Abstract
Similar to other yeasts, the human pathogen Candida glabrata ages when it undergoes asymmetric, finite cell divisions, which determines its replicative lifespan. We sought to investigate if and how aging changes resilience of C. glabrata populations in the host environment. Our data demonstrate that old C. glabrata are more resistant to hydrogen peroxide and neutrophil killing, whereas young cells adhere better to epithelial cell layers. Consequently, virulence of old compared to younger C. glabrata cells is enhanced in the Galleria mellonella infection model. Electron microscopy images of old C. glabrata cells indicate a marked increase in cell wall thickness. Comparison of transcriptomes of old and young C. glabrata cells reveals differential regulation of ergosterol and Hog pathway associated genes as well as adhesion proteins, and suggests that aging is accompanied by remodeling of the fungal cell wall. Biochemical analysis supports this conclusion as older cells exhibit a qualitatively different lipid composition, leading to the observed increased emergence of fluconazole resistance when grown in the presence of fluconazole selection pressure. Older C. glabrata cells accumulate during murine and human infection, which is statistically unlikely without very strong selection. Therefore, we tested the hypothesis that neutrophils constitute the predominant selection pressure in vivo. When we altered experimentally the selection pressure by antibody-mediated removal of neutrophils, we observed a significantly younger pathogen population in mice. Mathematical modeling confirmed that differential selection of older cells is sufficient to cause the observed demographic shift in the fungal population. Hence our data support the concept that pathogenesis is affected by the generational age distribution of the infecting C. glabrata population in a host. We conclude that replicative aging constitutes an emerging trait, which is selected by the host and may even play an unanticipated role in the transition from a commensal to a pathogen state.
Author summary
This study shows that aging makes Candida glabrata more resilient and virulent in vivo. Aging results in a thicker cell wall that is also qualitatively different with respect to sterol composition, which conveys enhanced resistance to azoles and cellular stresses. We observed greater accumulation of old cells in competent mice, indicating that selection pressures in the host preferentially kill younger cells. Depletion experiments and mathematical modeling confirm that neutrophils represent the predominant selection pressure, and their removal dramatically alters selection pressures, resulting in a younger pathogen population. We propose that replicative age is an unanticipated virulence trait that is selected in the host environment and may play an important role in the switch from a commensal to a pathogen state.
Introduction
Candida glabrata infections are common in immunocompromised patients and associated with prolonged treatment [1, 2], extended length of hospital stay, high costs and high mortality rates [3, 4]. Over the last decade, the incidence of C. glabrata infections has increased considerably due to higher numbers of immunocompromised patients, as well as broad empiric antifungal prophylaxis, which promotes colonization with azole-resistant C. glabrata [1]. C. glabrata is a very successful human pathogen because it has a high intrinsic stress tolerance, enabling it to withstand oxidative stress [5]. The yeast disseminates and attaches to host cells and indwelling devices, where it forms biofilms [6]. In vitro phagocytosed C. glabrata cells are able to survive and replicate inside human and murine macrophages [7, 8]. Imaging studies demonstrate that primary human neutrophils can kill or release phagocytosed C. glabrata [9]. Consequently, neutropenia constitutes a major risk factor for disseminated candidiasis in colonized patients [10, 11].
Most fungal infections are subacute or chronic [1, 10, 12]. A major barrier to improved antifungal drug therapy is a lack of understanding of how fungal populations change during chronic infection, and how this microevolution affects in vivo phenotypes and pathogenesis of the respective fungus [13, 14]. The pathogenic yeasts, Cryptococcus neoformans, Candida albicans and C. glabrata expand clonally in the host even though some of them retain the ability for mating and sexual reproduction [15]. Similar to other unicellular yeasts, C. glabrata is expected to undergo asymmetric mitotic divisions where original mother cells progressively age in a process referred to as replicative aging [16]. Saccharomyces cerevisiae has been the predominant model organism for investigating this process in yeasts. However, this fungus is rarely a pathogen in humans, and therefore research on it has historically focused on elucidating the molecular mechanisms that regulate aging [17].
Recent work in C. neoformans suggests that replicative aging can affect the pathogenesis of fungal populations [18, 19]. Specifically it was shown that old C. neoformans cells, referring to those with a higher replicative age, accumulate in spinal fluid during chronic infection, indicating the presence of a demographic shift within the infecting fungal population. Older cells manifested a thicker cell wall that rendered them more resistant to host response. This suggested that replicative age might be relevant in a clonally expanding C. neoformans population allowing it to gain resilience in the host environment. We proposed that older C. neoformans cells constitute the persister cells and contribute to the high treatment failure rate despite lack of antifungal resistance.
The intent of this study was to characterize the lifespan features of another important fungal pathogen, C. glabrata. Opposed to C. neoformans, this yeast can be a commensal, and it has been proposed that its resilience and high stress tolerance has evolved as a result of its intimate association with the mammalian host (for review [20]). The host is especially vulnerable for invasive disease, when the host becomes neutropenic and the major antifungal defenses are compromised. Then C. glabrata can shift from a commensal to an invasive pathogen. Therefore, we were particularly interested in studying the interplay between replicative aging of a C. glabrata population and the in vivo selection pressures due to neutrophil-mediated killing, which can be done by controlled depletion studies in the murine host. Our data demonstrate that aging leads to remodeling of the cell wall and that selection by neutrophils controls generational distribution within the C. glabrata population. We show that younger C. glabrata cells with more potential to adhere emerge in a neutropenic host, whereas older C. glabrata cells, which exhibit a thicker cell wall with an altered sterol composition mediate enhanced azole resistance and resilience and dominate in the immunocompetent host.
Results
Characterization of lifespan in clinical C. glabrata strains
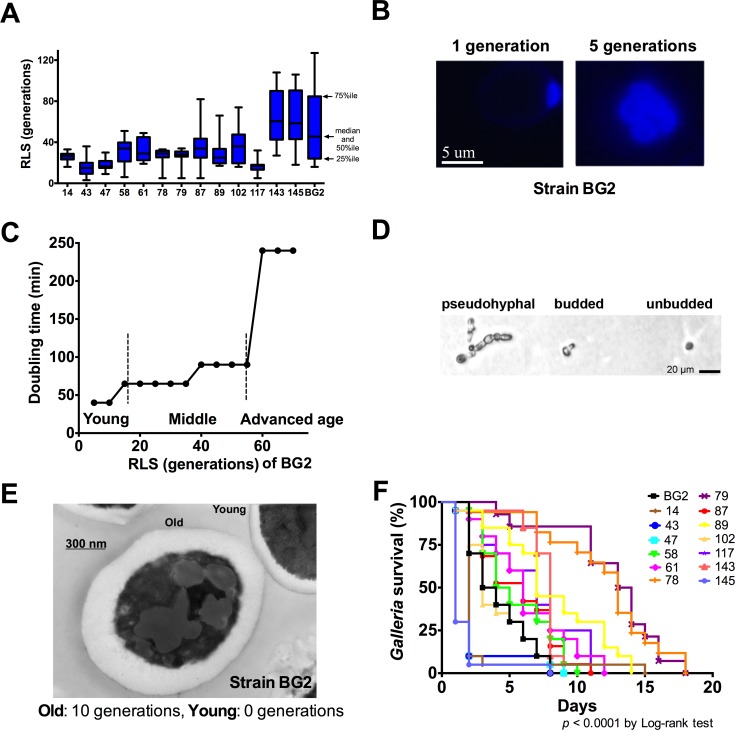
To investigate aging and its effects on various virulence-related phenotypes, we determined replicative lifespans (RLS) at 37°C on 14 clinical C. glabrata strains of different multilocus sequence types (MLST) and variable chronological lifespans (CLS) (Fig 1A and Table 1). The median RLS of C. glabrata strains varied from 15.0 (strain 43) to 61 generations (strain 143). Median RLS of cells across all strains was 29 generations and ranged from 3–127 generations, confirming the previous observation in C. neoformans [18, 19] that RLS is highly variable among strains, but a distinct and reproducible trait that defines the individual strain. High variability of RLS within a C. glabrata population was noted, where some cells exhibited a very short, and others a very long RLS. Neither prolonged nor shortened RLS was associated with a specific MLST type. In accordance with other publications [21], we found calcofluor reliably stains budscars, allowing us to identify the respective replicative age of older C. glabrata cells (Fig 1B and S1 Video). C. glabrata cells, regardless of their MLST, doubled every 45–50 min until they reached an advanced age when their doubling time slowed considerably from 90 to 240 min. Accordingly, the lifespan of C. glabrata strains could be divided into a young, middle, and advanced age phase (Fig 1C). Notably, at the end of their RLS, senescent C. glabrata cells arrested either as pseudohyphae, in a budded state, or as a single mother cell (unbudded) (Fig 1D). With every replication, the cell size and cell wall thickness increased (Fig 1E). Although considerable variability in relative cell body volume increase among strains was observed, there was no correlation between maximal volume and median RLS (Spearman r = -0.14, p = 0.78). Also despite variability, no correlation between virulence in the Galleria mellonella model and median RLS (Spearman r = -0.01, p = 0.96), or median CLS (Spearman r = -0.46, p = 0.10) was found. C. glabrata strains killed Galleria at markedly different rates (range 1–14 d, p < 0.0001) (Fig 1F). C. glabrata strains (14, 43, 47) that formed pseudohyphae were more virulent (median survival: 2 d).
Fig 1. Phenotypic characterization of RLS in C. glabrata.
(A) A box-and-whiskers plot demonstrates significant variability in RLSs of C. glabrata strains, shown as box plots with percentiles (n = 20 cells per experiment and 2–6 experiments compared by Log-Rank test). (B) Calcofluor stain reliably identifies generational age of C. glabrata (n = 100 cells). (C) Distinct doubling times during different lifespan phases, and (D) various cell morphologies were captured at time of death. (E) shows thicker cell wall in older BG2 cells by TEM (n = 100 cells compared by Student’s t-test). (F) C. glabrata demonstrated variable virulence in G. mellonella (n = 20 worms compared by Log-Rank test).
Table 1. Characterization of clinical C. glabrata isolates.
| Strain | MLST | Source of origin | Median CLS and range (days) |
|---|---|---|---|
| BG2 | 3 | Vaginal | 23 (20–25) |
| 14 | 8 | Urine | 24 (22–25) |
| 43 | 9 | Urine | 23 (20–25) |
| 47 | 8 | Urine | 22 (20–24) |
| 58 | 5 | Urine | 27 (25–30) |
| 61 | 10 | Urine | 22 (20–24) |
| 78 | 4 | Blood | 23 (20–25) |
| 79 | 14 | Blood | 24 (22–25) |
| 87 | 7 | Urine | 25 (23–27) |
| 89 | 17 | Urine | 30 (27–33) |
| 102 | 2 | Urine | 10 (7–13) |
| 117 | 9 | Urine | 26 (22–24) |
| 143 | 10 | Urine | 10 (7–13) |
| 145 | 10 | Urine | 10 (7–13) |
Higher age makes C. glabrata more resilient to host attack and increases switching to more virulent variants
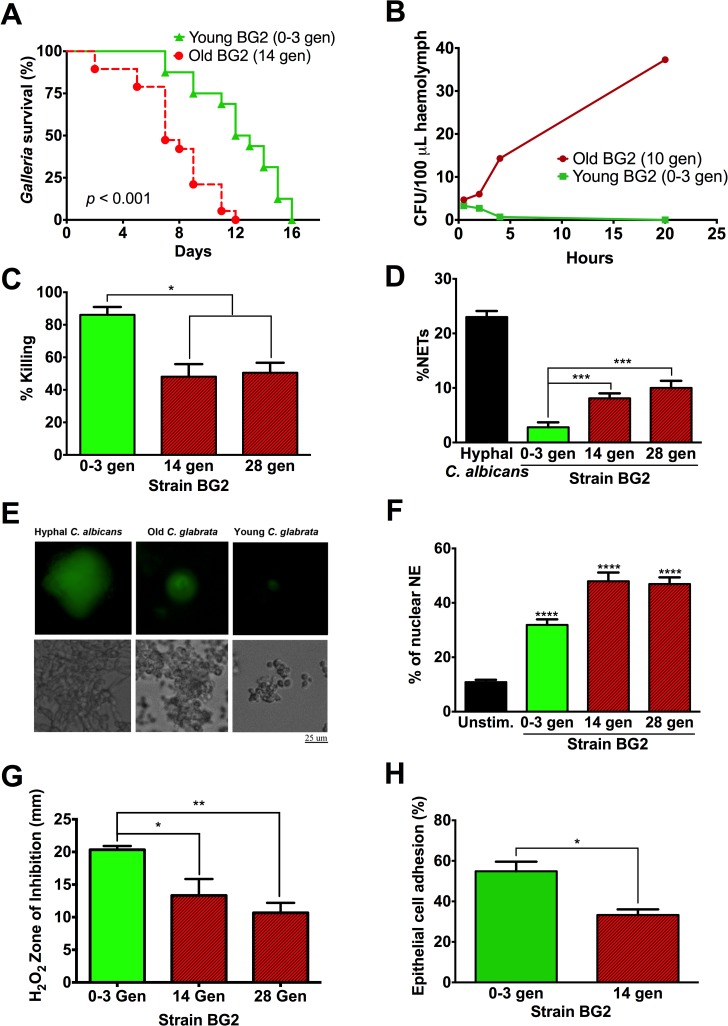
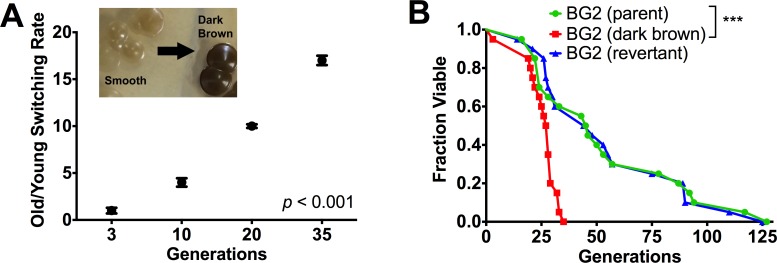
Similar to other yeast species, we hypothesized that replicative aging would augment C. glabrata’s resilience in vivo (Table 2). Old C. glabrata cells were isolated by biotin labeling a virgin population, allowing it ample time to divide and age, and then magnetically labeling it with streptavidin beads before sorting through a magnetic column. These cells, which henceforth we will label as “old,” are 10–14 generations, or for some experiments, 28 generations old. These are not young cells, but at this age are also far from senescence and thus have comparable growth dynamics to young cells (Fig 1C). The higher the replicative age of a cell, the rarer it becomes without stress: in a clonally expanding population a 14 generation old cell is rare and has a frequency of roughly 1:16,000 compared to other ages; this becomes 1:>108 for a 28 generation old cell. We compared the virulence of these old cells to young ones of the standard laboratory strain BG2 using the Galleria model. These experiments demonstrated that old cells (14 generations, average diameter 6–8 μm) were significantly (p < 0.05) more virulent compared to young cells (0–3 generations, average diameter 4.1 μm) (Fig 2A). It was determined that younger cells were cleared quicker than older cells, while older cells tended to accumulate in the first few hours during infection (Fig 2B). Consistent with that finding, older cells were phagocytosed at a lower rate by Galleria haemocytes (S1A Fig) and human neutrophils (S1B Fig), and more resistant to neutrophil-mediated killing (Fig 2C). Although older C. glabrata cells induce more neutrophil extracellular traps (NETs) than young C. glabrata cells (Fig 2D–2F), relative to C. albicans hyphae the overall NET induction is considerably lower, and it is unclear if NETs truly kill these C. glabrata cells. Similar enhancement of virulence (S2A and S2B Fig), and resistance to neutrophil-mediated phagocytosis and killing (S2C and S2D Fig) was also documented in older cells of C. glabrata strains 89 and 117. Consistent with augmented resistance to neutrophil killing, older cells exposed to H2O2 exhibited smaller zones of no growth in a disc diffusion assay (Fig 2G and S2E Fig). Interestingly, older cells exhibited decreased adhesion to epithelial cells compared with younger cells (Fig 2H) suggesting that they would more successfully invade after dissemination. Lastly, the effect of aging on switching was investigated, in C. glabrata, where the phenotypic switch from the parental smooth (S) variant to the dark brown (DB) variant, is associated with enhanced persistence in mice ([22] and S2F Fig). These experiments demonstrate that replicative aging of BG2 cells from 2 to 35 generations consistently resulted in higher phenotypic switching rate (Fig 3A), a finding confirmed also in strain 89 (S2G Fig). Interestingly, in both strains, S to DB switching (Fig 3B and S2H Fig) resulted in over 50% loss of median RLS, which was reversible and regained in the revertant colony. These findings further support the conception that RLS is a highly regulated trait in a fungus.
Table 2. Summary of the phenotypic characteristics of 14-generation-old C. glabrata cells compared to young cells (strain BG2).
| Phenotype | Effect in old cells |
|---|---|
| Cell volume | Increased (2.6 fold ± 1.0) |
| Cell wall thickness | Increased (2.3 fold ± 0.5)* |
| Doubling time | Increased (40 ± 10 min to 165 ± 75 min)* |
| Phagocytic killing | Decreased (32.3% ± 6.5%)** |
| NET induction | Increased (8.1% ± 1.0 from 2.7% ± 0.9)*** |
| Adherence | Decreased (40.0% ± 8.5%)* |
| Hydrogen peroxide resistance | Increased (2.3 fold ± 0.3)* |
| Phenotypic switching rate | Increased (4–17 fold ± 0.4)*** |
*P < 0.05
**P < 0.01
***P < 0.001.
Fig 2. Enhanced resilience of older C. glabrata cells.
(A) Older cells contributed to increased virulence in Galleria (n = 20 worms compared by Log-Rank test). (B) CFU counts continuously increased after Galleria mellonella infection with old BG2 cells (10 generations). Young BG2 cells (0–3 generations) were cleared before establishing infection within the first few hours post infection (n = 25 worms per time point). (C) Resistance to neutrophil-mediated killing was observed in older cells (experiments were run in duplicates, with six replicates, and compared by Student’s t-test). (D and E) NET induction was higher by old (14 and 28 generations) compared to young C. glabrata cells, but still significantly lower compared to that seen with hyphal C. albicans. % NETs indicates neutrophil nuclei >1,000 μm2 over total neutrophils, and upper panel in D shows sytox staining of nuclei (experiments were run in duplicates, with six replicates, and compared by Student’s t-test). (F) Neutrophil Elastase nuclear localization was higher in old (14 and 28 generations) compared to young C. glabrata cells and unstimulated neutrophils (10 neutrophils were analyzed per condition and compared by one-way ANOVA) (G) H2O2 disc diffusion assay shows smaller zone of inhibition for both 14 and 28 generation old cells (experiments were run in triplicates and compared by Student’s t-test). (H) Epithelial cell adhesion assays demonstrate decreased adhesion of older cells (experiments were run in triplicates and compared by Student’s t-test). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Fig 3. Switch variants have altered lifespan.
(A) Phenotypic switching in BG2 from S to DB colony morphology (inset) increased consistently with aging to 17 fold (experiments were run in duplicates). (B) RLS of DB was shortened over 50% relative to S and reconstituted in the revertant colony (n = 20 cells over two experiments; DB v. S RLS compared by Wilcoxon Rank Sum Test). ***P < 0.001.
C. glabrata resilience and enhanced resistance to antifungals is mediated by a cell wall remodeled in the process of aging
Given the above described striking phenotypes of resilience we pursued a transcriptome analysis comparing genes transcribed in young and 14-generation-old C. glabrata cells by RNA sequencing. Fungal cells were grown in YPD. RNAseq results were consistent with the observed distinct cell wall morphology on EM images of old cells and indicated that fundamental remodeling of the cell wall occurs throughout the process of aging. Specifically, several genes involved in ergosterol biosynthesis and sterol uptake (CAGL0I04246g, ERG1, ERG3, ERG5, ERG6, ERG11,) [23–27] were upregulated. ERG19 and ERG20 upstream in the “mevalonate pathway,” as well as CAGL0L09493g downstream in the “sito steryl glucoside synthesis” of ergosterol biosynthesis were also upregulated. Also CAGL0C04026g, a mannosyl transferase, which aids in protein glycosylation and uses farnesyl pyrophosphate, the endproduct of the mevalonate pathway was upregulated. In addition, several genes involved in cell wall remodeling (KNH1, SUN4) [28, 29], as well as the Hog pathway (CDC42, STE20, MSB2, CAGL0F03003g) [30–34] were significantly upregulated. Cell wall adhesins (PWP3 and PWP5) [21, 35–38] and adhesion proteins (AWP1, CAGL0J02530g, CAGL0L06424g) [21, 36, 39] were significantly downregulated (virulence-associated genes are included in Table 3, complete list is available at GSE85985), which was predicted by our functional adhesion assays. Interestingly EPA6, a cell wall adhesin [40–45], and FKS1, a β1,3-glucan synthase [46, 47], both implicated in azole resistance were upregulated in old cells.
Table 3. List of genes upregulated in old C. glabrata cells known to be involved in antifungal resistance.
| Gene Name | Gene ID | Fold Change | P-value | Function |
|---|---|---|---|---|
| CAGL0I04246g | 7.4 | 3.31E-16 | Transcription factor involved in sterol uptake | |
| ERG11 | CAGL0E04334g | 6.1 | 2.15E-17 | Putative cytochrome P-450 lanosterol 14-alpha-demethylase |
| CAGL0J07502g | 6.0 | 2.97E-13 | Putative protein similar to globins with a heme-binding domain | |
| CAGL0K04301g | 5.6 | 2.99E-15 | Mitochondrial Ser/Thr protein kinase | |
| TPO3 | CAGL0I10384g | 5.3 | 8.12E-17 | Predicted polyamine transporter of the major facilitator superfamily |
| FKS1 | CAGL0G01034g | 4.9 | 7.93E-14 | Putative 1,3-beta-glucan synthase component |
| KNH1 | CAGL0H07997g | 4.7 | 2.29E-14 | Involved in cell wall beta 1,6-glucan synthesis |
| AQR1 | CAGL0J09944g | 4.5 | 6.60E-15 | Plasma membrane drug:H+ antiporter |
| EPA6 | CAGL0C00110g | 4.1 | 6.72E-13 | Sub-telomerically encoded adhesin with a role in cell adhesion |
| ERG3 | CAGL0F01793g | 4.1 | 3.61E-16 | Delta 5,6 sterol desaturase |
| CAGL0M10219g | 3.9 | 1.92E-13 | Putative ceramide synthase component | |
| CAGL0M01870g | 3.8 | 2.02E-12 | Putative zinc finger protein | |
| SUR2 | CAGL0H01375g | 3.7 | 7.68E-16 | Predicted sphinganine hydroxylase with role in sphingolipid biosynthesis |
| ERG5 | CAGL0M07656g | 3.6 | 4.82E-16 | Putative C22 sterol desaturase |
| CAGL0F04917g | 3.6 | 1.82E-13 | Putative regulatory subunit for protein phosphatase | |
| YBT1 | CAGL0C03289g | 3.3 | 6.12E-11 | Putative ABC transporter involved in bile acid transport |
| QDR2 | CAGL0G08624g | 3.2 | 1.13E-15 | Drug:H+ antiporter of the Major Facilitator Superfamily |
| PFK1 | CAGL0F08041g | 3.1 | 1.49E-13 | Putative phosphofructokinase |
| PDR16 | CAGL0J07436g | 3.0 | 3.13E-11 | Putative ABC transporter |
| ERG6 | CAGL0H04653g | 2.9 | 5.59E-15 | C24 sterol methyltransferase |
| RSB1 | CAGL0L10142g | 2.2 | 2.36E-13 | Sphingolipid flippase |
| RTA1 | CAGL0K00715g | 2.2 | 3.25E-13 | Involved in 7-aminocholesterol resistance |
| NCP1 | CAGL0D04114g | 2.1 | 6.03E-13 | Has NADPH-hemoprotein reductase activity |
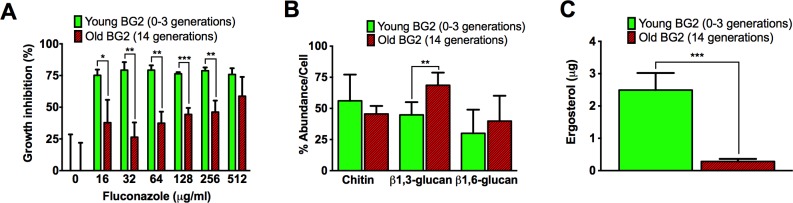
Given these results we predicted that old cells would be more resilient under the selection pressure of azole treatment. Older (5–7 generations) and young (0–2 generations) C. glabrata cells were grown and aged for 15 h under sub-therapeutic levels of fluconazole (32 μg/mL), and then subjected 4 h to increasing fluconazole concentrations (0–512 μg/mL) before plating on YPD plates. At fluconazole concentrations of 16–256 μg/mL, the younger cells were inhibited/killed at significantly higher rates than the older cells (Fig 4A). Specifically, failure of young cells to grow on fluconazole containing plates (16 to 64 μg/mL) ranged from 75–79%, respectively. Whereas failure of old cells to grow ranged from 26% (32 μg/mL) to 59% (512 μg/mL). Inhibition was relatively stable across various fluconazole levels for the young cells, whereas growth inhibition of older cells tended to slightly increase as the amount of fluconazole increased. These data suggest that older cells grown under selection pressure of sub-therapeutic fluconazole are significantly more resistant to subsequent fluconazole exposure compared to young cells exposed to the same fluconazole concentrations.
Fig 4. Enhanced resilience of older C. glabrata cells stems from cell wall remodeling.
(A) C. glabrata cells aged under sub-therapeutic fluconazole were more resistant to growth inhibition than younger cells when subjected to various concentrations of fluconazole for 4 h (experiments were run in triplicates and compared by Student’s t-test). (B) Older C. glabrata cells contain a slightly higher quantity of β1,3-glucans, but comparable amounts of β1,6-glucans and chitin (experiments were run in triplicates and compared by Student’s t-test). (C) Lipid analysis by GC-MS indicated that younger C. glabrata cells contain a higher amount of ergosterol compared to 14-generation-old cells (experiment was run in triplicates). *P < 0.05, **P < 0.01, ***P < 0.001.
Transcriptome analysis (GSE85985) identified upregulation of several β1,3-glucan enzymes: a synthase (FKS1) that is functionally redundant with Fks2, and in which “hot spot” mutations confer resistance [48], a exo-1,3,beta-glucosidase (CAGL0G09515g), a transglycosidase (GAS5) with an elongation role [21], and an ortholog with osmosensor and biosynthetic activity (CAGL0F03003g). Also, KNH1 [28], which is involved in 1,6-glucan synthesis, was upregulated, while a 1,4-alpha-glucosidase (CAGL0G02717g) was downregulated. In addition to the glucan signature, CHS3B, a Class IV chitin synthase that has a calcineurin pathway- and Tor1p-dependent role in cell wall integrity [49, 50] was upregulated. Nevertheless, major quantitative or qualitative differences for content of chitin, β1,3-, and β1,6-glucans could not be established by employed biochemical methods. Old C. glabrata cell walls contained both glucans, and only a slightly higher quantity of β1,3-glucans was noted, whereas chitin and β1,6-glucan content did not differ (Fig 4B).
Detailed transcriptome (Table 3) and gene ontology analysis (Table 4) identified enrichment of steroid metabolic and biosynthetic processes. Therefore, we analyzed the lipid composition of older C. glabrata cells to investigate if the upregulation of the ergosterol pathway leads to accumulation of sterol intermediates. The ergosterol content of old and young cells was quantified by GC-MS and normalized both to cell number as well as to lipid content. This quantitative analysis demonstrated approximately 10 fold lower ergosterol levels in old compared to young cells (Fig 4C), whereas analysis of the downstream sito steryl glucosides revealed slightly increased accumulation (10.5 vs 6.6 μg/ml). This latter quantification was done by LC-MS in pooled samples of 14-generation-old compared to young cells (0–3 generations old).
Table 4. Gene ontology enrichment analysis for genes upregulated in old C. glabrata cells.
| Gene ID | P-value |
|---|---|
| nucleobase transport | 8.35E-07 |
| organic hydroxy compound metabolic process | 0.0008 |
| rRNA processing | 0.00228 |
| thiamine-containing compound metabolic process | 0.00286 |
| thiamine metabolic process | 0.00286 |
| steroid metabolic process | 0.00349 |
| rRNA metabolic process | 0.00369 |
| organic hydroxy compound biosynthetic process | 0.00432 |
| sterol biosynthetic process | 0.00713 |
| sterol biosynthetic process | 0.01031 |
| sterol metabolic process | 0.01167 |
| alcohol metabolic process | 0.01393 |
| alcohol biosynthetic process | 0.01873 |
| maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 0.01907 |
| thiamine-containing compound biosynthetic process | 0.03803 |
| thiamine biosynthetic process | 0.03803 |
| ribosome biogenesis | 0.04021 |
| ribosomal small subunit biogenesis | 0.04025 |
| RNA phosphodiester bond hydrolysis, endonucleolytic | 0.04409 |
| ribosomal large subunit biogenesis | 0.04441 |
| cellular alcohol metabolic process | 0.04981 |
Taken together, the transcriptome data show that cell wall remodeling genes, several of which are already implicated in C. glabrata virulence and antifungal resistance are differentially regulated in older cells. Older cells exhibit markedly upregulated ergosterol pathway associated genes and greater resistance under increasing fluconazole selection. Furthermore, biochemical analysis of older C. glabrata cells show differences in ergosterol content and cell wall composition.
Host neutrophils select older cells in vivo
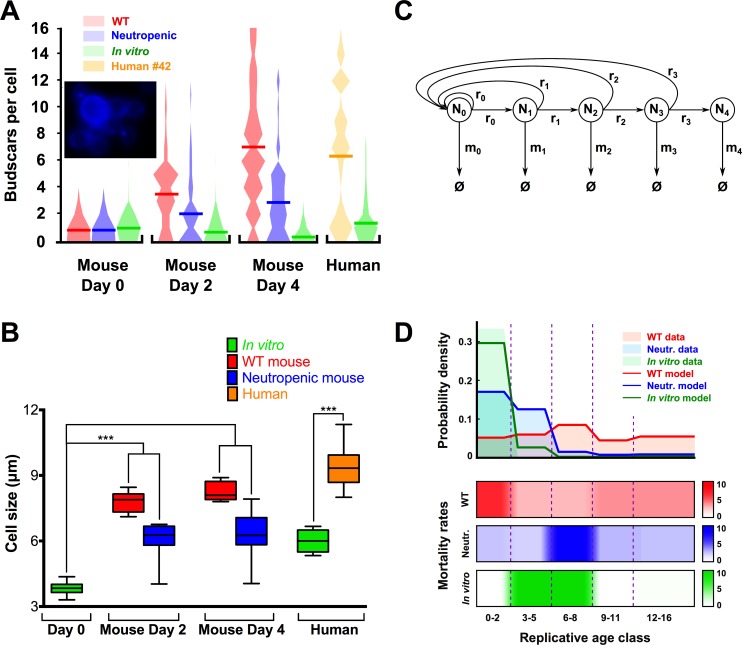
In a clonally expanding pathogen population, old cells statistically cannot dominate within the population unless forceful selection pressures are operative to promote their emergence and kill off younger cells. Based on the aforementioned in vitro assays, which suggest that old cells have a biological advantage under certain selection pressures, we hypothesized that the immune response of the host controls the generational distribution of a pathogen population by differential mortality. In support of this, larger C. glabrata cells with high budscar count were observed in human patients (n = 6) with urinary tract infections. Those patients were not neutropenic, and violin plots of the budscar counts indicate higher accumulation of old cells from in vivo derived yeast cells when compared to the same cells grown up without selection pressure with in vitro cultures (Fig 5A shows data from patient #42, and S3A Fig shows data from the remaining patients). Disseminated C. glabrata infections commonly occur in neutropenic patients, and thus we predicted that neutrophils constitute the selective force in mice, as they are the key effector cells that contain C. glabrata infection [51]. To test this hypothesis in vivo, the effect of neutrophil depletion was examined on the generational distribution of an infecting C. glabrata population with different strains (BG2 and 89) injected intravenously into wildtype (WT) or neutropenic mice. The latter immune status was achieved by injecting antibody RB6-8C5, which transiently depletes neutrophils in the mouse. Subsequently, C. glabrata was harvested from homogenized kidneys every 2 days, and yeast cells were directly stained with calcofluor to assess budscar count as a measure of replicative age. In addition, cell size was measured as increase in size correlates with older age. The violin plots demonstrate shift in generational distribution. Specifically, a significantly higher percentage of older cells, as demonstrated by increased budscars and cell size were found in WT mice when compared to neutropenic mice for both strains BG2 (Fig 5A and 5B, and S1 Table) and 89 (S3B Fig, and S1 Table). This supported the hypothesis that old cells are selected due to increased killing of cells by host neutrophils. Consequently, in neutropenic mice the pathogen population is younger, since there were fewer neutrophils to kill them, and the generational distribution was more similar to a population that had grown in vitro without exposure to selection forces.
Fig 5. Older C. glabrata cells accumulate in vivo.
(A) Calcofluor stain shows BG2 cells with increased budscars in mouse kidneys (inset). Significantly more BG2 cells with high budscar counts (mean budscars represented by line through plot) were found at day 2 and 4 in kidneys of WT compared to neutropenic mice injected intravenously, and also compared to day 0 in either host. Higher budscar counts were also found in C. glabrata from candiduric patient when compared to the same strain grown in vitro (#42). (B) BG2 cells were larger at days 2 and 4 in kidneys of WT mice compared to neutropenic mice and inoculum (day 0). (C) Model of expansion or contraction of a fungal cell population with five age classes: 0–2, 3–5, 6–8, 9–11 and 12–16 replications, whose populations are given by N0, N1, N2, N3 and N4, respectively. r0…r3 are replication rates, and m0…m4 are mortality rates of each age class. (D) Theoretical model can reproduce experimental results and confirm differential selection hypothesis. Upper panel: Probability distributions of replicative ages found experimentally (shaded areas) overlaid with results from the model parametrized with the most optimal parameters found using optimization (lines). Lower panel: Mortality rate profiles that yielded the best fits of the model to the data for each condition. Precise mortality rates were m = [6.9 3.0 2.8 4.1 4.0] (WT host); m = [2.8 2.3 9.3 2.9 2.8] (neutropenic host); m = [0.0, 10.4, 10.4, 0.0, 0.5] (in vitro).
Computational modeling supports differential selection hypothesis
To confirm the experimental results theoretically and gain additional insights, the selection pressure on cells of various ages was determined based on the distribution of replicative ages from the mouse experiment (Fig 5A). A system of ordinary differential equations was developed representing the time-dependent population sizes of five age classes (S4 Fig) to evaluate the age distributions resulting from various mortality rates on each age class (Fig 5C). Many initial mortality profiles (sets of mortality rates) were systematically explored, then local optimization was used to vary each mortality rate separately (S5 Fig). This returned the corresponding best-fit age distribution given the initial profile. Chosen from all local optima, the global-optimum mortality profile was robust to the initial profile, with the local optimizer converging on it for 38%, 52% and 63% (out of 3125 initial profiles) of the search space for WT, neutropenic, and in vitro populations, respectively; S6 Fig).
The experimental age distribution superimposed with the results from the model were parametrized with the most optimal mortality rates, as given by the global optimization (Fig 5D). The model fit the experimental age distribution closely for all three experimental populations. It was also able to capture the overall dynamics of the C. glabrata infections in vivo (S4 and S5 Figs). The estimated mortality rates took different forms for each condition (Fig 5D): high on the first age class and lower on the rest for WT mice; high on the middle age class and lower on the rest for neutropenic mice; very low on all age classes except the second and third for the in vitro population. Comparing the mortality rates with each other, the mortality profile of neutropenic mice was similar to that of in vitro cultures (Pearson r = 0.54, p = 0.35). Interestingly, mortalities in WT mice showed an opposite pattern; they were highest for young cells (r = -0.41, p = 0.49 between WT and neutropenic; r = -0.71, p = 0.18 between WT and in vitro). The lack of significance was a result of the comparison between two sets of five values (the number of age classes).
Overall, the computational results showed that in order to generate the replicative age distributions observed experimentally, a competent host must cause selection pressures on each age class that are different from those of a neutropenic host (as well as an in vitro population). This served as further confirmation of the hypothesis that neutrophils constitute the main selection pressure in a competent host, since the mortality profile with respect to age of a neutropenic host was more similar to that of an in vitro population than a competent host.
Discussion
To our knowledge, this is the first study to investigate replicative aging in clinical C. glabrata populations. Our data link replicative aging to pathogenesis in this important fungal pathogen. Specifically, we demonstrate that the natural process of aging results in a remodeling of the fungal cell wall leading to enhanced resilience. Furthermore, we show that neutrophil-mediated killing comprises the host’s selection pressure, which shifts the generational distribution of a C. glabrata population in vivo. Thus, this study supports a conceptually novel understanding of pathogenesis, where replicative aging in a yeast population facilitates demographic changes in the presence of a selective host response. Hence, the natural process of replication can be exploited as a unique mechanism of adaptation that contributes to phenotypic variation, resilience, and virulence. Since the phenotype of aging is not genetically inherited, but only seen through shifting selection pressures that promote persistence of older cells, the high risk of random permanent mutations is avoided. Instead constant rejuvenation through budding provides an assurance that all adaptive changes can be rapidly reversed as the daughter inherits asymmetrically [52, 53] and allows the emergence of a new generation.
Our original study indicated accumulation of old C. neoformans cells in chronic cryptococcosis [19, 54]. That work led to the hypothesis that aging is associated with pathogenesis and that the high number of C. neoformans cells with decreased remaining RLS, which we recovered from spinal fluid of infected rats or humans, represent old cells, which are statistically unlikely to occur in a clonally expanding population unless selected. To examine this hypothesis further and in depth, we chose C. glabrata, a commensal that can become pathogenic to cause invasive disease predominantly in neutropenic patients. Using this yeast we could study directly the association of aging and fungal pathogenesis, because opposed to C. neoformans, it maintains budscars on its cell surface when it continues to replicate, and therefore the actual generational age can be determined in real time. In addition, neutrophils, which are key host defense cells in several human and murine Candida spp. infections [12, 51, 55], can be studied in vitro and depleted in vivo. However, given the lack of prior studies on replicative aging in C. glabrata, baseline assumptions about replicative aging had to first be validated for this species.
We found considerable variability of RLS among clinical C. glabrata strains similar to that observed in other fungi [18, 19, 56, 57]. C. glabrata cell size increased consistently with each replication. In contrast to S. cerevisiae [58], maximal cell volume did not predict RLS, and future imaging studies will need to clarify whether time to first bud emergence correlates with length of RLS as recently demonstrated for S. cerevisiae [59]. Variable length of exposure to the intracellular environment of phagocytic cells has been associated with change in the virulence of several Candida spp. [13, 14]. Thus, it is conceivable that the intracellular environment of other phagocytic cells, especially macrophages, also contributes to variability in replicative age among the clinical strains.
C. glabrata can colonize the human host, and cause disease in humans only when host defenses fail and unchecked growth of the C. glabrata population ensues. We were therefore most interested in comparing resilience-associated phenotypes in young and old cells. Consequently, in vivo resilience of old and young C. glabrata cells was compared using a well-established infection model of G. mellonella [60–62]. These data demonstrate that aging indeed enhances resilience, and consequently virulence of this pathogen. Specifically, we found that older cells of both short and long-lived strains killed Galleria significantly faster than younger cells of the same C. glabrata strain. It is important to emphasize that 14-generation-old cells are far from senescence, and therefore fitness would not be significantly decreased. Accordingly, we show that 14-generation-old cells exhibit comparable doubling rates. Several important phenotypic differences between young and old cells explain the observed resilience of the older cells.
We demonstrated resistance of older cells to killing and phagocytic uptake. Young cells do not evade the basic immune response of Galleria, and they are therefore killed more rapidly and cannot establish infection as rapidly as the older C. glabrata cells. Galleria has an innate immune system composed of six types of phagocytic cells [60]. Consistent with our finding is the observation that larger C. neoformans cells are also more resistant to uptake by Galleria’s phagocytic cells [63].
Furthermore, our data shows that age confers resistance to intracellular killing after phagocytic uptake. In mammals, one contributing facilitator of killing in neutrophils are reactive oxygen species [55, 64]. Although C. glabrata is closely related to S. cerevisiae, higher resistance to ROS has evolved in this yeast as an adaptation to the intracellular environment of phagocytic cells, enabling the fungus to persist in the host [5]. Our data now indicate that aging augments this resistance, explaining the resistance of older C. glabrata cells to neutrophil-mediated killing. Transcriptome data indicate that genes in the Hog pathway, including CDC42, STE20, MSB2, were significantly upregulated in older cells, and this pathway is a well-known regulator of cell wall damage imparted by various stresses [65].
Hence, in order to be protected, older cells make a thicker cell wall through a concerted mechanism involving gene regulation, protein reorganization, and lipid remodeling. The observation that glucan is much more abundant in old cells compared to young cells is supported by recent studies in Candida tropicalis, where a thicker cell wall rendered strains less susceptible to amphotericin B and more resistant to stress responses [66]. Also in this instance, the authors found more beta-glucans in strains with thicker cell wall with no change in chitin content, similar to our findings. Our studies extend these observations to another fungus. We also documented a significant decrease in ergosterol in older cells, which could lead to a re-organization of the plasma membrane. In addition, sito steryl glucosides, a downstream product of ergosterol synthesis is known to accumulate in the plasma membrane [67, 68]. Data from Saccharomyces indicate that mutations of the ergosterol pathway affect the cell wall integrity [67]. Furthermore, vesicular trafficking and release of extracellular vesicles may be affected by ergosterol level, which may also modulate the immune response [69]. The increased azole resistance could be attributed to increased expression of the azole target, Erg11p. It is noteworthy that although ERG11 and several other ergosterol pathway genes are markedly upregulated in old cells, ergosterol levels were found to be significantly lower in old cells. Several factors could contribute to this finding. First the overexpression of upstream mevalonate pathway genes, ERG19 and ERG20, may redirect the sterol intermediates to the side pathway; as such we found a mannosyl transferase (CAGL0C04026g) upregulated. This enzyme aids in protein glycosylation [70]. Second, the accumulation of sito steryl glucosidase, which is downstream of ergosterol may lower the ergosterol levels. This may also promote overexpression of increased uptake. Future studies examining the azole influx and efflux in young and old cells may shed more light on how azole resistance is mediated [67]. Third, the transcriptional upregulation in older cells may be due to a compensatory response to ergosterol depletion. If older cells have mutations in the enzymes involved in ergosterol biosynthesis, it may explain the low ergosterol level and the resistance to fluconazole. Further studies are clearly needed to test these possibilities.
The cell wall constitutes the key defense against all external attacks. In addition it is also recognized by the host immune system, which can make both effective and ineffective harmful immune responses as a result of host-pathogen interaction [71, 72]. Lastly because cell walls of fungi are fundamentally different from the bi-lipid membranes of human cells, they are very attractive targets for antifungal therapy. The concept that cell walls are dynamic and that structural realignment occurs as a result of stress induced upregulation of signaling pathways is not novel and has been described for other yeast as well [73–75]. In our case, however, cell wall reorganization is not a response to an environmental change per se, but a process that is merely driven by an internal clock, namely the natural process of aging. This process is controlled by evolutionarily conserved signaling pathways similar to those of environmentally induced stress (Hog1), cell integrity (Mkc1, Cek1) and ergosterol metabolism (Erg11, Erg3, Erg5, Erg6, Sur2) [76].
A less obvious mechanism of how aging can affect pathogenesis is by augmenting phenotypic switching rates to more persistent or virulent variants. Older C. glabrata cells switch at a significantly higher rate to dark brown colony variants, which colonize the host more efficiently [22]. Interestingly, both the hypervirulent mucoid (C. neoformans) [19], and DB (C. glabrata) switch variants exhibit more than 30 generations shortened median RLS, a loss which is instantly recovered with reversal to the parent phenotype. This finding further supports the notion that isogenic strains with shortened RLS may be more virulent because of rapid aging as the more resilient “old age-associated” phenotypes are acquired earlier. In addition, it underscores the fact that lifespan, although variable among C. glabrata strains, is a highly regulated trait in the individual pathogenic yeast cell.
Lastly, it should be emphasized that staying young also has some benefits. Our results suggest that adherence to epithelial cells is enhanced in young cells. Adherence of C. glabrata to host cells is mediated in part by the Epa family of adhesins, which are encoded largely at subtelomeric loci and subject to transcriptional silencing of SIR genes [77]. EPA6 for instance is derepressed in response to limiting levels of nicotinamide adenine dinucleotide, which results in reduction of Sir2-mediated histone deacetylation and loss of silencing [41]. In S. cerevisiae, Sir2 levels decrease in old cells, and therefore one would predict higher adherence as a result of derepressed EPA expression [78]. However, it is conceivable that the thicker cell wall of older C. glabrata cells may affect surface exposure of Epa proteins, some of which reside in the C. glabrata cell wall [79]. Nonetheless in C. glabrata, adhesion induces actin-mediated phagocytosis and an increased inflammatory cytokine response [6]. Thus, the decreased adherence observed in the older cells in our studies is consistent with their decreased phagocytosis and ultimately decreased killing.
In C. glabrata, stainable budscars permitted us to assess in real-time the generational age distribution of an in vivo expanding C. glabrata population. This allowed us to design experiments that directly assessed the effect of host response on the emergence of old cells. First, we demonstrate by directly counting budscars that the in vivo derived C. glabrata population includes a large percentage of cells with many budscars, the hallmark of advanced generational age. Importantly this observation was confirmed with C. glabrata cells derived from patients with candiduria. Next, our hypothesis that the shift to an older population is the result of selection by a host response was tested by removing the selective force through depletion of neutrophils in mice. These cells were chosen as they represent the key inflammatory cells that contain the infection in vivo [51]. The results support our hypothesis and show that depletion of neutrophils shifts the age distribution of the pathogen population towards a younger population similar to that seen in exponential in vitro growth of C. glabrata. One concern is that budscar analysis maybe biased because the younger cells adhere better than the older cells. This is, however, unlikely as we homogenized the cells thoroughly and lysed away the murine tissue cells to achieve a single cell population for budscar staining. In addition, adhesion to epithelial cells would also be expected in neutropenic mice. Another concern is that budscar measurement of very old cells (30 generations) has not been experimentally confirmed. Although this concern may be valid, it would underestimate the age of old cells, and the result would be even more striking.
Further, by constructing and directly manipulating a mathematical model of an expanding or contracting cell population, we were able to generate replicative age distributions that matched our experimental data. This tells us that the different age distributions obtained in vivo were a direct result of differential selection (mortality) on each age class. This further supports our hypothesis that effective killing of young cells is the driving force behind the demographic shift in the fungal population derived from the host, and therefore that neutrophils are the cause of the shift. Future studies should aim to correlate the differential responses of neutrophils observed in our in vitro studies with the intracellular or extracellular locations of fungi in vivo. This might shed better insight into this particular selective force.
Conventional wisdom imparts that stress promotes aging [80]. Ironically, here we show that aging promotes stress resistance in the pathogen, C. glabrata. The most stressful environment for a commensal and a pathogen is the host, and we show how loss of host response results in a younger pathogen population. By contrast, in a competent host the younger cells are killed and the older yeast cells persist. Thus, while aging endows C. glabrata strains with increased resilience and promotes commensalism, youth selected in the neutropenic host may convey fitness, namely the ability to disseminate and adhere, and thus promote the pathogen state. Given that aging is a regulated process, it could potentially be targeted for antifungal drug development, which is in desperate need of new targets [81, 82]. It is conceivable that combination of antifungals and lifespan-altering drugs would result in synergy if they were tested in the host environment. We propose that replicative aging represents a virulence-associated trait of eukaryotic pathogen populations that contributes to microevolution of pathogens in the host. Importantly, this mechanism of microevolution may be relevant for other eukaryotic pathogens that either cause chronic infections, or alter between the commensal and pathogenic lifestyle.
Methods
Ethics statement
All animal experiments were carried out with the approval of the Stony Brook University Animal Institute. The protocol #628261 was approved by the Institutional Animal Care and Use Committee at Stony Brook. The study in strict accordance with federal, state, local and institutional guidelines that include “The Guide for the Care and Use of Laboratory Animals,” “The Animal Welfare Act,” and “Public Health Service Policy on Human Care and Use of Laboratory Animals.” Candida from patient urine samples was obtained with the approval of the Stony Brook Institutional Review Board (protocol #648612). No consent was required as the data was de-identified and anonymous. Neutrophils were isolated from donor blood (New York Blood Center) with the approval of the Stony Brook Institutional Review Board (protocol #646298).
C. glabrata strains
Clinical C. glabrata strains from the United States (Table 1) stored at -80°C were streaked for single colonies on Yeast Peptone Dextrose agar (Difco) at 37°C thrice before analysis. Multilocus Sequence Typing of C. glabrata strains was performed with primers as previously described [83].
Replicative and chronological lifespan
RLS was determined for C. glabrata strains by adaptation of S. cerevisiae methods [16]. Briefly, new buds from a virgin cell (n = 20) on YPD agar were separated at the end of each division using a 25 μm needle (CoraStyles) on a tetrad dissection microscope (Zeiss) at 250X magnification. Plates were returned to 37°C at the end of each division, and cells that had not divided for 24 h were declared dead. CLS was determined by adaptation of S. cerevisiae methods [17]. Briefly, cells were grown for 3 d in YPD broth (Difco) until they reached stationary phase, then transferred to dH2O, and their viability was determined by plating colony forming units (CFU) every 2 d until 99% of the cells were dead.
Isolation of C. glabrata cells with advanced replicative age
Counter-flow centrifugal elutriation was used to isolate old cells as previously described [18]. In the methods below, young describes cells that gave rise to fewer than 3 daughters, and old describes cells that are a third or more into their lifespan (approximately 14 generations old) or more (28 generations old) unless otherwise stated. With this method, the flow and/or the centrifugation speed can be adjusted to collect only those cells whose sedimentation velocity (the rate at which molecules move in response to the centrifugal force generated in a centrifuge) falls in a particular range. To avoid clumping, cells were washed twice with 30 mM EDTA, and then thrice with PBS. Briefly, newly budded C. glabrata cells were isolated by elutriation (Beckman JE-5.0 rotor in a Beckman J-6B centrifuge; Beckman Instruments Inc.), and labeled with Sulfo-NHS-LC-Biotin (Thermo Scientific). The newly budded and labeled cells were grown for several generations, and collected by first binding them to streptavidin-conjugated magnetic microbeads (Miltenyi Biotec), then isolating them on a magnetic column (Miltenyi Biotec). Unbound young yeast cells (0–3 generations old) that washed off the column and had been exposed to similar manipulations were used as controls. Purity of old cells was confirmed by Fluorescein isothiocyanate (FITC)-staining of the streptavidin-labeled cells.
Imaging
Cells suspended on YPD agar were imaged at 100X magnification on an Olympus AX70 microscope, and cell size was measured in Adobe Photoshop CS6 for Macintosh. For budscar stains, old C. glabrata cells were isolated as described earlier, or specimens from murine or human infection were directly stained with 100 μg/ml Calcofluor (Sigma) in dH2O for 5 min at room temperature in the dark, followed by washes in dH2O. Images were taken at 1000X magnification under oil immersion in the blue channel with a Qimaging Retiga 1300 digital camera and processed in the Qcapture Suite V2.46 software (Qimaging). Transmission electron microscopy was performed with the help of Stony Brook University microscopy facility. Briefly, virgin and 10-generation-old, and C. glabrata cells were harvested, resuspended in 5% glutaraldehyde, and fixed for 2 h at room temperature. Samples were postfixed in 1% OsO4 at 4 oC, and after several washing and dehydration steps, embedded in araldite. Ultrathin sections (70–74 nm) were cut using an ultramicrotome (Leica EM UC7), contrasted with lead citrate, and imaged using a JEOL JEM 1400 TEM.
Neutrophil-mediated killing and neutrophil extracellular trap assay
Neutrophils were isolated from donor blood (New York Blood Center) as previously described [84]. Neutrophil-mediated killing was determined on young or old C. glabrata cells as described [85]. Briefly, neutrophils were diluted in RPMI 1640 medium with 1% human sera for opsonization and allowed to adhere to 96 well plates for 30 min at 37°C and 5% CO2. C. glabrata cells were isolated as described earlier and added in a fungus-to-neutrophil ratio of 1:10 or 1:100. Wells with no neutrophils were included as controls. The plate was incubated for 30 min to allow phagocytosis, then incubated an additional hour to allow killing. Appropriate dilutions were plated on YPD plates and incubated at 37°C for 48 h. All experiments were performed in six replicates, and the killing percentage was calculated by comparison of CFU 1 h after phagocytosis. NET production and Nuclear Elastase (NE) were measured as previously described [86]. Briefly, for NET production, 5 x 104 neutrophils/ml were plated on 24 well plates in Hank’s balanced salt solution with 3% sera, and C. glabrata yeast (young or old), or preformed C. albicans hyphae were added at an MOI of 10. After 4 h of incubation, NETs were visualized by the addition of SYTOX (S702, Invitrogen), and imaged at 40x magnification in at least six random fields. The mean percentage and standard deviation of neutrophil nuclei larger than 1,000 μm2 over the total neutrophils was calculated. NE nuclear co-localization was determined after 5 x 104 neutrophils were plated on glass coverslips 1 h prior to stimulation. After 4 h of stimulation, cells were fixed with 2% paraformaldehyde for 20 min at 37°C. After immunofluorescence staining was performed with anti-neutrophil elastase (Abcam) and DAPI, confocal z-series (every 0.8 mm) covering the whole neutrophils were used to quantify total NE and nuclear localization using ImageJ. A mask of the DAPI channel was created to measure the NE nuclear localization, and the total area and percentage of co-localization in individual channels was used to yield the total NE signal (10 neutrophils per condition were used).
H2O2 disc diffusion
Young or old C. glabrata cells were isolated as described earlier and used to make a lawn on a YPD agar plate, to the center of which a 6 mm paper disc saturated with 10 μl of 30% H2O2 (Sigma) was added. A zone of inhibition (diameter of circle with no growth around the disc) was determined by measurement at 24 h. The zone of inhibition reflects the initial resistance of the plated yeast population prior to further growth.
Phenotypic switching and adhesion assays
Approximately 500 young or old C. glabrata cells of the parent phenotype were plated on 100 YPD agar plates with 1 mM CuSO4, grown for 2 d at 37°C, then stored at 4°C to enhance the switched colony morphology. The ratio of switched to parent colonies was determined over generational age and plotted as the phenotypic switching rate. The switched colonies were streaked for single colonies onto YPD plates three times to determine stability before RLS was determined as described above. Adhesion assays were performed using HeLa cells as described previously [6].
Antifungal resistance testing
Young or old (5–7 generations old) C. glabrata cells were isolated as described earlier. Both populations were biotin labeled to allow for isolation after they were allowed to grow in 32 μg/mL fluconazole (Sagent Pharmaceuticals) in YPD broth for 15 h, where old cells could age to 13–15 generations and young cells produced during these 15 hours could be separated from their older mothers. Cells were then washed and the original population of old cells (now 13–15 generations) and the new, young fraction from the originally labeled young population were used in a killing assay as previously described (45). Briefly, cells were diluted to a final concentration of 5 x 103 cells/mL in RPMI 1640 medium buffered with MOPS containing 0–512 μg/mL serial dilutions of fluconazole. 200 μL of each culture was plated in 6 replicates in a 96 well plate and was incubated at 37°C for 4 h. After 4 h, appropriate dilutions were made and plated on YPD plates. Percent inhibition was calculated by comparing CFUs normalized to the CFU from 0 μg/mL fluconazole.
Chitin and glucan analysis
Chitin composition was analyzed as described previously [87, 88]. For glucan analysis, SDS-treated young or old C. glabrata cells were separated into alkali-soluble and insoluble fractions, and each fraction was treated with recombinant glucanases and the digested reducing sugars were measured by p-aminohydroxybenzoic acid hydrazine method [89].
Lipid analysis
Lipids were extracted from young or old C. glabrata cells using a method adapted from [90]. Briefly, 108 cells for each replicate were pelleted in a glass tube in which mandala extraction buffer was added [91] and extraction was performed. Further extraction was performed per Bligh and Dyer [92] followed by base hydrolysis. The lipid-containing lower organic layer was evaporated to analyze lipids by liquid or gas chromatography-mass spectrometry. For GC-MS, the dried samples were derivatized using N,O-bis (trimethylsilyl) trifluoroacetamide /trimethylchlorosilane (Sigma-Aldrich) and analyzed using a 30 mt (0.25 μm) VF-5ms column on Agilent 7890 GC-MS (Agilent Technologies, Santa Clara, CA, USA). The retention time of the ergosterol standard (Matreya LLC, PA) was used as a reference. Cholesterol was added as an internal standard for these analyses prior to lipid extraction. LC-MS was performed by the Stony Brook Proteomics core on the samples to quantitate sito and glycosylated ergosterol. A standard curve was generated with sito (R2 = 0.991) and used to quantify glycosylated ergosterol.
Computational modeling of population dynamics
A system of ordinary differential equations was used to model the population dynamics of a C. glabrata population with distinct age classes over time. For simplicity, the full range of ages was grouped into several classes, namely 0–2, 3–5, 6–8, 9–11 and 12–16 replications (whose population sizes are represented by N0, N1, N2, N3 and N4, respectively). The free parameters of the model were the mortality rates of each age class, while the replication rates were fixed. Mortality rate profiles were tested for optimality by evaluating the solution of the differential equations analytically at 4 d and compared to the experimental age distribution. This procedure was repeated for many trial sets using a local optimizer, to obtain the mortality profile that gave the global best fit to the experimental data. Additional methods and results are described in Supplemental Information.
Infection models
Galleria mellonella infection was carried out as described previously [61] using a Hamilton syringe to deliver 106 C. glabrata cells per waxworm. CFU analysis was done by cleaning the waxworm surface with 70% ethanol, puncturing it with a 1 ml syringe, and collecting the haemolymph from 25 worms per time point. Appropriate dilutions were made, plated on YPD agar plates, and incubated at 37°C for 24 h to count CFU.
Murine infections were carried out by intravenously injecting 6–8 week old female Swiss/Webster mice (Taconic) with 7.5 x 108 young or old C. glabrata cells isolated as described above. One group (n = 15) was injected with the antibody RB6-8C5 [93] every 48 h to deplete neutrophils. Depletion of neutrophils was verified by peripheral cell counts. Mice from both groups were sacrificed (n = 3) at 2 d and 4 d. The fungal burden was collected by homogenizing the kidneys to determine CFU on YPD plates, and budscar count by calcofluor staining (Sigma) as described previously [94].
Galleria mellonella haemocyte collection and phagocytosis assay
Haemocyte collection was performed as previously described [95]. Briefly, haemolymph was collected and diluted 1:10 into Insect Physiological Saline (IPS) [150 mM sodium chloride, 5 mM potassium chloride, 10 mM Tris HCl, 10 mM EDTA and 30 mM sodium citrate] with 10 mM N-ethylmaleimide (anticoagulant). Cells were then centrifuged at 1200 rpm for 10 min, washed with IPS, and counted on a hemocytometer.
For the phagocytosis assay, cells were diluted in RPMI 1640 with 10% Fetal Bovine Serum (FBS). 105 haemocytes were plated in a 96-well plate and incubated at 37°C with 10% CO2 overnight to allow for adherence. The following day, non-adherent cells were washed off, and young or old C. glabrata cells were added to the wells in a 1:50 ratio. The well plate was spun at 1000 rpm for 1 min to ensure contact between haemocytes and yeast, incubated for 1 h to allow for phagocytosis, then washed to remove non-internalized yeast. Yeasts were fixed with cold ethanol for 30 min, and stained with Giemsa for 30 min followed by 3 washes of IPS. Yeasts were analyzed by averaging the number internalized in haemocytes (n = 100).
RNA sequencing and GO analysis
Three biological replicates of 108 young or old C. glabrata cells (strain BG2) were isolated, washed and resuspended in 0.5 mm zirconia beads and RLT buffer (Qiagen), then disrupted mechanically using a mini bead beater (Biospec) for 2 min for a total of 4 cycles with 1 min intervals on ice. Post lysis, total RNA was isolated using the RNeasy mini kit (Qiagen) per manufacturer’s instructions. The Genome Technology Access Centre of Washington University in St. Louis (GTAC-WUSTL) performed RNA hybridization, data acquisition and analysis. Briefly, RNA was polyA selected and sequenced on an Illumina HiSeq 2000. Raw sequence reads were converted to basecalls, demultiplexed, and aligned to a reference sequence using Tophat v2.0.9 and Bowtie2 v2.1.0 and gene abundances were derived by HTSeq. Differential expression was estimated by pair-wise negative binomial tests using EdgeR and DEXSeq. Any genes with more than a 2-fold change, a p < 0.05 by a hypergeometric test, and an FDR q < 0.05 were used to determine significance. Gene ontology (GO) enrichment analysis was done by entering significant genes into the Gene Ontology Term Finder software at http://candidagenome.org/cgi-bin/GO/goTermFinder. The data was deposited at NCBI and can be accessed on GEO (accession #GSE85985).
Statistics
Standard statistical analysis and non-parametric tests, such as Student’s t-test, Log-rank, one way ANOVA, and Wilcoxon rank sum tests were performed using Prism version 6 (Graphpad) or Microsoft Excel 2011 for Macintosh. Differences are marked with asterisks and considered significant if p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), p < 0.0001 (****).
Supporting information
(A) Younger cells were phagocytosed by Galleria mellonella haemocytes (B) and human neutrophils at a higher frequency compared to old cells. **P < 0.01, ****P < 0.0001.
(TIF)
Increased virulence in Galleria was observed with older cells of strains (A) 89 and (B) 117. (C) Younger cells were phagocytosed by Galleria mellonella haemocytes at a higher frequency compared to old cells of strain 89. (D) Increased resistance to neutrophil-mediated killing was observed in older cells of strains 89 and 117. (E) H2O2 disc diffusion assays showed smaller zone of inhibition in older cells of strain 89. (F) The DB variant exhibited a higher fungal burden than the parent (S) in strain 89. (G) Phenotypic switching in strain 89 from S to DB colony morphology increased consistently with age to 18 fold. (H) The RLS of DB was shortened over 50% relative to S and reconstituted in the revertant colony in strain 89. *P < 0.05, **P < 0.01, ***P < 0.001.
(TIF)
(A) Budscar staining of samples from urine of 5 patients showed a skewed distribution towards an increased frequency of older cells compared to their in vitro cultures. (B) Strain 89 cells with a proportionally high number of budscars (mean line) were found at days 2 and 4 in kidneys of WT compared to neutropenic mice, and also compared to day 0.
(TIF)
Solutions of ordinary differential system with optimum-fit mortality profile for (A) WT mouse host, (B) neutropenic mouse host, and (C) in vitro control population.
(TIF)
Mean of 1000 stochastic simulations of corresponding system of reactions with optimum-fit mortality profile for (A) WT mouse host, (B) neutropenic mouse host, and (C) in vitro control population.
(TIF)
Mortality profiles found using a local optimizer are shown for the entire search space of trial profiles. Each locally-optimized mortality profile is marked as being within +/- 20% of the global optimum or not, and its cost function (distance of model from data age distribution, as computed via Eq. (S2) is shown). (A) WT mouse host, (B) neutropenic mouse host, (C) in vitro control cell population.
(TIF)
(MP4)
(DOCX)
Supplemental methods and results for modeling are described separately.
(PDF)
Acknowledgments
We thank Christian Ruiz, Dr. Geoffrey Girnun, and Dr. Ashutosh Singh for their expertise with lipid analysis.
Data Availability
The data is deposited at NCBI and can be accessed on GEO (accession #GSE85985).
Funding Statement
This research was supported by NIH grants AI059681 and AI114259 to BCF; NIH grants AI56168 and AI125770, and a Merit Review grant I01BX002624 from the Veterans Affairs Program in Biomedical Laboratory Research and Development to MDP; and the Laufer Center for Physical & Quantitative Biology to GB. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Diekema D, Arbefeville S, Boyken L, Kroeger J, Pfaller M. The changing epidemiology of healthcare-associated candidemia over three decades. Diagnostic microbiology and infectious disease. 2012;73(1):45–8. Epub 2012/05/15. doi: 10.1016/j.diagmicrobio.2012.02.001 [DOI] [PubMed] [Google Scholar]
- 2.Wang E, Farmakiotis D, Yang D, McCue DA, Kantarjian HM, Kontoyiannis DP, et al. The ever-evolving landscape of candidaemia in patients with acute leukaemia: non-susceptibility to caspofungin and multidrug resistance are associated with increased mortality. J Antimicrob Chemother. 2015;70(8):2362–8. doi: 10.1093/jac/dkv087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silva S, Negri M, Henriques M, Oliveira R, Williams DW, Azeredo J. Candida glabrata, Candida parapsilosis and Candida tropicalis: biology, epidemiology, pathogenicity and antifungal resistance. FEMS microbiology reviews. 2012;36(2):288–305. Epub 2011/05/17. doi: 10.1111/j.1574-6976.2011.00278.x [DOI] [PubMed] [Google Scholar]
- 4.Fidel PL Jr., Vazquez JA, Sobel JD. Candida glabrata: review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clinical microbiology reviews. 1999;12(1):80–96. Epub 1999/01/09. PubMed Central PMCID: PMC88907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuellar-Cruz M, Briones-Martin-del-Campo M, Canas-Villamar I, Montalvo-Arredondo J, Riego-Ruiz L, Castano I, et al. High resistance to oxidative stress in the fungal pathogen Candida glabrata is mediated by a single catalase, Cta1p, and is controlled by the transcription factors Yap1p, Skn7p, Msn2p, and Msn4p. Eukaryotic cell. 2008;7(5):814–25. Epub 2008/04/01. PubMed Central PMCID: PMC2394966. doi: 10.1128/EC.00011-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cormack BP, Ghori N, Falkow S. An adhesin of the yeast pathogen Candida glabrata mediating adherence to human epithelial cells. Science. 1999;285(5427):578–82. Epub 1999/07/27. [DOI] [PubMed] [Google Scholar]
- 7.Otto V, Howard DH. Further studies on the intracellular behavior of Torulopsis glabrata. Infection and immunity. 1976;14(2):433–8. Epub 1976/08/01. PubMed Central PMCID: PMC420903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaur R, Ma B, Cormack BP. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc Natl Acad Sci U S A. 2007;104(18):7628–33. PubMed Central PMCID: PMCPMC1863504. doi: 10.1073/pnas.0611195104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Essig F, Hunniger K, Dietrich S, Figge MT, Kurzai O. Human neutrophils dump Candida glabrata after intracellular killing. Fungal Genet Biol. 2015;84:37–40. doi: 10.1016/j.fgb.2015.09.008 [DOI] [PubMed] [Google Scholar]
- 10.Perlroth J, Choi B, Spellberg B. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Medical mycology. 2007;45(4):321–46. Epub 2007/05/19. doi: 10.1080/13693780701218689 [DOI] [PubMed] [Google Scholar]
- 11.Pfaller MA, Castanheira M, Messer SA, Moet GJ, Jones RN. Variation in Candida spp. distribution and antifungal resistance rates among bloodstream infection isolates by patient age: report from the SENTRY Antimicrobial Surveillance Program (2008–2009). Diagnostic microbiology and infectious disease. 2010;68(3):278–83. Epub 2010/09/18. doi: 10.1016/j.diagmicrobio.2010.06.015 [DOI] [PubMed] [Google Scholar]
- 12.Hachem R, Hanna H, Kontoyiannis D, Jiang Y, Raad I. The changing epidemiology of invasive candidiasis: Candida glabrata and Candida krusei as the leading causes of candidemia in hematologic malignancy. Cancer. 2008;112(11):2493–9. Epub 2008/04/17. doi: 10.1002/cncr.23466 [DOI] [PubMed] [Google Scholar]
- 13.Brunke S, Seider K, Fischer D, Jacobsen ID, Kasper L, Jablonowski N, et al. One small step for a yeast—microevolution within macrophages renders Candida glabrata hypervirulent due to a single point mutation. PLoS Pathog. 2014;10(10):e1004478 PubMed Central PMCID: PMCPMC4214790. doi: 10.1371/journal.ppat.1004478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wartenberg A, Linde J, Martin R, Schreiner M, Horn F, Jacobsen ID, et al. Microevolution of Candida albicans in macrophages restores filamentation in a nonfilamentous mutant. PLoS Genet. 2014;10(12):e1004824 PubMed Central PMCID: PMCPMC4256171. doi: 10.1371/journal.pgen.1004824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garnacho-Montero J, Diaz-Martin A, Garcia-Cabrera E, Ruiz Perez de Pipaon M, Hernandez-Caballero C, Aznar-Martin J, et al. Risk factors for fluconazole-resistant candidemia. Antimicrob Agents Chemother. 2010;54(8):3149–54. ubMed Central PMCID: PMCPMC2916332. doi: 10.1128/AAC.00479-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steffen KK, Kennedy BK, Kaeberlein M. Measuring replicative life span in the budding yeast. Journal of visualized experiments: JoVE. 2009;(28). Epub 2009/06/27. PubMed Central PMCID: PMC2797481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burtner CR, Murakami CJ, Kennedy BK, Kaeberlein M. A molecular mechanism of chronological aging in yeast. Cell cycle. 2009;8(8):1256–70. Epub 2009/03/24. PubMed Central PMCID: PMC2746416. doi: 10.4161/cc.8.8.8287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain N, Cook E, Xess I, Hasan F, Fries D, Fries BC. Isolation and characterization of senescent Cryptococcus neoformans and implications for phenotypic switching and pathogenesis in chronic cryptococcosis. Eukaryotic cell. 2009;8(6):858–66. Epub 2009/05/05. PubMed Central PMCID: PMC2698302. doi: 10.1128/EC.00017-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bouklas T, Pechuan X, Goldman DL, Edelman B, Bergman A, Fries BC. Old Cryptococcus neoformans cells contribute to virulence in chronic cryptococcosis. mBio. 2013;4(4). Epub 2013/08/15. PubMed Central PMCID: PMC3747583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasper L, Seider K, Hube B. Intracellular survival of Candida glabrata in macrophages: immune evasion and persistence. FEMS Yeast Res. 2015;15(5):fov042 doi: 10.1093/femsyr/fov042 [DOI] [PubMed] [Google Scholar]
- 21.de Groot PW, Kraneveld EA, Yin QY, Dekker HL, Gross U, Crielaard W, et al. The cell wall of the human pathogen Candida glabrata: differential incorporation of novel adhesin-like wall proteins. Eukaryotic cell. 2008;7(11):1951–64. PubMed Central PMCID: PMC2583536. doi: 10.1128/EC.00284-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srikantha T, Daniels KJ, Wu W, Lockhart SR, Yi S, Sahni N, et al. Dark brown is the more virulent of the switch phenotypes of Candida glabrata. Microbiology. 2008;154(Pt 11):3309–18. Epub 2008/10/30. doi: 10.1099/mic.0.2008/020578-0 [DOI] [PubMed] [Google Scholar]
- 23.Geber A, Hitchcock CA, Swartz JE, Pullen FS, Marsden KE, Kwon-Chung KJ, et al. Deletion of the Candida glabrata ERG3 and ERG11 genes: effect on cell viability, cell growth, sterol composition, and antifungal susceptibility. Antimicrob Agents Chemother. 1995;39(12):2708–17. PubMed Central PMCID: PMC163017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marichal P, Vanden Bossche H, Odds FC, Nobels G, Warnock DW, Timmerman V, et al. Molecular biological characterization of an azole-resistant Candida glabrata isolate. Antimicrob Agents Chemother. 1997;41(10):2229–37. PubMed Central PMCID: PMC164098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogers PD, Vermitsky JP, Edlind TD, Hilliard GM. Proteomic analysis of experimentally induced azole resistance in Candida glabrata. J Antimicrob Chemother. 2006;58(2):434–8. doi: 10.1093/jac/dkl221 [DOI] [PubMed] [Google Scholar]
- 26.Healey KR, Katiyar SK, Raj S, Edlind TD. CRS-MIS in Candida glabrata: sphingolipids modulate echinocandin-Fks interaction. Molecular microbiology. 2012;86(2):303–13. PubMed Central PMCID: PMC3472958. doi: 10.1111/j.1365-2958.2012.08194.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakayama H, Nakayama N, Arisawa M, Aoki Y. In vitro and in vivo effects of 14alpha-demethylase (ERG11) depletion in Candida glabrata. Antimicrob Agents Chemother. 2001;45(11):3037–45. Central PMCID: PMC90779. doi: 10.1128/AAC.45.11.3037-3045.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagahashi S, Lussier M, Bussey H. Isolation of Candida glabrata homologs of the Saccharomyces cerevisiae KRE9 and KNH1 genes and their involvement in cell wall beta-1,6-glucan synthesis. J Bacteriol. 1998;180(19):5020–9. PubMed Central PMCID: PMC107535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bader O, Krauke Y, Hube B. Processing of predicted substrates of fungal Kex2 proteinases from Candida albicans, C. glabrata, Saccharomyces cerevisiae and Pichia pastoris. BMC Microbiol. 2008;8:116 PubMed Central PMCID: PMC2515848. doi: 10.1186/1471-2180-8-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roman E, Cottier F, Ernst JF, Pla J. Msb2 signaling mucin controls activation of Cek1 mitogen-activated protein kinase in Candida albicans. Eukaryotic cell. 2009;8(8):1235–49. PubMed Central PMCID: PMC2725568. doi: 10.1128/EC.00081-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cash CJ, Sardesai AM, Berman LH, Herrick MJ, Treece GM, Prager RW, et al. Spatial mapping of the brachial plexus using three-dimensional ultrasound. Br J Radiol. 2005;78(936):1086–94. doi: 10.1259/bjr/36348588 [DOI] [PubMed] [Google Scholar]
- 32.Lotz H, Sohn K, Brunner H, Muhlschlegel FA, Rupp S. RBR1, a novel pH-regulated cell wall gene of Candida albicans, is repressed by RIM101 and activated by NRG1. Eukaryotic cell. 2004;3(3):776–84. PubMed Central PMCID: PMC420143. doi: 10.1128/EC.3.3.776-784.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calcagno AM, Bignell E, Rogers TR, Canedo M, Muhlschlegel FA, Haynes K. Candida glabrata Ste20 is involved in maintaining cell wall integrity and adaptation to hypertonic stress, and is required for wild-type levels of virulence. Yeast. 2004;21(7):557–68. doi: 10.1002/yea.1125 [DOI] [PubMed] [Google Scholar]
- 34.Mirbod F, Nakashima S, Kitajima Y, Cannon RD, Nozawa Y. Molecular cloning of a Rho family, CDC42Ca gene from Candida albicans and its mRNA expression changes during morphogenesis. J Med Vet Mycol. 1997;35(3):173–9. [PubMed] [Google Scholar]
- 35.Thierry A, Dujon B, Richard GF. Megasatellites: a new class of large tandem repeats discovered in the pathogenic yeast Candida glabrata. Cell Mol Life Sci. 2010;67(5):671–6. doi: 10.1007/s00018-009-0216-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaudhuri R, Ansari FA, Raghunandanan MV, Ramachandran S. FungalRV: adhesin prediction and immunoinformatics portal for human fungal pathogens. BMC Genomics. 2011;12:192 PubMed Central PMCID: PMC3224177. doi: 10.1186/1471-2164-12-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sorgo AG, Heilmann CJ, Dekker HL, Bekker M, Brul S, de Koster CG, et al. Effects of fluconazole on the secretome, the wall proteome, and wall integrity of the clinical fungus Candida albicans. Eukaryotic cell. 2011;10(8):1071–81. PubMed Central PMCID: PMC3165447. doi: 10.1128/EC.05011-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, et al. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148(1–2):126–38. PubMed Central PMCID: PMC3266547. doi: 10.1016/j.cell.2011.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weig M, Jansch L, Gross U, De Koster CG, Klis FM, De Groot PW. Systematic identification in silico of covalently bound cell wall proteins and analysis of protein-polysaccharide linkages of the human pathogen Candida glabrata. Microbiology. 2004;150(Pt 10):3129–44. doi: 10.1099/mic.0.27256-0 [DOI] [PubMed] [Google Scholar]
- 40.Iraqui I, Garcia-Sanchez S, Aubert S, Dromer F, Ghigo JM, d'Enfert C, et al. The Yak1p kinase controls expression of adhesins and biofilm formation in Candida glabrata in a Sir4p-dependent pathway. Molecular microbiology. 2005;55(4):1259–71. doi: 10.1111/j.1365-2958.2004.04475.x [DOI] [PubMed] [Google Scholar]
- 41.Domergue R, Castano I, De Las Penas A, Zupancic M, Lockatell V, Hebel JR, et al. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science. 2005;308(5723):866–70. doi: 10.1126/science.1108640 [DOI] [PubMed] [Google Scholar]
- 42.Castano I, Pan SJ, Zupancic M, Hennequin C, Dujon B, Cormack BP. Telomere length control and transcriptional regulation of subtelomeric adhesins in Candida glabrata. Molecular microbiology. 2005;55(4):1246–58. doi: 10.1111/j.1365-2958.2004.04465.x [DOI] [PubMed] [Google Scholar]
- 43.Zupancic ML, Frieman M, Smith D, Alvarez RA, Cummings RD, Cormack BP. Glycan microarray analysis of Candida glabrata adhesin ligand specificity. Molecular microbiology. 2008;68(3):547–59. doi: 10.1111/j.1365-2958.2008.06184.x [DOI] [PubMed] [Google Scholar]
- 44.Kucharikova S, Tournu H, Lagrou K, Van Dijck P, Bujdakova H. Detailed comparison of Candida albicans and Candida glabrata biofilms under different conditions and their susceptibility to caspofungin and anidulafungin. J Med Microbiol. 2011;60(Pt 9):1261–9. doi: 10.1099/jmm.0.032037-0 [DOI] [PubMed] [Google Scholar]
- 45.Gomez-Molero E, de Boer AD, Dekker HL, Moreno-Martinez A, Kraneveld EA, Ichsan, et al. Proteomic analysis of hyperadhesive Candida glabrata clinical isolates reveals a core wall proteome and differential incorporation of adhesins. FEMS Yeast Res. 2015;15(8). [DOI] [PubMed] [Google Scholar]
- 46.Pfeiffer CD, Garcia-Effron G, Zaas AK, Perfect JR, Perlin DS, Alexander BD. Breakthrough invasive candidiasis in patients on micafungin. Journal of clinical microbiology. 2010;48(7):2373–80. PubMed Central PMCID: PMC2897493. doi: 10.1128/JCM.02390-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katiyar SK, Alastruey-Izquierdo A, Healey KR, Johnson ME, Perlin DS, Edlind TD. Fks1 and Fks2 are functionally redundant but differentially regulated in Candida glabrata: implications for echinocandin resistance. Antimicrob Agents Chemother. 2012;56(12):6304–9. PubMed Central PMCID: PMC3497156. doi: 10.1128/AAC.00813-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garcia-Effron G, Lee S, Park S, Cleary JD, Perlin DS. Effect of Candida glabrata FKS1 and FKS2 mutations on echinocandin sensitivity and kinetics of 1,3-beta-D-glucan synthase: implication for the existing susceptibility breakpoint. Antimicrob Agents Chemother. 2009;53(9):3690–9. PubMed Central PMCID: PMCPMC2737881. doi: 10.1128/AAC.00443-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ueno K, Namiki Y, Mitani H, Yamaguchi M, Chibana H. Differential cell wall remodeling of two chitin synthase deletants Deltachs3A and Deltachs3B in the pathogenic yeast Candida glabrata. FEMS Yeast Res. 2011;11(5):398–407. doi: 10.1111/j.1567-1364.2011.00728.x [DOI] [PubMed] [Google Scholar]
- 50.Namiki Y, Ueno K, Mitani H, Virtudazo EV, Ohkusu M, Shimizu K, et al. Scanning and negative-staining electron microscopy of protoplast regeneration of a wild-type and two chitin synthase mutants in the pathogenic yeast Candida glabrata. J Electron Microsc (Tokyo). 2011;60(2):157–65. [DOI] [PubMed] [Google Scholar]
- 51.Miramon P, Kasper L, Hube B. Thriving within the host: Candida spp. interactions with phagocytic cells. Med Microbiol Immunol. 2013;202(3):183–95. Epub 2013/01/29. doi: 10.1007/s00430-013-0288-z [DOI] [PubMed] [Google Scholar]
- 52.McFaline-Figueroa JR, Vevea J, Swayne TC, Zhou C, Liu C, Leung G, et al. Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging cell. 2011;10(5):885–95. Epub 2011/07/06. PubMed Central PMCID: PMC3173513. doi: 10.1111/j.1474-9726.2011.00731.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Henderson KA, Hughes AL, Gottschling DE. Mother-daughter asymmetry of pH underlies aging and rejuvenation in yeast. Elife. 2014;3:e03504 PubMed Central PMCID: PMCPMC4175738. doi: 10.7554/eLife.03504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bouklas T, Fries BC. Cryptococcus neoformans constitutes an ideal model organism to unravel the contribution of cellular aging to the virulence of chronic infections. Current opinion in microbiology. 2013;16(4):391–7. Epub 2013/05/02. PubMed Central PMCID: PMC3755025. doi: 10.1016/j.mib.2013.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Segal AW. How neutrophils kill microbes. Annu Rev Immunol. 2005;23:197–223. PubMed Central PMCID: PMCPMC2092448. doi: 10.1146/annurev.immunol.23.021704.115653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roux AE, Chartrand P, Ferbeyre G, Rokeach LA. Fission yeast and other yeasts as emergent models to unravel cellular aging in eukaryotes. J Gerontol A Biol Sci Med Sci. 2010;65(1):1–8. Epub 2009/10/31. doi: 10.1093/gerona/glp152 [DOI] [PubMed] [Google Scholar]
- 57.Qin H, Lu M. Natural variation in replicative and chronological life spans of Saccharomyces cerevisiae. Exp Gerontol. 2006;41(4):448–56. Epub 2006/03/07. doi: 10.1016/j.exger.2006.01.007 [DOI] [PubMed] [Google Scholar]
- 58.Yang J, Dungrawala H, Hua H, Manukyan A, Abraham L, Lane W, et al. Cell size and growth rate are major determinants of replicative lifespan. Cell cycle. 2011;10(1):144–55. PubMed Central PMCID: PMC3048081. doi: 10.4161/cc.10.1.14455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jo MC, Liu W, Gu L, Dang W, Qin L. High-throughput analysis of yeast replicative aging using a microfluidic system. Proc Natl Acad Sci U S A. 2015;112(30):9364–9. PubMed Central PMCID: PMCPMC4522780. doi: 10.1073/pnas.1510328112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fuchs BB, O'Brien E, Khoury JB, Mylonakis E. Methods for using Galleria mellonella as a model host to study fungal pathogenesis. Virulence. 2010;1(6):475–82. Epub 2010/12/24. [DOI] [PubMed] [Google Scholar]
- 61.Mylonakis E, Moreno R, El Khoury JB, Idnurm A, Heitman J, Calderwood SB, et al. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infection and immunity. 2005;73(7):3842–50. Epub 2005/06/24. PubMed Central PMCID: PMC1168598. doi: 10.1128/IAI.73.7.3842-3850.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bouklas T, Diago-Navarro E, Wang X, Fenster M, Fries BC. Characterization of the virulence of Cryptococcus neoformans strains in an insect model. Virulence. 2015;6(8):809–13. PubMed Central PMCID: PMCPMC4826120. doi: 10.1080/21505594.2015.1086868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garcia-Rodas R, Casadevall A, Rodriguez-Tudela JL, Cuenca-Estrella M, Zaragoza O. Cryptococcus neoformans capsular enlargement and cellular gigantism during Galleria mellonella infection. PloS one. 2011;6(9):e24485 Epub 2011/09/15. PubMed Central PMCID: PMC3168503. doi: 10.1371/journal.pone.0024485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reeves EP, Lu H, Jacobs HL, Messina CG, Bolsover S, Gabella G, et al. Killing activity of neutrophils is mediated through activation of proteases by K+ flux. Nature. 2002;416(6878):291–7. doi: 10.1038/416291a [DOI] [PubMed] [Google Scholar]
- 65.Gregori C, Schuller C, Roetzer A, Schwarzmuller T, Ammerer G, Kuchler K. The high-osmolarity glycerol response pathway in the human fungal pathogen Candida glabrata strain ATCC 2001 lacks a signaling branch that operates in baker's yeast. Eukaryotic cell. 2007;6(9):1635–45. PubMed Central PMCID: PMCPMC2043364. doi: 10.1128/EC.00106-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mesa-Arango AC, Rueda C, Roman E, Quintin J, Terron MC, Luque D, et al. Cell Wall Changes in Amphotericin B-Resistant Strains from Candida tropicalis and Relationship with the Immune Responses Elicited by the Host. Antimicrob Agents Chemother. 2016;60(4):2326–35. PubMed Central PMCID: PMCPMC4808153. doi: 10.1128/AAC.02681-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garcia R, Botet J, Rodriguez-Pena JM, Bermejo C, Ribas JC, Revuelta JL, et al. Genomic profiling of fungal cell wall-interfering compounds: identification of a common gene signature. BMC Genomics. 2015;16:683 PubMed Central PMCID: PMCPMC4560923. doi: 10.1186/s12864-015-1879-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Malinsky J, Tanner W, Opekarova M. Transmembrane voltage: Potential to induce lateral microdomains. Biochim Biophys Acta. 2016;1861(8 Pt B):806–11. [DOI] [PubMed] [Google Scholar]
- 69.Vargas G, Rocha JD, Oliveira DL, Albuquerque PC, Frases S, Santos SS, et al. Compositional and immunobiological analyses of extracellular vesicles released by Candida albicans. Cell Microbiol. 2015;17(3):389–407. doi: 10.1111/cmi.12374 [DOI] [PubMed] [Google Scholar]
- 70.Romero PA, Lussier M, Veronneau S, Sdicu AM, Herscovics A, Bussey H. Mnt2p and Mnt3p of Saccharomyces cerevisiae are members of the Mnn1p family of alpha-1,3-mannosyltransferases responsible for adding the terminal mannose residues of O-linked oligosaccharides. Glycobiology. 1999;9(10):1045–51. [DOI] [PubMed] [Google Scholar]
- 71.Casadevall A, Pirofski LA. Microbiology: Ditch the term pathogen. Nature. 2014;516(7530):165–6. doi: 10.1038/516165a [DOI] [PubMed] [Google Scholar]
- 72.Casadevall A, Fang FC, Pirofski LA. Microbial virulence as an emergent property: consequences and opportunities. PLoS Pathog. 2011;7(7):e1002136 PubMed Central PMCID: PMCPMC3141035. doi: 10.1371/journal.ppat.1002136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen YL, Konieczka JH, Springer DJ, Bowen SE, Zhang J, Silao FG, et al. Convergent Evolution of Calcineurin Pathway Roles in Thermotolerance and Virulence in Candida glabrata. G3. 2012;2(6):675–91. Epub 2012/06/13. PubMed Central PMCID: PMC3362297. doi: 10.1534/g3.112.002279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ene IV, Adya AK, Wehmeier S, Brand AC, MacCallum DM, Gow NA, et al. Host carbon sources modulate cell wall architecture, drug resistance and virulence in a fungal pathogen. Cell Microbiol. 2012;14(9):1319–35. PubMed Central PMCID: PMCPMC3465787. doi: 10.1111/j.1462-5822.2012.01813.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ene IV, Walker LA, Schiavone M, Lee KK, Martin-Yken H, Dague E, et al. Cell Wall Remodeling Enzymes Modulate Fungal Cell Wall Elasticity and Osmotic Stress Resistance. mBio. 2015;6(4):e00986 PubMed Central PMCID: PMCPMC4551979. doi: 10.1128/mBio.00986-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bahn YS, Jung KW. Stress signaling pathways for the pathogenicity of Cryptococcus. Eukaryotic cell. 2013;12(12):1564–77. PubMed Central PMCID: PMCPMC3889573. doi: 10.1128/EC.00218-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaur R, Domergue R, Zupancic ML, Cormack BP. A yeast by any other name: Candida glabrata and its interaction with the host. Current opinion in microbiology. 2005;8(4):378–84. Epub 2005/07/06. doi: 10.1016/j.mib.2005.06.012 [DOI] [PubMed] [Google Scholar]
- 78.Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459(7248):802–7. PubMed Central PMCID: PMCPMC2702157. doi: 10.1038/nature08085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Frieman MB, McCaffery JM, Cormack BP. Modular domain structure in the Candida glabrata adhesin Epa1p, a beta1,6 glucan-cross-linked cell wall protein. Molecular microbiology. 2002;46(2):479–92. Epub 2002/10/31. [DOI] [PubMed] [Google Scholar]
- 80.Saretzki G, Von Zglinicki T. Replicative aging, telomeres, and oxidative stress. Ann N Y Acad Sci. 2002;959:24–9. [DOI] [PubMed] [Google Scholar]
- 81.Bouklas T, Jain N, Fries BC. Modulation of Replicative Lifespan in Cryptococcus neoformans: Implications for Virulence. Front Microbiol. 2017;8:98 PubMed Central PMCID: PMCPMC5276861. doi: 10.3389/fmicb.2017.00098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Denning DW, Bromley MJ. How to bolster the antifungal pipeline. Science. 2015;347(6229):1414–6. doi: 10.1126/science.aaa6097 [DOI] [PubMed] [Google Scholar]
- 83.Dodgson AR, Pujol C, Denning DW, Soll DR, Fox AJ. Multilocus sequence typing of Candida glabrata reveals geographically enriched clades. Journal of clinical microbiology. 2003;41(12):5709–17. Epub 2003/12/10. PubMed Central PMCID: PMC309006. doi: 10.1128/JCM.41.12.5709-5717.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Oh H, Siano B, Diamond S. Neutrophil isolation protocol. Journal of visualized experiments: JoVE. 2008;(17). Epub 2008/12/11. PubMed Central PMCID: PMC3074468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Du C, Calderone RA. Phagocytosis and killing assays for Candida species. Methods in molecular biology. 2009;499:17–26. Epub 2009/01/20. doi: 10.1007/978-1-60327-151-6_3 [DOI] [PubMed] [Google Scholar]
- 86.Branzk N, Lubojemska A, Hardison SE, Wang Q, Gutierrez MG, Brown GD, et al. Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat Immunol. 2014;15(11):1017–25. PubMed Central PMCID: PMCPMC4236687. doi: 10.1038/ni.2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baker LG, Specht CA, Donlin MJ, Lodge JK. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryotic cell. 2007;6(5):855–67. PubMed Central PMCID: PMCPMC1899242. doi: 10.1128/EC.00399-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baker LG, Specht CA, Lodge JK. Chitinases are essential for sexual development but not vegetative growth in Cryptococcus neoformans. Eukaryotic cell. 2009;8(11):1692–705. Epub 2009/09/08. PubMed Central PMCID: PMC2772403. doi: 10.1128/EC.00227-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Beauvais A, Maubon D, Park S, Morelle W, Tanguy M, Huerre M, et al. Two alpha(1–3) glucan synthases with different functions in Aspergillus fumigatus. Appl Environ Microbiol. 2005;71(3):1531–8. PubMed Central PMCID: PMCPMC1065186. doi: 10.1128/AEM.71.3.1531-1538.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Singh A, Yadav V, Prasad R. Comparative lipidomics in clinical isolates of Candida albicans reveal crosstalk between mitochondria, cell wall integrity and azole resistance. PloS one. 2012;7(6):e39812 PubMed Central PMCID: PMCPMC3384591. doi: 10.1371/journal.pone.0039812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mandala SM, Thornton RA, Frommer BR, Curotto JE, Rozdilsky W, Kurtz MB, et al. The discovery of australifungin, a novel inhibitor of sphinganine N-acyltransferase from Sporormiella australis. Producing organism, fermentation, isolation, and biological activity. J Antibiot (Tokyo). 1995;48(5):349–56. [DOI] [PubMed] [Google Scholar]
- 92.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–7. doi: 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- 93.Hestdal K, Ruscetti FW, Ihle JN, Jacobsen SE, Dubois CM, Kopp WC, et al. Characterization and regulation of RB6-8C5 antigen expression on murine bone marrow cells. J Immunol. 1991;147(1):22–8. [PubMed] [Google Scholar]
- 94.Amberg DC, Burke DJ, Strathern JN. Yeast vital stains: calcofluor staining of chitin and bud scars. CSH Protoc. 2006;2006(1). [DOI] [PubMed] [Google Scholar]
- 95.Tomiotto-Pellissier F, Cataneo AH, Orsini TM, Thomazelli AP, Dalevedo GA, de Oliveira AG, et al. Galleria mellonella hemocytes: A novel phagocytic assay for Leishmania (Viannia) braziliensis. J Microbiol Methods. 2016;131:45–50. doi: 10.1016/j.mimet.2016.10.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Younger cells were phagocytosed by Galleria mellonella haemocytes (B) and human neutrophils at a higher frequency compared to old cells. **P < 0.01, ****P < 0.0001.
(TIF)
Increased virulence in Galleria was observed with older cells of strains (A) 89 and (B) 117. (C) Younger cells were phagocytosed by Galleria mellonella haemocytes at a higher frequency compared to old cells of strain 89. (D) Increased resistance to neutrophil-mediated killing was observed in older cells of strains 89 and 117. (E) H2O2 disc diffusion assays showed smaller zone of inhibition in older cells of strain 89. (F) The DB variant exhibited a higher fungal burden than the parent (S) in strain 89. (G) Phenotypic switching in strain 89 from S to DB colony morphology increased consistently with age to 18 fold. (H) The RLS of DB was shortened over 50% relative to S and reconstituted in the revertant colony in strain 89. *P < 0.05, **P < 0.01, ***P < 0.001.
(TIF)
(A) Budscar staining of samples from urine of 5 patients showed a skewed distribution towards an increased frequency of older cells compared to their in vitro cultures. (B) Strain 89 cells with a proportionally high number of budscars (mean line) were found at days 2 and 4 in kidneys of WT compared to neutropenic mice, and also compared to day 0.
(TIF)
Solutions of ordinary differential system with optimum-fit mortality profile for (A) WT mouse host, (B) neutropenic mouse host, and (C) in vitro control population.
(TIF)
Mean of 1000 stochastic simulations of corresponding system of reactions with optimum-fit mortality profile for (A) WT mouse host, (B) neutropenic mouse host, and (C) in vitro control population.
(TIF)
Mortality profiles found using a local optimizer are shown for the entire search space of trial profiles. Each locally-optimized mortality profile is marked as being within +/- 20% of the global optimum or not, and its cost function (distance of model from data age distribution, as computed via Eq. (S2) is shown). (A) WT mouse host, (B) neutropenic mouse host, (C) in vitro control cell population.
(TIF)
(MP4)
(DOCX)
Supplemental methods and results for modeling are described separately.
(PDF)
Data Availability Statement
The data is deposited at NCBI and can be accessed on GEO (accession #GSE85985).