Abstract
A molecular understanding of drug resistance mechanisms enables surveillance of the effectiveness of new antimicrobial therapies during development and deployment in the field. We used conventional drug resistance selection as well as a regime of limiting dilution at early stages of drug treatment to probe two antimalarial imidazolopiperazines, KAF156 and GNF179. The latter approach permits isolation of low-fitness mutants that might otherwise be out-competed during selection. Whole-genome sequencing of 24 independently-derived resistant P. falciparum clones revealed four parasites with mutations in the known cyclic amine resistance locus (pfcarl), and a further 20 with mutations in two previously unreported P. falciparum drug resistance genes, an acetyl-CoA transporter (pfact) and a UDP-galactose transporter (pfugt). Mutations were validated both in vitro by CRISPR editing in P. falciparum, and in vivo by evolution of resistant P. berghei mutants. Both PfACT and PfUGT were localized to the endoplasmic reticulum by fluorescence microscopy. As mutations in pfact and pfugt conveyed resistance against additional unrelated chemical scaffolds, these genes are likely to be involved in broad mechanisms of antimalarial drug resistance.
Introduction
Malaria is a global health menace with 3.2 billion people at risk of infection and approximately 438,000 deaths in 20151. In recent years, mounting evidence of resistance to the artemisinin component of artemisinin-based combination therapies (ACT), the current standard of care for P. falciparum malaria, has been observed2,3. As the emergence of artemisinin resistance threatens recent success in malaria control, it is critical that alternative drugs are developed.
Imidazolopiperazines are promising drug candidates with potential to aid in malaria elimination. These compounds, in particular KAF156 and the closely related probe compound, GNF179 (also known as KAF179), possess low nanomolar potency against P. falciparum liver stages, asexual blood stages and sexual stage gametocytes. KAF156 is currently in Phase II clinical trials (Fig. 1a)4–7 and could become the first new drug that can prophylactically protect against malaria, eliminate asexual blood stages that cause disease manifestations, and block transmission of intra-erythrocytic gametocytes to Anopheles mosquito vectors. Previous work7 demonstrated that KAF156 exposure can generate resistant parasites with a minimum inoculum for resistance (MIR)8 of 108 parasites, similar to other preclinical candidates that inhibit PfATP49 and PfeEF210.
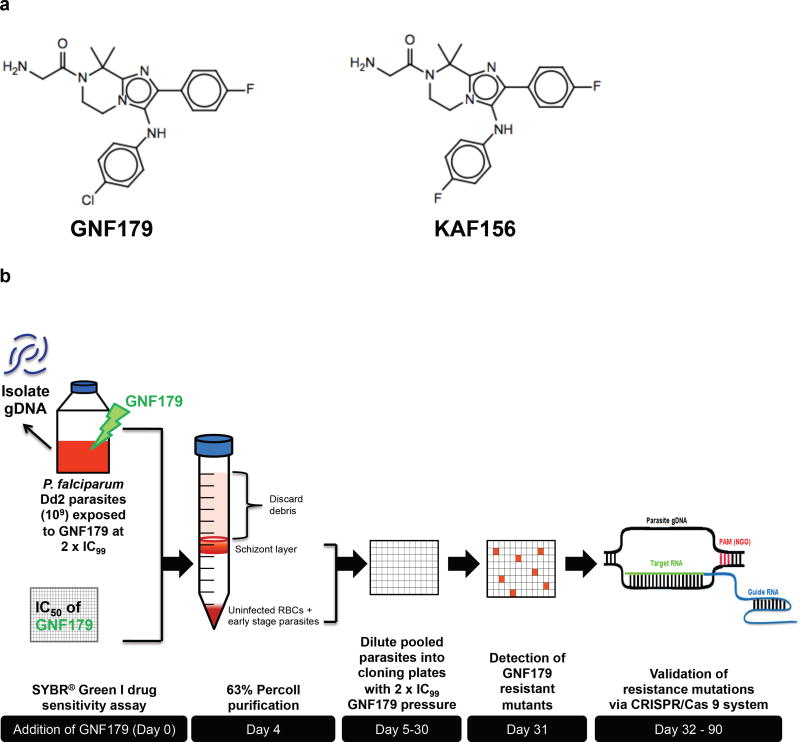
Figure 1. Selection of imidazolopiperazine-resistant mutants.
(a) Chemical structures of GNF179 and KAF156. (b) Schematic diagram showing the selection process for spontaneous GNF179-resistant mutants. Briefly, a large inoculum (109 parasites) of a clonal line of P. falciparum Dd2 was exposed to GNF179 at 2× the IC99 (38.4 nM) for two complete intraerythrocytic cycles (96 hr). After 25 days in culture, wells containing parasites were identified by Giemsa staining and light microscopy. The resistance mutations were then validated using CRISPR/Cas 9-based approaches.
Despite its potential to become a drug that can eliminate all stages of Plasmodium infection in human hosts, the mechanism of action of KAF156 remains poorly understood6,7,11,12. Previous work has shown that resistance to KAF156 can be conferred by mutations in the P. falciparum cyclic amine resistance locus (pfcarl)13. Here we describe an early dilution in vitro selection approach that enables the isolation of low-fitness resistant parasites. We use this method, as well as more conventional approaches, to discover two putative transporters14, pfugt and pfact. We also show, using genome editing, that variants of both genes confer resistance to KAF156 and GNF179 as well as to unrelated chemical scaffolds. These two transmembrane proteins constitute a previously unreported antimalarial resistance mechanism, distinct from those involving the multidrug resistance-1 (PfMDR1)15 and the chloroquine resistance transporter (PfCRT)15,16. The localization of PfUGT and PfACT to the endoplasmic reticulum offers insights into the mechanism of action of imidazolopiperazines.
Results
Selection of imidazolopiperazine-resistant mutants
To study the mechanisms of resistance to imidazolopiperazines, we employed a forward genetics approach that included aliquoting the cultures at single-parasite densities into 96-well plates immediately after selection with GNF179 (Fig. 1b). The rationale was that clonal isolation concurrent with compound exposure would allow the propagation of low-fitness mutants that might otherwise be outcompeted in a mixed culture. A total of 11 putatively resistant clones were obtained from two plates based upon this in vitro selection approach: six (A3, A9, B6, D6, E8 and B3) from plate A and five (H5, H8, A2, B5 and D8) from plate B. These initial 11 cloned lines were tested for drug sensitivity to both GNF179 and KAF156. All exhibited a dramatic (22 to 539-fold) increase in resistance compared to the parental Dd2 clone (Table 1).
Table 1. Compound susceptibility tests for GNF179-resistant P. falciparum mutants.
Mean IC50 values (nM) of mutants generated using an early dilution method, a high-concentration single-step method or a high-concentration ramp-up method against artemisinin and GNF179. Values were calculated from three biological experiments with technical duplicates (n=6) with 95% confidence intervals (C.I.).
| Artemisinin | KAF179 | |||||
|---|---|---|---|---|---|---|
|
|
||||||
| Sample | Putative Target | SNP | IC50 in nM (95% CI) | Fold Shift | IC50 in nM (95% CI) | Fold Shift |
| Early Dilution method | ||||||
| Parent Dd2 | - | - | 10.0 (7.7–13.0) | NA | 6.3 (4.7–8.6) | NA |
| Mutant H5 | PfCARL | P822L | 6.2 (3.9–9.8) | 0.6 | 274.4 (201.7–373.3) | 43 |
| Mutant D6 | PfCARL | S1076I | 9.3 (6.9–12.4) | 0.9 | 141.6 (125.2–160.1) | 22 |
| Mutant H8 | PfCARL | S1076I | 10.3 (7.0–15.3) | 1 | 185.6 (155.8–221.2) | 29 |
| Mutant A3 | PfUGT | F37V | 9.9 (7.3–13.3) | 1 | 2410 (1816–3198) | 382 |
| Mutant A9 | PfUGT | F37V | 10.6 (8.3–13.6) | 1.1 | 3074 (2749–3436) | 487 |
| Mutant B6 | PfUGT | F37V | 10.9 (8.4–14.1) | 1.1 | 3045 (2738–3387) | 483 |
| Mutant A2 | PfUGT | F37V | 8.2 (5.6–12.0) | 0.8 | 2858 (1936–4219) | 453 |
| Mutant B5 | PfUGT | F37V | 7.9 (5.3–11.8) | 0.8 | 2660 (2099–3371) | 422 |
| Mutant D8 | PfUGT | F37V | 10.9 (8.6–13.9) | 1.1 | 2210 (1710–2855) | 350 |
| Mutant E8 | PfUGT | F37V | 8.8 (6.2–12.5) | 0.9 | 3402 (2174–5324) | 539 |
| Mutant B3 | PfACT | S242* | 7.8 (6.0–10.1) | 0.8 | 2230 (1556–3196) | 353 |
| High-Concentration method | ||||||
| S1-Dd2 Parent | - | - | 10.0 (7.8–12.4) | - | 6.3 (4.2–8.3) | - |
| S1-179-Dd2-1A | PfACT | C193* | 21.7 (19.1–24.7) | 2.2 | 1898 (1492–2416) | 301 |
| S1-179-Dd2-1B | PfACT | G559R | 10.5 (8.5–13.0) | 1.0 | 1091 (854.7–1392) | 173 |
| S1-179-Dd2-2B | PfCARL | E834K | 13.7 (7.3–16.0) | 1.4 | 153.8 (137.7–171.6) | 24 |
| S1-179-Dd2-3A | PfACT | R108K | 12.3 (10.5–18.0) | 1.2 | 3141 (2594–3803) | 499 |
| S1-3D7Parent | - | - | 9.9 (6.9–13.5) | - | 3.1 (2.2–4.7) | - |
| S1-179-3D7-1B | PfACT | A94T | 7.6 (7.2–8.1) | 0.8 | 868.6 (534.4–1412) | 280 |
| S1-179-3D7-2A | PfACT | D165N | 8.9 (8.0–10.0) | 0.9 | 693.4 (451.1–1066) | 224 |
| S1-179-3D7-3A | PfACT | S110R | 7.3 (6.6–8.1) | 0.7 | 285 (130.7–619.9) | 92 |
| S1-179-3D7-3B | PfACT | S110R | 11.7 (8.1–15.9) | 1.2 | 3566 (3119–4076) | 1150 |
| Ramp-up method | ||||||
| S2-3D7Parent | - | - | 9.9 (6.9–13.5) | - | 3.1 | - |
| S2-179-3D7-1B | PfACT | Intronic | 28.8 (24.0–34.6) | 2.9 | 7962 (4152–15269) | 2568 |
| S2-179-3D7-2A | PfACT | Intronic | 30.7 (23.1–40.8) | 3.1 | 895.7 (316.2–2537) | 289 |
| S2-179-3D7-2C | PfACT | Intronic | 35.8 (27.0–47.4) | 3.6 | 3633 (2209–5975) | 1172 |
| S2-179-3D7-3A | PfACT | L253* | 27.4 (25.2–29.7) | 2.8 | 1613 (1121–2322) | 520 |
| S2-179-3D7-3C | PfACT | L253* | 29.2 (21.6–39.4) | 2.9 | 5185 (4076–6596) | 1673 |
|
| ||||||
| CRISPR lines | ||||||
|
| ||||||
| Dd2 Parent | - | - | 17.9 (13.1–24.4) | 5.1 (3.5–7.4) | NA | |
| CRISPR #1 | PfUGT | F37V | 18.6 (11.6–29.8) | 1 | 926.4 (707.4 to 1213) | 183 |
| CRISPR #2 | PfACT | S242* | 13.6 (9.6–19.3) | 0.8 | 4800 (2677 to 8604) | 947 |
Genotypic characterization of imidazolopiperazine-resistant mutants
Whole-genome sequencing was performed on the parental P. falciparum Dd2 clone and the 11 resistant clones selected with GNF179. These 12 genomes yielded 2.38×108 reads of 100 bp with 86% mapping to the P. falciparum reference genome. An average coverage of 59× was obtained with an average of 96.8% of the genome covered by five or more reads. No obvious copy number variants (CNV) were observed in any of the selected lines. Comparison of the called variants in the evolved lines to the reference Dd2 revealed that three lines had acquired single nucleotide variants (SNVs) in pfcarl (PlasmoDB ID: PF3D7_0321900) (Table 1), corroborating previous work that identified pfcarl as a gene associated with resistance to imidazolopiperazines7,12. Two out of the three pfcarl-mutated lines (D6 and H8) bore the same S1076I substitution, whereas pfcarl clone H5 encoded a P822L substitution. Interestingly, the remaining eight resistant lines possessed mutations in two genes not previously associated with imidazolopiperazine resistance, namely an UDP-galactose transporter (pfugt) (PlasmoDB ID: PF3D7_1113300) and an acetyl-CoA transporter (pfact) (PlasmoDB ID: PF3D7_1036800) (Fig. 2a). All seven lines mutated in pfugt possessed the same phenylalanine to valine change at position 37 (F37V). One line (B3) acquired a stop-gain mutation at position 242 of pfact (Table 1).
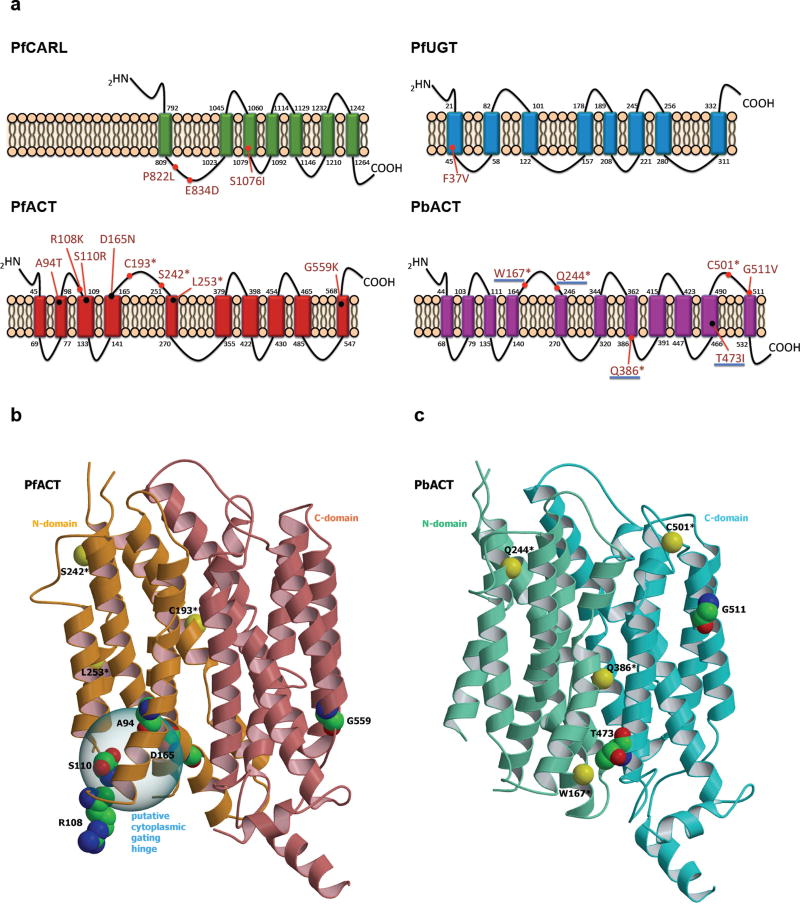
Figure 2. Characterization of resistance mutations.
(a) Annotated resistance mutations of PfCARL, PfUGT, PfACT and PbACT, with membrane topology predicted using TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). Coloured rectangle: transmembrane domain, PfCARL: P. falciparum Cyclic Amine Resistance Locus protein (PF3D7_0321900), PfUGT: P. falciparum UDP-galactose transporter (PF3D7_1113300), PfACT: P. falciparum acetyl-CoA transporter (PF3D7_1036800) and P. berghei acetyl-CoA transporter (PBANKA_051980). Underlined pbact mutations (W167*, Q244*, Q386* and T473I) indicate heterozygous mutations in polyclonal population of mutants. (b) Ribbon view of PfACT 3D model. The N- and C-terminal domains are colored in orange and coral, respectively. Residues of the resistance mutations obtained are labeled and shown with carbon in green, oxygen in red and nitrogen in blue. The carbon α of stop-gain mutations are represented as yellow ball and labeled. The putative cytoplasmic gating hinge (see text) is indicated. (c) Ribbon view of the PbACT 3D model. The N- and C-terminal domains are colored in aquamarine and cyan respectively. Residues of the resistance mutations are labeled and shown with carbon in green, oxygen in red and nitrogen in blue. The carbon α of stop-gain mutations are represented as yellow balls and labeled. The pictures (b) and (c) was obtained using MolScript56 and Raster3D57 programs successively.
The resistant clones selected with our early dilution cloning method were more resistant than the previously generated resistant clones to KAF156 and GNF179 bearing mutations in pfcarl7,12. Clones from previously reported imidazolopiperazine resistance studies were generated using a step-wise approach in which selection was stopped as soon as resistance emerged11. To determine whether mutations in pfact and pfugt would be found upon selection with higher concentrations of compound, we performed nine additional independent selections, using previously reported methodologies. In the first case, six single-step selections (3 each in 3D7 and Dd2 parasites) with 50 nM of GNF179 were performed, and in the second case, three independent step-wise selections were performed (in Dd2) over a period of three months until parasites resistant to 40 nM of GNF179 were recovered. Clones from these cultures were also all highly resistant to GNF179 - in some cases able to tolerate concentrations 1,000-fold greater than the IC50 of the parental line. Whole-genome sequencing to 90.4 fold coverage revealed seven additional mutations in pfact, as well as a previously observed resistance mutation (E834D) in pfcarl. Seven of the pfact SNVs were within the coding region (A94T, R108K, S110R, D165N, G559K, C183*, L253*) and two were located in the intron splice donor sequences immediately after exons 1 and 4, leading to intronic incorporation and a premature stop-gain mutation at “new codons” 78 and 198 respectively (Fig. S1). Overall, these data indicate that mutations in pfugt and pfact appeared when cultures were exposed to higher GNF179 concentrations.
In vivo selection of resistant mutants
To determine whether we would find similar mutations after in vivo growth, mice were infected with P. berghei ANKA parasites and once parasites were visible, were treated with a single GNF179 oral dose at two concentrations, 100 mg/kg or 30 mg/kg. After parasite recrudescence, mice were dosed a second time with the same concentration. Resistant parasites were inoculated into a new mouse and infected erythrocytes were collected. Eleven DNA samples (from two mice infected with the parental strain and nine mice harboring recrudescent parasites) were analyzed by whole-genome sequencing. On average, 22 million reads were collected for each sample. Of these an average of 31.1% aligned to the P. berghei ANKA reference genome (with the remaining mapping to the mouse). This sequencing yielded average coverage rates of 32.2 fold (range 6.0 to 92.9, excluding one failed sample). Alignment of the sequences and comparison to the parent reference sequence showed that six out of the seven P. berghei samples with good coverage had acquired mutations in the P. berghei homologue of pfact (PlasmoDB ID: PBANKA_051980). These changes included stop-gain mutations at positions W167, C501, Q244 and Q386, two non-synonymous changes (G511V and T473I), and an intronic change (Fig. 2a). These data suggest that mutations in the acetyl-CoA transporter may be more relevant in vivo than mutations in the cyclic amine resistance locus, and that the mechanism of resistance to imidazolopiperazines was conserved across parasite species (Fig. S2).
Resistance phenotypes of spontaneous mutants
GNF179 exhibited low nanomolar potency (5 to 9 nM) against the laboratory-adapted P. falciparum Dd2 strain that is multidrug-resistant (to chloroquine, quinine, pyrimethamine and sulfadoxine; Table 1) using a 72 hr proliferation assay17. Representative mutants in pfact, pfugt and pfcarl showed no cross-resistance with standard antimalarial drugs (Fig. 3a). The lines harboring mutations within either pfugt or pfact all demonstrated a >10-fold higher level of resistance to GNF179 than any of the observed pfcarl mutants.
Figure 3. CRISPR/Cas9 mutation validation and localization of PfUGT and PfACT.
(a) IC50 values of GNF179-resistant mutants against a panel of antimalarial compounds. (b) Schematic of the pfugt and pfact editing strategy. Cas9 and the target-specific gRNA were provided on a single hdhfr-marked plasmid. For pfugt editing (left panel), the gRNA was expressed from the T7 promoter, enabled by co-expression of T7 RNA polymerase. For pfact editing (right panel), the gRNA was expressed from the P. falciparum U6 promoter. The homologous donor sequence was provided on a plasmid expressing the blasticidin S-deaminase (bsd) selectable marker. The donor encoded the putative resistance mutation as well as silent mutations in the gRNA-binding site to prevent cleavage. (c) Comparison of GNF179 IC50 values for the drug-selected and CRISPR-generated parasite lines. (d–e) Live-cell imaging of parasites expressing (d) mRFP-PfUGT and the ER marker GFP-PfSec12, or (e) PfACT-GFP and stained with the ER marker ER-tracker red. Both PfUGT and PfACT were located in a perinuclear pattern characteristic of the parasite ER. Nuclei were stained with Hoechst 33342. Bar = 5 µm. For figure 3 (a) and (c) error bars represent standard deviation (s.d.) from three biological experiments with technical duplicates.
GNF179 exhibited micromolar potency against the seven pfugt F37V-mutant lines, a decrease in drug sensitivity in the range of 350 to 540 fold (Table 1). Despite acquiring a detrimental stop-gain mutation at position 242 in pfact, clone B3 remained viable and was highly resistant to GNF179, with an IC50 increase of 353-fold as compared to the parental strain (Table 1).
Membrane topology, homology modeling and distribution of genetic polymorphisms
TMHMM18 was used to predicted membrane topology for the PfCARL, PfUGT, PfACT and PbACT proteins (Fig. 2a). These models predicted all three proteins to be multispanning membrane proteins, with a subset of resistance mutations in PfACT and PbACT located primarily in cytosolic loops. The truncation mutations resulting from stop-gain codons in the acetyl-CoA transporter would result in the loss of several transmembrane domains and nearly half of the protein. ACT is likely a member of the Major Facilitator Superfamily (MFS), a large and ubiquitous secondary transporter family19. The crystal structure of a MFS transporter from the eukaryotic organism Piriformospora indica was used as the template for modeling PfACT and PbACT (Fig. 2b, c). All five non-synonymous amino acid substitutions (A94T, R108K, S110R, D165N, G559K) in PfACT were situated on the intracellular side of the membrane (Fig. 2b). Except for G559, the four other mutated residues were located at the site corresponding to the intracellular gate also known as the “cytosolic tunnel”20. Residue R108 belongs to the Motif-A (G-X3-(D/E)-(R/K)-X-G-[X]-(R/K)-(R/K)), a sequence motif known to play a role in intracellular gating across the MFS transporters21. In contrast to PfACT, the two non-synonymous substitutions in PbACT (G511V and T473I) localized to the C-terminus and their functional impact cannot be predicted (Fig. 2a, c). All mutated residues were located in highly conserved regions within the MFS transporters except for the G511 and G599 substitutions (Fig. S2, S3).
Mutant validation by CRISPR/Cas9 genome editing
Although mutations in the putative UDP-galactose and acetyl-CoA transporters were found at rates much higher than expected by chance, a few additional noncoding variants were found in the sequencing data. To demonstrate that pfugt and pfact mutations were sufficient to confer GNF179 resistance, we introduced the pfugt F37V and pfact S242* mutations into a wild-type Dd2 strain using the CRISPR/Cas9 system22 (Fig. 3b). Sanger sequencing of both edited loci revealed the presence of the resistance mutations as well as the silent mutations engineered into the Cas9-cleavage sites (Fig. S4). Introduction of the F37V mutation into the pfugt gene resulted in a 183-fold increase in resistance to GNF179 (926.4 nM vs 5.1 nM for the parental Dd2 strain) (Table 1, Fig. 3c), similar in magnitude to the fold-shift achieved with the evolved spontaneous mutants.
Evolved mutants with premature stop codons in pfact were able to tolerate high levels of GNF179 (IC50 values between 895 nM and 7.9 uM). Introduction of the S242* truncation mutation into pfact yielded a 947-fold decrease in sensitivity to GNF179 (IC50 = 4.8 µM versus 5.1 nM for the parental Dd2 line) similar to the expected value (2.3 µM) observed with the spontaneous pfact stop-gain mutant (Table 1, Fig. 3c). As with the evolved clones, the edited clones did not show cross-resistance to artemisinin (Table 1, Fig. 3a). Taken together our results showed that the SNVs observed in pfact and pfugt were sufficient to confer resistance to GNF179.
The proteins encoded by both pfugt and pfact are predicted by the Malaria Parasite Metabolic Pathways database23 to reside on the ER/Golgi membrane. To localize PfUGT, we generated a transgenic parasite line expressing a second copy of pfugt fused with mRFP. Live cell imaging revealed a perinuclear distribution of mRFP-PfUGT characteristic of the ER. This localization was confirmed by co-transfection of an ER marker, GFP-PfSec1224 which showed nearly perfect co-localization with mRFP-PfUGT (Fig. 3d). Live cell imaging also revealed a similar subcellular distribution of PfACT-GFP, which co-localized with the ER-specific cellular stain ER-tracker red (Fig. 3e).
PfUGT and PfACT mutations possess an associated fitness cost
One feature of the early dilution selection protocol (Fig. 1b) is that the serial dilution is upfront, limiting the probability of having fitter clones outgrow weak ones. We observed, during routine culturing, that parasites encoding a pfugt or pfact mutation grew at a slower rate compared to the parental strain or the pfcarl mutant lines. To document growth rates more quantitatively, we performed continuous culturing of mixed ratios of mutant and parental parasites over a period of ten generations. This competitive growth experiment revealed that pfugt-F37V and pfact-S242* mutants possessed a higher fitness cost compared to wild-type Dd2 (Fig. 4a–b). We performed individual growth kinetic analysis using flow cytometry to monitor the parasitemia of these lines in the presence and absence of GNF179. In the absence of compound pressure, the growth rates of the pfugt-F37V and pfact-S242* lines were similar to each other (Fig. 4c). The presence of GNF179 imposed a stress on the growth of the mutant lines, with parasitemia decreasing to a third to half that of the drug-free counterparts (Fig. 4c). Regardless of the presence of the GNF179, both the pfugt-F37V and the pfact-S242* mutants had impaired growth compared to the parental Dd2 strain. Despite this fitness cost, representative resistant lines subsequently cultured in the absence of GNF179 for four months retained similar levels of resistance, indicating these mutations still permit stable parasite growth in vitro (Fig. 4d). Examination of Giemsa-stained blood smears of the mutant parasite lines during routine culture, maintained in the presence of 38 nM GNF179 to prevent possible reversion, revealed morphological changes in both the pfugt and pfact mutant isolates, in particular during the late trophozoite to schizont stages (Fig. 4e). This suggests that PfUGT and PfACT could play an important role in late trophozoite and schizont development.
Figure 4. GNF179-resistant mutants display poor fitness.
(a–b). Competitive growth kinetics of (a) PfUGT mutant and (b) PfACT mutant, showing the ratio of the PfUGT or PfACT mutants versus the parental Dd2 parental strain over ten generations, as quantified using droplet digital polymerase chain reaction (DDPCR). Three different starting ratios of mutant:WT (1:10, 1:1 and 10:1) yielded similar results, with the WT line outgrowing the mutant lines. Error bars (s.e.) represent technical duplicates for a biological experiment. (c) Growth at 96 hr, expressed as mean parasitemias of the PfUGT and PfACT mutants, or the WT Dd2 strain, when grown in independent flasks, either in the absence of drug pressure (n = 4) or in the presence of GNF179 at 2× WT IC99, i.e. 38.4 nM (n = 2) for a biological experiment. Parasitemias were measured using flow cytometry. % parasitemias were significantly different in the presence of GNF179 for the PfUGT mutant (*p = 0.0166 < 0.05, t-test, two-sided) and PfACT mutant (**p = 0.0050 < 0.01, t-test, two sided), despite both mutants having an IC50 >50-fold higher than the concentration used. (d) PfUGT and PfACT mutants cultured in the absence of GNF179 for four months demonstrated no phenotypic reversion in drug susceptibility (p > 0.05) (n = 4). (e) Giemsa-stained morphology of PfUGT and PfACT mutants (GNF179 IC50 >2 µM for both lines), cultured in the presence of 38.4 nM GNF179 as in (c). Mutant parasites appeared less organized in the distribution of merozoites in the schizont stage but ring and early trophozoite stages remained similar to that of the wild-type strain. Bar = 5 µm. For Figure 4 (c) and (d) error bars represent the standard error (s.e.).
Cross-resistance against a panel of imidazolopiperazine analogs
Given that both pfugt and pfact appear to be transporters based upon their sequence homologies, we hypothesized that GNF179 resistance might be due to transporter activity, rather than representing direct targets of GNF179. To test this hypothesis, we examined whether the mutant forms of these genes also conferred resistance to additional chemical compounds. We first evaluated the pfugt-F37V and pfact-S242* evolved resistant lines for cross-resistance against a panel of 62 imidazolopiperazine chemical analogs. These analogs incorporated various substitutions at the R1 to R5 positions on the imidazolopiperazine core (Fig. 5a). Twenty-six imidazolopiperazine analogs were inactive on the wild-type strain and proved equally inactive against the pfugt and pfact mutants. A bulky substitution on both R1 and R2 generally resulted in lack of activity (IC50 > 10 µM) against all tested strains. Among the 36 analogs with activity against wild-type parasites, none could overcome the resistance phenotype of the pfugt and pfact mutant lines, although some variability was observed in the overall level of resistance. The stop-gain (S242*) mutation in the acetyl-CoA transporter rendered this parasite highly cross-resistant to all the chemical analogs tested, except for compound 13 and KAF156 that retained relatively good sub-micromolar potency (Fig. 5a). We note that the pfact-S242* mutant was slightly more sensitive to KAF156 than to GNF179, with a lower fold-shift in the 10–100× range.
Figure 5. Cross resistance against a panel of imidazolopiperazine analogs and unrelated antimalarial compounds.
(a) Heat maps of biological activity (pIC50) of 62 imidazolopiperazine analogs (median of two biological experiments with technical duplicates) against GNF179-resistant mutants. No association was observed between R-group descriptors (volume and lipophilicity) and the pIC50 activity profiles (defined as the -log10(IC50)) in the heat maps. (b) Structures of non-imidazolopiperazine scaffolds. (c) Activity profiles of non-imidazolopiperazine scaffolds in terms of fold-change in IC50 compared to wild type. Each colour represents a different compound class. The light grey lines correspond to compounds that show no shift in the mutants. Three biological experiments with technical duplicates were performed. Error bars represent standard deviation (s.d.).
Mutant lines show cross-resistance against non-imidazolopiperazine scaffolds
We next tested whether the pfugt-F37V (mutants A9 and B5) and pfact-S242* (mutant B3) resistant lines demonstrated cross-resistance against other antimalarials, defined as a greater than 5-fold increase in the mean IC50 value, against a panel of compounds of unrelated scaffolds (Fig. 5b–c). 69 compounds were evaluated, comprising a panel of 10 standard antimalarials and 59 additional Novartis compounds inclusive of KAF156 and GNF179. Thirteen compounds were active only or primarily against the parental strain, with evidence of cross-resistance in the mutant lines. These compounds included GNF156 and KAF179, as discussed below (Fig. 5b), and another nine that will form part of a future study. A further two compounds (compounds 5 and 6) showed increased potency against the mutant lines (Fig. 5b).
Four compounds displayed differential cross-resistance among the parasite lines. Compound 3 (Tyroscherin), a known inhibitor of insulin-like growth factor (IGF)-1 dependent growth25 and structurally unrelated to GNF179 and KAF156, demonstrated cross-resistance in both pfugt and pfact mutants with 30 to 50 fold IC50 increases (Fig. 5c). Compound 4, belonging to the oxazole class, was active against the pfact mutant but displayed a loss in potency against the pfugt mutant. In contrast, compounds 5 (KDU691) and 6, both lipid kinase inhibitors, demonstrated a modest increase in potency against both mutant lines (Fig. 5b and Table S1). Recent publications have also demonstrated pfcarl as a potential multidrug resistance mechanism13. The cross-resistance data presented here on pfugt-F37V and pfact-S242* mutants as well as on pfcarl13 suggest that these three proteins (PfCARL, PfUGT and PfACT) could participate in a common resistance mechanism.
Discussion
Although in vitro evolution and whole-genome analysis is often used to discover putative targets of small molecules identified in antimalarial cellular screens11, it is equally useful for characterizing potential pathways of resistance. In this study, we present pfugt and pfact as multidrug resistance genes in Plasmodium. Although their modification resulted in a significant loss of parasite fitness, their integrity should nevertheless be monitored in field studies involving imidazolopiperazines or other novel antimalarial compounds undergoing development that do not act against traditional antimalarial targets such as PfDHFR, β-hematin, or cytochrome bc1.
Here we provide definitive evidence that mutations in pfact and pfugt confer resistance to imidazolopiperazines, as well as other scaffolds. More work will be needed to establish their cellular role. The fact that both are predicted to encode transporters and that pfact is nonessential argues that neither is a target of KAF156. Both predicted proteins are members of the MFS26, the largest and most ubiquitous secondary transporter family responsible for the translocation of small molecules including metabolites, nucleosides, oligosaccharides, amino acids, oxyanions and drugs19. MFS transporters have been established as drug-specific or multidrug efflux pumps in bacteria and fungi27–29.
All members of the SLC35A2 (UDP-galactose translocator) family bear Pfam domains belonging to eukaryotic triose phosphate transporter (TPT), the nucleotide-sugar transporter (NST) families30,31 and drug/metabolite transporter superfamily (DMT)32. Interestingly, the P. falciparum chloroquine resistance transporter (PfCRT) is also a member of DMT33,34 but is localized to the digestive vacuole. NSTs have been associated with drug resistance in organisms such as Candida albicans and Saccharomyces cerevisiae31. In humans, mutations in SLC35A2 cause a congenital disorder of glycosylation35. Although further work is required to understand the native function of PfUGT in Plasmodium, the ER localization is consistent with a function in transporting UDP-galactose from the cytosol into the ER where it may play a role in post-translational modification of proteins14,36,37.
The function of PfACT in P. falciparum is predicted only by homology. Evolutionarily it is orthologous to a human AT-1 protein (also known as SLC33A1). Heterologous expression of its cDNA in COS-1 cells allows these cells to transport radiolabeled acetyl-coA and to form O-acetylated (Ac) gangliosides38 a process that occurs in Golgi vesicles39. In addition, AT-1 regulates the acetylation status of ER-transiting proteins in human cells40 and is also associated with neurodegenerative disorders41. Many of the identified mutations are predicted to localize to the cytoplasmic side of PfACT, suggesting that they prevent KAF156 from entering the ER or Golgi.
The observations that mutations in pfugt and pfact, as well as pfcarl, which encodes a Golgi-localized protein13, convey resistance to a variety of imidazolopiperazine compounds as well as unrelated scaffolds provides clues about the mechanism of action of KAF156/GNF179 and other small molecules whose activity can be modulated through mutations in these genes. Our hypothesis is that PfACT/PfUGT may be involved in transporting compounds into the ER or Golgi, where they may act on cellular processes such as membrane trafficking or posttranslational modification. Interestingly KDU691, another antimalarial candidate that targets PfPI4K, a phosphatidlyinositol-4-kinase, affects membrane trafficking from the Golgi by depleting phosphatidylinositol-4-phosphate lipid pools42. Furthermore the PfPI4K ortholog in Saccharomyces, Pik1, is localized to the Golgi where it regulates secretion, and yeast temperature-sensitive Pik1 mutants can be complemented by the Plasmodium enzyme43,44. The cellular targets of other small molecules, including KAF156 are not known but tyroscherin contains a hydrocarbon tail and in C. albicans down-regulation of ergosterol biosynthesis genes sensitize cells to its activity45. Although our dataset is small it is interesting to note that compounds that seem to be affected by these pathways are more likely to have activity in other stages of the lifecycle—both KAF156 and KDU691 have potent activity against stage V gametocytes and parasite liver stages, indicating that they do not act through hemoglobin digestion, or DNA replication. These data suggest that pfact and pfugt could become more important as drug discovery efforts shift away from compounds that act mostly against asexual blood stage parasites, for example, by inhibiting hemoglobin digestion.
Although the dramatic decrease in drug sensitivity caused by these mutations raises concerns that imidazolopiperazines might succumb to drug resistance in the field, parasites that harbor mutations in these genes proliferate at a reduced rate. These low fitness mutants might not survive the competition for nutrients in a polyclonal environment such as the human body, or may be more readily cleared by the immune system. Furthermore, while pfact is not essential for blood stages, it could be essential for other stages of the lifecycle and the mutations might not be transmitted, as has been recently shown for atovaquone resistance mutations46. Nevertheless, it will be critical to select an adequate combination partner for KAF156 as it enters phase III clinical trials.
Materials and Methods
P. falciparum culture
The parental P. falciparum Dd2 strain and all GNF179-resistant mutants were cultured under standard conditions47. The Dd2 strain is resistant to chloroquine, quinine, pyrimethamine and sulfadoxine and replicates at a rate of 5–6 fold per 48-hr generation in vitro48. Single donor human O+ whole blood (Innovative Research) was obtained from Novi, Mississippi. Leukocyte-free erythrocytes were stored at 50% hematocrit in RPMI-1640 washing media.
Determination of parasitemia
A thin blood smear was fixed with 100% methanol (Fisher Scientific) for one min before staining in 10% v/v Giemsa (Sigma-Aldrich) in phosphate-buffered saline for 15 minutes. Giemsa is used to visually distinguish P. falciparum parasites from surrounding uninfected erythrocytes. Parasitemia was determined from Giemsa-stained slides under the light microscope (Nikon YS100) with 100× magnification.
Selection of imidazolopiperazine-resistant P. falciparum mutants
A clonal P. falciparum Dd2 parasite line was cultured at 37°C in complete RPMI media at 4% hematocrit in an incubator with reduced oxygen environment47. The half maximal inhibitory concentration (IC50) of GNF179 was determined to be 9.0 nM using a 72 hr SYBR® Green I drug sensitivity assay17. Prior to selection, an aliquot of the parental line was stocked as a reference for subsequent whole-genome sequencing analysis. A single-step selection for pre-existing resistant mutants was employed by the addition of GNF179 at greater than fully inhibitory concentration (2 × IC99, 38.4 nM) to a population of 109 parasites (Fig. 1b). Excessive debris produced by the death of drug-sensitive parasites was removed using a 63% Percoll purification step49, performed four days post exposure to GNF179. The schizont-containing interface and pellet fraction containing ring and trophozoite-stage parasites were pooled together and re-suspended in 100 mL of culture media, adjusted to 2.5% hematocrit, dispensed into 96-well plates and maintained under 2 × IC99 drug pressure for 25 days. Negative control wells that contained culture at 2.5% hematocrit without any parasite were also present in each plate. Parasites were kept under constant drug pressure with replacement of complete media, washed blood and drug every other day. After 25 days in culture, mutants were selected microscopically using Giemsa-stained slides.
Additional selections were performed using either the traditional “ramp-up” selection method or the high-concentration selection method. For the ramp-up selection method, representing selection S2 in Table 1, 3D7 strain parasites were cultured under normal conditions while simultaneously treated with gradually increasing doses of GNF179 for 2 months (starting at 5 nM and ending at 40 nM), with cultures monitored via Giemsa-strained thin smears every day, until parasite cultures exhibited at least 5× resistance versus wild-type. Parasites were cloned by limiting dilution and DNA was extracted for whole-genome sequencing. For the high-concentration selections (selection S1 3D7 and Dd2 in Table 1), both 3D7 and Dd2 parasites were exposed to 50 nM GNF179 for 10 days. GNF179 was removed and parasite cultures were allowed to recover under normal culturing conditions. Parasites were then cloned via limiting dilution.
Isolation and determination of splicing variants in pfact
Both wild-type 3D7 and S2-3D7-1B and S2-3D7-2C parasites were cultured as indicated above. Parasites were washed with 1× cold PBS, treated with 0.15% saponin, washed twice more with 1× cold PBS, and total RNA extracted from asynchronous parasites using the Qiagen RNeasy mini kit. cDNA was synthesized using random hexamers and superscript II (Life technologies) at 42°C for 2 hours. Regions of PfACT were amplified with primers indicated in Table S3 via PCR, then examined via Sanger sequencing.
Selection of imidazolopiperazines-resistant P. berghei-ANKA mutants
The P. berghei ANKA strain was a gift from Dr. C. Jensen, Leiden University Medical Center, Netherlands. GNF179-resistant P. berghei ANKA mutants were selected in 4–8 week old female BALB/c mice (n=18). Briefly, 10 mice were infected through the tail vein with 107 P. berghei ANKA parasites from a single batch and treated with either 30 mg/kg (n=4) or 100 mg/kg (n=4) of GNF179 or were untreated (n=2). Parasites extracted from all 8 treated mice were re-infected into 8 naïve mice for propagation and isolation of infected reticulocytes for DNA preparation. For the purpose of in vivo mutant selection of P. berghei parasites we did not require a statistically significant number of animals. Animals were neither randomized nor blinded. Parasites were enumerated by thin blood smear and staining in 10% Giemsa as described above. The formulation in terms of v/v used for GNF179 was 10% ethanol, 30% PEG400 and 60% Vitamin E TPGS (10% solution) and was administered by gavage. The first treatment was initiated at a level of parasitemia reached 4–10%, left to drop below the limit of detection and repeated only once recrudescent parasites reached once again 4–10% parasitemia. This was repeated until the level of parasitemia no longer decreased. Surviving parasites were transfused into naive mice under either 30 or 100 mg/kg daily treatment with GNF179 and the growth kinetics monitored every two days by sampling and Giemsa staining. Parasites were harvested by cardiac puncture at 4% parasitemia, passed through a filter to remove white blood cells prior to DNA isolation and sequencing as described for P. falciparum. Ethical approval for the study was obtained from the Animal Care and Use Committee at Novartis Institute for Tropical Diseases.
Drug sensitivity assay using SYBR® Green I
Drug susceptibility was measured using the malaria SYBR® Green I-based fluorescence assay17. Each compound was tested in duplicate on a ten-point concentration curve prepared by a three-fold dilution from 10 µM to 0.5 nM. At least five independent experiments were carried out for IC50 determinations of KAF156 and GNF179-resistant mutants. IC50 values were calculated using a nonlinear regression curve fit in GraphPad Prism 5.0 (California, USA).
Parasite extraction and genomic DNA isolation
Cultures were scaled up to at least 4–5% parasitemia in 100 mL of RPMI media at 2.5% hematocrit prior to parasite extraction from the red blood cells. The cultures are reconstituted to 10% hematocrit with fresh RPMI media and transferred to a 15 mL falcon tube. Lysis buffer (1.5% w/v saponin and 1.0% w/v bovine serum albumin fraction V in PBS) was added on ice in 10 pellet volumes. Upon lysis of red blood cells, indicated by a clear red supernatant, the lysed cultures were centrifuged at 2700 × g for 5 minutes at 4°C. The supernatant was removed by aspiration and the cells were washed twice using chilled PBS in microcentrifuge tubes. Cell pellets were stored in −80°C until genomic DNA was isolated using a Blood and Cell Culture DNA mini extraction kit (Cat. No. 13323, Qiagen).
Whole-Genome Sequencing
DNA libraries for each gDNA sample were prepared for sequencing using the Nextera XT kit (Cat. No. FC-131-1024, Illumina), following standard dual index protocols. Libraries were clustered and run on the Illumina HiSeq 2500 using the RapidRun mode, sequencing 100 base pairs in depth on either end. Paired-end reads were either aligned to the P. falciparum 3D7 reference genome (PlasmoDB v. 11.0) or the P. berghei ANKA reference genome (PlasmoDB v 9.0), as previously described50. Because P. berghei samples were unable to be cloned using limiting dilution methods, the sample analysis for these samples included mutations with mixed read ratios (pbact mutations W167*, Q244*, Q386* and T473I). To identify possible CNVs, interval depth for each gene was calculated using GATK DiagnoseTargets. To reduce noise levels, interval depths were normalized across each sample, and then calibrated against four 3D7 parent samples sequenced previously in our laboratory. Finally, samples were compared to their respective parents, and normalized interval depth ratios ≥2 were flagged. All SNV were visually confirmed using the Broad Institute web-based sequence browser IGV. The NCBI short read archive accession number, for the P. falciparum samples isolated using the early limiting dilution as well as several P. berghei mutant lines, is SRP068203. The higher dose ramp-up and high selection mutants resistant to GNF179 were uploaded under the study accession number SRP072010. The additional P. berghei samples, which were treated with 30 or 100 mg/kg as well as their parent, were uploaded under the study accession number SRP075559. Files are named according to Table 1.
CRISPR/Cas9 Genome Editing
Individual mutations that were identified via in vitro selection were confirmed by introducing those mutations into the parental Dd2 line using a CRISPR/Cas9 system. Briefly, Cas9 was expressed from a pDC2-based human dhfr plasmid, along with a sequence encoding the specific guide RNA. Expression of the latter was driven either by a short T7 promoter (for pfugt), via co-expression of the T7 RNA polymerase from a pDC2-bsd plasmid, or via a U6 promoter (pfact)51. A donor template with homology to the target site was also supplied on a pDC2-bsd plasmid, and contained both the desired nucleotide replacement and also silent mutations in the Cas9 cut site to prevent cleavage of the donor or the modified genome. These plasmids were electroporated (310kV, 950uF) into sorbitol-synchronized ring-stage Dd2 parasites. Parasites were first selected for 6 days with 1.5 nM WR99210 and 2 µg/ml blasticidin, and then with 5 × IC50 GNF179. The time to recover polyclonal parasite lines was approximately 30 and 60 days for pfugt and pfact respectively. Transformed parasite lines were then cloned by limiting dilution. Editing was validated by PCR and Sanger sequencing. Validated parasite lines with the desired mutation were then assayed for resistance to GNF179 using the SYBR® Green I assay described previously.
The guide RNA sequence (listed as gRNA+PAM) used for PF3D7_1113300 was GATTTTACAAGTACTTGTGTAGG, while the guide RNA sequence for PF3D7_1036800 was GAAAAAGTTTTTAAAATTCTGG. Donor templates consisted of the full-length coding region (1032 bp) for the UDP-galactose transporter, and a 1 kb fragment centered on the mutation for the Acetyl-CoA transporter. Mutations in the donor template, both for the gRNA target site and the desired SNV, were introduced via the Quikchange II kit (Agilent Technologies). Primers for the donor templates are indicated in Table S3.
Cross resistance studies
The P. falciparum Dd2 parental strain and 11 resistant mutants obtained with the early dilution method were tested for cross-resistance against 62 imidazolopiperazine chemical analogs and 10 standard antimalarial drugs using the SYBR® Green I drug sensitivity assay as described above. Representative resistant lines were tested for cross-resistance against a panel of 59 compounds13 of various scaffolds using the SYBR® Green I drug sensitivity assay as described above. For the chemogenomics analysis, the median IC50 value was used for mutants that shared the same SNV. The biological activity data were converted to pIC50 values and fold-shifts were calculated for the mutants relative to the wild type. The data were clustered using software R 3.2.0 using the following method: hclust (d, method = "complete", members = NULL). This method finds similar clusters with Euclidean distance between activity profiles.
Competitive growth kinetics of resistant mutants
P. falciparum Dd2 parental (WT) parasites were mixed separately with two mutant lines harboring mutations in the UDP-galactose transporter (Mutant A9, F37V) or acetyl-CoA transporter (Mutant B3, S242*). Mutant: WT ratios were started at 1:10, 1:1 and 10:1. The mutant and WT mixed cultures were adjusted to have an initial parasitemia of 1% in a 12 mL culture at 4% hematocrit. The parasites were passaged in a 1 in 6 dilution every 2nd day for 10 generations. Genomic DNA was also harvested every other day using the QIAamp DNA Blood Mini Kit (Cat. No.51106, Qiagen). The genomic DNA was subsequently used for digital PCR to amplify an amplicon between 75–150 base pairs within the target of interest containing the SNV. The results from droplet digital PCR (DDPCR; see below) were used to analyze the frequencies of WT and mutant strain alleles, which in turn reflects the growth kinetics of the WT and mutant strains (Fig. 4a, b).
Individual growth kinetics of resistant mutants
P. falciparum Dd2 WT and three mutant lines (see preceding paragraph) were synchronized with 5% v/v sorbitol and MACS purification and adjusted to an initial parasitemia of 0.1% and hematocrit of 3%. Quadruplicates were performed for mutant lines in the absence of GNF179 and replicates for mutant lines in the presence of 38.4 nM GNF179. Six replicates of the parental Dd2 strain were cultured in the absence of drug. A 20 µL sample was taken daily for flow cytometry analysis throughout the course of the experiment, namely at 0 hr, 24 hr, 48 hr, 72 hr and 96 hr. Media was changed at 24, 48, 60, 72 and 84 hr. Each sample was stained with 5 µg/ml dihydroethidium (Sigma) and 8 mM of Hoechst 33342 (Sigma) as previously described52. Flow cytometry analysis used FlowJo software (Tree Star). Two-sided t-tests at α=0.05 with equal variances were performed to compare parasitemia ± GNF179. F-tests were performed to test for equality of variance (PfUGT: p = 0.375 > 0.05, PfACT: 0.230 > 0.05).
Droplet Digital PCR experiments (DDPCR)
DDPCR reactions were performed with 2 ng of genomic DNA template with 2× droplet PCR supermix for probes (no dUTP). The mix was supplemented with an additional 50 µM dATP/dTTP. Primer and probe concentrations were 900 nM and 250 nM, respectively. Primers and probes were synthesized by Integrated DNA Technologies. Wild type probes were 5’-HEX labeled and mutant probes were 5’FAM labeled. Both probes were 3’-labeled with an Iowa Black Quencher. Emulsions were generated in the DDPCR droplet generator and then transferred to an Eppendorf TwinTec PCR plate and sealed with a pierceable foil seal. Emulsions underwent 40 cycles of amplification in a Bio-Rad C1000 under the following condition: 95°C for 10 minutes, a two-step amplification at 94°C for 30 seconds and annealing temperature for 1 minute for 40 cycles with a 2.5°C/s ramp rate, 98°C for 10 minutes, and finally a 12°C hold. For the UDP experiments, extension time was increased to 1.5 minutes. Optimal annealing temperatures determined by gradient PCR were 57.6°C, 54°C, and 60°C for pfugt, and pfact, respectively. Droplets were read on a QX150 reader. Droplet populations were manually gated into double-negative, single-positive, and double-positive populations using the QuantaSoft software.
Morphology of resistant mutants
The morphology of the resistant mutants was examined and imaged using Giemsa-stained slides under light microscopy (Nikon TS100 and DS-Fi2 camera) with 100× magnification. The cultures were under drug pressure (38.4 nM GNF179).
Phenotypic reversion of mutations to wild-type
All 11 resistant mutants were cultured without drug pressure for 4 months. Phenotypic reversion was defined by a change in IC50 values and morphology. IC50 assays using the SYBR® Green I drug sensitivity assay were carried out at the 2nd and 4th month to evaluate changes in IC50 values. Two-sided t-tests at α=0.05 with unequal variances were performed to compare IC50 at 2nd and 4th month. F-tests were performed to test for equality of variance (PfUGT: p = 0.778 > 0.05, PfACT: 0.03 < 0.05).
Localization of the UDP-galactose and the Acetyl CoA transporter
The full-length coding sequence for pfugt (PF3D7_1113300) was amplified from genomic DNA using primers p4794 / p4795 (see Table S3) and subcloned into a pDC2-based expression vector downstream of the P. berghei EF1alpha promoter and mRFP, yielding the expression plasmid pDC2-PbEF1pro-mRFP-PfUGT-attP-BSD. This expression plasmid was transfected into the NF54-attB parasite line together with the pINT plasmid to achieve attP×attB integrase-mediated recombination53. A plasmid (pDC2-GFP-Sec12-hDHFR) expressing the ER marker, GFP-PfSec12 was co-transfected with mRFP-PfUGT and pINT, and the parasites were selected with 2.5 nM WR99210 / 2 µg/ml blasticidin / 250 µg/ml G41824. In a similar manner to PfUGT, the full-length coding sequence for pfact (PF3D7_1036800) was amplified from genomic DNA using primers indicated in Table S3 and subcloned into a pDC2-based expression vector downstream of the P. falciparum calmodulin promoter and downstream of GFP, yielding the expression plasmid pDC2-PfCAMpro-PfACT-GFP-attP-BSD. This plasmid was transfected into the Dd2-attB parasite line together with the pINT plasmid to yield attP×attB integrase-mediated recombination, and selected for as above53. The ER was stained with ER-tracker-red at 1ug/ml for 15 min. Cultures were then washed twice with 1× PBS and imaged.
Fluorescence microscopy imaging
Live-cell imaging of parasites expressing GFP- and mRFP-tagged proteins was performed on a Nikon TiE PFS inverted microscope equipped with a CoolSNAP HQ2 monochrome camera. Aliquots (100 µL) of parasite culture were washed once with RPMI-1640 media lacking Albumax II, transferred to MatTek poly-l-lysine coated glass-bottom culture dishes, and overlaid with 1 ml of media containing 2 µg/ml of the nuclear stain Hoechst 33342 (Sigma).
PfACT and PbACT homology modeling
Considering similarities in protein length (> 500 residues) and in sequence (33%), the MFS phosphate transporter from the fungi Piriformospora indica (PDB id 4J05) was used as the 3D template to model PfACT and PbACT. The sequence alignment between PfACT and the template was calculated using ClustalW, followed by manually refining gaps based on the transmembrane regions both observed in the crystal template structure and predicted using TMpred server (http://www.ch.embnet.org/software/TMPRED_form.html). The alignment is shown in Fig. S2. In addition 846 sequences similar to PfACT were retrieved by BlastP, and aligned automatically with ClustalW and refined manually. The WebLogo server (http://weblogo.berkeley.edu/logo.cgi) was used to plot the amino acid distribution in a 5-residue window around the mutated residue in PfACT and PbACT as shown in Fig S3. The conservation score were also shown below each position. These score were computed with AL2CO program54 and were linearly rescaled from 0 (no conservation at this position) to 9 (highest conservation in the alignment). 3D models of PfACT and PbACT were constructed with MODELLER-9v1655. Among the 20 models generated for each of both proteins, the one with the lowest energy was selected as the final model.
Data Availability
All evolved GNF179 and GNF156 resistant strains will be made available upon request to P.B. (early limiting dilution evolved Dd2 strains, including those used for all assays, and all P. berghei strains) and/or E.A.W. (engineered Dd2 parasite lines and all GNF179-resistant strains obtained by previously reported methods). All relevant whole-genome sequencing data have been uploaded to Accession numbers SRP068203 (P. berghei and early limiting dilution P. falciparum), SRP072010 (the higher dose ramp-up and high selection GNF179-resistant P. falciparum clones) and SRP075559 (additional P. berghei samples, which were treated with either 30 or 100 mg/kg GNF179), as indicated in the methods section. The authors declare that all other data supporting the findings of this study are available within the article and its supplementary information.
Supplementary Material
Acknowledgments
M.Y.L. is supported by the Economic Development Board-Industrial Postgraduate Programme (EDB-IPP) scholarship. G.L. is supported by an A.P. Giannini Post-Doctoral Fellowship. Work at UCSD was supported by grants from NIH (R01 AI090141 and R01 AI103058) to EAW. DAF gratefully acknowledges support from the Medicines for Malaria Venture. B.M. and L.R. are supported by Singapore Immunology Network under the Agency for Science, Technology and Research (A*STAR, Singapore). R.W. is a Research Associate at the National Fund for Scientific Research FNRS-FRS (Belgium). The authors would like to thank Dr. C. Jensen (Leiden University Medical Center, Netherlands) for the donation of P. berghei ANKA strain.
Footnotes
Supplementary information is available online.
Author Contributions
MYL, GL, MCSL, EAW and PB designed the experiments. MYL, GL, MCSL, CR, BH, VC, BT, AC, MN, BM, EDC and LL performed the experiments. Modeling work was performed by RW. MYL, GL, MCSL, CR, VC, MN, EC and PG analyzed the data. GB, PH, LR and DAF contributed support. MYL, GL, MCSL, BY, DF, EAW and PB wrote and proofread the manuscript. EAW and PB gave technical support and conceptual advice. The manuscript was edited by all authors.
Competing Financial Interests
The authors declare no competing financial interest.
References
- 1.WHO. World Malaria Report 2015. WHO Press; 2015. pp. 1–280. [Google Scholar]
- 2.Dondorp AM. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashley EA, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagle A, et al. Imidazolopiperazines: lead optimization of the second-generation antimalarial agents. J. Med. Chem. 2012;55:4244–4273. doi: 10.1021/jm300041e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu T, et al. Imidazolopiperazines: hit to lead optimization of new antimalarial agents. J. Med. Chem. 2011;54:5116–5130. doi: 10.1021/jm2003359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leong FJ, et al. A first-in-human randomized, double-blind, placebo-controlled, single- and multiple-ascending oral dose study of novel Imidazolopiperazine KAF156 to assess its safety, tolerability, and pharmacokinetics in healthy adult volunteers. Antimicrob. Agents Chemother. 2014;58:6437–6443. doi: 10.1128/AAC.03478-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhen KL, et al. KAF156 is an antimalarial clinical candidate with potential for use in prophylaxis, treatment, and prevention of disease transmission. Antimicrob. Agents Chemother. 2014;58:5060–5067. doi: 10.1128/AAC.02727-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding XC, Ubben D, Wells TN. A framework for assessing the risk of resistance for anti-malarials in development. Malaria J. 2012;11:292. doi: 10.1186/1475-2875-11-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jimenez-Diaz MB, et al. (+)-SJ733, a clinical candidate for malaria that acts through ATP4 to induce rapid host-mediated clearance of Plasmodium. Proc. Natl. Acad. Sci. U S A. 2014;111:E5455–5462. doi: 10.1073/pnas.1414221111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baragana B, et al. A novel multiple-stage antimalarial agent that inhibits protein synthesis. Nature. 2015;522:315–320. doi: 10.1038/nature14451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flannery EL, Fidock DA, Winzeler EA. Using genetic methods to define the targets of compounds with antimalarial activity. J. Med. Chem. 2013;56:7761–7771. doi: 10.1021/jm400325j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meister S, et al. Imaging of Plasmodium liver stages to drive next-generation antimalarial drug discovery. Science. 2011;334:1372–1377. doi: 10.1126/science.1211936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LaMonte G, et al. Mutations in the Plasmodium falciparum Cyclic Amine Resistance Locus (PfCARL) Confer Multidrug Resistance. MBio. 2016;7 doi: 10.1128/mBio.00696-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin RE, Henry RI, Abbey JL, Clements JD, Kirk K. The 'permeome' of the malaria parasite: an overview of the membrane transport proteins of Plasmodium falciparum. Genome Biol. 2005;6:R26. doi: 10.1186/gb-2005-6-3-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valderramos SG, Fiddock DA. Transporters involved in resistance to antimalarial drugs. Trends Pharmacol. Sci. 2006;27:594–601. doi: 10.1016/j.tips.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ecker A, Lehane AM, Clain J, Fidock DA. PfCRT and its role in antimalarial drug resistance. Trends Parasitol. 2012;28:504–514. doi: 10.1016/j.pt.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson JD, et al. Assessment and continued validation of the malaria SYBR green I-based fluorescence assay for use in malaria drug screening. Antimicrob. Agents Chemother. 2007;51:1926–1933. doi: 10.1128/AAC.01607-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 19.Yan N. Structural Biology of the Major Facilitator Superfamily Transporters. Annu. Rev. Biophys. 2015;44:257–283. doi: 10.1146/annurev-biophys-060414-033901. [DOI] [PubMed] [Google Scholar]
- 20.Pedersen BP, et al. Crystal structure of a eukaryotic phosphate transporter. Nature. 2013;496:533–536. doi: 10.1038/nature12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quistgaard EM, Low C, Guettou F, Nordlund P. Understanding transport by the major facilitator superfamily (MFS): structures pave the way. Nature Rev. Mol. Cell Biol. 2016;17:123–132. doi: 10.1038/nrm.2015.25. [DOI] [PubMed] [Google Scholar]
- 22.Lee MC, Fidock DA. CRISPR-mediated genome editing of Plasmodium falciparum malaria parasites. Genome Med. 2014;6:63. doi: 10.1186/s13073-014-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ginsburg H. Malaria Parasite Metabolic Pathways. 2015 doi: 10.1016/j.pt.2015.10.003. http://mpmp.huji.ac.il/maps/ERGolgiVacuole.html. [DOI] [PubMed]
- 24.Lee MC, Moura PA, Miller EA, Fidock DA. Plasmodium falciparum Sec24 marks transitional ER that exports a model cargo via a diacidic motif. Mol. Microbiol. 2008;68:1535–1546. doi: 10.1111/j.1365-2958.2008.06250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayakawa Y, et al. Structure of tyroscherin, an antitumor antibiotic against IGF-1-dependent cells from Pseudallescheria sp. J. Antibio. 2004;57:634–638. doi: 10.7164/antibiotics.57.634. [DOI] [PubMed] [Google Scholar]
- 26.Hediger MA, et al. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteins. Pflugers Archiv : Eur. J. Physiol. 2004;447:465–468. doi: 10.1007/s00424-003-1192-y. [DOI] [PubMed] [Google Scholar]
- 27.Prasad R, Rawal MK. Efflux pump proteins in antifungal resistance. Front. Pharmac. 2014;5:202. doi: 10.3389/fphar.2014.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar S, et al. Bacterial Multidrug Efflux Pumps of the Major Facilitator Superfamily as Targets for Modulation. Infect. Disorders Drug Targets. 2016;16:28–43. doi: 10.2174/1871526516666160407113848. [DOI] [PubMed] [Google Scholar]
- 29.Dos Santos SC, Teixeira MC, Dias PJ, Sa-Correia I. MFS transporters required for multidrug/multixenobiotic (MD/MX) resistance in the model yeast: understanding their physiological function through post-genomic approaches. Front. Physiol. 2014;5:180. doi: 10.3389/fphys.2014.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aurrecoechea C, et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perlin MH, Andrews J, Toh SS. Essential letters in the fungal alphabet: ABC and MFS transporters and their roles in survival and pathogenicity. Adv. Genet. 2014;85:201–253. doi: 10.1016/B978-0-12-800271-1.00004-4. [DOI] [PubMed] [Google Scholar]
- 32.Jack DL, Yang NM, Saier MH., Jr The drug/metabolite transporter superfamily. Eur. J. Biochem. 2001;268:3620–3639. doi: 10.1046/j.1432-1327.2001.02265.x. [DOI] [PubMed] [Google Scholar]
- 33.Tran CV, Saier MH., Jr The principal chloroquine resistance protein of Plasmodium falciparum is a member of the drug/metabolite transporter superfamily. Microbiology. 2004;150:1–3. doi: 10.1099/mic.0.26818-0. [DOI] [PubMed] [Google Scholar]
- 34.Martin RE, Kirk K. The malaria parasite's chloroquine resistance transporter is a member of the drug/metabolite transporter superfamily. Mol. Biol. Evol. 2004;21:1938–1949. doi: 10.1093/molbev/msh205. [DOI] [PubMed] [Google Scholar]
- 35.Ng BG, et al. Mosaicism of the UDP-galactose transporter SLC35A2 causes a congenital disorder of glycosylation. Am. J. Hum. Gent. 2013;92:632–636. doi: 10.1016/j.ajhg.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sprong H, et al. Association of the Golgi UDP-galactose transporter with UDP-galactose:ceramide galactosyltransferase allows UDP-galactose import in the endoplasmic reticulum. Mol. Biol. Cell. 2003;14:3482–3493. doi: 10.1091/mbc.E03-03-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorre K, et al. A new case of UDP-galactose transporter deficiency (SLC35A2-CDG): molecular basis, clinical phenotype, and therapeutic approach. J. Inher. Met. Dis. 2015;38:931–940. doi: 10.1007/s10545-015-9828-6. [DOI] [PubMed] [Google Scholar]
- 38.Kanamori A, et al. Expression cloning and characterization of a cDNA encoding a novel membrane protein required for the formation of O-acetylated ganglioside: a putative acetyl-CoA transporter. Proc. Natl. Acad. Sci. U S A. 1997;94:2897–2902. doi: 10.1073/pnas.94.7.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varki A, Diaz S. The transport and utilization of acetyl coenzyme A by rat liver Golgi vesicles. O-acetylated sialic acids are a major product. J. Biol. Chem. 1985;260:6600–6608. [PubMed] [Google Scholar]
- 40.Jonas MC, Pehar M, Puglielli L. AT-1 is the ER membrane acetyl-CoA transporter and is essential for cell viability. J. Cell Sci. 2010;123:3378–3388. doi: 10.1242/jcs.068841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirabayashi Y, Nomura KH, Nomura K. The acetyl-CoA transporter family SLC33. Mol. Aspects. Med. 2013;34:586–589. doi: 10.1016/j.mam.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 42.McNamara CW, et al. Targeting Plasmodium PI(4)K to eliminate malaria. Nature. 2013;504:248–253. doi: 10.1038/nature12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walch-Solimena C, Novick P. The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nature Cell Biol. 1999;1:523–525. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
- 44.Kruger T, Sanchez CP, Lanzer M. Complementation of Saccharomyces cerevisiaepik1ts by a phosphatidylinositol 4-kinase from Plasmodium falciparum. Mol. Biochem. Parasitol. 2010;172:149–151. doi: 10.1016/j.molbiopara.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 45.Roemer T, et al. Confronting the challenges of natural product-based antifungal discovery. Chem. Biol. 2011;18:148–164. doi: 10.1016/j.chembiol.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 46.Goodman CD, et al. Parasites resistant to the antimalarial atovaquone fail to transmit by mosquitoes. Science. 2016;352:349–353. doi: 10.1126/science.aad9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1979;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 48.Fidock DA, Rosenthal PJ, Croft SL, Brun R, Nwaka S. Antimalarial drug discovery: efficacy models for compound screening. Nature Rev. Drug Disc. 2004;3:509–520. doi: 10.1038/nrd1416. [DOI] [PubMed] [Google Scholar]
- 49.Kariuki MM, et al. Plasmodium falciparum: purification of the various gametocyte developmental stages from in vitro-cultivated parasites. Am. J. Trop.. Med. Hyg. 1998;59:505–508. doi: 10.4269/ajtmh.1998.59.505. [DOI] [PubMed] [Google Scholar]
- 50.Manary MJ, et al. Identification of pathogen genomic variants through an integrated pipeline. BMC Bioinformatics. 2014;15:63. doi: 10.1186/1471-2105-15-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ng CL, et al. CRISPR-Cas9-modified pfmdr1 protects Plasmodium falciparum asexual blood stages and gametocytes against a class of piperazine-containing compounds but potentiates artemisinin-based combination therapy partner drugs. Mol. Microbiol. 2016;101:381–393. doi: 10.1111/mmi.13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malleret B, et al. A rapid and robust tri-color flow cytometry assay for monitoring malaria parasite development. Sci. Rep. 2011;1:118. doi: 10.1038/srep00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nkrumah LJ, et al. Efficient site-specific integration in Plasmodium falciparum chromosomes mediated by mycobacteriophage Bxb1 integrase. Nature Meth. 2006;3:615–621. doi: 10.1038/nmeth904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fiser A, Sali A. Modeller: generation and refinement of homology-based protein structure models. Meth. Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- 56.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 1991;24:946–950. [Google Scholar]
- 57.Merritt EA, Bacon DJ. Raster3D: Photorealistic molecular graphics. Meth. Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All evolved GNF179 and GNF156 resistant strains will be made available upon request to P.B. (early limiting dilution evolved Dd2 strains, including those used for all assays, and all P. berghei strains) and/or E.A.W. (engineered Dd2 parasite lines and all GNF179-resistant strains obtained by previously reported methods). All relevant whole-genome sequencing data have been uploaded to Accession numbers SRP068203 (P. berghei and early limiting dilution P. falciparum), SRP072010 (the higher dose ramp-up and high selection GNF179-resistant P. falciparum clones) and SRP075559 (additional P. berghei samples, which were treated with either 30 or 100 mg/kg GNF179), as indicated in the methods section. The authors declare that all other data supporting the findings of this study are available within the article and its supplementary information.