Abstract
Biocatalysts, especially enzymes, have the ability to catalyze reactions with high product selectivity, utilize a broad range of substrates, and maintain activity at low temperature and pressure. Therefore, they represent a renewable, environmentally friendly alternative to conventional catalysts. Most current industrial-scale chemical production processes using biocatalysts employ soluble enzymes or whole cells expressing intracellular enzymes. Cell surface display systems differ by presenting heterologous enzymes extracellularly, overcoming some of the limitations associated with enzyme purification and substrate transport. Additionally, coupled with directed evolution, cell surface display is a powerful platform for engineering enzymes with enhanced properties. In this review, we will introduce the molecular and cellular principles of cell surface display and discuss how it has been applied to engineer enzymes with improved properties as well as to develop surface-engineered microbes as whole-cell biocatalysts.
1. Introduction to Biocatalysis
Biocatalysis is the subfield of reaction engineering concerned with the application of enzymes for chemical production.1,2 Similar to conventional catalysts, enzymes are not consumed by a chemical reaction; instead, both accelerate the reaction rate by providing an alternative reaction path characterized by a lower activation energy. While the fundamental thermodynamics governing the action of enzymes and conventional catalysts are identical, several properties associated with enzymes are considered to be advantageous for industrial processes,3 such as higher catalytic efficiency,2 product selectivity,3–6 ability to catalyze chemical reactions under mild conditions,7 and biodegradability.3,4,6 As such, the global market for industrial biocatalysis is expected to grow to 5.5 billion USD through 2015 with increasing applications in the production of detergents, alcohols, textiles, and pharmaceuticals.8
Current industrial biocatalysis is based largely on the use of cell-free or intracellular enzyme systems (Figure 1). Cell-free systems are typically used for cofactor-independent reactions1 and require purification of the enzymes from high-density cell cultures using liquid chromatography or liquid–liquid extraction to minimize downstream separation difficulties and competing reactions from contaminant enzymes in the cell lysate.9 Cell-free enzymes have direct access to substrate in solution and, as a result, often exhibit reaction-rate limited kinetics. However, these systems lack the important ability to piggyback on the natural metabolic network of the cell, limiting their application to simple reactions. In contrast to cell-free systems, intracellular enzyme systems first require internalization of the substrate via active or passive transport mechanisms. Because the rate of substrate internalization is almost always lower than the rate of reaction, the kinetics governing intracellular enzyme biocatalysts are mass-transport limited and typically 10- to 100-fold slower10,11 than cell-free enzymes. Despite slower kinetics, intracellular enzyme expression systems are commonly used in chemical synthesis.1 This is primarily due to their ability to utilize cofactor-dependent enzymes, as living cells can couple metabolic reactions to regenerate oxidized and reduced forms of cofactors. Additionally, these systems are less expensive to prepare than purified cell-free enzymes, and the tightly regulated intracellular environment protects enzymes from toxic and potentially inhibitory reaction conditions.12,13
Figure 1.
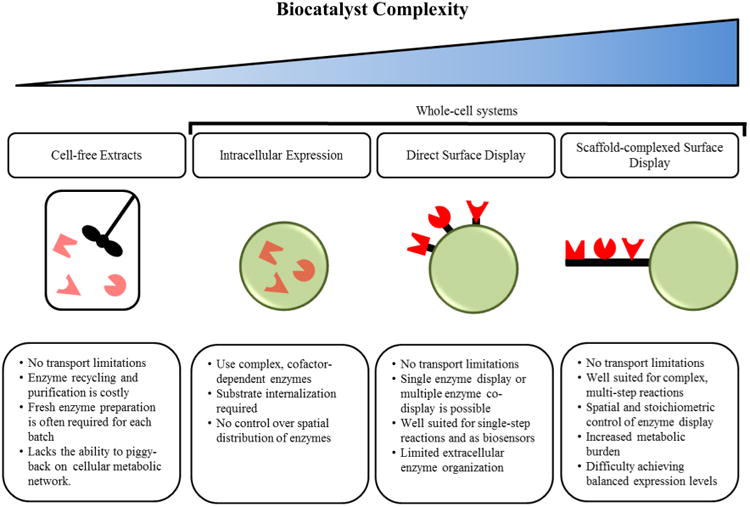
Comparison of biocatalytic systems ranging in complexity. The pros and cons of each system are summarized.
Despite demonstrated success, many industries have been reluctant to adopt enzyme biocatalysts in large-scale production processes. The most commonly cited drawbacks associated with enzyme biocatalysts have included process condition limitations, susceptibility to substrate/product inhibition, decreased activity in non-aqueous environment, low production capacity, and the lack of naturally occurring enzymes capable of mediating desired chemical transformations.14 Addressing some of these shortcomings, the breakthroughs in our understanding of cellular biology and protein engineering have helped to rapidly advance the field of biocatalysis. This evolution has been described by Bornscheuer et al. as the “three waves of biocatalysis”15 (Figure 2). In particular, the third wave, characterized by the implementation of directed evolution strategies, pioneered by Frances Arnold and Willem Stemmer around the turn of the 21st century,16 has been the most crucial of all. In addition to enhancing existing enzymatic properties, directed evolution has empowered researchers to develop enzymes with novel capabilities and utilize heterologous reaction pathways.15 The maturation of cell surface display technology at the turn of the second and third waves enabled researchers to harness its natural ability to link genotype and phenotype as a powerful library-screening tool for protein engineering, especially via directed evolution. Moreover, novel whole-cell biocatalysts have been engineered using surface-displayed enzymes. Enzyme cell surface display allows whole cells to readily access soluble substrate while retaining the metabolic potential of intracellular enzyme systems (e.g., extracellular degradation of cellulose and internalization of the major product, glucose, to produce ethanol). In this review, we will compare commonly used surface display systems and discuss how they, coupled with breakthroughs in the field of protein engineering, have generated new excitement about expanding the use of engineered enzyme biocatalysts in industry.
Figure 2.
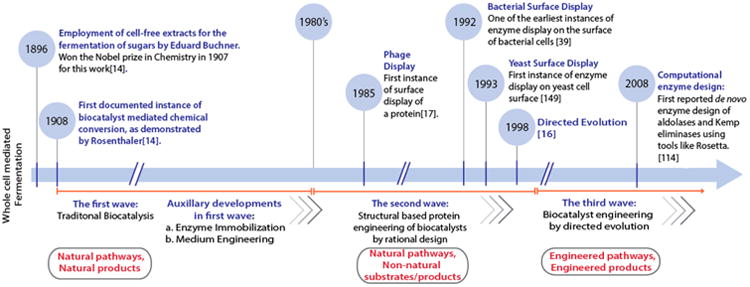
Timeline depicting the synergistic overlap of biocatalysis evolution and maturation of surface display39,149 as a tool for biocatalyst engineering. The evolution of biocatalysis, detailed in three waves,15 transitions from the utilization of natural metabolic pathways for the production of natural products (first wave) to the exploitation of natural pathways and enzymes for the generation of non-natural products (second wave) to engineering non-natural pathways for novel product generation (third wave).
2. Molecular and Cellular Principles of Cell Surface Display
Cell surface display systems have explored a variety of host organisms with a varying degree of complexity ranging from viral capsids17,18 to bacterial spores19 to prokaryotes20 to eukaryotes.21 Despite differences in the host organisms, surface display systems often have three core features in common: (a) a signal peptide to direct the protein of interest toward the secretory pathway, (b) an endogenous surface protein amenable to recombination (i.e., insertion, deletion, and fusion) to enable stable surface anchorage of the target protein, and (c) an epitope tag to facilitate the detection of successful surface display. Currently, no one-size-fits-all surface display platform exists, and the most suitable host system depends on several factors, including the complexity of the recombinant enzyme being displayed and its intended application (Table 1). In this section, we will introduce and compare some of the more widely used surface display systems pertinent to biocatalysis.
Table 1. Comparison of Cell Surface Display Systems with Significance in Biocatalysis.
| phage | prokaryote | eukaryote | |
|---|---|---|---|
| Commonly used host organisms | Filamentous phages fd, M13; Enterobacteria phages T4, T7, lambda, phagemid | E. coli, B. subtilis, L. bacillus, S. carnosus | S. cerevisiae, P. pastoris |
| Typical library size | 1 × 1010 to 1 × 1012 | 1 × 108 to 1 × 1010 | 1 × 106 to 1 × 107 |
| Post-translational machinery | Capable of simple post- translational modifications | Capable of moderate post-translational modifications | Capable of sophisticated post-translational modifications |
| Surface anchorage | Capsid proteins | Lpp-OmpA, autotransporter proteins, ice nucleation proteins | Agglutination proteins, flocculation proteins |
| Main applications | Peptide affinity maturation, peptide–DNA interactions, phage assisted continuous evolution (PACE) | Directed evolution for modification of nonmammalian proteins, biocatalysis, genotype–phenotype studies | Directed evolution of mammalian proteins, biocatalysis, genotype–phenotype studies |
2.1. Phage Display Systems
Bacteriophages (phages) comprise a diverse group of viruses that infect prokaryotic organisms. One of the first successful demonstrations of the surface display of peptides was accomplished using a phage display system.17 Phage display has been, and continues to be, used for many diverse applications, including antibody affinitymaturation,22–24 selection of molecular imaging probes,25,26 antigen identification for vaccine development,27,28 and the discovery of novel enzyme substrates and inhibitors.29–31 However, phage display is inherently limited by the post-translational machinery of its bacterial host as well as the size of the target protein. As a result, phage display systems lack the ability to present diverse libraries of large, full-length heterologous enzymes. Nevertheless, they still play an important role in improving the properties of enzyme biocatalysts. For example, phage display has been used to select improved enzyme subunits,32 to study substrate binding and enzyme inhibition,33 and to assess the interactions between small molecules and enzymes.33
2.2. Prokaryotic Display Systems
In order to account for some of the limitations associated with phage display, a number of prokaryotic cell-surface display systems have been developed. The greatest advantage of prokaryotic display over phage display is the ability to screen and sort cells using fluorescence activated cell sorting (FACS).34 Prokaryotic systems are also capable of presenting larger, more complex polypeptides than phage display systems.35 Furthermore, most bacteria express a high number of native anchor proteins. This allows for high-level surface display of foreign enzymes without exhibiting avidity effects associated with the use of some abundant phage capsid proteins.36 Escherichia coli has emerged as the most widely used host primarily due to its well-established genetic toolbox and its ability to achieve high-density surface display of full-length recombinant proteins. A detailed review of diverse prokaryotic surface display systems can be found elsewhere.37,38 Here, we will provide an overview that will serve to emphasize the significance of prokaryotic cell surface display in engineering novel and improved biocatalysts.
E. coli cell surface display is accomplished by fusing a target protein (the passenger) to a native surface protein (the carrier) anchored in the outer membrane. The carrier protein mediates the transport of the passenger through the E. coli inner membrane and periplasm for stable anchoring in the outer membrane. The properties of the carrier protein dictate the orientation (i.e., N- or C-terminal fusion) and the size limit of the target gene. For example, complications in outer membrane localization using outer membrane protein A (OmpA) as the carrier were observed when attempting to surface display large target proteins.39 This issue was addressed by fusing the signal and the first nine amino acids of a lipoprotein (Lpp) to the N-terminal region of OmpA and has been used to display many functionally diverse heterologous enzymes.40,41
While Lpp-OmpA is among the most widely used carrier proteins for prokaryotic surface display, recent efforts have focused on two carrier proteins: autotransporter proteins42 and ice nucleation proteins (INPs).43 Autotransporter protein display (autodisplay) has two significant advantages over traditional prokaryotic surface display systems including Lpp-OmpA. First, autotransporter proteins are relatively simple, modular proteins.44 This modular structure allows the native passenger protein (typically, a virulence factor) to be readily exchanged to display the desired heterologous enzyme on the bacterial cell surface.45 The second advantage of the autodisplay systems lies in the mobility of the β-barrel anchor,45 which allows the autodisplayed proteins to form multimers by associating in the membrane after expression. This has been demonstrated for several dimeric enzymes46,47 and could be adapted to display functional heterodimers and multimers.48 Autodisplay systems are also capable of displaying >180 000 copies of autotransporter-fused enzyme per cell, rivaling the expression of endogenous E. coli surface proteins (100 000–200 000 copies).44 Similar to autodisplay, INP-based display systems are capable of achieving higher surface display levels than Lpp-OmpA.49 In addition, INPs contain a highly degenerate internal region that has been used as a scalable linker to display heterologous enzymes at varying distances from the cell surface.50 This has allowed large fusion proteins to be surface displayed without prokaryotic cell-surface interference.
While greatly expanding the potential of cell surface display beyond phage display systems, prokaryotic systems also have their own limitations. Prokaryotes are capable of performing only relatively simple post-translational modifications and are known to exhibit expression bias against more complex mammalian proteins.21 Additionally, most prokaryotes lack the innate machinery to produce and tolerate high levels of toxic metabolites, such as ethanol.51
2.3. Eukaryotic Display Systems
To address some of the limitations of prokaryotic display, several eukaryotic surface display systems have been explored, ranging from yeast to insect52 to mammalian cell display.53 These eukaryotic display systems, particularly yeast display, offer several distinct advantages, including the ability to fold, process, and present complex heterologous proteins,21 the availability of an established genetic toolbox, and the ability to tolerate harsh conditions by exhibiting cross-protection54–56 against environmental stresses. A detailed review of yeast surface display mechanisms can be found elsewhere.57,58 Here, we will introduce some of the more common yeast surface display systems with demonstrated relevance in biocatalysis.
The mating-type-specific agglutination proteins, a- and α-agglutinin, have been among the most widely used anchor proteins for yeast surface display.57 The a-agglutinin protein consists of two units linked by a disulfide bridge: the Aga1 anchor and an Aga2 carrier. This disulfide bridge is a prominent feature of a-agglutinin-based display, as it provides a natural spacer between the target protein and the cell surface. In contrast, the α-agglutinin protein consists of a single anchor unit, Agα1. Another important difference between the two agglutinin-based surface display systems is the orientation of the target protein. a-Agglutinin is more commonly used for enzymes with a C-terminal functional domain, whereas α-agglutinin is more commonly used for those with a functional domain near the N-terminus.58,59 The agglutinin-based display systems have been adapted to display a diverse array of peptides, enzymes, and structural proteins on the yeast cell surface for a plethora of applications ranging from antibody affinity maturation to biofuel production.58–64 Another group of anchor proteins used for yeast surface display are flocculation proteins. The yeast flocculation protein, Flo1,65 has been widely used to display functional lipases on the surface of both Saccharomyces cerevisiae66 and Pichia pastoris.67,68 Additionally, it has been shown in some cases that the fusion of the target enzyme to the flocculation protein does not abolish its native function.69,70 The resulting flocculation phenotype is often desirable because it can serve as an indicator of nutrient depletion in media, and the yeast flocs can be easily separated from a liquid suspension.71,72
While yeast surface display has helped to advance the field of protein engineering, it is not without its limitations. First, yeast cell growth is significantly slower than that of most bacteria, which may hamper the economic viability of processes utilizing yeast-based whole-cell biocatalysts. Another commonly cited drawback associated with yeast surface display is its low transformation efficiency (Table 1), which limits the enzyme mutant library size of directed-evolution-based engineering strategies in yeast, as discussed below.
3. Cell Surface Display for Engineering Industrial Biocatalysts
While nature has provided a plethora of functionally diverse enzymes, most have evolved to operate within a narrow range of physiological conditions. As a result, many wild-type enzymes cannot function in the harsh environments associated with industrial processes. To address this issue, researchers have used directed evolution and cell surface display to engineer and select mutant enzymes better suited to handle industrial processing (Section 3.1). In addition to serving as a powerful tool for protein engineering, cell surface display of modified or wild-type enzymes can be used to create novel whole-cell biocatalysts for use in chemical processes ranging from biofuel production and petroleum refining to environmental pollutant detection.58,73–76 Two surface-display strategies are currently being explored for this purpose: (a) direct surface display of multiple enzymes in a noncomplexed form (Section 3.2) and (b) complexed display of enzymes organized on surface-displayed protein scaffolds (Section 3.3) as depicted in Figure 1.
3.1. Direct Cell Surface Display of Single Enzymes
The simplest form of cell surface display employs the direct genetic linkage of the target enzyme to a transmembrane anchorage protein of the cell. Coupling this direct cell surface display of single enzymes with directed evolution has emerged as a powerful approach for engineering and high-throughput screening of biocatalysts with improved properties. A typical application of this approach consists of five steps (Figure 3): (i) selection of an enzyme as the engineering target, (ii) generation of a library of enzyme mutants by random or semirational mutagenesis, (iii) cellular expression of the mutant library, (iv) cell surface display of the expressed enzymes for selection of mutants exhibiting enhanced properties, and (v) reiteration of these four steps to enrich the selected mutant populations with improved properties.76–78 In this section, we will describe how cell surface display of single enzymes has been used in conjunction with directed evolution to engineer novel and improved biocatalysts. Specific properties with importance in industrial processes include, but are not limited to, thermostability, tolerance to organic solvents, activity, substrate specificity, and enantioselectivity of reaction products.79-81 Some of the recent applications of the five-step engineering approach have been summarized in Table 2. In the discussion below, we will focus on studies that have introduced new aspects to the five-step engineering approach.
Figure 3.
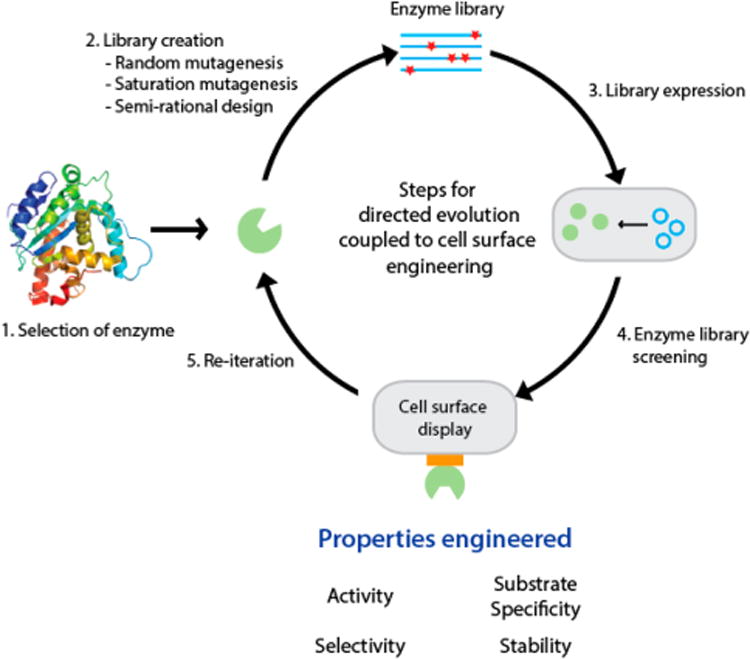
Overview of the application of cell surface display as a high-throughput screening tool for biocatalyst engineering via directed evolution.
Table 2. Recent Progress in Biocatalyst Engineering Based on the Use of Cell Surface Engineering as a Tool for Directed Enzyme Evolution.
| enzyme | enzyme characteristics | anchor protein | host organism | property engineered | |
|---|---|---|---|---|---|
| Oxidoreductase | Horseradish peroxidase | 44 kDa; glycoprotein metalloenzyme, heme group, 2 calcium atoms; single chain polypeptide, 4 disulfide bridges | Aga2 | S. cerevisiae | Activity104 Enatioselecivity110 |
| Glucose dehydrogenase | 112 kDa; short chain dehydrogenase reducatase enzyme; homotetramer | INP | E. coli | Substrate specificity and stability96 | |
| Transferase | Sfp phosphor-pantetheinyl transferase | 26 kDa; protein posttranslational modification (PTM) enzyme; monomer, two domains, pseudo 2-fold symmetry | pIII | M13 phage | Substrate specificity97 |
| Hydrolase | Lipase B from Candida antarctica | 33 kDa; catalytic triad of Ser-His-Asp; no hydrophobic lid = no interfacial activation; α/β hydrolytic fold | Cwp Flo1p |
H. polymorpha P. pastoris |
Activity94 Thermostability85 |
| Human β-glucuronidase | 312 kDa; glycoprotein, TIM barrel; homotetramer | B7-1 | Mouse fibroblasts | Activity93 | |
| ApeE esterase | 67 kDa; GDSL family outer membrane serine esterase/lipase | AIDA-I | E. coli | Activity150 | |
| Rhizomucor miehei lipase | 31.6 kDa; single polypeptide chain, singly wound β sheet domain; catalytic triad Ser-His-Asp, amphipathic helix lid | Flo1p | P. pastoris | Stability89,90 | |
| Cel5A hydrolase | 38 kDa; single catalytic domain, compact 8-fold β/α-barrel; Cys–Cys disulfide bridge | INP | E. coli | Thermostability84 | |
| Tobacco Etch Virus protease | 27 kDa; Cys endopeptidase; cleavage product of single massive polyprotein, two domain antiparallel β-barrel fold | Aga2 | S. cerevisiae | Substrate specificity100 | |
| Bacillus subtilis lipase A | 19 kDa; catalytic triad Ser-His-Asp; no interfacial activation; compact minimal α/β hydrolytic fold | pIII | M13 phage | Enantioselectivity112 | |
| Lyase | Bacillus subtilis endoxylanase XynA | 21.2 kDa; glycosyl hydrolase; β-jelly roll fold | pIII | M13 phage | Thermostability83 |
| Ligase | Non-ribosomal peptide synthetase | Large multimodular enzyme | Aga2 | S. cerevisiae | Substrate specificity92 |
3.1.1. Enzyme Stability and Solvent Tolerance
Many industrial processes operate at temperatures, pH ranges, and chemical/solvent conditions not optimal for maintaining the stability of wild-type enzymes. Therefore, a considerable effort has been made to engineer more robust enzyme biocatalysts with improved stability under non-ideal conditions. One of the most industrially relevant properties of biocatalysts is their thermostability. To date, only a few studies have reported the use of cell surface display to improve the thermostability of enzymes (Table 2). This may be due to the intrinsic thermal instability of unadapted living cells and protective mechanisms triggered (upregulation of heat shock proteins, suspension of protein transcription and translation, etc.) under extreme temperature conditions82 that can detrimentally affect the expression level of the enzyme library. Studies have evaluated the thermostability profile of mutated enzymes using cell surface display up to moderately high temperatures (70 °C) and showed greater than 50% residual activity for a relatively short time duration (a few hours to 4–5 days).83–86
Lipases are one class of enzymes that have demonstrated the potential to generate biofuels by the esterification of free fatty acids. However, many lipases can be inactivated not only at high temperatures but also in the presence of organic solvents such as methanol, both of which are required for efficient biodiesel production.87 For this reason, considerable research has been focused on improving the methanol tolerance of biodiesel-producing lipases, in addition to thermostability.85,88,89 However, the aqueous conditions of high-throughput screening systems designed to identify hydrolytic enzyme activity is not suitable for selecting lipases, of which the catalytic activity requires a non-aqueous environment. To address this issue, Zhang et al.90 developed a new screening technique to identify thermostable mutants of Rhizomucor miehei lipase (RML). Saturation mutagenesis was used to generate the mutant libraries, and the resulting RML variants were displayed on the surface of yeast cells. Unlike the conventional surface screening techniques applied in most studies described in Table 2, screening for thermostable RML mutants was performed at 70 °C by combining yeast surface display with pH monitoring and colony plating using organic media into a comprehensive high-throughput screening system.90 Thermostable enzymes were identified by heating the plates with yeast colonies grown on BMMG (buffered minimal methanol media)/BMMY (buffered methanol complex media) prior to the addition of the reaction substrate. Active and thermostable mutants then utilized the substrate, tributyrin, to form a halo around the corresponding yeast colony. This technique enabled the visual identification of improved lipases via non-aqueous screening, allowing for the rapid selection of thermostable mutants.
3.1.2. Enzyme Activity and Substrate Specificity
Being crucial parameters for the overall economy of a process, extensive efforts have been made to improve the activity and substrate specificity of enzymes using cell surface display60,75,91–97 (Table 2). However, the majority of such studies have relied on conventional low-throughput assays for mutant identification, similar to those used for soluble proteins, indicating the employment of cell surface display to bypass protein purification rather than a high-throughput screening tool. For protein affinity engineering, cell surface display has been routinely used in combination with FACS, which allows the screening of a few thousand cells per second.98 To harness its power in high-throughput screening of improved enzymes, the main challenge lies in establishing a stable link between the activity or substrate specificity of a mutant enzyme with the clone that harbors that particular mutant, i.e., preventing dissociation of the reaction product from the cell that expresses that enzyme. Thus, there is a need for a more elegant design that can help to overcome this particular drawback, and two recent studies on protease engineering using cell surface display have done just that. Several industries, including sanitation, food, pharmaceuticals, biomedical, and waste management, have seen a growing interest in employing protease-based biocatalysts in their processes.99 Although similar in their approaches, which involved FACS detection of the products generated from selective proteolysis of a multidomain protein substrate, the two studies differed in the location where the proteolytic cleavage took place.
In the first system,100 named yeast endoplasmic reticulum sequestration screening (YESS), proteolysis occurred intra-cellularly in the endoplasmic reticulum (ER) due to the addition of an ER retention signal peptide that directed both the enzyme variant and multiepitope tagged substrate to the ER. Protease mutants with altered substrate specificities cleaved at different residues, generating products with unique epitope tag combinations, but each proteolytic event resulted in the removal of the ER retention sequence. As a result, each uniquely tagged product was displayed on the yeast surface as an Aga2 fusion protein. Surface staining using epitope-specific antibodies coupled with FACS analysis enabled the detection of unique fluorescent profiles, which indicated selective cleavage by the protease variant. Additionally, improved activity of the enzyme mutant was identified based on the intensity of the fluorescence.
In the second study,101 enzymatic cleavage took place extracellularly by co-displaying the enzyme variant and the substrate on the cell surface. The authors showed that a protease variant with altered substrate specificity would cleave at amino acid residues other than a basic residue, such as hydrophobic, aromatic, polar, or negatively charged residues. This differential cleavage specificity was linked to the release or retention of an autoinhibitory peptide on the substrate. Removal of the inhibitor peptide by an altered protease would enable binding of a fluorescent dye, whereas its retention would not allow fluorescence labeling. The difference in fluorescence profiles could then be detected by high-throughput screening using FACS. Besides detection of altered specificity profiles, the designs used in these two systems also demonstrated the additional advantage of quantitative evaluation of enzyme activity based on the intensity of the fluorescence profiles.
Recent years have seen an increased interest in developing highly sensitive screening techniques based on microfluidics.102,103 By combining in vitro compartmentalization, microfluidics, and cell surface display, Agresti et al.104 demonstrated a new screening system that takes advantage of physical separation at the single-cell level, an advantage that is otherwise lost by evaluating a mixed population of cells displaying different variants of the enzyme on the surface. Specifically, yeast cells were engineered to display libraries of horseradish peroxidase (HRP) mutants as Aga-2 fusion proteins. The two libraries were generated by error-prone PCR (epPCR) followed by saturation mutagenesis and staggered expression process, respectively. In this system, a distinctive shift from conventional selection process was the compartmentalization of the recombinant yeast cells in picoliter droplets, with each droplet containing at most one yeast cell displaying one particular enzyme mutant. Using a microfluidics device, each picoliter droplet was sorted based on the presence or absence of a fluorescent signal. Droplets with desired enzyme mutants appeared as fluorescent bubbles, whereas those with no yeast cells or inactive enzymes did not emit light. With a sorting rate of 1000 events/s, this ultra-high-throughput system was capable of screening a library size of ∼108 in about 10 h,104 thus offering an efficiency comparable to that of current high-throughput screening techniques, such as FACS and microtiter plate screening.
3.1.3. Enzyme Enantioselectivity
Establishing a stable genotype-phenotype link is vital to the success of any directed evolution process.105 Historically, this has proven to be a significant challenge in developing effective screening methods for selection of enantioselective mutants, as the property of enantioselectivity is a kinetic one. Thus, linking high enantioselectivity of a beneficial mutant to the cell expressing that mutant is difficult.106 A number of studies have been devoted to the development of high-throughput screening systems with the ability to elucidate the relationship between enzyme mutations and the resulting product selectivity107–112 (Table 2). One of the more promising studies regarding enantioselectivity engineering was performed by developing a novel, high-throughput screening system called enzyme screening by covalent attachment of products via enzyme display (ES-CAPED).113 Traditional screening systems allow for the immediate detachment of the enantiomer product following the enzymatic reaction. ESCAPED differs by maintaining a covalent attachment of the reaction product with the enzyme on the host cell surface following the reaction and is therefore amenable to flow cytometric analysis. This system was validated by displaying an error-prone PCR mutant library of the Pseudomonas aeruginosa esterase (EstA) on the surface of E. coli.114 The highly selective mutant enzyme presented on the cell surface catalyzed a peroxide radical mediated reaction for the hydrolysis of a tyramide ester substrate (S- or R-2-MDA). The reaction product was immediately covalently attached to the surface of the bacterial cell due to radical transfer from the tyramide group to a tyrosine residue of an endogenous surface protein.115 The surface-displayed enzyme–product complexes were stained with enantiospecific antibodies and screened using FACS to identify mutants exhibiting a high degree of enantioselectivity. This elegant study provided a turning point in simplifying surface engineering of enantioselectivity of biocatalysts.
3.2. Direct Cell Surface Display of Multiple Enzymes
In addition to serving as a powerful screening platform for enzyme engineering, direct surface display of single enzymes has shown some biocatalytic potential. For example, whole-cell biocatalysts displaying single enzymes have been used to degrade toxic pollutants116 and exhibited greater activity in biodiesel production than enzymes immobilized on resin beads.117 However, these systems are largely restricted to simple, one-step reactions. In contrast, direct surface display of multiple enzymes has been used to engineer novel biocatalysts capable of acting on multiple substrates. One area where direct cell surface display of multiple enzymes has demonstrated success is in the detection or degradation of some pollutants (a process known as bioremediation). One common target of bioremediation is highly toxic organophosphorus pesticides (OPs). OPs have been widely employed in the agricultural industry for decades, and the development of biocatalysts that degrade toxic OPs is an active area of research.11,75,118 A number of successful whole-cell biocatalysts using direct display of OP-degrading enzymes have been developed,75,119,120 including a system utilizing co-display of two enzymes for simultaneous OP detection and degradation.118 In this system, organophosphorus hydrolase (OPH) and a methyl parathion hydrolase (MPH)–GFP fusion were co-displayed using truncated INP and Lpp-OmpA anchors, respectively. Because GFP fluorescence intensity is sensitive to environmental pH and the degradation of OPs by OPH and MPH generates protons, the surface-engineered E. coli strain was able to detect and degrade a broad range of OPs by monitoring changes in GFP fluorescence.118
Whole-cell biocatalysts utilizing surface-displayed enzymes are also being explored for the production of biofuels. For example, direct co-display of multiple cellulases has shown potential in the degradation of crystalline cellulose, a major bottleneck in the conversion of biomass to biofuels.121 One of the most recent breakthroughs in utilizing direct co-display of cellulases for the degradation of crystalline cellulose has been the incorporation of small, non-enzymatic proteins called expansins.122 As their name suggests, expansins are believed to act on crystalline cellulose by disrupting hydrogen bonding between cellulose microfibrils, disrupting the structure and increasing the surface area accessible to cellulase enzymes.123 In a recent study, Nakatani et al.124 co-expressed three cellulases on the S. cerevisiae cell surface in addition to a fungal expansin-like protein as Agα;1-fusions. The resulting strain was able to increase ethanol production from phosphoric acid swollen cellulose (PASC) from 2.3 to 3.4 g/L. This is one of the first examples of using direct surface display of both enzymatic and non-enzymatic proteins to harness enzyme synergy.
Whole-cell biocatalysts utilizing direct surface display have received increasing attention as potential food-grade biocatalysts. Innocuous whole-cell biocatalysts not only represent an environmentally friendly alternative to traditional catalytic processes but also have the potential to act on foodstuffs without the use of harmful materials in the production process. For example, the most common method for chemical synthesis of vanillin requires the toxic precursors phenol and guaiacol.125 E. coli whole-cell biocatalysts expressing intracellular enzymes have been engineered to produce vanillin using innocuous precursors glucose125 and ferulic acid.126 Another advantage associated with food-grade biocatalysts is the relatively low energy requirement for activation. Direct display of lipases on P. pastoris has resulted in the generation of safe, whole-cell biocatalysts capable of efficiently enriching the fatty acids EPA and DHA in fish oil with high selectivity, reducing the energy costs associated with downstream separation.127 P. pastoris displaying lipases have also been used to produce the fruit-flavored esters, isoamyl acetate and cis-3-hexenyl, exceeding 95% conversion.128,129
3.3. Scaffold-Complexed Multienzyme Surface Display
The most recent development in cell surface display has been the engineering of highly specific, modular protein scaffolds. Owing to the ability of scaffold-complexed systems to surface display multiple heterologous enzymes in highly organized patterns, they can be used to catalyze multistep reactions with a higher catalytic efficiency than the direct co-display of multiple enzymes due to enzyme–proximity synergy.130,131 In nature, protein scaffolds are found in some anaerobic bacteria and are made up of binding modules called cohesins.132 These cohesin-containing scaffolds, termed scaffoldins, are loaded with hydrolytic enzymes, each of which has a dockerin domain. The dockerin binds to cohesin in a species-dependent manner to form a cellulosome complex. Novel designer cellulosomes can be engineered by recombining cohesins from different species into a single construct. Dockerins from species corresponding to the desired cohesins can then be fused to target enzymes to create sophisticated enzyme networks in the extracellular space.
Yeast surface display of designer cellulosomes has been demonstrated as a promising platform for ethanol production from lignocellulosic biomass. Because yeast have the innate machinery to produce high levels of ethanol, yeast with surface displayed scaffoldin-complexed cellulases have the potential to directly ferment sugars freed from cellulose to ethanol.133–135 This feature is critical to the development of consolidated bioprocessing (CBP),136,137 a highly integrated process configuration with great potential to reduce the cost of lignocellulosic biofuel production. Therefore, yeast surface display of designer cellulosomes for biomass saccharification is an active research area, and several groups have investigated different methods of organizing enzymes in the extracellular space to harness synergistic effects. We have developed one such system using an Aga2-fused trifunctional scaffoldin complexed with three cellulases: endoglucanase, cellobiohydrolase, and β-glucosidase. The resulting yeast strain showed a 1.6-fold increase in PASC hydrolysis as a result of enzyme–proximity synergy and an additional 5.5-fold increase as a result of enzyme–enzyme synergy.135 This idea has been expanded to include self-surface assembly of a two-member miniscaffoldin on the S. cerevisiae cell surface, where one miniscaffoldin contained an Aga2 fusion and the other contained a dockerin fusion.138 This elegant design enabled the yeast surface display of multiple minicellulosomes with the same three enzymatic activities on a secondary scaffold to achieve hydrolysis of crystalline cellulose. The engineered yeast biocatalyst was able to achieve an ethanol yield of 1.412 g/L. The complexity of yeast-displayed scaffolds has been expanded further to include multivalent scaffold structures containing scaffold chains branching off from a surface-displayed Aga2-fused scaffold139 and even ill-defined amyloid-like oligomeric cohesin scaffolds.140
Another recent study demonstrating improved efficiency of scaffoldin-complexed biocatalysts was performed by Liang et al.,141 in which a pentavalent scaffold was displayed on the surface of S. cerevisiae. The Aga2-fused cohesin scaffold was complexed with an endoglucanase, a cellobiohydrolase, a β-glucosidase, a cellulose dehydrogenase (CDH), and a lytic polysaccharide monooxygenase (LPMO). The LPMO is believed to cleave cellulose in the presence of CDH, increasing the number of active sites available upon which the other scaffoldin-complexed cellulases can act. The addition of the CDH and LPMO enzymes increased the ethanol titers from PASC from 1.8 to 2.7 g/L.
Yeast surface display is not the only system used for scaffoldin-complexed surface display. Bacterial systems utilizing Bacillus subtilis and E. coli have also demonstrated potential to surface display designer cellulosomes.142 In the context of biofuel production and CBP, the optimal host for scaffoldin-complexed surface display of enzymes depends on a number of factors including metabolite tolerance, the innate fermentation machinery, process stability, and the amenability to scale-up. Due to the well-established genetic toolbox and its long history of industrial ethanol production, S. cerevisiae is among the most popular host systems being explored for the production of fuels from cellulosic biomass.51,143
3.4. Biocatalytic Process Considerations for Surface-Displayed Enzymes
Despite success on the laboratory scale, there are several considerations that arise for the industrial use of multicomponent whole-cell biocatalysts (direct co-display and scaffoldin-complexed cell surface display). Like intracellular expression systems, surface-display-based biocatalysts are derived from living organisms and therefore require special processing steps to maintain activity (removal of dead cells, selectable markers to remove wild-type contaminating strains, etc.). Furthermore, complex multicomponent biocatalysts using cell surface display often require the expression of multiple proteins and therefore special attention should be given to balancing protein expression. Additionally, as the number of enzymes expressed in a single whole-cell biocatalysts increases, plasmid stability can become a serious issue when attempting to scale-up biocatalytic processes from the laboratory scale.144 Enzyme expression can be controlled in part by careful selection of the type and strength of the promoters that initiate protein expression. Cells harboring biocatalytic proteins under an inducible promoter will not express that protein until they are given a specific substrate to unlock the target gene for transcription. Surface-displayed proteins regulated by an inducible promoter often express the target protein at high concentrations; however, in many applications, the high levels of foreign protein expression are toxic to cells.145 Therefore, biocatalysts utilizing high-density surface display of target enzymes under inducible promoters are more suited for batch processing. On the contrary, cells harboring enzymes under a constitutive promoter will express the enzyme continuously, albeit at a lower level than enzymes under inducible promoters. This is advantageous in that it does not require the use of expensive inducing agents or separate growth and induction media that can be problematic on a large scale. As a result, constitutive expression is more suited for continuous processes.146 However, it should be noted that there is a lack of strong constitutive promoters in many prokaryotic hosts.145 Some studies have suggested that the combined use of inducible and constitutive promoters enhances the overall expression of the target enzyme.147,148
4. Coclusion and Outlook
While engineering the properties of biocatalysts using cell surface display and directed evolution has greatly expanded their industrial applicability, the majority of these studies have focused on altering one enzyme property at a time. This approach does not take into consideration the consequences of sequence mutations on other enzyme properties. Therefore, future engineering effort should include strategies for multiparameter screening to evaluate several properties of an enzyme simultaneously. Smart design of enzyme libraries is likely to be the key to the success of this effort, underscoring the importance of predictive modeling and rational design in engineering an optimal biocatalyst for a set of conditions. Real-time evaluation of the effect of mutations on multiple enzyme properties could prove to be a more holistic approach to identifying enzyme species optimized for the chemical industry. Cell surface display combined with multiplexed single-cell screening technologies like FACS offers a clear advantage over other library screening techniques in that it offers the potential for multiparameter screening. The development of such a well-designed system is exemplified by the YESS system,100 where the protease was screened for both improved activity and altered selectivity.
Additionally, most biocatalyst engineering strategies have been focused on the local optimization of individual enzymes in isolation. To date, there has been little research focused on the engineering of individual enzymes in the context of a multienzyme system toward a global optimum. Cell surface display, especially co-display and scaffoldin-complexed display systems, represents a great platform to integrate protein engineering with microbial surface engineering for the development of superior industrial whole-cell biocatalysts. As the application of scaffoldin-complexed enzyme biocatalysts continues to advance, collective enzyme engineering strategies will likely be developed to enhance the overall performance of the multienzyme system. This research venue will require rapid, high-dimensional analysis of many enzyme properties, with each property corresponding to different enzymes used for a particular industrial application.
Biocatalytic processes represent a safe, environmentally friendly alternative to traditional catalytic processes. While chemicals produced in biocatalytic processes currently account for 6.2% of all chemical sales, they are expected to grow to 19.2% of all chemical sales by 2020.13 The transition toward intracellular whole-cell biocatalysis has been ongoing for the past decade, and several large chemical companies including DSM, Lonza, and BASF are developing processes utilizing whole-cell biocatalysts for the production of insecticides, antibiotics, and drug precursors.1 While no large-scale production processes utilizing cell surface display are used in industry today, this increasing prevalence of biocatalytic systems in industry coupled with advances in cell surface display will likely accelerate the adoption of whole-cell biocatalysts displaying enzymes.
Acknowledgments
The authors would like to acknowledge the financial support provided by the Department of Chemical Engineering and MCubed at the University of Michigan and NIH/NCI grant CA191952. We would also like to thank members of the Wen research group for insightful feedback and comments on this review.
Footnotes
Notes: The authors declare no competing financial interest.
References
- 1.Schmid A, Dordick JS, Hauer B, Kiener A, Wubbolts M, Witholt B. Industrial biocatalysis today and tomorrow. Nature. 2001;409:258–268. doi: 10.1038/35051736. [DOI] [PubMed] [Google Scholar]
- 2.Rothenberg G. Kirk-Othmer Encyclopedia of Chemical Technology. John Wiley & Sons, Inc.; Hoboken, NJ: 2000. [Google Scholar]
- 3.Johannes TW, Simurdiak MR, Zhao H. Biocatalysis. In: Lee S, editor. Encyclopedia of Chemical Processing. Taylor & Francis; New York: 2006. pp. 101–110. [Google Scholar]
- 4.Schoemaker HE, Mink D, Wubbolts MG. Dispelling the myths—biocatalysis in industrial synthesis. Science. 2003;299:1694–1697. doi: 10.1126/science.1079237. [DOI] [PubMed] [Google Scholar]
- 5.Faber K. Biotransformations in Organic Chemistry. Springer-Verlag; Berlin: 2011. pp. 9–10. [Google Scholar]
- 6.Bommarius A, Riebel B. Biocatalysis: Fundamentals and Applications. Wiley-VCH; Weinheim, Germany: 2004. Introduction to biocatalysis; pp. 1–18. [Google Scholar]
- 7.Frock AD, Kelly RM. Extreme thermophiles: moving beyond single-enzyme biocatalysis. Curr Opin Chem Eng. 2012;1:363–372. doi: 10.1016/j.coche.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jemli S, Ayadi-Zouari D, Hlima HB, Bejar S. Biocatalysts: application and engineering for industrial purposes. Crit Rev Biotechnol. 2014;8551:1–13. doi: 10.3109/07388551.2014.950550. [DOI] [PubMed] [Google Scholar]
- 9.Straathof AJ, Adlercreutz P, editors. Applied Biocatalysis. 2nd. CRC Press; Boca Raton, FL: 2000. [Google Scholar]
- 10.Ni Y, Chen RR. Accelerating whole-cell biocatalysis byreducing outer membrane permeability barrier. Biotechnol Bioeng. 2004;87:804–811. doi: 10.1002/bit.20202. [DOI] [PubMed] [Google Scholar]
- 11.Yang J, Liu R, Jiang H, Yang Y, Qiao C. Selection of a whole-cell biocatalyst for methyl parathion biodegradation. Appl Microbiol Biotechnol. 2012;95:1625–1632. doi: 10.1007/s00253-011-3792-3. [DOI] [PubMed] [Google Scholar]
- 12.Ishige T, Honda K, Shimizu S. Whole organism biocatalysis. Curr Opin Chem Biol. 2005;9:174–180. doi: 10.1016/j.cbpa.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Schüürmann J, Quehl P, Festel G, Jose J. Bacterial whole-cell biocatalysts by surface display of enzymes: toward industrial application. Appl Microbiol Biotechnol. 2014;98:8031–8046. doi: 10.1007/s00253-014-5897-y. [DOI] [PubMed] [Google Scholar]
- 14.Reetz MT. Biocatalysis in organic chemistry and biotechnology: past, present, and future. J Am Chem Soc. 2013;135:12480–12496. doi: 10.1021/ja405051f. [DOI] [PubMed] [Google Scholar]
- 15.Bornscheuer UT, Huisman GW, Kazlauskas RJ, Lutz S, Moore JC, Robins K. Engineering the third wave of biocatalysis. Nature. 2012;485:185–194. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- 16.Arnold F. Design by directed evolution. Acc Chem Res. 1998;31:125–131. [Google Scholar]
- 17.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 18.Makela A, Oker-Blom C. The baculovirus display technology—an evolving instrument for molecular screening and drug delivery. Comb Chem High Throughput Screening. 2008;11:86–98. doi: 10.2174/138620708783744525. [DOI] [PubMed] [Google Scholar]
- 19.Xu X, Gao C, Zhang X, Che B, Ma C, Qiu J, Tao F, Xu P. Production of N-acetyl-d-neuraminic acid by use of an efficient spore surface display system. Appl Environ Microbiol. 2011;77:3197–3201. doi: 10.1128/AEM.00151-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freudl R, MacIntyre S, Degen M, Henning U. Cell surfaceexposure of the outer membrane protein OmpA of Escherichia coli K-12. J Mol Biol. 1986:491–494. doi: 10.1016/0022-2836(86)90171-3. [DOI] [PubMed] [Google Scholar]
- 21.Boder E, Wittrup K. Yeast surface display for screeningcombinatorial polypeptide libraries. Nat Biotechnol. 1997:15. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 22.Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, Finkle D, Venook R, Wu X, Ridgway J, Schahin-Reed D, Dow GJ, Shelton A, Stawicki S, Watts RJ, Zhang J, Choy R, Howard P, Kadyk L, Yan M, Zha J, Callahan CA, Hymowitz SG, Siebel CW. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 23.Shi L, Wheeler JC, Sweet RW, Lu J, Luo J, Tornetta M, Whitaker B, Reddy R, Brittingham R, Borozdina L, Chen Q, Amegadzie B, Knight DM, Almagro JC, Tsui P. De novo selection of high-affinity antibodies from synthetic fab libraries displayed on phage as pIX fusion proteins. J Mol Biol. 2010;397:385–396. doi: 10.1016/j.jmb.2010.01.034. [DOI] [PubMed] [Google Scholar]
- 24.Pershad K, Pavlovic JD, Gräslund S, Nilsson P, Colwill K, Karatt-Vellatt A, Schofield DJ, Dyson MR, Pawson T, Kay BK, McCafferty J. Generating a panel of highly specific antibodies to 20 human SH2 domains by phage display. Protein Eng, Des Sel. 2010;23:279–288. doi: 10.1093/protein/gzq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen K, Sun X, Niu G, Ma Y, Yap LP, Hui X, Wu K, Fan D, Conti PS, Chen X. Evaluation of 64Cu labeled GX1: a phage display peptide probe for PET imaging of tumor vasculature. Mol Imaging Biol. 2012;14:96–105. doi: 10.1007/s11307-011-0479-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deutscher SL. Phage display in molecular imaging and diagnosis of cancer. Chem Rev. 2010;110:3196–3211. doi: 10.1021/cr900317f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khurana S, Chearwae W, Castellino F, Manischewitz J, King LR, Honorkiewicz A, Rock MT, Edwards KM, Del Giudice G, Rappuoli R, Golding H. Vaccines with MF59 adjuvant expand the antibody repertoire to target protective sites of pandemic avian H5N1 influenza virus. Sci Transl Med. 2010;2:15ra5. doi: 10.1126/scitranslmed.3000624. [DOI] [PubMed] [Google Scholar]
- 28.Khurana S, Verma N, Yewdell JW, Hilbert AK, Castellino F, Lattanzi M, Del Giudice G, Rappuoli R, Golding H. MF59 adjuvant enhances diversity and affinity of antibody-mediated immune response to pandemic influenza vaccines. Sci Transl Med. 2011;3:85ra48. doi: 10.1126/scitranslmed.3002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szabó A, Héja D, Szakács D, Zboray K, Kékesi Ka, Radisky ES, Sahin-Tóth M, Pál G. High affinity small protein inhibitors of human chymotrypsin C (CTRC) selected by phage display reveal unusual preference for P4′ acidic residues. J Biol Chem. 2011;286:22535–22545. doi: 10.1074/jbc.M111.235754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Héja D, Harmat V, Fodor K, Wilmanns M, Dobó J, Kékesi KA, Závodszky P, Gál P, Pál G. Monospecific inhibitors show that both mannan-binding lectin-associated serine protease-1 (MASP-1) and -2 are essential for lectin pathway activation and reveal structural plasticity of MASP-2. J Biol Chem. 2012;287:20290–20300. doi: 10.1074/jbc.M112.354332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piotukh K, Geltinger B, Heinrich N, Gerth F, Beyermann M, Freund C, Schwarzer D. Directed evolution of sortase A mutants with altered substrate selectivity profiles. J Am Chem Soc. 2011;133:17536–17539. doi: 10.1021/ja205630g. [DOI] [PubMed] [Google Scholar]
- 32.Gasparian ME, Bobik TV, Kim YV, Ponomarenko NA, Dolgikh DA, Gabibov AG, Kirpichnikov MP. Heterogeneous catalysis on the phage surface: display of active human enteropeptidase. Biochimie. 2013;95:2076–2081. doi: 10.1016/j.biochi.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 33.Fernandez-Gacio A, Uguen M, Fastrez J. Phage display as a tool for the directed evolution of enzymes. Trends Biotechnol. 2003;21:408–414. doi: 10.1016/S0167-7799(03)00194-X. [DOI] [PubMed] [Google Scholar]
- 34.Levin AM, Weiss GA. Optimizing the affinity and specificity of proteins with molecular display. Mol BioSyst. 2006;2:49–57. doi: 10.1039/b511782h. [DOI] [PubMed] [Google Scholar]
- 35.Lee SY, Choi JH, Xu Z. Microbial cell-surface display. Trends Biotechnol. 2003;21:45–52. doi: 10.1016/s0167-7799(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 36.Knez K, Noppe W, Geukens N, Janssen KPF, Spasic D, Heyligen J, Vriens K, Thevissen K, Cammue BPa, Petrenko V, Ulens C, Deckmyn H, Lammertyn J. Affinity comparison of p3 and p8 peptide displaying bacteriophages using surface plasmon resonance. Anal Chem. 2013;85:10075–10082. doi: 10.1021/ac402192k. [DOI] [PubMed] [Google Scholar]
- 37.Van Bloois E, Winter RT, Kolmar H, Fraaije MW. Decorating microbes: surface display of proteins on Escherichia coli. Trends Biotechnol. 2011;29:79–86. doi: 10.1016/j.tibtech.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 38.Schneewind O, Missiakas DM. Protein secretion and surface display in Gram-positive bacteria. Philos Trans R Soc, B. 2012;367:1123–1139. doi: 10.1098/rstb.2011.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Francisco J. Transport and anchoring of beta-lactamase to the external surface of Escherichia coli. Proc Natl Acad Sci USA. 1992;89:2713–2717. doi: 10.1073/pnas.89.7.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang C, Zhao Q, Liu Z, Li Q, Qiao C, Mulchandani A, Chen W. Cell surface display of functional macromolecule fusions on Escherichia coli for development of an autofluorescent whole-cell biocatalyst. Environ Sci Technol. 2008;42:6105–6110. doi: 10.1021/es800441t. [DOI] [PubMed] [Google Scholar]
- 41.Wei W, Liu X, Sun P, Wang X, Zhu H, Hong M, Mao ZW, Zhao J. Simple whole-cell biodetection and bioremediation of heavy metals based on an engineered lead-specific operon. Environ Sci Technol. 2014;48:3363–3371. doi: 10.1021/es4046567. [DOI] [PubMed] [Google Scholar]
- 42.Maurer J, Jose J, Meyer TF. Autodisplay: one-component system for efficient surface display and release of soluble recombinant proteins from Escherichia coli. J Bacteriol. 1997;179:794–804. doi: 10.1128/jb.179.3.794-804.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jung HC, Lebeault JM, Pan JG. Surface display of Zymomonas mobilis levansucrase by using the ice-nucleation protein of Pseudomonas syringae. Nat Biotechnol. 1998;16:576–580. doi: 10.1038/nbt0698-576. [DOI] [PubMed] [Google Scholar]
- 44.Jose J, Meyer TF. The autodisplay story, from discovery to biotechnical and biomedical applications. Microbiol Mol Biol Rev. 2007;71:600–619. doi: 10.1128/MMBR.00011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicolay T, Vanderleyden J, Spaepen S. Autotransporter-based cell surface display in Gram-negative bacteria. Crit Rev Microbiol. 2013;7828:1–15. doi: 10.3109/1040841X.2013.804032. [DOI] [PubMed] [Google Scholar]
- 46.Jose J, von Schwichow S. Autodisplay of active sorbitol dehydrogenase (SDH) yields a whole cell biocatalyst for the synthesis of rare sugars. ChemBioChem. 2004;5:491–499. doi: 10.1002/cbic.200300774. [DOI] [PubMed] [Google Scholar]
- 47.Detzel C, Maas R, Tubeleviciute A, Jose J. Autodisplay of nitrilase from Klebsiella pneumoniae and whole-cell degradation of oxynil herbicides and related compounds. Appl Microbiol Biotechnol. 2013;97:4887–4896. doi: 10.1007/s00253-012-4401-9. [DOI] [PubMed] [Google Scholar]
- 48.Jose J, Maas RM, Teese MG. Autodisplay of enzymes—molecular basis and perspectives. J Biotechnol. 2012;161:92–103. doi: 10.1016/j.jbiotec.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 49.Karami A, Latifi AM, Khodi S. Comparison of theorganophosphorus hydrolase surface display using InaVN and Lpp-OmpA systems in Escherichia coli. J Microbiol Biotechnol. 2014;24:379–385. doi: 10.4014/jmb.1309.09066. [DOI] [PubMed] [Google Scholar]
- 50.Li L, Kang DG, Cha HJ. Functional display of foreign proteinon surface of Escherichia coli using N-terminal domain of ice nucleation protein. Biotechnol Bioeng. 2004;85:214–221. doi: 10.1002/bit.10892. [DOI] [PubMed] [Google Scholar]
- 51.Fischer CR, Klein-Marcuschamer D, Stephanopoulos G. Selection and optimization of microbial hosts for biofuels production. Metab Eng. 2008;10:295–304. doi: 10.1016/j.ymben.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 52.Ernst W, Grabherr R, Wegner D, Borth N, Grassauer A, Katinger H. Baculovirus surface display: Construction and screening of a eukaryotic epitope library. Nucleic Acids Res. 1998;26:1718–1723. doi: 10.1093/nar/26.7.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beerli RR, Bauer M, Buser RB, Gwerder M, Muntwiler S, Maurer P, Saudan P, Bachmann MF. Isolation of human monoclonal antibodies by mammalian cell display. Proc Natl Acad Sci USA. 2008;105:14336–14341. doi: 10.1073/pnas.0805942105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martínez-Pastor MT, Marchler G, Schüller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 55.Marchler G, Schüller C, Adam G, Ruis H. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 1993;12:1997–2003. doi: 10.1002/j.1460-2075.1993.tb05849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dhar R, Sägesser R, Weikert C, Wagner A. Yeast adapts to a changing stressful environment by evolving cross-protection and anticipatory gene regulation. Mol Biol Evol. 2013;30:573–588. doi: 10.1093/molbev/mss253. [DOI] [PubMed] [Google Scholar]
- 57.Pepper L, Cho Y. A decade of yeast surface display technology: where are we now? Comb Chem High Throughput Screening. 2008;11:127–134. doi: 10.2174/138620708783744516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanaka T, Yamada R, Ogino C, Kondo A. Recent developments in yeast cell surface display toward extended applications in biotechnology. Appl Microbiol Biotechnol. 2012;95:577–591. doi: 10.1007/s00253-012-4175-0. [DOI] [PubMed] [Google Scholar]
- 59.Liu Y, Zhang R, Lian Z, Wang S, Wright AT. Yeast cell surface display for lipase whole cell catalyst and its applications. J Mol Catal B: Enzym. 2014;106:17–25. [Google Scholar]
- 60.Blazic M, Kovacevic G, Prodanovic O, Ostafe R, Gavrovic-Jankulovic M, Fischer R, Prodanovic R. Yeast surface display for the expression, purification and characterization of wild-type and B11 mutant glucose oxidases. Protein Expression Purif. 2013;89:175–180. doi: 10.1016/j.pep.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 61.Yeasmin S, Kim CH, Park HJ, Sheikh MI, Lee JY, Kim JW, Back KK, Kim SH. Cell surface display of cellulase activity-free xylanase enzyme on Saccharomyces cerevisiae EBY100. Appl Biochem Biotechnol. 2011;164:294–304. doi: 10.1007/s12010-010-9135-5. [DOI] [PubMed] [Google Scholar]
- 62.Gera N, Hussain M, Rao BM. Protein selection using yeast surface display. Methods. 2013;60:15–26. doi: 10.1016/j.ymeth.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 63.Birnbaum ME, Mendoza JL, Sethi DK, Dong S, Glanville J, Dobbins J, Özkan E, Davis MM, Wucherpfennig KW, Garcia KC. Deconstructing the peptide-MHC specificity of T cell recognition. Cell. 2014;157:1073–1087. doi: 10.1016/j.cell.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wen F, Sethi DK, Wucherpfennig KW, Zhao H. Cell surface display of functional human MHC class II proteins: yeast display versus insect cell display. Protein Eng, Des Sel. 2011;24:701–709. doi: 10.1093/protein/gzr035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bony M, Thines-Sempoux D, Barre P, Blondin B. Localization and cell surface anchoring of the Saccharomyces cerevisiae flocculation protein Flo1p. J Bacteriol. 1997;179:4929–4936. doi: 10.1128/jb.179.15.4929-4936.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matsumoto T, Fukuda H, Ueda M, Tanaka A, Kondo A. Construction of yeast strains with high cell surface lipase activity by using novel display systems based on the Flo1p flocculation functional domain. Appl Environ Microbiol. 2002;68:4517–4522. doi: 10.1128/AEM.68.9.4517-4522.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiang ZB, Song HT, Gupta N, Ma LX, Wu ZB. Cell surface display of functionally active lipases from Yarrowia lipolytica in Pichia pastoris. Protein Expression Purif. 2007;56:35–39. doi: 10.1016/j.pep.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 68.Han Z, Han S, Zheng S, Lin Y. Enhancing thermostability of a Rhizomucor miehei lipase by engineering a disulfide bond and displaying on the yeast cell surface. Appl Microbiol Biotechnol. 2009;85:117–126. doi: 10.1007/s00253-009-2067-8. [DOI] [PubMed] [Google Scholar]
- 69.Kondo A, Shigechi H, Abe M, Uyama K, Matsumoto T, Takahashi S, Ueda M, Tanaka A, Kishimoto M, Fukuda H. High-level ethanol production from starch by a flocculent Saccharomyces cerevisiae strain displaying cell-surface glucoamylase. Appl Microbiol Biotechnol. 2002;58:291–296. doi: 10.1007/s00253-001-0900-9. [DOI] [PubMed] [Google Scholar]
- 70.Kondo A, Ueda M. Yeast cell-surface display—applications of molecular display. Appl Microbiol Biotechnol. 2004;64:28–40. doi: 10.1007/s00253-003-1492-3. [DOI] [PubMed] [Google Scholar]
- 71.Vallejo JA, Sánchez-Pérez A, Martínez JP, Villa TG. Cell aggregations in yeasts and their applications. Appl Microbiol Biotechnol. 2013;97:2305–2318. doi: 10.1007/s00253-013-4735-y. [DOI] [PubMed] [Google Scholar]
- 72.Bauer FF, Govender P, Bester MC. Yeast flocculation and its biotechnological relevance. Appl Microbiol Biotechnol. 2010;88:31–39. doi: 10.1007/s00253-010-2783-0. [DOI] [PubMed] [Google Scholar]
- 73.Mohebali G, Ball AS. Biocatalytic desulfurization (BDS) of petrodiesel fuels. Microbiology. 2008;154:2169–2183. doi: 10.1099/mic.0.2008/017608-0. [DOI] [PubMed] [Google Scholar]
- 74.Wang H, Lang Q, Li L, Liang B, Tang X, Kong L, Mascini M, Liu A. Yeast surface displaying glucose oxidase as whole-cell biocatalyst: construction, characterization, and its electrochemical glucose sensing application. Anal Chem. 2013;85:6107–6112. doi: 10.1021/ac400979r. [DOI] [PubMed] [Google Scholar]
- 75.Tang X, Liang B, Yi T, Manco G, Palchetti I, Palchetti I, Liu A. Cell surface display of organophosphorus hydrolase for sensitive spectrophotometric detection of p-nitrophenol substituted organo-phosphates. Enzyme Microb Technol. 2014;55:107–112. doi: 10.1016/j.enzmictec.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 76.Wang M, Si T, Zhao H. Biocatalyst development by directed evolution. Bioresour Technol. 2012;115:117–125. doi: 10.1016/j.biortech.2012.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martínez R, Schwaneberg U. A roadmap to directed enzyme evolution and screening systems for biotechnological applications. Biol Res. 2013;46:395–405. doi: 10.4067/S0716-97602013000400011. [DOI] [PubMed] [Google Scholar]
- 78.Wen F, Mclachlan M, Zhao H. Wiley Encyclopedia of Chemical Biology. Wiley; Hoboken, NJ: 2008. Directed evolution: novel and improved enzymes. [Google Scholar]
- 79.Kumar A, Singh S. Directed evolution: tailoring biocatalysts for industrial applications. Crit Rev Biotechnol. 2013;33:365–378. doi: 10.3109/07388551.2012.716810. [DOI] [PubMed] [Google Scholar]
- 80.Turner NJ. Directed evolution drives the next generation of biocatalysts. Nat Chem Biol. 2009;5:567–573. doi: 10.1038/nchembio.203. [DOI] [PubMed] [Google Scholar]
- 81.Rubin-Pitel SB, Zhao H. Recent advances in biocatalysis by directed enzyme evolution. Comb Chem High Throughput Screening. 2006;9:247–257. doi: 10.2174/138620706776843183. [DOI] [PubMed] [Google Scholar]
- 82.Fulda S, Gorman AM, Hori O, Samali A. Cellular stress responses: cell survival and cell death. Int J Cell Biol. 2010;2010:214074. doi: 10.1155/2010/214074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Beliën T, Verjans P, Courtin CM, Delcour Ja. Phage display based identification of novel stabilizing mutations in glycosyl hydrolase family 11 Bsubtilis endoxylanase XynA. Biochem Biophys Res Commun. 2008;368:74–80. doi: 10.1016/j.bbrc.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 84.Liu W, Zhang XZ, Zhang Z, Zhang YHP. Engineering of Clostridium phytofermentans endoglucanase Cel5A for improved thermostability. Appl Environ Microbiol. 2010;76:4914–4917. doi: 10.1128/AEM.00958-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Peng XQ. Improved thermostability of lipase B from Candida antarctica by directed evolution and display on yeast surface. Appl Biochem Biotechnol. 2012;169:351–358. doi: 10.1007/s12010-012-9954-7. [DOI] [PubMed] [Google Scholar]
- 86.Chen YP, Hwang IE, Lin CJ, Wang HJ, Tseng CP. Enhancing the stability of xylanase from Cellulomonas fimi by cell-surface display on Escherichia coli. J Appl Microbiol. 2012;112:455–463. doi: 10.1111/j.1365-2672.2012.05232.x. [DOI] [PubMed] [Google Scholar]
- 87.Korman TP, Sahachartsiri B, Charbonneau DM, Huang GL, Beauregard M, Bowie JU. Dieselzymes: development of a stable and methanol tolerant lipase for biodiesel production by directed evolution. Biotechnol Biofuels. 2013;6:70. doi: 10.1186/1754-6834-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reetz MT, Soni P, Fernández L, Gumulya Y, Carballeira JD. Increasing the stability of an enzyme toward hostile organic solvents by directed evolution based on iterative saturation mutagenesis using the B-FIT method. Chem Commun. 2010;46:8657–8658. doi: 10.1039/c0cc02657c. [DOI] [PubMed] [Google Scholar]
- 89.Han S, Zhang J, Han Z, Zheng S, Lin Y. Combination of site-directed mutagenesis and yeast surface display enhances Rhizomucor miehei lipase esterification activity in organic solvent. Biotechnol Lett. 2011;33:2431–2438. doi: 10.1007/s10529-011-0705-6. [DOI] [PubMed] [Google Scholar]
- 90.Zhang J, Lin Y, Sun Y, Ye Y, Zheng S, Han S. High-throughput screening of B factor saturation mutated Rhizomucor miehei lipase thermostability based on synthetic reaction. Enzyme Microb Technol. 2012;50:325–330. doi: 10.1016/j.enzmictec.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 91.Tan LT, Hiraishi T, Sudesh K, Maeda M. Directed evolution of poly[(R)-3-hydroxybutyrate] depolymerase using cell surface display system: functional importance of asparagine at position 285. Appl Microbiol Biotechnol. 2013;97:4859–4871. doi: 10.1007/s00253-012-4366-8. [DOI] [PubMed] [Google Scholar]
- 92.Zhang K, Nelson KM, Bhuripanyo K, Grimes KD, Zhao B, Aldrich CC, Yin J. Engineering the substrate specificity of the DhbE adenylation domain by yeast cell surface display. Chem Biol. 2013;20:92–101. doi: 10.1016/j.chembiol.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen KC, Wu CH, Chang CY, Lu WC, Tseng Q, Prijovich ZM, Schechinger W, Liaw YC, Leu YL, Roffler SR. Directed evolution of a lysosomal enzyme with enhanced activity at neutral pH by mammalian cell-surface display. Chem Biol. 2008;15:1277–1286. doi: 10.1016/j.chembiol.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 94.Kim SY, Sohn JH, Pyun YR, Kim KH, Choi ES. In vitro evolution of lipase B from Candida antarctica using surface display in Hansenula polymorpha. J Microbiol Biotechnol. 2007;17:1308–1315. [PubMed] [Google Scholar]
- 95.Gupta N, Farinas ET. Directed evolution of CotA laccase for increased substrate specificity using Bacillus subtilis spores. Protein Eng, Des Sel. 2010;23:679–682. doi: 10.1093/protein/gzq036. [DOI] [PubMed] [Google Scholar]
- 96.Liang B, Lang Q, Tang X, Liu A. Simultaneously improving stability and specificity of cell surface displayed glucose dehydrogenase mutants to construct whole-cell biocatalyst for glucose biosensor application. Bioresour Technol. 2013;147:492–498. doi: 10.1016/j.biortech.2013.08.088. [DOI] [PubMed] [Google Scholar]
- 97.Sunbul M, Marshall NJ, Zou Y, Zhang K, Yin J. Catalytic turnover-based phage selection for engineering the substrate specificity of Sfp phosphopantetheinyl transferase. J Mol Biol. 2009;387:883–898. doi: 10.1016/j.jmb.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 98.Bonner WA, Hulett HR, Sweet RG, Herzenberg LA. Fluorescence activated cell sorting. Rev Sci Instrum. 1972;43:404–409. doi: 10.1063/1.1685647. [DOI] [PubMed] [Google Scholar]
- 99.Gupta R, Beg QK, Lorenz P. Bacterial alkaline proteases: molecular approaches and industrial applications. Appl Microbiol Biotechnol. 2002;59:15–32. doi: 10.1007/s00253-002-0975-y. [DOI] [PubMed] [Google Scholar]
- 100.Yi L, Gebhard MC, Li Q, Taft JM, Georgiou G, Iverson BL. Engineering of TEV protease variants by yeast ER sequestration screening (YESS) of combinatorial libraries. Proc Natl Acad Sci USA. 2013;110:7229–7234. doi: 10.1073/pnas.1215994110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yoo TH, Pogson M, Iverson BL, Georgiou G. Directed evolution of highly selective proteases using a novel FACS based screen that capitalizes on the p53 regulator MDM2. ChemBioChem. 2012;13:649–653. doi: 10.1002/cbic.201100718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Paegel BM, Joyce GF. Microfluidic compartmentalized directed evolution. Chem Biol. 2010;17:717–724. doi: 10.1016/j.chembiol.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Griffiths AD, Tawfik DS. Man-made enzymes—from design to in vitro compartmentalisation. Curr Opin Biotechnol. 2000;11:338–353. doi: 10.1016/s0958-1669(00)00109-9. [DOI] [PubMed] [Google Scholar]
- 104.Agresti JJ, Antipov E, Abate AR, Ahn K, Rowat AC, Baret JC, Marquez M, Klibanov AM, Griffiths AD, Weitz DA. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc Natl Acad Sci USA. 2010;107:4004–4009. doi: 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Acevedo-Rocha CG, Agudo R, Reetz MT. Directed evolution of stereoselective enzymes based on genetic selection as opposed to screening systems. J Biotechnol. 2014;191:3–10. doi: 10.1016/j.jbiotec.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 106.Reetz MT. Combinatorial and evolution-based methods in the creation of enantioselective catalysts. Angew Chem, Int Ed. 2001;40:284–310. [PubMed] [Google Scholar]
- 107.Liebeton K, Zonta A, Schimossek K, Nardini M, Lang D, Dijkstra BW, Reetz MT, Jaeger KE. Directed evolution of an enantioselective lipase. Chem Biol. 2000;7:709–718. doi: 10.1016/s1074-5521(00)00015-6. [DOI] [PubMed] [Google Scholar]
- 108.Reetz MT, Becker MH, Klien HW, Stockigt D. A method for high-throughput screening of enantioselective catalysts. Angew Chem, Int Ed. 1999;38:1758–1761. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1758::AID-ANIE1758>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 109.Reetz MT, Zonta A, Schimossek K, Liebeton K, Jaeger KE. Creation of enantioselective biocatalysts for organic chemistry by in vitro evolution. Angew Chem, Int Ed Engl. 1997;36:2830–2832. [Google Scholar]
- 110.Lipovsek D, Antipov E, Armstrong KA, Olsen MJ, Klibanov AM, Tidor B, Wittrup KD. Selection of horseradish peroxidase variants with enhanced enantioselectivity by yeast surface display. Chem Biol. 2007;14:1176–1185. doi: 10.1016/j.chembiol.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 111.Antipov E, Cho AE, Wittrup KD, Klibanov AM. Highly l and d enantioselective variants of horseradish peroxidase discovered by an ultrahigh-throughput selection method. Proc Natl Acad Sci USA. 2008;105:17694–17699. doi: 10.1073/pnas.0809851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dröge MJ, Boersma YL, van Pouderoyen G, Vrenken TE, Rüggeberg CJ, Reetz MT, Dijkstra BW, Quax WJ. Directed evolution of Bacillus subtilis lipase A by use of enantiomeric phosphonate inhibitors: crystal structures and phage display selection. ChemBioChem. 2006;7:149–157. doi: 10.1002/cbic.200500308. [DOI] [PubMed] [Google Scholar]
- 113.Becker S, Schmoldt HU, Adams TM, Wilhelm S, Kolmar H. Ultra-high-throughput screening based on cell-surface display and fluorescence-activated cell sorting for the identification of novel biocatalysts. Curr Opin Biotechnol. 2004;15:323–329. doi: 10.1016/j.copbio.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 114.Becker S, Höbenreich H, Vogel A, Knorr J, Wilhelm S, Rosenau F, Jaeger KE, Reetz MT, Kolmar H. Single-cell high-throughput screening to identify enantioselective hydrolytic enzymes. Angew Chem, Int Ed. 2008;47:5085–5088. doi: 10.1002/anie.200705236. [DOI] [PubMed] [Google Scholar]
- 115.Becker S, Michalczyk A, Wilhelm S, Jaeger KE, Kolmar H. Ultrahigh-throughput screening to identify E. coli cells expressing functionally active enzymes on their surface. ChemBioChem. 2007;8:943–949. doi: 10.1002/cbic.200700020. [DOI] [PubMed] [Google Scholar]
- 116.Richins RD, Kaneva I, Mulchandani A, Chen W. Biodegradation of organophosphorus pesticides by surface-expressed organophosphorus hydrolase. Nat Biotechnol. 1997;15:984–987. doi: 10.1038/nbt1097-984. [DOI] [PubMed] [Google Scholar]
- 117.Kim S, Song JK, Kim HK. Cell surface display of Staphylococcus haemolyticus L62 lipase in Escherichia coli and its application as a whole cell biocatalyst for biodiesel production. J Mol Catal B: Enzym. 2013;97:54–61. [Google Scholar]
- 118.Liu R, Yang C, Xu Y, Xu P, Jiang H, Qiao C. Development of a whole-cell biocatalyst/biosensor by display of multiple heterologous proteins on the Escherichia coli cell surface for the detoxification and detection of organophosphates. J Agric Food Chem. 2013;61:7810–7816. doi: 10.1021/jf402999b. [DOI] [PubMed] [Google Scholar]
- 119.Shimazu M, Mulchandani A, Chen W. Cell surface display of organophosphorus hydrolase using ice nucleation protein. Biotechnol Prog. 2001;17:76–80. doi: 10.1021/bp0001563. [DOI] [PubMed] [Google Scholar]
- 120.Takayama K, Suye S, Kuroda K, Ueda M, Kitaguchi T, Tsuchiyama K, Fukuda T, Chen W, Mulchandani A. Surface display of organophosphorus hydrolase on Saccharomyces cerevisiae. Biotechnol Prog. 2006;22:939–943. doi: 10.1021/bp060107b. [DOI] [PubMed] [Google Scholar]
- 121.Matano Y, Hasunuma T, Kondo A. Simultaneous improvement of saccharification and ethanol production from crystalline cellulose by alleviation of irreversible adsorption of cellulase with a cell surface-engineered yeast strain. Appl Microbiol Biotechnol. 2013;97:2231–2237. doi: 10.1007/s00253-012-4587-x. [DOI] [PubMed] [Google Scholar]
- 122.Chen X, Ishida N, Todaka N, Nakamura R, Maruyama J, Takahashi H, Kitamoto K. Promotion of efficient saccharification of crystalline cellulose by Aspergillus fumigatus Swo1. Appl Environ Microbiol. 2010;76:2556–2561. doi: 10.1128/AEM.02499-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Arantes V, Saddler JN. Access to cellulose limits the efficiency of enzymatic hydrolysis: the role of amorphogenesis. Biotechnol Biofuels. 2010;3:4. doi: 10.1186/1754-6834-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nakatani Y, Yamada R, Ogino C, Kondo A. Synergetic effect of yeast cell-surface expression of cellulase and expansin-like protein on direct ethanol production from cellulose. Microb Cell Fact. 2013;12:66. doi: 10.1186/1475-2859-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li K, Frost JW. Synthesis of vanillin from glucose. J Am Chem Soc. 1998;120:10545–10546. [Google Scholar]
- 126.Barghini P, Di Gioia D, Fava F, Ruzzi M. Vanillin production using metabolically engineered Escherichia coli under non-growing conditions. Microb Cell Fact. 2007;6:13. doi: 10.1186/1475-2859-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pan XX, Xu L, Zhang Y, Xiao X, Wang XF, Liu Y, Zhang H, Yan YJ. Efficient display of active Geotrichum sp. lipase on Pichia pastoris cell wall and its application as a whole-cell biocatalyst to enrich EPA and DHA in fish oil. J Agric Food Chem. 2012;60:9673–9679. doi: 10.1021/jf301827y. [DOI] [PubMed] [Google Scholar]
- 128.Jin Z, Liang S, Zhang X, Han S, Ren C, Lin Y, Zheng S. Synthesis of fructose laurate esters catalyzed by a CALB-displaying Pichia pastoris whole-cell biocatalyst in a non-aqueous system. Biotechnol Bioprocess Eng. 2013;18:365–374. [Google Scholar]
- 129.Jin Z, Ntwali J, Han SY, Zheng SP, Lin Y. Production of flavor esters catalyzed by CALB-displaying Pichia pastoris whole-cells in a batch reactor. J Biotechnol. 2012;159:108–114. doi: 10.1016/j.jbiotec.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 130.Resch MG, Donohoe BS, Baker JO, Decker SR, Bayer Ea, Beckham GT, Himmel ME. Fungal cellulases and complexed cellulosomal enzymes exhibit synergistic mechanisms in cellulose deconstruction. Energy Environ Sci. 2013;6:1858. [Google Scholar]
- 131.Hyeon JE, Jeon SD, Han SO. Cellulosome-based, Clostridium-derived multi-functional enzyme complexes for advanced biotechnology tool development: advances and applications. Biotechnol Adv. 2013;31:936–944. doi: 10.1016/j.biotechadv.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 132.Schwarz WH. The cellulosome and cellulose degradation by anaerobic bacteria. Appl Microbiol Biotechnol. 2001;56:634–649. doi: 10.1007/s002530100710. [DOI] [PubMed] [Google Scholar]
- 133.Tsai SL, Oh J, Singh S, Chen R, Chen W. Functional assembly of minicellulosomes on the Saccharomyces cerevisiae cell surface for cellulose hydrolysis and ethanol production. Appl Environ Microbiol. 2009;75:6087–6093. doi: 10.1128/AEM.01538-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tsai SL, Goyal G, Chen W. Surface display of a functional minicellulosome by intracellular complementation using a synthetic yeast consortium and its application to cellulose hydrolysis and ethanol production. Appl Environ Microbiol. 2010;76:7514–7520. doi: 10.1128/AEM.01777-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wen F, Sun J, Zhao H. Yeast surface display of trifunctional minicellulosomes for simultaneous saccharification and fermentation of cellulose to ethanol. Appl Environ Microbiol. 2010;76:1251–1260. doi: 10.1128/AEM.01687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lynd LR, van Zyl WH, McBride JE, Laser M. Consolidated bioprocessing of cellulosic biomass: an update. Curr Opin Biotechnol. 2005;16:577–583. doi: 10.1016/j.copbio.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 137.Olson DG, McBride JE, Shaw AJ, Lynd LR. Recent progress in consolidated bioprocessing. Curr Opin Biotechnol. 2012;23:396–405. doi: 10.1016/j.copbio.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 138.Fan L, Zhang Z, Yu X. Self-surface assembly of cellulosomes with two miniscaffoldins on Saccharomyces cerevisiae for cellulosic ethanol production. Proc Natl Acad Sci USA. 2012:1–6. doi: 10.1073/pnas.1209856109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tsai SL, DaSilva NA, Chen W. Functional display of complex cellulosomes on the yeast surface via adaptive assembly. ACS Synth Biol. 2013;2:14–21. doi: 10.1021/sb300047u. [DOI] [PubMed] [Google Scholar]
- 140.Han Z, Zhang B, Wang YE, Zuo YY, Su WW. Self-assembled amyloid-like oligomeric-cohesin scaffoldin for augmented protein display on the Saccharomyces cerevisiae cell surface. Appl Environ Microbiol. 2012;78:3249–3255. doi: 10.1128/AEM.07745-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liang Y, Si T, Ang EL, Zhao H. An engineered pentafunctional minicellulosome for simultaneous saccharification and ethanol fermentation in Saccharomyces cerevisiae. Appl Environ Microbiol. 2014;80:6677–6684. doi: 10.1128/AEM.02070-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.You C, Zhang XZ, Sathitsuksanoh N, Lynd LR, Zhang YHP. Enhanced microbial utilization of recalcitrant cellulose by an ex vivo cellulosome-microbe complex. Appl Environ Microbiol. 2012;78:1437–1444. doi: 10.1128/AEM.07138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lin Y, Tanaka S. Ethanol fermentation from biomass resources: current state and prospects. Appl Microbiol Biotechnol. 2006;69:627–642. doi: 10.1007/s00253-005-0229-x. [DOI] [PubMed] [Google Scholar]
- 144.Ensley BD. Stability of recombinant plasmids in industrial microorganisms. Crit Rev Biotechnol. 1986;4:263–277. [Google Scholar]
- 145.Xu Y, Tao F, Ma C, Xu P. New constitutive vectors: useful genetic engineering tools for biocatalysis. Appl Environ Microbiol. 2013;79:2836–2840. doi: 10.1128/AEM.03746-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Goodrick JC, Xu M, Finnegan R, Schilling BM, Schiavi S, Hoppe H, Wan NC. High-level expression and stabilization of recombinant human chitinase produced in a continuous constitutive Pichia pastoris expression system. Biotechnol Bioeng. 2001;74:492–497. doi: 10.1002/bit.1140. [DOI] [PubMed] [Google Scholar]
- 147.He X, Liu N, Li W, Zhang Z, Zhang B, Ma Y. Inducible and constitutive expression of a novel thermostable alkaline β-mannanase from alkaliphilic Bacillus sp. N16-5 in Pichia pastoris and characterization of the recombinant enzyme. Enzyme Microb Technol. 2008;43:13–18. [Google Scholar]
- 148.Wu JM, Lin JC, Chieng LL, Lee CK, Hsu TA. Combined use of GAP and AOX1 promoter to enhance the expression of human granulocyte-macrophage colony-stimulating factor in Pichia pastoris. Enzyme Microb Technol. 2003;33:453–459. [Google Scholar]
- 149.Schreuder MP, Brekelmans S, van den Ende H, Klis FM. Targeting of a heterologous protein to the cell wall of Saccharomyces cerevisiae. Yeast. 1993;9:399–409. doi: 10.1002/yea.320090410. [DOI] [PubMed] [Google Scholar]
- 150.Schultheiss E, Weiss S, Winterer E, Maas R, Heinzle E, Jose J. Esterase autodisplay: enzyme engineering and whole-cell activity determination in microplates with pH sensors. Appl Environ Microbiol. 2008;74:4782–4791. doi: 10.1128/AEM.01575-07. [DOI] [PMC free article] [PubMed] [Google Scholar]


