Abstract
Emerging evidence indicates that nuclear factor I/B (NFIB), a transcription factor required for proper development and regulation of cellular differentiation in several tissues, also plays critical roles in cancer. Despite being a metastatic driver in small cell lung cancer and melanoma, it has become apparent that NFIB also exhibits tumour suppressive functions in many malignancies. The contradictory contributions of NFIB to both the inhibition and promotion of tumour development and progression, corroborates its diverse and context-dependent roles in many tissues and cell types. Considering the frequent involvement of NFIB in cancer, a better understanding of its multifaceted nature may ultimately benefit the development of novel strategies for the management of a broad spectrum of malignancies. Here we discuss recent findings which bring to light NFIB as a crucial and paradoxical player in cancer.
Keywords: Cancer, Oncogene, Tumour suppressor, Cellular differentiation, NFIB, Developmental transcription factor
Highlights
-
•
NFIB, a versatile regulator of cell differentiation, is emerging as a crucial driver of cancer metastasis.
-
•
Paradoxically, NFIB also exhibits tumour suppressive functions in several cancer types.
-
•
A deeper understanding of the multifaceted and context-dependent nature of NFIB has the potential to improve the clinical management of a variety of cancers.
1. NFIB in Development and Cell Physiology
The Nuclear Factor I (NFI) family of site-specific DNA binding proteins functions in adenoviral DNA replication and in the regulation of transcription of a large variety of cellular and viral genes (Gronostajski, 2000). This family is comprised of four genes in vertebrates (NFIA, NFIB, NFIC and NFIX), whose encoded proteins interact with DNA as homo- or hetero-dimers. They bind to the palindromic sequence TTGGC(N5)GCCAA with high affinity, resulting in transcriptional activation or repression, depending on the cellular context and regulatory region (Gronostajski, 2000). Binding sites for these factors have been identified in promoter, enhancer and silencer regions of a plethora of genes expressed in almost every organ and tissue (Kruse and Sippel, 1994, Gronostajski, 2000). Reflecting their important roles, NFIs are essential for the development of a number of organ systems and show non-redundant functions during murine development (Chaudhry et al., 1997, das Neves et al., 1999, Steele-Perkins et al., 2005, Barry et al., 2008).
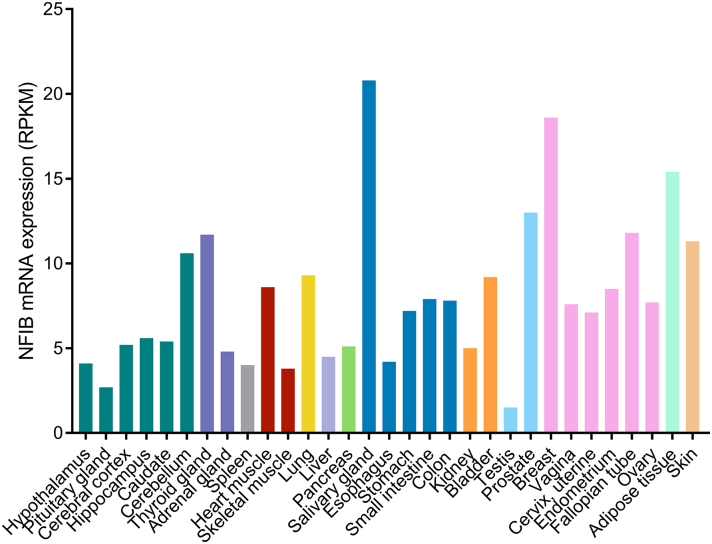
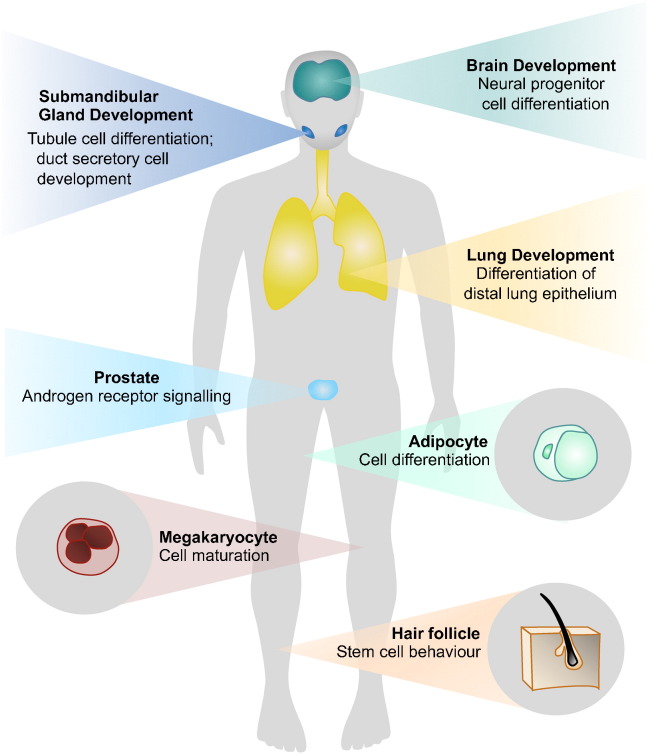
Transcriptome and proteome analyses reveal that NFIB is commonly expressed throughout the human body (Fig. 1; GTEx Consortium, 2015, Uhlen et al., 2015). Consistent with this ubiquitous expression pattern and the apparent abundance of target genes, data supports that NFIB plays a fundamental role in a range of biological processes (Fig. 2). Mice lacking this gene present with a very severe phenotype, marked by the death of all animals shortly after birth due to lung dysfunction (Steele-Perkins et al., 2005). Loss of Nfib results in an undifferentiated primordial respiratory system in addition to major neuroanatomic defects, including corpus callosum dysgenesis and delayed glial and neuronal differentiation (Steele-Perkins et al., 2005, Barry et al., 2008, Piper et al., 2014). Notably, some Nfib heterozygous animals show related phenotypes, suggesting haploinsufficiency at the Nfib locus (Steele-Perkins et al., 2005). Besides being essential to lung and brain development, Nfib has also been shown to be required for tubule cell differentiation during development of mouse submandibular glands (Mellas et al., 2015).
Fig. 1.
NFIB expression overview in human tissues. NFIB is expressed in a range of tissues. RNA-sequencing data from 31 tissues generated by the Genotype-Tissue Expression Project (GTEx; https://www.gtexportal.org/) are reported as median RPKM (Reads Per Kilobase of transcript per Million mapped reads). Colour-coding is based on 13 tissue groups, each consisting of tissues with common functional features (adapted from the Human Protein Atlas: http://www.proteinatlas.org/ENSG00000147862-NFIB/tissue, available from v16.1.proteinatlas.org).
Fig. 2.
NFIB functions in development and physiology. NFIB is required for the development of the lung, brain and submandibular glands. It is also required for the maintenance of a range of physiological processes in several tissues, including adipocyte differentiation, megakaryocyte maturation, regulation of androgen receptor signaling in the prostate, and epithelial-melanocyte stem cell behaviour in the hair follicle niche. Note: Experiments assessing the diverse functions of NFIB were performed mainly using mouse models, and in some cases human cell lines.
In addition to these roles in development, NFIB has been implicated in a range of physiological processes, such as, adipocyte differentiation (Waki et al., 2011), megakaryocyte maturation (Chen et al., 2014), and in the regulation of androgen receptor signaling in the prostate (Grabowska et al., 2014). Furthermore, NFIB functions as a gatekeeper, governing activity within the quiescent stem cell niche of hair follicles, where its loss enhances melanocyte stem-cell self-renewal, disturbing epithelial-melanocyte stem cell synchrony (Chang et al., 2013). Recently, NFIB has also been shown to regulate hippocampal neural stem cell fate (Rolando et al., 2016).
Corroborating its functions in regulating a variety of developmental and physiological processes, NFIB has become increasingly implicated in a range of malignancies (Table 1), which is the focus of this review article.
Table 1.
Summary of alterations in NFIB reported in cancer.
| Type of aberration | Organ/site | Tumour type | Function/potential role | Reference(s) |
|---|---|---|---|---|
| Amplification/overexpression | Lung | Small cell lung cancer | Oncogene |
Denny et al., 2016 Dooley et al., 2011, Semenova et al., 2016, Wu et al., 2016 |
| Overexpression | Skin | Melanoma | Oncogene | Fane et al., 2017 |
| Amplification/overexpression | Breast | Triple negative; ER negative | Oncogene | Han et al., 2008 |
| Amplification | Esophagus | Squamous cell carcinoma | Unknown | Yang et al., 2001 |
| Amplification | Submandibular gland | Large cell neuroendocrine carcinoma | Unknown | Andreasen et al., 2016 |
| Amplification | Bone | Metastatic giant cell tumour | Unknown | Quattrini et al., 2015 |
| Underexpression | Lung | Non-small cell lung cancer | Tumour suppressor | Becker-Santos et al., 2016 |
| Loss of heterozygosity/underexpression | Brain | Glioma, Glioblastoma | Tumour suppressor *Oncogenic behaviour in some subtypes of glioblastoma |
Stringer et al., 2016, Suzuki et al., 2015 |
| Germline mutation | Bone | Osteosarcoma | Tumour suppressor | Mirabello et al., 2015 |
| Underexpression | Skin | Cutaneous squamous cell carcinoma | Tumour suppressor | Zhou et al., 2014 |
| Gene fusions (MYB-NFIB, MYBL-NFIB, NFIB-AIG1, NFIB-MAN1A1, NFIB-NKAIN2, NFIB-PTPRD, NFIB-XRCC4) | Salivary, lacrimal & ceruminal glands; breast; vulva | Adenoid cystic carcinomas | Unknown |
Marchio et al., 2010 Xing et al., 2016 Persson et al., 2009 Mitani et al., 2010 Brayer et al., 2016 Mitani et al., 2011 Mitani et al., 2016 |
| Gene fusions (HMGA2-NFIB) | Head & neck | Pleomorphic adenoma | Unknown | Geurts et al., 1998 |
| Gene fusions (HMGA2-NFIB) | Colon & retroperitoneal space; intramuscular | Lipoma | Unknown | Italiano et al., 2008, Pierron et al., 2009 |
2. NFIB as an Oncogene
2.1. Small Cell Lung Cancer
Using genetically engineered mouse model systems of small cell lung cancer (SCLC) in combination with analyses of human SCLC specimens, a number of studies have defined NFIB as an oncogene. In 2011, Dooley et al., identified Nfib amplification/overexpression within murine tumour tissue, showed that Nfib regulated cell viability and proliferation during transformation of murine SCLC, and reported recurrent amplification of NFIB in ~ 15% of primary human SCLC (Dooley et al., 2011). More recently, a major oncogenic role was assigned to NFIB in this class of lung tumours. In a series of experiments, Denny et al. implicated NFIB in critical molecular events that drive metastasis in SCLC (Denny et al., 2016). They showed that Nfib is both necessary and sufficient to promote multiple steps of the metastatic cascade in vivo, through the reconfiguration of chromatin accessibility in SCLC cells. Chromatin in metastatic lesions displayed a widespread increase in accessibility at gene distal regions that were enriched for NFI motifs, resembling regions found in neural tissue. NFIB was associated with the newly open chromatin sites, maintained the hyper-accessible chromatin state in these regions, and was also proposed to alter a variety of gene expression programs by influencing the combinatorial binding of other transcription factors to these open chromatin regions (Denny et al., 2016). In addition, a different study using another mouse model of SCLC also showed that NFIB promotes metastatic spread, and that it is highly overexpressed in human metastatic high-grade neuroendocrine lung tumours (Semenova et al., 2016). Moreover, an additional report demonstrated oncogenic properties of Nfib in a related model system of SCLC, supporting its role as a metastatic driver, and identifying target gene networks including those related to axon guidance, focal adhesion and extracellular matrix-receptor interactions (Wu et al., 2016).
2.2. Melanoma
Most recently NFIB has been shown to mediate a highly invasive and migratory phenotype in melanoma, where it directly promotes EZH2 expression, also leading to changes in the chromatin state of tumour cells to facilitate this aggressive behaviour (Fane et al., 2017). This study showed that the direct regulation of NFIB expression by BRN2 in melanoma cells, leads to increased cell migration and potentially invasion through the positive and negative regulation of EZH2 and MITF respectively. In melanoma, heterogeneous expression of the MITF and BRN2 transcription factors has been proposed to constitute a key switching mechanism between phenotypic states essential to tumour development and progression (Goodall et al., 2008, Hoek and Goding, 2010). While MITF is a driver of a highly proliferative, less invasive cell state, BRN2 promotes an invasive and less differentiated state crucial to drive tumour progression towards metastasis. NFIB seems to be a key mediator of this phenotype switching.
2.3. Other Cancers
Increased copy number and expression of NFIB have also been reported in triple negative breast cancer (Han et al., 2008) consistent with an oncogenic role in estrogen receptor-negative breast tumours (Moon et al., 2011). Furthermore, NFIB amplifications within squamous cell carcinoma of the esophagus (Yang et al., 2001), large cell neuroendocrine carcinoma of the submandibular gland (Andreasen et al., 2016), and metastatic giant cell tumour of the bone (Quattrini et al., 2015) have also been reported, although, the role of NFIB in these cancers is unknown.
3. NFIB and Tumour Suppressive Characteristics
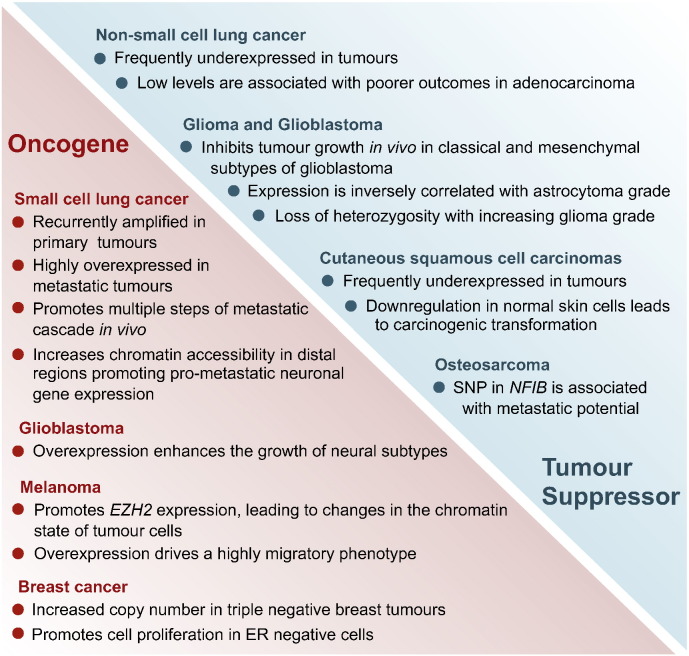
Despite NFIB's established role as an oncogene in SCLC, and most recently in melanoma (and potentially in other malignancies as well), several lines of evidence suggest a tumour suppressor function in other cancer types (Fig. 3).
Fig. 3.
Paradoxical roles of NFIB in cancer. NFIB has shown both oncogenic and tumour suppressive functions in different cancer types and subtypes.
3.1. Non-small Cell Lung Cancer
We have shown that NFIB is underexpressed in 40–70% of non-small cell lung cancers (NSCLC), and that higher NFIB expression is associated with favourable prognosis in lung adenocarcinoma, but not in squamous cell carcinoma patients (Becker-Santos et al., 2016). This lineage-specific phenotype, likely reflects the role of NFIB in regulating the differentiation of cell types comprising the terminal respiratory units of the lung (Steele-Perkins et al., 2005) where lung adenocarcinomas, but not squamous cell carcinomas, typically develop. Accordingly, we observed that tumours presenting low levels of NFIB, displayed less differentiated phenotypes, accompanied by the repression of lung differentiation markers involved in the development of type II pneumocytes, which are thought to be the progenitor cells for lung adenocarcinomas (Becker-Santos et al., 2016).
3.2. Glioma and Glioblastoma
NFIB has shown tumour suppressor activity in glioblastoma, where its expression is inversely correlated with astrocytoma grade, and ectopic expression significantly inhibits tumour growth in vivo. Similar to the findings in NSCLC versus SCLC, NFIB appears to exert a context-dependent role in glioblastoma, whereby its expression induced differentiation and inhibited proliferation and self-renewal of classical and mesenchymal glioblastoma subtypes, while enhancing the growth of neural subtypes (Stringer et al., 2016). Furthermore, a tumour suppressive function for NFIB in brain is also supported by a genome-wide study of genetic alterations associated with gliomas, which revealed NFIB loss of heterozygosity with increasing glioma grade (Suzuki et al., 2015).
3.3. Cutaneous Squamous Cell Carcinoma
Contrary to the findings in melanoma, expression of NFIB has been proposed as a barrier for the development of cutaneous squamous cell carcinoma. Underexpression of NFIB has been reported as a general feature in tumours from patients with this type of skin cancer, and its downregulation in keratinocytes led to carcinogenic transformation. While suppression of NFIB led to upregulation of CDK6 and Bcl-2, it also decreased p53 levels, suggesting that NFIB may mediate G1 arrest and consequently apoptosis in cutaneous squamous cell carcinoma (Zhou et al., 2014).
3.4. Osteosarcoma
NFIB underexpression has been associated with aggressive osteosarcoma phenotypes. A multistage genome wide association study assessing the connection between germline genetic variation and osteosarcoma metastasis, identified a common SNP in NFIB (9p24.1), which is associated with a decrease in NFIB expression and metastasis at diagnosis (Mirabello et al., 2015). Decreased NFIB levels led to increased osteosarcoma cell line proliferation, migration and colony formation, supporting its contribution to susceptibility to metastasis.
4. Gene Fusions Involving NFIB
4.1. Adenoid Cystic Carcinomas
NFIB has been linked to other malignancies through gene fusions, which is frequently the case in adenoid cystic carcinomas (tumours that most commonly arise from salivary and lachrymal glands, although they can also occur in other tissues containing secretory glands such as breast, cervix and vulva) (Persson et al., 2009, Mitani et al., 2010, Brayer et al., 2016, Marchio et al., 2010, Xing et al., 2016). These cancers are often characterized by a recurrent translocation t(6;9)(q22–23;p23–24) involving MYB and NFIB, which leads to high expression of a functional MYB and truncation of NFIB – in the majority of cases only exon 9 of NFIB (encoding the last 5 amino acids) is present in the chimeric mRNA transcripts. The MYB-NFIB gene fusion was reported in 23–86% of adenoid cystic carcinomas arising from different anatomical sites (Wysocki et al., 2016). NFIB may have a tumour suppressive role in these tumours independent of MYB, as rearrangements leading to truncation of NFIB, and presumably loss of its function also occur with other partners (e.g.: NFIB-AIG1, NFIB-MAN1A1, NFIB-NKAIN2, NFIB-PTPRD, NFIB-XRCC4) (Mitani et al., 2011, Mitani et al., 2016). Further supporting a tumour suppressor role in adenoid cystic carcinomas, truncating mutations and homozygous deletions affecting NFIB have also been reported in these tumours (Ho et al., 2013).
4.2. Lipomas and Pleomorphic Adenomas
Other chromosomal translocations involving NFIB include HMGA2-NFIB fusions in lipomas and pleomorphic adenomas, which lead to upregulation of HMGA2 and truncation of NFIB (as in the MYB-NFIB rearrangement, in many cases only five amino acid residues encoded by NFIB were shown to replace the carboxyterminal portion of HMGA2) (Geurts et al., 1998, Italiano et al., 2008, Pierron et al., 2009). Similar to adenoid cystic carcinomas, NFIB most likely plays a role independently of HMGA2, as it is also present in rearrangements with other partners in these malignancies.
Despite the high frequency of rearrangements involving NFIB in adenoid cystic carcinomas, lipomas and pleomorphic adenomas, apart from the fact that the relocation of NFIB regulatory elements has been proposed to contribute to high expression of its fusion partners (Wysocki et al., 2016), little attention has been focused on a potential direct role for NFIB in these cancers – this highlights the need for studies assessing the direct contribution of NFIB to these malignancies, where disruption resulting in its loss of function might play a key role.
5. NFIB Gene Regulation and Downstream Targets
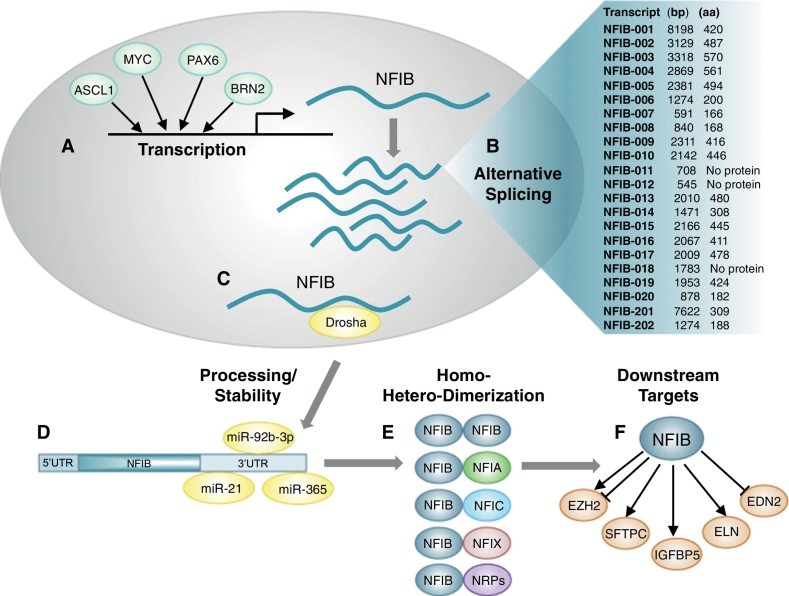
An understanding of the seemingly paradoxical nature of NFIB to display both oncogenic properties and tumour suppressor activity, is hampered by the paucity of information regarding regulation of its expression in various cancer types/subtypes, and the identification of downstream effectors. Moreover, the NFIB locus (9p23–9p22.3) is very complex, where at least 20 spliced variants have been identified (Ensembl version 88; Yates et al., 2016), although their possible distinct functions remain to be explored. The presence of a large 3′UTR extending up to 7.8 kb, suggests that NFIB levels may be commonly regulated by miRNAs. Indeed, we and others have reported that miRNAs such as miR-92b-3p, miR-21 and miR-365, which are frequently deregulated in cancers where NFIB is altered, can bind to the 3′UTR of NFIB, leading to its downregulation (Becker-Santos et al., 2016, Fujita et al., 2008, Zhou et al., 2014). Recently, NFIB has also been shown to be directly repressed by Drosha, independent of miRNAs, representing a novel cell-intrinsic mechanism that regulates the fate of adult hippocampal stem cells (Rolando et al., 2016). Nevertheless, the context-dependent and cell type-specific mechanisms by which NFIB levels are regulated remain largely unknown, and only a few transcription factors have been shown to directly regulate its expression. These include ASCL1 and MYC in a SCLC context (Borromeo et al., 2016, Mollaoglu et al., 2016), PAX6 in neural progenitor cells (Ninkovic et al., 2013), and BRN2 in melanoma (Fane et al., 2017).
Similarly, for most tissue and cell types where NFIB is expressed, only a few direct downstream targets have been identified. Examples include EZH2, which is repressed by NFIB during cortical development (Piper et al., 2014) but activated by NFIB in melanoma (Fane et al., 2017), IGFBP5 which is activated by NFIB in osteoblasts (Perez-Casellas et al., 2009), and SFTPC (Bachurski et al., 2003) and ELN (Hsu et al., 2011) both of which are activated by NFIB in lung development. NFIB also regulates EDN2 in a context related to epithelial-melanocyte stem cell proliferation and differentiation in hair follicles, where it was also linked to the regulation of expression of 1449 target genes by ChIP (Chang et al., 2013). Since NFI members encode proteins with highly homologous DNA-binding domains with similar DNA-binding specificities, it is possible that there are common downstream targets for all NFI genes. In contrast to the highly conserved N-terminal DNA binding domain, the C-terminal region of NFIB, which encodes a transactivation domain, diverges extensively between other NFI members as well as between isoforms, and therefore might promote interactions with different nuclear regulatory proteins depending on the cellular context (Gronostajski, 2000). Adding to this complexity, post-translational modifications (phosphorylation, O- or N-glycosylation) can significantly influence the activity of NFI proteins (Sabova et al., 2013). Fig. 4 summarizes the regulation of NFIB activity at multiple levels.
Fig. 4.
NFIB activity is regulated at multiple levels. A) Cell-type and context-specific transcription of NFIB in various tissues is regulated by multiple transcription factors including ASCL1, MYC, PAX6 and BRN2. B) At least 20 alternatively-spliced isoforms of NFIB have been identified. C & D) Drosha has been shown to destabilize NFIB nuclear transcripts (C) and miRNAs, such as, miR-92b-3p, miR-21 and miR-365 have been shown to affect cytosolic NFIB transcript stability (D). E) NFIB protein products of alternatively-spliced transcripts can form homo- and hetero-dimers with other NFI gene products. In addition, NFIB activity can also be influenced by its binding to other nuclear regulatory proteins (NRPs), and by post-translational modifications (not shown). F) NFIB regulates the expression levels of multiple downstream targets in various tissues, including EZH2, SFTPC, IGFBP5, ELN and EDN2.
It is also noteworthy that NFIB is located in close proximity to the fragile site FRA9G at 9p22.2, which is present in a large fraction of the population (Sawinska et al., 2007), and coincides with recurrent chromosomal breakpoints in cancer cells (Arlt et al., 2006). This location could explain the high frequency of chromosomal rearrangements involving NFIB described in several cancers (Table 1), although this hypothesis needs to be further explored.
6. Conclusions and Perspectives
A number of transcription factors that induce lineage-specific differentiation during embryonic and fetal development, play crucial, and sometimes paradoxical roles in the malignant transformation of adult cells. Although the roles of some of these transcription factors have been well studied in cancer, which is the case for NKX2-1 and SOX members (Yamaguchi et al., 2013, Thu et al., 2014), much remains to be deciphered in terms of understanding the oncogenic and tumour suppressive functions of the vast majority of such important players. The first NFI transcription factor was identified over thirty years ago as a protein required for efficient initiation of adenovirus replication (Nagata et al., 1982). Since then, this family of transcription factors has been implicated in the transcriptional regulation of a variety of viral and cellular genes, and is critically important for the proper development of a number of organs (Chaudhry et al., 1997). Thus, as is the case with other transcription factors involved in developmental processes, it is not surprising that NFIB has become increasingly appreciated as an important player in tumour development and progression.
Recent studies have ascertained a potent oncogenic role for NFIB in SCLC. By increasing chromatin accessibility similar to open regions found in neural tissue, NFIB has emerged as a driver of metastasis in these highly aggressive neuroendocrine lung tumours (Denny et al., 2016). These findings suggest that this mechanism may be relevant to other cancers, especially to other neuroendocrine tumours, which similar to SCLC, also rely on the activation of neuron-like programs, and therefore, might depend on NFIB to drive their metastatic behaviour. In addition to SCLC, a vital role for NFIB in triggering invasive behaviours that drive metastatic spread in melanoma was recently shown (Fane et al., 2017). In this type of skin cancer, NFIB is capable of propagating the acquisition of a more invasive phenotype through broad changes in chromatin status, in large part by increasing expression and function of the histone methyl-transferase enzyme EZH2. Taken together, both studies reveal that NFIB has the ability to promote dynamic changes in the chromatin state of tumour cells to facilitate migration, invasion, and ultimately, metastasis. While in melanoma a key conduct of these effects is the regulation of EZH2 by NFIB, it remains to be determined if a similar epigenetic mechanism could be involved in other tumour types to drive metastatic progression.
Despite a clear oncogenic role for NFIB in SCLC and melanoma, tumour suppressive functions have been demonstrated or strongly suggested in other cancers. Although the molecular mechanisms behind its anti-oncogenic functions are not well understood, the fact that NFIB is widely expressed in multiple normal tissues and cell types, supports a potential role in the maintenance of cellular homeostasis, and conceivably as a barrier to malignant transformation. Accordingly, we and others have shown that downregulation of NFIB leads to the activation of less differentiated tumour phenotypes, culminating in increased cancer aggressiveness and ultimately poorer patient survival. The concept that cells undergo a process of dedifferentiation to a more progenitor like state frequently associated with metastasis, has been documented in various cancer models (Friedmann-Morvinski and Verma, 2014). Moreover, NFIB appears to play a critical role in maintaining stem cell quiescence in some adult tissues (Harris et al., 2015, Chang et al., 2013, Rolando et al., 2016). Further supporting a link between NFIB and the modulation of cellular fate, this NFI member has been shown to be regulated/interact with two key pluripotent transcription factors, MYC and SOX2, respectively (Mollaoglu et al., 2016, Engelen et al., 2011).
Although cancer-related studies have focused mostly on cell-intrinsic changes caused by NFIB, it is worth noting that tumour microenvironment changes might also contribute to drive NFIB-induced cancer aggressiveness. Supporting this hypothesis, NFIB has been shown to regulate endothelins – secreted factors with the ability to mediate intercellular crosstalk – which can promote tumour angiogenesis, migration and invasion (Chang et al., 2013, Rosano et al., 2013). Corroborating a potential role in the tumour microenvironment, NFIB also appears to be expressed in the stroma surrounding tumours (Grabowska et al., 2016; unpublished observations).
Major questions that remain to be answered pertain to the understanding of how NFIB's diverse functions – which promote cell differentiation during late stages of development in a range of tissues, and regulate the maintenance of populations of stem cells in adult tissues – contribute to its significant and paradoxical roles in tumourigenesis. Some specific questions that need to be addressed are: 1) What are the important upstream regulators of NFIB in different cancer-related contexts? 2) Do the many alternatively-spliced isoforms of NFIB play cooperative, or perhaps antagonist, roles during tumourigenesis? 3) What factors might mediate stabilization or enhanced degradation of NFIB transcripts in tumours? 4) How does the expression of NFIB binding partners, including other NFI family members and other transcription factors, influence its activity in cancer? Lastly, 5) What are the direct downstream targets of NFIB in different tumour types and stages, and are they the same or different from those identified during normal development and maintenance of tissue homeostasis? The development of quantitative pull-down assays with NFIB partner proteins and in vivo FRET measurements of NFI-partner protein interactions, combined with ChIP-seq, ATAC-seq, single cell RNA-seq and the use of conditional knock-out alleles of NFIB and other NFI family members, should allow us to answer these important questions.
In closing, the paradoxical involvement of NFIB in both the inhibition and promotion of tumour development and progression in different malignancies; especially between different tumour subtypes within a single organ system, such as in lung, brain and skin, corroborates its diverse and distinct roles in specific tissues and cell types. This follows from the fact that NFI-mediated transcriptional activation or repression of specific gene promoters, varies depending on cell type and upon details of the cellular context (Chaudhry et al., 1998, Chaudhry et al., 1999), resulting in the modulation of the expression of a plethora of diverse tissue-specific genes (Gronostajski, 2000). Consequently, caution must be exercised in the development of any future therapies aimed to manipulate NFIB levels – which may result in unexpected/undesired effects, with the potential for exacerbation of tumour aggressiveness. Further insights into the fascinating role of this enigmatic transcription factor in cancer will certainly open new doors to clinical translation of these findings.
Conflict of Interest Statement
The authors disclose no potential conflicts of interest.
Search Strategy and Selection Criteria
Data for this review were identified by searches of PubMed, using the following search terms in various combinations: “NFIB”, “cancer”, “development”, “cellular differentiation”, “NFI transcription factors”. Articles resulting from these searches and references cited in those articles were selected based on their relevance to the topic covered in the review. Only articles published in English between 1982 and 2017 were included. The majority of the articles reported were published in the last 5 years.
Acknowledgments
We are grateful to Dr. William W. Lockwood for critical reading of the manuscript and stimulating discussions, and to Sonia H.Y. Kung for assistance with illustrations. This work was supported by grants from the Canadian Institutes of Health Research (CIHR FDN-143345 to WLL) and NYSTEM (C030133 to RMG). DDBS was supported by a Vanier Canada Graduate Scholarship.
References
- Andreasen S., Persson M., Kiss K., Homoe P., Heegaard S., Stenman G. Genomic profiling of a combined large cell neuroendocrine carcinoma of the submandibular gland. Oncol. Rep. 2016;35:2177–2182. doi: 10.3892/or.2016.4621. [DOI] [PubMed] [Google Scholar]
- Arlt M.F., Durkin S.G., Ragland R.L., Glover T.W. Common fragile sites as targets for chromosome rearrangements. DNA Repair. 2006;5:1126–1135. doi: 10.1016/j.dnarep.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Bachurski C.J., Yang G.H., Currier T.A., Gronostajski R.M., Hong D. Nuclear factor I/thyroid transcription factor 1 interactions modulate surfactant protein C transcription. Mol. Cell. Biol. 2003;23:9014–9024. doi: 10.1128/MCB.23.24.9014-9024.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry G., Piper M., Lindwall C., Moldrich R., Mason S., Little E., Sarkar A., Tole S., Gronostajski R.M., Richards L.J. Specific glial populations regulate hippocampal morphogenesis. J. Neurosci. 2008;28:12328–12340. doi: 10.1523/JNEUROSCI.4000-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker-Santos D.D., Thu K.L., English J.C., Pikor L.A., Martinez V.D., Zhang M., Vucic E.A., Luk M.T., Carraro A., Korbelik J. Developmental transcription factor NFIB is a putative target of oncofetal miRNAs and is associated with tumour aggressiveness in lung adenocarcinoma. J. Pathol. 2016;240:161–172. doi: 10.1002/path.4765. [DOI] [PubMed] [Google Scholar]
- Borromeo M.D., Savage T.K., Kollipara R.K., He M., Augustyn A., Osborne J.K., Girard L., Minna J.D., Gazdar A.F., Cobb M.H. ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep. 2016;16:1259–1272. doi: 10.1016/j.celrep.2016.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brayer K.J., Frerich C.A., Kang H., Ness S.A. Recurrent fusions in MYB and MYBL1 define a common, transcription factor-driven oncogenic pathway in salivary gland adenoid cystic carcinoma. Cancer Discov. 2016;6:176–187. doi: 10.1158/2159-8290.CD-15-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.Y., Pasolli H.A., Giannopoulou E.G., Guasch G., Gronostajski R.M., Elemento O., Fuchs E. NFIB is a governor of epithelial-melanocyte stem cell behaviour in a shared niche. Nature. 2013;495:98–102. doi: 10.1038/nature11847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry A.Z., Lyons G.E., Gronostajski R.M. Expression patterns of the four nuclear factor I genes during mouse embryogenesis indicate a potential role in development. Dev. Dyn. 1997;208:313–325. doi: 10.1002/(SICI)1097-0177(199703)208:3<313::AID-AJA3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Chaudhry A.Z., Vitullo A.D., Gronostajski R.M. Nuclear factor I NFI isoforms differentially activate simple versus complex NFI-responsive promoters. J. Biol. Chem. 1998;273:18538–18546. doi: 10.1074/jbc.273.29.18538. [DOI] [PubMed] [Google Scholar]
- Chaudhry A.Z., Vitullo A.D., Gronostajski R.M. Nuclear factor I-mediated repression of the mouse mammary tumor virus promoter is abrogated by the coactivators p300/CBP and SRC-1. J. Biol. Chem. 1999;274:7072–7081. doi: 10.1074/jbc.274.11.7072. [DOI] [PubMed] [Google Scholar]
- Chen L., Kostadima M., Martens J.H., Canu G., Garcia S.P., Turro E., Downes K., Macaulay I.C., Bielczyk-Maczynska E., Coe S. Transcriptional diversity during lineage commitment of human blood progenitors. Science. 2014;345:1251033. doi: 10.1126/science.1251033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny S.K., Yang D., Chuang C.H., Brady J.J., Lim J.S., Gruner B.M., Chiou S.H., Schep A.N., Baral J., Hamard C. Nfib promotes metastasis through a widespread increase in chromatin accessibility. Cell. 2016;166:328–342. doi: 10.1016/j.cell.2016.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley A.L., Winslow M.M., Chiang D.Y., Banerji S., Stransky N., Dayton T.L., Snyder E.L., Senna S., Whittaker C.A., Bronson R.T. Nuclear factor I/B is an oncogene in small cell lung cancer. Genes Dev. 2011;25:1470–1475. doi: 10.1101/gad.2046711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelen E., Akinci U., Bryne J.C., Hou J., Gontan C., Moen M., Szumska D., Kockx C., van Ijcken W., Dekkers D.H. Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat. Genet. 2011;43:607–611. doi: 10.1038/ng.825. [DOI] [PubMed] [Google Scholar]
- Fane M.E., Chhabra Y., Hollingsworth D.E., Simmons J.L., Spoerri L., Oh T.G., Chauhan J., Chin T., Harris L., Harvey T.J. NFIB mediates BRN2 driven melanoma cell migration and invasion through regulation of EZH2 and MITF. EBioMedicine. 2017;16:63–75. doi: 10.1016/j.ebiom.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann-Morvinski D., Verma I.M. Dedifferentiation and reprogramming: origins of cancer stem cells. EMBO Rep. 2014;15:244–253. doi: 10.1002/embr.201338254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita S., Ito T., Mizutani T., Minoguchi S., Yamamichi N., Sakurai K., Iba H. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J. Mol. Biol. 2008;378:492–504. doi: 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- Geurts J.M., Schoenmakers E.F., Roijer E., Astrom A.K., Stenman G., van de Ven W.J. Identification of NFIB as recurrent translocation partner gene of HMGIC in pleomorphic adenomas. Oncogene. 1998;16:865–872. doi: 10.1038/sj.onc.1201609. [DOI] [PubMed] [Google Scholar]
- Goodall J., Carreira S., Denat L., Kobi D., Davidson I., Nuciforo P., Sturm R.A., Larue L., Goding C.R. Brn-2 represses microphthalmia-associated transcription factor expression and marks a distinct subpopulation of microphthalmia-associated transcription factor-negative melanoma cells. Cancer Res. 2008;68:7788–7794. doi: 10.1158/0008-5472.CAN-08-1053. [DOI] [PubMed] [Google Scholar]
- Grabowska M.M., Elliott A.D., DeGraff D.J., Anderson P.D., Anumanthan G., Yamashita H., Sun Q., Friedman D.B., Hachey D.L., Yu X. NFI transcription factors interact with FOXA1 to regulate prostate-specific gene expression. Mol. Endocrinol. 2014;28:949–964. doi: 10.1210/me.2013-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowska M.M., Kelly S.M., Reese A.L., Cates J.M., Case T.C., Zhang J., DeGraff D.J., Strand D.W., Miller N.L., Clark P.E. Nfib regulates transcriptional networks that control the development of prostatic hyperplasia. Endocrinology. 2016;157:1094–1109. doi: 10.1210/en.2015-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronostajski R.M. Roles of the NFI/CTF gene family in transcription and development. Gene. 2000;249:31–45. doi: 10.1016/s0378-1119(00)00140-2. [DOI] [PubMed] [Google Scholar]
- GTEx Consortium Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W., Jung E.M., Cho J., Lee J.W., Hwang K.T., Yang S.J., Kang J.J., Bae J.Y., Jeon Y.K., Park I.A. DNA copy number alterations and expression of relevant genes in triple-negative breast cancer. Genes Chromosom. Cancer. 2008;47:490–499. doi: 10.1002/gcc.20550. [DOI] [PubMed] [Google Scholar]
- Harris L., Genovesi L.A., Gronostajski R.M., Wainwright B.J., Piper M. Nuclear factor one transcription factors: divergent functions in developmental versus adult stem cell populations. Dev. Dyn. 2015;244:227–238. doi: 10.1002/dvdy.24182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho A.S., Kannan K., Roy D.M., Morris L.G., Ganly I., Katabi N., Ramaswami D., Walsh L.A., Eng S., Huse J.T. The mutational landscape of adenoid cystic carcinoma. Nat. Genet. 2013;45:791–798. doi: 10.1038/ng.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek K.S., Goding C.R. Cancer stem cells versus phenotype-switching in melanoma. Pigment Cell Melanoma Res. 2010;23:746–759. doi: 10.1111/j.1755-148X.2010.00757.x. [DOI] [PubMed] [Google Scholar]
- Hsu Y.C., Osinski J., Campbell C.E., Litwack E.D., Wang D., Liu S., Bachurski C.J., Gronostajski R.M. Mesenchymal nuclear factor I B regulates cell proliferation and epithelial differentiation during lung maturation. Dev. Biol. 2011;354:242–252. doi: 10.1016/j.ydbio.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiano A., Ebran N., Attias R., Chevallier A., Monticelli I., Mainguene C., Benchimol D., Pedeutour F. NFIB rearrangement in superficial, retroperitoneal, and colonic lipomas with aberrations involving chromosome band 9p22. Genes Chromosom. Cancer. 2008;47:971–977. doi: 10.1002/gcc.20602. [DOI] [PubMed] [Google Scholar]
- Kruse U., Sippel A.E. Transcription factor nuclear factor I proteins form stable homo- and heterodimers. FEBS Lett. 1994;348:46–50. doi: 10.1016/0014-5793(94)00585-0. [DOI] [PubMed] [Google Scholar]
- Marchio C., Weigelt B., Reis-Filho J.S. Adenoid cystic carcinomas of the breast and salivary glands or 'The strange case of Dr Jekyll and Mr Hyde' of exocrine gland carcinomas. J. Clin. Pathol. 2010;63:220–228. doi: 10.1136/jcp.2009.073908. [DOI] [PubMed] [Google Scholar]
- Mellas R.E., Kim H., Osinski J., Sadibasic S., Gronostajski R.M., Cho M., Baker O.J. NFIB regulates embryonic development of submandibular glands. J. Dent. Res. 2015;94:312–319. doi: 10.1177/0022034514559129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabello L., Koster R., Moriarity B.S., Spector L.G., Meltzer P.S., Gary J., Machiela M.J., Pankratz N., Panagiotou O.A., Largaespada D. A genome-wide scan identifies variants in NFIB associated with metastasis in patients with osteosarcoma. Cancer Dis. 2015;5:920–931. doi: 10.1158/2159-8290.CD-15-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani Y., Li J., Rao P.H., Zhao Y.J., Bell D., Lippman S.M., Weber R.S., Caulin C., El-Naggar A.K. Comprehensive analysis of the MYB-NFIB gene fusion in salivary adenoid cystic carcinoma: incidence, variability, and clinicopathologic significance. Clin. Cancer Res. 2010;16:4722–4731. doi: 10.1158/1078-0432.CCR-10-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani Y., Rao P.H., Futreal P.A., Roberts D.B., Stephens P.J., Zhao Y.J., Zhang L., Mitani M., Weber R.S., Lippman S.M. Novel chromosomal rearrangements and break points at the t6;9 in salivary adenoid cystic carcinoma: association with MYB-NFIB chimeric fusion, MYB expression, and clinical outcome. Clin. Cancer Res. 2011;17:7003–7014. doi: 10.1158/1078-0432.CCR-11-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani Y., Liu B., Rao P.H., Borra V.J., Zafereo M., Weber R.S., Kies M., Lozano G., Futreal P.A., Caulin C. Novel MYBL1 gene rearrangements with recurrent MYBL1-NFIB fusions in salivary adenoid cystic carcinomas lacking t(6;9) translocations. Clin. Cancer Res. 2016;22:725–733. doi: 10.1158/1078-0432.CCR-15-2867-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollaoglu G., Guthrie M.R., Bohm S., Bragelmann J., Can I., Ballieu P.M., Marx A., George J., Heinen C., Chalishazar M.D. MYC drives progression of small cell lung cancer to a variant neuroendocrine subtype with vulnerability to aurora kinase inhibition. Cancer Cell. 2016 doi: 10.1016/j.ccell.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon H.G., Hwang K.T., Kim J.A., Kim H.S., Lee M.J., Jung E.M., Ko E., Han W., Noh D.Y. NFIB is a potential target for estrogen receptor-negative breast cancers. Mol. Oncol. 2011;5:538–544. doi: 10.1016/j.molonc.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K., Guggenheimer R.A., Enomoto T., Lichy J.H., Hurwitz J. Adenovirus DNA replication in vitro: identification of a host factor that stimulates synthesis of the preterminal protein-dCMP complex. Proc. Natl. Acad. Sci. U. S. A. 1982;79:6438–6442. doi: 10.1073/pnas.79.21.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- das Neves L., Duchala C.S., Tolentino-Silva F., Haxhiu M.A., Colmenares C., Macklin W.B., Campbell C.E., Butz K.G., Gronostajski R.M. Disruption of the murine nuclear factor I-A gene Nfia results in perinatal lethality, hydrocephalus, and agenesis of the corpus callosum. Proc. Natl. Acad. Sci. U. S. A. 1999;96:11946–11951. doi: 10.1073/pnas.96.21.11946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninkovic J., Steiner-Mezzadri A., Jawerka M., Akinci U., Masserdotti G., Petricca S., Fischer J., von Holst A., Beckers J., Lie C.D. The BAF complex interacts with Pax6 in adult neural progenitors to establish a neurogenic cross-regulatory transcriptional network. Cell Stem Cell. 2013;13:403–418. doi: 10.1016/j.stem.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Casellas L.A., Wang X., Howard K.D., Rehage M.W., Strong D.D., Linkhart T.A. Nuclear factor I transcription factors regulate IGF binding protein 5 gene transcription in human osteoblasts. Biochim. Biophys. Acta. 2009;1789:78–87. doi: 10.1016/j.bbagrm.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson M., Andren Y., Mark J., Horlings H.M., Persson F., Stenman G. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18740–18744. doi: 10.1073/pnas.0909114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierron A., Fernandez C., Saada E., Keslair F., Hery G., Zattara H., Pedeutour F. HMGA2-NFIB fusion in a pediatric intramuscular lipoma: a novel case of NFIB alteration in a large deep-seated adipocytic tumor. Cancer Genet. Cytogenet. 2009;195:66–70. doi: 10.1016/j.cancergencyto.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Piper M., Barry G., Harvey T.J., McLeay R., Smith A.G., Harris L., Mason S., Stringer B.W., Day B.W., Wray N.R. NFIB-mediated repression of the epigenetic factor Ezh2 regulates cortical development. J. Neurosci. 2014;34:2921–2930. doi: 10.1523/JNEUROSCI.2319-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrini I., Pollino S., Pazzaglia L., Conti A., Novello C., Ferrari C., Pignotti E., Picci P., Benassi M.S. Prognostic role of nuclear factor/IB and bone remodeling proteins in metastatic giant cell tumor of bone: a retrospective study. J. Orthop. Res. 2015;33:1205–1211. doi: 10.1002/jor.22873. [DOI] [PubMed] [Google Scholar]
- Rolando C., Erni A., Grison A., Beattie R., Engler A., Gokhale P.J., Milo M., Wegleiter T., Jessberger S., Taylor V. Multipotency of adult hippocampal NSCs in vivo is restricted by drosha/NFIB. Cell Stem Cell. 2016;19:653–662. doi: 10.1016/j.stem.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Rosano L., Spinella F., Bagnato A. Endothelin 1 in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer. 2013;13:637–651. doi: 10.1038/nrc3546. [DOI] [PubMed] [Google Scholar]
- Sabova L., Kretova M., Luciakova K. New insights into the role of NF1 in cancer. Neoplasma. 2013;60:233–239. doi: 10.4149/neo_2013_031. [DOI] [PubMed] [Google Scholar]
- Sawinska M., Schmitt J.G., Sagulenko E., Westermann F., Schwab M., Savelyeva L. Novel aphidicolin-inducible common fragile site FRA9G maps to 9p22.2, within the C9orf39 gene. Genes Chromosom. Cancer. 2007;46:991–999. doi: 10.1002/gcc.20484. [DOI] [PubMed] [Google Scholar]
- Semenova E.A., Kwon M.C., Monkhorst K., Song J.Y., Bhaskaran R., Krijgsman O., Kuilman T., Peters D., Buikhuisen W.A., Smit E.F. Transcription factor NFIB is a driver of small cell lung cancer progression in mice and marks metastatic disease in patients. Cell Rep. 2016;16:631–643. doi: 10.1016/j.celrep.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele-Perkins G., Plachez C., Butz K.G., Yang G., Bachurski C.J., Kinsman S.L., Litwack E.D., Richards L.J., Gronostajski R.M. The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol. Cell. Biol. 2005;25:685–698. doi: 10.1128/MCB.25.2.685-698.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer B.W., Bunt J., Day B.W., Barry G., Jamieson P.R., Ensbey K.S., Bruce Z.C., Goasdoue K., Vidal H., Charmsaz S. Nuclear factor one B NFIB encodes a subtype-specific tumour suppressor in glioblastoma. Oncotarget. 2016;7:29306–29320. doi: 10.18632/oncotarget.8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Aoki K., Chiba K., Sato Y., Shiozawa Y., Shiraishi Y., Shimamura T., Niida A., Motomura K., Ohka F. Mutational landscape and clonal architecture in grade II and III gliomas. Nat. Genet. 2015;47:458–468. doi: 10.1038/ng.3273. [DOI] [PubMed] [Google Scholar]
- Thu K.L., Becker-Santos D.D., Radulovich N., Pikor L.A., Lam W.L., Tsao M.S. SOX15 and other SOX family members are important mediators of tumorigenesis in multiple cancer types. Oncoscience. 2014;1:326–335. doi: 10.18632/oncoscience.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C.A., Odeberg J., Djureinovic D., Takanen J.O., Hober S., Alm T., Edqvist P.H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J.M., Hamsten M., Von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., Von Heijne G., Nielsen J., Ponten F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Waki H., Nakamura M., Yamauchi T., Wakabayashi K., Yu J., Hirose-Yotsuya L., Take K., Sun W., Iwabu M., Okada-Iwabu M. Global mapping of cell type-specific open chromatin by FAIRE-seq reveals the regulatory role of the NFI family in adipocyte differentiation. PLoS Genet. 2011;7:e1002311. doi: 10.1371/journal.pgen.1002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N., Jia D., Ibrahim A.H., Bachurski C.J., Gronostajski R.M., MacPherson D. NFIB overexpression cooperates with Rb/p53 deletion to promote small cell lung cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocki P.T., Izumchenko E., Meir J., Ha P.K., Sidransky D., Brait M. Adenoid cystic carcinoma: emerging role of translocations and gene fusions. Oncotarget. 2016 doi: 10.18632/oncotarget.11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing D., Bakhsh S., Melnyk N., Isacson C., Ho J., Huntsman D.G., Gilks C.B., Ronnett B.M., Horlings H.M. Frequent NFIB-associated gene rearrangement in adenoid cystic carcinoma of the vulva. Int. J. Gynecol. Pathol. 2016 doi: 10.1097/PGP.0000000000000324. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Hosono Y., Yanagisawa K., Takahashi T. NKX2-1/TTF-1: an enigmatic oncogene that functions as a double-edged sword for cancer cell survival and progression. Cancer Cell. 2013;23:718–723. doi: 10.1016/j.ccr.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Yang Z.Q., Imoto I., Pimkhaokham A., Shimada Y., Sasaki K., Oka M., Inazawa J. A novel amplicon at 9p23-24 in squamous cell carcinoma of the esophagus that lies proximal to GASC1 and harbors NFIB. Jpn. J. Cancer Res. 2001;92:423–428. doi: 10.1111/j.1349-7006.2001.tb01112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates A., Akanni W., Amode M.R., Barrell D., Billis K., Carvalho-Silva D., Cummins C., Clapham P., Fitzgerald S., Gil L., Giron C.G., Gordon L., Hourlier T., Hunt S.E., Janacek S.H., Johnson N., Juettemann T., Keenan S., Lavidas I., Martin F.J., Maurel T., Mclaren W., Murphy D.N., Nag R., Nuhn M., Parker A., Patricio M., Pignatelli M., Rahtz M., Riat H.S., Sheppard D., Taylor K., Thormann A., Vullo A., Wilder S.P., Zadissa A., Birney E., Harrow J., Muffato M., Perry E., Ruffier M., Spudich G., Trevanion S.J., Cunningham F., Aken B.L., Zerbino D.R., Flicek P. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–D716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Zhou L., Zheng L., Guo L., Wang Y., Liu H., Ou C., Ding Z. miR-365 promotes cutaneous squamous cell carcinoma CSCC through targeting nuclear factor I/B NFIB. PLoS One. 2014;9:e100620. doi: 10.1371/journal.pone.0100620. [DOI] [PMC free article] [PubMed] [Google Scholar]