Abstract
We report the generation-characterization of a fetal liver (FL) B-cell progenitor (BCP)-derived human induced pluripotent stem cell (hiPSC) line CRISPR/Cas9-edited to carry/express a single copy of doxycycline-inducible Cas9 gene in the “safe locus” AAVS1 (iCas9-FL-BCP-hiPSC). Gene-edited iPSCs remained pluripotent after CRISPR/Cas9 genome-edition. Correct genomic integration of a unique copy of Cas9 was confirmed by PCR and Southern blot. Cas9 was robustly and specifically expressed on doxycycline exposure. T7-endonuclease assay demonstrated that iCas9 induces robust gene-edition when gRNAs against hematopoietic transcription factors were tested. This iCas9-FL-BCP-hiPSC will facilitate gene-editing approaches for studies on developmental biology, drug screening and disease modeling.
Resource Table
| Unique stem cell line identifier | JCLRIi001-A-1 |
| Alternative name of stem cell line | iCas9-FL-BCP-hiPSC |
| Institution | Josep Carreras Leukemia Research Institute |
| Contact information of distributor | Julio Castaño, jcastano@carrerasresearch.org |
| Type of cell line | iPSC |
| Origin | human |
| Additional origin info | Age: 19–22 weeks of human fetal development Sex: XX |
| Cell source | Fetal liver B-cell progenitors |
| Method of reprogramming | Non-integrative (Sendai virus) |
| Associated disease | Non applicable |
| Gene/locus | Cas9 inserted in AAVS1 locus |
| Method of modification | CRISPR-Cas9 |
| Gene correction | NO |
| Name of transgene or resistance | Cas9 |
| Inducible/constitutive system | Doxycycline inducible system |
| Date archived/stock date | December 2016 |
| Cell line repository/bank | |
| Ethical approval | Patient's informed consent obtained. Institutional Review Board approval obtained (CMRB-CEIC-26/2013) |
Resource utility
This iCas9-FL-BCP-iPSC constitutes a unique tool facilitating the screening on multiple sgRNAs (and libraries) for the generation of locus-specific genetic-edited (knock-in, knock-out, codon substitution, structural rearrangements, etc.) hiPSC for developmental biology, compound screening and disease modeling.
Resource details
Fresh fetal liver (FL) was collected from developing human embryos aborted at 19–22 weeks of pregnancy. Human tissue was provided by The Vrelinghuis abortion clinic (Utrecht, The Netherlands) upon signed informed consent and approval by our local Ethics and Biozahard Board Committees (CMRB-CEIC-26/2013) through a formal collaboration with the Erasmus-Medical Centre, Rotterdam, The Netherlands). Mononuclear cells (MNCs) were isolated using Ficoll-Hypaque and CD34 + CD19 + B-cell progenitors (BCP) were FACS-purified and reprogrammed by infection with non-integrative tetracistronic SeV vectors encoding the transcription factors OCT4, SOX2, KLF4, and MYC (OSKM) (Munoz-Lopez et al., 2016a, Munoz-Lopez et al., 2016b). Resulting FL CD34 + CD19 + − iPSC lines (FL-BCP-hiPSC) were established as previously reported (Munoz-Lopez et al., 2016a, Munoz-Lopez et al., 2016b, Bueno et al., 2016). This FL-BCP-hiPSC was genome-edited to harbor the Cas9 coding sequence controlled by a doxycycline-inducible cassette in the genomic “safe harbor” AAVS1 (iCas9-FL-BCP-hiPSC). A single cassette containing both the rTetR activator under CAG promoter and the Tetracycline Response Element (TRE) promoter driving the expression of Cas9, was inserted in the AAVS1 locus by homologous recombination using the Cas9 nuclease and a guide RNA (gRNA) sequence (Mali et al., 2013) against intron 1 of AAVS1 locus (Fig. 1 panel A). The Fig. 1 panel A shows a schematic representation of the donor vector used for insertion of the iCas9 cassette into the AAVS1 locus (HA, homology arm; puro, puromycin; SA, splice acceptor; T2A, self-cleaving 2A peptide; CAG, CMV early enhancer/chicken β actin promoter. rTetR, reverse Tet repressor; pA, poly A signal; TRE, Tet response element). Correct genomic integration of a unique copy of Cas9 was confirmed by both genomic PCR (not shown) and Southern blot analysis (Fig. 1 panel B) in several iPSC clones, using a 5′-internal probe (left panel) and a 3′-external probe (right panel). Red asterisks indicate clones with the desired targeted insertions of the iCas9. Three iCas9-FL-BCP-hiPSC clones were induced for 72 h with 2 μg/ml of doxycycline and analyzed for Cas9 expression by qPCR (Fig. 1 panel C), showing a robust, non-leaky expression of Cas9. To functionally validate the Cas9 expression, iCas9-FL-BCP-hiPSCs were nucleofected with different gRNAs against three hematopoietic transcription factors (MLL, GATA2 and AF4) in presence/absence of doxycycline. The T7 endonuclease I assay confirmed a high percentage (25%–62%) of cleavage (Fig. 1 panel D). Red asterisks depict the expected T7EI-specific fragments used to quantify indel frequency. The in silico-predicted (crispr.mit.edu) top off-targets of AAVS1 gRNA (RNF4, RHOT2, FAIM2, RPL8, BTNL8, MYBL2) were sequence-verified in iCas9-FL-BCP-hiPSCs and they were consistently found unaltered, demonstrating the high specificity of the approach used (data not shown).
Fig. 1.
CRISPR/Cas9-mediated generation of iCas9-FL-BCP-hiPSCs by gene targeting at the AAVS1 locus.
Importantly, iPSCs remained pluripotent after CRISPR/Cas9 gene editing. iCas9-FL-BCP-hiPSCs retained hESC-like morphology and expressed the pluripotency markers alkaline phosphatase (AP) (Fig. 1 panel E), OCT4, NANOG, SOX2, REX1, CRIPTO, and DNMT3B (Fig. 1 panel F). Endogenous expression of NANOG and OCT4 was accompanied by the extensive loss of CpG methylation in their promoters (Fig. 1 panel G). By flow cytometry, gene-edited iPSCs consistently expressed SSEA-3, SSEA-4, TRA-1-60, and TRA-1-81 (Fig. 1 panel H). In vivo, the differentiation capacity was confirmed by teratoma formation in NSG mice comprising tissue representing all three germ layers (Fig. 1 panel I).
Materials and methods
iPSC generation, maintenance and characterization
iPSCs were generated using OKSM polycistronic SeV vector (Munoz-Lopez et al., 2016a, Munoz-Lopez et al., 2016b) and were fully characterized before and after gene edition as previously described (Bueno et al., 2016).
Promoter demethylation
Bisulfite pyrosequencing of OCT4 and NANOG promoters was done as described (Bueno et al., 2016).
Immunophenotyping
Antibodies used to check by flow cytometry the pluripotency-associated markers are detailed in Table 2.
Table 2.
Antibodies and primers used in this study.
| Antibodies used for immunocytochemistry/flow-citometry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # and RRID | |
| SSEA-3-PE | Rat anti-SSEA-3 | 1:100 | BD Bioscience Cat#560237, RRID:AB_1645542 |
| SSEA-4-v450 | Mouse anti-SSEA-4 | 1:100 | BD Bioscience Cat#561156, RRID:AB_10896140 |
| TRA-1-60-BV510 | Mouse anti-TRA-1-60 | 1:100 | BD Bioscience Cat#563188, RRID:AB_2637036 |
| TRA-1-81-AlexaFlour647® | Mouse anti-TRA-1-81 | 1:100 | BD Bioscience Cat#560793, RRID:AB_10550550 |
| Primers | |||
| Target | Forward/Reverse primer (5′–3′) | ||
| Genomic PCR | 5′ junction | CTGCCGTCTCTCTCCTGAGT/GTGGGCTTGTACTCGGTCAT | |
| 3′ junction | GGCGATCTGACGGTTCACTAAAC/GAATCCACCCAAAAGGCAGC | ||
| Southern blot | 5′ probe | AGGTTCCGTCTTCCTCCACT/GTCCAGGCAAAGAAAGCAAG | |
| 3′ probe | ACAGGTACCATGTGGGGTTC/CTTGCCTCACCTGGCGATAT | ||
| T7 assay | MLL | CAGCACTCTCTCCAATGGCA/TAAGCCTCCCATCTCCCACA | |
| AF4 | GGGGAAAAAAAACATTTCGGCGACATG/CTACCATTTCCCTCATTCCAATTCACTCC | ||
| GATA2 | CGTGTCGCTGGGATCAAG/TCCCCAAAGAAAGCCAGAAAC | ||
| RNA in vitro transcription | AAVS1 IVT | GAAATTAATACGACTCACTATAGGGGGCCACTAGGGACAGGATGTTTTAGAGCTAGAAA/AAAAGCACCGACTCGGTGCC | |
| MLL IVT | GAAATTAATACGACTCACTATAGTTAGCAGGTGGGTTTAGCGCGTTTTAGAGCTAGAAA/AAAAGCACCGACTCGGTGCC | ||
| AF4 IVT | GAAATTAATACGACTCACTATAGGTCTCATTCCAGCAACACGTGTTTTAGAGCTAGAAA/AAAAGCACCGACTCGGTGCC | ||
| GATA2 IVT | CATGTAGTTGTGCGCCGTTTTAGAGCTAGA/AAAAGCACCGACTCGGTGCC | ||
| Pluripotency Markers (qPCR) | OCT4 | GGGTTTTTGGGATTAAGTTCTTCA/GCCCCCACCCTTTGTGTT | |
| NANOG | ACAACTGGCCGAAGAATAGCA/GGTTCCCAGTCGGGTTCAC | ||
| SOX2 | CAAAAATGGCCATGCAGGTT/AGTTGGGATCGAACAAAAGCTATT | ||
| REX1 | CCTGCAGGCGGAAATAGAAC/GCACACATAGCCATCACATAAGG | ||
| CRIPTO | CGGAACTGTGAGCACGATGT/GGGCAGCCAGGTGTCATG | ||
| DNMT3B | GCTCACAGGGCCCGATACTT/GCAGTCCTGCAGCTCGAGTTTA | ||
| Housekeeping gene | GAPDH | GCACCGTCAAGGCTGAGAAC/AGGGATCTCGCTCCTGGAA | |
| Off-target genomic PCR | RNF4 | CAGACCGTGACTCCCGAAA/GTCAGCGGGGAACAAAAACC | |
| RHOT2 | TGTTACTGGGCGAGGGTAGG/CTACGGCCGCTACCTGAGTA | ||
| FAIM2 | AGGCTCGTCCCATCCTTTTG/CACATCCCCATTTGCTCCCT | ||
| RPL8 | GCAGGCAGTTCTAGAAGCCA/CCTTAGTTATCTGGATTTCCAGAAC | ||
| BTNL8 | TAGGAGTCTTGGTGGTGTTCAT/ATATCGTGGCACCTGGCTAC | ||
| MYBL2 | GCAGTCGGAGGAAGTGACAA/CTCCTGGCCCCTCTTAGACT | ||
| Mycoplasma PCR | Nature | TGCACCATCTGTCACTCTGTTAACCTC/GGGAGCAAACAGGATTAGATACCCT | |
| M1 | ACACCATGGGAGCTGGTAAT/CTTCATCGACTTTCAGACCCAAGGCAT | ||
CRISPR/Cas9-edition of hiPSCs expressing a doxycycline-inducible Cas9
The CAG-rTetR cassette was PCR amplified from AAVS1-Neo-M2rtTA (Addgene #60843) with primers containing restriction sites for SalI and ClaI. After amplification and enzyme-digestion, the CAG-rTetR cassette was cloned in the Puro-Cas9 donor vector (Addgene #58409). Finally, a gBlock fragment (IDT Technologies) designed with two opposite poly-A sequences was cloned using MluI and ClaI enzymes. A gRNA sequence targeting the AAVS1 intron 1 (5′-GGGGCCACTAGGGACAGGAT-3′) was in vitro transcribed (IVT). To edit the iPSCs, 200.000 cells were electroporated with 100 pmol Cas9 nuclease (IDT - Integrated DNA Technologies), 120 pmol the IVT-gRNA against AAVS1 intron 1, and 5 μg of linearized donor vector. Electroporation was performed using Neon Transfection System (ThermoFisher) at 1400 V, 5 ms and 3 pulses in a 100 μl tip. Cells were then selected with 1 μg/ml puromycin.
Southern blot
Genomic DNA from each cell line was isolated with Maxwell® RSC Cultured Cells DNA Kit (Promega). 6 μg of DNA from each clone was digested with SphI (for 5′ probe) or BglII (for 3′ probe) (New England Biolabs), separated on a 1% agarose gel and transferred to a nylon membrane (RPN303B, Amersham). Membranes were hybridized with DIG-dUTP labeled probes. Probes were detected by an AP-conjugated DIG-Antibody (Roche Diagnostics) using CDP-Star (Sigma-Aldrich) as a substrate for chemiluminescence. Probes were synthesized by PCR using the PCR DIG Probe Synthesis Kit (Roche Diagnostics). 5′ probe was generated using plasmidic DNA and 3′ probe using genomic DNA as a templates. Primers used for probes are detailed in Table 2.
T7 endonuclease assay
iCas9-FL-BCP-hiPSCs were treated with doxycycline (2 μg/ml) for two days before and during transfection. Cells were dissociated with Accutase (Stem Cell Technologies) and 200.000 cells were electroporated with 120 pmol of a single IVT-gRNA (MLL, GATA2 or AF4). Genomic DNA was extracted four days after gRNA transfection. Genomic regions flanking the CRISPR target sites were PCR amplified (Table 1). PCRs were denatured and re-annealed and then PCRs were treated with 5U of T7EI at 37 °C for 1 h.
Table 1.
Summary of quality control testing and results for iCas9-FL-BCP-hiPSC.
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography | hESC-like morphology | Fig. 1 panel E top |
| AP staining | Positive | Fig. 1 panel E bottom | |
| Phenotype | qPCR | Expression of pluripotency markers: OCT4, NANOG, SOX2, CRIPTO, REX, DNMT3B | Fig. 1 panel F |
| Promoter demethylation | loss of CpG methylation in OCT4 and NANOG promoters | Fig. 1 panel G | |
| Flow cytometry | SSEA-3 (76%), SSEA-4 (100%), TRA-1-60 (100%) and TRA-1-81 (99%) | Fig. 1 panel H | |
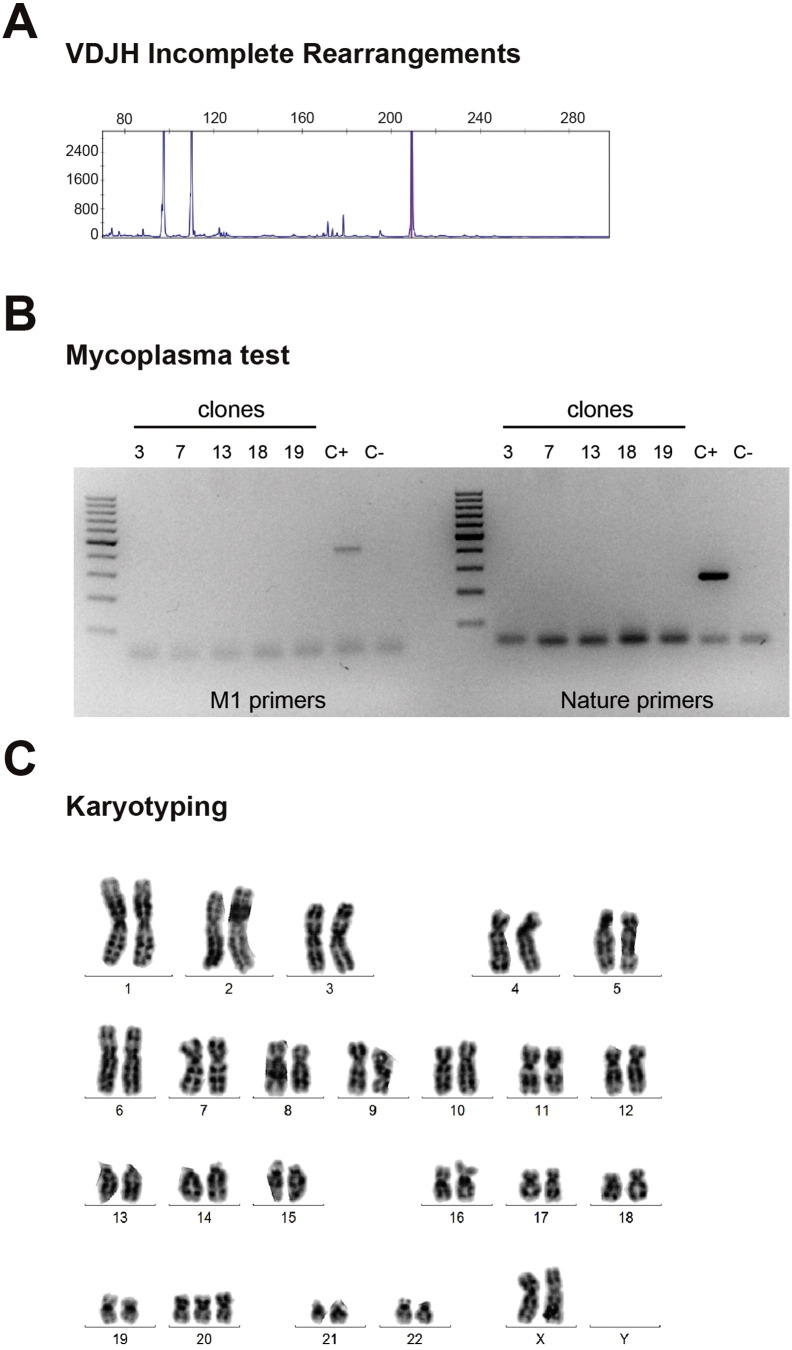
| Genotype | Karyotype | 47 (XX) + 20 Resolution: 400-band level |
Fig. 1S panel C |
| Identity | VDJH (BCR) rearrangement | Incomplete VDJH rearrangement (progenitor B cell) | Fig. 1S panel A |
| Mutation analysis | Southern blot | One specific insertion at AAVS1 locus | Fig. 1 panel B |
| Microbiology and virology | Mycoplasma | Mycoplasma tested by PCR: negative | Fig. 1S panel B |
| Differentiation potential | Teratoma formation | Representation of all three germ layers | Fig. 1 panel I |
| Donor screening | N/A | ||
| Genotype additional info | N/A |
In vitro transcription
T7 RNA polymerase promoter was added to gRNA sequences by PCR using as a template the pSpCas9(BB)-2A-GFP (PX458) plasmid (Addgene #48138) containing the guide RNA sequences for MLL, GATA2 or AF4. PCR amplification were performed using specific forward primers and a universal reverse primer (Table 2). PCR products were used as templates for IVT using the HiScribe™ T7 High Yield RNA Synthesis Kit (New England Biolabs). The resulting gRNAs were purified using the MEGAclear kit (Life Technologies), eluted in RNase-free water and stored at − 80 °C until use.
Mycoplasma test
Primers used are listed in Table 2. PCR conditions were:
94 °C: 20″
- × 40 cycles
- 94ªC: 20″
- 55 °C: 20″ (Nature primer) or 58 °C: 10″ (M1 primer)
- 65ªC: 20–40″
The following are the supplementary data related to this article.
Supplementary Fig. 1.
Supplementary quality control testing and results for iCas9-FL-BCP-hiPSC.
Supplementary material
References
- Muñoz-López A., Romero-Moya D., Prieto C., Ramos-Mejia V., Agraz-Doblas A., Varela I., Buschbeck M., Palau A., Carvajal-Vergara X., Giorgetti A., Ford A., Lako M., Granada I., Ruiz-Xiville N., Rodriguez-Perales S., Torres-Ruiz R., Stam R.W., Fuster J.L., Fraga M.F., Nakanishi M., Cazzaniga G., Bardini M., Cobo I., Bayon G.F., Fernandez A.F., Bueno C., Menéndez P. Development refractoriness of MLL-rearranged human B cell acute leukemias to reprogramming into pluripotency. Stem Cell Rep. 2016;7:602–618. doi: 10.1016/j.stemcr.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-López A., van Roon E.H., Romero-Moya D., Lopez-Millan B., Stam R.W., Colomer D., Nakanishi M., Bueno C., Menéndez P. Cellular ontogeny and hierarchy influence the reprogramming efficiency of human B cells into induced pluripotent stem cells. Stem Cells. 2016;34:581–587. doi: 10.1002/stem.2303. [DOI] [PubMed] [Google Scholar]
- Bueno C., Sardina J.L., Di Stefano B., Romero-Moya D., Muñoz-López A., Ariza L., Chillon M.C., Balanzategui A., Castaño J., Herreros A., Fraga M.F., Fernandez A., Granada I., Quintana-Bustamante O., Segovia J.C., Nishimura K., Ohtaka M., Nakanishi M., Graf T., Menéndez P. Reprogramming human B cells into induced pluripotent stem cells and its enhancement by C/EBPalpha. Leukemia. 2016;30:674–682. doi: 10.1038/leu.2015.294. [DOI] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material