Abstract
Defining direct targets of transcription factors and regulatory pathways is key to understanding their roles in physiology and disease. Here we combine SLAM-seq, a method for direct quantification of newly synthesized mRNAs, with pharmacological and chemical-genetic perturbation to define regulatory functions of two transcriptional hubs in cancer, BRD4 and MYC, and to interrogate direct responses to BET bromodomain inhibitors (BETi). We find that BRD4 acts as general co-activator of RNA polymerase II (Pol2)-dependent transcription, which is broadly repressed upon high-dose BETi treatment. At doses triggering selective effects in leukemia, BETi deregulate a small set of hypersensitive targets including MYC. In contrast to BRD4, MYC primarily acts as a selective transcriptional activator controlling metabolic processes such as ribosome biogenesis and de-novo purine synthesis. Our study establishes a simple and scalable strategy to identify direct transcriptional targets of any gene or pathway.
Transcription factors (TFs) and chromatin regulators govern the identity and fate of a cell, and their mutation or dysregulation drives cancer and other human diseases (1). Epigenetic regulators maintaining aberrant cell states have emerged as accessible entry points for targeted therapies (2). Among these, BETi have shown activity in pre-clinical models of leukemia and other cancers (2, 3), yet underlying mechanisms remain poorly understood. While BETi interfere with multiple BET proteins, therapeutic effects have mainly been attributed to displacement of BRD4 from acetylated histones and repression of its target genes. In hematopoietic malignancies, BETi commonly trigger repression of MYC (4–6), an oncogenic TF that is overexpressed in up to 70% of human cancers (7).
Defining direct targets of transcriptional regulators such as BRD4 and MYC is critical, both for understanding their cellular function and for therapy development. However, deciphering direct regulatory relationships remains challenging, since genomic binding of a factor does not predict regulatory functions on neighboring genes, while conventional expression analyses following gene perturbation preclude a clear distinction between direct and indirect effects due to vast differences in mRNA and protein half-lives (8, 9) (fig. S1A). An ideal strategy for defining direct transcriptional targets would combine rapid protein perturbation and subsequent measurement of changes in mRNA output at time-scales that preclude secondary effects.
Thiol(SH)-linked alkylation for the metabolic sequencing of RNA (SLAM-seq) enables the direct quantification of 4-thiouridine (4sU) labeled mRNAs within the total mRNA pool (10). This is achieved through alkylation of the thiol-group in 4sU (fig. S1B), which prompts misincorporation of G during reverse transcription, enabling the detection of 4sU as thymine-to-cytosine (T>C) conversion in 3’-end mRNA-sequencing. To test the suitability of SLAM-seq for detecting immediate and global changes in mRNA production, we measured responses to inhibition of CDK9, a cyclin dependent kinase globally required for releasing Pol2 from promoter-proximal pausing (11). To this end, we treated human K562 leukemia cells with the CDK inhibitor flavopiridol and performed SLAM-seq following 45 min of 4sU labeling (fig. S1C). As expected, only few transcripts showed deregulation at the total mRNA level, while transcripts containing T>C conversions were broadly repressed (fig. S1, D and E). We further optimized the setup to eliminate noise introduced by PCR and sequencing errors (fig. S1F) and to maximize the recovery of labeled reads (fig. S1G). To test whether SLAM-seq captures more specific transcriptional responses, we treated K562 cells with small-molecule inhibitors of their driving oncogene BCR/ABL, as well as MEK and AKT, which act in distinct signaling cascades downstream of BCR/ABL (fig. S2A) (12). SLAM-seq revealed prominent immediate responses to these inhibitors (fig. S2, B and C) that were not biased by mRNA half-lives (fig. S2D). Combined inhibition of MEK and AKT approximated to effects of BCR/ABL inhibition, recapitulating their function as key effector pathways of BCR/ABL (fig. S2, E and F). Together, these pilot studies establish SLAM-seq as a rapid and scalable approach to probe direct transcriptional responses to drug treatment.
To generalize this approach for investigating the vast number of regulators for which, as in the case of BRD4, no selective inhibitors are available, we sought to combine SLAM-seq with chemical-genetic protein degradation (Fig. 1A). To achieve sufficiently rapid kinetics, we employed the auxin-inducible degron (AID) system reported to degrade AID-tagged proteins within less than 1h (13). Specifically, we introduced a minimal AID-tag into the BRD4 locus of K562 cells (Fig. 1B), and transduced homozygous knock-in clones with a lentiviral vector expressing the rice F-box protein Tir1, which mediates ubiquitination of AID-tagged proteins upon treatment with IAA (indole-3-acetic acid). IAA treatment of edited cells triggered a highly specific and near complete degradation of BRD4 within 30 min (Fig. 1B, fig. S3, A to C, and table S1). While introduction of the tag or Tir1 expression and IAA treatment were well tolerated, prolonged BRD4 degradation strongly suppressed cell proliferation (fig. S3, D and E) in line with its essential function (14).
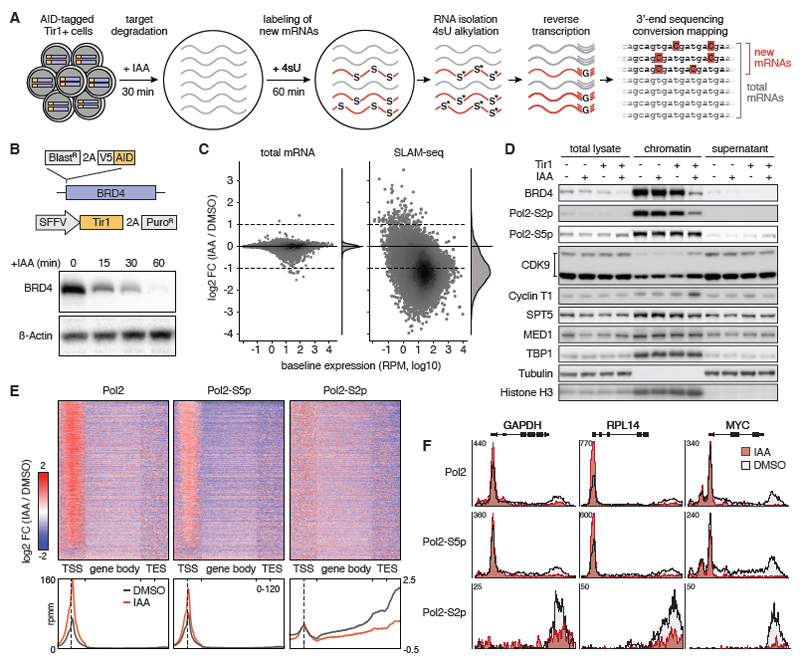
Fig. 1. Global transcriptional control by BRD4.
(A) Sample workflow of a SLAM-seq experiment mapping direct transcriptional responses to degradation of auxin-inducible degron (AID)-tagged proteins. (B) Schematic of the AID-BRD4 knock-in allele and Tir1 delivery vector SOP. Immunoblotting of BRD4 in K562AID-BRD4 + Tir1 cells treated with 100μM IAA for the indicated time points. (C) Changes in the abundance of total and newly synthesized mRNAs (detected in SLAM-seq based on T>C conversions) in K562AID-BRD4+Tir1 cells treated with IAA for 30 min followed by 4sU labeling over 60 min. FC, fold-change. (D) Immunoblotting of indicated transcriptional core regulators and controls in total cell lysate, chromatin fraction and supernatant of K562AID-BRD4+Tir1 cells treated with IAA for 60 min. (E) Spike-in controlled ChIP-seq of hypo-phosphorylated, S2-phosphorylated and S5-phosphorylated Pol2 in K562AID-BRD4+Tir1 cells treated with IAA for 60 min. Heatmaps and density diagrams show change of signals across genes at transcription start sites (TSS, +/- 1kb), gene-bodies (scaled) and transcription end sites (TES, +/- 1kb). A density scale from low (blue) to high (red) is shown. (F) Changes of Pol2 occupancy upon BRD4 degradation shown in (E) for indicated genes.
SLAM-seq following acute BRD4 degradation and 60 min of 4sU labelling revealed a global downregulation of transcription (Fig. 1C and fig. S3F), similar to effects of CDK9 inhibition. These effects are not due to displacement of core transcriptional machinery as loss of BRD4 did not impair chromatin binding of factors involved in transcriptional initiation (TBP1, MED1) or pause-release and elongation (CDK9, Cyclin T1, SPT5) (Fig. 1D). While initiation-associated phosphorylation of Pol2 at serine 5 (S5) of its C-terminal repeat domain was unaffected, BRD4 degradation led to a marked reduction of elongation-associated serine 2 (S2)-phosphorylated Pol2, indicating a defect in promoter proximal pause release. Indeed, spike-in controlled ChIP-sequencing upon BRD4 degradation showed an accumulation of total and S5-phosphorylated Pol2 levels at active transcription start sites (TSS), while total, S5- and S2-phophorylated Pol2 were reduced throughout gene bodies (Fig. 1, E and F, and fig. S4). These results are in line with a recent report showing a widespread reduction of transcription upon pan-BET protein degradation independent of CDK9 recruitment (15), and convincingly show that BRD4 globally controls transcription by promoting the release of stalled Pol2.
While these findings are consistent with the promiscuous binding of BRD4 to active TSS (16), they contrast with selective effects of BETi, which have been widely reported based on results of conventional expression analyses. To define immediate transcriptional responses to BETi, we performed SLAM-seq following treatment with different doses of the BETi JQ1 (17) in K562 and human MV4-11 acute myeloid leukemia (AML) cells. In both cell types, high-dose JQ1 treatment (1 or 5 μM) broadly suppressed transcription (Fig. 2A, fig. S5A) and globally reduced Pol2-S2 phosphorylation (fig. S5B) similar to effects observed after BRD4 degradation, showing that global transcriptional functions of BRD4 are BET bromodomain-dependent. Importantly, effects of high-dose BETi on Pol2-S2 phosphorylation were recapitulated after knockdown of BRD4, but not BRD2 or BRD3 (fig. S5, C and D), indicating that global effects of BETi are primarily mediated by BRD4 inhibition and cannot be compensated by other BET proteins.
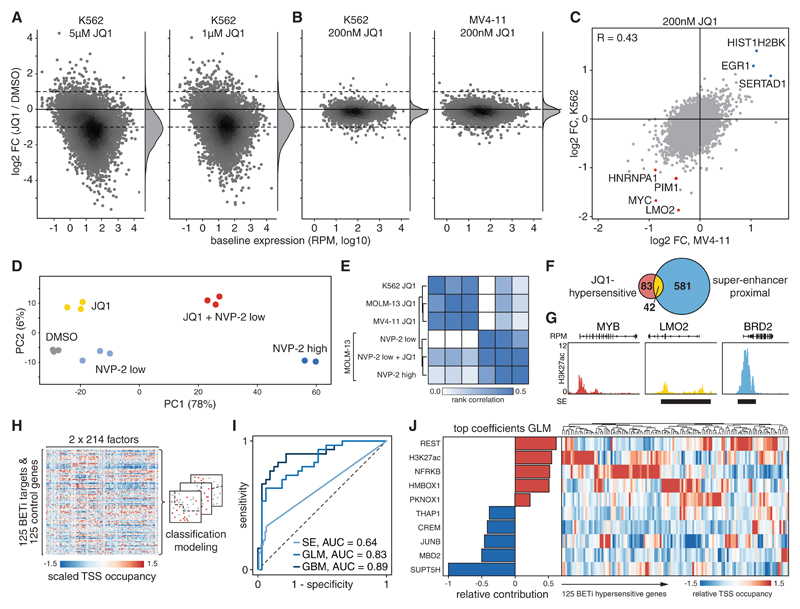
Fig. 2. Dose dependency and determinants of responses to BETi.
(A) SLAM-seq responses of K562 cells treated with indicated doses of JQ1 for 30 min before 4sU labeling for 60 min. (B) SLAM-seq responses of K562 and MV4-11 cells treated with 200nM JQ1 as in (A). (C) Pairwise comparison of SLAM-seq responses to JQ1 shown in (B). R, Pearson correlation coefficient. (D) Principal component analysis of SLAM-seq profiles from MOLM-13 cells treated with JQ1 or NVP-2 as in (A). (E) Heatmap and hierarchical clustering of Spearman’s rank correlations between SLAM-seq responses to JQ1 and NVP-2 in indicated cell lines. (F) Venn diagram showing overlap between BETi-hypersensitive genes and published super-enhancer targets in K562 cells. (G) Sample tracks of H3K27ac ChIP-seq with super-enhancer (SE) annotation exemplifying categories in (F). (H) Simplified model generation workflow for classifying BETi-hypersensitive genes based on 214 chromatin signatures. (I) ROC curve for classification of BETi-hypersensitive genes by super-enhancer assignment or two independent chromatin signature-based models assessed on a held-out test set. (J) Relative contribution of the strongest positive and negative predictors to the GLM shown in (I) based on normalized model coefficients. Heatmap shows relative ChIP-seq densities of these factors at TSS of 125 BETi-hypersensitive genes.
As JQ1 doses above 1μM vastly exceed growth-inhibitory concentrations in AML and other sensitive cell lines, we explored direct transcriptional responses to a more selective dose of 200nM, which triggers strong anti-leukemic effects in a wide range of AML models (4). In K562 cells, one of few BETi insensitive leukemia cell lines, 200nM JQ1 induced a selective deregulation of a small number of transcripts (Fig. 2B). Treatment of two highly sensitive AML cell lines with the same dose triggered transcriptional responses that were comparable in scale (Fig. 2B and fig. S6, A and B) and affected a similar set of BETi-hypersensitive transcripts, including MYC and other genes known to be essential in myeloid leukemia cells (Fig. 2C and fig. S6, C and D) (14). These findings are in line with the notion that sensitivity to BETi at the cellular level is determined by secondary adaptation rather than differences in the primary transcriptional response (18, 19). We also noted a small set of genes that were commonly upregulated following BET inhibition or BRD4 degradation (fig. S6E) through mechanisms that remain elusive. Collectively, our results reveal a profound dose-dependency of direct responses to BETi and show that therapeutically active doses trigger anti-leukemic effects by deregulating a small set of hypersensitive genes.
We next explored whether the BETi hypersensitivity of certain transcripts simply reflects a pronounced sensitivity to interference with general Pol2 pause-release machinery. To test this, we used SLAM-seq to compare transcriptional responses to BET inhibition (200nM JQ1) with effects triggered by different doses of the selective CDK9 inhibitor NVP-2 (20). While high-dose CDK9 inhibition (60nM NVP-2) globally suppressed transcription, an intermediate dose (6nM NVP-2) triggered selective transcriptional responses that were distinct from the conserved response to BETi (Fig. 2, D and E and fig S7, A and B). As CDK9 and BET inhibitors display strong synergistic effects (20) (fig. S7, C and D), we sought to investigate transcriptional responses underlying this phenomenon. In contrast to selective effects seen after single-agent treatment, combining intermediate doses of JQ1 and NVP-2 triggered a global loss of transcription similar to high-dose CDK9 inhibition (Fig. 2, D and E and fig. S7A). These observations hold true in a genetically distinct AML cell line (fig. S7, E and F), suggesting that the therapeutic synergy between BETi and CDK9i is largely based on synergistic suppression of global transcription, raising concerns about toxicities of this combination. These results also suggest that therapeutically active doses of CDK9 and BET inhibitors exploit different bottlenecks in Pol2 pause-release to trigger selective transcriptional responses.
To investigate whether BETi hypersensitivity is determined by specific chromatin features at target genes, we first tested whether BRD4 occupancy levels at TSS or their accessibility to BETi could distinguish direct BETi targets (FDR ≤ 0.1, log2FC ≤ -0.7) from an equally sized cohort of unresponsive genes with identical baseline expression (FDR ≤ 0.1, -0.1 ≤ log2FC ≤ 0.1; fig. S8A). While chromatin occupancy of BRD4 did not predict BETi hypersensitive target genes (AUC 0.52, fig. S8B), recently reported chromatin binding levels of BETi measured by Click-seq could partly account for BETi responses (AUC 0.63; fig. S8C), suggesting that differences in drug accessibility contribute to selective BETi effects. Another model attributes transcriptional and therapeutic effects of BETi to their ability to selectively suppress super-enhancers (16). This notion has been challenged by a recent study identifying H3K27ac-based regulatory potential as a superior predictor of BETi targets (21). As these studies relied on conventional RNA-seq after prolonged drug treatment, we re-evaluated both models using SLAM-seq profiles. Both, the H3K27ac-based regulatory potential of genes, as well as their association with super-enhancers (22), predicted hypersensitivity to BETi with modest accuracy (AUC 0.66 and 0.64, respectively, fig. S8B). However, two-thirds of BETi-sensitive genes could not be assigned to super-enhancers, and the vast majority of expressed super-enhancer-associated genes did not respond to BETi treatment (Fig. 2, F and G). These observations hold true in other leukemia cell lines (fig. S8D) and show that the sensitivity to BET inhibition is associated with, but not determined by, the presence of super-enhancers, suggesting that more complex factors underlie this phenomenon.
To further explore determinants of BETi hypersensitivity, we took advantage of extensive profiling data available for K562 cells (23, 24) and devised an unbiased approach for modeling combinatorial modes of gene regulation. Specifically, we extracted signals of 214 ChIP- and methylome sequencing experiments within 500 and 2000 bp around the TSS of BETi-sensitive and unresponsive genes, and used this data to train various classification models that were later evaluated based on held-out test genes (Fig. 2H and fig. S8E). This approach yielded multiple classifiers predicting BETi sensitivity with high fidelity (AUC > 0.8, Fig 2I and fig. S8F), among them a generalized linear model (GLM) derived by elastic net regression. Re-analyzing coefficients of this model revealed that several factors including high levels of TSS-proximal REST and H3K27ac are associated with BETi hypersensitivity, while high occupancy of SUPT5H, itself a regulator of elongation (25, 26), was the strongest negative predictor (Fig. 2J and fig. S9A). Unsupervised clustering revealed that predictive TFs and co-factors are enriched only at distinct sub-clusters of BETi sensitive or unresponsive genes (Fig. 2J and fig. S9B), suggesting that the transcriptional response to BETi is determined by locus-specific regulators and cannot be predicted based on a single unifying chromatin factor.
Therapeutic effects of BETi are likely mediated through deregulation of multiple hypersensitive genes. While repression of MYC has been identified as a common and relevant effector mechanism in leukemia (4), direct regulatory functions of MYC remain under debate. Previous reports have described activating or repressive effects of MYC on specific target genes, whereas other studies suggest that MYC acts as a general transcriptional amplifier (27–31). To test these models, we sought to measure direct changes in mRNA output following acute loss of endogenous MYC. We therefore engineered an AID-tag into the endogenous MYC locus of K562 cells (Fig. 3A and fig. S10), which in homozygous Tir1-expressing clones allowed for rapid degradation of MYC within less than 30 min (Fig. 3B). We then used SLAM-seq to quantify the output of newly synthesized mRNAs over 60 min following MYC degradation. In contrast to degradation of BRD4, acute loss of MYC resulted in highly specific rather than global changes in mRNA production (Fig. 3C). These were dominated by repressive effects on 712 genes, while only 15 mRNAs were strongly up-regulated (fig. S11A). Hence, in K562 cells, MYC does not act as a direct repressor or general amplifier of transcription, but predominantly functions as a transcriptional activator of specific target genes.
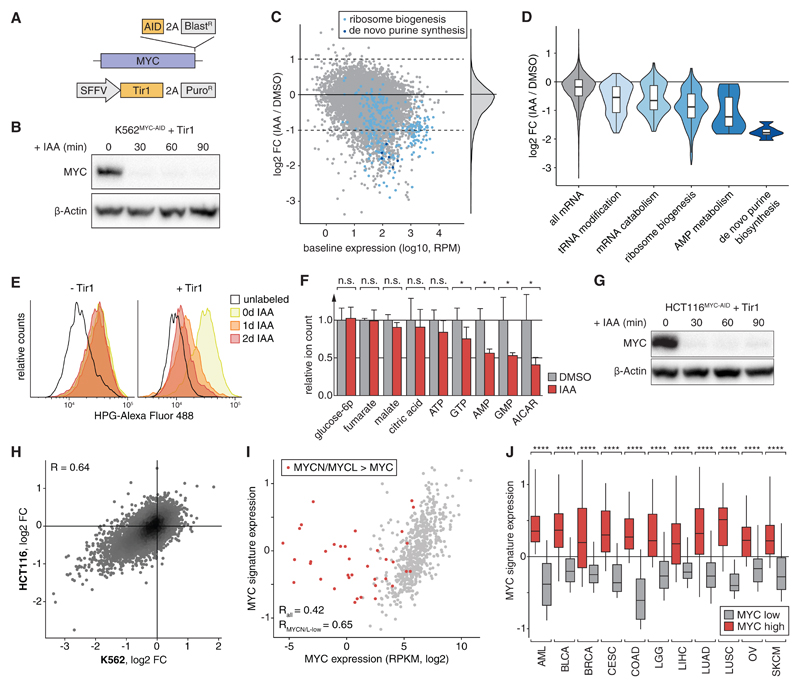
Fig. 3. MYC is a selective transcriptional activator of genes involved in biosynthesis processes.
(A) Schematic of the MYC-AID knock-in allele and Tir1 delivery-vector. (B) Immunoblotting of MYC in K562MYC-AID+Tir1 cells treated with IAA. (C) SLAM-seq profile following MYC-degradation in K562MYC-AID+Tir1 cells (30 min IAA treatment, 60 min 4sU-labeling). Highlighted are ribosome biogenesis factors (light blue) and de-novo purine synthesis enzymes (dark blue). (D) Violin plots depicting SLAM-seq responses of significantly enriched gene ontology classes. (E) Measurement of global protein synthesis by L-homopropargylglycine (HPG)-incorporation and flow cytometry in K562MYC-AID cells treated with IAA. (F) Targeted mass spectrometry quantification of indicated metabolites in K562MYC-AID+Tir1 cells after 48h of IAA treatment. Bars show means of 3 independent experiments. Error bars indicate one standard deviation. (G) MYC-immunoblotting in HCT116MYC-AID+Tir1 cells as in (B). (H) Comparison of SLAM-seq responses in K562MYC-AID+Tir1 and HCT116MYC-AID+Tir1 cells. (I) Expression of MYC compared with a signature of the top 100 common MYC-dependent transcripts in (H) across 672 cancer cell lines. (J) MYC-target signature expression across 5583 patient samples separated based on high (top 20%) or low (bottom 20%) MYC-expression and cancer type. ****, p<0.0001 (Wilcoxon’s rank-sum test).
Since MYC is known to occupy most active promoters (27), we next investigated how MYC exerts selective transcriptional activation despite ubiquitous binding. To this end, we trained classification models to predict MYC-dependent transcripts (FDR ≤ 0.1, log2FC ≤ -1) based on different ChIP-seq signals at their promoter. Elastic net regression yielded a simple GLM that was highly predictive of MYC-dependent gene regulation (AUC 0.91, fig. S11B). The strongest contributor in this model was the abundance of MYC itself (fig. S11C). Indeed, while the presence of MYC at promoters determined by conventional peak calling fails to identify MYC-sensitive transcripts, binding levels of MYC or its co-factor MAX predict MYC-dependent gene regulation with intermediate accuracy (AUC 0.76 and 0.74, respectively; fig. S11, D and E). These results suggest that genes directly activated by MYC are defined by strong binding of MYC and by further modulation through additional factors such as MNT, NKRF, TBL1XR1, EP300 and YY1.
To investigate the cellular function of MYC-dependent gene regulation, we performed gene ontology analysis of direct MYC target genes in K562 cells. Strikingly, acute MYC degradation predominantly led to down-regulation genes associated with protein and nucleotide biosynthesis, including 36% of all ribosome biogenesis factors, key regulators in AMP metabolism, and all six enzymes of the de-novo purine synthesis pathway (Fig. 3, C and D and table S2). Indeed, MYC degradation progressively impaired protein synthesis (Fig. 3E) and led to a strong reduction in cellular AMP and GMP levels as well as their upstream intermediate AICAR prior to the onset of cell proliferation defects (Fig. 3F and fig. S12, A and B). MYC’s role as a direct regulator of key enzymes in protein and nucleotide biosynthesis, as well as several subunits of polymerases I, II and III (fig. S12C), provide an explanation for the reported increase in total cellular RNA upon MYC overexpression and support the notion that these effects are secondary rather than due to global transcriptional effects (32).
To test whether direct transcriptional functions of MYC are conserved in other cellular contexts, we introduced homozygous AID-tags into the MYC locus of HCT116 human colon carcinoma cells. As with K562 cells, IAA treatment of TIR1-expressing HCT116MYC-AID cells triggered complete degradation of MYC within less than 30 min (Fig. 3G). SLAM-seq profiling revealed a selective transcriptional response (Fig. 3H and fig. S12D) that affected the same cellular processes (Fig. S12E) and correlated with effects observed in K562 cells (R = 0.64, Fig 3H). To test whether the conservation of MYC targets extends to other cancer types, we derived a signature of the 100 most strongly downregulated genes in SLAM-seq (table S3) and compared its expression with MYC levels in a panel of 672 cancer cell lines (33). Indeed, expression of MYC and our MYC target signature correlated well (Fig. 3I), except for a small fraction of outliers. Notably, all of these express high levels of MYCN or MYCL (fig. S13A), indicating that MYC paralogs have redundant functions in the regulation of core MYC targets. Our signature of direct MYC targets was also strongly correlated with MYC levels in publicly available RNA-seq profiles from 5583 primary patient samples across 11 major human cancers (Fig. 3J, fig. S13B) (34). Together, these findings suggest that MYC drives expression of a conserved set of transcriptional targets, which should be considered as entry points for blocking its oncogenic functions.
In summary, combining rapid chemical-genetic perturbation and SLAM-seq establishes a simple yet powerful strategy for probing specific and global direct functions of transcription factors and co-factors. Using this approach, we functionally characterize BRD4, a protein widely studied as a regulator of lineage- and disease-associated expression programs, as a general co-factor in transcriptional pause-release. We also find that MYC, which has previously been implicated as a global transcriptional amplifier, activates a confined and conserved set of target genes to fuel basic anabolic processes, particularly protein and nucleotide biosynthesis. More generally, by enabling the direct quantification of changes in mRNA output, SLAM-seq provides a simple, robust and scalable method for defining direct transcriptional responses to any perturbation, and thereby explore the regulatory wiring of a cell.
Supplementary Material
Acknowledgments
We are grateful to all members of the Zuber and Ameres labs, A. Stark, F. Muerdter, M. Rath and D. Kaiser for experimental advice and helpful discussions. We thank A. Sommer, I. Tamir and the VBCF-NGS team (www.vbcf.ac.at) for deep sequencing services, G. Dürnberger, K. Mechtler, E. Roitinger and M. Schutzbier at the IMP/IMBA protein biochemistry core facility for performing MS-based proteomics, and the IMP/IMBA bio-optics and molecular biology services for continuous support.
Funding: This work was funded by Starting Grants of the European Research Council to J.Z. (ERC-StG-336860) and S.L.A. (ERC-StG-338252) and the Austrian Science Fund (SFB grants F4710 and F4322, Y-733-B22 START and W127-B09). M.M. is recipient of a DOC fellowship of the Austrian Academy of Sciences. Research at the IMP is generously supported by Boehringer Ingelheim. VBCF is funded by the City of Vienna through the Vienna Business Agency.
Footnotes
Author contributions: M.M., S.L.A. and J.Z. conceived and planned this project. M.M., A.E., C.U., C.W., T.K. and J.P. designed and conducted experiments. M.M., A.E., T.N., S.L.A. and J.Z. analyzed and interpreted original and publicly available data. T.N. established and performed deep sequencing data analyses. J.J., P.R., V.A.H., B.R., D.A.C., T.H., P.B. and M.F.S. established critical reagents and methodology. J.J.L., A.v.H., A.C.O. and J.P. provided critical input on experimental designs and data analyses. M.M. and J.Z. co-wrote the manuscript with input from co-authors.
Competing interests: S.L.A., B.R., V.A.H., J.Z. and M.M. are inventors on patent application EU17166629.0-1403 submitted by the Institute of Molecular Biotechnology (IMBA) that covers methods for the modification and identification of nucleic acids, which have been licensed to Lexogen GmbH.
Data and materials availability: All deep sequencing data is available through GEO (https://www.ncbi.nlm.nih.gov/geo) under the accession code GSE111463. Materials will be provided upon request under a material transfer agreement with the IMP.
References and Notes
- 1.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones PA, Issa J-PJ, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17:630–641. doi: 10.1038/nrg.2016.93. [DOI] [PubMed] [Google Scholar]
- 3.Shi J, Vakoc CR. The Mechanisms behind the Therapeutic Activity of BET Bromodomain Inhibition. Mol Cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dawson MA, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–33. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Housden BE, et al. Loss-of-function genetic tools for animal models: cross-species and cross-platform differences. Nat Rev Genet. 2017;18:24–40. doi: 10.1038/nrg.2016.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 10.Herzog VA, et al. Thiol-linked alkylation of RNA to assess expression dynamics. Nat Methods. 2017;14:1198–1204. doi: 10.1038/nmeth.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rahl PB, et al. C-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren R. Mechanisms of BCR–ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer. 2005;5:172–183. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- 13.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–22. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 14.Wang T, et al. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winter GE, et al. BET Bromodomain Proteins Function as Master Transcription Elongation Factors Independent of CDK9 Recruitment. Mol Cell. 2017;67:5–18.e19. doi: 10.1016/j.molcel.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lovén J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–34. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rathert P, et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature. 2015;525:543–547. doi: 10.1038/nature14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fong CY, et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature. 2015;525:538–42. doi: 10.1038/nature14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu H, et al. Compensatory induction of MYC expression by sustained CDK9 inhibition via a BRD4-dependent mechanism. Elife. 2015;4:1–26. doi: 10.7554/eLife.06535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S, et al. Modeling cis-regulation with a compendium of genome-wide histone H3K27ac profiles. Genome Res. 2016;26:1417–1429. doi: 10.1101/gr.201574.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mei S, et al. Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res. 2017;45:D658–D662. doi: 10.1093/nar/gkw983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wada T, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–56. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shetty A, et al. Spt5 Plays Vital Roles in the Control of Sense and Antisense Transcription Elongation. Mol Cell. 2017;66:77–88.e5. doi: 10.1016/j.molcel.2017.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin CY, et al. Transcriptional Amplification in Tumor Cells with Elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nie Z, et al. c-Myc Is a Universal Amplifier of Expressed Genes in Lymphocytes and Embryonic Stem Cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sabò A, et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014;511:488–92. doi: 10.1038/nature13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walz S, et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014;511:483–487. doi: 10.1038/nature13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorenzin F, et al. Different promoter affinities account for specificity in MYC-dependent gene regulation. Elife. 2016;5:1–35. doi: 10.7554/eLife.15161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kress TR, Sabò A, Amati B. MYC: connecting selective transcriptional control to global RNA production. Nat Rev Cancer. 2015;15:593–607. doi: 10.1038/nrc3984. [DOI] [PubMed] [Google Scholar]
- 33.Klijn C, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol. 2015;33:306–12. doi: 10.1038/nbt.3080. [DOI] [PubMed] [Google Scholar]
- 34.Grossman RL, et al. Toward a Shared Vision for Cancer Genomic Data. N Engl J Med. 2016;375:1109–1112. doi: 10.1056/NEJMp1607591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen B, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–91. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wutz G, et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 2017;36:3573–3599. doi: 10.15252/embj.201798004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fellmann C, et al. An Optimized microRNA Backbone for Effective Single-Copy RNAi. Cell Rep. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 38.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramírez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orlando DA, et al. Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep. 2014;9:1163–70. doi: 10.1016/j.celrep.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 42.Kuhn M. Building Predictive Models in R Using the caret Package. J Stat Softw. 2008;28:1–27. [Google Scholar]
- 43.Dorfer V, et al. MS Amanda, a universal identification algorithm optimized for high accuracy tandem mass spectra. J Proteome Res. 2014;13:3679–84. doi: 10.1021/pr500202e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923–5. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 45.Smyth GK. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Stat Appl Genet Mol Biol. 2004;3:1–25. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.