Abstract
Nanotechnology holds many advantages. Here we report another advantage of applying RNA nanotechnology for directional control. The orientation of arrow-shaped RNA was altered to control ligand-display on extracellular vesicle (EV) membranes for specific cell targeting, or to regulate intracellular trafficking of siRNA/miRNA. Placing membrane-anchoring cholesterol at the arrow-tail results in display of RNA aptamer or folate on EV outer surface. In contrast, placing the cholesterol at the arrow-head results in partial loading of RNA nanoparticles into the EVs. Taking advantage of the RNA ligand for specific targeting and EVs for efficient membrane fusion, the resulting ligand-displaying EVs were competent for specific delivery of siRNA to cells, and efficiently block tumor growth in three cancer models. PSMA aptamer-displaying EVs loaded with survivin siRNA inhibited prostate cancer xenograft. The same EV but displaying EGFR aptamer inhibited orthotopic breast cancer models. Likewise, survivin-loaded and folate-displaying EVs inhibited patient derived colorectal cancer xenograft.
Design and construction of arrow-shaped RNA nanostructures
The three-way junction (3WJ)1, 2 of the bacteriophage phi29 motor pRNA3, 4 folds by its intrinsic nature into a planar arrangement with three angles of 60°, 120°, and 180° between helical regions (Fig. 1a–b).2 The pRNA-3WJ was extended into an arrow-shaped structure by incorporating an RNA aptamer serving as a targeting ligand for binding to specific receptors overexpressed on cancer cells. The engineered pRNA-3WJ was used to decorate EVs purified from HEK293T cell culture supernatants to create ligand-decorated EVs. HEK293T EVs were used as they contain minimal intrinsic biological cargos compared to EVs generated by other cells.5 As shown in Western blots (Fig. S1a), HEK293T isolated EVs showed negative staining for several common integrin markers as seen on EVs for cancerous origins,6, 7 with only positive staining for TSG101. Additional steps were taken to remove EVs from FBS used in the HEK293T cell culture; although, centrifugation might not completely remove the FBS EVs.8, 9 Ultracentrifugation using OptiPrep was used to purify EVs (see Methods).10 The addition of iso-osmotic OptiPrep cushion layer greatly enhanced reproducibility of EVs purification in purity (Fig. S1c), and also minimized physical disruption of EVs by ultracentrifugation pelleting as shown by Electron Microscopy (EM) imaging (Fig. 1c). The presence of the OptiPrep cushion layer did not change the EVs particle size distribution or zeta potential significantly (Fig. 1d–e), but rather preserved the native shape of EVs. The EVs purified without the OptiPrep cushion appear as flattened spheres (Fig. 1c right), while the majority of EVs purified with the cushion appear as full spheres (Fig. 1c left). The size of EVs from EM image might not always represent its particle size distribution in the population. Nanoparticle Tracking Analysis (NTA) and Dynamic Light Scattering (DLS) revealed that the isolated native EVs were physically homogeneous, with a narrow size distribution centered around 96 nm (Fig. 1d) and a negative zeta potential (Fig. 1e). The purified EVs were further identified by the presence of EV specific marker TSG10111 by Western blot (Fig. S1a). The yield of purified EVs from HEK293T cell culture supernatant was about 10–15 μg (measured as protein concentration), or 0.1 ‒ 1.9×109 EV particles (measured by NTA) per 106 cells. A single steroid molecule, cholesterol-triethylene glycol (TEG), was conjugated into the arrow-tail of the pRNA-3WJ to promote the anchorage of the 3WJ onto the EV membrane (Fig. 1b). Cholesterol spontaneously inserts into the membrane of EVs via its hydrophobic moiety.12, 13 Display of RNA nanoparticles on surface of purified EVs was achieved by simply incubating the cholesterol-modified RNA nanoparticles with EVs at 37 °C for one hour.
Figure 1. RNA nanotechnology for decorating native EVs.

(a) AFM image of extended 3WJ of the motor pRNA of bacteriophage phi29. (b) Illustration of the location for cholesterol labeling of the arrow-head or arrow-tail of 3WJ. (c) Negative-stained EM image of EVs from HEK293T cells purified with differential ultracentrifugation method and cushion modified ultracentrifugation method. (d–g) NTA for size analysis and DLS for Zeta potential measurements. (h) 2D structure (left panel) and native PAGE for testing 3WJ assembly from three component strands, as indicated. (i). EVs loading and RNA aptamer display.
EVs hold great promise as emerging therapeutic carriers given their role in intercellular communication. They can enter cells through multiple routes including membrane fusion, tetraspanin and integrin receptor-mediated endocytosis, lipid raft mediated endocytosis, or micropinocytosis. However, there is limited specificity regarding the recipient cells.14, 15 In order to confer specific targeting of EVs to cancer cells, three classes of targeting ligands, folate, PSMA RNA aptamer, or EGFR RNA aptamer were conjugated to the 3WJ for displaying on the EVs surface. Folate is an attractive targeting ligand since many cancers of epithelial origin, such as colorectal cancers, overexpress folate receptors.16 PSMA is expressed at an abnormally high level in prostate cancer cells, and its expression is also associated with more aggressive diseases.17 A PSMA-binding 2′-Fluoro (2′-F) modified RNA aptamer A9g18, 19 was displayed on EVs to enhance targeting efficiency to prostate cancer cells. The PSMA aptamer A9g is a 43-mer truncated version of A9, which binds PSMA specifically with Kd 130nM18 and used as RNA based ligand. EGFR is highly overexpressed in triple negative breast cancer (TNBC) tumors and metastatic TNBC tumors.20 An EGFR specific 2′F-RNA aptamer21, 22 was incorporated to one end of pRNA-3WJ and thereby displayed on EVs for enhanced targeting of breast cancer cells. For imaging, one of the pRNA-3WJ strands was end-labeled with a fluorescent dye Alexa647 (Fig. 1h). The size distribution and zeta potential of RNA nanoparticle-decorated EVs did not change significantly compared with native EVs as measured by NTA and DLS (Fig. 1f–g).
Survivin, an inhibitor of cell apoptosis, is an attractive target for cancer therapy, since its knockdown can decrease tumorigenicity and inhibit metastases.23, 24 In combination with the survivin siRNA loaded in the EVs (Fig. 1i), siRNA loaded EVs with targeting moieties were prepared to evaluate in vivo prostate, breast, and colon cancer inhibition efficacy (see section 5). To improve the stability of siRNA in vivo, the passenger strand was 2′-F modified on pyrimidines to provide RNase resistance, while the guide strand was kept unmodified.25, 26 For tracking siRNA loading efficiency in EVs, the survivin siRNA was fused to an Alexa647-labeled 3WJ core and assembled into RNA nanoparticles (Fig. S1b). After loading siRNA into EVs and decorating EVs with PSMAapt/3WJ/Cholesterol RNA nanoparticles, there was not a significant change in the EVs size, as measured by NTA with two peaks at 103 and 120 nm (Fig. 1f). Treating survivin-3WJ RNA nanoparticles in PBS with ExoFect, without EVs, showed a different particle size distribution profile (PBS/siSurvivin) and about 40-times lower particle concentration (Fig. S1e). The loading efficiency for siRNA-3WJ RNA nanoparticles was around 70% (Fig. S1d) as measured by fluorescent intensity of the free RNA nanoparticles. Controls without EVs or with only the ExoFect reagent showed as low as 15% pelleting.
Arrow-head or arrow-tail for RNA loading or membrane display
Serum digestion assay was used to differentiate between entry and surface display on EVs. The orientation and angle of the arrow-shaped pRNA-3WJ nanostructure was used to control RNA loading or surface display of EVs. Serum digestion was performed to confirm the localization of 2′-F RNA nanoparticles with EVs. Although 2′-F 3WJ RNA nanoparticles are relatively resistant to RNaseA (Fig. S2a), they can be digested in 67 % fetal bovine serum (FBS) and incubated at 37 °C for 2 hours (Fig. S2b). Alexa647-2′F RNA nanoparticle-displaying EVs were purified from free RNA nanoparticles by ultracentrifugation, then subjected to serum digestion. Alexa647-2′F RNA with cholesterol on the arrow-tail for EVs decoration were degraded (31.6 ± 8.8 %) much more than the arrow-head cholesterol-decorated counterparts (9.5 ± 11.9 %) after 37 °C FBS incubation (Fig. 2a–d). These results indicate that cholesterol on the arrow-tail promoted display of either folate-3WJ or RNA aptamers on the surface of the EVs and were therefore degraded. While cholesterol on the arrow-head promoted RNA nanoparticles entering EVs, as evidenced by the protection of the Alexa647-2′F RNA nanoparticles against serum digestion. In the arrow-tail configuration, it seems as if the two arms that form a 60° angle can act as a hook to lock the RNA nanoparticle in place. If this was the case, the effect would prevent the hooked RNA from passing through the membrane (Fig. 2a). The concentration of FBS used in the serum digestion experiment was kept extremely high purposefully to degrade the externally displayed RNA on EVs. The decorated PSMAapt-3WJ 2′F RNA nanoparticles have been shown to remain stable and intact under physiological conditions.19, 22
Figure 2. Comparison of the role between arrow-head and arrow-tail 3WJ.

(a–b) Illustration showing the difference between arrow-head and arrow-tail display. (c) Syner gel to test arrow-head and arrow-tail Alexa647-3WJ/EV degradation by RNase in FBS. The gel was imaged at Alexa647 channel (d) and the bands were quantified by Image J. (e–i) Assay to compare cell binding of folate-3WJ arrow-tail (e–g) and arrow-head (h–i) on folate receptor positive and negative cells.
Competition assay was used to differentiate between entry and surface display on EVs. As described above, when cholesterol was attached to the arrow-tail of pRNA-3WJ, the RNA nanoparticles were anchored on the membrane of EVs, and the incorporated ligands were displayed on the outer surface of the EVs (Fig. 2a). An increase in the binding of EVs to folate receptor-overexpressing KB cells was detected by displaying folate on the EV surface using arrow-tail cholesterol RNA nanoparticles (Fig. 2e,f). When incubating with low folate receptor-expressing MDA-MB-231 breast cancer cells, arrow-tail-shaped FA-3WJ/EV did not enhance its cell binding compared to arrow-tail ligand free 3WJ/EV (Fig. 2g). The surface display of folate was further confirmed by free folate competition assay, in which a baseline of binding by the cholesterol arrow-tail FA-3WJ/EVs to KB cells was established. A decrease (48.3 ± 0.6 %) in the cellular binding to KB cells was detected when 10 μM of free folate was added to compete with the cholesterol-arrow-tail FA-3WJ/EV for folate receptor binding (Fig. 2f). In contrast, competition by free folate in arrow-head FA-3WJ/EV (Fig. 2h) binding to KB cells was much lower (24.8 ± 0.6 %) (Fig. 2i), which is possibly due to partial internalization of the arrow-head-shaped FA-3WJ nanoparticle into the EVs, which resulted in a lower display intensity of folate on the surface of the EVs.
EVs can mediate intercellular communication by transporting mRNA, siRNA, miRNA or proteins and peptides between cells. They internalize into recipient cells through various pathways, including micropinocytosis, receptor-mediated endocytosis, or lipid raft-mediated endocytosis.14 Although the natural process for the uptake of EVs is not ligand-dependent, the arrow-tail cholesterol RNA-3WJ allows for displaying ligand onto the surface of EVs, and increasing its targeting efficiency to the corresponding receptor overexpressing cancer cells.
Cancer-targeting and gene silencing of RNA-displaying EVs
Specific cancer cell-targeting is an important prerequisite for applying nano-vesicles to cancer therapy. To generate cancer cell-targeting EVs, approaches to express cancer cell-specific ligands on EVs have been explored. One way to increase the specificity of EVs to target cells is to overexpress peptide ligands fused to EV membrane proteins.27 Neuron acetylcholine receptor specific peptide RVG has been fused to EV membrane protein Lamp2b to be overexpressed on dendritic cells.27 GE11 peptide, which is a ligand to EGFR (Epidermal Growth Factor Receptor), was fused to the transmembrane domain of the platelet-derived growth factor receptor to be overexpressed on EV donor HEK293T cells.28 RGD peptide was fused to EV protein Lamp2b; thus, the EVs can deliver the chemical drug doxorubicin specifically to tumor cells.29 One problem in using fusion peptide for targeted exosomal delivery is that the displayed peptide can be degraded during EV biogenesis.30 We explored a method of displaying ligands onto the EVs surface post-biogenesis to enhance its specificity.
The targeting, delivery and gene silencing efficiency of the PSMA aptamer displaying EVs were examined in PSMA-positive LNCaP prostate cancer cells. To confer RNase resistance, 2′-F modifications were applied to the RNA nanoparticles placed on the surface of EVs,1 while the thermodynamic stability of pRNA-3WJ provided a rigid structure to ensure the correct folding of RNA aptamers.1, 31 PSMA aptamer-displaying EVs showed enhanced binding and apparent uptake to PSMA(+) LNCaP cells compared to EVs without PSMA aptamer by flow cytometry and confocal microscopy analysis, but not to the PC-3 cells, which is a low PSMA receptor expressing cell line (Fig. 3a). Upon incubation with LNCaP cells, PSMAapt/EV/siSurvivin was able to knock down the survivin expression at the mRNA level as demonstrated by real-time PCR (37.73 ± 11.59%, p<0.05) (Fig. 3b) and protein level as shown by Western Blot (62.89 ± 8.5 %, p<0.05) (Fig. S3). Cell viability by MTT assays indicated that the viability of LNCaP cells were decreased as a result of survivin siRNA delivery (70.98 ± 6.46 %, p<0.05) (Fig. 3c).
Figure 3. Specific binding and siRNA delivery to cells in vitro using PSMA aptamer-displaying EVs.

(a) Flow cytometry (left) and confocal images (right) showing the binding of PSMA RNA aptamer-displaying EVs to PSMA-receptor positive and negative cells. Nucleus (Blue), cytoskeleton (Green), and RNA (Red) in confocal images. (b) RT-PCR assay for PSMA aptamer-mediated delivery of survivin siRNA by EVs to PSMA(+) prostate cancer cells. Statistics: n=4; experiment was run in four biological replicates and two to four technical repeats with an ANOVA analysis; holm adjusted p = 0.0120, 0.0067 comparing PSMAapt/EV/siSurvivin to PSMAapt/EV/siScramble and 3WJ/EV/siSurvivin, respectively. (c) MTT assay showing reduced cellular proliferation. n=3, p = 0.003, 0.031 comparing PSMAapt/EV/siSurvivin to PSMAapt/EV/siScramble and 3WJ/EV/siSurvivin respectively. *p<0.05, **p<0.01.
The ligand displaying EVs target tumors
The tumor targeting and biodistribution properties of ligand-displaying EVs were evaluated. FA-3WJ/EVs were systemically administered via the tail vein into KB subcutaneous xenograft mice model. 3WJ/EVs and PBS treated mice were tested as a control. Ex vivo images of healthy organs and tumors taken from mice after 8 hrs showed that the FA-3WJ/EVs mainly accumulated in tumors, with low accumulation in vital organs in comparison with PBS control mice, and with more accumulation in tumors in comparison with 3WJ/EVs control mice (Fig. 4a). Normal EVs without surface modification usually showed accumulation in liver after systemic delivery.28 Both RNA and cell membranes are negatively charged. The electrostatic repulsion effect has been shown to play a role in reducing the accumulation of RNA nanoparticles in healthy organs.19, 22, 32 We hypothesize that displaying targeting RNAs on the EVs surface reduces their accumulation in normal organs, and the ideal nano-scale size of RNA displaying EVs facilitates tumor targeting via Enhance Permeability and Retention (EPR) effects, thereby avoiding toxicity and side effects.
Figure 4. Animal trials using ligands displaying EV for tumor inhibition.
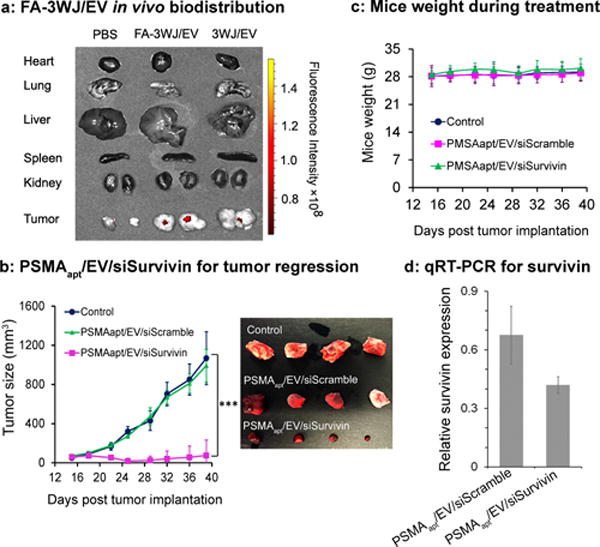
(a) Organ images showing specific tumor targeting 8 hrs after systemic injection of folate displaying EVs to mice with subcutaneous KB cell xenografts. n = 2, two independent experiments. (b) Intravenous treatment of nude mice bearing LNCaP-LN3 subcutaneous xenografts with PSMAapt/EV/siSurvivin or PSMAapt/EV/siScramble (both with 0.6 mg/kg, siRNA/mice body weight), and PBS, injected twice per week for three weeks. n=10 biological replicates, 2 independent experiments, and statistics were calculated using a two-sided t-test expressed as averages and with standard deviation. p = 0.347, 0.6–2, 1.5e–6, 8.2e–8, 2.1e–7, 1.0e–7, 1.9e–7, 1.8e–6 for days 15, 18, 22, 25, 29, 32, 36, and 39 respectively for PSMAapt/EV/siSurvivin compared to control. (c) Body weight of mice during the time course of EVs treatment. (d) RT-PCR showing the trend of knockdown survivin mRNA expression in prostate tumors after EV treatment.
Inhibition of tumor growth by ligand-3WJ-displaying EVs
The PSMA aptamer displaying EVs completely inhibits prostate cancer growth in mice. The therapeutic effect of PSMA aptamer-displaying EVs for prostate cancer treatment was evaluated using LNCaP-LN3 tumor xenografts.33, 34 Treatment with PSMAapt/EV/siSurvivin (1 dose every 3 days; total 6 doses) completely suppressed in vivo tumor growth, compared to control groups (Fig. 4b). EVs are biocompatible and well tolerated in vivo, we did not observe any significant toxicity as indicated by body weights of the mice, assessed over 40 days post-treatment (Fig. 4c). Analyzing the survivin mRNA expression levels in the tumor by real time PCR using GAPDH as internal control showed a trend of knocking down survivin by PSMAapt/EV/siSurvivin (Fig. 4d). Taken together, PSMA aptamer displaying EVs is a promising vector for delivering survivin siRNA in vivo and systemic injection of PSMAapt/EV/siSurvivin might achieve desired therapeutic efficacy.
The in vivo cancer growth inhibition effect was more pronounced than in vitro MTT assays in prostate cancer studies. The displaying of PSMA aptamer on the surface of EVs slightly enhanced its targeting to PSMA receptor overexpressing cancer cells in vitro, while the negatively charged RNA on EV surface might have minimized its nonspecific distribution to healthy cells as seen in the FA-3WJ/EVs biodistribution test. The EPR effect could also promote the homing of nanoscale EVs into tumors in vivo; although the biodistribution presented in Fig. 4a may not apply to the functional evaluation presented in Fig. 4b. All these results suggest that RNA aptamer displaying EVs are suitable for in vivo applications.
The EGFR aptamer displaying EVs inhibited breast cancer growth in mice. Overexpression of EGFR in breast cancer cells is associated with high proliferation, and risk of relapse in patients receiving treatment.35 We constructed pRNA-3WJ nanoparticles harboring EGFR aptamer (Fig. S4a) for display on EV surface and loaded the EVs with survivin siRNA. The resulting EGFRapt/EV/siSurvivin particles were administered via tail vein into the MDA-MB-468 orthotopic xenograft tumor bearing mice. 3WJ/EV/siSurvivin (without targeting ligand) and PBS treated mice served as controls. The analysis was completed with three mice per group. Ex vivo images taken after 8 hrs showed that the EGFRapt/EV/siSurvivin accumulated more in tumors than the control groups (Fig. 5a), indicating that displaying EGFR aptamer on the surface of EVs greatly enhanced its tumor targeting capabilities in vivo. Treatment with EGFRapt/EV/siSurvivin at a dose of 0.5 mg siRNA/kg of mice body weight (6 doses weekly) significantly suppressed in vivo tumor growth as monitored by tumor volume, compared to controls (Fig. 5b). The specific knockdown of survivin was validated from three representative tumors from each group by both Western blot (Fig. 5c) and quantitative real-time PCR (Fig. 5d), where GAPDH was used as an internal normalization control. The results indicate that successful delivery of survivin siRNA to breast tumor cells inhibited survivin expression at both protein and mRNA levels.
Figure 5. EGFR aptamer displaying EVs can deliver survivin siRNA to breast cancer orthotopic xenograft mouse model.

(a) EGFR aptamer displaying EVs showed enhanced targeting effect to breast tumor in orthotopic xenograft mice models. (b) Intravenous treatment of nude mice bearing breast cancer orthotopic xenografts with EGFRapt/EV/siSurvivin and controls (n=5). After 6 weeks, EGFRapt/EV/siSurvivin treated group had significantly smaller tumor size than other controls. p = 0.008 comparing EGFRapt/EV/siSurvivin to EGFRapt/EV/siScramble. (c) Analysis of the protein expression in tumor extracts showed that EGFRapt/EV/siSurvivin treatment significantly reduced the expression of Survivin. p=0.0004 comparing EGFRapt/EV/siSurvivin to EGFRapt/EV/siScramble. (d) Quantitative real-time PCR on extracted RNA from tumors showed the reduction of Survivin mRNA in the EGFRapt/EV/siSurvivin treated mice compared to controls. p=0.024 comparing EGFRapt/EV/siSurvivin to EGFRapt/EV/siScramble. Error bars indicate s.e.m. * p < 0.05, ** p < 0.01, *** p<0.001.
Folate displaying EVs inhibited colorectal cancer growth in mice. Survivin gene, an anti-apoptotic protein, is upregulated in most colorectal cancers, as tested by immunohistochemistry (IHC) imaging of tumor tissues from 9 colorectal cancer patients (Fig. S5). Utilizing a similar strategy, we constructed pRNA-3WJ nanoparticles harboring folate (Fig. S4b) for display on EV surface and loaded the EVs with survivin siRNA. The functionalized EVs were then evaluated in a clinically relevant patient derived CRC xenograft (PDX-CRC) mouse model. Treatment with FA/EV/siSurvivin at a dose of 0.5 mg siRNA/kg of mice body weight (6 doses weekly) significantly suppressed in vivo tumor growth as measured by tumor volume and tumor weight, compared to control group (Fig. 6a–b). The data suggests that folate displaying EVs can be used as a vector for delivering siRNA for colorectal cancer treatment.
Figure 6. Folate displaying EVs can deliver survivin siRNA to patient derived colorectal cancer xenograft (PDX-CRC) mouse model.

(a) Intravenous treatment of nude mice bearing PDX-CRC xenografts with FA/EV/siSurvivin and controls (n=4). After 6 weeks, FA/EV/siSurvivin treated group had significantly smaller tumor size, p = 0.0098 and 0.0387 comparing FA/EV/siSurvivin to FA/EV/siScramble at week 4 and week 5 respectively. (b) Lower tumor weight than controls. p = 0.0024 comparing FA/EV/siSurvivin to FA/EV/siScramble. Error bars indicate s.e.m. * p < 0.05, ** p < 0.01.
The application of RNA interference technology, such as siRNA, to knockdown gene expression has been of great interest.36 The nanometer-scale EVs37–40 can deliver biomolecules into cells by direct fusion with the cell membrane through tetraspanin domains, or back-fusion with endosomal compartment membranes for endosome escape. Therapeutic payloads, such as siRNA, can fully function after delivery to cells by EVs.37–40 However, EVs lack selectivity and can also randomly fuse to healthy cells. To generate specific cell-targeting EVs, approaches by in vivo expression of cell specific peptide ligands on the surface of EVs have been explored.27, 28 However, in vivo expression of protein ligands is limited to the availability of ligands in their producing cell types.37, 40,41 It would be desirable for in vivo cancer cell targeting using in vitro surface display technology to display nucleic acid-based or chemical targeting ligands on EVs.
This article reports the in vitro application of RNA nanotechnology42 to reprogram natural EVs for specific delivery of siRNA to cancer models in vitro and in animal models (Fig. 1a–c). Taking advantage of the thermodynamically stable properties of pRNA-3WJ,1, 31, 43 multifunctional RNA nanoparticles harboring membrane-anchoring lipid domain, imaging modules and targeting modules were generated. The arrow-shaped pRNA-3WJ offered the opportunity to control either partial loading of RNA into EVs or decoration of ligands on the surface of EVs. With cholesterol placed on the arrow-tail of the 3WJ, the RNA-ligand was prevented from trafficking into EVs, ensuring oriented surface display of targeting modules for cancer receptor binding. This was explicitly demonstrated by serum digestion and folate competition assays (Fig. 2f), as well as by enhanced binding to LNCaP cells after PSMA aptamer display (Fig. 3a) and during in vivo breast cancer by the EGFR aptamer display (Fig. 5a). Additionally, the placement of cholesterol on the arrow-head allowed for partial internalization of the RNA nanoparticle within the EVs (Fig. 2b, h). The incorporation of arrow-tail 3WJ-RNA nanoparticles to the surface of the EVs not only provided a targeting ligand to the EVs, but also added a negative charge on the EVs surface. Displaying negatively charged RNA nanoparticles on EV surface might assist in the reduction of non-specific binding of EV to normal cells. We have noticed previously that negatively charged RNA nanoparticles with a proper ligand tend to accumulate into tumors specifically after systemic administration.19, 22, 32 The cholesterol-TEG-modified RNA nanoparticles should preferentially anchor onto the raft-forming domains of the lipid bilayer of EVs,12 and further studies will be necessary to illustrate this process. EVs have the intrinsic ability to back-fuse with endosomal compartment membranes following receptor mediated endocytosis.37–39 Our in vitro decoration approach preserved the favorable endogenous composition of EVs as delivery vectors, thus eliminating the need to build artificial endosome-escape strategies into the EV vectors compared to using other synthetic nanovectors for siRNA delivery.44, 45
CONCLUSION
This study demonstrates the effective reprogramming of native EVs using RNA nanotechnology. RNA nanoparticles orientation was used to controls siRNA and miRNA loading or surface display on EVs for efficient cell targeting, siRNA and miRNA delivery and cancer regression. The reprogrammed EVs displayed robust physiochemical properties, enhanced cancer cell specific targeting, and efficient intracellular release of siRNA to suppress tumor growth in three animal models.
Supplementary Material
Acknowledgments
We thank Huang-Ge Zhang for his valuable communication during the investigation of the exosome project; Jianying Zhang, Heidi Weiss, Di Wu, and Di Gao for assistance with statistical analysis. The research was supported mainly by NIH grants UH3TR000875 and U01CA207946 (P. Guo), and partially by R01CA186100 (B. Guo), R35CA197706 (C.M.C.), P30CA177558 and R01CA195573 (B.M.E.).
Footnotes
CONFLICT OF INTEREST
P. Guo’s Sylvan G. Frank Endowed Chair position in Pharmaceutics and Drug Delivery is funded by the CM Chen Foundation, and he is a consultant of Oxford Nanopore, Nanobio Delivery Pharmaceutical Co., Ltd, and the cofounder of P&Z Biological Technology LLC. Pi now works for Nanobio Delivery Pharmaceutical Co., Ltd, S. Wang and F. Haque now work for P&Z Biological Technology LLC.
AUTHOR CONTRIBUTIONS
P. Guo generated the original arrow-head and arrow-tail idea in using the 3WJ structure orientation to control cell entry or cell surface anchoring, respectively, and designed the Arrow-Head and Arrow-Tail technology. P. Guo and F. Pi conceived and designed the experiments. H. Li and S. Wang developed the method for RNA insertion to EV. F. Pi, H. Li, D. Binzel and Z. Li performed the experiments. M. Sun and B. Guo performed the prostate cancer mouse studies. T. Lee performed the breast cancer mouse studies. P. Rychahou performed the colorectal cancer mouse studies. P. Guo and F. Haque supervised the project. P. Guo, C. Croce and M. Evers provided the funding and resources. P. Guo, F. Pi, F. Haque, D. Binzel co-wrote the manuscript and all authors refined the manuscript.
ADDITIONAL INFORMATION
Supplementary information is available in the online version of the paper.
References
- 1.Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. Thermodynamically stable RNA three-way junctions for constructing multifuntional nanoparticles for delivery of therapeutics. Nature Nanotechnology. 2011;6:658–667. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang H, et al. Crystal Structure of 3WJ Core Revealing Divalent Ion-promoted Thermostability and Assembly of the Phi29 Hexameric Motor pRNA. RNA. 2013;19:1226–1237. doi: 10.1261/rna.037077.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo P, Erickson S, Anderson D. A small viral RNA is required for in vitro packaging of bacteriophage phi29 DNA. Science. 1987;236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 4.Guo P, Zhang C, Chen C, Trottier M, Garver K. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 5.Lamichhane TN, Raiker RS, Jay SM. Exogenous DNA Loading into Extracellular Vesicles via Electroporation is Size-Dependent and Enables Limited Gene Delivery. Mol Pharm. 2015;12:3650–3657. doi: 10.1021/acs.molpharmaceut.5b00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rak J. Cancer: Organ-seeking vesicles. Nature. 2015;527:312–314. doi: 10.1038/nature15642. [DOI] [PubMed] [Google Scholar]
- 7.Melo SA, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Witwer KW, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shelke GV, Lasser C, Gho YS, Lotvall J. Importance of exosome depletion protocols to eliminate functional and RNA-containing extracellular vesicles from fetal bovine serum. J Extracell Vesicles. 2014;3 doi: 10.3402/jev.v3.24783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006 doi: 10.1002/0471143030.cb0322s30. Chapter 3, Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 11.Kumar D, Gupta D, Shankar S, Srivastava RK. Biomolecular characterization of exosomes released from cancer stem cells: Possible implications for biomarker and treatment of cancer. Oncotarget. 2015;6:3280–3291. doi: 10.18632/oncotarget.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bunge A, et al. Lipid membranes carrying lipophilic cholesterol-based oligonucleotides–characterization and application on layer-by-layer coated particles. J Phys Chem B. 2009;113:16425–16434. doi: 10.1021/jp9067747. [DOI] [PubMed] [Google Scholar]
- 13.Pfeiffer I, Hook F. Bivalent cholesterol-based coupling of oligonucletides to lipid membrane assemblies. J Am Chem Soc. 2004;126:10224–10225. doi: 10.1021/ja048514b. [DOI] [PubMed] [Google Scholar]
- 14.Marcus ME, Leonard JN. FedExosomes: Engineering Therapeutic Biological Nanoparticles that Truly Deliver. Pharmaceuticals (Basel) 2013;6:659–680. doi: 10.3390/ph6050659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Dongen HM, Masoumi N, Witwer KW, Pegtel DM. Extracellular Vesicles Exploit Viral Entry Routes for Cargo Delivery. Microbiol Mol Biol Rev. 2016;80:369–386. doi: 10.1128/MMBR.00063-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker N, et al. Folate receptor expression in carcinomas and normal tissues determined by a quantitative radioligand binding assay. Anal Biochem. 2005;338:284–293. doi: 10.1016/j.ab.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 17.Dassie JP, et al. Targeted inhibition of prostate cancer metastases with an RNA aptamer to prostate-specific membrane antigen. Mol Ther. 2014;22:1910–1922. doi: 10.1038/mt.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rockey WM, et al. Rational truncation of an RNA aptamer to prostate-specific membrane antigen using computational structural modeling. Nucleic Acid Ther. 2011;21:299–314. doi: 10.1089/nat.2011.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Binzel D, et al. Specific Delivery of MiRNA for High Efficient Inhibition of Prostate Cancer by RNA Nanotechnology. Molecular Therapy. 2016;24:1267–1277. doi: 10.1038/mt.2016.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 21.Esposito CL, et al. A neutralizing RNA aptamer against EGFR causes selective apoptotic cell death. PLoS ONE. 2011;6:e24071. doi: 10.1371/journal.pone.0024071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shu D, et al. Systemic delivery of anti-miRNA for suppression of triple negative breast cancer utilizing RNA nanotechnology. ACS Nano. 2015;9:9731–9740. doi: 10.1021/acsnano.5b02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paduano F, et al. Silencing of survivin gene by small interfering RNAs produces supra-additive growth suppression in combination with 17-allylamino-17-demethoxygeldanamycin in human prostate cancer cells. Molecular Cancer Therapeutics. 2006;5:179–186. doi: 10.1158/1535-7163.MCT-05-0132. [DOI] [PubMed] [Google Scholar]
- 24.Khaled A, Guo S, Li F, Guo P. Controllable Self-Assembly of Nanoparticles for Specific Delivery of Multiple Therapeutic Molecules to Cancer Cells Using RNA Nanotechnology. Nano Letters. 2005;5:1797–1808. doi: 10.1021/nl051264s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui D, et al. Regression of gastric cancer by systemic injection of RNA nanoparticles carrying both ligand and siRNA. Scientific reports. 2015;5:10726. doi: 10.1038/srep10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee TJ, et al. RNA nanoparticles as a vector for targeted siRNA delivery into glioblastoma mouse model. Oncotarget. 2015;6:14766–14776. doi: 10.18632/oncotarget.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.varez-Erviti L, et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 28.Ohno S, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21:185–191. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian Y, et al. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014;35:2383–2390. doi: 10.1016/j.biomaterials.2013.11.083. [DOI] [PubMed] [Google Scholar]
- 30.Hung ME, Leonard JN. Stabilization of exosome-targeting peptides via engineered glycosylation. J Biol Chem. 2015;290:8166–8172. doi: 10.1074/jbc.M114.621383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Binzel DW, Khisamutdinov EF, Guo P. Entropy-driven one-step formation of Phi29 pRNA 3WJ from three RNA fragments. Biochemistry. 2014;53:2221–2231. doi: 10.1021/bi4017022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haque F, et al. Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers. Nano Today. 2012;7:245–257. doi: 10.1016/j.nantod.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Tian Z, Rizvi SM, Bander NH, Allen BJ. In vitro and preclinical targeted alpha therapy of human prostate cancer with Bi-213 labeled J591 antibody against the prostate specific membrane antigen. Prostate Cancer Prostatic Dis. 2002;5:36–46. doi: 10.1038/sj.pcan.4500543. [DOI] [PubMed] [Google Scholar]
- 34.Pettaway CA, et al. Selection of highly metastatic variants of different human prostatic carcinomas using orthotopic implantation in nude mice. Clin Cancer Res. 1996;2:1627–1636. [PubMed] [Google Scholar]
- 35.Rimawi MF, et al. Epidermal growth factor receptor expression in breast cancer association with biologic phenotype and clinical outcomes. Cancer. 2010;116:1234–1242. doi: 10.1002/cncr.24816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK. RNA interference in the clinic: challenges and future directions. Nat Rev Cancer. 2011;11:59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.EL-Andaloussi S, Mager I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 38.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 39.El-Andaloussi S, Lakhal S, Mager I, Wood MJ. Exosomes for targeted siRNA delivery across biological barriers. Adv Drug Deliv Rev. 2013;65:391–397. doi: 10.1016/j.addr.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 40.van Dommelen SM, et al. Microvesicles and exosomes: opportunities for cell-derived membrane vesicles in drug delivery. J Control Release. 2012;161:635–644. doi: 10.1016/j.jconrel.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 41.Wiklander OP, et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J Extracell Vesicles. 2015;4:26316. doi: 10.3402/jev.v4.26316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo P. The emerging field of RNA nanotechnology. Nature Nanotechnology. 2010;5:833–842. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shu D, Khisamutdinov E, Zhang L, Guo P. Programmable folding of fusion RNA complex driven by the 3WJ motif of phi29 motor pRNA. Nucleic Acids Res. 2013;42:e10. doi: 10.1093/nar/gkt885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Varkouhi AK, Scholte M, Storm G, Haisma HJ. Endosomal escape pathways for delivery of biologicals. J Control Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 45.Kilchrist KV, Evans BC, Brophy CM, Duvall CL. Mechanism of Enhanced Cellular Uptake and Cytosolic Retention of MK2 Inhibitory Peptide Nano-polyplexes. Cell Mol Bioeng. 2016;9:368–381. doi: 10.1007/s12195-016-0446-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


