Abstract
Group 2 innate lymphoid cells (ILC2s) regulate inflammation, tissue repair and metabolic homeostasis1. ILC2 activation is driven by host-derived cytokines and alarmins1. While discrete immune cell subsets integrate nervous system cues2–4, it remains unclear whether neuronal-derived signals control ILC2s. Here we show that Neuromedin U (NMU) is a uniquely fast and potent regulator of type 2 innate immunity in the context of a novel neuron-ILC2 unit. We found that ILC2s selectively express Neuromedin U receptor 1 (Nmur1), while mucosal neurons express NMU. ILC2-autonomous activation with NMU resulted in immediate and strong production of innate inflammatory and tissue repair cytokines, in a NMUR1-dependent manner. NMU controlled ILC2s downstream of extracellular signal–regulated kinase (ERK) and calcium (Ca2+)-influx-dependent activation of Calcineurin and nuclear factor of activated T cells (NFAT). NMU treatment in vivo resulted in immediate protective type 2 responses. Accordingly, ILC2-autonomous ablation of Nmur1 led to impaired type 2 responses and poor worm infection control. Strikingly, mucosal neurons were found adjacent to ILC2s, directly sensed worm products and alarmins to induce NMU and to control innate type 2 cytokines. Our work reveals that neuron-ILC2 cell units are poised to confer a first-line of immediate tissue protection via coordinated neuro-immune sensory responses.
ILC2s are abundant at mucosal barriers and act as key initiators of type 2 inflammation and tissue repair1. ILC2s are activated by cell-extrinsic cytokines, including interleukin 25 (IL-25), IL-33 and thymic stromal lymphopoietin (TSLP)1. Previous reports indicated that discrete lymphocyte subsets and haematopoietic progenitors are controlled by dietary signals and neuroregulators2,4–11, suggesting that ILC2s may exert their function in the context of neuro-immune cell units (NICUs)4.
To interrogate whether ILC2s directly and selectively perceive neuronal-derived molecules, we employed genome-wide transcriptional profiling of ILC2s versus their adaptive (T helper cells) and innate (ILC1 and ILC3) counterparts12 (Fig.1a,b). This analysis identified the gene Nmur1 as being selectively enriched in ILC2s when compared to ILC1s, ILC3s and T helper cells (Fig.1a,b and Extended Data Fig.1a,b). This finding was confirmed by independent quantitative expression assays in multiple subsets of immune cells, including ILC1s, ILC3s, natural killer (NK) cells, eosinophils, mast cells, macrophages, neutrophils, dendritic cells, T cell subsets and B cells (Fig.1c and Extended Data Fig.1c,d). Noteworthy, human ILC2 also expressed NMUR1 (Fig.1d). This gene encodes for a transmembrane receptor for NMU. The latter is a secreted neuropeptide found in the brain and highly expressed in the gastrointestinal tract13–16. As such, NMU acts as a neuronal-derived regulator in diverse physiologic processes16. NMU was shown to be produced by cholinergic enteric neurons, which also express the neurotrophic factor receptor RET13–15,17. In agreement, neurons in the lamina propria were main expressers of the NMU gene (Nmu), while these transcripts were not detectable in enteric neuroglia and epithelial cells (Fig.1e). Similarly, all analysed immune cell subsets, including dendritic cells, macrophages and B cells, had no significant Nmu expression (Fig.1e and Extended Data Fig.1e). Strikingly, neuronal reporter mice (RetGFP and Chat-Cre.Rosa26RFP)18–20 revealed that CD3-KLRG1+ candidate ILC2s are adjacent to the intestinal neuronal network (4.716μm±0.656), and lung ILC2 could also be found in the vicinity of local neurons (Fig.1f,g and Extended Data Fig.1f-h). Taken together these data suggest a paracrine neuron-ILC2 crosstalk orchestrated by NMU-NMUR1 interactions.
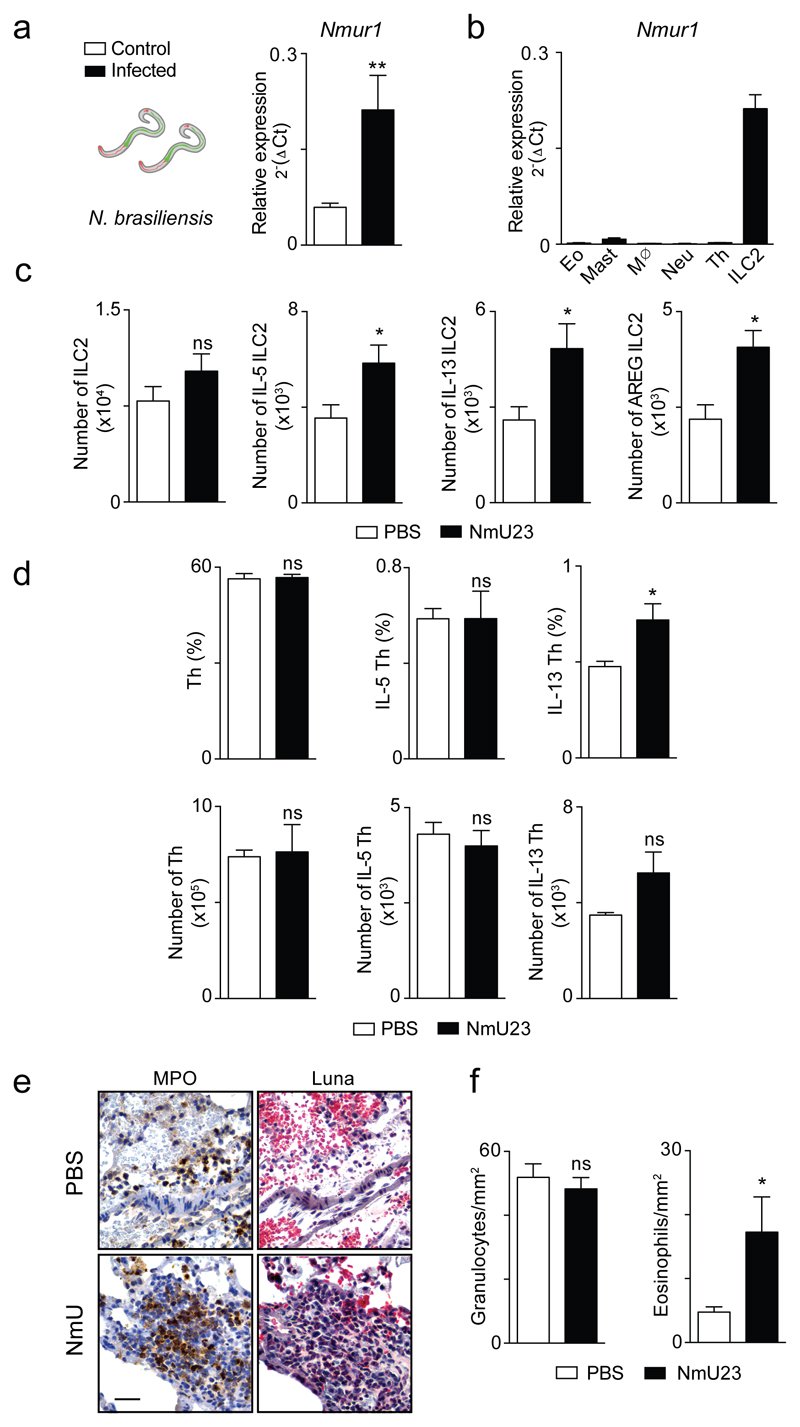
Figure 1. ILC2s express NMUR1 and closely locate with NMU-expressing neurons.
a, Heat map for 40 neuronal-related mRNA transcripts in T helper cells (Th), ILC1s, ILC2s, NCR- (CD4+ and CD4-) and NCR+ ILC3s subsets. b, Comparison of ILC2 gene expression with ILC1, ILC3 NCR+ and Th cells12, by volcano plots. c, Nmur1 quantitative RT-PCR analysis in intestinal lamina propria cells unless stated otherwise. Bone marrow common lymphoid progenitor (CLP); Bone Marrow common helper innate lymphoid progenitor (CHILP); Bone marrow ILC2 progenitor (ILC2P); Mature ILC subsets; Eosinophils (Eo); Mast cells (Mast); Macrophages (Mø); Neutrophils (Neu); Dendritic cells (DC); Naïve helper T cells (Thn); Polarised Th2 cells (Th2); Memory helper T cells (Thm); B cells (B); Lamina propria glial cells (G) and Neurons (N); Epithelial cells (Ep). n=6. d, NMUR1 expression in human T helper cells and ILC2 from blood. n=3. e, Nmu expression in intestinal populations. n=6. f-g Confocal analysis of intestinal lamina propria. f, Left: Green: neurons (RetGFP); Red: CD3; Cyan: KLRG1. Right: details of the left panel. g, Red: neurons (Chat-Cre.Rosa26RFP); Green: CD3; Cyan: KLRG1. Arrows: examples of ILC2s. Scale bars: 30μm. Data are representative of 2-3 independent experiments. Error bars show s.e.m. **P<0.01.
To explore this hypothesis, intestine and lung-derived ILC2s were purified and activated with recombinant mouse peptide NMU (NmU23) (Fig.2a-e). Astonishingly, cell-autonomous activation of ILC2s with NmU23 resulted in prompt and very potent expression of the pro-inflammatory and tissue-protective type 2 cytokines genes Il5, Il13, amphiregulin (Areg) and colony stimulating factor 2 (Csf2) (Fig.2a,b and Extended Data Fig.2a). NmU23-dependent activation of ILC2s also increased ILC2 proliferation as measured by Ki67 expression (Extended Data Fig.2b-d). NMU was shown to bind with similar affinity to two orphan class A G-protein-coupled receptors, NMUR1 and NMUR216. Formal definition that NMUR1 activation is the molecular link between NMU-dependent ILC2 activation and cytokine production was provided by genetic ablation of Nmur1. Activation of purified ILC2s with NmU23 led to potent expression of the type 2 cytokine proteins IL-5 and IL-13 in a NMUR1-dependent manner (Fig.2c,d and Extended Data Fig.2e-h). Strikingly, and in contrast to the canonical ILC2-activating cytokines (IL-33 and IL-25), NmU23 led to an immediate strong expression of innate IL-5, IL-13 and AREG, indicating that NMU is a uniquely fast and potent regulator of ILC2-derived type 2 cytokines (Fig.2e and Extended Data Fig.3a,b). In agreement, in vivo administration of the neuropeptide NmU23 resulted in immediate and selective type 2 cytokine production from ILC2s, while their adaptive T helper cell-derived counterparts were unperturbed (Fig.2f,g and Extended Data Fig.3c). Noteworthy, while in the absence of IL-33 and IL-25 receptor signals NMU restored ILC2 numbers in vivo, this neuropeptide failed to efficiently induce innate cytokines in the similar contexts (Extended Data Fig.4a-d). This suggests that NMU signals critically fine-tune immediate innate type 2 cytokines, while the canonical IL-25/IL-33 circuit regulates ILC2 homeostasis and function with comparatively delayed kinetics (Extended Data Fig.4a-d). To formally establish the link between ILC2-autonomous NMUR1 activation and innate type 2 cytokine production, we generated mixed bone-marrow (BM) chimeras with Nmur1 sufficient and deficient BM progenitors. While NMUR1 signals were dispensable for ILC2 homeostasis (Extended Data Fig 5a-f), an intact NMU-NMUR1 axis was critical for ILC2-derived cytokines. Notably, NmU23 administration to BM chimeras revealed that Nmur1 deficient ILC2 had reduced innate IL-5 and IL-13 expression when compared to their Nmur1 sufficient counterparts (Fig.2h,i). In contrast, T helper cell-derived cytokines were unperturbed in Nmur1 deficient cells (Extended Data Fig.5g,h). Together, these data indicate that NMU is a uniquely fast and potent cell-autonomous regulator of innate type 2 inflammatory and tissue repair cytokines, via NMUR1 activation.
Figure 2. NMU is a potent ILC2-intrinsic regulator of type 2 cytokines, via NMUR1 activation.
a-b, Type 2 cytokine gene expression in ILC2s after NmU23 activation. n=6. a, Gut ILC2s. n=6. b, Lung ILC2s. n=6. c-d, Type 2 cytokine protein expression in ILC2s after NmU23 activation. c, Intestinal ILC2s. IL-5 n=6; IL-13 n=6; AREG n=3. d, IL-5 and IL-13 expression in enteric Nmur1 competent and deficient ILC2s. e, Enteric ILC2-derived cytokines upon activation with increasing concentrations (10, 50 and 100ng/mL) of IL-33, IL-25, NmU23 and PMA+Ionomycin (P+I). n=3. f-g, in vivo administration of NmU23. f, Gut ILC2-derived type 2 cytokines. n=6. g, Intestinal Th cell-derived type 2 cytokines. n=6. h-i, BM mixed chimeras with Nmur1-/- and their Nmur1+/+ WT littermate controls upon NmU23 administration. h, Percentage of lung ILC2s. Nmur1+/+ n=5; Nmur1-/- n=4. i, Number of Iung ILC2. Nmur1+/+ n=5; Nmur1-/- n=4. Data are representative of 2-6 independent experiments. Error bars show s.e.m. *P<0.05; **P<0.01; ***P<0.001; ns not significant.
To further examine how NMU controls innate type 2 responses, we investigated the signalling cues provided by activated NMUR1 in ILC2s. In neurons, activation of NMU receptors leads to increased Ca2+ influx and ERK1/2 activation, while NFAT activity is required for type 2 cytokine production21–23. Stimulation of ILC2s with NMU led to immediate and efficient ERK1/2 activation, while inhibition of ERK activity upon NmU23-induced ILC2 activation resulted in impaired type 2 cytokine gene expression (Fig.3a,b). Analysis of NMU-induced activation of ILC2s also led to immediate and robust Ca2+ influx, suggesting a role of the calcium-dependent serine/threonine protein phosphatase Calcineurin in NmU23-induced type 2 responses (Fig.3c). Accordingly, inhibition of Calcineurin upon NmU23 activation led to impaired innate Il5, Il13 and Csf2 expression, while NmU23 activation led to rapid NFAT nuclear translocation in ILC2 (Fig.3d-f and Extended Data Fig.6). Finally, inhibition of NFAT activity upon NmU23-induced NMUR1 activation led to a similar decreased in Il5, Il13 and Csf2 (Fig.3g). Thus, we conclude that the neuropeptide NMU can operate in an ILC2-intrinsic manner by activating NMUR1, which regulates innate type-2 cytokines downstream of a Ca2+/Calcineurin/NFAT cascade and ERK1/2 phosphorylation.
Figure 3. NMU regulates ILC2-derived cytokines via ERK1/2 and a Ca2+/Calcineurin/NFAT cascade.
Intestinal ILC2 activation by NMU. a, Top: p-ERK. Bottom: Percentage of p-ERK cells n=4. Mean fluorescence intensity (MFI) of p-ERK expression. n=4. b, Il5, Il13 and Csf2 expression in ILC2s cultured with medium (control), NmU23 or NmU23 and ERK inhibitor PD98059. n=3. c, Left and centre: Ca2+ influx, represented by Fluo-4 AM intensity. NmU23 was added 60 seconds after ILC2 baseline acquisition (arrow). Right: Mean intensity of Ca2+ influx. n=3. d, Il5, Il13 and Csf2 expression in ILC2s cultured with medium (control), NmU23 or NmU23 and Calcineurin inhibitor FK506. n=12. e, Nuclear translocation of NFAT (red) upon NmU23 activation in ILC2. f, Left: Percentage of ILC2 with nuclear NFAT. n=3. Right: NFAT nuclear fluorescence intensity. Control n=16; NmU23 n=7; P+I n=8. g, Il5, Il13 and Csf2 expression in ILC2s cultured with medium (control), NmU23 or NmU23 and NFAT inhibitor 11R-VIVIT. n=6. Scale bars: 5μm. Data are representative of 2-4 independent experiments. Error bars show s.e.m. *P<0.05; **P<0.01; ***P<0.001; ns not significant.
To interrogate whether neuronal peptides can regulate mucosal defence we tested the impact of varying degrees of NMUR1 signals during infection with the helminth parasite Nippostrongylus brasiliensis24. Strikingly, infection of wild-type (WT) mice with N. brasiliensis resulted in strongly increased and sustained Nmu expression in infected tissues (Fig.4a), while expression of high levels of Nmur1 was still restricted to ILC2 (Extended Data Fig.7a,b). These observations suggest that NMU may regulate in vivo responses to worm infection. Accordingly, administration of the neuropeptide NmU23 in N. brasiliensis infected mice resulted in a robust and selective ILC2 response (Fig.4b and Extended Data Fig.7c,d), decreased tissue haemorrhage (Fig.4c and Extended Data Fig.7e,f), with subsequent increased eosinophil and mast cell infiltrates in the lung and reduced infection burden when compared to their vehicle (PBS) treated counterparts (Fig.4d,e). To further explore the role of NMUR1 in innate type 2 responses we infected Nmur1 deficient mice and their WT littermate controls with N. brasiliensis (Fig.4f-i). Strikingly, when compared to their WT littermate counterparts, Nmur1 deficient mice had decreased type 2 responses, notably reduced ILC2-derived cytokines (Fig.4f and Extended Data Fig.8a), increased tissue haemorrhage (Fig.4g and Extended Data Fig.8b-c) and impaired eosinophil and mastocyte infiltrates (Fig.4h). In line with these findings, Nmur1 deficient mice had increased N. brasiliensis infection burden at the peak of lung and gut infection phases, while late worm expulsion was intact (Fig.4i and Extended Data Fig.8d). To more specifically define the link between ILC2, the NMU-NMUR1 signalling axis and protection to infection, we performed chimeras of Nmur1 sufficient and deficient ILC2 into alymphoid host mice. Infection of such chimeras with N. brasiliensis revealed that Nmur1 knockout ILC2 had decreased type 2 cytokine responses when compared to their WT counterparts (Fig.4j). In line with these findings, Nmur1 deficient ILC2-chimeras had increased N. brasiliensis infection burden (Fig.4k). Thus, we conclude that NMU regulates innate type 2 responses to provide immediate mucosal protection against early phases of worm infection. To interrogate whether enteric neurons support innate type 2 cytokine production, we have generated neurosphere-derived neuronal organoids (Fig.4l). Neuronal stimulation with the alarmin IL-33 or N. brasiliensis excretory/secretory products (NES) efficiently induced neuron-derived Nmu (Fig.4m). In line with this findings, ILC2 stimulation with NES-activated neuronal supernatants increased innate type 2 cytokines and intranasal challenge with NES led to similar increase of ILC2-derived cytokines in the lung (Extended Data Fig.9a,b). Enteric neurons have been previously shown to express Toll-like receptors2,4. Strikingly, activation of neuronal organoids with NES or IL-33 induced Nmu expression in a myeloid differentiation primary response gene 88 (MYD88)-dependent manner (Fig.4m). To formally demonstrate the physiological importance of MYD88-dependent neuronal sensing on ILC2 function, we deleted Myd88 in cholinergic neurons by breeding Chat-Cre to Myd88fl/fl mice. Strikingly, cholinergic-neuron-intrinsic deletion of Myd88 resulted in impaired cytokine-producing ILC2 during N. brasiliensis infection (Fig.4n). Taken together, these data indicate that neurons can orchestrate innate type 2 cytokines through direct sensing of worm products and alarmins.
Figure 4. Neuron-derived NMU sets ILC2-autonomous protection against worm infection.
a, Nmu expression in total lung and gut after infection. Lung n=3; Gut n=6. b, Lung ILC2s at day 1 after infection in NmU23 treated and control animals. n=5. c, Left: Lung haemorrhage; Right: Haematoxylin and eosin (HE) lung sections at day 2 after infection. d, Eosinophil and mast cell frequencies in the bronchoalveolar lavage (BAL) at day 6 after infection. n=5. e, N. brasiliensis infection burden in: Lung phase. n=5; Intestinal phase. n=5. f-i, Nmur1-/- and their Nmur1+/+ littermate controls were infected with N. brasiliensis. f, Lung ILC2s at day 6 after infection. Nmur1+/+ n=6; Nmur1-/- n=8. g, HE lung sections 2 days after infection. h, Eosinophil and mast cell frequencies in the BAL at day 6 after infection. Nmur1+/+ n=6; Nmur1-/- n=7. i, N. brasiliensis infection burden in: Lung phase. n=3; Intestinal phase. Nmur1+/+ n=6; Nmur1-/- n=7. j-k. Nmur1-/- and Nmur1+/+ WT littermate control ILC2 chimeras. j. Lung ILC2. Nmur1+/+ n=5; Nmur1-/- n=6. k, N. brasiliensis infection burden in the intestinal phase. Nmur1+/+ n=5; Nmur1-/- n=6. l, Neurosphere-derived neuronal organoids. m, Neuron activation with NES, LPS and IL-33. WT: NES n=12; LPS n=18; IL-33 n=17. Myd88-/-: NES n=10; LPS n=15; IL-33 n=12. n, Myd88wt/fl n=3; Chat-Cre.Myd88fl/fl n=6 Scale bar: 50μm. Data are representative of 1-3 independent experiments. Error bars show s.e.m. *P<0.05; **P<0.01; ***P<0.001; ns not significant.
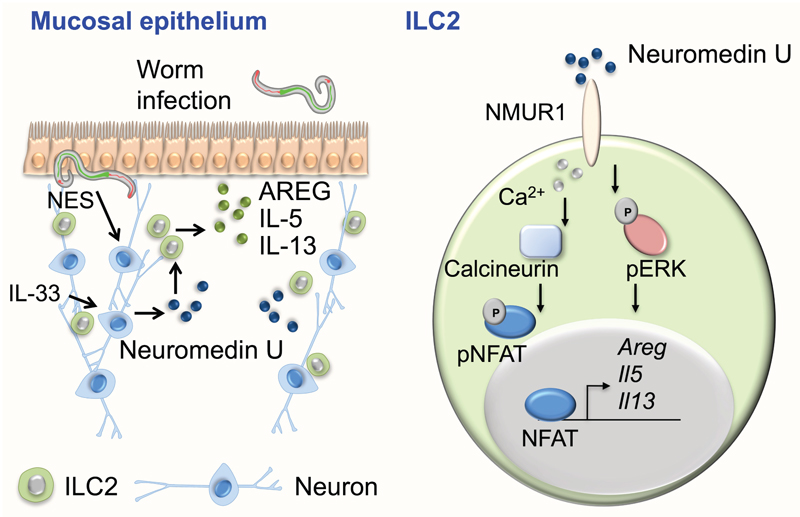
Deciphering the mechanisms by which ILC2s perceive, integrate and respond to environmental signals is critical to understand tissue and organ homeostasis. Our work establishes unexpected relationships between ILC2s and their environment. We deciphered a novel neuron-ILC2 cell unit orchestrated by NMU (Extended Data Fig.10). Mucosal neurons can directly sense alarmin and worm products to produce NMU. NMU activates ILC2s, via NMUR1, resulting in a uniquely potent and immediate production of innate type 2 cytokine downstream of ERK phosphorylation and activation of a Ca2+/Calcineurin/NFAT cascade (Extended Data Fig.10).
While it is well established that ILC2s integrate cytokine signals, including IL-25, IL-33 and TSLP1,11, here we demonstrate that ILC2s can integrate signals from different germ-layer-derived tissues to immediately trigger inflammatory and tissue repair type 2 responses. Thus, neuron-ILC2 cell units might be poised to confer a first-line of immediate tissue protection via coordinated neuro-immune sensory responses (Extended Data Fig.10). While afferent extrinsic nervous cues might also fine-tune local NICUs, future studies utilising emergent technology platforms2,4 are required to interrogate these putative interactions.
Previous studies demonstrated that ILC2s contribute to multiple homeostatic processes, including nutrient sensing, metabolism, tissue repair and infection control1,11,24–28. Here we reveal that NMU is the molecular link between neuronal sensing, innate type 2 responses and mucosal protection. Coupling neuronal activity and ILC2-dependent immune regulation may have ensured potent, efficient and integrated multi-tissue responses to environmental challenges throughout evolution. Notably, coordinated NMU-induced smooth muscle contraction16 and type 2 innate immunity may have coevolved to control worms that have been intimate evolution partners of mammals. In line with this hypothesis, NMU is highly conserved across mammalian, amphibian, avian and fish species16. Finally, our current data and other independent studies indicate that the mucosal nervous system partners with immune cells to ensure local tissue regulation2,3,29,30; thus, it is tempting to speculate the existence of neuro-immune sensory units that regulate physiology and homeostasis at an organismic level.
Methods
Mice
C57BL/6J (B6) mice were purchased from Charles River. Nod/Scid/Gamma (NSG) mice were bought from The Jackson Laboratory. Sperm from the strain C57BL/6-Nmur1tm1.1(KOMP)Vlcg, which contains a Nmur1 deletion, was obtained from the KOMP Repository, University of California Davis and Children’s Hospital Oakland Research Institute, US. Nmur1-/- mice were generated by in vitro fertilisation at the Champalimaud Centre for the Unknown, Portugal. Chat-Cre20, Rosa26RFP 19, Rag2-/- Il2rg-/- 31,32, Myd88-/- 33 and RetGFP 18 and Myd88fl/fl 33 mice were on a C57BL/6J background. Mice were bred and maintained at the Champalimaud Centre for the Unknown and iMM Lisboa animal facilities under specific pathogen free conditions. Il1rl1-/- Il17rb-/- mice 34,35 and their WT controls were on a BALB/c background and were bred and maintained at École Polytechnique Fédérale de Lausanne (EPFL), Switzerland. Mice were systematically compared with co-housed littermate controls unless stated otherwise. 7-12 weeks old males and females were used in this study. All animal experiments were approved by national and institutional ethical committees, respectively Direção Geral de Veterinária and Service de la Consummation et des Affaires Vétérinaires Federal (Canton Vaud, Switzerland), Champalimaud Centre for the Unknown, iMM Lisboa and EPFL ethical committees. Randomisation and blinding were not used unless stated otherwise. Power analysis was performed to estimate the number of experimental mice.
Analysis of gene expression microarray data
The expression profile of 239 genes related to neural pathways was performed in mouse lymphoid cells based on the Affymetrix Mouse Gene 1.0 ST Array dataset (GEO accession number GSE37448)12. Preprocessing of microarray data (including background correction and normalisation) was performed applying the robust multiarray (RMA) method36, included in the Bioconductor package affy37 for the statistical software environment R38. Linear models and the B (empirical Bayes) statistic were employed in differential gene expression analysis, using Bioconductor package limma39. Plots associated with the microarray data analyses were generated in R.
In vitro and in vivo ILC2 activation
For in vitro experiments, purified lung and small intestine lamina propria ILC2s were cultured in complete RPMI (supplemented with 10% foetal bovine serum (FBS), 1% hepes, sodium pyruvate, glutamine, streptomycin and penicillin) at 37ºC. ILC2s were stimulated overnight for 2, 4, 6 and 20 hours with recombinant mouse Neuromedin U 23 peptide (NmU23) (100ng/mL unless stated otherwise; Phoenix Pharmaceuticals), recombinant mouse IL-25 and IL-33 (10ng/mL, unless stated otherwise) (R&D Systems). Control and activated ILC2s were cultured in the presence of IL-2 and IL-7 (10ng/mL; PeproTech), unless stated otherwise. ILC2s were lysed using RLT buffer (Qiagen). For cytokine protein analysis in vitro, ILC2s were incubated with brefeldin A (eBioscience) for 2, 4, 6 or 20 hours prior to intracellular staining. For in vivo experiments, mice were injected intraperitoneal (i.p.) with NmU23 peptide (8μg/day) during Nippostrongylus brasiliensis infection or with a single dose of NmU23 (20μg) and analysed after 12 hours. Control mice were treated with PBS alone. For cytokine protein analysis ex vivo, ILC2s were incubated with PMA (50ng/mL), ionomycin (500ng/mL) (Sigma) and brefeldin A (eBioscience) for 4 hours prior to intracellular staining.
Bone marrow transplantation
Bone marrow cells were flushed out from femurs and tibiae of Nmur1-/-, Il1rl1-/-.Il17rb-/- mice and their respective WT controls. Bone marrow cells were CD3-depleted using Dynabeads Biotin Binder (Thermo Fisher Scientific) according to the manufacturer’s instructions. 106 cells of each genotype (CD45.2) were injected intravenously alone or in direct competition with a third-party WT competitor (CD45.1/CD45.2), in a 1:1 ratio, into non-lethally irradiated (150 Rad) NSG mice (CD45.1). Mice were analysed at 8 weeks after transplantation.
ILC2 adoptive transfer
For adoptive cell transfer, small intestine ILC2s from Nmur1-/- and Nmur1+/+ littermate control mice were purified and expanded in vitro in supplemented RPMI in the presence of recombinant mouse IL-2, IL-7 (10ng/mL; Peprotech) and IL-33 (10ng/mL; R&D Systems) for 1 week. 2×105 expanded ILC2s were injected intravenously into Rag2-/-Il2rg-/- recipients. Mice were infected with 400 Nippostrongylus brasiliensis larvae 1 week after ILC2 transplantation.
Parasite infection
Nippostrongylus brasiliensis was maintained by monthly passages in Lewis rats as previously described40. Infective (iL3) worms were kindly provided by Nicola Harris (EPFL, Switzerland). iL3 larvae were treated for 15 minutes with streptomycin and penicillin (300U/mL; Thermo Fisher Scientific), gentamicin (1.5mg/mL; Sigma) and tetracyclin (30μg/mL; Sigma), washed with PBS and counted under a stereomicroscope. Mice were injected subcutaneously with 400 iL3 in 200μL of sterile PBS using a 21G needle. Mice were sacrificed at day 1, 2 and 6 post-infection and lungs, bronchoalveolar lavage (BAL) and small intestine were collected and analysed.
Infection burden
Lung and small intestine parasite burden was quantified in minced lungs and small intestine as previously described40. Briefly, lungs and small intestine were placed on sterile cheesecloth and suspended in a 50mL tube containing PBS at 37°C for at least 4 hours. Viable worms that migrate out into the bottom of the tube were counted under a stereomicroscope (steREO Lumar V12; Zeiss).
Airway challenge
C57BL/6J mice were anesthetised by inhalation of isoflurane. Mice were challenged with a single intranasal (i.n.) dose of Nippostrongylus brasiliensis excretory/secretory products (NES) (5μg) (provided by Nicola Harris (EPFL)) or with PBS in a volume of 30μL for 4 consecutive days. 24 hours after the last challenge lung and BAL were recovered for analysis.
Differentiation of naïve T helper cells
Enteric naïve T helper cells were purified and cultured in an anti-CD3 (2μg/mL; 17A2; Biolegend) pre-coated plate. For Th2 differentiation, cells were stimulated in the presence of soluble anti-CD28 (1μg/mL; 37.51), recombinant mouse IL-2 (10ng/mL) (Peprotech), IL-4 (50ng/mL), anti-IFN-γ (1μg/mL; R4-6A2) and anti-IL-12 (1μg/mL; C17.8) (Biolegend). Polarised Th2 cells were analysed after 4 days of differentiation.
Cell isolation
Lungs were perfused with a solution of cold PBS with 2% heparin through the right ventricle of the heart and were subsequently finely minced and digested in complete RPMI supplemented with collagenase D (0.1mg/mL; Roche) and DNase I (20U/mL; Affymetrix) for 1h at 37°C under gentle agitation. For isolation of small intestine lamina propria cells, intestines were thoroughly rinsed with PBS, cut in 1cm pieces, and shaken for 30 minutes in PBS containing 2% FBS, 1% hepes and 5mM EDTA to remove intraepithelial and epithelial cells. Intestines were then digested with collagenase D (0.5mg/mL; Roche) and DNase I (20U/mL; Affymetrix) in complete RPMI for 30 minutes at 37°C, under gentle agitation. Enteric neurons and glial cells were isolated as previously described3,41. Briefly, isolated tissues were digested with Liberase TM (7.5μg/mL; Roche) and DNase I (20U/mL; Affymetrix) in complete RPMI for 30 minutes at 37°C, under gentle agitation. Digested organs were disrupted by passage through a 100μm cell strainer (BD Biosciences). A 40-80% percoll gradient centrifugation was used for additional leukocyte purification from lung and small intestine cell suspensions. Erythrocytes from lung, small intestine and bone marrow preparations were lysed with RBC lysis buffer (eBioscience).
Flow cytometry and cell sorting
For cytokine protein analysis ex vivo, cells were incubated with PMA (50ng/mL), ionomycin (500ng/mL) (Sigma) and brefeldin A (eBioscience) for 4 hours prior to intracellular staining. Intracellular staining was performed using IC fixation/permeabilisation kit (eBioscience). Cell suspensions were stained with anti-CD45 (30-F11), anti-TER119 (TER-119), TCRβ (H57-597), anti-CD3ε (eBio500A2), anti-CD19 (eBio1D3), anti-NK1.1 (PK136), anti-CD11c (N418), anti-Gr1 (RB6-8C5), anti-CD11b (Mi/70), anti-CD127 (IL-7Rα; A7R34), anti-α4β7 (DATK32), anti-Flt3 (A2F10), anti-CD25 (PC61.5), anti-c-Kit (2B8), anti-Thy1.2 (53-2.1), anti-CD49b (DX5), anti-TCRγδ (GL3), anti-NKp46 (29A1.4), anti-CD4 (GK1.5), anti-CD31 (390), anti-IL-13 (eBio13A), anti-F4/80 (BM8), anti-FcεR1 (MAR-1), anti-CD44 (IM7), anti-CD62L (MEL-14), 7AAD viability dye, anti-CD16/CD32 (93) from eBioscience; anti-CD8α (53-6.7), anti- KLRG1 (2F1/KLRG1), anti-Sca1 (D7), anti-CCR3 (J073E), anti-MHC-II (M5/114.15.2), anti-Ki67 (16A8), anti-streptavidin and anti-CD326 (G8.8) from Biolegend, anti-IL-5 (MH9A3) from BD Biosciences, anti-amphiregulin (R&D Systems). LIVE/DEAD Fixable Aqua Dead Cell Stain Kit was purchased from Invitrogen. Cell populations were defined as: Common Lymphoid Progenitor (CLP) - Lin−CD127+Flt3+Sca1intc-Kitint; Common Helper Innate Lymphoid Progenitor (CHILP) - Lin−CD127+α4β7+Flt3−CD25−; ILC2 precursor (ILC2P) - Lin−CD127+α4β7+Flt3−CD25+; ILC2 - CD45+Lin-Thy1.2+KLRG1+Sca1+; ILC1 - CD45+Lin-NKp46+NK1.1+CD49b-CD127+; ILC3 - CD45+Lin-Thy1.2hiKLRG1-; for ILC3 subsets additional markers were employed: ILC3 CD4+ - NKp46-CD4+; ILC3 NCR- - NKp46-CD4-; ILC3 NCR+ - NKp46+CD4-; NK cells - CD45+Lin-NKp46+NK1.1+CD49b+CD127-; Lineage was composed by CD3ε, CD8α, TCRβ, TCRγδ, CD19, Gr1, CD11c and TER119; Eosinophils (Eo) - MHC-II-CCR3hiGR1int; Mast cells (Mast) - CD3-FcεR1+; Macrophages (MØ) - CD3-MHC-II+F4/80+; Neutrophils (Neu) - MHC-II-CCR3-GR1hi; Dendritic cells (DC) – CD45+MHC-II+F4/80-CD11c+; T cells - CD45+CD3+TCRβ+; T helper (Th) cells - CD45+CD3+TCRβ+CD4+; Th naïve (Thn) cells - CD45+CD3+CD4+CD44loCD62Lhi; memory Th (Thm) cells - CD45+CD3+CD4+CD44hiCD62Llo; B cells - CD45+CD19+; enteric glial cells (G) - CD45-CD31-TER119-CD49b+; enteric neurons (N) - CD45-CD31-TER119-RETGFP/+; Epithelial cells (Ep) – CD45-CD31-CD326+. Flow cytometry analysis and cell sorting were performed using LSRFortessa, FACSAria and FACSFusion flow cytometers (BD Biosciences). The percentage of ILC2s is gated in live CD45+Lin-Thy1.2+ cells, unless stated otherwise. Sorted populations were >95% pure. Data analysis was done using FlowJo software (Tristar).
Enteric neurosphere-derived neurons
Enteric neurosphere-derived neurons were obtained as previously described29. Briefly, total intestines from Embryonic day 14.5 (E14.5) to E18.5 Myd88−/− and control embryos were digested with collagenase D (0.5mg/mL; Roche) and DNase I (20U/mL; Affymetrix) in supplemented DMEM/F-12, GlutaMAX (1% hepes, penicillin and streptomycin and 0.1% β-mercaptoethanol) (Gibco) for 1 hour at 37 °C under gentle agitation. Cells were mechanically disrupted, washed and cultured in a non-adherent plate for 1 week in a CO2 incubator at 37 °C in supplemented DMEM/F-12, GlutaMAX with B27 (Gibco), EGF and FGF2 (20ng/mL; R&D Systems). After neurosphere formation, cells were dissociated using NeuroCult™ (STEMCELL Technologies) chemical dissociation kit, according to the manufacturer's instructions. Dissociated neurospheres were plated on a 24-well plate previously coated with PDL (100μg/mL; Sigma) and cultured in supplemented NEUROBASAL media (1% hepes, glutamine, penicillin and streptomycin) (Gibco) with B27 for neuronal differentiation. After 7 days, neuronal organoids were used either for activation or immunostaining. Enteric neurosphere-derived neurons were activated with LPS (5μg/mL; Invivogen), recombinant mouse IL-33 (100ng/mL; R&D Systems) and NES (10μg/mL), for 24 hours followed by RNA extraction. For ILC2 activation experiments, conditioned media from NES activated enteric neurons was collected and applied overnight to purified ILC2s.
Purification of human ILC2s and T helper cells
For purification of human peripheral ILC2s and T helper cells, peripheral blood mononuclear cells (PBMCs) were isolated from human buffy coats by diluting the blood 1:2 in PBS FBS 2%. The blood was layered on top of Lymphoprep™ (STEMCELL Technologies) and centrifuge at 800x g for 30 minutes at room temperature without brake. Human PBMCs were then washed and stained with antibodies against human lineage markers namely, CD3 (OKT3), αβTCR (IP26), γδTCR (B1.1), CD19 (HIB19), CD14 (61D3), CD11c (3.9), CD16 (eBioCB16) (eBioscience); and CD45 (HI30), CD127 (A019D5), CD161 (HP-3G10) (Biolegend), CRTH2 (BM16) (BD Biosciences) and CD4 (OKT4; Biolegend). Human ILC2s were defined as CD45+Lin-CRTH2+CD127+CD161+ and human helper T cells as CD45+Lin+CD4+.
Quantitative RT-PCR
Total RNA was extracted using RNeasy micro kit (Qiagen) according to the manufacturer’s protocol. When indicated, total brain, lung and proximal duodenum was collected for RNA extraction using Trizol (Invitrogen) and zirconia/silica beads (BioSpec) in a bead beater (MIDSCI), according to the manufacturer’s protocol. RNA concentration was determined using Nanodrop Spectrophotometer (Nanodrop Technologies). Quantitative real-time RT–PCR was performed as previously described5,8. Hprt, Gapdh and Eef1a1 were used as housekeeping genes. For neurospheres-derived neurons Hprt, Gapdh, Tubb3 and Rbfox3 were used as housekeeping genes. For TaqMan assays (Applied Biosystems) RNA was retro-transcribed using a High Capacity RNA-to-cDNA Kit (Applied Biosystems), followed by a pre-amplification PCR using TaqMan PreAmp Master Mix (Applied Biosystems). TaqMan Gene Expression Master Mix (Applied Biosystems) was used in real-time PCR. TaqMan Gene Expression Assays (Applied Biosystems) were the following: Hprt Mm00446968_m1; Gapdh Mm99999915_g1; Eef1a1 Mm01973893_g1; Il5 Mm00439646_m1; Il13 Mm00434204_m1; Areg Mm1354339_m1; Csf2 Mm01290062_m1; Gata3 Mm00484683_m1; Rora Mm01173766_m1; Nmu Mm00479868_m1; Nmur1 Mm04207994_m1; Nmur2 Mm00600704_m1; Tubb3 Mm00727586_s1 and Rbfox3 Mm01248771_m1. Real-time PCR analysis was performed using StepOne Real-Time PCR system (Applied Biosystems). For the analysis of human blood cells the TaqMan Gene Expression Assays were the following: HPRT1 Hs02800695_m1; GAPDH Hs02786624_g1; and NMUR1 Hs00173804_m1. Samples were normalised using HPRT1 and GAPDH as housekeeping genes. The mRNA analysis was done using the comparative CT method (2−ΔCT). When comparison or fold change between samples was required, the comparative ΔCT method (2−ΔΔCT) was applied.
Cell signalling
Purified ILC2s from small intestine and lung were FBS starved for 2 hours before in vitro activation with NmU23 at 37°C. To test for ERK phosphorylation (Cell Signaling Technology), purified ILC2s were activated with NmU23 (100ng/mL; Phoenix Pharmaceuticals) in the presence of IL-2 and IL-7 (10ng/mL; PeproTech) for 10 minutes prior to intracellular staining. To test ERK, calcineurin and NFAT activation, ILC2s were cultured for 1 hour with their respective inhibitor and then stimulated with NmU23 overnight before mRNA expression analysis. ERK inhibitor - PD98059 (40μM; Sigma); Calcineurin inhibitor - FK506 (100nM) and CsA (100nM) (Tocris Bioscience); NFAT inhibitor - 11R-VIVIT (10μM) (Tocris Bioscience).
Calcium signalling
Purified ILC2s from the small intestine were cultured with IL-2 and IL-7 (10ng/mL) and FBS deprived for 6 hours prior to calcium signaling experiments. ILC2s were stained with Fluo-4 Direct Calcium Assay Kit (Thermo Fisher Scientific) according to manufacturer’s protocol. Calcium (Ca2+) influx, represented by the Fluo-4 AM, was recorded over time on a BD Accuri C6 (BD Biosciences) flow cytometer as previously reported42. The recombinant mouse NmU23 was added 60 seconds after ILC2 baseline recording. Data was represented by the mean values of Ca2+ influx kinetics between the ILC2 baseline response and the peak of response after recombinant mouse NmU23 addition.
Histopathology analysis
Mice were sacrificed by cervical dislocation. The caudal lobe of the right lung was harvested, fixed in 10% neutral buffered formalin and processed for paraffin embedding. Serial 4μm sections were stained for hematoxylin and eosin (H&E), Luna and immunohistochemistry for myeloperoxidase (MPO) was performed. Briefly, using standard protocols, antigen heat-retrieval was performed at low pH43 in Dako PT module, followed by incubation with the primary antibody (polyclonal rabbit anti-human Myeloperoxidase, Dako Corp). Incubation with ENVISION kit (Peroxidase/DAB detection system, Dako Corp) was followed by Harri’s hematoxylin counterstaining (Bio Otica). Negative control included the absence of primary antibodies. Slides were analysed by a pathologist blinded to experimental groups and images were acquired in a Leica DM2500 microscope, coupled with a Leica MC170 HD microscope camera. Quantification of inflammatory cell infiltration of the lung was performed in MPO-stained sections by manual counting of MPO-positive cells at 20x original magnification, corresponding to 0.2mm2 per field. Quantification of pulmonary eosinophils was performed in Luna-stained slides by manual counting the number of granulocytes with eosinophilic granular cytoplasm in low power fields (1mm2 per field).
Microscopy
Sections from lungs were obtained from NGS and irradiated (400 rad) Chat-Cre.Rosa26RFP mice after reconstitution with WT ILC2s. Sections of small intestine were obtained from Chat-Cre.Rosa26RFP and RetGFP mice. Sections of 100μm were obtained with a Leica VT1200/VT1200 S vibratome. Briefly, samples were fixed with 4% PFA at 4°C overnight and were then incorporated in 4% low-melting temperature agarose (Invitrogen). Samples were blocked and permeabilised with PBS containing 0.6% Triton X-100 and 2% BSA and incubated for 1–2 days at room temperature with the following antibodies: mouse monoclonal anti-KLRG1 (2F1/KLRG1; Biolegend); anti-CD3 (17A2; Biolegend) and anti-Tubulin β 3 (TUBB3) (TUJ1; Covance) for small intestine imaging and anti-Thy1.2 (53-2.1; eBioscience), anti-CD45.1 (A20; Biolegend) and anti-TUBB3 (TUJ1; Covance) for lung imaging. Alexa Fluor 647 goat anti-hamster, Alexa Fluor 568 goat anti-rat and Alexa Fluor 633 goat anti-mouse (Invitrogen) were used as secondary antibodies overnight at room temperature. Distances between neurons and ILC2 were measured using Imaris software. For NFAT2 imaging ILC2s were sorted from small intestine of C57Bl/6J mice. ILC2s were FBS starved for 2 hours and treated for 90 minutes with NmU23 (100ng/mL). After stimulation, cells were plated in a pre-coated 0.01% of Poly-L-Lysine (Sigma) coverslip and let to adhere. Cells were fixed with 2% PFA for 30 minutes at room temperature. Coverslips were blocked and permeabilised at room temperature for 30 minutes with a PBS solution containing 0.3% Triton X-100 and 1% BSA. Cells were stained at room temperature for 150 minutes with mouse monoclonal anti-NFAT2 antibody (7A6; Abcam). ILC2s were incubated subsequently at room temperature in the dark for 30 minutes with Alexa Fluor 488 goat anti–mouse antibody (Invitrogen) and then counterstained for 5 minutes with DAPI. For quantification of NFAT2 fluorescence intensity single-cell ILC2 nuclei were identified by DAPI staining. Regions of interest (ROIs) were defined from each cell nucleus and NFAT signal was measured using ImageJ software. Enteric neurosphere-derived neurons were fixed in 2% PFA for 30 minutes at room temperature. Cells were blocked and permeabilised at room temperature for 30 minutes. Differentiated neurons were stained at room temperature for 150 minutes with anti-TUBB3 (TUJ1; Covance). Neuronal organoids were sequentially incubated at room temperature in the dark for 30 minutes with Alexa Fluor 488 goat anti–mouse antibody (Invitrogen) and then counterstained for 5 minutes with DAPI. Samples were mounted in Mowiol and were acquired on a Zeiss LSM710 confocal microscope using Plan Apochromat 20x/0.8 M27 objective and EC Plan-Neofluar 40x/1.30 and Plan-Apochromat 63x/1.4 oil immersion objectives.
Statistics
Results are shown as mean ± s.e.m. Statistical analysis was performed with GraphPad Prism software (GraphPad Software, La Jolla, Calif). Student’s t-test was performed on homoscedastic populations. Unpaired t-test was applied on samples with different variances. Results were considered significant at *P<0.05, **P<0.01, ***P<0.001.
Extended Data
Extended Data Figure 1. Genome-wide ILC2 transcriptional profiling and neuron-ILC2 interactions.
a, Weighted Unifrac PCoA analysis of ILC2s, Th cells, ILC1s and ILC3s. b, Levels of Nmur1 expression in ILC2s, Th cells, ILC1 and ILC3 populations. c, Nmur1 expression in lung ILC2s, Eosinophils (Eo); Mast cells (Mast); Macrophages (Mø); Neutrophils (Neu); Dendritic cells (DC); helper T (Th); B cells (B) and glial cells (G). n=3. d, Nmur2 expression in intestinal ILC2s, Eosinophils (Eo); Mast cells (Mast); Macrophages (Mø); Neutrophils (Neu); Dendritic cells (DC); helper T (Th); B cells (B), Glial cells (G); Neurons (N) and Brain (Br). n=3. e, Nmu expression in lung immune cell subsets. n=3. f, Distance of T cells and ILC2 to adjacent enteric neurons. T cells n=22; ILC2 n=28. g, Confocal analysis of lung. Red: neurons (TUBB3); Green: Thy1.2, Blue: DAPI. Green arrows: candidate ILC2s. Red arrow: neuron. Scale bar: 5μm. h, Confocal analysis of lung. Red: neurons (Chat-Cre.Rosa26RFP); Green: CD45.1, Blue: DAPI. Green arrows: candidate ILC2s. Red arrow: neuron. Scale bar: 5μm. Error bars show s.e.m. *P<0.05.
Extended Data Figure 2. Neuromedin U is potent regulator of innate type 2 cytokines, via NMUR1 activation.
a, ILC2 and Th2 were activated with NmU23. ILC2 n=6; Th2 n=3. b, Proliferation (as measured by Ki67 expression) of gut ILC2 upon NmU23 activation in the presence or absence of IL-2 and IL-7 in vitro. n=3. c, Percentage ofKi67 expression in enteric ILC2 upon NmU23 administration in vivo. n=5. d, Dot plots representing Ki67 expression in gut ILC2 upon NmU23 administration in vivo. e, Lung innate type 2 cytokines after NmU23 in vitro stimulation. n=3. f, Dot plots representing lung ILC2-derived type 2 cytokines after NmU23 in vitro activation. g, Enteric ILC2-derived type 2 cytokines upon NmU23 stimulation over different incubation times or PMA+Ionomycin (P+I) activation for 4h. h, Dot plots representing gut ILC2-derived type 2 cytokines upon NmU23 stimulation over different incubation time periods. Error bars show s.e.m. *P<0.05; **P<0.01; ***P<0.001; ns not significant.
Extended Data Figure 3. NMU is a fast and potent inducer of ILC2 cytokines.
a, Dot plots representing enteric ILC2-derived type 2 cytokines upon activation with increasing concentrations (10, 50 and 100ng/mL) of IL-33, IL-25 and NmU23 for 20 hours. b, Gut ILC2-derived cytokines after stimulation with increasing concentrations (10, 50 and 100ng/mL) of IL-33, IL-25, NmU23 and PMA+Ionomycin (P+I) for 4h. n=3. c, Lung ILC2- and Th cell-derived type 2 cytokines after in vivo administration of NmU23. n=3. Error bars show s.e.m. *P<0.05; **P<0.01; ns not significant.
Extended Data Figure 4. Activation of ILC2s by NMU and IL-25/IL-33 signals.
a, Lung ILC2s from Il1rl1-/-Il17rb-/- (DKO) and their WT controls after NmU23 stimulation. n=6. b, Type 2 cytokines in Nmur1 sufficient and deficient ILC2s after IL-33 and IL-25 (10ng/mL) activation for 24 hours. n=3. c, Intestinal ILC2-derived cytokines after NmU23 administration in Il1rl1-/-Il17rb-/- (DKO) and their WT controls. Left panel represents ILC2 percentage gated in total live cells. n=5. d, Lung ILC2-derived cytokines in WT BALB/c and Il1rl1-/-Il17rb-/- bone marrow chimeras upon NmU23 administration. n=5. Error bars show s.e.m. *P<0.05; **P<0.01; ***P<0.001; ns not significant.
Extended Data Figure 5. ILC2-autonomous NMUR1 signals.
a-d, ILC2-derived type 2 cytokines in Nmur1-/- and in their Nmur1+/+ WT littermate controls. a, Percentage of intestinal ILC2s and their signature cytokines. Nmur1+/+ n=6; Nmur1-/- n=9. b, Number of intestinal ILC2s and their signature cytokines. Nmur1+/+ n=6; Nmur1-/- n=9. c, Percentage of lung ILC2s and their signature cytokines. Nmur1+/+ n=6; Nmur1-/- n=9. d, Number of lung ILC2s and their signature cytokines. Nmur1+/+ n=6; Nmur1-/- n=9. e-h, Competitive bone marrow chimeras. 106 cells of each genotype (CD45.2) were injected intravenously in direct competition with a third-party WT competitor (CD45.1/CD45.2), in a 1:1 ratio, into non-lethally irradiated (150Rad) NSG mice (CD45.1). e, Percentage and number of donor ILC2s in the intestine. Nmur1+/+ n=8; Nmur1-/- n=6. f, Percentage and number of donor ILC2s in the lung. Nmur1+/+ n=12; Nmur1-/- n=13. g-h, Bone marrow mixed chimeras upon NmU23 administration. g, Percentage of lung Th cell-expressing type 2 cytokines. Nmur1+/+ n=5; Nmur1-/- n=4. h, Number of lung Th cell-expressing type 2 cytokines. Nmur1+/+ n=5; Nmur1-/- n=4. Error bars show s.e.m. ns not significant.
Extended Data Figure 6. Calcineurin inhibition during NMU-dependent ILC2 activation.
Intestinal ILC2 activation with NmU23. Il5, Il13 and Csf2 expression in ILC2s cultured with medium (control), NmU23 or NmU23 and calcineurin inhibitor cyclosporine (CsA) (n=3). Error bars show s.e.m. ***P<0.001.
Extended Data Figure 7. Neuromedin U administration during worm infection.
a-f, Mice were infected with N. brasiliensis larvae and treated with NmU23. a, Nmur1 expression in lung ILC2 at day 6 post-infection. n=6. b, Nmur1 expression in lung immune populations. Eosinophils (Eo); Mast cells (Mast); Macrophages (Mø); Neutrophils (Neu); T helper cells (Th) and ILC2. n=3. c, Number of lung ILC2s at day 1 after infection in NmU23 treated and control animals. n=5. d, Lung T helper cells at day 1 after infection in NmU23 treated and control animals. n=5. e, Myeloperoxidase (MPO)- (granulocytes) and Luna-stained (eosinophils) lung sections at day 2 after infection. f, Lung granulocyte and eosinophilic cell counts (cells/mm2) at day 2 after infection. n=8. Scale bars: 50μm. Error bars show s.e.m. *P<0.05; **P<0.01; ns not significant.
Extended Data Figure 8. Worm infection in Nmur1 deficient mice.
Nmur1-/- and in their Nmur1+/+ WT littermate control mice were infected with N. brasiliensis. a, Number of lung ILC2s and their cytokines at day 6 after infection. Nmur1+/+ n=6; Nmur1-/- n=8. b, Myeloperoxidase (MPO)- (granulocytes) and Luna-stained (eosinophils) lung sections at day 2 after infection. Scale bars: 50μm. c, Lung granulocyte and eosinophil cell counts (cells/mm2) at day 2 after infection. n=8. d, Worm infection burden at day 6 and 9 post-infection in the small intestine of Nmur1 sufficient and deficient mice. D6 n=6; D9 n=5. Error bars show s.e.m. *P<0.05; **P<0.01; ***P<0.001; ns not significant.
Extended Data Figure 9. Secretory worm products induce ILC2-derived type 2 cytokines.
a, ILC2-derived cytokines after stimulation with Nippostrongylus brasiliensis excretory/secretory products (NES) alone or with NES activated neurosphere-derived enteric neurons conditioned media (SN NES). Control n=3; NES n=3; SN NES n=3. b, Percentage and number of lung ILC2s and their signature cytokines after intranasal NES administration to WT mice. PBS n=5, NES n=5. Error bars show s.e.m. *P<0.05; **P<0.01; ns not significant.
Extended Data Figure 10. A novel neuron-ILC2 unit orchestrated by Neuromedin U.
Mucosal neurons can directly sense worm products (NES) and the host alarmin (IL-33) to control Neuromedin U expression. Neuromedin U activates ILC2s in a cell-autonomous and NMUR1 dependent manner, resulting in a fast and potent production of inflammatory and tissue repair cytokines that confer immediate protection to worm infection. Neuromedin U activates NMUR1 inducing type 2 cytokine expression downstream of ERK phosphorylation and activation of a Ca2+/Calcineurin/NFAT cascade. This model indicates that neuron-ILC2 cell units are poised to uniquely ensure potent and immediate type 2 responses in a Neuromedin U-dependent manner.
Supplementary Material
Acknowledgements
We thank the Histology and Bioimaging services at iMM Lisboa. We thank the Vivarium and Flow Cytometry platforms at iMM Lisboa and at the Champalimaud Centre for the Unknown. We thank A. McKenzie for kindly providing Il1rl1-/- and Il17rb-/- mice; D. Fonseca-Pereira, V. Fonseca, S. Xapelli and L. Lopes for helpful discussions and M. Rendas for technical assistance. V.C was supported by Fundação para a Ciência e Tecnologia (FCT), Portugal. J.C. by Fondation pour la Recherche Médicale (FRM), France, and by Marie Skłodowska-Curie fellowship (750030), EU. B.G.-C. by FP7 (289720), EU. N.L.B.-M. is supported by FCT, Portugal, and European Molecular Biology Organisation (EMBO). N.H. by Swiss National Science Foundation (310030_156517). H.V.-F. by ERC (647274), EU; Kenneth Rainin Foundation, US; Crohn’s and Colitis Foundation of America, US; and FCT, Portugal.
Footnotes
Author contribution
V.C. and J.C designed, performed and analysed the experiments in Fig.1-4; and Extended Data Fig.1-10. H.R. provided technical assistance in Fig.4a and managed the animal colony. B. G.-C. contributed to experiments in Fig.1f,g, Fig.3e,f and Extended Data Fig.1f-g. T.C. analysed the experiments in Fig.4c,g and Extended Data Fig.7e,f and Extended Data Fig.8b,c. N.L.B.-M. analysed the experiments in Fig.1a,b and Extended Data Fig.1a,b.; T.B., K.S. and N.H. contributed to the design of the experiments in Fig.4, Extended Data Fig.4c and provided N. brasiliensis larvae and NES. H.V.-F. supervised the work, planned the experiments and wrote the manuscript.
Author information
The authors declare no competing financial interests.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author [H.V.-F.]. The data are not publicly available due to patent application request.
References
- 1.Cording S, Medvedovic J, Aychek T, Eberl G. Innate lymphoid cells in defense, immunopathology and immunotherapy. Nat Immunol. 2016;17:755–757. doi: 10.1038/ni.3448. [DOI] [PubMed] [Google Scholar]
- 2.Veiga-Fernandes H, Mucida D. Neuro-Immune Interactions at Barrier Surfaces. Cell. 2016;165:801–811. doi: 10.1016/j.cell.2016.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ibiza S, et al. Glial-cell-derived neuroregulators control type 3 innate lymphoid cells and gut defence. Nature. 2016;535:440–443. doi: 10.1038/nature18644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veiga-Fernandes H, Pachnis V. Neuroimmune regulation during intestinal development and homeostasis. Nat Immunol. 2017;18:116–122. doi: 10.1038/ni.3634. [DOI] [PubMed] [Google Scholar]
- 5.van de Pavert SA, et al. Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature. 2014;508:123–127. doi: 10.1038/nature13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel A, et al. Differential RET signaling pathways drive development of the enteric lymphoid and nervous systems. Sci Signal. 2012;5:ra55. doi: 10.1126/scisignal.2002734. [DOI] [PubMed] [Google Scholar]
- 7.Veiga-Fernandes H, et al. Tyrosine kinase receptor RET is a key regulator of Peyer's Patch organogenesis. Nature. 2007;446:547–551. doi: 10.1038/nature05597. [DOI] [PubMed] [Google Scholar]
- 8.Fonseca-Pereira D, et al. The neurotrophic factor receptor RET drives haematopoietic stem cell survival and function. Nature. 2014;514:98–101. doi: 10.1038/nature13498. [DOI] [PubMed] [Google Scholar]
- 9.Veldhoen M, Veiga-Fernandes H. Feeding immunity: skepticism, delicacies and delights. Nat Immunol. 2015;16:215–219. doi: 10.1038/ni.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiss EA, et al. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011;334:1561–1565. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- 11.Spencer SP, et al. Adaptation of innate lymphoid cells to a micronutrient deficiency promotes type 2 barrier immunity. Science. 2014;343:432–437. doi: 10.1126/science.1247606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinette ML, et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol. 2015;16:306–317. doi: 10.1038/ni.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Augood SJ, Keast JR, Emson PC. Distribution and characterisation of neuromedin U-like immunoreactivity in rat brain and intestine and in guinea pig intestine. Regul Pept. 1988;20:281–292. doi: 10.1016/0167-0115(88)90063-8. [DOI] [PubMed] [Google Scholar]
- 14.Ballesta J, et al. Occurrence and developmental pattern of neuromedin U-immunoreactive nerves in the gastrointestinal tract and brain of the rat. Neuroscience. 1988;25:797–816. doi: 10.1016/0306-4522(88)90037-1. [DOI] [PubMed] [Google Scholar]
- 15.Honzawa M, Sudoh T, Minamino N, Kangawa K, Matsuo H. Neuromedin U-like immunoreactivity in rat intestine: regional distribution and immunohistochemical study. Neuropeptides. 1990;15:1–9. doi: 10.1016/0143-4179(90)90153-p. [DOI] [PubMed] [Google Scholar]
- 16.Martinez VG, O'Driscoll L. Neuromedin U: a multifunctional neuropeptide with pleiotropic roles. Clin Chem. 2015;61:471–482. doi: 10.1373/clinchem.2014.231753. [DOI] [PubMed] [Google Scholar]
- 17.Mulligan LM. RET revisited: expanding the oncogenic portfolio. Nat Rev Cancer. 2014;14:173–186. doi: 10.1038/nrc3680. [DOI] [PubMed] [Google Scholar]
- 18.Hoshi M, Batourina E, Mendelsohn C, Jain S. Novel mechanisms of early upper and lower urinary tract patterning regulated by RetY1015 docking tyrosine in mice. Development. 2012;139:2405–2415. doi: 10.1242/dev.078667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossi J, et al. Melanocortin-4 receptors expressed by cholinergic neurons regulate energy balance and glucose homeostasis. Cell Metab. 2011;13:195–204. doi: 10.1016/j.cmet.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hermann-Kleiter N, Baier G. NFAT pulls the strings during CD4+ T helper cell effector functions. Blood. 2010;115:2989–2997. doi: 10.1182/blood-2009-10-233585. [DOI] [PubMed] [Google Scholar]
- 22.Howard AD, et al. Identification of receptors for neuromedin U and its role in feeding. Nature. 2000;406:70–74. doi: 10.1038/35017610. [DOI] [PubMed] [Google Scholar]
- 23.Raddatz R, et al. Identification and characterization of two neuromedin U receptors differentially expressed in peripheral tissues and the central nervous system. J Biol Chem. 2000;275:32452–32459. doi: 10.1074/jbc.M004613200. [DOI] [PubMed] [Google Scholar]
- 24.Fallon PG, et al. Identification of an interleukin (IL)-25-dependent cell population that provides IL-4, IL-5, and IL-13 at the onset of helminth expulsion. The Journal of experimental medicine. 2006;203:1105–1116. doi: 10.1084/jem.20051615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monticelli LA, et al. Arginase 1 is an innate lymphoid-cell-intrinsic metabolic checkpoint controlling type 2 inflammation. Nat Immunol. 2016;17:656–665. doi: 10.1038/ni.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monticelli LA, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol. 2011;12:1045–1054. doi: 10.1031/ni.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brestoff JR, et al. Group 2 innate lymphoid cells promote beiging of white adipose tissue and limit obesity. Nature. 2015;519:242–246. doi: 10.1038/nature14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nussbaum JC, et al. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature. 2013;502:245–248. doi: 10.1038/nature12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gabanyi I, et al. Neuro-immune Interactions Drive Tissue Programming in Intestinal Macrophages. Cell. 2016;164:378–391. doi: 10.1016/j.cell.2015.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muller PA, et al. Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell. 2014;158:300–313. doi: 10.1016/j.cell.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shinkai Y, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 32.Cao X, et al. Defective lymphoid development in mice lacking expression of the common cytokine receptor gamma chain. Immunity. 1995;2:223–238. doi: 10.1016/1074-7613(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 33.Adachi O, et al. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 34.Neill DR, et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 2010;464:1367–1370. doi: 10.1038/nature08900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Townsend MJ, Fallon PG, Matthews DJ, Jolin HE, McKenzie AN. T1/ST2-deficient mice demonstrate the importance of T1/ST2 in developing primary T helper cell type 2 responses. The Journal of experimental medicine. 2000;191:1069–1076. doi: 10.1084/jem.191.6.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 37.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 38.Huber W, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nature methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bouchery T, et al. ILC2s and T cells cooperate to ensure maintenance of M2 macrophages for lung immunity against hookworms. Nat Commun. 2015;6:6970. doi: 10.1038/ncomms7970. [DOI] [PubMed] [Google Scholar]
- 41.Joseph NM, et al. Enteric glia are multipotent in culture but primarily form glia in the adult rodent gut. The Journal of clinical investigation. 2011;121:3398–3411. doi: 10.1172/JCI58186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doherty TA, et al. Lung type 2 innate lymphoid cells express cysteinyl leukotriene receptor 1, which regulates TH2 cytokine production. J Allergy Clin Immunol. 2013;132:205–213. doi: 10.1016/j.jaci.2013.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu JD. Myeloid cell-lineage and premylocytic-stage-specific- expression of themouse myeloperoxidase gene is controlled at initiation as well as elongation levels of transcription. Cell Res. 1999;9:107–134. doi: 10.1038/sj.cr.7290010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.