presence of both SCS and ASCT strongly suggest that these deeper-branching CLOs can generate ATP in their MROs.
Two types of ACS were identified in metamonads, and at least one of these types of ACS was found in Trimastix marina, Ca. membranifera, A. paluster, Chilomastix spp., E. cyprinoides, K. bialata, D. brevis, and diplomonad species. Although some ACSs function in MROs4,27, ACSs in Chilomastix spp., K. bialata and D. brevis were closely related to the cytosolically functioning ACSs of diplomonads (Giardia spp., S. salmonicida, and Trepomonas sp.; henceforth, ACS1), and only distantly related to the MRO-localised homologue of S. salmonicida (Supplementary Figs. 17 and 18). This suggests that the cytosolic ACS1 was acquired through a single LGT event into the last common ancestor of Chilomastix, Kipferlia, Dysnectes and diplomonads. The other metamonad ACS homologues are of different origin(s) (ACS2), although because of the lack of resolution in this phylogeny it is unclear whether or not ACS2 was ancestrally present in metamonads (Figs. S17 and S18). No predicted MRO-localising homologues of ACS, SCS, or ASCT were identified in the transcriptome of D. brevis, nor in any diplomonads, with the exception of S. salmonicida4.
As described above, GCS, NuoE and NuoF are metabolically linked enzyme systems that are only known to function within mitochondria and MROs. However, NuoE and NuoF require an electron sink in order to regenerate NAD+ (required by the GCS), for example via reduction of protons to hydrogen. We recovered these proteins, and [FeFe]-hydrogenase maturases with predicted MTS, from the transcriptome of D. brevis, strongly suggesting that [FeFe]-hydrogenase is active in its MRO. Yet, despite the high transcriptome coverage in D. brevis, we have identified only the cytosolic-type ATP production machinery in D. brevis (ACS1, see above). We therefore hypothesise that D. brevis harbours a new type of mitochondrion-related organelle that evolves molecular hydrogen, but does not produce ATP (Fig. 3).
ATP synthesis was lost and regained in metamonad MROs
The extremely patchy distribution of the ACS gene in the eukaryotic tree of life has been explained by multiple lateral gene transfers4,28,29. Our phylogenetic analyses placed the hydrogenosome-localising ACS2 homologue of S. salmonicida separately from all other metamonad homologues (Supplementary Fig. 17 and 18). Meanwhile, since only cytosolic ACS1 was identified in other diplomonads and in their close relative D. brevis, a parsimonious explanation is that the common ancestor of diplomonads and D. brevis possessed an MRO incapable of ATP production. This would imply that the MROs of S. salmonicida acquired the capacity for ATP production secondarily, through LGT of the hydrogenosomal ACS2.
Complex functions in the last common ancestor of metamonads
The hydrogenosomes in T. vaginalis function in pyruvate metabolism and amino acid metabolism30, whereas in Giardia spp. these pathways occur in the cytosol5,17. We found homologues of enzymes of pyruvate metabolism and amino acid metabolism in CLOs (data not shown), Trimastix and Paratrimastix; but, in the absence of any predicted MTS, we could not conclude that they function in the MROs.
In addition to the proteins discussed above, we identified other possible MRO-localising proteins that are not present in the hydrogenosomes of Trichomonas spp. or in the mitosomes of Giardia spp., but that are broadly conserved in canonical mitochondria. These include cardiolipin synthase in Ca. membranifera (Supplementary Fig. 19) and aminoadipate-semialdehyde dehydrogenase in Trim. marina (Supplementary Fig. 20). The metamonad homologues showed affinity to other eukaryotic homologues in phylogenetic analyses (Supplementary Figs. 19–20), indicating that they have likely been vertically inherited from the last eukaryotic common ancestor. These findings further support our inference that the metabolic pathways and functions in MROs of the last metamonad common ancestor were more complex than those of the hydrogenosomes of Trichomonas spp.
Discussion
As a primarily transcriptomic analysis, our work is necessarily vulnerable to the possibility that the culture conditions under which the samples were generated affected the transcripts recovered. As a result, complete genome-based analyses will be helpful to cross-check our observations. Our transcriptome analyses indicate that the MROs of the last common ancestor of Fornicata resembled those of T. vaginalis in that they generated ATP with the associated production of hydrogen (Fig. 1B; Fig. 3). In contrast, the MROs of the last common ancestor of Diplomonadida had already secondarily lost the ATP synthesis machinery, but had retained the capacity for hydrogen production (Fig. 1B; Fig. 3). Hydrogenosomes are generally characterised as functioning in anaerobic ATP production, with the concomitant production of hydrogen1,3. Our analyses suggest that in some CLOs, such as Dysnectes, these functions are spatially uncoupled within the cell. The MROs of these organisms resemble hydrogenosomes in their involvement in hydrogen production, but are more similar to mitosomes in their inability to synthesise ATP.
In organisms (such as Dysnectes) that possess this type of organelle, ATP is likely synthesised through the cytosolic ACS-based reaction, as in Giardia. Nevertheless, we hypothesise that there is a constraint against loss of hydrogen production from these MROs because hydrogen production functions as an electron acceptor, not for pyruvate metabolism, but for amino acid metabolism. As parasites are able to rely on the host in terms of metabolites, the evolutionary transition to parasitism might lead to further functional reduction, allowing loss of amino acid metabolism pathways from MROs without affecting cell viability. As a result, MROs originally resembling those of Dysnectes might then lose both amino acid metabolism pathways and hydrogen production, resulting in MROs like those found in Giardia, unable to produce either ATP or hydrogen. Thus, this type of MRO may represent an evolutionarily intermediate state between hydrogenosomes and mitosomes. In any case, mitosomes in G. intestinalis were apparently shaped by stepwise losses from an organelle similar to the hydrogenosome of T. vaginalis, but which possessed a richer set of metabolic pathways, including a complete GCS. The MROs of Dysnectes are important snapshots of an intermediate step in this process.
Methods
Culturing, RNA isolation and sequencing
Cultures were maintained as described in ref. 32 (in the case of Trimastix marina, isolate PCT), as described in ref. 7 (in the case of Chilomastix caulleryi), following the procedures set forth by the American Type Culture Collection (ATCC; http://www.atcc.org/products/all/50927.aspx#culturemethod) (in the case of Chilomastix cuspidata), or as described in ref. 33 (all other Carpediemonas-like organisms). RNA was isolated from ~1–3L of culture using TRIzol Reagent (Invitrogen, USA; Life Technologies, USA) according to the manufacturer’s instructions. Sanger and 454 sequencing libraries were prepared by Agencourt (Germany), and Illumina libraries by Genome Quebec (Canada). 454 (GS FLX Titanium, single-end) and Illumina (HiSeq2000, paired-end) sequencing were carried out by Genome Quebec. For Aduncisulcus paluster and Dysnectes brevis, cDNA synthesis and sequencing by Illumina (HiSeq2000, paired-end) were instead carried out by Eurofins Genetic Services (Eurofins, Germany). In the case of Ergobibamus cyprinoides, an initial Sanger transcriptome was sequenced: mRNA was double purified from total RNA using magnetic beads (Ambion, USA), a Sanger sequencing cDNA library was prepared, and Expressed Sequence Tag (EST) reads were sequenced. This cDNA library was subsequently modified for 454 sequencing by Agencourt, and subjected to 454 sequencing by Genome Quebec. The number of reads per species is as follows: Ca. membranifera – 57.6 thousand 454 reads and 16.4 million Illumina reads; E. cyprinoides –15.9 thousand 454 reads and 5220 Sanger reads; A. paluster – 147 million Illumina reads; K. bialata – 28.1 thousand 454 reads; D. brevis – 148.6 million Illumina reads; Ch. cuspidata – 183.4 million Illumina reads; Ch. caulleryi – 28.3 thousand reads; and Trimastix marina PCT – 35.2 million Illumina reads. Raw sequencing data were deposited into the NCBI Short Reads Archive (SRA) database under accession number SRP077666.
Assembly
Sanger and 454 sequence data were assembled using the MIRA3 assembler with miraEST34, using default assembly parameters. Where both types of data were available, a hybrid assembly was performed. Adapter (SSAHA scan output) and quality clipping were performed by MIRA3. RNASeq data were quality trimmed using an in-house script (25 minimum quality score over a 20 nucleotide sliding window; minimal length of read after trimming 50 nucleotides) and assembled using the program inchworm from the Trinity software package with default parameters35.
Phylogenomic dataset assembly
Data were added to our previously published phylogenomic dataset36 and the new taxa were added to the dataset using the procedures described in ref. 36. Briefly, each reference protein was used as a query in tblastn searches of each metamonad species, and the five top hits (with an e-value < 1×10−10) were retained. These were then added to the alignments, and preliminary single-gene trees were constructed using MAFFT37 for alignment, BMGE38 for trimming of ambiguously aligned positions, and FastTree39 for tree construction. The single gene trees were inspected by eye, and paralogues were removed (in the case of multiple in-paralogues, the best coverage or shortest branching sequence was selected). Final single-gene datasets were constructed using MAFFT L-INS-I, and trimmed with BMGE. Final concatenation was carried out using the program alvert from Barrel-O-Monkeys (http://rogerlab.biochemistryandmolecularbiology.dal.ca/monkeybarrel.php). The final alignment used for the phylogenomic analysis consisted of 94 taxa and 39089 sites. The phylogenomic tree was constructed using RAxML40 under the PROTGAMMALGF model, with 25 independent searches and 500 regular non-parametric bootstraps; and using IQ-TREE41 under the LG+C60+F+Gamma model, with 1000 ultrafast bootstrap replicates42. A Bayesian phylogeny was constructed using the CAT-GTR model incorporating among-site rate variation approximated by a discrete gamma distribution with four categories (CAT-GTR + Γ model), implemented in PhyloBayes-MPI 1.5a43, after the exclusion of constant sites. Four independent Markov Chain Monte Carlo (MCMC) chains were run for over 10,000 cycles, sampling every two cycles. After this time, 3/4 chains converged on the topology for metamonads shown in Fig. 1 with a 20% burnin, but failed to converge on a single topology for several other parts of the tree.
Alignments available from the Dryad Digital Repository: doi 10.5061/dryad.34qd7.
Mitochondrial gene tree construction
A custom metamonad protein database was assembled from our newly sequenced datasets, as well as from publicly available genomes and transcriptomes of other metamonads (Paratrimastix pyriformis, Trichomonas vaginalis, Pentatrichomonas hominis, Tritrichomonas foetus, Spironucleus vortens, S. barkhanus, S. salmonicida, Giardia intestinalis, and Trepomonas sp.). For each gene an initial tree was assembled as follows:
A seed sequence was used as a blast query against the NCBI nr database with an evalue < 1×10−10, and the top 250 hits were retrieved.
The same seed sequence was used as a blast query against the custom metamonad protein database, recovering all sequence hits with an e-value threshold of 1×10−5, creating the raw Metamonada dataset.
The raw Metamonada dataset sequences were clustered using CD-HIT44 at 99% identity, creating the clustered Metamonada dataset.
For each sequence in the clustered Metamonada dataset, the top three hits were recovered by blast searches against each major group of Bacteria and Eukaryotes in the nr taxonomy database. Duplicate sequences were removed.
The sequences from steps 1, 3 and 4 were grouped to make a final dataset. Sequences were aligned using MAFFT, ambiguously aligned positions were removed using trimAl45 and initial trees were constructed using FastTree.
Each initial tree was then visually inspected to select orthologues of interest. After removal of paralogues, each alignment was refined using MAFFT L-INS-I46, ambiguously aligned positions were removed using BMGE and the final phylogenies were constructed in RAxML with 100 rapid bootstrap replicates under the PROTGAMMALGF model. In the cases of MPP and ACS, removal of paralogues or manual addition of sequences was performed several times in order to ensure that the correct orthologue was present in the final tree.
The cardiolipin synthase phylogeny was constructed by adding the Ca. membranifera sequence to a dataset previously assembled and published by Noguchi et al. (ref. 47). Alignments available from the Dryad Digital Repository: doi 10.5061/dryad.34qd7.
Mitochondrial targeting prediction
For each protein sequence of interest, the presence of a Mitochondrial Targeting Sequence was predicted using MitoProt48, TargetP49 (with default parameters for non-plant sequences) and MitoFates50 (with default parameters for fungal sequences).
Data availability statement
Sequence data that support the findings of this study have been deposited in the NCBI Short Reads Archive (SRA) database under accession number SRP077666. The alignment data that support the findings of this study have been made available through the Dryad Digital Repository: doi: 10.5061/dryad.34qd7
Supplementary Material
Acknowledgments
This work and M.K., M.M.L. and C.W.S. were supported by a grant (MOP-142349) from the Canadian Institutes of Health Research awarded to A.J.R. This work was also supported, in part, by a grant from the JSPS Strategic Young Researcher Overseas Visits Program (awarded to R.K.), by NSERC Grant 298366-2009 to AGBS, by a Czech Science Foundation grant to I. Č. (project GA14-14105S) and by grants from the Japan Society for the Promotion of Science (JSPS; nos 15H05606 and 15K14591 awarded to R.K., 23117005 and 15H05231 awarded to T.H., 23117006 awarded to Y.I.). We would like to thank Dr. Aaron A. Heiss for his help with Trimastix marina data generation, and Núria Ros for helpful comments on the manuscript.
Footnotes
Author contributions
MK, RK, JOA, YI, AGBS, TH and AJR conceived and designed the experiments; MK, KK, IČ, JDS, FX, AY, QZ performed the experiments; MML, MK, RK, CWS, KK, LE, YI analyzed the data; RK, CWS, JDS, KT, YI, AGBS, TH and AJR contributed materials and/or analysis tools; and MML, MK, RK, AGBS and AJR wrote the paper.
Competing Financial Interests Statement
The authors declare that they have no competing financial interests.
References
- 1.Stairs CW, Leger MM, Roger AJ. Diversity and origins of anaerobic metabolism in mitochondria and related organelles. Philos Trans R Soc L B Biol Sci. 2015:370. doi: 10.1098/rstb.2014.0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindmark DG, Müller M. Hydrogenosome, a cytoplasmic organelle of the anaerobic flagellate Tritrichomonas foetus, and its role in pyruvate metabolism. J Biol Chem. 1973;248:7724–7728. [PubMed] [Google Scholar]
- 3.Müller M, et al. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev. 2012;76:444–95. doi: 10.1128/MMBR.05024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jerlstrom-Hultqvist J, et al. Hydrogenosomes in the diplomonad Spironucleus salmonicida. Nat Commun. 2013;4:2493. doi: 10.1038/ncomms3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jedelsky PL, et al. The minimal proteome in the reduced mitochondrion of the parasitic protist Giardia intestinalis. PLoS One. 2011;6:e17285. doi: 10.1371/journal.pone.0017285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karnkowska A, et al. A eukaryote without a mitochondrial organelle. Curr Biol. 2016;26:1274–1284. doi: 10.1016/j.cub.2016.03.053. [DOI] [PubMed] [Google Scholar]
- 7.Takishita K, et al. Multigene phylogenies of diverse Carpediemonas-like organisms identify the closest relatives of ‘amitochondriate’ diplomonads and retortamonads. Protist. 2012;163:344–355. doi: 10.1016/j.protis.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Simpson AG, Patterson DJ. On core jakobids and excavate taxa: the ultrastructure of Jakoba incarcerata. J Eukaryot Microbiol. 2001;48:480–492. doi: 10.1111/j.1550-7408.2001.tb00183.x. [DOI] [PubMed] [Google Scholar]
- 9.Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- 10.Martincova E, et al. Probing the biology of Giardia intestinalis mitosomes using in vivo enzymatic tagging. Mol Cell Biol. 2015;35:2864–2874. doi: 10.1128/MCB.00448-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–49. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 12.Yaffe MP, Ohta S, Schatz G. A yeast mutant temperature-sensitive for mitochondrial assembly is deficient in a mitochondrial protease activity that cleaves imported precursor polypeptides. EMBO J. 1985;4:2069–2074. doi: 10.1002/j.1460-2075.1985.tb03893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris SR, Matus A, Hrdy I, Kute E. Reductive evolution of the mitochondrial processing peptidases of the unicellular parasites Trichomonas vaginalis and Giardia intestinalis. PLoS Pathog. 2008;4:e1000243. doi: 10.1371/journal.ppat.1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garg S, et al. Conservation of transit peptide-independent protein import into the mitochondrial and hydrogenosomal matrix. Genome Biol Evol. 2015;7:2716–2726. doi: 10.1093/gbe/evv175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kikuchi G. The glycine cleavage system: composition, reaction mechanism, and physiological significance. Mol Cell Biochem. 1973;1:169–187. doi: 10.1007/BF01659328. [DOI] [PubMed] [Google Scholar]
- 16.Mukherjee M, Brown MT, McArthur AG, Johnson PJ. Proteins of the glycine decarboxylase complex in the hydrogenosome of Trichomonas vaginalis. Eukaryot Cell. 2006;5:2062–2071. doi: 10.1128/EC.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrison HG, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–6. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- 18.Zubacova Z, et al. The mitochondrion-like organelle of Trimastix pyriformis contains the complete glycine cleavage system. PLoS One. 2013;8:e55417. doi: 10.1371/journal.pone.0055417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hampl V, et al. Genetic evidence for a mitochondriate ancestry in the ‘amitochondriate’ flagellate Trimastix pyriformis. PLoS One. 2008;3:e1383. doi: 10.1371/journal.pone.0001383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nývltová E, Smutná T, Tachezy J, Hrdý I. OsmC and incomplete glycine decarboxylase complex mediate reductive detoxification of peroxides in hydrogenosomes of Trichomonas vaginalis. Mol Biochem Parasitol. 2016;206:29–38. doi: 10.1016/j.molbiopara.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 21.Xu F, et al. On the reversibility of parasitism: adaptation to a free-living lifestyle via gene acquisitions in the diplomonad Trepomonas sp. PC1. BMC Biol. 2016;14:62. doi: 10.1186/s12915-016-0284-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hampson RK, Barron LL, Olson MS. Regulation of the glycine cleavage system in isolated rat liver mitochondria. J Biol Chem. 1983;258:2993–2999. [PubMed] [Google Scholar]
- 23.Steinbüchel A, Müller M. Anaerobic pyruvate metabolism of Tritrichomonas foetus and Trichomonas vaginalis hydrogenosomes. Mol Biochem Parasitol. 1986;20:57–65. doi: 10.1016/0166-6851(86)90142-8. [DOI] [PubMed] [Google Scholar]
- 24.Van Hellemond JJ, Klockiewicz M, Gaasenbeek CP, Roos MH, Tielens AG. Rhodoquinone and complex II of the electron transport chain in anaerobically functioning eukaryotes. J Biol Chem. 1995;270:31065–31070. doi: 10.1074/jbc.270.52.31065. [DOI] [PubMed] [Google Scholar]
- 25.Tielens AGM, van Grinsven KWA, Henze K, van Hellemond JJ, Martin W. Acetate formation in the energy metabolism of parasitic helminths and protists. Int J Parasitol. 2010;40:387–397. doi: 10.1016/j.ijpara.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez LB, Müller M. Purification and characterization of the acetate forming enzyme, acetyl-CoA synthetase (ADP-forming) from the amitochondriate protist, Giardia lamblia. FEBS Lett. 1996;378:240–244. doi: 10.1016/0014-5793(95)01463-2. [DOI] [PubMed] [Google Scholar]
- 27.Noguchi F, et al. Metabolic capacity of mitochondrion-related organelles in the free-living anaerobic stramenopile Cantina marsupialis. Protist. 2015;166:534–550. doi: 10.1016/j.protis.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Field J, Rosenthal B, Samuelson J. Early lateral transfer of genes encoding malic enzyme, acetyl-CoA synthetase and alcohol dehydrogenases from anaerobic prokaryotes to Entamoeba histolytica. Mol Microbiol. 2000;38:446–455. doi: 10.1046/j.1365-2958.2000.02143.x. [DOI] [PubMed] [Google Scholar]
- 29.Nývltová E, et al. Lateral gene transfer and gene duplication played a key role in the evolution of Mastigamoeba balamuthi hydrogenosomes. Mol Biol Evol. 2015;32:1039–1055. doi: 10.1093/molbev/msu408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider RE, et al. The Trichomonas vaginalis hydrogenosome proteome is highly reduced relative to mitochondria, yet complex compared with mitosomes. Int J Parasitol. 2011;41:1421–1434. doi: 10.1016/j.ijpara.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murcha MW, Narsai R, Devenish J, Kubiszewski-Jakubiak S, Whelan J. MPIC: A mitochondrial protein import components database for plant and non-plant species. Plant Cell Physiol. 2015;56:e10. doi: 10.1093/pcp/pcu186. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Q, et al. Marine isolates of Trimastix marina form a plesiomorphic deep-branching lineage within Preaxostyla, separate from other known Trimastigids (Paratrimastix n. gen.) Protist. 2015;166:468–491. doi: 10.1016/j.protis.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 33.Kolisko M, et al. A wide diversity of previously undetected free-living relatives of diplomonads isolated from marine/saline habitats. Env Microbiol. 2010;12:2700–2710. doi: 10.1111/j.1462-2920.2010.02239.x. [DOI] [PubMed] [Google Scholar]
- 34.Chevreux B, et al. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 2004;14:1147–1159. doi: 10.1101/gr.1917404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gentekaki E, et al. Large-scale phylogenomic analysis reveals the phylogenetic position of the problematic taxon Protocruzia and unravels the deep phylogenetic affinities of the ciliate lineages. Mol Phylogenet Evol. 2014;78:36–42. doi: 10.1016/j.ympev.2014.04.020. [DOI] [PubMed] [Google Scholar]
- 37.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minh BQ, Nguyen MA, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lartillot N, Rodrigue N, Stubbs D, Richer J. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 2013;62:611–615. doi: 10.1093/sysbio/syt022. [DOI] [PubMed] [Google Scholar]
- 44.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Br Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 47.Noguchi F, Tanifuji G, Brown MW, Fujikura K, Takishita K. Complex evolution of two types of cardiolipin synthase in the eukaryotic lineage stramenopiles. Mol Phylogenet Evol. 2016;101:133–141. doi: 10.1016/j.ympev.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 48.Claros MG. MitoProt, a Macintosh application for studying mitochondrial proteins. Comput Appl Biosci. 1995;11:441–447. doi: 10.1093/bioinformatics/11.4.441. [DOI] [PubMed] [Google Scholar]
- 49.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 50.Fukasawa Y, et al. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol Cell Proteomics. 2015;14:1113–1126. doi: 10.1074/mcp.M114.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data that support the findings of this study have been deposited in the NCBI Short Reads Archive (SRA) database under accession number SRP077666. The alignment data that support the findings of this study have been made available through the Dryad Digital Repository: doi: 10.5061/dryad.34qd7