Abstract
Type I interferons (IFN-α/β) and the more recently identified type III IFNs (IFN-λ) function as the first line of defense against virus infection and regulate the development of both innate and adaptive immune responses. Type III IFNs were originally identified as a novel ligand-receptor system acting in parallel with type I IFNs, but subsequent studies have provided increasing evidence for distinct roles for each IFN family. In addition to their compartmentalized antiviral actions, these two systems appear to have multiple levels of cross-regulation and act coordinately to achieve effective antimicrobial protection with minimal collateral damage to the host.
Keywords: antiviral agent, interferon, mucosal immunology, viral immunology, virology
Interferon kingdom
Interferons (IFNs)3 represent a group of cytokines that are able to induce resistance to virus infections in treated cells. Most vertebrates possess three types (or families) of IFNs, based on similarities in gene and protein sequences and structures, receptor utilization, and biological functions, which are shared among the members of each IFN type but are distinct between the three IFN types. Type I IFN (IFN-α/β) and type III IFN (IFN-λ) families generally have multiple members, whereas type II IFN (IFN-γ) is encoded by a single gene in most species (1).
All vertebrates appear to possess type I IFN genes, which are generally numerous and intronless. The human genome contains 13 IFNA genes encoding 12 distinct, but similar, IFN-α proteins (IFN-α1 and IFN-α13 have identical sequences), and one gene each for IFN-β, IFN-ω, IFN-κ, and IFN-ϵ, which share only limited amino acid similarities with each other and with IFN-α. Only the IFNK gene contains an intron in the 3′-UTR; all other human type I IFN genes lack introns. In contrast, type III IFN genes do have introns (Fig. 1A) in most species. Amphibian genomes contain both intronless genes and genes with introns, which encode either type I or type III IFNs (2, 3). Thus far, all IFN genes identified in fish contain introns but, based on their sequences, are more related to type I IFNs found in other species rather than to type III IFNs (4). The intron-containing genes encoding either type I or type III IFNs share a common intron/exon structure that is also conserved in genes encoding IL-10-related cytokines (5, 6). IFN-λs also share limited similarity with both type I IFNs and the IL-10-related cytokines (Fig. 1B). In addition, all IFNs and IL-10-related cytokines signal through receptors belonging to the same class II cytokine receptor family (5, 7). Therefore, all these cytokines are likely to have arisen from a common ancestor by gene duplication and expansion. The divergence into two IFN families is likely to have happened relatively early in the tetrapod evolution, because amphibians, reptiles (including birds), and mammals have distinctive type I and type III IFNs. Although fish IFNs resemble type I IFNs, they utilize two heterodimeric receptor complexes, in which one receptor subunit is shared (8). Therefore, it appears that IFNs began to diverge into two subfamilies during the evolution of teleosts.
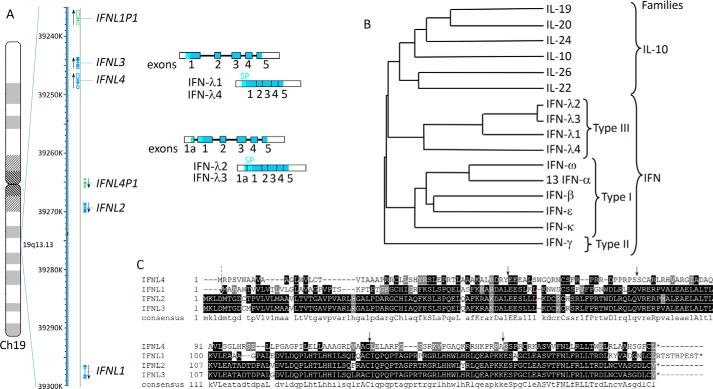
Figure 1.
Human type III IFNs. A, schematic representations of the chromosomal localization and intron/exon organization of the genes encoding human IFN-λs. The genes are transcribed in the direction indicated by the arrows. Genes encoding functional proteins are shown in blue and pseudogenes in green. Unspliced transcripts are schematically shown as strings of filled or open boxes (exons) joined by intervening solid lines (introns). Spliced transcripts are also shown as shaded/open boxes with vertical lines indicating the relative positions of former introns. The coding regions of exons are shaded, and the segments corresponding to the 5′- and 3′-untranslated regions are open (not shaded). The position of the signal peptide (SP) is shown in light blue. IFNL1 and IFNL4 genes are composed of five exons, whereas IFNL2 and IFNL3 genes have an additional upstream exon 1a. There are in-frame Met codons in both exons 1 and 1a; exon 1a extends the signal peptide by four additional amino acids. B, phylogenetic tree for IFN-λs with other IFNs and IL-10-related cytokines, (IL-10, IL-19, IL-22, IL-24, and IL-26) encoded in the human genome. Only one IFN-α was used in this alignment because the 13 human IFN-α subtypes have nearly identical sequences. C, sequence alignment of human IFN-λ proteins with the consensus sequence shown on the bottom. Arrows indicate positions of common introns (solid arrows) and of an additional intron, which is present only in IFNL2 and IFNL3 genes (dashed arrow).
Although designated as an IFN, and present in fish, IFN-γ has relatively weak intrinsic antiviral activity and, unlike type I and type III IFNs, its production is not directly triggered by viral infection. The sequence of IFN-γ is substantially divergent from IFNs and IL-10-related cytokines (Fig. 1B), and the IFNG gene has a distinct intron/exon structure.
The term IFN was originally devised for a substance produced by influenza A virus-infected chick embryo chorioallantoic membranes that can “interfere” with virus replication. Because chickens have type I and type III IFN-based systems, it is likely that, in the original experiments, both types of IFNs were present (9). However, because of the design of cloning strategies, the first IFNs to be characterized happened to be type I IFNs. IFN-β and several IFN-α subtypes were cloned in the 1980s based on their IFN-specific antiviral activities. Type III IFNs were first discovered in silico as novel cytokines that were shown to possess antiviral activities similar to those of type I IFNs. The type III IFN family in humans consists of four functional IFN-λ proteins. IFN-λ1, IFN-λ2, and IFN-λ3, also designated as IL-28/29, were discovered in 2002/2003 by two independent groups (10, 11). IFN-λ4 was identified in 2013 based on RNA sequence data from poly(I-C)-treated primary hepatocytes (12), and it shares ∼30% identity with other IFN-λs (Fig. 1C). The genes encoding type III IFNs are clustered on human chromosome 19 (Fig. 1A), whereas all type I IFN genes map to chromosome 9. In addition to four IFNL genes encoding functional proteins, there are three pseudogenes (Fig. 1A). Mice have only two genes encoding functional type III IFNs, IFN-λ2 and IFN-λ3, and two pseudogenes (13).
IFN-λ receptor complex and signaling
All IFN-λs engage a heterodimeric receptor complex for signaling. This complex is composed of IFN-λR1 (also known as IL-28R), a receptor subunit unique to the IFN-λ receptor (IFNLR), and the IL-10R2 chain that is shared with receptor complexes for IL-10, IL-22, and IL-26. There are three splice variants of the human IFNLR1 gene, encoding either the full-length functional IFN-λR1, a soluble IFN-λR1, or an IFN-λR1 variant lacking a membrane-proximal region of the intracellular domain and expected to be signal-incapable (5). The expression pattern of the splice variants and their functional importance are not well studied, although it has been suggested that soluble IFN-λR1 can partially antagonize IFN-λ activities (14). All type I IFNs signal through the IFNAR complex composed of IFN-αR1 and IFN-αR2 subunits. Interestingly, although type I and type III IFNs signal through distinct non-overlapping receptors, there is a poxvirus-encoded IFN antagonist that neutralizes the action of all human type I and type III IFNs, with the exception of IFN-λ4 (15). This viral glycoprotein binds type I and type III IFNs in a competitive manner, suggesting the existence of conserved epitopes between these two IFN family members, despite the minimal amino acid sequence similarity (15).
Signaling cascades triggered downstream of the IFN-λ receptor complex are very similar to those activated in response to type I IFNs binding to their specific receptor complex (Fig. 2) (16). The predominant signaling from either IFNLR or IFNAR is transmitted by the JAK-STAT pathway. Although the activation and involvement of JAK2 kinase in IFN-λ signaling has been reported (17, 18), STAT activation downstream of IFN-λR1 proceeded normally in JAK2-deficient cells (19). Because Tyk2 associates with IL-10R2 (20, 21), and JAK1 is expected to interact with IFN-λR1 (19), the importance of JAK2 for IFN-λ signaling requires further investigation. The hallmark of IFN signaling is the activation of a transcription complex designated IFN-stimulated gene factor 3 (ISGF3), which is highly specific to type I and type III IFN-induced signaling. ISGF3 is composed of STAT1, STAT2, and IRF9 and induces transcription of hundreds of ISGs. As expected, the sets of ISGs induced by either type I or type III IFNs are very similar if not identical. However, the kinetics of ISG induction by type I and type III IFNs differ. In general, the levels of ISG expression peak earlier and decline faster in response to type I IFNs and demonstrate a delayed and prolonged induction pattern in response to IFN-λs (22–24).
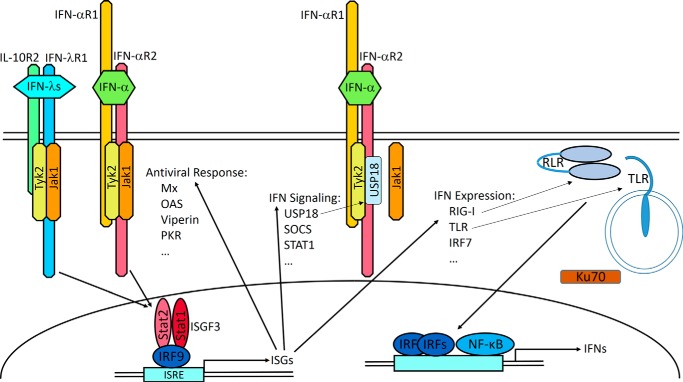
Figure 2.
IFN receptor complexes and signaling. IFN-λs and type I IFNs use distinct heterodimeric receptor complexes. The IFN-λs engage the unique IFN-λR1 and IL-10R2, a subunit also used by the IL-10, IL-22, and IL-26 receptor complexes. IFN-αR1 and IFN-αR2 form the active type I IFN receptor complex. IFN-λ binding to its receptor complex leads to the activation of receptor-associated JAK1 and Tyk2 kinases, which phosphorylate Tyr residues within the IFN-λR1 intracellular domain. These phosphotyrosine-based motifs serve as docking or recruitment sites for the latent transcriptional factors of the STAT family. The main STATs that become recruited and activated are STAT1 and STAT2, although activation of STAT3, STAT4, and STAT5 can be also detected (10, 19, 97). Phosphorylated STAT1 and STAT2 heterodimerize and interact with another transcription factor, IFN regulatory factor 9 (IRF9), leading to the formation of a transcription complex designated IFN-stimulated gene factor 3 (ISGF3). After translocation to the nucleus, ISGF3 binds to the IFN-specific response element (ISRE) that is commonly present in the promoter regions of hundreds of IFN-stimulated genes (ISGs), culminating in their transcriptional expression. The many ISGs encode a variety of antiviral mediators (98) enabling the establishment of an antiviral state effective against a broad spectrum of viruses. Subsets of ISGs encode proteins involved in virus recognition and IFN induction such as TLR, RIG-I-like receptors (RLR), and IRF7 as well as modulators of IFN signaling such as STAT1, USP18, and SOCS1 (99). USP18 binds to the IFN-αR2 intracellular domain, displaces JAK1, and suppresses IFN-α signaling. Ku70 is a DNA sensor that selectively triggers expression of IFN-λs (100).
One of the ISGs encodes a negative regulator of IFN signaling, USP18 (Fig. 2), which competes with JAK1 for binding to IFN-αR2 (25). USP18 has a stronger inhibitory effect toward IFN-αs than toward IFN-β (26), which may reflect differential affinities of IFN subtypes for their shared receptor subunits. The action of USP18 is of particular interest, because it provides a mechanism for selective inhibition and for cross-regulation of type I and type III IFN-induced signaling. USP18 is induced by either type of IFN (Fig. 2), but it exerts stronger inhibition of IFN-α- than IFN-λ-mediated activities (26, 27). Through USP18 induction, IFN-λs can selectively constrain IFN-α actions in cells responsive to both types of IFN.
Little is known about IFN-λ subtypes regarding their receptor binding and differential signaling. It came as a surprise that human IFN-λ3 has much higher antiviral potency than IFN-λ2, despite the fact that they have 96% amino acid identity (28). Although IFN-λ1, IFN-λ3, and IFN-λ4 seem to have comparable biological activities (28, 29), it remains to be seen whether the kinetics of ISG induction, mode of receptor engagement, or sensitivity to negative regulators may differ among these IFN-λ subtypes.
Expression of IFN-λ
The presence of regulatory elements, similar to those found in the promoters of type I IFN genes, led to the initial characterization of IFN-λs as antiviral cytokines (11). Indeed, IRF-, NF-κB-, and AP1-binding sites are commonly present in the promoters of both type I and type III IFN genes (16, 30, 31). As predicted, initial experiments with cell lines or primary cells demonstrated that type I and type III IFNs are co-produced in response to virus infection or small molecular mimetics that trigger activation of extracellular or intracellular sensors, which recognize pathogen-associated molecular pattern molecules, particularly nucleic acids. However, experiments carried out in vivo are revealing a much more nuanced and complex picture. Mucosal infections appear to trigger predominantly IFN-λ expression and a low level of IFN-β (32, 33). The source of these IFNs is often epithelial cells, which preferentially express IFN-λs (34–41). For example, epithelial cells were the primary source of IFN-λ production in the gastrointestinal tract of mice infected with reovirus and rotavirus (35, 41). IFN-αs are produced later and require the presence of immune cells, such as plasmacytoid dendritic cells (pDCs), which produce both IFN-αs and IFN-λs (42–44). BDCA3+ dendritic cells (DCs) can also produce IFN-λs in response to poly(I-C) or co-culture with HCV-infected cells (45–47). Overall, the expression patterns and levels of production of type I and type III IFNs vary depending on the site of the infection and the specific viral pathogen. Although not surprising, given the many different mechanisms by which viruses stimulate and suppress IFN production (48), this variability is often underappreciated. It appears that production of IFN-λs can be induced to substantial levels by either IRF or NF-κB pathways (49–51), whereas type I IFN production requires activation of both pathways for efficient production. Because many viruses target the IRF pathway, the preferential expression of IFN-λs by epithelial cells during virus infection may be occurring via an unaffected NF-κB pathway.
Another mechanism favoring IFN-λ production by epithelial cells is the increased number of peroxisomes found in polarized cells at mucosal surfaces (18). MAVS, a critical mediator of a signaling cascade leading to IFN production downstream of RIG-I-like receptors, can be found localized to mitochondria or peroxisomes (52). Activation of peroxisome-associated MAVS triggers the production of IFN-λs, whereas type I IFNs are induced by mitochondrion-associated MAVS (18, 52). IRF1, IRF3, and NF-κB, but not AP1, mediate the signaling leading to IFN-λ production from peroxisome-bound MAVS (18, 52). This pathway was initially assumed to be a novel, IFN-independent, antiviral pathway, as no expression of type I IFNs could be detected (52), but it was subsequently shown to lead to selective IFN-λ production (18). Bacterial infections of epithelial cells also triggered IFN-λ production mediated by peroxisome-associated MAVs (18, 53, 54). Increased polarization status of intestinal epithelial cells not only augments their ability to produce IFN-λs (18) but also enhances their sensitivity to IFN-λ treatment (41) emphasizing the importance of type III IFNs in intestinal mucosal immunity (Fig. 3). Thus, type III IFNs appear to play an important role in the protection of barrier surfaces against a variety of microbes and are likely to contribute to the maintenance of tissue homeostasis.
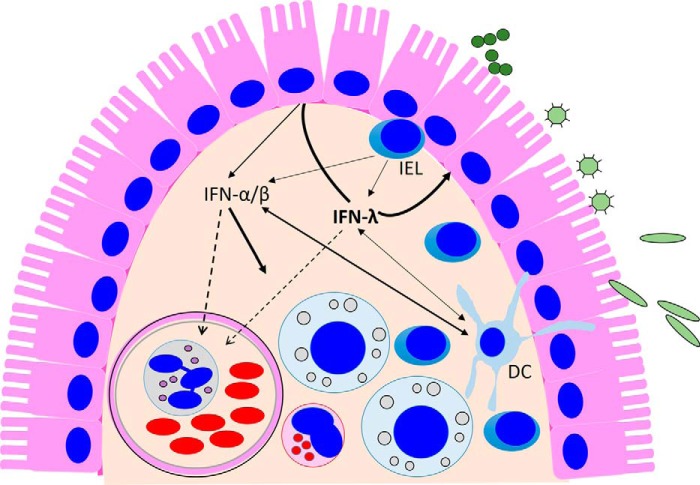
Figure 3.
IFN-λ-centric model on the production and action of IFNs at the epithelial barrier. IFN production in the intestine is induced by invading bacterial and viral pathogens and likely by the many microbial by-products present in the intestinal lumen. These stimuli trigger predominantly IFN-λ expression from epithelial cells, and these cells preferentially respond to type III IFNs. Therefore, it appears that the antiviral protection of IECs relies on the IFN-λ-based autocrine system. In addition to IECs, DCs present in the lamina propria beneath the epithelial surface produce and respond to both types of IFNs, but pDCs preferentially produce IFN-α. Intraepithelial T lymphocytes (IELs), which are in continuous contact with the epithelial layer, also secrete both type I and type III IFNs upon antigen stimulation (101). IFN-α/β secreted into the submucosa (solid arrows) act on lamina propria cells. Both IFN types can also act systemically through entry into the blood stream (dashed line).
IFN-λ induction at low levels, and with delayed kinetics, was detected in HepG2 cells following treatment by either type I or type III IFNs (55) and in IFN-α-treated pDCs (43), demonstrating that the IFNL genes are ISGs. ISGs encode many proteins, such as TLRs, RIG-I-like receptors, IRF7, STAT1, SOCS, and USP18, that can positively or negatively modulate IFN production or signaling (Fig. 2). This provides a basis for cross-talk between these two IFN systems. It has been shown that treatment of DCs and macrophages with IFN-α prior to, or together with, virus infection or TLR agonists potentiates IFN-λ production (46, 56–58). Priming of IFN-λ expression by type I IFNs is also suspected because of the decreased IFN-λ expression in response to virus infection observed in IFNAR-deficient mice (33, 59). The extent to which type I IFN expression is potentiated by IFN-λs is less clear. This was reported for macrophages and DCs following exposure to virus (46, 56), but pretreatment of airway epithelial cells with IFN-λ had no effect on influenza A virus-induced IFN-β expression (37). Because immune cells are thought to be the major source of type I IFN production in vivo, and only a restricted set of immune cells respond to IFN-λ, priming by IFN-λ is unlikely to affect type I IFN expression.
IFN-λs and HCV
In 2009, several independent groups discovered that polymorphisms in the vicinity of the IFNL3 gene predict response to IFN-α-based antiviral therapies in patients chronically infected with HCV, as well as spontaneous clearance of HCV infection (60–62). Subsequent studies reported the association of these single nucleotide polymorphisms (SNPs) with other chronic viral infections and other diseases, including non-alcoholic fatty liver disease and allergy. It was proposed that the SNPs affect the level of IFN-λ3 expression; however, this was not conclusively demonstrated. Sets of SNPs were found to be in linkage disequilibrium, revealing the existence of several distinct haplotypes in humans. In 2013, a novel IFNL gene, designated IFNL4, was discovered upstream of the IFNL3 gene (Fig. 1) (12). Two SNPs have been identified in this gene in the human population; one of these causes a frameshift mutation resulting in non-functional IFN-λ4 and another generates an amino acid substitution (12, 63). IFN-λ4 possesses strong antiviral activity, comparable with that of IFN-λ3 and IFN-λ1, and signals through the same receptor complex, despite being only ∼30% identical to other IFN-λs (29). The P70S amino acid substitution reduces IFN-λ4 activity (63). Paradoxically, the presence of an allele encoding fully active IFN-λ4, the protein that, in vitro, shares anti-HCV activities with other IFN-λs, is strongly associated with unfavorable outcomes from HCV infection. The principles underlying this seeming contradiction remain elusive. One possible explanation (61, 62) arises from previous observations: (i) the increased expression levels of ISGs, including USP18 (Fig. 2), in patients refractory to anti-HCV therapies (62), and (ii) selective suppression of IFN-α but not IFN-λ signaling by HCV (64, 65). This suggests a scenario in which continuously produced IFN-λ4 (66) is responsible for the chronic up-regulation of ISG expression, including negative regulators such as USP18 that maintain the refractoriness to IFN-α-based therapy. However, it remains unclear what makes IFN-λ4 so different from other IFN-λs, because transcription from all four IFNL genes is simultaneously induced by the HCV mimetic poly(I-C) in primary hepatocytes, but the establishment of chronic infection is most strongly associated with the presence of the functional IFNL4 gene. One distinction conserved among mammalian IFN-λ4 proteins is the fact that, unlike other IFN-λs, this cytokine is poorly secreted from the producer cells (12, 29, 67), but how and whether this feature may translate into a unique function during HCV infection remain unclear. Rodents lack the IFNL4 gene, but noteworthy is the report that T cell responses were opposite in IFNLR−/− mice infected with acute versus persistent strains of lymphocytic choriomeningitis virus (68). Therefore, even in rodents, the effects of IFN-λs may be tuned by chronic inflammation.
The conundrum of IFN-λ4 in HCV infection will be difficult to untangle as this phenomenon cannot be studied in the mouse model. Mouse hepatocytes are not responsive to type III IFNs in vivo, despite their ability to respond to these cytokines ex vivo (69). Human hepatocytes are sensitive to type III IFNs in vivo as shown by successful clinical trials of IFN-λ1 for the treatment of patients with chronic HCV infection (70), patterns of human IFN-λR1 expression in liver biopsies (71, 72), and IFN-λ responsiveness of human primary hepatocytes. Nonetheless, IFN-λR1 expression varies in human hepatocytes in vivo (72), indicating that hepatocyte responsiveness to IFNs, and the factors and pathways affecting this, deserves additional examination.
Role of IFN-λs in vivo
The existence of the IFN-λ family was not suspected prior to 2003, largely because type I and type III IFNs are induced by many of the same stimuli and mediate the same effects despite their distinct receptors. Thus, one of most interesting questions facing IFN researchers is understanding how and whether type III IFNs have a distinct biological role. One clear distinction is the essentially non-overlapping distribution of type I and type III IFN receptors.
Utilization of different receptors allows type I and type III IFNs to target distinct compartments in vivo. Despite the fact that IFNAR has long been thought to be expressed on all cell types, it is now becoming clear that, at least in the gastrointestinal tract, intestinal epithelial cells (IECs) respond almost exclusively to type III IFNs, whereas type I IFNs are mostly active on immune and endothelial cells (Fig. 3) (35, 41, 73–76). Accordingly, IFN-λ limits rotavirus and reovirus replication in IECs, whereas type I IFNs control virus spread beyond the epithelial compartment (35, 41). Interestingly, this difference is observed only in adult animals, with IECs in neonatal mice responsive to both IFN types (41). It is important to emphasize that isolated intestinal epithelial cells respond to either type I or type III IFNs ex vivo, as do a variety of cell lines derived from the GI epithelium (10, 76). The basis of this age-dependent restriction in vivo is not yet understood. It has been suggested that the level of IFN-λR1 expression is down-regulated in IECs in vivo (35), but the mechanism(s) determining this altered responsiveness is currently unknown. Also unresolved is the localization of IFN receptors on polarized epithelial surfaces in vivo. Polarized rat IECs were found to respond to type I IFNs added to the apical surface only, whereas type III IFNs could signal from either the apical or the basolateral surface (76). This is contradicted by an in vivo study showing both IFN-α/β and IFN-λ responses by neonatal murine IECs following subcutaneous administration of either cytokine (41). It is important that receptor orientation, in mouse and man, be determined if we are to understand innate immune responses at the mucosal surface.
Acting on epithelial cells, IFN-λs may be important for limiting virus release into the lumina of the gastrointestinal and respiratory tracts. Indeed, fecal shedding of rotavirus and reovirus, as well as murine norovirus (MNV) that primarily replicates in B cells, was increased in IFNLR-deficient mice (35, 76, 77). Although rotavirus and reovirus replicate in IECs, MNV replicates in hematopoietic cells, where IFN-λ is inactive. Therefore, indirect action of IFN-λs was proposed for controlling persistent MNV infection (77). Moreover, depletion of microbiota by antibiotics prevented the establishment of persistent infection in mice infected with low MNV doses, an effect that was not observed in IFNLR-deficient animals (78). However, MNV replicates to higher titers in IFNLR-deficient mice, and higher MNV doses lead to persistent MNV infection even in antibiotic-treated animals (78). Thus, the precise mechanisms of IFN-λ-mediated protection against MNV, and the cells responding to this cytokine in MNV infection, require further investigation.
The MNV example raises an additional consideration. It is possible that type III IFNs reduce MNV shedding by acting to strengthen the epithelial barrier even in the absence of epithelial infection. Of note, tightening of the blood-brain barrier by IFN-λ decreased neuroinvasion of West Nile virus (79). Effects of IFNs on the permeability of the GI epithelial barrier may also explain increased sensitivity of mice to the dextran sulfate sodium-containing water in the mouse model of acute colitis (80).
Current data suggest that type I and type III IFNs are redundant in the protection against respiratory viruses where replication of the virus is limited to the epithelial compartment. In the case of influenza virus (73, 81) or severe acute respiratory syndrome (SARS) virus (82) infection, either type I or type III IFNs confer the same level of protection seen in wild-type mice, with significantly enhanced virus titers in the absence of both pathways. Consistent with these observations, influenza A virus infection induces the same set of ISGs in murine tracheal epithelial cells obtained from either wild-type, IFNAR-, or IFNLR-deficient mice (83), confirming the redundancy of type I and type III IFN-based antiviral responses in airway epithelium. Enhanced respiratory syncytial virus replication is present only in mice deficient in both type I and type III IFN receptors (73). However, it was recently reported that IFN-λs are more important in controlling the spread of respiratory viruses from the upper respiratory tract to the lungs in mice and virus transmission between animals (84). Therefore, it appears that there may be non-redundant IFN-based functions within the respiratory as well as the GI tract.
It has been particularly challenging to define immune cell subsets sensitive to type III IFNs due to the lack of verified research reagents, and the fact that the assessment of levels of transcriptional or protein expression of IFN-λR1 does not equate to responsiveness of the cells to IFNs. As an example, B cells may express IFN-λR1 but are barely responsive to IFN-λs (14, 85). NK cells, which are important mediators of anti-tumor activities of both type I and type III IFNs (86–88), are sensitive to type I IFNs, but whether IFN-λs can directly affect NK functions is controversial (86–90). There are numerous and sometimes contradictory reports on this topic, but in aggregate it appears that T cells, NK cells, and monocytes are not directly responsive to type III IFNs. Nonetheless, this does not rule out the possibility that subsets of these cells may become IFN-λ-responsive in some settings. It has recently been shown, for example, that the responsiveness of neutrophils to IFN-λ depends on their activation or differentiation status; neutrophils isolated from peripheral blood do not respond to this cytokine, whereas the neutrophils isolated from bone marrow are very sensitive to IFN-λs (91). Plasmacytoid dendritic cells isolated from human subjects are also responsive to both IFN types (43, 92).
Overall, it can be concluded that the type III IFN system evolved to serve as a first line of defense at the mucosal barrier, which is the initial target of most invasive pathogens. Mainly, IFN-λs protect these epithelial layers, but some subsets of immune cells, particularly those involved in innate rather than adaptive immune responses, are responsive to IFN-λs constitutively or upon activation (Fig. 3). Acting on DCs, IFN-λs can indirectly affect the development of adaptive immune responses (43, 92–94). Neutrophil and DC responses to type I IFNs might also be down-regulated by prolonged IFN-λ-mediated induction of USP18, adding another level of immune regulation. This is an important consideration as type I IFNs generally promote inflammatory as well as antiviral responses (95). Consistent with the idea that type III IFNs may act as a brake on the proinflammatory effects of type I IFNs, it was reported that IFN-λs acting on DCs promote the development of T regulatory cells (93) and dampened inflammation in mouse models of asthma (94) and rheumatoid arthritis (91). In addition, in vivo studies also suggest a role for type III IFNs in wound healing and anti-tumor defense (61), although the mechanisms of these additional functions are not well understood.
Clinical applications
The most compelling aspect of our current knowledge of type III IFN biology is the possibility that IFN-λs could act as potent antivirals against multiple infections without the side effects associated with IFN-α treatment. This hypothesis was tested in clinical trials of IFN-λ1 therapy for HCV infection, which confirmed that this cytokine had antiviral effects equivalent to IFN-α without the same level of associated toxicity (70). Studies of IFN-λ treatment of influenza A virus-infected mice have shown similar results (96). Exogenous IFN-λ was also able to prevent or cure murine norovirus infection (77) and reduce replication of rotavirus (35, 41, 76). Taken together, these data suggest that IFN-λ can potentially be developed as a broad-spectrum antiviral agent for prophylaxis and treatment of a large number of viral infections without the unwanted proinflammatory effects of IFN-α. This is an exciting possibility and would represent a major clinical advance. In addition, type III IFNs have now been shown to play an important role in protecting the lung from fungal infection,4 suggesting applications beyond the treatment and prophylaxis of viral infection. The potential use of IFN-λ in the treatment of malignancy is also an area of active research (61).
Conclusion
The host response to invading pathogens requires well-tuned defense mechanisms that eradicate harmful microbes while limiting tissue damage and systemic inflammation. Most species evolved two ligand-receptor systems, based on either type I IFNs (IFN-α/β) or type III IFNs (IFN-λs), which function as the first line of innate defense against virus infection, but they also act as regulators of both innate and adaptive immune responses. Although type III IFNs were originally shown to provide antiviral protection in parallel with and independent of the type I IFNs, a growing body of evidence indicates distinct roles for each IFN family, as well as non-overlapping functions for subtypes within each family. Of equal importance is the realization that, in addition to their compartmentalized antimicrobial actions, these two antiviral systems appear to have multiple levels of cross-regulation and act coordinately during both the innate and adaptive phases of the immune response. As our understanding of these complementary systems grows, so will opportunities for more nuanced therapeutic interventions in infectious and inflammatory disease.
Acknowledgments
We are thankful to Russell Durbin and Jerome Langer for careful reading and editing the manuscript.
This work was supported in part by National Institutes of Health Grants RO1 AI057468 (to S. V. K.) and AI104669 (to S. V. K. and J. E. D.). S. V. K. is an inventor of patents and patent applications related to IFN-λs, which have been licensed for commercial development. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
A. Rivera, J. E. Durbin, and S. V. Kotenko, unpublished data.
- IFN
- interferon
- ISG
- IFN-stimulated gene
- IEC
- intestinal epithelial cell
- IFNAR
- IFN-α/β receptor
- IFNLR
- IFN-λ receptor
- DC
- dendritic cell
- pDC
- plasmacytoid dendritic cell
- HCV
- hepatitis C virus
- TLR
- Toll-like receptor
- SNP
- single nucleotide polymorphism
- GI
- gastrointestinal
- MNV
- murine norovirus
- MAVS
- mitochondrial antiviral signaling protein.
References
- 1. Krause C. D., and Pestka S. (2015) Cut, copy, move, delete: the study of human interferon genes reveals multiple mechanisms underlying their evolution in amniotes. Cytokine 76, 480–495 [DOI] [PubMed] [Google Scholar]
- 2. Qi Z., Nie P., Secombes C. J., and Zou J. (2010) Intron-containing type I and type III IFN coexist in amphibians: refuting the concept that a retroposition event gave rise to type I IFNs. J. Immunol. 184, 5038–5046 [DOI] [PubMed] [Google Scholar]
- 3. Sang Y., Liu Q., Lee J., Ma W., McVey D. S., and Blecha F. (2016) Expansion of amphibian intronless interferons revises the paradigm for interferon evolution and functional diversity. Sci. Rep. 6, 29072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boudinot P., Langevin C., Secombes C. J., and Levraud J. P. (2016) The peculiar characteristics of fish type I interferons. Viruses 8, E298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kotenko S. V., and Langer J. A. (2004) Full house: 12 receptors for 27 cytokines. Int. Immunopharmacol. 4, 593–608 [DOI] [PubMed] [Google Scholar]
- 6. Kotenko S. V. (2002) The family of IL-10-related cytokines and their receptors: related, but to what extent? Cytokine Growth Factor Rev. 13, 223–240 [DOI] [PubMed] [Google Scholar]
- 7. Langer J. A., Cutrone E. C., and Kotenko S. (2004) The class II cytokine receptor (CRF2) family: overview and patterns of receptor-ligand interactions. Cytokine Growth Factor Rev. 15, 33–48 [DOI] [PubMed] [Google Scholar]
- 8. Aggad D., Mazel M., Boudinot P., Mogensen K. E., Hamming O. J., Hartmann R., Kotenko S., Herbomel P., Lutfalla G., and Levraud J. P. (2009) The two groups of zebrafish virus-induced interferons signal via distinct receptors with specific and shared chains. J. Immunol. 183, 3924–3931 [DOI] [PubMed] [Google Scholar]
- 9. Isaacs A., and Lindenmann J. (1957) Virus interference: 1. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 147, 258–267 [PubMed] [Google Scholar]
- 10. Kotenko S. V., Gallagher G., Baurin V. V., Lewis-Antes A., Shen M., Shah N. K., Langer J. A., Sheikh F., Dickensheets H., and Donnelly R. P. (2003) IFN-λs mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 4, 69–77 [DOI] [PubMed] [Google Scholar]
- 11. Sheppard P., Kindsvogel W., Xu W., Henderson K., Schlutsmeyer S., Whitmore T. E., Kuestner R., Garrigues U., Birks C., Roraback J., Ostrander C., Dong D., Shin J., Presnell S., Fox B., et al. (2003) IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 4, 63–68 [DOI] [PubMed] [Google Scholar]
- 12. Prokunina-Olsson L., Muchmore B., Tang W., Pfeiffer R. M., Park H., Dickensheets H., Hergott D., Porter-Gill P., Mumy A., Kohaar I., Chen S., Brand N., Tarway M., Liu L., Sheikh F., et al. (2013) A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 45, 164–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lasfar A., Lewis-Antes A., Smirnov S. V., Anantha S., Abushahba W., Tian B., Reuhl K., Dickensheets H., Sheikh F., Donnelly R. P., Raveche E., and Kotenko S. V. (2006) Characterization of the mouse IFN-λ ligand-receptor system: IFN-λs exhibit antitumor activity against B16 melanoma. Cancer Res. 66, 4468–4477 [DOI] [PubMed] [Google Scholar]
- 14. Witte K., Gruetz G., Volk H. D., Looman A. C., Asadullah K., Sterry W., Sabat R., and Wolk K. (2009) Despite IFN-λ receptor expression, blood immune cells, but not keratinocytes or melanocytes, have an impaired response to type III interferons: implications for therapeutic applications of these cytokines. Genes Immun. 10, 702–714 [DOI] [PubMed] [Google Scholar]
- 15. Huang J., Smirnov S. V., Lewis-Antes A., Balan M., Li W., Tang S., Silke G. V., Pütz M. M., Smith G. L., and Kotenko S. V. (2007) Inhibition of type I and type III interferons by a secreted glycoprotein from Yaba-like disease virus. Proc. Natl. Acad. Sci. U.S.A. 104, 9822–9827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kotenko S. V. (2011) IFN-λs. Curr. Opin. Immunol. 23, 583–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee S. J., Kim W. J., and Moon S. K. (2012) Role of the p38 MAPK signaling pathway in mediating interleukin-28A-induced migration of UMUC-3 cells. Int. J. Mol. Med. 30, 945–952 [DOI] [PubMed] [Google Scholar]
- 18. Odendall C., Dixit E., Stavru F., Bierne H., Franz K. M., Durbin A. F., Boulant S., Gehrke L., Cossart P., and Kagan J. C. (2014) Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat. Immunol. 15, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dumoutier L., Lejeune D., Hor S., Fickenscher H., and Renauld J. C. (2003) Cloning of a new type II cytokine receptor activating signal transducer and activator of transcription (STAT)1, STAT2 and STAT3. Biochem. J. 370, 391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kotenko S. V., Izotova L. S., Pollack B. P., Muthukumaran G., Paukku K., Silvennoinen O., Ihle J. N., and Pestka S. (1996) Other kinases can substitute for Jak2 in signal transduction by interferon-γ. J. Biol. Chem. 271, 17174–17182 [DOI] [PubMed] [Google Scholar]
- 21. Kotenko S. V., Krause C. D., Izotova L. S., Pollack B. P., Wu W., and Pestka S. (1997) Identification and functional characterization of a second chain of the interleukin-10 receptor complex. EMBO J. 16, 5894–5903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marcello T., Grakoui A., Barba-Spaeth G., Machlin E. S., Kotenko S. V., MacDonald M. R., and Rice C. M. (2006) Interferons α and λ inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology 131, 1887–1898 [DOI] [PubMed] [Google Scholar]
- 23. Bolen C. R., Ding S., Robek M. D., and Kleinstein S. H. (2014) Dynamic expression profiling of type I and type III interferon-stimulated hepatocytes reveals a stable hierarchy of gene expression. Hepatology 59, 1262–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jilg N., Lin W., Hong J., Schaefer E. A., Wolski D., Meixong J., Goto K., Brisac C., Chusri P., Fusco D. N., Chevaliez S., Luther J., Kumthip K., Urban T. J., Peng L. F., et al. (2014) Kinetic differences in the induction of interferon stimulated genes by interferon-α and interleukin 28B are altered by infection with hepatitis C virus. Hepatology 59, 1250–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Malakhova O. A., Kim K. I., Luo J. K., Zou W., Kumar K. G., Fuchs S. Y., Shuai K., and Zhang D. E. (2006) UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 25, 2358–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. François-Newton V., Magno de Freitas Almeida G., Payelle-Brogard B., Monneron D., Pichard-Garcia L., Piehler J., Pellegrini S., and Uzé G. (2011) USP18-based negative feedback control is induced by type I and type III interferons and specifically inactivates interferon α response. PLoS ONE 6, e22200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Burkart C., Arimoto K., Tang T., Cong X., Xiao N., Liu Y. C., Kotenko S. V., Ellies L. G., and Zhang D. E. (2013) Usp18 deficient mammary epithelial cells create an antitumour environment driven by hypersensitivity to IFN-λ and elevated secretion of Cxcl10. EMBO Mol. Med. 5, 1035–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dellgren C., Gad H. H., Hamming O. J., Melchjorsen J., and Hartmann R. (2009) Human interferon-λ3 is a potent member of the type III interferon family. Genes Immun. 10, 125–131 [DOI] [PubMed] [Google Scholar]
- 29. Hamming O. J., Terczyńska-Dyla E., Vieyres G., Dijkman R., Jørgensen S. E., Akhtar H., Siupka P., Pietschmann T., Thiel V., and Hartmann R. (2013) Interferon λ4 signals via the IFNλ receptor to regulate antiviral activity against HCV and coronaviruses. EMBO J. 32, 3055–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Donnelly R. P., and Kotenko S. V. (2010) Interferon-λ: a new addition to an old family. J. Interferon Cytokine Res. 30, 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Iversen M. B., and Paludan S. R. (2010) Mechanisms of type III interferon expression. J. Interferon Cytokine Res. 30, 573–578 [DOI] [PubMed] [Google Scholar]
- 32. Nakagawa S., Hirata Y., Kameyama T., Tokunaga Y., Nishito Y., Hirabayashi K., Yano J., Ochiya T., Tateno C., Tanaka Y., Mizokami M., Tsukiyama-Kohara K., Inoue K., Yoshiba M., Takaoka A., and Kohara M. (2013) Targeted induction of interferon-λ in humanized chimeric mouse liver abrogates hepatotropic virus infection. PLoS ONE 8, e59611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jewell N. A., Cline T., Mertz S. E., Smirnov S. V., Flaño E., Schindler C., Grieves J. L., Durbin R. K., Kotenko S. V., and Durbin J. E. (2010) λ interferon is the predominant interferon induced by influenza A virus infection in vivo. J. Virol. 84, 11515–11522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sato S., Li K., Kameyama T., Hayashi T., Ishida Y., Murakami S., Watanabe T., Iijima S., Sakurai Y., Watashi K., Tsutsumi S., Sato Y., Akita H., Wakita T., Rice C. M., et al. (2015) The RNA sensor RIG-I dually functions as an innate sensor and direct antiviral factor for hepatitis B virus. Immunity 42, 123–132 [DOI] [PubMed] [Google Scholar]
- 35. Mahlakõiv T., Hernandez P., Gronke K., Diefenbach A., and Staeheli P. (2015) Leukocyte-derived IFN-α/β and epithelial IFN-λ constitute a compartmentalized mucosal defense system that restricts enteric virus infections. PLoS Pathog. 11, e1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thomas E., Gonzalez V. D., Li Q., Modi A. A., Chen W., Noureddin M., Rotman Y., and Liang T. J. (2012) HCV infection induces a unique hepatic innate immune response associated with robust production of type III interferons. Gastroenterology 142, 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang J., Oberley-Deegan R., Wang S., Nikrad M., Funk C. J., Hartshorn K. L., and Mason R. J. (2009) Differentiated human alveolar type II cells secrete antiviral IL-29 (IFN-λ1) in response to influenza A infection. J. Immunol. 182, 1296–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marukian S., Andrus L., Sheahan T. P., Jones C. T., Charles E. D., Ploss A., Rice C. M., and Dustin L. B. (2011) Hepatitis C virus induces interferon-λ and interferon-stimulated genes in primary liver cultures. Hepatology 54, 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Okabayashi T., Kojima T., Masaki T., Yokota S., Imaizumi T., Tsutsumi H., Himi T., Fujii N., and Sawada N. (2011) Type-III interferon, not type-I, is the predominant interferon induced by respiratory viruses in nasal epithelial cells. Virus Res. 160, 360–366 [DOI] [PubMed] [Google Scholar]
- 40. Khaitov M. R., Laza-Stanca V., Edwards M. R., Walton R. P., Rohde G., Contoli M., Papi A., Stanciu L. A., Kotenko S. V., and Johnston S. L. (2009) Respiratory virus induction of α-, β- and λ-interferons in bronchial epithelial cells and peripheral blood mononuclear cells. Allergy 64, 375–386 [DOI] [PubMed] [Google Scholar]
- 41. Lin J. D., Feng N., Sen A., Balan M., Tseng H. C., McElrath C., Smirnov S. V., Peng J., Yasukawa L. L., Durbin R. K., Durbin J. E., Greenberg H. B., and Kotenko S. V. (2016) Distinct roles of type I and type III interferons in intestinal immunity to homologous and heterologous rotavirus infections. PLoS Pathog. 12, e1005600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Megjugorac N. J., Gallagher G. E., and Gallagher G. (2010) IL-4 enhances IFN-λ1 (IL-29) production by plasmacytoid DCs via monocyte secretion of IL-1Ra. Blood 115, 4185–4190 [DOI] [PubMed] [Google Scholar]
- 43. Yin Z., Dai J., Deng J., Sheikh F., Natalia M., Shih T., Lewis-Antes A., Amrute S. B., Garrigues U., Doyle S., Donnelly R. P., Kotenko S. V., and Fitzgerald-Bocarsly P. (2012) Type III IFNs are produced by and stimulate human plasmacytoid dendritic cells. J. Immunol. 189, 2735–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Coccia E. M., Severa M., Giacomini E., Monneron D., Remoli M. E., Julkunen I., Cella M., Lande R., and Uzé G. (2004) Viral infection and Toll-like receptor agonists induce a differential expression of type I and λ interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur. J. Immunol. 34, 796–805 [DOI] [PubMed] [Google Scholar]
- 45. Yoshio S., Kanto T., Kuroda S., Matsubara T., Higashitani K., Kakita N., Ishida H., Hiramatsu N., Nagano H., Sugiyama M., Murata K., Fukuhara T., Matsuura Y., Hayashi N., Mizokami M., and Takehara T. (2013) Human blood dendritic cell antigen 3 (BDCA3) dendritic cells are a potent producer of interferon-λ in response to hepatitis C virus. Hepatology 57, 1705–1715 [DOI] [PubMed] [Google Scholar]
- 46. Zhang S., Kodys K., Li K., and Szabo G. (2013) Human type 2 myeloid dendritic cells produce interferon-λ and amplify interferon-α in response to hepatitis C virus infection. Gastroenterology 144, 414–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lauterbach H., Bathke B., Gilles S., Traidl-Hoffmann C., Luber C. A., Fejer G., Freudenberg M. A., Davey G. M., Vremec D., Kallies A., Wu L., Shortman K., Chaplin P., Suter M., O'Keeffe M., and Hochrein H. (2010) Mouse CD8α+ DCs and human BDCA3+ DCs are major producers of IFN-λ in response to poly IC. J. Exp. Med. 207, 2703–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hoffmann H. H., Schneider W. M., and Rice C. M. (2015) Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol. 36, 124–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee H. C., Narayanan S., Park S. J., Seong S. Y., and Hahn Y. S. (2014) Transcriptional regulation of IFN-λ genes in hepatitis C virus-infected hepatocytes via IRF-3.IRF-7.NF-κB complex. J. Biol. Chem. 289, 5310–5319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Iversen M. B., Ank N., Melchjorsen J., and Paludan S. R. (2010) Expression of type III interferon (IFN) in the vaginal mucosa is mediated primarily by dendritic cells and displays stronger dependence on NF-κB than type I IFNs. J. Virol. 84, 4579–4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thomson S. J., Goh F. G., Banks H., Krausgruber T., Kotenko S. V., Foxwell B. M., and Udalova I. A. (2009) The role of transposable elements in the regulation of IFN-λ1 gene expression. Proc. Natl. Acad. Sci. U.S.A. 106, 11564–11569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dixit E., Boulant S., Zhang Y., Lee A. S., Odendall C., Shum B., Hacohen N., Chen Z. J., Whelan S. P., Fransen M., Nibert M. L., Superti-Furga G., and Kagan J. C. (2010) Peroxisomes are signaling platforms for antiviral innate immunity. Cell 141, 668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lebreton A., Lakisic G., Job V., Fritsch L., Tham T. N., Camejo A., Matteï P. J., Regnault B., Nahori M. A., Cabanes D., Gautreau A., Ait-Si-Ali S., Dessen A., Cossart P., and Bierne H. (2011) A bacterial protein targets the BAHD1 chromatin complex to stimulate type III interferon response. Science 331, 1319–1321 [DOI] [PubMed] [Google Scholar]
- 54. Bierne H., Travier L., Mahlakõiv T., Tailleux L., Subtil A., Lebreton A., Paliwal A., Gicquel B., Staeheli P., Lecuit M., and Cossart P. (2012) Activation of type III interferon genes by pathogenic bacteria in infected epithelial cells and mouse placenta. PLoS ONE 7, e39080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ank N., West H., Bartholdy C., Eriksson K., Thomsen A. R., and Paludan S. R. (2006) λ interferon (IFN-λ), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J. Virol. 80, 4501–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Melchjorsen J., Sirén J., Julkunen I., Paludan S. R., and Matikainen S. (2006) Induction of cytokine expression by herpes simplex virus in human monocyte-derived macrophages and dendritic cells is dependent on virus replication and is counteracted by ICP27 targeting NF-κB and IRF-3. J. Gen. Virol. 87, 1099–1108 [DOI] [PubMed] [Google Scholar]
- 57. Sirén J., Pirhonen J., Julkunen I., and Matikainen S. (2005) IFN-α regulates TLR-dependent gene expression of IFN-α, IFN-β, IL-28, and IL-29. J. Immunol. 174, 1932–1937 [DOI] [PubMed] [Google Scholar]
- 58. Osterlund P., Veckman V., Sirén J., Klucher K. M., Hiscott J., Matikainen S., and Julkunen I. (2005) Gene expression and antiviral activity of α/β interferons and interleukin-29 in virus-infected human myeloid dendritic cells. J. Virol. 79, 9608–9617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ank N., Iversen M. B., Bartholdy C., Staeheli P., Hartmann R., Jensen U. B., Dagnaes-Hansen F., Thomsen A. R., Chen Z., Haugen H., Klucher K., and Paludan S. R. (2008) An important role for type III interferon (IFN-λ/IL-28) in TLR-induced antiviral activity. J. Immunol. 180, 2474–2485 [DOI] [PubMed] [Google Scholar]
- 60. Wack A., Terczyńska-Dyla E., and Hartmann R. (2015) Guarding the frontiers: the biology of type III interferons. Nat. Immunol. 16, 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lazear H. M., Nice T. J., and Diamond M. S. (2015) Interferon-λ: immune functions at barrier surfaces and beyond. Immunity 43, 15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Heim M. H., Bochud P. Y., and George J. (2016) Host-hepatitis C viral interactions: the role of genetics. J. Hepatol. 65, S22–32 [DOI] [PubMed] [Google Scholar]
- 63. Terczyńska-Dyla E., Bibert S., Duong F. H., Krol I., Jørgensen S., Collinet E., Kutalik Z., Aubert V., Cerny A., Kaiser L., Malinverni R., Mangia A., Moradpour D., Müllhaupt B., Negro F., et al. (2014) Reduced IFNλ4 activity is associated with improved HCV clearance and reduced expression of interferon-stimulated genes. Nat. Commun. 5, 5699. [DOI] [PubMed] [Google Scholar]
- 64. Friborg J., Ross-Macdonald P., Cao J., Willard R., Lin B., Eggers B., and McPhee F. (2015) Impairment of type I but not type III IFN signaling by hepatitis C virus infection influences antiviral responses in primary human hepatocytes. PLoS ONE 10, e0121734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chandra P. K., Bao L., Song K., Aboulnasr F. M., Baker D. P., Shores N., Wimley W. C., Liu S., Hagedorn C. H., Fuchs S. Y., Wu T., Balart L. A., and Dash S. (2014) HCV infection selectively impairs type I but not type III IFN signaling. Am. J. Pathol. 184, 214–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Amanzada A., Kopp W., Spengler U., Ramadori G., and Mihm S. (2013) Interferon-λ4 (IFNL4) transcript expression in human liver tissue samples. PLoS ONE 8, e84026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Paquin A., Onabajo O. O., Tang W., and Prokunina-Olsson L. (2016) Comparative functional analysis of 12 mammalian IFN-λ4 orthologs. J. Interferon Cytokine Res. 36, 30–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Misumi I., and Whitmire J. K. (2014) IFN-λ exerts opposing effects on T cell responses depending on the chronicity of the virus infection. J. Immunol. 192, 3596–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dickensheets H., Sheikh F., Park O., Gao B., and Donnelly R. P. (2013) Interferon-λ (IFN-λ) induces signal transduction and gene expression in human hepatocytes, but not in lymphocytes or monocytes. J. Leukocyte Biol. 93, 377–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Muir A. J., Arora S., Everson G., Flisiak R., George J., Ghalib R., Gordon S. C., Gray T., Greenbloom S., Hassanein T., Hillson J., Horga M. A., Jacobson I. M., Jeffers L., Kowdley K. V., et al. (2014) A randomized phase 2b study of peginterferon λ-1a for the treatment of chronic HCV infection. J. Hepatol. 61, 1238–1246 [DOI] [PubMed] [Google Scholar]
- 71. Doyle S. E., Schreckhise H., Khuu-Duong K., Henderson K., Rosler R., Storey H., Yao L., Liu H., Barahmand-pour F., Sivakumar P., Chan C., Birks C., Foster D., Clegg C. H., Wietzke-Braun P., Mihm S., and Klucher K. M. (2006) Interleukin-29 uses a type 1 interferon-like program to promote antiviral responses in human hepatocytes. Hepatology 44, 896–906 [DOI] [PubMed] [Google Scholar]
- 72. Duong F. H., Trincucci G., Boldanova T., Calabrese D., Campana B., Krol I., Durand S. C., Heydmann L., Zeisel M. B., Baumert T. F., and Heim M. H. (2014) IFN-λ receptor 1 expression is induced in chronic hepatitis C and correlates with the IFN-λ3 genotype and with nonresponsiveness to IFN-α therapies. J. Exp. Med. 211, 857–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mordstein M., Neugebauer E., Ditt V., Jessen B., Rieger T., Falcone V., Sorgeloos F., Ehl S., Mayer D., Kochs G., Schwemmle M., Günther S., Drosten C., Michiels T., and Staeheli P. (2010) λ interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J. Virol. 84, 5670–5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sommereyns C., Paul S., Staeheli P., and Michiels T. (2008) IFN-λ (IFN-λ) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 4, e1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pulverer J. E., Rand U., Lienenklaus S., Kugel D., Zietara N., Kochs G., Naumann R., Weiss S., Staeheli P., Hauser H., and Köster M. (2010) Temporal and spatial resolution of type I and III IFN responses in vivo. J. Virol. 84, 8626–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pott J., Mahlakõiv T., Mordstein M., Duerr C. U., Michiels T., Stockinger S., Staeheli P., and Hornef M. W. (2011) IFN-λ determines the intestinal epithelial antiviral host defense. Proc. Natl. Acad. Sci. U.S.A. 108, 7944–7949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nice T. J., Baldridge M. T., McCune B. T., Norman J. M., Lazear H. M., Artyomov M., Diamond M. S., and Virgin H. W. (2015) Interferon-λ cures persistent murine norovirus infection in the absence of adaptive immunity. Science 347, 269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Baldridge M. T., Nice T. J., McCune B. T., Yokoyama C. C., Kambal A., Wheadon M., Diamond M. S., Ivanova Y., Artyomov M., and Virgin H. W. (2015) Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science 347, 266–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lazear H. M., Daniels B. P., Pinto A. K., Huang A. C., Vick S. C., Doyle S. E., Gale M. Jr., Klein R. S., and Diamond M. S. (2015) Interferon-λ restricts West Nile virus neuroinvasion by tightening the blood-brain barrier. Sci. Transl. Med. 7, 284ra259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Rauch I., Rosebrock F., Hainzl E., Heider S., Majoros A., Wienerroither S., Strobl B., Stockinger S., Kenner L., Müller M., and Decker T. (2015) Noncanonical effects of IRF9 in intestinal inflammation: more than type I and type III interferons. Mol. Cell. Biol. 35, 2332–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mordstein M., Kochs G., Dumoutier L., Renauld J. C., Paludan S. R., Klucher K., and Staeheli P. (2008) Interferon-λ contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog. 4, e1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Mahlakõiv T., Ritz D., Mordstein M., DeDiego M. L., Enjuanes L., Müller M. A., Drosten C., and Staeheli P. (2012) Combined action of type I and type III interferon restricts initial replication of severe acute respiratory syndrome coronavirus in the lung but fails to inhibit systemic virus spread. J. Gen. Virol. 93, 2601–2605 [DOI] [PubMed] [Google Scholar]
- 83. Crotta S., Davidson S., Mahlakoiv T., Desmet C. J., Buckwalter M. R., Albert M. L., Staeheli P., and Wack A. (2013) Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog. 9, e1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Klinkhammer J. S., D., Schwanderlapp M;, Mahlakoiv T., and Staeheli P. (2016) Interferon-λ controls the spread of influenza viruses from the upper respiratory tract to the lungs and restricts virus transmission in mice. Cytokine 76, 74 [Google Scholar]
- 85. Kelly A., Robinson M. W., Roche G., Biron C. A., O'Farrelly C., and Ryan E. J. (2016) Immune cell profiling of IFN-λ response shows pDCs express highest level of IFN-λR1 and are directly responsive via the JAK-STAT pathway. J. Interferon Cytokine Res. 36, 671–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Souza-Fonseca-Guimaraes F., Young A., Mittal D., Martinet L., Bruedigam C., Takeda K., Andoniou C. E., Degli-Esposti M. A., Hill G. R., and Smyth M. J. (2015) NK cells require IL-28R for optimal in vivo activity. Proc. Natl. Acad. Sci. U.S.A. 112, E2376–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Abushahba W., Balan M., Castaneda I., Yuan Y., Reuhl K., Raveche E., de la Torre A., Lasfar A., and Kotenko S. V. (2010) Antitumor activity of type I and type III interferons in BNL hepatoma model. Cancer Immunol. Immunother. 59, 1059–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lasfar A., de laTorre A., Abushahba W., Cohen-Solal K. A., Castaneda I., Yuan Y., Reuhl K., Zloza A., Raveche E., Laskin D. L., and Kotenko S. V. (2016) Concerted action of IFN-α and IFN-λ induces local NK cell immunity and halts cancer growth. Oncotarget 7, 49259–49267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. de Groen R. A., Boltjes A., Hou J., Liu B. S., McPhee F., Friborg J., Janssen H. L., and Boonstra A. (2015) IFN-λ-mediated IL-12 production in macrophages induces IFN-γ production in human NK cells. Eur. J. Immunol. 45, 250–259 [DOI] [PubMed] [Google Scholar]
- 90. Morrison M. H., Keane C., Quinn L. M., Kelly A., O'Farrelly C., Bergin C., and Gardiner C. M. (2014) IFNL cytokines do not modulate human or murine NK cell functions. Hum. Immunol. 75, 996–1000 [DOI] [PubMed] [Google Scholar]
- 91. Blazek K., Eames H. L., Weiss M., Byrne A. J., Perocheau D., Pease J. E., Doyle S., McCann F., Williams R. O., and Udalova I. A. (2015) IFN-λ resolves inflammation via suppression of neutrophil infiltration and IL-1β production. J. Exp. Med. 212, 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Megjugorac N. J., Gallagher G. E., and Gallagher G. (2009) Modulation of human plasmacytoid DC function by IFN-λ1 (IL-29). J. Leukocyte Biol. 86, 1359–1363 [DOI] [PubMed] [Google Scholar]
- 93. Mennechet F. J., and Uzé G. (2006) Interferon-λ-treated dendritic cells specifically induce proliferation of FOXP3-expressing suppressor T cells. Blood 107, 4417–4423 [DOI] [PubMed] [Google Scholar]
- 94. Koltsida O., Hausding M., Stavropoulos A., Koch S., Tzelepis G., Ubel C., Kotenko S. V., Sideras P., Lehr H. A., Tepe M., Klucher K. M., Doyle S. E., Neurath M. F., Finotto S., and Andreakos E. (2011) IL-28A (IFN-λ2) modulates lung DC function to promote Th1 immune skewing and suppress allergic airway disease. EMBO Mol. Med. 3, 348–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Davidson S., Crotta S., McCabe T. M., and Wack A. (2014) Pathogenic potential of interferon αβ in acute influenza infection. Nat. Commun. 5, 3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Davidson S., McCabe T. M., Crotta S., Gad H. H., Hessel E. M., Beinke S., Hartmann R., and Wack A. (2016) IFNλ is a potent anti-influenza therapeutic without the inflammatory side effects of IFNα treatment. EMBO Mol. Med. 8, 1099–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Dumoutier L., Tounsi A., Michiels T., Sommereyns C., Kotenko S. V., and Renauld J. C. (2004) Role of the interleukin (IL)-28 receptor tyrosine residues for antiviral and antiproliferative activity of IL-29/interferon-λ1: similarities with type I interferon signaling. J. Biol. Chem. 279, 32269–32274 [DOI] [PubMed] [Google Scholar]
- 98. Schoggins J. W., and Rice C. M. (2011) Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 1, 519–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Brand S., Zitzmann K., Dambacher J., Beigel F., Olszak T., Vlotides G., Eichhorst S. T., Göke B., Diepolder H., and Auernhammer C. J. (2005) SOCS-1 inhibits expression of the antiviral proteins 2′,5′-OAS and MxA induced by the novel interferon-λs IL-28A and IL-29. Biochem. Biophys. Res. Commun. 331, 543–548 [DOI] [PubMed] [Google Scholar]
- 100. Zhang X., Brann T. W., Zhou M., Yang J., Oguariri R. M., Lidie K. B., Imamichi H., Huang D. W., Lempicki R. A., Baseler M. W., Veenstra T. D., Young H. A., Lane H. C., and Imamichi T. (2011) Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J. Immunol. 186, 4541–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Swamy M., Abeler-Dörner L., Chettle J., Mahlakõiv T., Goubau D., Chakravarty P., Ramsay G., Reis e Sousa C., Staeheli P., Blacklaws B. A., Heeney J. L., and Hayday A. C. (2015) Intestinal intraepithelial lymphocyte activation promotes innate antiviral resistance. Nat. Commun. 6, 7090. [DOI] [PMC free article] [PubMed] [Google Scholar]