Abstract
High-throughput preparation of plasmid DNA libraries for next-generation sequencing (NGS) is an important capability for molecular biology laboratories. In particular, it is an essential quality control (QC) check when large numbers of plasmid variants are being generated. Here, we describe the use of the Design of Experiments (DOE) methodology to optimise the miniaturised preparation of plasmid DNA libraries for NGS, using the Illumina® Nextera XT technology and the Labcyte Echo® acoustic liquid dispensing system. Furthermore, we describe methods which can be implemented as a QC check for identifying the presence of genomic DNA (gDNA) in plasmid DNA samples and the subsequent shearing of the gDNA, which otherwise prevents the acoustic transfer of plasmid DNA. This workflow enables the preparation of plasmid DNA libraries which yield high-quality sequencing data.
1. Introduction
The rapid advancement of next-generation sequencing (NGS) technologies has revolutionised genomics research [1]. In order to reduce the cost of sequencing large numbers of samples simultaneously, methods have been developed for barcoding and miniaturising the preparation of libraries for NGS [[2], [3], [4]]. However, there has predominantly been a focus on the preparation of genomic DNA (gDNA) libraries, rather than plasmid DNA. Here, we describe the development of a method for the high-throughput preparation of plasmid DNA libraries for NGS, using the Illumina® Nextera XT technology and the Labcyte Echo® acoustic liquid dispensing system. This method prepares libraries directly from purified plasmid DNA, without the use of rolling circle amplification (RCA), as has been described previously [3]. The use of the Labcyte Echo® enables reactions to be miniaturised, decreasing reagent requirements and reducing the cost per reaction [3,5]. The sequencing of complete plasmids is an important capability in molecular biology laboratories, particularly those producing large numbers of construct variants, the identity of which need to be confirmed for quality control. High-throughput laboratories, such as the London DNA Foundry [6,7], generate large numbers of DNA assemblies with different gene combinations and permutations for which the sequence must be confirmed, particularly if these are to be used in further rounds of DNA assembly.
DNA assembly methods, such as golden gate [8], bring multiple DNA parts together before an enzymatic reaction creates larger, contiguous DNA plasmids. Completing such DNA assemblies on a large scale requires a high number of liquid transfers. For example, a full 384-well plate of six-part assemblies would require 2688 individual liquid transfers. This number of transfers is impractical using tip-based systems due to the requirement to change tips in between each transfer. Due to the tip change, each transfer can take up to one minute, resulting in reagent evaporation concerns. Tip-free acoustic dispensing methods, for example using the Labcyte Echo® system, are preferable. Acoustic dispensing is contactless which greatly reduces the overall processing time, with the added advantage that using nanoliter volumes reduces reagent costs and sample requirements. During acoustic dispensing, droplets are physically ejected from the meniscus of a liquid, however, in the case of highly concentrated RCA DNA droplets cannot be transferred at concentrations greater than 10 ng/μl [3]. Due to our use of the Echo® acoustic liquid handling system to dispense DNA, the successful acoustic transfer of DNA samples was something which needed to be monitored and quality controlled.
The Nextera technology can be used to prepare indexed libraries from DNA samples for sequencing on multiple NGS platforms, including Illumina® sequencing systems [9]. The preparation involves an enzymatic reaction termed ‘tagmentation’, in which the DNA is fragmented and adaptor sequences are added. Sequencing primers and indices are then added by reduced-cycle PCR amplification, allowing for the unique barcoding of the DNA fragments in preparation for sequencing. The Nextera XT kit is typically used for smaller genomes than the Nextera kit and one major advantage is the requirement of a very low amount of input DNA (1 ng for Nextera XT compared to 50 ng for Nextera), as well as the removal of a column purification step [10]. The preparation of plasmid DNA using RCA and the Nextera kit, as well as the processing of gDNA samples using the Nextera XT kit, has previously been described [2,3]; however, to our knowledge there has been no large scale exploration of applying Nextera XT to whole plasmid sequencing. Potentially due to the supercoiled nature of bacterial plasmid DNA, the preparation of libraries is likely to differ from the optimised protocols developed for gDNA sequencing [2]. Here, we use Design of Experiments (DOE) [11] to develop a high-throughput library preparation method for plasmid DNA, ensuring the correct mean fragment size was suitable for NGS. The method was developed using the Nextera XT kit on the Labcyte Echo® acoustic liquid dispensing system, which enabled a significant reduction in reagent use resulting in cost- and DNA sample-saving benefits. Plasmid DNA libraries were multiplexed on an Illumina® MiSeq [12,13] and were shown to generate high quality sequencing data. The methods described here, and the DOE approach used, could aid the field of synthetic biology, and others, looking to work with plasmid DNA on acoustic dispensers, prepare plasmid DNA for NGS, or those interested in using DOE for the optimisation of biological assays.
2. Material and methods
2.1. DOE optimisation of Nextera XT NGS library preparation
JMP® Pro 13 (SAS, UK) was used to generate custom designs for the optimisation of the library preparation of plasmid DNA samples for NGS, using the Nextera XT library preparation kit (FC-131-1096, Illumina Inc., USA). Plasmid DNA samples were subjected to a series of three multi-factorial experiments in which the reaction conditions were optimised for three input variables: tagmentation incubation time (5–20 min), DNA sample volume (50–1000 nl) and magnetic bead concentration (0.6–1.8x sample volume). Libraries were prepared from plasmid DNA samples using the various reaction conditions defined by the DOE models and then run on the Fragment Analyzer (Advanced Analytical Technologies Inc., USA) and/or quantified in a dsDNA DNA quantification assay (see below). The data were input into the DOE model and analysed using the JMP® Pro 13 software.
2.2. Nextera XT library preparation of plasmid DNA for NGS
All reagents used were provided in the Illumina® Nextera XT DNA Library Preparation Kit (FC-131-1096, Illumina Inc., USA) and Nextera XT Index Kit v2 Set A (FC-131-2001, Illumina, Inc., USA). Prior to library preparation, all samples were diluted using the Echo® acoustic liquid dispensing system, to 0.4 ng/μl in ddH2O in a final volume of 5 μl/well in an Echo® Qualified 384-well, Low Dead Volume microplate (Labcyte Inc., USA). For the tagmentation reaction, the Echo® was used to combine 500 nl/well Tagment DNA (TD) buffer with 250 nl/well Amplicon Tagment Mix (ATM) in a 384-well PCR plate. Next, 50 nl/well normalised plasmid DNA (0.4 ng/μl) was added to the PCR plate and the samples were incubated at 55 °C for 12.5 min, using the C1000 Touch™ Thermal Cycler (Bio-Rad Laboratories, Inc., USA). The samples were then allowed to cool to 4 °C before the immediate addition of 250 nl/well of the Neutralize Tagment (NT) buffer. The NT buffer was incubated with the samples for 5 min at room temperature, to terminate the tagmentation reaction.
Using the Echo®, 1875 nl/well of the Nextera PCR Master Mix (NPM) was combined with an equivalent volume of ddH2O in a 384-well PCR plate. A unique combination of an i5 and an i7 index primer (625 nl each primer per well) was then added to each tagmented DNA sample. The primers used are listed in Supplementary Table S1. The index primers were attached to each sample via 12 PCR cycles. The thermocycling protocol was 72 °C for 3 min followed by 30 s at 90 °C. There were then 12 cycles of heating to 98 °C for 10 s then 55 °C for 30 s, followed by 72 °C for 30 s. Following the PCR cycles, the samples were held at 72 °C for 5 min before being cooled and held at 4 °C. The PCR products were purified using an automated, magnetic bead purification method (see below). The concentration of the purified DNA samples was quantified in a miniaturised dsDNA quantification assay using the PicoGreen® reagent (see below), with an expected concentration range of 0.5–5 ng/μl, prior to being pooled for sequencing.
2.3. Purification of DNA samples
To remove small DNA fragments and unbound index primers, the PCR products were purified using a plate based magnetic bead purification protocol. Using the CyBio® FeliX pipetting platform (Analytik Jena, Germany), PCR products were combined with 18 μl/well AmpiClean™ Magnetic Bead solution (Nimagen, Netherlands) in a round-bottom, 96-well plate. The mixture was incubated at room temperature for 5 min to allow the DNA to bind to the beads. The plate was then transferred to a 96-well magnet and incubated for 2 min for rings of beads to form. The supernatant was then removed from all wells. The beads were washed twice with 70% Ethanol before removal of all liquid and air drying of the magnetic bead/DNA complexes. The 96-well plate was then removed from the magnet and the DNA eluted from the beads with 20 μl/well ddH2O. After placing the 96-well plate back on the magnet, the supernatant was removed from the beads and then contained the purified PCR product.
2.4. Next generation sequencing of plasmid DNA samples using the Illumina® MiSeq system
Plasmid DNA libraries, prepared using the Nextera XT library preparation kit (see above), were pooled to give a final concentration of 4–10 nM in a minimum volume of 15 μl. Normalisation of the pooled libraries according to concentration and size was not necessary. The average fragment size of the pooled library was determined by testing the sample on the Fragment Analyzer™ (Advanced Analytical Technologies Inc., USA). The library was sequenced using the Illumina® MiSeq system, with the MiSeq Micro reagent kit v2 (2 × 150 bp reads; Illumina, Inc., USA).
The data analyses were performed on the generated Illumina® FASTQ files using FastQC, to determine if each of the sample reads were of reliable quality. The Velvet de novo assembler [14] was used to build the reads into contiguous sequences (contigs). The contigs from the de novo assembly were then aligned to the reference sequence, built in silico, using Clustal Omega [15] to determine if the plasmids had assembled correctly. The aligned contigs were modified to account for circular DNA by cleaving the unaligned trailing nucleotides, if matched with the opposite end of the assembled contig, producing a more accurate final consensus sequence.
2.5. Fragment analysis
Using the dsDNA 915 reagent kit on the Fragment Analyzer™ Automated CE System (Advanced Analytical Technologies Inc., USA), purified PCR products were analysed to measure the mean size of the DNA fragments in each sample. Samples were tested neat, according to the manufacturer's instructions. Data was analysed using the PROsize® analysis software (Advanced Analytical Technologies Inc., USA).
2.6. dsDNA quantification assay
The PicoGeen® dsDNA Quantification Reagent (Thermo Fisher, USA) was used to detect and quantify dsDNA. The reagent was used in a miniaturised, high-throughput assay developed in the London DNA Foundry and implemented using the Labcyte Echo® acoustic dispensing system (Labcyte Inc., USA). The protocol can be used to determine the concentration of dsDNA samples, as well as indicating if plasmid DNA samples are contaminated with gDNA. If a DNA sample is contaminated with gDNA, it will either not transfer at all or transfer variably on the Echo®. Therefore, the quantification of dsDNA in the destination plate informs on the presence or absence of gDNA in a given DNA sample. If a DNA sample failed to transfer, it was identified as having gDNA contamination. The PicoGreen® reagent was added to a 384-well plate at 50 nl/well. The reagent was diluted to the appropriate concentration through the addition of 20 μl/well 1x Tris-EDTA (TE) buffer using a bulk dispenser, prior to the transfer of 100 nl/well DNA sample on the Echo®. The assay plate was incubated for 3 min at room temperature, prior to the measurement of the fluorescent readout on a microplate reader (485ex/520em). The concentration of the DNA samples was interpolated from a standard curve, prepared using a plasmid DNA sample of a known concentration.
2.7. Shearing of gDNA in plasmid DNA samples
If a plasmid DNA sample was found to contain gDNA using the dsDNA quantification assay (see above), the gDNA was sheared by sonication, which retains the plasmid DNA intact. Bath sonication was performed using a Q700 sonicator and a Microplate Horn (Qsonica, USA). To shear gDNA, 100 μl/well of the DNA sample was added to a 96-well PCR plate and sonicated floating in the water-filled reservoir of the Microplate Horn. Sonication was performed at room temperature for a total of 3 min with a 2 s pulse, followed by a 2 s pause, at an amplitude of 10%.
2.8. Agarose gel electrophoresis
Agarose gel electrophoresis was used to test plasmid DNA samples to confirm the presence of gDNA, during assay development. Samples were run in a 1% agarose gel prepared with SYBR™ Safe (Thermo Fisher, USA), at either 60 V or 80 V, for 90/60 min respectively. The GelDoc (BioRad) was used to image the gels and the size of the bands was determined by comparison to a 1 Kb Plus DNA ladder (Thermo Fisher, USA).
3. Results
3.1. Use of a dsDNA quantification assay as a QC check for the contamination of plasmid DNA samples with gDNA
The miniaturisation of plasmid DNA library preparation for NGS, involves the use of the Labcyte Echo® to transfer small volumes of plasmid DNA samples. It is known that the transfer of long linear DNA, such as gDNA, at concentrations greater than 10 ng/μl, is not successful using acoustic dispensing [3] (Fig. 1A). However, we demonstrate that plasmid DNA samples, tested up to a concentration of 80 μg/ml, transfer acoustically with success, likely due to their supercoiled nature (Fig. 1B). During assay development, we observed variable results when transferring plasmid DNA samples acoustically. We hypothesised that gDNA contamination was preventing the transfer of the affected plasmid DNA samples on the Echo®. Therefore, it became important to ensure that plasmid DNA samples, prepared using a high-throughput plasmid isolation method [16], were free from gDNA contamination, prior to their transfer on the Echo®. For this, we developed a miniaturised assay, using the Echo®, for the quantification of dsDNA, using the PicoGreen® dsDNA quantitation reagent (Thermo Fisher, USA). In the presence of dsDNA, the dye emits fluorescence which can be quantified using a standard curve [17].
Fig. 1.
Long linear DNA fails to transfer by acoustic dispensing. PicoGreen® dsDNA dye was used to measure the fluorescent signal from long, linear lamda DNA (A) or supercoiled plasmid DNA (B) at a range of concentrations after 1 μl sample was transferred either by manual pipetting or acoustically using the Echo®, in triplicate. At concentrations above 5 μg/ml, lamda DNA does not transfer well on the Echo®. However, supercoiled plasmid DNA transfers efficiently at all tested concentrations. The concentration of 7 plasmid DNA samples was measured using the PicoGreen® reagent. The plasmid DNA samples were either transferred by manual pipetting (C) or using acoustic dispensing on the Labcyte Echo® 550 (D). All samples were tested in 3 replicates, as represented by different coloured bars. The data show that 3 samples failed to transfer by acoustic dispensing (samples 1–3) while one transfers with variable amounts (sample 5). (E) The plasmid DNA samples were run in 1% w/v agarose gel (in 1× TAE) at 80 V for 1 h; M = 1 Kb Plus DNA Ladder (ThermoFisher Scientific). Due to the large size of gDNA, it remains above the range of the ladder, as indicated by the red arrow. The gel shows the presence of gDNA in samples 1–3, which are those that did not transfer acoustically. (F) The concentrations of plasmid DNA samples in (C) and (D) were interpolated from a standard curve, generated using a plasmid DNA sample of a known concentration.
We tested a selection of plasmid DNA samples in the miniaturised dsDNA quantification assay, with samples transferred either manually or by acoustic dispensing (Fig. 1Cand D). The samples were also run on an agarose gel (Fig. 1E). When the concentration of the samples was interpolated from the standard curve in the assay (Fig. 1F), we observe that all of the tested samples have the expected concentration when transferred manually (Fig. 1C). However, some samples have no fluorescent readout when the transfer of the DNA was carried out acoustically (Fig. 1D). The agarose gel shows that those samples which failed to transfer acoustically on the Echo® are contaminated with gDNA (Fig. 1E, lanes 1–3). The miniaturised PicoGreen® dsDNA quantitation assay provides a quality control (QC) check for the presence of gDNA in plasmid DNA samples. The lack of a fluorescence signal indicates a failed transfer of a DNA sample. This failed transfer could be due either to a lack of DNA in the well or gDNA contamination. By testing the sample with manual transfer the presence of DNA can be confirmed, indicating such a sample contains gDNA. In the London DNA Foundry, all plasmid DNA samples are run in the gDNA QC assay prior to their addition to our DNA parts library, to ensure that they will transfer as required. The variability of DNA concentration across replicates is hypothesised to indicate an intermediate level of gDNA contamination, which is undesirable in an acoustic DNA library workflow (Fig. 1Cand D, sample 5).
3.2. Shearing of gDNA in plasmid DNA samples
When plasmid DNA samples are found to contain gDNA, they cannot be used (Fig. 1). We developed a method to shear gDNA by sonication, while maintaining an intact plasmid (Fig. 2A). The gDNA QC assay data show that while an untreated, gDNA contaminated sample transfers variably by acoustic dispensing, the sonicated sample transfers successfully, with the expected concentration consistently measured (Fig. 2B). The resulting DNA was also transformed into DH5α E. coli cells to determine the plasmid viability, giving rise to antibiotic resistant clones and indicating that the plasmid DNA is intact (Supplementary Fig. S1).
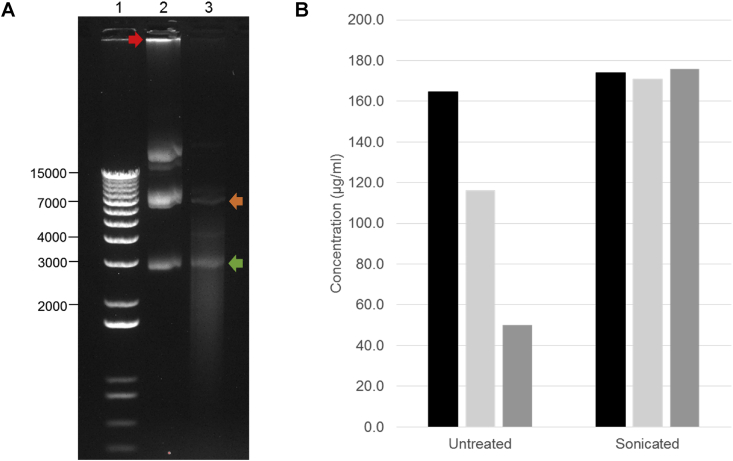
Fig. 2.
Sonication of DNA samples fragments gDNA while maintaining the plasmid DNA intact. (A) Samples were run in 1% w/v agarose gel (in 1× TAE) at 60 V for 90 min; Lane 1: 1 Kb Plus DNA Ladder (ThermoFisher Scientific); Lane 2: DNA plasmid part, 50 ng/μl/kbp; Lane 3: DNA plasmid part, 50 ng/μl/kbp, after sonication (3 min, 2 s pulse, 10% amplitude). The red arrow indicates gDNA visible in the untreated sample (lane 2). The green arrow indicates the supercoiled plasmid DNA, seen in both the untreated (lane 2) and the sonicated (lane 3) samples. The orange arrow indicates the open circular form of the plasmid DNA. (B) The untreated and sonicated plasmid DNA samples were tested in triplicate in the miniaturised gDNA QC assay and the concentration of dsDNA was calculated, according to the standard curve. In the untreated sample, variable concentrations were measured in each of the three replicates, likely due to contamination of the plasmid DNA sample by gDNA. In the sonicated sample, all replicates gave a similar quantification, indicating successful acoustic dispense and shearing of gDNA.
These data indicate that shearing by sonication is appropriate for the fragmentation of gDNA in plasmid DNA samples, leaving the plasmid DNA intact and ready for use in downstream applications. Treated samples can also be successfully transferred acoustically, which is essential for miniaturised methods utilising this method of dispensing, such as the NGS library preparation protocol described here.
3.3. Optimisation of plasmid DNA library preparation for NGS using Design of Experiments
For sequencing of plasmid DNA using the Illumina® MiSeq platform, a library of fragments with a mean size of between 200 and 400 bp, each with a concentration of 0.5–5 ng/μl is required. Using an existing method for gDNA library preparation [2], the processing of plasmid DNA samples yielded fragments of variable size which were generally too large and had low relative fluorescence units (RFU), indicative of low concentration (Fig. 3A–C).
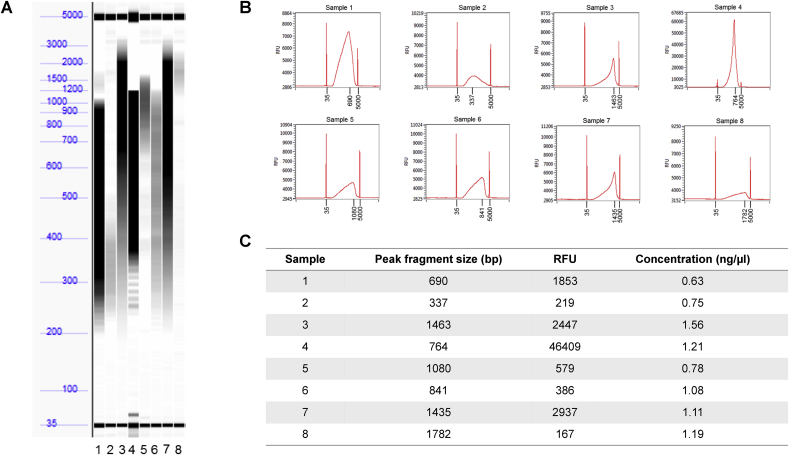
Fig. 3.
Preparation of plasmid DNA libraries for NGS using miniaturised gDNA method. (A) Eight plasmid DNA samples were prepared according to Labcyte's miniaturised NGS gDNA library preparation method [2], using the Nextera XT kit. After purification, the samples were run on the Fragment Analyzer to determine the size of the fragments. (B) Plots of the Fragment Analyzer data show the peak fragment size for 7/8 of the samples is greater than 400 bp, which is the maximum limit required for sequencing. (C) Summary of the data shows variable relative fluorescence units (RFU) for the 8 samples. The concentration of each purified sample, as measured using the PicoGreen dsDNA assay, is within the range required (0.5–5 ng/μl).
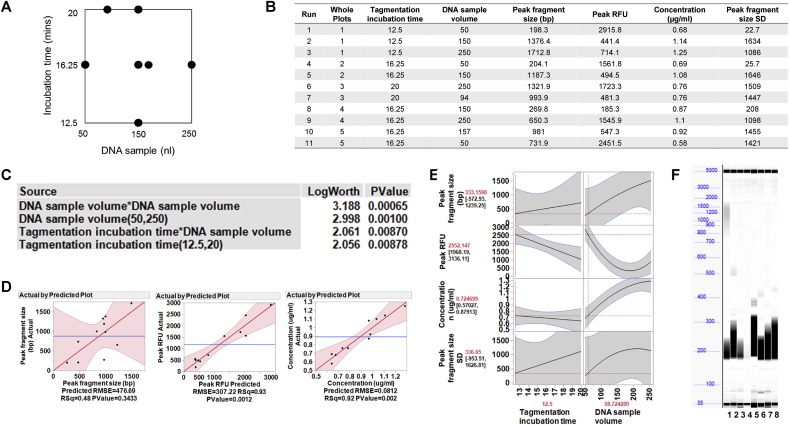
We used DOE and modelling to optimise the miniaturised preparation of a plasmid DNA library for NGS, using the Nextera XT kit. Three sequential DOE models were tested in order to optimise the conditions for the library preparation. In the first experiment, the input variables included in the model were the tagmentation incubation time, the DNA sample volume and the concentration of the magnetic beads used during the DNA purification step (Fig. 4A). The ratio of the magnetic beads to the sample volume determines the size of the DNA fragments bound by the beads; use of a lower ratio (and therefore lower concentration) of magnetic beads allows larger DNA fragments to be bound, and subsequently removed, from the DNA sample. We needed to generate and retain fragments of at least 300 bp, ideally removing fragments of 200 bp and below. The DOE model had 15 runs which were divided into 5 whole plots, within each of which the tagmentation incubation time remained constant (Fig. 4B).
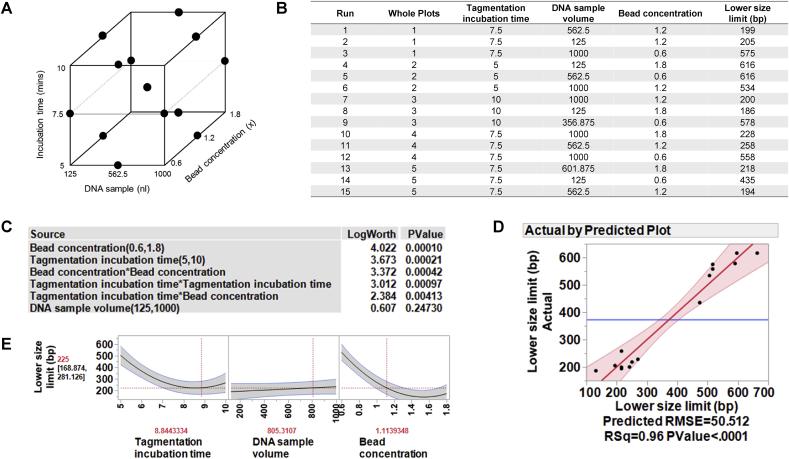
Fig. 4.
Design of experiments (DOE) model number 1 for NGS plasmid DNA library preparation: Optimisation of the lower size limit of fragments. (A) Three factors were evaluated using a custom designed model generated with JMP® software: tagmentation incubation time, plasmid DNA sample volume, and the concentration of magnetic beads used in the automated DNA purification method. The evaluated range for each variable is shown. (B) There was a total of 15 runs in random order from 5 whole plots. Each whole plot represents the same condition for the tagmentation incubation time and was performed in a separate plate. The response variable to be optimised was the lower size limit (the lowest size of DNA fragment detected after magnetic bead purification). A plasmid DNA sample was prepared with the Nextera XT library preparation kit using the miniaturised method, according to each set of conditions defined in the DOE model. After magnetic bead purification, samples were run neat on the Fragment Analyzer. (C) The data were modelled according to the DOE design, using JMP® software. Both bead concentration and tagmentation incubation time has a significant effect on the lower size limit of the fragmented DNA (Log Worth >2). (D) The data predicted by the DOE model correlated with the actual data with an R2 value of 0.96, indicating a very good correlation. (E) Visualisation of the optimal lower size limit model using the prediction profiler tool in the JMP® software, shows that the desired lower size limit of less than 300 bp is achieved with a tagmentation incubation time of >7.5 min and a magnetic bead concentration of between 1.1–1.8x sample volume.
The data were generated using a single plasmid DNA sample, prepared according to the conditions of each of the different runs in the model. When the data were fitted to the model, it was demonstrated that to achieve the desired lower size limit of <300 bp, two variables were significant factors: the bead concentration and the tagmentation incubation time, with a significant interaction observed between these two variables (Fig. 4C). The model fit shows an excellent correlation between the predicted and the actual data (R2 = 0.96; Fig. 4D), with a tagmentation incubation time of >7.5 min and a magnetic bead ratio of between 1.1–1.8x sample volume, giving a lower fragment size limit of <300 bp, as required (Fig. 4E). The DNA sample volume did not have a significant effect on the lower fragment size limit.
In the second experiment, there were two response variables: the RFU (to be maximised); and the peak fragment size (200–400 bp required). The input variables remained the same as in the initial model with the tagmentation incubation time and the magnetic bead concentration adjusted according to the results of the first model (Fig. 5A). The model had 15 runs and 5 whole plots, within which the tagmentation incubation time remained constant (Fig. 5B). The modelled data show two significant factors, namely the DNA sample volume and the tagmentation incubation time, with a significant interaction observed between these two variables (Fig. 5C). There was a good correlation between the actual data and the values predicted by the models for both the peak fragment size and the RFU, with R2 values of 0.98 and 0.93 respectively (Fig. 5D). The DOE model indicates that a long tagmentation incubation time and a low DNA volume give the highest RFU with fragments within the required range (Fig. 5E). A low volume of DNA sample yields smaller peak fragment sizes, while a long tagmentation incubation time significantly increases the peak RFU. The magnetic bead concentration was not a significant factor in this model. A concentration of 1.8x magnetic beads was used in all future experiments, as this is consistent with existing protocols. The experimental conditions optimised in this model were tested across a 96-well plate of different plasmid DNA preparations. However, after purification, the prepared sample libraries had highly variable fragment sizes (Fig. 5F), making them unsuitable for sequencing.
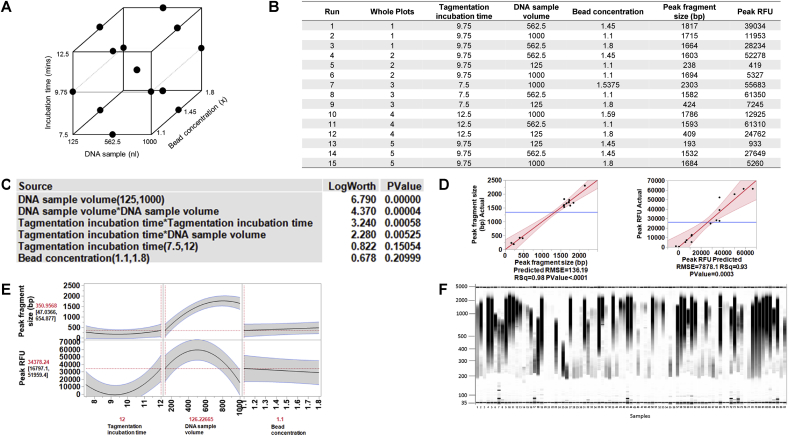
Fig. 5.
Design of experiments (DOE) model number 2 for NGS plasmid DNA library preparation: Optimisation of fragment size and concentration. (A) Three factors were evaluated using a custom designed model generated with JMP® software: tagmentation incubation time, plasmid DNA sample volume, and the concentration of magnetic beads used in the automated DNA purification method. The evaluated range for each variable is shown. (B) There was a total of 15 runs in random order from 5 whole plots. Each whole plot represents the same condition for the tagmentation incubation time and was performed in a separate plate. There were two response variables to be optimised: the peak fragment size (representing the size of most DNA fragments); and the peak relative fluorescence unit (RFU), indicative of concentration. A plasmid DNA sample was prepared with the Nextera XT library preparation kit using the miniaturised method, according to each of the conditions defined in the DOE model. After magnetic bead purification, samples were run neat on the Fragment Analyzer. (C) The data were modelled according to the DOE design using JMP® software. The effect summary shows that the tagmentation incubation time and DNA sample volume had a significant effect individually on the output variables. There was also a significant interaction between these two input variables. (D) The peak fragment size predicted by the DOE model correlated with the actual data with an R2 value of 0.98, while the predicted peak RFU data correlated with the actual data with an R2 value of 0.93, both indicative of a very good correlation. (E) The prediction profiler tool in the JMP® software was used to visualise the data. When the desirability was maximised (peak fragment size of 200–400 bp and maximum RFU), the optimised conditions suggested by the model are a DNA sample volume of 126 nl and a tagmentation incubation time of 12 min. (F) When the optimised conditions were tested on a multiwell plate of 96 samples, some samples had the desired peak fragment size (200–400 bp), however, many had larger fragment sizes than desired. Therefore, although the conditions optimised here are applicable to some plasmid DNA samples, further optimisation was required to establish the correct conditions for all plasmid DNA preparations.
The aim of the third DOE model was to reduce the variability between different plasmid DNA preparations. The experimental goals were mean fragment sizes between 200 and 400 bp for all samples, a high RFU and for the magnetic bead purified samples to be at an appropriate concentration (0.5–5 ng/μl), as measured in the dsDNA quantification assay. To investigate the reproducibility of the tested conditions, 8 different plasmid DNA preparations were used for each run condition and the mean data were analysed in the model, in addition to the standard deviation of the fragment sizes. Two input variables were tested in this design, the tagmentation incubation time and the DNA sample volume (Fig. 6A). Based on the conclusions from the previous experiment, the range of the tagmentation incubation time was extended while the range for the DNA sample volume was lowered. There were 11 runs in the design and 5 whole plots, within each of which the tagmentation time remained constant (Fig. 6B).
Fig. 6.
Design of experiments (DOE) model number 3 for NGS plasmid DNA library preparation: Optimising the reproducibility between different plasmid DNA preparations. (A) Two factors were evaluated using a custom designed model generated with JMP® software: tagmentation incubation time and plasmid DNA sample volume. The evaluated range for each variable is shown. (B) There was a total of 11 runs in random order from 5 whole plots. Each whole plot represents the same condition for the tagmentation incubation time and was performed in a separate plate. There were three response variables: the peak fragment size (representing the size at which most DNA fragments are); the peak relative fluorescence unit (RFU); and the concentration of the sample, after magnetic bead purification. 8 plasmid DNA samples were prepared with the Nextera XT library preparation kit using the miniaturised method, according to each of the conditions defined in the DOE model. After magnetic bead purification, samples were run neat on the Fragment Analyzer. The concentration of the purified samples was also determined using the PicoGreen® dsDNA quantification assay. (C) The data were modelled according to the DOE design using JMP® software. The effect summary shows that tagmentation incubation time and DNA sample volume each had a significant effect individually. There was also a significant interaction between the two variables. (D) The data predicted by the DOE model correlated with the actual data with an R2 value of 0.48, 0.93 and 0.92 for the peak fragment size, peak RFU and concentration respectively. (E) The prediction profiler tool in the JMP® software was used to visualise the data. When the desirability was maximised (peak fragment size of 200–400 bp, maximum RFU, a concentration of 0.5–5 ng/μl and minimised peak fragment size standard deviation), the optimised conditions suggested by the model are a DNA sample volume of 58.7 nl and a tagmentation incubation time of 12.5 min. (F) The Fragment Analyzer outputs for 8 samples, run with a 12.5 min tagmentation incubation time and 50 nl DNA, show that 7/8 of the samples have a peak fragment size of between 200 and 300 bp and an average concentration of 0.68 ng/μl (±0.33 SD). The undetected sample (sample 4) had a concentration below the limit of detection of the Fragment Analyzer (<0.5 ng/μl). These optimised conditions give reproducible results across multiple plasmid DNA samples.
The data show that both the DNA sample volume and the tagmentation incubation time had a significant effect in the model (Fig. 6C). The correlation between the peak fragment size predicted by the model and the actual peak fragment size was low, with an R2 value of 0.48 (Fig. 6D), although there is an observable trend in the data. The correlation was high however for the peak RFU and the purified sample concentration, with R2 values of 0.93 and 0.92 respectively (Fig. 6D). The model shows that an optimal peak fragment size of 333 bp, with a high RFU was achieved with a low DNA sample volume (∼50 nl) and a 12.5 min tagmentation incubation time (Fig. 6E). The concentration of the prepared samples was not affected by the tagmentation incubation time. However, the DNA sample volume did have a significant effect on output concentration, with higher sample volumes conferring a higher concentration of library preparation product, as expected (Fig. 6E). However, as an increase in the concentration of DNA also reduces the RFU and increases the peak fragment size to outside of the target range, a low DNA volume has to be used. The model therefore suggests using a low DNA volume and a tagmentation time of 12.5 min (Fig. 6E). For simplification, the DNA volume was set at 50 nl. These conditions achieve fragment sizes of 200–400 bp, with sufficiently high RFU and an average concentration of 0.68 ng/μl (±0.33 SD), which is within the required range (Fig. 6F).
3.4. Next generation sequencing of plasmid DNA library
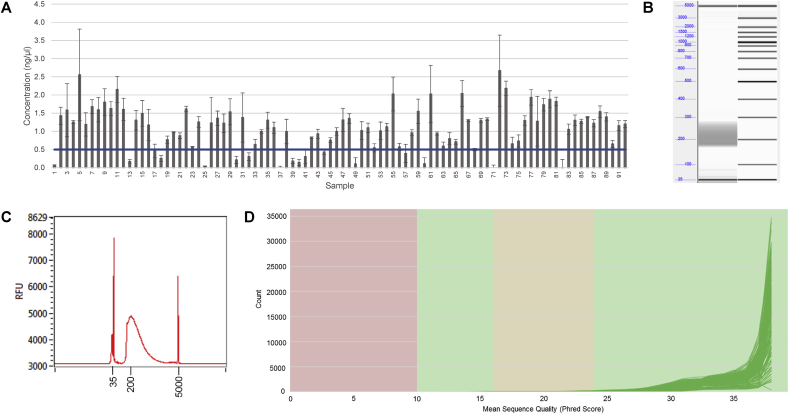
Using the optimised conditions for library preparation, a 96-well plate of plasmid samples, including positive and negative controls, were processed. After purification, the average concentration of samples was 1.1 ng/μl (±0.6 SD), which is within the desired range of 0.5–5 ng/μl (Fig. 7A). Samples were pooled, with a pooled library concentration of 6.64 nM and a DNA library mean fragment size in the desired range of 200–400 bp (Fig. 7Band C). Samples were sequenced using the Illumina® MiSeq platform (2 × 150 method) and the mean quality scores for all samples passed the QC criteria (>30 Phred score; Fig. 7D). All the positive control plasmid DNA samples matched the reference sequence with 99.87% similarity. In addition, a DNA part which was sonicated prior to its use in DNA assembly due to gDNA contamination, had the expected sequence when mapped to the appropriate reference sequence (Supplementary Fig. S2). These data indicate that the library preparation method described yields high-quality sequencing data, appropriate for use in high-throughput applications.
Fig. 7.
Next generation sequencing of plasmid DNA libraries, prepared using a miniaturised method with the Nextera XT library preparation kit. 96 plasmid DNA libraries were prepared for NGS with the Nextera XT library preparation kit, using optimised conditions (12.5 min incubation, 50 nl sample, 1.8x magnetic bead solution). (A) After purification, the samples were quantified in the PicoGreen® dsDNA quantification assay. The data show that 75/92 samples have a concentration within the desired range (0.5–5 ng/μl), with an average concentration of 1.1 ng/μl. These samples were pooled, with a final concentration of 6.64 nM and run on the Fragment Analyzer (B) and (C). All fragments in the pooled libraries are of the desired size (200–400 bp). (D) The pooled library was sequenced on the Illumina® MiSeq system (2 × 150 method). The mean sequence quality (Phred) scores are plotted for each sample. For all samples, the sequence quality (Phred) score was >30 for more than 85% of the base pairs, indicating that all samples passed the QC criteria.
4. Discussion
Plasmid DNA is used widely in the field of molecular biology, in particular during DNA assembly workflows for engineering biology. The use of plasmid DNA isolated directly from bacterial cultures, for sequencing and DNA assembly, offers an advantage over DNA obtained from rolling circle amplification (RCA) as it is possible to transfer it acoustically at high concentrations if it is free of gDNA contamination (Fig. 1) [18]. We describe the importance of testing plasmid DNA samples for gDNA contamination prior to the use of acoustic dispensing systems, due to the presence of gDNA (or other long linear DNA) preventing the acoustic transfer of samples. The implementation of an assay using the PicoGreen® dsDNA quantification reagent on the Labcyte Echo® provides a gDNA contamination QC test for plasmid DNA samples. We have also described a method to recover gDNA contaminated samples using sonication, which enables DNA to be used in an acoustic liquid handling workflow reliably for synthetic biology applications without the need for re-preparation of plasmid DNA samples.
We have used a DOE approach to develop a miniaturised high-throughput workflow for the preparation of plasmid DNA libraries for NGS, using the Nextera XT kit on the Labcyte Echo® acoustic dispensing system (Fig. 8). The sequencing of the full length of DNA plasmids is an important capability in molecular biology laboratories, in particular for the QC of DNA constructs. Existing methods for the preparation of DNA libraries for sequencing have either focused on gDNA libraries or plasmid DNA obtained from RCA. To our knowledge, this is the first method described for the preparation of plasmid DNA libraries using the Nextera XT kit on the Labcyte Echo®. The use of the Echo® allowed the miniaturisation of the method, significantly reducing reagent requirements and therefore cost. The method described here, costs approximately £10.90 per plasmid sample, inclusive of library preparation and sequencing, as compared to services currently on the market which cost in the region of $165 per sample for full length plasmid sequencing [19]. Using DOE, three sequential experiments were rapidly executed in which a total of only 41 data points were required in order to model the entire experimental space and to accurately predict the optimal conditions for the protocol. This rapid assay development approach using DOE has both time and cost-saving benefits.
Fig. 8.
High-throughput workflow for the preparation of plasmid DNA libraries for NGS. Plasmid DNA samples are isolated from bacteria cells using a high-throughput plasmid isolation method on the CyBio® FeliX robot. All steps performed using the FeliX platform are highlighted with a grey outline. The isolated plasmid samples are tested for the presence of genomic DNA (gDNA), prior to library preparation, using the Labcyte Echo®. All steps performed using the Labcyte Echo® are highlighted with a shaded grey box. If samples are free from gDNA, they are diluted to 0.4 ng/μl in H2O. If gDNA is detected, the samples are sonicated prior to re-testing in the gDNA QC assay. Using reagents from the Nextera XT kit, a tagmentation reaction is performed on all samples under the optimised conditions, followed by neutralization of the reaction. Unique combinations of index primers are added to all samples via 12 PCR cycles, followed by magnetic bead purification of the PCR products. The concentration of the purified dsDNA is then determined using the PicoGreen® reagent assay and the libraries are pooled to give a final concentration of 4–10 nM, in a minimum volume of 15 μl. The average fragment size of the pooled libraries is measured using the Fragment Analyzer before being sequenced on the Illumina® MiSeq system.
We opted to use Illumina's Nextera XT library preparation technology. One problem with many NGS library preparation methods is the loss of sample material during the procedure, meaning a high amount of input DNA is required. The Nextera technology has significantly reduced the loss of sample material, enabling a lower amount of input DNA [1]. The reduction in sample loss has been achieved both by combining the DNA fragmentation, end-polishing and adapter ligation steps into a single reaction [9] and also through replacing gel and column purification steps with magnetic beads [20]. The Nextera XT method was also preferable over the original Nextera method due to the requirement of a very low amount of input DNA (1 ng for Nextera XT compared to 50 ng for Nextera) [10]. For the sequencing of our prepared DNA libraries, we used the Illumina® bench-top MiSeq sequencer. Illumina® provide market leading sequencing platforms with high-throughput and low per-base cost [1,21]. In addition, Illumina® sequencing platforms have very low error rates, as compared to other platforms such as the Ion Torrent and PacBio sequencers [12]. Although the MiSeq instrument has a lower throughput than other Illumina® instruments, such as the HiSeq 2000, it has a significantly faster turnaround time and with a lower instrument cost, is more accessible to smaller laboratories [12].
4.1. Conclusions
The utilisation of the Echo® dispensing system enabled the miniaturisation of both a PicoGreen® dsDNA quantification assay and an NGS plasmid DNA library preparation method. In both cases, this reduces the amount of sample DNA required and significantly lowers the cost of the methods by minimising reagent volumes. We demonstrate that the use of the developed protocols enables the high-throughput preparation of plasmid DNA libraries which yield high quality sequencing data, at a reduced cost compared to existing methods. We believe that these methods will aid those working in the field of molecular biology and in particular synthetic biology where plasmid DNA construction at scale is widely used to enable the testing of multiple genetic designs.
Funding
This work was supported by the Engineering and Physical Sciences Research Council (EPSRC) [EP/L011573/1, EP/M006700/1, EP/N023854/1]; and the DNA Synthesis Capital Award [BB/M025632/1].
Acknowledgements
We thank the Imperial BRC Genomics Facility and Alex Foster for technical assistance.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2019.01.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Table S1. Nextera XT Index Primers used for library preparation of plasmid DNA samples. Unique combinations of the i7 and i5 index primers were added to tagmented DNA samples. 12 i7 (N7XX) and 8 i5 primers (S5XX) were used, enabling the unique labelling of 96 samples at one time.
Fig. S1. Plasmid DNA samples remain viable after sonication under optimised conditions. Plasmid DNA samples with ampicillin resistance were sonicated under 5 different conditions (S1-S5), to shear gDNA contaminants, and subsequently transformed into DH5α E.coli cells. When sonicated using the conditions described in this study (S4, labelled with ‘A’), the plasmid DNA was still intact and viable as shown by the successful growth of antibiotic resistant E.coli colonies. The original unsonicated, gDNA contaminated plasmid DNA sample was also transformed into DH5α cells, for comparison (‘B’).
Fig. S2. Alignment of a sequenced promoter part to a reference sequence. Sequence 1 is a reference sequence for the inducible tac promoter, from an in silico assembly. Sequence 2 is the de novo assembled sequence from the NGS data for a DNA part encoding the tac promoter. The two sequences show a good alignment.
References
- 1.van Dijk E.L., Auger H., Jaszczyszyn Y., Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 2014;30:418–426. doi: 10.1016/j.tig.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Technical note: effective miniaturization of Illumina Nextera XT library prep for multiplexed whole genome sequencing and microbiome applications [internet] Labcyte Inc; San Jose, CA: 2017. http://learn.labcyte.com/rs/450-DEG-211/images/APP-G121-Miniaturization-Nextera-XT-Library-Prep.pdf Available: [Google Scholar]
- 3.Shapland E.B., Holmes V., Reeves C.D., Sorokin E., Durot M., Platt D. Low-cost, high-throughput sequencing of DNA assemblies using a highly multiplexed Nextera process. ACS Synth Biol. 2015;4:860–866. doi: 10.1021/sb500362n. [DOI] [PubMed] [Google Scholar]
- 4.Tan S.J., Phan H., Gerry B.M., Kuhn A., Hong L.Z., Ong Y.M. A microfluidic device for preparing next generation DNA sequencing libraries and for automating other laboratory protocols that require one or more column chromatography steps. PLoS One. 2013;8 doi: 10.1371/journal.pone.0064084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agrawal S., Cifelli S., Johnstone R., Pechter D., Barbey D.A., Lin K. Utilizing low-volume Aqueous acoustic transfer with the Echo 525 to enable miniaturization of qRT-PCR assay. J Lab Autom. 2016;21:57–63. doi: 10.1177/2211068215609315. [DOI] [PubMed] [Google Scholar]
- 6.Chambers S., Kitney R., Freemont P. The Foundry: the DNA synthesis and construction Foundry at imperial college. Biochem Soc Trans. 2016;44:687–688. doi: 10.1042/BST20160007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McClymont D.W., Freemont P.S. With all due respect to Maholo, lab automation isn't anthropomorphic. Nat Biotechnol. 2017;35:312–314. doi: 10.1038/nbt.3795. [DOI] [PubMed] [Google Scholar]
- 8.Engler C., Gruetzner R., Kandzia R., Marillonnet S. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005553. journals.plos.org. e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caruccio N. Preparation of next-generation sequencing libraries using NexteraTM technology: simultaneous DNA fragmentation and adaptor tagging by in vitro transposition. Methods Mol Biol. 2011;733:241–255. doi: 10.1007/978-1-61779-089-8_17. [DOI] [PubMed] [Google Scholar]
- 10.Technical note: Nextera XT DNA library prep kit [internet] Illumina, Inc.; San Diego, CA: 2018. https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/samplepreps_nextera/nextera-xt/nextera-xt-library-prep-reference-guide-15031942-03.pdf Available: [Google Scholar]
- 11.Anderson M.J., Whitcomb P.J. Kirk-othmer encyclopedia of chemical technology. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2000. Design of experiments; p. 16. John Wiley & Sons, Inc. [Google Scholar]
- 12.Quail M.A., Smith M., Coupland P., Otto T.D., Harris S.R., Connor T.R. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012;13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bentley D.R., Balasubramanian S., Swerdlow H.P., Smith G.P., Milton J., Brown C.G. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Technical Note . SynbiCITE, UK & Analytik Jena; Germany: 2018. Automated high-throughput plasmid DNA isolation from bacteria cells [internet]https://www.analytik-jena.de/en/lab-automation/products-lab-automation/liquid-handling/cybior-felix.html Available: [Google Scholar]
- 17.Ahn S. PicoGreen quantitation of DNA: effective evaluation of samples pre- or post-PCR. Nucleic Acids Res. 1996;24:2623–2625. doi: 10.1093/nar/24.13.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dean F.B., Nelson J.R., Giesler T.L., Lasken R.S. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Applied Biological Materials Inc. Full Length Plasmid Sequencing Service ($125.00) | ABM Inc [Internet]. [cited 14 Jan 2019]. Available: https://www.abmgood.com/Full-Length-Plasmid-Sequencing-Service.html.
- 20.DeAngelis M.M., Wang D.G., Hawkins T.L. Solid-phase reversible immobilization for the isolation of PCR products. Nucleic Acids Res. 1995;23:4742–4743. doi: 10.1093/nar/23.22.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu L., Li Y., Li S., Hu N., He Y., Pong R. Comparison of next-generation sequencing systems. J Biomed Biotechnol. 2012;2012:251364. doi: 10.1155/2012/251364. downloads.hindawi.com. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.