Abstract
During embryogenesis, cells acquire distinct fates by transitioning through transcriptional states. To uncover these transcriptional trajectories during zebrafish embryogenesis, we sequenced 38,731 cells and developed URD, a simulated diffusion-based computational reconstruction method. URD identified the trajectories of 25 cell types through early somitogenesis, gene expression along them, and their spatial origin in the blastula. Analysis of Nodal signaling mutants revealed that their transcriptomes were canalized into a subset of wild-type transcriptional trajectories. Some wild-type developmental branchpoints contained cells expressing genes characteristic of multiple fates. These cells appeared to trans-specify from one fate to another. These findings reconstruct the transcriptional trajectories of a vertebrate embryo, highlight the concurrent canalization and plasticity of embryonic specification, and provide a framework to reconstruct complex developmental trees from single-cell transcriptomes.
One Sentence Summary:
The first specification tree of vertebrate embryogenesis constructed by combining scRNA-seq with a new computational technique, URD.
During embryogenesis, a single totipotent cell gives rise to numerous cell types with distinct functions, morphologies, and spatial positions. Since this process is primarily controlled through transcriptional regulation, the identification of the transcriptional states underlying cell fate acquisition is paramount to understanding and manipulating development. Previous studies have presented different views of cell fate specification. For example, artificially altering transcription factor expression (e.g. in reprogramming) has revealed remarkable plasticity of cellular fates (1-3). Conversely, classic embryological studies have indicated that cells are canalized to adopt perduring fates separated by epigenetic barriers. Technological limitations necessitated that traditional embryological studies focus on specific fate decisions with selected marker genes, but the advent of single-cell RNA sequencing (scRNA-seq) raises the possibility of fully defining the transcriptomic states of embryonic cells as they acquire their fates (4-8). However, the large number of transcriptional states and branchpoints, as well as the asynchrony in developmental processes, pose major challenges to the comprehensive identification of cell types and the computational reconstruction of their developmental trajectories. Pioneering computational approaches to uncover developmental trajectories (5-7, 9-11) were either designed to address stationary or steady-state processes or accommodate only small numbers of branchpoints, and thus are insufficient for addressing the complex branching structure of time-series developmental data. Here, we address these challenges by combining large-scale single-cell transcriptomics during zebrafish embryogenesis with the development of a novel simulated diffusion-based computational approach to reconstruct developmental trajectories, called URD (named after the Norse mythological figure who nurtures the world tree and decides all fates).
High-throughput scRNA-seq from Zebrafish Embryos
We profiled 38,731 cells from 694 embryos across 12 closely spaced stages of early zebrafish development using Drop-seq, a massively parallel scRNA-seq method (12). Samples spanned from high blastula stage (3.3 hours post-fertilization, just after transcription from the zygotic genome begins), when most cells are pluripotent, to 6-somite stage (12 hours post-fertilization, shortly after the completion of gastrulation), when many cells have differentiated into specific cell types (Fig. 1A, table S1). In a t-distributed Stochastic Neighbor Embedding (tSNE) plot (13) of the entire dataset based on transcriptional similarity, it is evident that developmental time was a strong source of variation in the data, but the underlying developmental trajectories were not readily apparent (Fig. 1B). Consistent with the understanding that cell types become more transcriptionally divergent over time, cells from early stages formed large continuums in the tSNE plot, while more discrete clusters emerged at later stages (Fig. 1C).
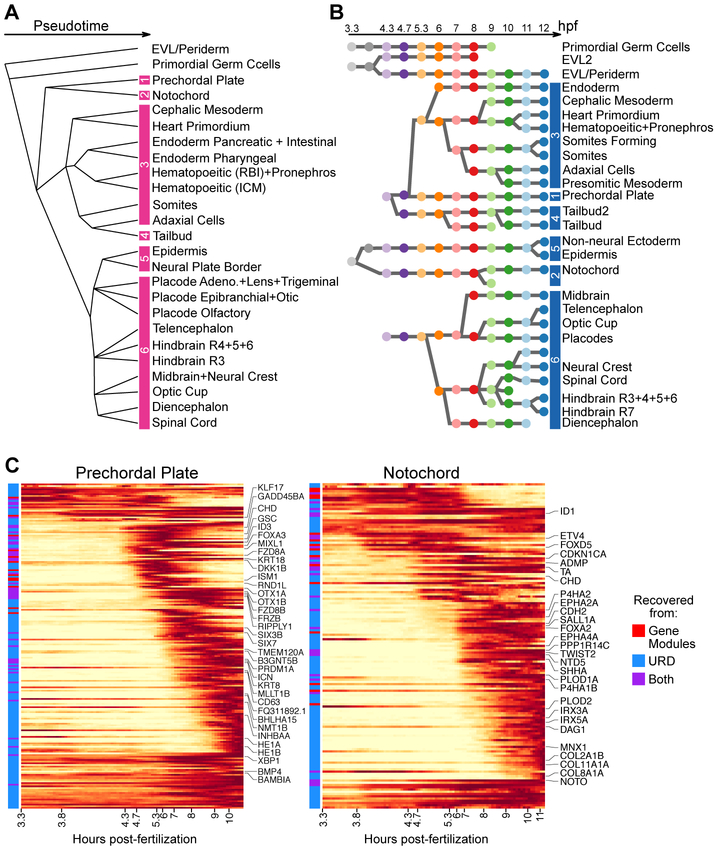
Fig 1. Generation of a developmental specification tree for early zebrafish embryogenesis using URD.
(A) Single-cell transcriptomes were collected from zebrafish embryos at 12 developmental stages (colored dots) spanning 3.3–12 hours post-fertilization (hpf). (B) tSNE plot of the entire data, colored by stage (as in Fig. 1A). Developmental time is a strong source of variation, and the underlying developmental trajectories are not immediately apparent. (C) tSNE plot of data from two stages (top: 50% epiboly, bottom: 6-somite). Clusters are more discrete at the later stage. (D) URD’s approach for finding developmental trajectories: (1) Transition probabilities are computed from the distances between transcriptomes and used to connect cells with similar gene expression. (2) From a user-defined ‘root’ (e.g. cells of the earliest timepoint), pseudotime is calculated as the average number of transitions required to reach each cell from the root. (3) Trajectories from user-defined ‘tips’ (e.g. cell clusters in the final timepoint) back to the root are identified by simulated random walks that are biased towards transitioning to cells younger or equal in pseudotime. (4) To recover an underlying branching tree structure, trajectories are joined agglomeratively at the point where they contain cells that are reached from multiple tips. (5) The data is visualized using a force-directed layout based on cells’ visitation frequency by the random walks from each tip. (E) Force-directed layout of early zebrafish embryogenesis, optimized for 2D visualization (fig. S2, Methods, movie S1), colored by stage (as in Fig. 1A) with terminal populations labeled. Abbreviations: EVL (Enveloping Layer), P (Placode), Adeno. (Adenohypophyseal), Trig. (Trigeminal), Epi. (Epibranchial), Panc.+Int. (Pancreatic + Intestinal), RBI (Rostral Blood Island), ICM (Intermediate Cell Mass). (F–I) Cell populations downstream of early and intermediate branchpoints recovered by URD.
URD Reconstructs Complex Branching Developmental Trajectories
Acquisition of many single-cell embryonic transcriptomes with high temporal resolution created the possibility of reconstructing developmental trajectories through similarity in gene expression profiles. Such an approach would allow the investigation of the gene expression dynamics and the timing of molecular specification — when progenitor populations become transcriptionally distinct from each other and begin to express the regulators that will drive their future fates. Therefore, we developed URD, a new approach to uncover complex developmental trajectories as a branching tree. URD extends diffusion maps (originally presented for single-cell differentiation analysis in pioneering work by Haghverdi et al. and their R package, destiny (9, 10)) through several advances: it introduces new ways to order cells in pseudotime, finds developmental trajectories, discovers an underlying branching tree that abstracts specification, and visualizes the data (Fig. 1D, Methods).
In general, URD operates by “simulating diffusion”, using discrete random walks and graph searches to approximate the continuous process of diffusion (Methods). Briefly, URD constructs a k-nearest-neighbor graph between transcriptomes in gene expression space; graph edges are assigned transition probabilities that are used as weights in later simulations and describe the chance a random walk would move along each edge (9, 10) (Fig. 1D, 1). The user identifies the root(s) (starting points) and tips (end points) of the developmental process. Cells are next assigned a pseudotime—an ordering that should reflect their developmental progress rather than absolute time—in order to compensate for developmental asynchrony. URD calculates pseudotime by simulating diffusion from the root to determine each cell’s distance from the root (as the average number of diffusive transitions needed to reach it across several simulations) (Fig. 1D, 2). Next, the developmental trajectory (the path in gene expression) between each tip and the root is determined by identifying which cells are visited by simulated biased random walks initiated in that tip; the walks are biased to only transition to cells of equal or earlier pseudotime, so that when they reach developmental branchpoints they proceed toward the root and do not explore other cell types (Fig. 1D, 3). Then, URD reconstructs a branching tree structure by joining pairs of trajectories where they pass through the same cells (Fig. 1D, 4; e.g. black and purple edges). Finally, the data is visualized using a force-directed layout based on cells’ visitation frequency by the random walks from each tip (14) (Fig. 1D, 5). The developmental trajectories identified by URD are akin to cell lineages, but they differ from classical definitions of cell lineage, because they are reconstructed from observed gene expression, and do not measure mother-daughter relationships between cells. Importantly, URD does not require any prior knowledge of the developmental trajectories it seeks to find (i.e. the number of branchpoints, or any definition of intermediate states).
Reconstructed Developmental Tree Recapitulates Molecular Specification During Zebrafish Embryogenesis
Application of URD to the early zebrafish embryogenesis scRNA-seq data generated a tree whose branches reflected embryonic specification trajectories. To define the final cell populations (i.e. the tips of the tree), we clustered cells from the final stage of our timecourse and determined cluster identity through the expression of known marker genes (fig. S1). The recovered tree followed the specification of 25 final cell populations across 16 branchpoints (Fig. 1E, fig. S2, movie S1). The reconstructed tree substantially recapitulated the developmental trajectories expected from classical embryological studies (15-17). For example, the primordial germ cells (PGCs), the enveloping layer cells (EVL), and the deep layer blastomeres already formed separate trajectories by high stage (Fig. 1F). Unexpectedly, the first branchpoint within the blastoderm not only separated the ectoderm from the mesendoderm, but also divided the axial mesoderm from the remainder of the mesendoderm (Fig. 1G). Later branching events were also recovered, such as the separation of paraxial, lateral, and intermediate mesoderm (Fig. 1H), the separation of the non-neural and neural ectoderm (Fig. 1I), and the eventual branching of the non-neural ectoderm into epidermis, neural plate border, and multiple preplacodal ectoderm trajectories (Fig. 1E, Fig. 2 ‘Trajectory’, fig. S3). Displaying the expression of classic marker genes on the developmental tree highlighted expected trajectories and confirmed their annotation (Fig. 2 ‘Gene Expression’, fig. S3). For example, consistent with its known expression, the notochord marker gene, noto, was restricted primarily to two trajectories (the entire notochord trajectory and the later stages of the tailbud trajectory) and confirmed that the branchpoint between the notochord and prechordal plate was correctly placed (18). These results show that URD can reconstruct the highly complex branching trajectories of early zebrafish embryogenesis solely based on large-scale scRNA-seq data.
Fig 2. Developmental trajectories, genes, and connected gene modules overlaid on the force-directed layout.
From top to bottom: (1) the trajectories identified by URD from the root to a given population (or group of populations), (2) gene expression of a classical marker of that population, and (3) expression of a 6-somite gene module active in the population and its connected modules from earlier stages. (The remainder are presented in fig. S3).
Connected Gene Modules Support Reconstructed Developmental Trajectories
To complement the specification tree and in order to find groups of genes that are coexpressed within cell populations, we applied non-negative matrix factorization (NMF) to the single-cell transcriptomes (19). This approach produced modules of co-varying genes and described cells in terms of module expression (which is more robust than individual gene expression measurements) (4, 20). Modules were annotated post hoc according to their highly ranked classic marker genes, and similar modules from adjacent stages were linked to each other according to the overlap of their most highly ranked genes. This approach created chains of connected gene modules that provided an alternative way to track developmental trajectories (fig. S4, table S2). For example, the prechordal plate chain of connected gene modules extended from 50% epiboly to 6-somite stage, during which the top ranking genes gradually changed from early to late markers for the prechordal plate (table S3).
The URD-generated developmental tree and the chains of connected gene modules provided two different ways to analyze the scRNA-seq data and define developmental trajectories. To determine how congruent these approaches are, we highlighted cells in the developmental tree based on their expression of connected gene modules. Notably, cells that express connected gene modules occupied specific URD-recovered developmental trajectories, further supporting the structure of the developmental tree reconstructed by URD (Fig 2 ‘Module Expression’, fig. S3).
Gene Cascades Reveal Expression Dynamics Along Developmental Trajectories
Gene expression and gene module analysis were combined to find candidate regulators and markers along each trajectory uncovered by URD. Genes and connected gene modules were associated with developmental trajectories by testing for their differential expression downstream of each branchpoint of URD’s recovered branching structure (Fig. 3A–B, Methods). Gene expression dynamics were then fit with an impulse model (21) to determine the onset and offset time of their expression, which was then used to order genes. As an example, sequential expression was observed for 197 genes enriched in the prechordal plate during its specification, including several well-known transcription factors or signaling molecules that confirm the validity of our approach (e.g. gsc, foxa3, klf17, frzb) (22-25) (Fig. 3C, fig. S5–S7). This cascade contained several regulatory factors (e.g. fzd8a, fzd8b, mllt1b, inhbaa) without described roles in the prechordal plate that would be candidates for reverse genetic screens. This cascade also contained over 40 genes that were not previously annotated as associated with the prechordal plate (fig. S6), and those tested by in situ hybridization were indeed expressed in the prechordal plate (fig. S7). Thus, combining URD and gene module analysis uncovered the transcriptional cascades that accompanied the development of progenitors into differentiated cells and highlighted both previously characterized and newly identified trajectory-enriched genes.
Fig 3. Association of developmental trajectories with temporal gene expression patterns.
(A) The underlying branching structure found by URD. Pink bars demarcate collections of cell types used in Fig. 4A. (B) The structure of connected gene modules. Each circular node represents a module and is colored by the developmental stage the module was computed from (as in Fig. 1A). Blue bars demarcate collection of modules downstream of each 50% epiboly (5.3 hpf) gene module used in Fig. 4B. (C) Gene expression cascades during specification of the prechordal plate and notochord. Expression is displayed as a moving-window average in pseudotime (along the x-axis), scaled to the maximum observed expression. Selected genes are labeled along the y-axis. Genes are annotated with whether they were identified as a differentially expressed gene, as a top ranking member of a differentially expressed connected gene module, or both. Cascades for all trajectories (with all genes labeled) are presented in Fig. S5.
Combining Developmental Trajectories With Spatial Analysis Infers Progenitor Locations
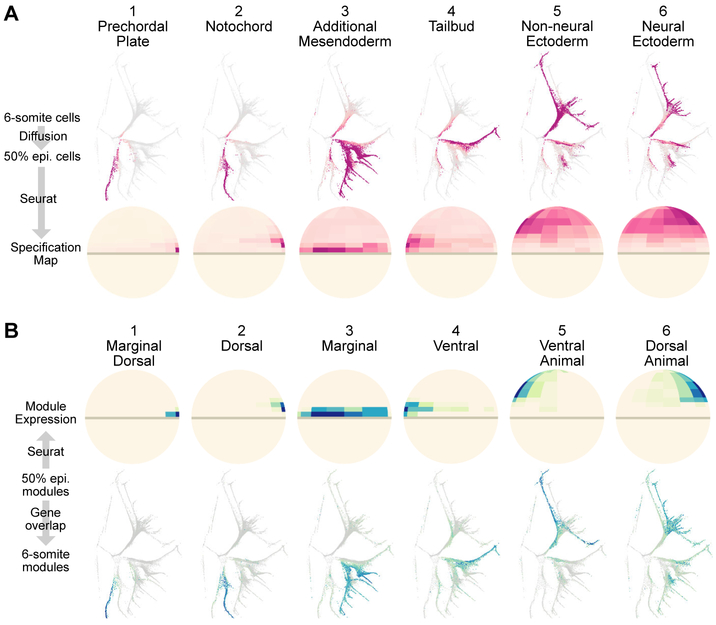
The URD-generated tree is a powerful way to visualize developmental trajectories, but it lacks spatial information. We therefore asked if the trajectories could be traced to their spatial origin at blastula stage (16, 17). First, a spatial map of the Drop-seq 50% epiboly transcriptomes was generated using Seurat, a method we previously developed to infer the spatial locations of single cell transcriptomes by comparing the genes expressed in each transcriptome to the spatial expression patterns of a few landmark genes obtained from RNA in situ hybridization (20). Second, Seurat’s spatial map was combined with either URD or connected gene module analysis (as parallel, independent approaches) to associate cell populations at 6-somite stage with the location of their ‘pseudoprogenitors’ at 50% epiboly. In one approach, we used URD’s simulated random walks from cell populations at 6-somite (Fig. 3A, pink bars) to infer their ‘pseudoprogenitor’ cells, and then plotted the spatial location of the 50% epiboly ‘pseudoprogenitors’ using Seurat (Fig. 4A). In the other approach, we plotted the spatial expression of each 50% epiboly gene module and identified its connected gene modules at later stages (Fig. 3B, blue bars on right); the cells that express these connected gene modules are highlighted on the developmental tree (Fig. 4B) and are the ‘pseudodescendents’ that arise from the spatial domain identified by the 50% epiboly gene module.
Fig 4. Molecular specification maps relate cell position at 50% epiboly to cell fate at 6-somite.
(A) Visitation by random walks from given tip(s) (as proportion of visitation from all tips), and the spatial location of visited 50% epiboly cells (ventral side to the left). The six tip groups are marked in Fig. 3A. (B) Spatial expression of 50% epiboly gene modules; expression of connected gene modules plotted on the force-directed layout highlight populations that will emerge from the 50% epiboly module’s expression domain. The six groups of connected gene modules are marked in Fig. 3B.
The results from the two approaches were highly concordant, and agreed with classic fate mapping experiments (16, 17). For instance, both approaches associated the dorsal margin of the 50% epiboly embryo with axial mesodermal fates (prechordal plate and notochord). Likewise, in both cases, the animal pole was associated with ectodermal fates, with the ventral animal side biased to non-neural fate and the dorsal animal side to neural fate. Overall, these results show that scRNA-seq data can be used not only to reconstruct specification trajectories and their associated gene cascades, but also to connect the earlier spatial position of progenitors to the later fate of their descendants.
Nodal Signaling Mutant Cells are Canalized into a Subset of Wild-Type Transcriptional States
Previous studies of mutant embryos have provided important insights into embryonic fate specification, but scRNA-seq raises the possibility of rapidly phenotyping mutants both genome-wide and at single-cell resolution. To test this idea, we profiled maternal-zygotic one-eyed pinhead mutants (MZoep), which lack the coreceptor for the mesendoderm inducer Nodal (26, 27). We first asked whether previous MZoep embryological results could be reconstructed simply from scRNA-seq data. Transcriptomes were generated with SMART-seq2 (28), with deeper mRNA coverage than Drop-seq (fig. S8), to collect 325 MZoep and 1,047 wild-type transcriptomes at 50% epiboly, when Nodal signaling is normally active at the blastoderm margin. Using Seurat, we inferred the spatial origin of both wild-type and MZoep transcriptomes based on a wild-type landmark map (Fig. 5A). As predicted, no MZoep cells mapped to the margin of a wild-type embryo, where mesendodermal progenitors arise. Applying NMF revealed that in MZoep mutants, expression of the marginal dorsal, dorsal, and marginal gene modules is greatly reduced or absent, but no mutant-specific modules were found (Fig. 5B). NMF recovered the same 50% epiboly gene modules from SMART-seq and Drop-seq data (Fig. 5B, table S4), which allowed us to use the chains of connected Drop-seq gene modules to predict the MZoep mutant phenotype at later stages. Namely, our approach successfully predicted the loss of paraxial mesoderm, ventrolateral mesoderm, axial mesoderm, and endoderm (Fig. 5C), as well as the continued presence of the tailbud, as found in previous embryological studies (26). Analysis of additional single cell transcriptomes from wild-type and MZoep mutants at 6-somite stage largely verified our predictions (fig. S9, table S5). Together, these results show that combining modest scale scRNA-seq in mutants with the large-scale developmental reference tree constructed for wild type can rapidly provide phenotypic insights: in the absence of Nodal signaling, marginal blastomeres that would normally become mesendodermal progenitors instead become ectodermal and tail progenitors, resulting in the absence of mesendodermal cell types and an altered fate map, as shown in previous cell tracing experiments (27).
Fig 5. Characterization of Nodal signaling mutant by scRNA-seq and developmental specification tree.
(A) Spatial assignment of wild-type and MZoep transcriptomes using a wild-type landmark map indicates an absence of wild-type marginal fates in MZoep (ventral, left). 311 wild-type transcriptomes are shown at random (to match MZoep cell number). (B) Top: wild-type expression domain of spatially restricted gene modules identified in Smart-seq data (ventral to left). Bottom: Violin plot of the maximum-scaled gene module levels in wild-type and MZoep mutant cells. The marginal dorsal, dorsal, and marginal gene modules are absent or strongly reduced in MZoep. (C) Expression of gene modules connected to those missing in MZoep (marginal dorsal, dorsal, and marginal, red) and connected to those remaining in MZoep (blue). (D) Hierarchical clustering of wild-type and MZoep mutant transcriptomes, based on the scaled expression of gene modules. Number of clusters is determined by the Davies-Bouldin index. Genotype is indicated beneath the heatmap (wild type, green; MZoep, red). Clusters 3 and 8 contain only wild-type cells. All other clusters contain a mixture of wild-type and MZoep cells. This clustering analysis was sufficiently sensitive to detect computationally simulated altered states (Fig. S10).
We next asked if the mutant scRNA-seq dataset could provide novel insights into cell identity in the absence of Nodal signaling. Morphological analysis and fate mapping of MZoep mutants has found that all cells appear to adopt wild-type fates (26, 27). However, intricate interactions exist between the several signaling pathways active during early development, and the elimination of Nodal signaling changes levels and domains of other developmental signals in the embryo. Thus, MZoep mutant cells could potentially perceive novel signaling input combinations, which may result in novel gene expression states. We therefore wondered whether mutant cells expressed novel combinations of gene modules under this altered signaling landscape, or were transcriptionally equivalent to a subset of wild-type states. Coclustering of wild-type and MZoep transcriptomes by gene module expression revealed that while some mesendormal cell types were absent in MZoep at 50% epiboly, the remaining cells clustered with wild-type states (Fig. 5D, fig. S10). This result indicates that even on the whole transcriptomic and single-cell level, mutant cells are canalized into a subset of wild-type fates after the loss of an essential signaling pathway.
Hybrid Gene Expression States Reveal Developmental Plasticity
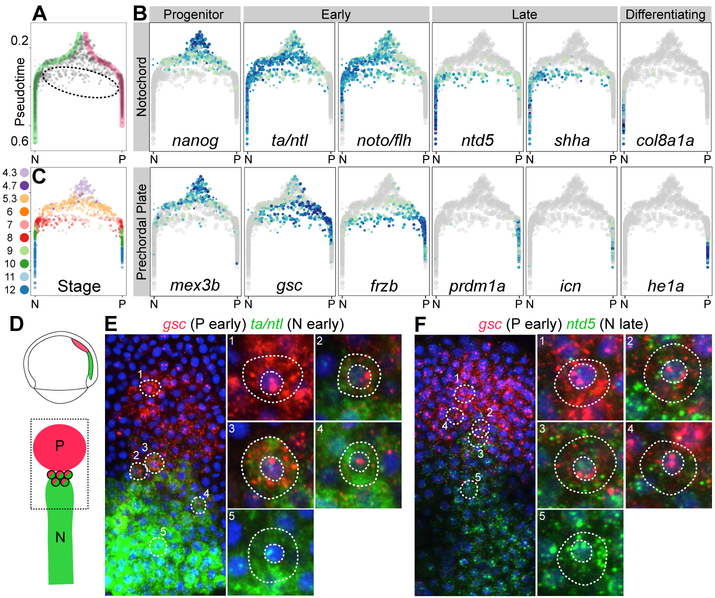
Inspection of the developmental tree revealed that most cells fell along tight trajectories but some cells were located in intermediate zones between branches. This observation seemed at odds with the view that embryonic cells traverse a developmental trajectory until a branchpoint funnels them cleanly and irreversibly into one of multiple downstream branches. For instance, at the axial mesoderm branchpoint, most cells fell along the classic bifurcation from progenitor into notochord or prechordal plate fates (15, 29), with waves of gene expression corresponding to their specification and differentiation status (Fig. 6A, highlighted). However, ~5.4% were intermediate cells that expressed both notochord and prechordal plate markers (Fig. 6A–B). The intermediate cells and the completely bifurcated axial mesoderm cells with similar pseudotimes came from embryos at the same developmental stage (Fig. 6C); moreover, they no longer expressed genes characteristic of the progenitors (e.g. nanog, mex3b). These observations eliminated models where intermediate cells retained their progenitor state and delayed specification. Instead, we noticed that these cells expressed early markers of both programs (gsc, frzb, ta, noto) but later markers of only the notochord program (ntd5, shha). This observation raised two possible models: (1) Cells initially express both programs, then shut off the prechordal plate program, and produce only notochord markers later (“dual-specification” model); or (2) Cells specified as notochord first express early and late notochord markers, and then change their specification by shutting off notochord expression and initiating early prechordal plate marker expression (“trans-specification” model).
Fig 6. Hybrid state of cells in the axial mesoderm.
(A) Branchpoint plot, showing pseudotime (y-axis) and random walk visitation preference from the notochord (N, left) and prechordal plate (P, right) tips (x-axis), defined as the difference in visitation from the two tips divided by the sum of visitation from the two tips. Direct trajectories to notochord (green) and prechordal plate (pink) are highlighted, and intermediate cells are circled. (B) Gene expression of notochord markers (top row) and prechordal plate markers (bottom row) at the axial mesoderm branchpoint. Intermediate cells express early (ta/ntl, noto) and late (ntd5, shha) notochord markers, but only early prechordal plate markers (gsc, frzb). (C) Cells at the branchpoint, colored by developmental stage. Intermediate cells have the same developmental stage as fully bifurcated cells with similar pseudotimes. (D) Cartoon of the prechordal plate (P) and notochord (N) in the 75% epiboly embryo. (E–F) Double fluorescent in situ expression of the early prechordal plate marker gsc (red) and either the early notochord marker ta/ntl (E, green) or the late notochord marker ntd5 (F, green) at 75% epiboly (8 hpf). Most cells contain only prechordal plate marker mRNA (e.g. 1) or notochord marker mRNA (e.g. 5). Cells with both prechordal plate and notochord marker mRNA are observed at the boundary of the two tissues, with red nuclear transcription dots, indicating active transcription of gsc (e.g. 2–4) (fig. S11). Cells that contained both ntd5 and gsc mRNA were identified and scored for their nuclear transcription foci, which indicate active transcription (see Methods); 56% had 1 or more observable nuclear transcription dots, of which 80% showed only active gsc transcription, 7% showed both active gsc and active ntd5 transcription, and 13% showed active ntd5 transcription alone.
To distinguish between these two models, we investigated their properties and location using fluorescent RNA in situ hybridization. Indeed, some 75% epiboly cells located in the border region between the two tissues co-expressed an early prechordal plate marker (gsc) with either an early (ta/ntl) or late (ntd5) notochord marker (Fig. 6D–F, fig. S11). The existence of these intermediate cells in situ confirms they are not an artifact of scRNA-seq (e.g. cell doublets), and their defined spatial localization near the border of the two tissues suggests that they are not merely biological or technical noise. Instead, the location of co-expressing cells raises the possibility that the boundary between the notochord and prechordal plate territories is refined during gastrulation, after the two populations become transcriptionally distinct. Many of the co-expressing cells exhibited bright nuclear foci that denoted sites of active transcription of the probed genes. Strikingly, most of the cells with nuclear transcription foci for a single gene exhibited transcription of the early prechordal plate marker, gsc. In contrast, cells with active transcription of the notochord markers ta/ntl or ntd5 were rare (Fig. 6E–F). The active transcription of the prechordal plate program supports the “trans-specification” model—the intriguing possibility that some axial mesoderm cells move down the notochord specification path but then trans-specify into prechordal plate cells.
Discussion
We describe the first molecular specification tree of an early vertebrate embryo by exploring large-scale single-cell transcriptomic data with URD, a novel computational approach to reveal developmental trajectories in transcriptional space. Our study lays the foundation for multiple areas of future exploration.
First, URD is a powerful tool to reveal the transcriptional trajectories of complex developmental processes such as zebrafish embryogenesis. Combining URD with connected gene module analysis uncovered the transcriptional cascades underlying fate specification. Moreover, combining URD with Seurat enabled the anchoring of developmental trajectories to their spatial origins. We anticipate that similar augmentation of URD with information about lineage relationships (30, 31), chromatin dynamics (32), and signaling will further deepen insights into developmental processes. Additionally, some of URD’s limitations could be addressed by future enhancements. For instance, branching events driven by single genes (e.g. sox32 in the endoderm) (33, 34) are not captured by URD until additional transcriptional differences arise, which could potentially be improved with more aggressive, iterative branch calling. Also, improvements to the throughput and quality of scRNA-seq may drive improved performance from URD—more closely spaced timepoints could enable detection of rapidly changing fate decisions and could reveal bifurcations at branch sites currently thought to enter multiple trajectories; larger numbers of cells could enable reconstruction of rare, transcriptionally indistinct populations (e.g. the floor plate and hypochord); and improved sequencing depth could enable detection of important, but lowly expressed regulators that are not found in the current data (e.g. npas4l/cloche). In the future, URD could be used to analyze additional systems, such as in vitro differentiation, tissue regeneration, cancer and disease progression.
Second, the scRNA-seq data and developmental tree provide a rich resource for future studies of zebrafish embryogenesis. The presented data describe embryonic gene expression with unparalleled temporal and cellular resolution, and the reconstructed tree thus provides an atlas of the expression pattern and dynamics for nearly all genes. This allows inspection of gene co-expression with much greater ease than by multi-color in situ hybridization, reveals new markers for cell types of interest, and associates uncharacterized genes with particular cell types. Moreover, the data reveal the progressive nature of cell fate specification and suggest potentially redundant regulators of developmental decisions with overlapping spatial and temporal expression, which would be missed by forward genetic screens.
Third, this work begins to illustrate the trajectories, canalization, and potential plasticity of fate specification. With respect to trajectories, our analysis suggests that not all cell fate decisions are binary—multiple trajectories can arise simultaneously from a pool of equipotent progenitors. For example, at the molecular level, the earliest specification of blastomeres separates the axial mesoderm, non-axial mesendoderm, and ectoderm, rather than just separating the germ layers. This seemingly surprising conclusion is compatible with the observation that the axial mesoderm progenitors reside at the dorsal blastula margin, where maternal factors that specify the Mangold-Spemann organizer simultaneously affect the fate of those progenitors (15). Concerning canalization, the analysis of a Nodal signaling mutant reveals that, even on the whole transcriptomic level, mutant cells adopt a subset of wild-type transcriptomic states, but not new ones, demonstrating the extensive canalization during embryogenesis even after abnormal developmental signaling. With respect to plasticity, the identification of intermediate cells at the axial mesoderm branchpoint demonstrates hybrid developmental states. In situ hybridization experiments suggest that these cells trans-specify from one cell type (notochord) to another (prechordal plate), and their border zone spatial localization suggests refinement of the boundary between these two regions. These results support an alternative view of developmental fate choice to the common interpretation of Waddington’s model: downstream of some branchpoints, some cells still transition across the ‘ridges’ between different lineages even after they are canalized into well-defined transcriptional states. This suggests a developmental plasticity that has been observed after perturbation (1-3, 35), but has not been well established in normal developmental processes. We propose that, in a continuous morphogen gradient that resolves into discrete states, some cells will receive an amount of signal that is on the cusp between the two states; these cells would end up at the boundary between the two tissues. It is conceivable that continued signaling could provide a ‘push’ over a shallow ‘ridge’, such as continued Nodal signaling in the axial mesoderm (22) or continued retinoic acid signaling in the hindbrain (36, 37). Future studies combining lineage tracing and in vivo imaging of gene expression will be needed to directly observe such transitions.
Collectively, our approach provides a framework to reconstruct the specification trajectories of many developmental systems without the need for prior knowledge of gene expression patterns, fate maps, or lineage trajectories. The generation of developmental trees for different species will enable comparative studies, and developmental statistics generated from such comparisons will help reveal conserved pathways and species-specific idiosyncracies.
Supplementary Material
Acknowledgements:
We thank B. Raj and J. Gagnon for assistance collecting samples when two hands were insufficient, M. Rabani for assistance with the impulse model, M. Haesemeyer for the critical suggestions of directly simulating diffusion and of using visitation frequency as a method of dimensionality reduction, the Harvard Center for Biological Imaging and Bauer Core Facility for support, and members of the Schier and Regev labs for helpful discussions. We thank B. Raj, J. Gagnon, S. Pandey, N. Lord, M. Rabani, A. Carte, M. Haesemeyer, I. Whitney, and T. Montague for comments on the manuscript.
Funding: This research was supported by the NIH (A.F.S., J.A.F., A.R.), the Allen Discovery Center for Cell Lineage Tracing (A.F.S.), Jane Coffin Childs Memorial Fund (J.A.F.), Charles A. King Trust (J.A.F.), Howard Hughes Medical Institute (A.R.), and the Klarman Cell Observatory (A.R.).
Footnotes
Competing Interests: A.R. is an SAB member of ThermoFisher Scientific, Syros Pharmaceuticals, and Driver Group.
Data availability: The raw data reported in this paper are archived at NCBI GEO (Accession Number GSE106587) and in processed and interactively browseable forms in the Broad Single-Cell Portal (https://portals.broadinstitute.org/single_cell/study/single-cell-reconstruction-of-developmental-trajectories-during-zebrafish-embryogenesis). URD is available from GitHub (https://github.com/farrellja/URD).
References:
- 1.Davis RL, Weintraub H, Lassar AB, Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 51, 987–1000 (1987). [DOI] [PubMed] [Google Scholar]
- 2.Tapscott SJ et al. , MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 242, 405–411 (1988). [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Yamanaka S, Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell. 126, 663–676 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Wagner A, Regev A, Yosef N, Revealing the vectors of cellular identity with single-cell genomics. Nat Biotechnol. 34, 1145–1160 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannoodt R, Saelens W, Saeys Y, Computational methods for trajectory inference from single-cell transcriptomics. Eur. J. Immunol 46, 2496–2506 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Bendall SC et al. , Single-Cell Trajectory Detection Uncovers Progression and Regulatory Coordination in Human B Cell Development. Cell. 157, 714–725 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trapnell C et al. , The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jang S et al. , Dynamics of embryonic stem cell differentiation inferred from single-cell transcriptomics show a series of transitions through discrete cell states. eLife Sciences. 6, e20487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haghverdi L, Buettner F, Theis FJ, Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics. 31, 2989–2998 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Haghverdi L, Büttner M, Wolf FA, Buettner F, Theis FJ, Diffusion pseudotime robustly reconstructs lineage branching. Nat Methods (2016), doi:10.1038/nmeth.3971. [DOI] [PubMed] [Google Scholar]
- 11.Setty M et al. , Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat Biotechnol. 34, 637–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macosko EZ et al. , Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Maaten L, Hinton G, Visualizing Data using t-SNE. Journal of Machine Learning Research. 9, 2579–2605 (2008). [Google Scholar]
- 14.Fruchterman TMJ, Reingold EM, Graph drawing by force-directed placement. Software: Practice and Experience. 21, 1129–1164 (1991). [Google Scholar]
- 15.Schier AF, Talbot WS, Molecular genetics of axis formation in zebrafish. Annu. Rev. Genet 39, 561–613 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Kimmel CB, Warga RM, Schilling TF, Origin and organization of the zebrafish fate map. Development. 108, 581–594 (1990). [DOI] [PubMed] [Google Scholar]
- 17.Woo K, Shih J, Fraser SE, Fate maps of the zebrafish embryo. Curr. Opin. Genet. Dev 5, 439–443 (1995). [DOI] [PubMed] [Google Scholar]
- 18.Talbot WS et al. , A homeobox gene essential for zebrafish notochord development. Nature. 378, 150–157 (1995). [DOI] [PubMed] [Google Scholar]
- 19.Brunet J-P, Tamayo P, Golub TR, Mesirov JP, Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci USA. 101, 4164–4169 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Satija R, Farrell JA, Gennert D, Schier AF, Regev A, Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chechik G et al. , Activity motifs reveal principles of timing in transcriptional control of the yeast metabolic network. Nat Biotechnol. 26, 1251–1259 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thisse C, Thisse B, Halpern ME, Postlethwait JH, Goosecoid expression in neurectoderm and mesendoderm is disrupted in zebrafish cyclops gastrulas. Dev Biol. 164, 420–429 (1994). [DOI] [PubMed] [Google Scholar]
- 23.Seiliez I, Thisse B, Thisse C, FoxA3 and goosecoid promote anterior neural fate through inhibition of Wnt8a activity before the onset of gastrulation. Dev Biol. 290, 152–163 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Gardiner MR, Gongora MM, Grimmond SM, Perkins AC, A global role for zebrafish klf4 in embryonic erythropoiesis. Mech Dev. 124, 762–774 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Swindell EC et al. , Regulation and function offoxe3 during early zebrafish development. Genesis. 46, 177–183 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Gritsman K et al. , The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell. 97, 121–132 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Carmany-Rampey A, Schier AF, Single-cell internalization during zebrafish gastrulation. Curr Biol. 11, 1261–1265 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Picelli S et al. , Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 10, 1096–1098 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Gritsman K, Talbot WS, Schier AF, Nodal signaling patterns the organizer. Development. 127, 921–932 (2000). [DOI] [PubMed] [Google Scholar]
- 30.McKenna A et al. , Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science. 353, aaf7907 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raj B et al. , Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat Biotechnol. 40, 181 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cusanovich DA et al. , The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature. 555, 538–542 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dickmeis T et al. , A crucial component of the endoderm formation pathway, CASANOVA, is encoded by a novel sox-related gene. Genes Dev. 15, 1487–1492 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kikuchi Y et al. , casanova encodes a novel Sox-related protein necessary and sufficient for early endoderm formation in zebrafish. Genes Dev. 15, 1493–1505 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halpern ME et al. , Cell-autonomous shift from axial to paraxial mesodermal development in zebrafish floating head mutants. Development. 121, 4257–4264 (1995). [DOI] [PubMed] [Google Scholar]
- 36.Moens CB, Prince VE, Constructing the hindbrain: Insights from the zebrafish. Developmental Dynamics. 224, 1–17 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Zhang L et al. , Noise drives sharpening of gene expression boundaries in the zebrafish hindbrain. Mol Syst Biol. 8, 933 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pandey S, Shekhar K, Regev A, Schier AF, Comprehensive Identification and Spatial Mapping of Habenular Neuronal Types Using Single-cell RNA-seq. Current Biology. 28 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marchenko VA, Pastur LA, Distribution of eigenvalues in certain sets of random matrices. Mat. Sb. (N.S.) 72 (114), 507–536 (1967). [Google Scholar]
- 40.Shekhar K et al. , Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell. 166, 1308–1323.e30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Maaten L, Accelerating t-SNE using Tree-Based Algorithms. Journal of Machine Learning Research. 15, 3221–3245 (2014). [Google Scholar]
- 42.Levine JH et al. , Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 162, 184–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosvall M, Bergstrom CT, Maps of random walks on complex networks reveal community structure. Proceedings of the National Academy of Sciences. 105, 1118–1123 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haghverdi L, Lun ATL, Morgan MD, Marioni JC, Correcting batch effects in single-cell RNA sequencing data by matching mutual nearest neighbours. bioRxiv, 165118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holm S, A Simple Sequentially Rejective Multiple Test Procedure. Scandinavian Journal of Statistics. 6, 65–70 (1979). [Google Scholar]
- 46.Pedregosa F, Varoquaux GE, Gramfort A, al E, Scikit-learn: machine learning in Python. J. Mach. Learn. Res 12, 2825–2830 (2011). [Google Scholar]
- 47.Hammerschmidt M et al. , Mutations affecting morphogenesis during gastrulation and tail formation in the zebrafish, Danio rerio. Development. 123, 143–151 (1996). [DOI] [PubMed] [Google Scholar]
- 48.Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nat Methods. 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li B, Dewey CN, RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2014 15:1. 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davies DL, Bouldin DW, A Cluster Separation Measure. IEEE Transactions on Pattern Analysis and Machine Intelligence. PAMI-1, 224–227 (1979). [PubMed] [Google Scholar]
- 51.van der Walt S et al. , scikit-image: image processing in Python. PeerJ. 2, e453 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.