Significance
CRISPR-Cas is an adaptive immunity system that protects bacteria and archaea from mobile genetic elements. We present comparative genomic and phylogenetic analysis of minimal CRISPR-Cas variants associated with distinct families of transposable elements and develop the hypothesis that such repurposed defense systems contribute to the transposable element propagation by facilitating transposition into specific sites. Thus, these transposable elements are predicted to propagate via RNA-guided transposition, a mechanism that has not been previously described for DNA transposons.
Keywords: CRISPR-Cas systems, Tn7 transposon, transposition strategy, crRNA guide, target-site selection
Abstract
A survey of bacterial and archaeal genomes shows that many Tn7-like transposons contain minimal type I-F CRISPR-Cas systems that consist of fused cas8f and cas5f, cas7f, and cas6f genes and a short CRISPR array. Several small groups of Tn7-like transposons encompass similarly truncated type I-B CRISPR-Cas. This minimal gene complement of the transposon-associated CRISPR-Cas systems implies that they are competent for pre-CRISPR RNA (precrRNA) processing yielding mature crRNAs and target binding but not target cleavage that is required for interference. Phylogenetic analysis demonstrates that evolution of the CRISPR-Cas–containing transposons included a single, ancestral capture of a type I-F locus and two independent instances of type I-B loci capture. We show that the transposon-associated CRISPR arrays contain spacers homologous to plasmid and temperate phage sequences and, in some cases, chromosomal sequences adjacent to the transposon. We hypothesize that the transposon-encoded CRISPR-Cas systems generate displacement (R-loops) in the cognate DNA sites, targeting the transposon to these sites and thus facilitating their spread via plasmids and phages. These findings suggest the existence of RNA-guided transposition and fit the guns-for-hire concept whereby mobile genetic elements capture host defense systems and repurpose them for different stages in the life cycle of the element.
Mechanisms for recognizing specific nucleic acid sequences are essential to accessing and maintaining the genome in all life forms. The most widespread molecular systems based on sequence recognition involve dedicated nucleic acid-binding proteins (1, 2). In particular, promoter recognition by transcription factors and recognition of chromosomal replication origins by initiation proteins are fundamental, universal processes central to normal cell function (3, 4). Additionally, recognition of nucleic acids by proteins is the basis of self vs. nonself discrimination that is essential for defense functions, such as restriction modification in prokaryotes (5, 6). However, there is a growing appreciation of how nucleic acids themselves are harnessed for the task of sequence recognition. A key advantage of these systems is their flexibility whereby a guide nucleic acid molecule can be adapted to recognize any target sequence with high specificity. Thanks to this inherent capacity, nucleic acid based-machinery has been exploited extensively in the evolution for defense of the genome against mobile genetic elements (MGE) as well as regulatory functions (7). A major case in point is the vast RNAi network, apparently the most conserved, ancestral innate immunity system in eukaryotes (8–10). The RNAi machinery takes advantage of dsRNA produced by viruses and transposons to generate specific guide RNAs for defense and has also spawned a variety of regulatory mechanisms.
Prokaryotes possess a system of innate immunity centered around the Argonaute proteins that appears to be the evolutionary antecedent of eukaryotic RNAi (7, 11, 12) as well as CRISPR-Cas systems of adaptive immunity (13–15). The CRISPR-Cas systems provide guide RNA-based defense against viruses and other MGE in nearly all archaea and about one third of bacteria (16).
CRISPR-Cas systems possess modular organization which roughly reflects the three main functional stages of the CRISPR immune response: (i) spacer acquisition (known as “adaptation”), (ii) pre–CRISPR RNA (precrRNA) processing, and (iii) interference (14). CRISPR-Cas systems are highly diverse but can be partitioned into two distinct classes based on the organization of the effector module that is responsible for processing and adaptation (15, 16). Class 1 CRISPR-Cas systems are further divided into three types and 12 subtypes in all of which the effector modules are multisubunit complexes of Cas proteins (16). In contrast, in the currently identified three types and 12 subtypes of class 2, the effector modules are represented by a single multidomain protein, such as the thoroughly characterized Cas9 (15, 17, 18).
At the adaptation stage, the Cas1–Cas2 protein complex, in some instances with additional involvement of accessory adaptation proteins and/or effector module proteins, captures a segment of the target DNA (known as the “protospacer”) and inserts it at the 5′ end of a CRISPR array (19–23). In the second processing stage, a CRISPR array is transcribed into a long transcript known as “precrRNA” that is bound by Cas proteins and processed into mature, small crRNAs. In most class 1 systems, the precrRNA processing is catalyzed by the Cas6 protein that, in some cases, is loosely associated with the effector complex (14, 24). The final interference step involves binding of the mature crRNA by the effector complex, scanning a DNA or RNA molecule for a sequence matching the crRNA guide and containing a protospacer adjacent motif (PAM), and cleavage of the target by a dedicated nuclease domain(s) (14, 24–26). The identity of the nuclease(s) differs between type I and type III CRISPR-Cas systems. In type I, the protein responsible for target cleavage is Cas3, which typically consists of a superfamily II helicase and HD-family nuclease domains (27). After the effector complex, which is denoted “Cascade” [CRISPR-associated complex for antiviral defense (28)] in type I systems, recognizes the cognate protospacer in the target DNA, it recruits Cas3, after which the helicase unwinds the target DNA duplex, and the HD nuclease cleaves both strands (29, 30). Type III systems lack Cas3, and the protein responsible for target cleavage is Cas10, which contains polymerase-cyclase and HD-nuclease domains that are both required for the target degradation (31, 32).
In some of the CRISPR-Cas systems the adaptation genes are encoded separately or even are missing from the genome containing effector complex genes. Among these nonautonomous CRISPR-Cas systems, those of type III have been characterized in most detail (14). It has been shown that type III effector complexes can use crRNA originating from CRISPR arrays associated with type I systems and thus do not depend on their own adaptation modules (33–37). Furthermore, the CRISPR-Cas systems of type IV, which are often encoded on plasmids, typically consist of the effector genes only (16). No adaptation genes and no associated nuclease domains could be found in the type IV loci, although occasionally CRISPR arrays and cas6-like genes are present. The type IV systems have not yet been studied experimentally, so their mode of action remains unknown. Finally, several variants of type I systems, similarly to type IV, lack adaptation genes and genes for proteins involved in DNA cleavage. A “minimal” variant of subtype I-F has been identified in the bacterium Shewanella putrefaciens, with an effector module that consists only of Cas5f, Cas6f, and Cas7f proteins and lacks the large and small subunits present in other Cascade complexes (38). Even more dramatic minimization of subtype I-F has been observed in another variant of subtype I-F that lacks the adaptation module and consists solely of three effector genes, namely a fusion of cas8f (large subunit) with cas5f, that is unique for this variant, cas7f, and cas6f (Fig. 1A) (16). Given the composition of their Cascade complex, these Cas1-less minimal subtype I-F systems can be predicted to process precrRNA, yielding mature crRNAs, and to recognize the target. However, they lack the Cas3 protein and therefore cannot be expected to be competent for target cleavage. Here we report a comprehensive in silico analysis of this system showing that it is linked to a specialized group of transposons related to the well-studied Tn7.
Fig. 1.
Schematic representation of the complete and minimal type I-F CRISPR-Cas systems and Tn7 transposition. (A) Gene organization of a complete and a minimal type I-F CRISPR-Cas system lacking the genes for proteins responsible for adaptation and target cleavage. Minimal I-F systems contain fused cas8f and cas5f genes that are characteristic of this group (16). Together, these proteins can be predicted to be subunits of a minimal Cascade complex. (B) Gene structure of the Tn7 genes flanked by left (L) and right (R) end sequences. Transposition catalyzed by the TnsABC+TnsD proteins directs the transposon into a single chromosomal site (attTn7) in bacterial genomes. Transposition catalyzed by the TnsABC+TnsE proteins preferentially directs transposition into actively conjugating DNA and filamentous bacteriophage (shown by a red circle with arrows). The transposon is denoted by a rectangle in the attachment site. The DNA sequence omitted in the graphic is denoted by two slashes. See text for details.
As genomic parasites, transposons have evolved to limit the negative effects they exert on the host. A variety of regulatory mechanisms are used to maintain transposition at a low frequency and sometimes coordinate transposition with various cell processes. Some prokaryotic transposons also can mobilize functions that benefit the host or otherwise help maintain the element. Certain transposons also evolved mechanisms of tight control over target site selection, the most notable example being the Tn7 family (39).Three transposon-encoded proteins form the core transposition machinery of Tn7: a heteromeric transposase (TnsA and TnsB) and a regulator protein (TnsC) (Fig. 1B). In addition to the core TnsABC transposition proteins, Tn7 elements encode dedicated target site-selection proteins, TnsD and TnsE. In conjunction with TnsABC, the sequence-specific DNA-binding protein TnsD directs transposition into a conserved site referred to as the “Tn7 attachment site,” attTn7 (40). TnsD is a member of a large family of proteins that also includes TniQ, a protein found in other types of bacterial transposons. TniQ is incompletely characterized at the molecular level but has been shown to target transposition into the resolution sites of plasmids (41). Transposition into the attTn7 site shows no negative impact on the host, providing a “safe haven” for these elements that appear to be universally maintained in bacteria. Transposition mediated by TnsABC + TnsE is preferentially directed into plasmids and bacteriophages owing to the ability of TnsE to recognize complexes formed during specific types of DNA replication (42–45). The TnsE-mediated transposition that preferentially directs insertion into other MGE is likely responsible for the wide distribution of Tn7 elements among bacteria.
Here we show that minimal subtype I-F CRISPR-Cas systems are specifically associated with a distinct group of Tn7-like elements. These transposons encode TnsD(TniQ)-like proteins and use previously uncharacterized attachment sites but lack TnsE-like proteins that normally promote horizontal transfer of the elements. Several identified matches for the spacers from the transposon-associated CRISPR arrays suggest that this system might function by targeting transposition to target sites enabled by guide crRNAs. We hypothesize that the CRISPR-Cas machinery recruited by these elements facilitates their horizontal dissemination, mostly via plasmids and/or phages. Thus, this group of MGE is likely to possess a functionality that has not been described previously for DNA transposons, namely, RNA-guided transposition.
Results and Discussion
A Variant of the Type I-F CRISPR-Cas System Is Specifically Associated with a Distinct Family of Tn7-Like Elements.
For the purpose of comprehensive identification of type I-F CRISPR-Cas loci, we chose the Cas7f protein as the probe, given that it is the most conserved component in all systems of this subtype including the minimal variant lacking cas1, cas2, and cas3 genes. Using a PSI-BLAST search started with Cas7f profiles, we obtained 2,905 Cas7f protein sequences, mapped them onto the respective genomes, and annotated the genes in the neighborhoods 10 kb up- and downstream of the cas7f genes using PSI-BLAST against the conserved domain database (CDD). These 20-kb loci are long enough to cover a typical complete I-F system that consists of six genes (16). We then reconstructed a phylogenetic tree from all identified Cas7f protein sequences (Fig. 2A and Dataset S1; see the respective Newick tree at ftp://ftp.ncbi.nih.gov/pub/makarova/supplement/Peters_et_al_2017). Mapping gene neighborhoods on the tree revealed a single, monophyletic, strongly supported branch that included all cas1-less I-F variants. As of this analysis, the branch encompassed 423 sequences from 19 genera of Gammaproteobacteria and appears to derive from a typical, complete I-F system (Figs. 1A and 2A). Indeed, all other branches in the tree consist of Cas7f homologs from complete I-F systems containing a cas1 gene within the locus. A few exceptions that are scattered in the tree are from either small contigs or disrupted cas loci. In the vast majority of the loci corresponding to the cas1-less branch, a tnsD(tniQ) gene is located next to the cas genes (Fig. 3).
Fig. 2.
Schematic evolutionary trees for the Cas7f, TnsA, and TnsD(TniQ) protein families. (A) The dendrogram was built using 2,905 Cas7f proteins as described in Methods (see the complete tree at ftp://ftp.ncbi.nih.gov/pub/makarova/supplement/Peters_et_al_2017). The major subtrees are collapsed and shown by triangles. The branch corresponding to the minimal I-F variant is colored in orange, and the bootstrap value for this subtree is shown. (B) The dendrogram was built using 7,023 TnsA protein sequences (see the complete tree at ftp://ftp.ncbi.nih.gov/pub/makarova/supplement/Peters_et_al_2017). The branch corresponding to TnsA in the loci containing I-F variant cas genes is colored in orange, and I-B subtype cas genes are colored in green. The CRISPR-Cas subtypes are indicated next to the respective branches. Distinct cyanobacterial strains are indicated next to the respective I-B systems. The bootstrap value for the TnsA branch associated with I-F cas genes is shown. (C) The dendrogram was built using 7,963 TnsD(TniQ) proteins (see the complete tree at ftp://ftp.ncbi.nih.gov/pub/makarova/supplement/Peters_et_al_2017). The outgroup consists of the TnsD(TniQ)-like proteins that form the sister group of those associated with the type I-F CRISPR-Cas systems but encoded by Tn6022 elements lacking CRISPR-Cas (see the complete tree for the full information). The designations are as in B.
Fig. 3.
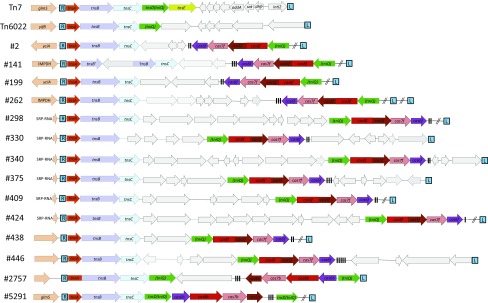
Schematic representation of Tn7, Tn6022, and selected Tn7-like transposons containing cas genes. Genomic features recognized by the transposon-encoded TniQ protein are indicated on the left (glmS, yifB, IMPDH, yciA, and SRP-RNA). Color coding and labeling are as in Fig. 1. Elements other than Tn7 and Tn6022 are denoted by the respective TnsA tree leaves (#XX) (Tn6022 = Tree node #582) (Dataset S2). Other genes are shown in gray, and known Tn7 cargo genes are indicated. Black vertical bars indicate repeats in the element-encoded arrays. DNA sequences omitted in the graphic are indicated by two slashes. See text for details.
To determine whether the association of the Cas1-less I-F systems with Tn7-like elements was unique or emerged independently on several occasions, we analyzed the TnsD(TniQ) and TnsA families. The TnsA protein is the most highly conserved gene of the Tn7-like elements and is responsible for the unique behavior of the elements with heteromeric transposases (46–49). We collected and annotated 10,349 loci containing at least tniQ/tnsD or tnsA (Dataset S2) and reconstructed a tree for both protein families (Fig. 2 B and C and see respective Newick trees at ftp://ftp.ncbi.nih.gov/pub/makarova/supplement/Peters_et_al_2017). In both trees, the loci containing cas genes of the cas1-less I-F variant mapped to strongly supported clades (Fig. 2 B and C). Thus, phylogenetic analysis of both Cas7f and the associated transposon-encoded proteins reveals a strong link between a specific group of Tn7-like elements and a distinct variant of the subtype I-F CRISPR-Cas systems. The Tn7-like elements in the clade that includes Tn6022 were identified as the outgroups to the respective branches in both the TnsA and TnsD(TniQ) trees, suggesting that a member of the Tn6022 family is the ancestor of the CRISPR-associated variety of Tn7-like transposons (Fig. 2 B and C). Both clades include multiple, deep branches that are not associated with cas genes in the respective loci, indicating that the link with the I-F system evolved relatively late in the history of this group of Tn7-like elements (see respective Newick trees at ftp://ftp.ncbi.nih.gov/pub/makarova/supplement/Peters_et_al_2017). In several cases, however, distribution of the cas genes among the tree branches implies that these were lost from the vicinity of the conserved transposon genes (e.g., Shewanella baltica OS678 and Thiomicrospira crunogena XCL_2), implying that the CRISPR-Cas system is not essential for the transposon survival. Notably, however, the converse is not the case: We detected no intact cas1-less I-F systems outside this transposon neighborhood, with the implication that this CRISPR-Cas variant is functional only when associated with a Tn7-like element.
We further investigated the tnsD and tnsA loci to identify any other CRISPR-Cas systems that might be linked to Tn7-like transposons. Only a few such instances were detected, mostly complete loci containing the adaptation genes. The respective tnsA and/or tniQ/tnsD genes are scattered in the phylogenetic trees, suggesting that most of these associations are effectively random and might be transient (Dataset S2). However, some such loci do show a degree of evolutionary coherence. Specifically, they form two small, unrelated branches in both the TnsA and the TnsD(TniQ) trees (see I-B in Fig. 2 B and C). All these CRISPR-cas loci are present in different cyanobacteria, belong to the I-B subtype, and lack adaptation genes as well as the cas3 gene that is required for DNA cleavage in type I systems. Thus, to a large extent, these type I-B variants mimic the organization of the more common transposon-associated, cas1-less I-F variant (see below).
The cas1-Less Type I-F CRISPR-Cas System Is Mobilized Together with Conserved Transposition Genes.
We analyzed the transposon end sequences in the loci containing the I-F and I-B CRISPR-Cas variants to determine whether the cas genes were located within the boundaries of these elements or are simply adjacent to the transposon. The structure of the left and right ends of canonical Tn7 has been defined previously (Fig. S1). Tn7 ends are marked by a series of 22-bp TnsB-binding sites (50–52). Flanking the most distal TnsB-binding sites is an 8-bp terminal sequence ending with 5′-TGT-3′/3′-ACA-5′. Tn7 contains four overlapping TnsB-binding sites in the ∼90-bp right end of the element and three dispersed sites in the ∼150-bp left end of the element, but the number and distribution of TnsB-binding sites can vary among Tn7-like elements (39, 49). End sequences of Tn7-related elements can be determined by identifying the directly repeated 5-bp target site duplication, the terminal 8-bp sequence, and 22-bp TnsB-binding sites (Fig. S1). Compared with the canonical Tn7 and Tn6022, Tn7-like elements show extensive variation in size and gene complements as illustrated by a representative set of 12 complete elements ranging in size from 22 kb to almost 120 kb (Fig. 3 and Table S1) (53, 54). One of these elements has been previously identified in Vibrio parahaemolyticus RIMD2210633 as a member of the Tn7 superfamily and encodes the Vibrio pathogenicity determinant, thermostable direct hemolysin (TDH) (55). It should be emphasized that in closely related bacterial genomes (e.g., different strains of V. parahaemolyticus), CRISPR-Cas–carrying Tn7-like elements are often inserted in different sites (Fig. 4), which is indicative of recent mobility of these elements.
Fig. S1.
Schematic representation of the end structure of Tn7-like elements: anatomy of a Tn7 insertion. Typically, the insertion occurs at a single site about 25 bp downstream from the last codon of glmS. The Tn7 end proximal to tnsA is closest to glmS (by convention it is referred to as the “right” end). Transposition generates a target site duplication (shown in red) of the chromosomal sequence that now forms a direct repeat on either side the element. In the case of insertion of the canonical Tn7 element at the attTn7 site, this sequence is GCGGG; an 8-bp end sequence starts with TGT/CAC, and immediately after this end sequence is the first 22-bp binding site for TnsB.
Table S1.
Characteristics of selected Tn7 loci associated with cas genes
 |
TnsA node number for each element cross-lists as indicated in Figs. 3 and 4 and Datasets S1 and S2.
Fig. 4.
Phylogenetic tree of selected representatives of type I-F-associated TnsD(TniQ)-like proteins. A maximum likelihood phylogenetic tree was built as described in Methods for a selected set of TnsD(TniQ)-like proteins associated with the type I-F CRISPR-Cas variant and the same outgroup as in Fig. 2C. The numbers at internal branches indicate percent bootstrap support; only values greater than 70% are indicated. Elements located in one of the three attachment sites identified in this work are shown by color as indicated (yciA, IMPDH, and SRP-RNA); random sites are in black. The leaves of the tree for the TnsD(TniQ)-like proteins (#XX) (Dataset S2) are shown in green.
In our analysis of CRISPR-Cas systems, two groups of type I-B variants were identified in association with Tn7-like elements (Fig. 2 B and C). Similar to the type I-F CRISPR-Cas variant, these I-B systems are expected to be functional for maturing CRISPR transcripts and forming crRNA complexes at protospacers but lack adaptation genes and Cas3 and, accordingly, are likely to be defective for interference. Furthermore, these type I-B CRISPR-Cas variants are associated with short CRISPR arrays (Fig. 3).
Taken together, these findings indicate that the type I-F and I-B CRISPR-Cas variants identified in this work are part of the core gene repertoire in multiple clades of Tn7-like elements.
Chromosomal Insertions of the I-F CRISPR-Cas–Associated Elements Show Three Recognizable Attachment Sites Likely Accessed By Dedicated TnsD(TniQ) Proteins.
The canonical Tn7 element and especially the transposition pathway that directs the element into the attTn7 site located downstream of the conserved glmS gene have been studied extensively. The Tn7 TnsD(TniQ) protein is a sequence-specific DNA-binding protein that recognizes a highly conserved 36-bp sequence in the downstream region of the glmS gene-coding sequence (40, 56). Transposition events promoted by TnsABC+D are directed into a position 23 bp downstream of the region bound by TnsD. Tn7 transposition is orientation specific in all transposition pathways; the transposon end proximal to the tnsA gene (the “right” end of the element) is adjacent to the DNA sequence or a specific protein complex recognized in each pathway (44, 56–58).
We analyzed the region adjacent to the point of insertion of the Tn7-like elements and identified three previously uncharacterized attachment sites for the cas1-less, type I-F–associated transposons. Similar to Tn7 insertions, one subgroup of the elements occurred downstream of a gene, but instead of glmS, these insertions were found downstream of an inosine-5′-monophosphate dehydrogenase gene (Figs. 3 and 4 and Table S1). The configurations found with the other recognizable attachment sites have not been described previously for Tn7-like elements. In one case, the attachment site was located upstream of the yciA gene, which encodes an acyl-CoA thioester hydrolase (Figs. 3 and 4 and Table S1). The third attachment site identified for the cas1-less type I-F–associated elements is in a non–protein-encoding gene, namely, the gene for the signal-recognition particle RNA (SRP-RNA), another configuration not reported previously (Figs. 3 and 4 and Table S1). The concordance between the phylogeny of the TnsD(TniQ) proteins and the attachment site used by the element is consistent with the hypothesis that each attachment site is recognized by a cognate TnsD(TniQ) protein (Fig. 4). However, many transposons appear to be inserted in random sites (Fig. 4). It remains unclear how insertions were directed into these sites because they are unlikely to be specifically recognized by TnsD(TniQ) proteins encoded by these elements, and these elements lack a homolog of the TnsE protein found in typical Tn7 transposons.
Analysis of CRISPR Arrays Associated with the cas1-Less I-F Systems.
The great majority of the transposon-associated I-F and I-B systems encompass a CRISPR array downstream of the cas6 gene (Fig. 3 for examples). In most cases, this array contains only one or two spacers, suggesting that spacer acquisition in these arrays occurs only rarely (Fig. 3 and Table S2). Nevertheless, the spacers are typically unrelated, even in closely related bacterial genomes, indicating that, occasionally, new spacers are incorporated, and old ones are lost. Obviously, only adaptation genes acting in trans can insert new spacers into these arrays. Among the 14 complete bacterial genomes containing Tn7-like elements with the I-F CRISPR-Cas, only two encompass other CRISPR-Cas loci containing adaptation genes, namely, Vibrio fluvialis ATCC 33809 and Pseudoalteromonas rubra SCSIO6842, which possess I-F and I-C systems, respectively. Among draft genomes, there are more cases where additional, complete CRISPR-Cas systems, mostly I-F and I-E, are present in the same genomes. Nevertheless, most of the genomes that contain the Tn7-associated I-F lack other CRISPR-Cas systems that would be able to provide for adaptation, which might account for the short CRISPR arrays. All four complete genomes containing elements associated with I-B systems encompass additional CRISPR-Cas loci containing adaptation genes, often of subtype I-D, which is abundant in cyanobacteria (16).
Table S2.
Spacers with identified matches
 |
The TnsA nodes are as in Table S1.
Spacers were identified as described in the Materials and Methods section. Spacers (and 3 bp located 3′ in the repeat) are indicated for comparison with the match identified in the database (presumed PAM sequences). Mismatches between the spacer (3′ sequence) and protospacer (PAM) are indicated in red.
Altogether, more than 800 spacers were identified in the transposon-associated I-F and I-B CRISPR arrays (see automatically and manually identified spacers at ftp://ftp.ncbi.nih.gov/pub/makarova/supplement/Peters_et_al_2017). As in most analyses of CRISPR spacers, including a recent comprehensive survey (59–62), only a small fraction of these spacers yielded significant matches to sequences in public databases. However, the matches that could be detected were informative because they were to plasmids and bacteriophages associated with the same bacterial genera in which the respective elements are found (Table S2). We identified two cases (in Photobacterium kishitanii and Photobacterium leiognathi) of special interest, in which spacers matched the region adjacent to the tnsA-gene–proximal side of the element (Table S2), i.e., the specific region where complexes involved in targeting transposition events interact with the target DNA (44, 56, 58). An additional spacer match was found inside the transposon boundaries in several V. parahaemolyticus strains (Table S2). A similar situation might have also occurred in a Tn7-like transposon associated with a type I-B CRISPR-Cas variant in a Cyanothece PCC 7822 plasmid, although end sequences could not be unambiguously defined for this element (Table S2).
A Potential Role for CRISPR-Cas in Targeting Transposition.
Taking into account all the observations on the transposon-associated CRISPR-Cas systems and previous studies on the mechanism of target site activation, we propose a model for the involvement of Cas1-less CRISPR-Cas systems in targeting transposition to facilitate cell–cell transfer of the element (Fig. 5). Canonical Tn7 encodes two targeting pathways that are both mediated by the same set of TnsABC proteins (Fig. 1B). The TnsABC+TnsD(TniQ) pathway appears to be broadly conserved, allowing high-frequency transposition into an attachment site recognized by a cognate TnsD(TniQ) protein (Figs. 1 and 4 and Table S1) (49). The cas1-less I-F CRISPR-Cas variant is encoded in the same location where the tnsE gene that promotes transposition into conjugal plasmids and filamentous bacteriophages is typically located (Fig. 3). Thus, it appears likely that the CRISPR-Cas system functionally replaces TnsE as a mechanism facilitating horizontal transfer of the element. Support for this possibility comes from the observation that the transposon-associated CRISPR arrays largely carry plasmid and phage-specific spacers and could direct the transposon to the respective elements (Table S2).
Fig. 5.
Model of the two targeting pathways for Tn7 elements containing CRISPR-Cas system. Designations are as in Fig. 1.
Distortions in B-Form DNA Induced By Cas-crRNA Could Play a Role in Recruiting Transposition.
Transposition into attTn7 is well understood at the molecular level; the DNA structure in the vicinity of the attachment site plays a central role in transposition (Fig. 6 A–C). TnsD binding induces an asymmetric distortion in the attTn7 target DNA that is essentially responsible for attracting TnsC for target site selection during transposition (Fig. 6A) (56, 63). The TnsABC proteins are normally insufficient for Tn7 transposition in vivo or in vitro (64); however, certain gain-of-function mutations in the regulator protein TnsC (TnsC*) allow untargeted transposition in the absence of TnsD or TnsE (47, 65, 66). Notably, transposition in this case is attracted to a specific location adjacent to a short segment of triplex-forming DNAs (58, 67). Analogous to transposition events found in attTn7, these events are targeted to a position on one side of the triplex-forming DNA in a unique orientation owing to the ability of TnsC to recognize the distortion formed at the triplex–to–B-form DNA transition (Fig. 6C). Distortions induced in the target DNA are also implicated in transposition targeting by TnsABC+E (Fig. 6B) (68). Given that distortions in B-form DNA are also expected adjacent to crRNA-bound effector complexes that generate R-loops through duplex formation between the crRNA and the protospacer (26, 69), there could be a mechanistic link between the well-understood Tn7 targeting process and DNA targeting by the CRISPR-Cas effector complexes (Fig. 6D).
Fig. 6.
Models of the three previously described Tn7 targeting pathways and the proposed CRISPR-Cas–facilitated transposition pathway. Representations of TnsABC+TnsD (A) and TnsABC+TnsE (B) transposition pathways, the synthetic transposition pathway that targets triplex DNA complexes with a mutant form of TnsC, TnsABC* (C), and the proposed targeting pathway mediated by Cas interference complexes (D) are shown. Known host factors that participate in the TnsD (ACP, L29) and TnsE (DnaN) pathways are also shown. See text for details and references.
Evolution of the Association Between CRISPR-Cas Variants and Tn7-Like Elements.
Given that type I CRISPR-Cas systems have been shown to selectively integrate spacers from plasmids and phages (19, 70), an attractive hypothesis is that the CRISPR-cas loci that randomly became associated with the transposon were fixed through selection for their ability to facilitate dissemination of transposons. As discussed above, because changes in DNA structure play a key role in target site selection by Tn7, relatively little evolutionary adaptation might be needed to allow the core TnsABC machinery to recognize crRNA-bound effector complexes for targeting. In this light, it is intriguing that association between CRISPR-Cas systems and Tn7-like elements occurred on multiple, independent occasions. The consistent minimalist features in the organization of the transposon-associated type I-F and I-B variants imply that they coevolved with Tn7-like elements along parallel paths of reductive evolution. Both type I-F and type I-B systems have lost the adaptation module (cas1 and cas2) and the cas3 gene, which is required to cleave the target DNA in other type I systems (14). The absence of Cas3 implies that these CRISPR-Cas systems recognize but do not cleave the target, a mode of action that would allow the targeted DNA to serve as a vehicle for horizontal transfer of the respective Tn7-like transposon.
The transposon-associated CRISPR arrays are short, and the respective bacterial genomes often lack CRISPR adaptation modules. Thus, the majority of the CRISPR-containing transposons are likely to be relatively recent arrivals to the respective genomes, conceivably, brought about by the plasmid or phage against which they carry a spacer. Once integrated into a new host attachment site, such transposons could “lie in wait” for a horizontal transfer vehicle, either as a result of in trans acquisition of a new spacer that is specific to an endogenous plasmid or prophage or via the entry of an element that is already represented by a cognate spacer in the transposon-encoded CRISPR array. In some cases, an incoming plasmid or phage recognized by the CRISPR-Cas system and targeted for transposition would be incapacitated by the integration event. Nevertheless, even such unproductive integrations would still benefit the CRISPR-carrying transposon by protecting the host. In such cases, CRISPR-directed integration that is in keeping with a selfish behavior for the transposon would also qualify as altruistic behavior toward the host. Occasionally, the Tn7-encoded CRISPR-Cas systems appear to acquire spacers from the host chromosome, conceivably stimulating ectopic transposition within the same genome. This mechanism could provide for transposition in hosts that lack attachment sites recognized by the element-encoded TnsD(TniQ) protein.
Concluding Remarks
Here we identify three distinct groups of Tn7-like transposons that encode minimal variants of type I CRISPR-Cas systems. The transposon-encoded CRISPR-Cas variants lack the interference nucleases, whereas the transposons themselves lack the TnsE protein that directs transposition to MGE. Therefore, we hypothesize that these CRISPR-Cas systems functionally replace TnsE and comprise an RNA-guided transposition machinery. To the best of our knowledge, such a mechanism has not been identified or proposed previously for DNA transposons. However, homology between the MGE RNA and the integration region in the host genome is exploited during group II intron retrohoming (71, 72), suggesting that RNA-guided target recognition evolved more than once in MGE evolution.
Many questions remain regarding the functioning of the CRISPR-Cas in Tn7-like transposons, including the possibility of direct interaction between the CRISPR effector complexes and TnsD(TniQ), TnsABC, or other transposon-encoded accessory proteins. It is also unclear if these CRISPR-Cas variants might perform alternative or additional functions beyond facilitation of transposition, such as gene silencing or protection of the transposon.
From the evolutionary standpoint, the transposon-associated CRISPR-Cas systems fit the guns-for-hire paradigm (73). Under this concept, MGE genes are often recruited by host defense systems, and, conversely, defense systems or components thereof can be captured by MGE and repurposed for counter defense or other roles in the life cycle of the element. Recruitment of MGE apparently was central to the evolution of CRISPR-Cas, contributing to the origin of both the adaptation module and the class 2 effector modules (15, 17, 74). On the other side of the equation, virus-encoded CRISPR-Cas systems have been identified and implicated in inhibition of host defense (75). The observations described here, if validated experimentally, seem to “close the circle” by demonstrating recruitment of CRISPR-Cas systems by transposons, conceivably for a role in targeting transposition, a key step in transposon propagation.
This work also raises the possibility that other, complicated molecular machines may be identified that use RNA or DNA guides to recognize specific nucleotide sequences in different functional contexts. Finally, it has not escaped our notice that the transposon-encoded CRISPR-Cas systems described here potentially could be harnessed for genome-engineering applications, namely, precise targeting of synthetic transposons encoding selectable markers and other genes of interest.
Methods
Prokaryotic Genome Database and ORF Annotation.
Archaeal and bacterial complete and draft genome sequences were downloaded from the National Center for Biotechnology Information (NCBI) FTP site (ftp://ftp.ncbi.nlm.nih.gov/genomes/all/) in March 2016. For incompletely annotated genomes (coding density less than 0.6 coding DNA segments/kbp), the existing annotation was discarded and replaced with the Meta-GeneMark 1 (76) annotation using the standard model MetaGeneMark_v1.mod (Heuristic model for genetic code 11 and GC 30). Altogether, the database includes 4,961 completely sequenced and assembled genomes and 43,599 partially sequenced genomes.
Profiles for three protein families, namely Cas7f (cd09737, pfam09615), TnsA (pfam08722, pfam08721), and TnsD(TniQ) (pfam06527), which are available in the NCBI CDD database (77), were used as queries for PSI-BLAST searches (E-value: 10−4; other parameters were default) to find respective homologs. All ORFs within 10-kb regions up- and downstream of cas7f genes (to cover the potential complete I-F system) and 20-kb regions up- and downstream of tnsD(tniQ) and tnsA (to cover potential Tn7-like elements) were further annotated using RPS-BLAST searches with 30,953 profiles (COG, pfam, cd) from the NCBI CDD database and 217 custom Cas protein profiles (16). The CRISPR-Cas system (sub)type identification for all loci was performed using previously described procedures (16).
Protospacer Analysis.
The CRISPRfinder (78) and PILER-CR (79) programs were used with default parameters to identify CRISPR arrays in Cas7f and TnsA/TnsD loci. The MEGABLAST program (80) (word size 18; otherwise default parameters) was used to search for protospacers in the virus subset of the NR (nonredundant) database and the prokaryotic genome database. Matches were considered only if they showed at least 95% identity and at least 95% length coverage in the case of the NR database and 80% identity and 80% length coverage for the self-hits (hits were classified as “self” if they matched the same genomes or genome of the same species disregarding the strain information). Because the automatic approach missed several short CRISPR arrays, loci initially found to lack CRISPR were analyzed manually by examining the intergenic region downstream of the cas6f gene for repeats and using the BLASTN program with the default parameters to find matches to the spacer identified.
Clustering and Phylogenetic Analysis.
To construct a nonredundant, representative sequence set, protein sequences within families of interest were clustered using the NCBI BLASTCLUST program (ftp://ftp.ncbi.nih.gov/blast/documents/blastclust.html) with the sequence identity threshold of 90% and length coverage threshold of 0.9. Short fragments or disrupted sequences were discarded. Multiple alignments of protein sequences were constructed using MUSCLE (81) or MAFFT (82) programs. Sites with the gap character fraction values >0.5 and homogeneity <0.1 were removed from the alignment. Phylogenetic analysis was performed using the FastTree program (83), with the WAG evolutionary model and the discrete gamma model with 20 rate categories. The same program was used to compute bootstrap values.
Relationships within diverse sequence families were established using the following procedure: Initial sequence clusters were obtained using UCLUST (84) with the sequence similarity threshold of 0.5; sequences were aligned within clusters using MUSCLE (81). Then, cluster-to-cluster similarity scores were obtained using HHsearch (85) (including trivial clusters consisting of a single sequence each), and an unweighted pair-group method with arithmetic mean (UPGMA) dendrogram was constructed from the pairwise similarity scores. Highly similar clusters (pairwise score to self-score ratio >0.1) were aligned to each other using HHALIGN (85), and the procedure was repeated iteratively. At the last step, sequence-based trees were reconstructed from the cluster alignments using the FastTree program (83) as described above and rooted by midpoint; these trees were grafted onto the tips of the profile similarity-based UPGMA dendrogram.
Analysis of Tn7-Like Elements.
End sequences of Tn7-like elements were determined by identifying the directly repeated 5-bp target site duplication, the terminal 8-bp sequence, and 22-bp TnsB-binding sites as described in the text using Gene Construction Kit 4.0 to manipulate DNA sequences and search for DNA repeats. Sequence files were derived from matches to cas7f, tnsA, and tniQ as described above.
Supplementary Material
Acknowledgments
J.E.P. was supported by US Department of Agriculture National Institute of Food and Agriculture Hatch Project NYC-189438. K.S.M., S.S., and E.V.K. are supported by the intramural program of the US Department of Health and Human Services (to the National Library of Medicine).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1709035114/-/DCSupplemental.
References
- 1.Trifonov EN. The multiple codes of nucleotide sequences. Bull Math Biol. 1989;51:417–432. doi: 10.1007/BF02460081. [DOI] [PubMed] [Google Scholar]
- 2.Parker SC, Tullius TD. DNA shape, genetic codes, and evolution. Curr Opin Struct Biol. 2011;21:342–347. doi: 10.1016/j.sbi.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burton ZF, Opron K, Wei G, Geiger JH. A model for genesis of transcription systems. Transcription. 2016;7:1–13. doi: 10.1080/21541264.2015.1128518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bleichert F, Botchan MR, Berger JM. Mechanisms for initiating cellular DNA replication. Science. 2017;355:eaah6317. doi: 10.1126/science.aah6317. [DOI] [PubMed] [Google Scholar]
- 5.Makarova KS, Wolf YI, Koonin EV. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res. 2013;41:4360–4377. doi: 10.1093/nar/gkt157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pingoud A, Wilson GG, Wende W. Type II restriction endonucleases–a historical perspective and more. Nucleic Acids Res. 2014;42:7489–7527. doi: 10.1093/nar/gku447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koonin EV. Evolution of RNA- and DNA-guided antivirus defense systems in prokaryotes and eukaryotes: Common ancestry vs convergence. Biol Direct. 2017;12:5. doi: 10.1186/s13062-017-0177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerutti H, Casas-Mollano JA. On the origin and functions of RNA-mediated silencing: From protists to man. Curr Genet. 2006;50:81–99. doi: 10.1007/s00294-006-0078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shabalina SA, Koonin EV. Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol. 2008;23:578–587. doi: 10.1016/j.tree.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swarts DC, et al. The evolutionary journey of Argonaute proteins. Nat Struct Mol Biol. 2014;21:743–753. doi: 10.1038/nsmb.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hur JK, Olovnikov I, Aravin AA. Prokaryotic Argonautes defend genomes against invasive DNA. Trends Biochem Sci. 2014;39:257–259. doi: 10.1016/j.tibs.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrangou R, Horvath P. A decade of discovery: CRISPR functions and applications. Nat Microbiol. 2017;2:17092. doi: 10.1038/nmicrobiol.2017.92. [DOI] [PubMed] [Google Scholar]
- 14.Mohanraju P, et al. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016;353:aad5147. doi: 10.1126/science.aad5147. [DOI] [PubMed] [Google Scholar]
- 15.Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makarova KS, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shmakov S, et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. 2017;15:169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burstein D, et al. New CRISPR-Cas systems from uncultivated microbes. Nature. 2017;542:237–241. doi: 10.1038/nature21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amitai G, Sorek R. CRISPR-Cas adaptation: Insights into the mechanism of action. Nat Rev Microbiol. 2016;14:67–76. doi: 10.1038/nrmicro.2015.14. [DOI] [PubMed] [Google Scholar]
- 20.Heler R, et al. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature. 2015;519:199–202. doi: 10.1038/nature14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei Y, Terns RM, Terns MP. Cas9 function and host genome sampling in type II-A CRISPR-Cas adaptation. Genes Dev. 2015;29:356–361. doi: 10.1101/gad.257550.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vorontsova D, et al. Foreign DNA acquisition by the I-F CRISPR-Cas system requires all components of the interference machinery. Nucleic Acids Res. 2015;43:10848–10860. doi: 10.1093/nar/gkv1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson SA, et al. CRISPR-Cas: Adapting to change. Science. 2017;356:eaal5056. doi: 10.1126/science.aal5056. [DOI] [PubMed] [Google Scholar]
- 24.Charpentier E, Richter H, van der Oost J, White MF. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity. FEMS Microbiol Rev. 2015;39:428–441. doi: 10.1093/femsre/fuv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson RN, Wiedenheft B. A conserved structural chassis for mounting versatile CRISPR RNA-guided immune responses. Mol Cell. 2015;58:722–728. doi: 10.1016/j.molcel.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsui TK, Li H. Structure principles of CRISPR-Cas surveillance and effector complexes. Annu Rev Biophys. 2015;44:229–255. doi: 10.1146/annurev-biophys-060414-033939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: Computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hochstrasser ML, et al. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc Natl Acad Sci USA. 2014;111:6618–6623. doi: 10.1073/pnas.1405079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huo Y, et al. Structures of CRISPR Cas3 offer mechanistic insights into cascade-activated DNA unwinding and degradation. Nat Struct Mol Biol. 2014;21:771–777. doi: 10.1038/nsmb.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samai P, et al. Co-transcriptional DNA and RNA cleavage during type III CRISPR-Cas immunity. Cell. 2015;161:1164–1174. doi: 10.1016/j.cell.2015.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, Graham S, Tello A, Liu H, White MF. Multiple nucleic acid cleavage modes in divergent type III CRISPR systems. Nucleic Acids Res. 2016;44:1789–1799. doi: 10.1093/nar/gkw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Majumdar S, et al. Three CRISPR-Cas immune effector complexes coexist in Pyrococcus furiosus. RNA. 2015;21:1147–1158. doi: 10.1261/rna.049130.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng L, Garrett RA, Shah SA, Peng X, She Q. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol Microbiol. 2013;87:1088–1099. doi: 10.1111/mmi.12152. [DOI] [PubMed] [Google Scholar]
- 35.Staals RHJ, et al. Structure and activity of the RNA-targeting type III-B CRISPR-Cas complex of Thermus thermophilus. Mol Cell. 2013;52:135–145. doi: 10.1016/j.molcel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staals RH, et al. RNA targeting by the type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol Cell. 2014;56:518–530. doi: 10.1016/j.molcel.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elmore JR, et al. Bipartite recognition of target RNAs activates DNA cleavage by the type III-B CRISPR-Cas system. Genes Dev. 2016;30:447–459. doi: 10.1101/gad.272153.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gleditzsch D, et al. Modulating the cascade architecture of a minimal Type I-F CRISPR-Cas system. Nucleic Acids Res. 2016;44:5872–5882. doi: 10.1093/nar/gkw469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peters JE. Tn7. Microbiol Spectr. 2014;2:1–20. doi: 10.1128/microbiolspec.MDNA3-0010-2014. [DOI] [PubMed] [Google Scholar]
- 40.Mitra R, McKenzie GJ, Yi L, Lee CA, Craig NL. Characterization of the TnsD-attTn7 complex that promotes site-specific insertion of Tn7. Mob DNA. 2010;1:18. doi: 10.1186/1759-8753-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minakhina S, Kholodii G, Mindlin S, Yurieva O, Nikiforov V. Tn5053 family transposons are res site hunters sensing plasmidal res sites occupied by cognate resolvases. Mol Microbiol. 1999;33:1059–1068. doi: 10.1046/j.1365-2958.1999.01548.x. [DOI] [PubMed] [Google Scholar]
- 42.Wolkow CA, DeBoy RT, Craig NL. Conjugating plasmids are preferred targets for Tn7. Genes Dev. 1996;10:2145–2157. doi: 10.1101/gad.10.17.2145. [DOI] [PubMed] [Google Scholar]
- 43.Finn JA, Parks AR, Peters JE. Transposon Tn7 directs transposition into the genome of filamentous bacteriophage M13 using the element-encoded TnsE protein. J Bacteriol. 2007;189:9122–9125. doi: 10.1128/JB.01451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parks AR, et al. Transposition into replicating DNA occurs through interaction with the processivity factor. Cell. 2009;138:685–695. doi: 10.1016/j.cell.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi Q, et al. Conformational toggling controls target site choice for the heteromeric transposase element Tn7. Nucleic Acids Res. 2015;43:10734–10745. doi: 10.1093/nar/gkv913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.May EW, Craig NL. Switching from cut-and-paste to replicative Tn7 transposition. Science. 1996;272:401–404. doi: 10.1126/science.272.5260.401. [DOI] [PubMed] [Google Scholar]
- 47.Choi KY, Li Y, Sarnovsky R, Craig NL. Direct interaction between the TnsA and TnsB subunits controls the heteromeric Tn7 transposase. Proc Natl Acad Sci USA. 2013;110:E2038–E2045. doi: 10.1073/pnas.1305716110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hickman AB, et al. Unexpected structural diversity in DNA recombination: The restriction endonuclease connection. Mol Cell. 2000;5:1025–1034. doi: 10.1016/s1097-2765(00)80267-1. [DOI] [PubMed] [Google Scholar]
- 49.Peters JE, Fricker AD, Kapili BJ, Petassi MT. Heteromeric transposase elements: Generators of genomic islands across diverse bacteria. Mol Microbiol. 2014;93:1084–1092. doi: 10.1111/mmi.12740. [DOI] [PubMed] [Google Scholar]
- 50.Arciszewska LK, Craig NL. Interaction of the Tn7-encoded transposition protein TnsB with the ends of the transposon. Nucleic Acids Res. 1991;19:5021–5029. doi: 10.1093/nar/19.18.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gary PA, Biery MC, Bainton RJ, Craig NL. Multiple DNA processing reactions underlie Tn7 transposition. J Mol Biol. 1996;257:301–316. doi: 10.1006/jmbi.1996.0164. [DOI] [PubMed] [Google Scholar]
- 52.Holder JW, Craig NL. Architecture of the Tn7 posttransposition complex: An elaborate nucleoprotein structure. J Mol Biol. 2010;401:167–181. doi: 10.1016/j.jmb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rose A. TnAbaR1: A novel Tn7-related transposon in Acinetobacter baumannii that contributes to the accumulation and dissemination of large repertoires of resistance genes. Bioscience Horizons. 2010;3:40–48. [Google Scholar]
- 54.Hamidian M, Hall RM. AbaR4 replaces AbaR3 in a carbapenem-resistant Acinetobacter baumannii isolate belonging to global clone 1 from an Australian hospital. J Antimicrob Chemother. 2011;66:2484–2491. doi: 10.1093/jac/dkr356. [DOI] [PubMed] [Google Scholar]
- 55.Sugiyama T, Iida T, Izutsu K, Park KS, Honda T. Precise region and the character of the pathogenicity island in clinical Vibrio parahaemolyticus strains. J Bacteriol. 2008;190:1835–1837. doi: 10.1128/JB.01293-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bainton RJ, Kubo KM, Feng JN, Craig NL. Tn7 transposition: Target DNA recognition is mediated by multiple Tn7-encoded proteins in a purified in vitro system. Cell. 1993;72:931–943. doi: 10.1016/0092-8674(93)90581-a. [DOI] [PubMed] [Google Scholar]
- 57.McKown RL, Orle KA, Chen T, Craig NL. Sequence requirements of Escherichia coli attTn7, a specific site of transposon Tn7 insertion. J Bacteriol. 1988;170:352–358. doi: 10.1128/jb.170.1.352-358.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rao JE, Miller PS, Craig NL. Recognition of triple-helical DNA structures by transposon Tn7. Proc Natl Acad Sci USA. 2000;97:3936–3941. doi: 10.1073/pnas.080061497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mojica FJ, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 60.Shah SA, Hansen NR, Garrett RA. Distribution of CRISPR spacer matches in viruses and plasmids of crenarchaeal acidothermophiles and implications for their inhibitory mechanism. Biochem Soc Trans. 2009;37:23–28. doi: 10.1042/BST0370023. [DOI] [PubMed] [Google Scholar]
- 61.Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. Self-targeting by CRISPR: Gene regulation or autoimmunity? Trends Genet. 2010;26:335–340. doi: 10.1016/j.tig.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shmakov SA, et al. 2017 The CRISPR spacer space is dominated by sequences from the species-specific mobilome. Available at biorxiv.org/content/early/2017/05/12/137356. Accessed July 18, 2017.
- 63.Kuduvalli PN, Rao JE, Craig NL. Target DNA structure plays a critical role in Tn7 transposition. EMBO J. 2001;20:924–932. doi: 10.1093/emboj/20.4.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waddell CS, Craig NL. Tn7 transposition: Two transposition pathways directed by five Tn7-encoded genes. Genes Dev. 1988;2:137–149. doi: 10.1101/gad.2.2.137. [DOI] [PubMed] [Google Scholar]
- 65.Stellwagen AE, Craig NL. Gain-of-function mutations in TnsC, an ATP-dependent transposition protein that activates the bacterial transposon Tn7. Genetics. 1997;145:573–585. doi: 10.1093/genetics/145.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Biery MC, Stewart FJ, Stellwagen AE, Raleigh EA, Craig NL. A simple in vitro Tn7-based transposition system with low target site selectivity for genome and gene analysis. Nucleic Acids Res. 2000;28:1067–1077. doi: 10.1093/nar/28.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rao JE, Craig NL. Selective recognition of pyrimidine motif triplexes by a protein encoded by the bacterial transposon Tn7. J Mol Biol. 2001;307:1161–1170. doi: 10.1006/jmbi.2001.4553. [DOI] [PubMed] [Google Scholar]
- 68.Peters JE, Craig NL. Tn7 recognizes transposition target structures associated with DNA replication using the DNA-binding protein TnsE. Genes Dev. 2001;15:737–747. doi: 10.1101/gad.870201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rutkauskas M, et al. Directional R-loop formation by the CRISPR-Cas surveillance complex cascade provides efficient off-target site rejection. Cell Rep. 2015;10:1534–1543. doi: 10.1016/j.celrep.2015.01.067. [DOI] [PubMed] [Google Scholar]
- 70.Levy A, et al. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015;520:505–510. doi: 10.1038/nature14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cousineau B, et al. Retrohoming of a bacterial group II intron: Mobility via complete reverse splicing, independent of homologous DNA recombination. Cell. 1998;94:451–462. doi: 10.1016/s0092-8674(00)81586-x. [DOI] [PubMed] [Google Scholar]
- 72.Ichiyanagi K, et al. Retrotransposition of the Ll.LtrB group II intron proceeds predominantly via reverse splicing into DNA targets. Mol Microbiol. 2002;46:1259–1272. doi: 10.1046/j.1365-2958.2002.03226.x. [DOI] [PubMed] [Google Scholar]
- 73.Koonin EV, Krupovic M. 2015 A moveable defense. The Scientist. Available at www.the-scientist.com/?articles.view/articleNo/41702/title/A-Movable-Defense/. Accessed July 18, 2017.
- 74.Krupovic M, Makarova KS, Forterre P, Prangishvili D, Koonin EV. Casposons: A new superfamily of self-synthesizing DNA transposons at the origin of prokaryotic CRISPR-Cas immunity. BMC Biol. 2014;12:36. doi: 10.1186/1741-7007-12-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seed KD, Lazinski DW, Calderwood SB, Camilli A. A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature. 2013;494:489–491. doi: 10.1038/nature11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marchler-Bauer A, et al. CDD: Conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;41:D348–D352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Edgar RC. PILER-CR: Fast and accurate identification of CRISPR repeats. BMC Bioinformatics. 2007;8:18. doi: 10.1186/1471-2105-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morgulis A, et al. Database indexing for production MegaBLAST searches. Bioinformatics. 2008;24:1757–1764. doi: 10.1093/bioinformatics/btn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 85.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.