Summary
Tumors display profound changes in cellular metabolism, yet how these changes aid the development and growth of tumors is not fully understood. Here we use a multi-omic approach to examine liver carcinogenesis and regeneration, and find that progressive loss of branched-chain amino acid (BCAA) catabolism promotes tumor development and growth. In human hepatocellular carcinomas and animal models of liver cancer, suppression of BCAA catabolic enzyme expression led to BCAA accumulation in tumors, though this was not observed in regenerating liver tissues. The degree of enzyme suppression strongly correlated with tumor aggressiveness, and was an independent predictor of clinical outcome. Moreover, modulating BCAA accumulation regulated cancer cell proliferation in vitro, and tumor burden and overall survival in vivo. Dietary BCAA intake in humans also correlated with cancer mortality risk. In summary, loss of BCAA catabolism in tumors confers functional advantages, which could be exploited for therapeutic interventions in certain cancers.
Graphical Abstract

Using a multi-omic approach to characterize normal, regenerating, and cancerous liver tissues in mice and humans, Ericksen et. al. find that BCAA catabolism is lost during tumor development and progression, but maintained in normal proliferating cells. Tissue BCAA content is also influenced by dietary BCAA intake and affects mTORC1 activity.
Introduction
The metabolic reprogramming of cancer has been observed for decades (Pavlova and Thompson, 2016; Vander Heiden et al., 2009), yet whether this reprogramming is a general aspect of proliferation, an unintended consequence of aberrant signaling pathways, or functionally involved in the oncogenic processes remains poorly understood. Previous reports have examined numerous metabolic alterations, demonstrating that they support tumor growth by meeting the energetic, biosynthetic, and redox needs of rapidly dividing cancer cells (Pavlova and Thompson, 2016). However, further examination has frequently shown that many of the same pathways are also used by normal proliferating cells (Pearce and Pearce, 2013), and therefore targeting these may not provide therapeutic effects without unintended consequences. The liver is one of the few organs that have the capacity to completely regenerate after partial resection. Thus, analysis of liver regeneration and oncogenesis provides a unique opportunity to understand both benign and malignant proliferative processes, respectively.
Liver cancer itself is a major health burden as the second leading cause of cancer-related death (Kladney et al., 2010). Predominantly of the hepatocellular carcinoma (HCC) subtype, tumors can develop due to a range of etiological factors, including Hepatitis B (HBV) or Hepatitis C (HCV) infection, alcohol consumption, as well as metabolic perturbations leading to non-alcoholic fatty liver disease (NAFLD) and non-alcoholic steatohepatitis (NASH). HCC is typically diagnosed late in the disease progression where treatment options are limited, causing disease-free survival rates to remain dismal (Yang and Roberts, 2010). Recent genetic profiling has shown that HCCs display a preponderance of mutations affecting p53, the Wnt-β-catenin pathway, and the PI3K-AKT-mTOR pathway (Ally et al., 2017; Schulze et al., 2015). However, targeting these for effective preventive or therapeutic interventions has remained difficult and largely unsuccessful. Thus, identification of additional factors or alternative methods for intervention remains a high priority. In an attempt to identify targetable tumor-specific pathways, we used unbiased comprehensive transcriptomic and metabolomic analyses to characterize human tumors, cancer cell lines, and animal models of liver cancer and regeneration.
Results
Progressive Loss of BCAA Catabolism in Hepatocellular Carcinoma Development and Progression
First, we analyzed a cohort of 48 HCC patients seen at Singapore General Hospital (Figure S1A). Transcriptomic profiling and differential expression analysis of matching tumor and adjacent nontumor liver tissue samples returned 3624 significant genes (Figure S1B). To help focus the large gene set, as well as validate the findings and ensure robustness, we compared expression changes to an independent, well-characterized HCC cohort from The Cancer Genome Atlas (TCGA-LIHC) (Ally et al., 2017). As expected, there was an overall direct correlation between the two datasets, and use of a stringent significance threshold (p<1×10−8) returned 1405 genes that were potently altered in both cohorts (Figures 1A and 1B). KEGG pathway analysis of this gene set identified branched-chain amino acid (BCAA) degradation as the most significant pathway (Figure 1C). Approximately 40 enzymes are involved in BCAA catabolism, and with exception of the reversible transamination step performed by BCAT1 and BCAT2 (Figures S1C–S1E), we found that the transcripts were broadly suppressed in tumors (Figure 1D). The first two critical irreversible steps of BCAA catabolism involve the branched-chain ketoacid dehydrogenase (BCKDH) complex and acyl-CoA dehydrogenase (ACAD) enzymes (Shin et al., 2014). Western blot and immunohistochemistry analysis demonstrated that tumors had a potent and robust downregulation of proteins involved in these catalytic steps, namely BCKDHA, ACADS, and ACADSB (Figure 1E). Biopsies from an additional independent cohort immunostained by The Human Protein Atlas (Uhlen et al., 2015) confirmed that the BCAA catabolic enzymes are broadly downregulated at the protein level (Figures 1F and S1F). Accordingly, tumors displayed a sharp reduction of BCKDH complex activity in an ex vivo enzymatic assay (Figure 1G).
Figure 1. BCAA Catabolism Is Suppressed During Hepatocellular Carcinoma Development and Progression.
(A) Comparison of gene expression changes in hepatocellular carcinomas (HCCs) relative to adjacent nontumor liver tissues, collected at Singapore General Hospital (SGH) or characterized by the cancer genome atlas (TCGA-LIHC).
(B) 1405 genes were similarly altered in both cohorts with high significance (p<1×10−8).
(C) KEGG pathway analysis of the 1405 gene set.
(D) Summary of branched-chain amino acid (BCAA) catabolic enzyme transcript levels across HCCs and nontumor liver tissues of both cohorts.
(E) Immunoblots of selected BCAA catabolic enzymes from paired HCCs and nontumor liver tissues from patients of the SGH cohort.
(F) Representative immunohistochemical micrographs from nontumor liver tissue and HCC biopsies, as profiled by The Human Protein Atlas.
(G) Ex vivo tissue BCKDH complex activity from paired HCCs and nontumor liver tissues from patients of the SGH cohort (n=6 per group).
(H) Summary of all significantly different metabolites from targeted metabolomics analyses from paired HCCs and nontumor liver tissues from patients of the SGH cohort (n=7 per group).
(I) Summary of BCAA catabolic enzyme transcript levels in HCCs from the SGH and TCGA cohorts, sorted by stage, grade, metastasis, vascular invasion, and local invasion.
(J) Kaplan-Meier survival estimate curves for patients ranked by a combined index of tumor BCKDHA, ACADS, and ACADSB expression. P-values for log-rank test and cox proportional hazard ratios (HR, with 95% percent confidence intervals) adjusted for age, sex, tumor stage and grade, and radiation, prescription and additional therapies shown.
*P<0.05, compared to respective controls. Data are shown as mean ± s.e.m.
See also Figure S1.
Next, we performed targeted metabolomic analyses for amino acids, organic acids, and acylcarnitines on the paired tumor and adjacent nontumor liver tissues (Figures 1H and S1G). Among the noteworthy changes in tumors, we observed a significant increase in lactate, acetyl-carnitine, and numerous long chain acylcarnitines, as well as a significant decrease in multiple medium chain acylcarnitines (Figure 1H). Importantly, there was also a significant increase of all three BCAAs, and a significant decrease of downstream BCAA-derived metabolites in tumors (e.g. C3 acylcarnitine), which is consistent with a decreased BCAA catabolic flux.
Notably, the transcript levels of BCAA catabolic enzymes were not only suppressed in tumors when compared to adjacent normal tissue, but the degree of suppression correlated with multiple indicators of disease progression and tumor aggressiveness, including stage, grade, vascular invasion, lymph node invasion, and distant metastasis (Figure 1I). We also examined whether tumor etiology (e.g. Hepatitis B-, Hepatitis C-, alcohol-, and NAFLD-associated), liver inflammation, and patient race/ethnicity had any influence on BCaA catabolic enzyme expression. However, after factoring in differences in tumor grade, stage, and invasion, all groups had comparable changes (Figure S1H).
Expression levels of the catabolic enzymes were also strongly associated with clinical outcome, even when adjusting for patient and tumor characteristics (Figure S1I). Maintaining high expression of these enzymes, therefore allowing efficient catabolism of BCAAs, was associated with significantly better patient outcome. Surveying the prognostic utility of aggregating multiple (up to 5) genes identified the combined expression index of BCKDHA, ACADS, and ACADSB as the most robust predictor of patient survival in both cohorts (Figure 1J). The bifurcation in patient survival was most dramatic among high-grade tumors, with a 54-point difference in survival at 5 years (88% versus 34%; Figure 1J). Thus, the transcriptomic, enzymatic, and metabolomic analyses suggest that suppression of catabolic enzymes in HCC spare the BCAAs from degradation, leading to their accumulation in tumors. The data also suggest that the loss of BCAA catabolic enzyme expression in tumors is positively selected for during disease progression, and that it has a significant impact on patient outcome.
BCAA Catabolism is Suppressed in Tumors, but Not Regenerating Liver Tissues
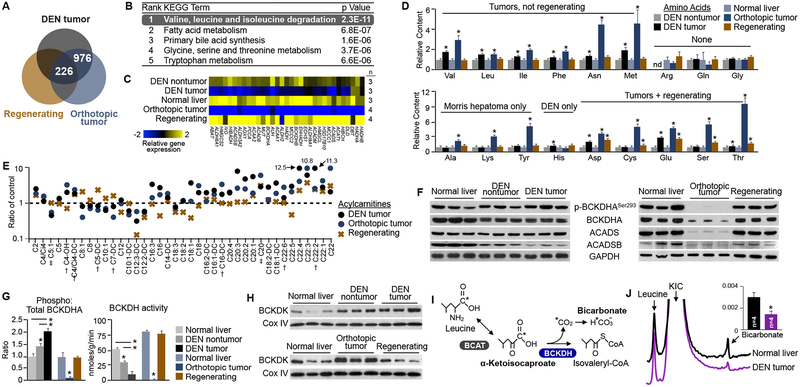
Given that the metabolic needs of proliferating cancer cells are potentially similar to normal proliferating hepatocytes, we questioned whether the suppression of BCAA catabolism was unique to tumors, or also observed in liver regeneration. Thus, we examined multiple in vivo animal models and performed similar comprehensive transcriptomic and metabolomic analyses. Tumor and nontumor tissues were harvested from animals administered diethylnitrosamine (DEN), as this compound generates endogenous tumors resembling poor-prognosis HCC by causing multiple stochastic DNA mutations in the liver (Heindryckx et al., 2009). We also used the syngeneic orthotopic Morris Hepatoma 3924a model to represent aggressive, fast-growing tumors. KEGG analysis of all significant, differentially expressed genes shared among the tumor models confirmed that BCAA catabolism ranked as the top pathway (Figure S2A). Importantly, BCAA catabolism remained the most significant pathway even after omitting genes differentially expressed in proliferating hepatocytes after partial hepatectomy (Figures 2A, 2B and S2B–F). Indeed, while tumors had reduced expression, regenerating tissues actually enhanced the expression of BCAA catabolic enzymes modestly (Figure 2C). An initial non-targeted screen of over 200 metabolites identified five that had accumulated in liver tumors with high significance, but not regenerating tissues, and among these were all three BCAAs (Figure S2G). Subsequent targeted analyses confirmed that the BCAAs and only three other amino acids (phenylalanine, methionine, and asparagine) had accumulated in both tumor models but not regenerating tissues (Figure 2D). Accordingly, there was a significant decrease of downstream BCAA-derived metabolites (e.g. C5:1 acylcarnitine) in tumors but not regenerating tissues (Figure 2E).
Figure 2. Loss of BCAA Catabolism Occurs in Liver Tumors, but Not Regenerating Liver Tissues.
(A-E) Transcriptomic and metabolomic characterization of animal tumor models. DEN-induced tumors are compared to DEN nontumor tissue, and orthotopic tumors (Morris Hepatoma) and regenerating liver tissue after partial hepatectomy are compared to normal liver tissue. (A) Summary of significant, differentially-expressed genes. (B) KEGG pathway analysis of the 976 genes differentially expressed in tumors but not regenerating tissues. (C) Summary of BCAA catabolic enzyme transcript levels across normal, tumor, and regenerating tissues. (D) Quantification of tissue amino acid content, normalized to respective controls. nd=not detected. (E) Quantification of tissue acylcarnitine content, normalized to respective controls. Only acylcarnitines that trended in the same direction in both tumor models are shown. †P<0.05, in 1 of 2 tumor models, ‡P<0.05 in both tumor models, compared to respective controls, while not significantly different in regenerating tissue.
(F) Immunoblots of selected BCAA catabolic enzymes in normal, tumor, and regenerating liver tissues.
(G) Quantification of phospho:total BCKDHA ratio, and corresponding ex vivo tissue BCKDH complex enzymatic activity.
(H) Immunoblots of mitochondrial fractions from normal, tumor, and regenerating liver tissues.
(I) Schematic of magnetic resonance spectroscopy (MRS)-based in vivo BCKDH complex activity assay. Hyperpolarized [1-C13] α-ketoisocaproate was injected by tail vein, and enzyme activity was assessed by detection of labeled bicarbonate in the liver in live, anesthetized animals.
(J) Representative liver MRS spectra after intravenous injection of hyperpolarized [1-C13]α-ketoisocaproate (KIC). Baselines are shifted to display the difference in bicarbonate peak. Quantification of relative bicarbonate levels over a 30 second interval inset.
*P<0.05, compared to respective controls; Data are shown as mean ± s.e.m.
See also Figure S2.
We next examined the protein levels and enzymatic activity of normal, tumor and regenerating liver tissues. Of note, in addition to suppression of total protein levels, inhibition of BCKDH complex’s activity can also be achieved by phosphorylation of the BCKDHA subunit by the kinase BCKDK (Lynch and Adams, 2014; Shin et al., 2014). We observed that while Morris Hepatoma tumors had sharp reductions in total protein levels of the BCAA catabolic enzymes, DEN-induced tumors displayed more overt changes in BCKDHA phosphorylation (Figure 2F). Relative to normal (DEN-free) liver tissues, the phospho/total BCKDHA ratios were elevated in DEN-exposed pre-tumor tissues, and even further increased in tumors (Figures 2G and S2H). Correspondingly, BCKDK was overexpressed in pre-tumor and tumor tissues of the animal tumor models, as well as human HCC (Figures 2H and S2I). BCKDH enzyme activity assays confirmed that regardless of the primary method of suppression, all animal tumor tissues tested had significantly reduced catabolic capacity (Figures 2G and S2J). Importantly, phospho/total BCKDHA ratios, BCKDH activity, and BCKDK expression remained unchanged in regenerating tissues (Figures 2G and 2H). Based on the concept of the ex vivo BCKDH assay, we also developed a hyperpolarized 13C magnetic resonance spectroscopy method (Lee et al., 2013) to quantify enzyme activity in vivo (Figure 2I). Using this platform, we detected significantly reduced BCKDH activity in the livers of rats bearing DEN-induced tumors (Figures 2J and S2K), indicating that this method can be used to noninvasively monitor enzyme activity in live subjects. Overall, these data demonstrate that in contrast to liver tumorigenesis, proliferating hepatocytes of the regenerating liver do not display decreased BCAA catabolism or an increase in BCAA content.
BCAA Catabolic Enzyme Expression in Tumors is Associated with Copy Number and Transcription Factor Changes
To understand what may govern the broad and robust change in BCAA catabolic enzymes, we next examined potential factors regulating their expression. No significant difference in promoter methylation of the BCAA catabolic enzymes or associations with changes in microRNA expression was observed (data not shown). In contrast, we noted that the most potent decrease in BCAA catabolic enzyme mRNA expression was from tumors with somatic copy number variation (CNV) loss (Figures 3A and 3B). The frequency of CNV loss varied by gene, but on average affected approximately 20% of HCCs (Figure 3B). However, tumors with normal CNVs still displayed reduced gene expression relative to normal tissues, suggesting that additional regulatory mechanisms are possibly involved (Figure 3A). Thus, to identify potential regulating transcription factors, we performed upstream analysis using all significant, differentially expressed genes in human liver cancers and animal tumor models, as well as specifically the BCAA catabolic enzymes. Ingenuity Pathway Analysis (IPA) predicted significant changes in the activity of multiple metabolic transcription factors, most notably PPAR α (Figure S3A). PPAR α was also among a subset of transcription factors whose expression correlated with tumor aggressiveness and patient survival (Figures S3B and S3C). TRANSFAC analysis demonstrated that PPAR α binding motifs were enriched in the promoters of genes differentially expressed in liver cancers, in particular the BCAA catabolic enzymes (Figure S3D). ENCODE ChIP-seq data also confirmed that PPAR α bound to these promoters in the basal state (Figure S3E).
Figure 3. Loss of BCAA Catabolic Enzyme Expression Is Associated with Changes in Copy Number Variation and Transcription Factor Expression/Activity.
(A) Summary of BCAA catabolic enzyme transcript levels in nontumor liver tissues, and tumors with or without CNV loss of indicated gene.
(B) Distribution of CNV losses of the BCAA catabolic enzymes in the TCGA-LIHC cohort, sorted by unsupervised hierarchical clustering.
(C) Summary of BCAA catabolic enzyme expression in nontumorigenic (HepG2) and tumorigenic (remaining) hepatoma/HCC-derived cell lines.
(D) Immunoblots of selected BCAA catabolic enzymes across the cell line panel. (E) RT-PCR analysis of BCAA catabolic enzymes from HepG2 cells treated with the PPARα antagonist GW6471 for 48 hours.
*P<0.05, compared to respective controls. Data are shown as mean ± s.e.m.
See also Figure S3.
To further investigate expression changes, we examined a panel of human liver-derived cell lines. Of note, expression patterns of the BCAA catabolic enzymes in the cell lines were consistent with human and animal liver cancers. Specifically, relative to the well-differentiated, nontumorigenic cell line HepG2, the tumorigenic HCC cell lines had reduced expression of the BCAA catabolic enzymes at both the mRNA and the protein level (Figures 3C and 3D). Consistent with the upstream analyses, knocking down PPAR α, or repressing its transactivation with the antagonist GW6471 significantly suppressed BCAA catabolic enzyme expression in HepG2 cells (Figures 3E and S3F). Collectively, these data suggest that changes in somatic copy number and transcription factor function likely account for the observed downregulation of BCAA catabolic enzyme expression in liver tumors.
BCAA Catabolism Regulates mTOR Activity and In Vitro Cell Proliferation
Like numerous other cancers, HCCs frequently harbor mutations in proteins that mimic chronic growth factor stimulation to the mammalian target of rapamycin complex 1(mTORC1) (Ally et al., 2017; Sanchez-Vega et al., 2018; Schulze et al., 2015). Previous reports have shown that activation of mTORC1 potently enhances cell growth and tumorigenesis in numerous human cancers and animal tumors models, including liver cancer (Kenerson et al., 2013; Menon et al., 2012; Villanueva et al., 2008). Consistent with this, we observed that the human HCCs and animal tumor models we analyzed overexpressed and/or displayed hyperactivation (i.e. hyperphosphorylation) of the downstream mTORC1 effectors S6K and S6 (Figures 4A and S4A). Moreover, gene set enrichment analysis (GSEA) demonstrated that active Mtorc1-related signatures were enriched in tumors of both human HCCs and the animal tumors models (Figure S4B).
Figure 4. BCAA Catabolism Regulates mTORC1 Activity and in vitro Cell Proliferation.
(A) Immunoblots of mTORCl downstream effectors (S6K and S6) and their activation states (p-S6KThr389 and p-S6ser235/236) in paired HCCs and nontumor liver tissues from patients of the SGH cohort.
(B) Real-time proliferation curves, immunoblots detailing knockdown efficiency (BCKDHA) and mTORC1 activity, and intracellular BCAA content of AML12 cells expressing a tet-inducible BCKDHA shRNA in the absence or presence of doxycycline and/or the mTOR inhibitors rapamycin (0.05nM) or Torin 1(0.5nM).
(C) Real-time proliferation curves and immunoblots of Hep3B cells grown in media with reduced levels of BCAAs.
(D) Real-time proliferation curves and immunoblots of Hep3B cells overexpressing Flag-tagged BCKDHA, ACADS, or ACADSB.
(E) Real-time proliferation curves and immunoblots of control and CRISPR-Cas9-mediated BCKDK null Hep3B clones.
(F) Real-time proliferation curves of Hep3B cells treated with the BCKDK inhibitor BT2, and immunoblots after 2 hours of BT2 treatment.
(G) Calculation of proliferation rates (number of divisions per day) for HCC cell lines treated with BT2.
(H) Colocalization of mTOR and the lysosomal marker LAMP2 in Hep3B cells grown in nutrient sufficient (control) media, after 1 hour of amino acid withdrawal, or after 2 hours of BT2 treatment (150μM). Quantification displays Manders overlap of 20 random cells.
(I) Immunoblots of 293T cells expressing control vectors (pLOC or Flag-Raptor) or constitutively active components of mTORCbrelated nutrient sensing complexes (HA-RagBS75L/HA-RagCQ99L or Flag-Raptor-Rheb 15) treated with vehicle or indicated concentration of BT2 for 2 hours.
(J) Immunoblots of 293T cells with GFP or Sestrin 1, 2, and 3 knockdown, treated with vehicle or indicated concentration of BT2 for 2 hours.
*P<0.05, compared to respective controls. Data are shown as mean ± s.e.m.
See also Figure S4.
mTORC1 integrates signals from both growth factors and nutrient abundance, and removal of either input is sufficient to block its activity (Sancak et al., 2008). Significant progress has been made recently in identifying the protein complexes and signaling cascades involved in mTORC1-related nutrient sensing (Shimobayashi and Hall, 2016). Specifically, key metabolites lead to the activation of the Rag proteins, which recruit mTORC1 to the lysosome so that Rheb can activate mTOR (Sancak et al., 2008). Leucine in particular has a unique role in regulating this process in that its removal, even in the presence of other amino acids, inactivates the Rag proteins (Sancak et al., 2008). While mutations activating the growth factor arm of mTORC1 are relatively common, whether the nutrient sensing arm of mTORC1 is altered in tumors has not been thoroughly explored previously. Examining the TCGA datasets, we observed that mutations impacting the currently identified proteins of mTORC1’s nutrient sensing arm are extremely rare in HCC and other cancers (Figure S3G). Thus, we hypothesized that reduced BCAA catabolism and enhanced tissue BCAA accumulation may be a primary mechanism utilized by tumors to facilitate the chronic activation of mTORC1. To examine this possibility, we undertook a series of in vitro experiments manipulating the BCAA catabolic and mTORC1 pathways. First, to recapitulate the loss of BCAA catabolic enzyme activity in tumors, we targeted a key subunit of the BCKDH complex in the immortalized hepatocyte cell line AML12. Consistent with our hypothesis, knocking down BCKDHA with inducible shRNAs raised intracellular BCAA content, enhanced mTORC1 activity, and increased cellular proliferation rates (Figures 4B and S4C). Moreover, normalizing mTORC1 activity with the inhibitors Rapamycin or Torin 1 returned proliferation rates to baseline levels (Figures 4B and S4C).
BCAAs are essential amino acids and therefore not produced endogenously. Thus, we examined whether restricting the supply or restoring the catabolism of BCAAs influenced liver cancer cell proliferation in vitro. Typical cell culture media preparations have BCAAs in far excess of physiological levels, and reducing or removing BCAAs significantly suppressed mTORC1 activity and proliferation rates of the HCC cell line Hep3B (Figure 4C). In contrast to Kras-driven pancreatic cancer cells (Palm et al., 2015), supplementing Leucine- or BCAA-starved liver cancer cells with albumin or other protein sources (either alone or in combination with the mTOR inhibitors Rapamycin or Torin 1)failed to rescue proliferation rates (Figure S4D). Similar to restricting the supply of BCAAs, restoring expression of the BCAA catabolic enzymes BCKDHA, ACADS, or ACADSB also caused mTORC1 activity and cell proliferation to significantly decrease (Figure 4D).
Given the enhanced expression of BCKDK in animal tumor models and human HCC, and the attractiveness of kinases as drug targets, we also examined whether inhibiting the activity of this kinase could restore BCAA catabolism and suppress cancer cell proliferation. Indeed, CRISPR-Cas9-mediated knockout of BCKDK in Hep3B cells reduced proliferation rates and mTORC1 activity (Figures 4E and S4E). As a complementary approach, we utilized BT2, an established compound that specifically inhibits BCKDK (Tso et al., 2014). As expected, treating Hep3B cells with BT2 also caused a dose-dependent decrease in cell proliferation and mTORC1 activity (Figure 4F). The suppression of mTORC1 activity and cell proliferation was also consistent across the panel of HCC cell lines, and cell line sensitivity to BT2-mediated growth inhibition was analogous to Rapamycin and Torin 1 sensitivity (Figures 4G and S4F–I).
Removal of the nutrient sufficiency signal, such as after total amino acid withdrawal, causes mTOR to move from an active, lysosomal localization to an inactive, diffuse cytosolic localization (Sancak et al., 2010) (Figure 4H). We observed that BT2 treatment induced a similar dispersion of mTOR, suggesting it suppressed mTORC1 by modulating the nutrient sensing arm (Figure 4H). Accordingly, expressing constitutively active mutant Rag proteins (RagBS75L/RagCQ99L) (Sancak et al., 2008), or a key component of the mTORC1 complex constitutively targeted to the lysosome (Raptor-Rheb 15) (Sancak et al., 2010), rendered cells resistant to BT2-mediated mTORC1 inhibition (Figure 4I). Sestrin 2 is believed to be a primary leucine sensor that regulates the Rag proteins through the GATOR1 and GATOR2 complexes (Chantranupong et al., 2014). Consistent with this role, a similar resistance to BT2-mediated mTORC1 inhibition was also observed in Sestrin 1/2/3 knockdown cells (Figure 4J). Collectively, these data demonstrate that BCAA supply and catabolism regulate cancer cell proliferation and mTORC1 activity, and suggest that loss of BCAA catabolism in tumors can help sustain chronic mTORC1 activation.
High Dietary BCAA Intake Enhances Tumor Development and Growth In Vivo
We next explored how manipulating BCAA levels may influence the development of tumors in vivo. Because essential amino acids are derived from dietary sources in vivo, we fed DEN-injected mice purified diets with standard or high levels of BCAAs (Table S1). Given the influence of fatty acids on BCAA accumulation (Newgard et al., 2009), we examined the effects in diets with either standard low (LFD) or high (HFD) fat content. Five months after DEN injection, at a time when no tumors developed in mice fed the control LFD, mice fed diets with high fat and/or BCAAs had a tumor incidence of 20–40% (Figure 5A). Moreover, of the tumors that did develop, the largest were from mice fed high BCAA diets (Figure 5B). By 8 months post-injection, mice fed high BCAA diets had a potent increase in both tumor number and size (Figures 5D–F). Notably, at both time points, the liver masses of DEN-injected mice fed high BCAA diets were elevated, while this was not observed in uninjected mice (Figures 5C and 5G). This suggests that the dietary BCAAs may influence liver tissues predisposed to tumorigenesis, but have minimal impact on healthy liver tissues. Accordingly, normal liver tissues of uninjected mice responded to diets high in BCAAs or fat by enhancing the expression of BCAA catabolic enzymes (Figure 5H). In contrast, BCAA catabolic enzymes were significantly suppressed in nontumor tissues of DEN-injected mice, even further suppressed in tumors, and largely failed to increase in response to high BCAAs or fat (Figures 5H, and S5A).
Figure 5. High Dietary BCAA Intake Enhances Tumor Development and Growth in vivo.
(A-C) Analysis of mice 5 months after DEN injection, fed either a low fat diet (LFD, 10% kcal from fat) or high fat diet (HFD, 45% kcal from fat) with normal or high (+150%, +BCAA) levels of BCAAs. (A) Representative livers from DEN-injected mice. (B) Quantification of tumor incidence and tumor sizes of DEN-injected mice. (C) Liver masses of DEN-injected and control uninjected mice, normalized to total body weight.
(D-M) Analysis of mice 8 months after DEN injection fed indicated diets. (D) Representative livers from DEN-injected mice. (E) Quantification of tumor incidence of DEN-injected groups (F) Quantification of the number of tumors (≥3mm) and size of the largest tumor per mouse from DEN-injected mice. (G) Liver masses of DEN-injected and uninjected mice, normalized to total body weight. (H) RT-PCR analysis of BCAA catabolic enzymes of normal liver tissue (from uninjected mice), and nontumor and tumor liver tissues (from DEN-injected mice). Results normalized to normal liver tissue of LFD-fed mice. Statistical analyses summarized in Figure S5A. (I) Quantification of BCAA content in nontumor liver tissues of DEN-injected mice, and its correlation with tumor multiplicity. (J) Quantification of BCAA content in tumors of DEN-injected mice. (K) Representative histological and immunohistochemical analyses of livers from DEN-injected mice. (L) Immunoblots of nontumor liver tissues from DEN-injected mice and quantification of phospho:total p70S6K ratios.
*P<0.05 vs. LFD, §P<0.05 vs HFD. Data are shown as mean ± s.e.m.
See also Figure S5.
Metabolomic analyses of liver tissues revealed that significant BCAA accumulation had occurred in nontumor liver tissues of mice fed high BCAA diets at both time points, while this was not consistently observed for other amino acids or metabolites (Figures 5I and S5B–D). Interestingly, the BCAA content of nontumor liver tissues directly correlated with tumor multiplicity (Figure 5I). High dietary BCAAs also enhanced the accumulation of BCAAs in tumors, which was associated with tumor sizes of the experimental groups (Figures 5F and 5J). As expected, inflammation was enhanced in livers developing tumors and/or exposed to a HFD, although dietary BCAA content appeared to have minimal impact on overall immune responses (Figure S5E). Moreover, high dietary BCAAs did not enhance DNA damage, fibrosis, or cell death leading to compensatory proliferation (Figures 5K and S5F,G), additional factors that influence liver tumorigenesis. Rather, in agreement with in vitro data, the mTORC1 pathway was hyperactivated in liver tissues of mice fed high BCAA diets (Figures 5L). Thus, tissue BCAA content is directly related to liver tumor development and size, which is dependent on dietary intake of the BCAAs.
Enhancing BCAA Catabolism or Restricting Dietary BCAAs Limits Tumor Burden
We then investigated whether limiting BCAA accumulation through enhanced catabolism or dietary restriction could limit tumor burden. Administration of the BCKDK inhibitor BT2 at low doses through the diet (200mg per kg of diet) was able to significantly reduce the tumor burden of DEN-injected mice fed high BCAA diets (Figures 6A and 6B). Similar trends were also observed in DEN-injected mice fed chow diets with BT2 (Figure S5H). Notably, this compound was well tolerated for the duration of the study and did not lead to any observable adverse side effects. Next, to restrict the supply of BCAAs in vivo, we fed mice diets with a 50% reduction in BCAA content (as essential amino acids they cannot be completely removed long term; Table S1). All DEN-injected mice fed a low BCAA diet survived the study duration, while this was true for only 70% of mice fed normal or high levels of BCAAs (Figure 6C). Upon sacrifice, mice fed low BCAA diets also had significantly smaller tumors and reduced liver BCAA content (Figures 6D–6F). Importantly, normal lean body mass and liver function was maintained in both DEN-injected and control mice fed low BCAA diets (Figures 6G–I and S5I,J). The effects of the high and low BCAA diets were not due to differences in protein intake, as the tumor burden of mice fed diets adjusted for total protein content were similar (Figure 6J). Overall, these data show that BCAA accumulation is critical for the development and growth of liver tumors in vivo, and that dietary interventions influence tumor progression and overall survival.
Figure 6. Enhancing BCAA Catabolism or Restricting Dietary BCAAs Limits Tumor Burden in vivo.
(A,B) Analysis of mice 8 months after DEN injection, fed either LFD+BCAA or HFD+BCAA diets without/with 0.02% BT2. (A) Representative livers. (B) Quantification of the number of tumors (≥3mm) and size of the largest tumor per mouse.
(C) Kaplan-Meier survival curves of DEN-injected mice fed LFDs with low (−50%, -lowBCAA), standard, or high (+150%, +BCAA) levels of BCAAs.
(D-J) Analysis of mice 12 months after DEN injection, fed indicated diets. (D) Representative livers. (E) Average tumor sizes. (F) Quantification of BCAA content in nontumor liver tissues. (G) Representative histological and immunohistochemical analyses of livers from DEN-injected mice. (H) Liver masses (normalized to total body weight) and nonliver lean body mass of DEN-injected and uninjected mice. (I) RT-PCR analysis of BCAA catabolic enzymes of normal liver tissue (from uninjected mice), and nontumor and tumor liver tissues (from DEN-injected mice). Results normalized to normal liver tissue of LFD-fed mice. Statistical analyses summarized in Figure S5H. (J) Average tumor sizes of DEN-injected mice, based on % kcal from BCAAs and % kcal from total protein. Non-BCAA amino acids were adjusted proportionally to match the total protein content of low- or high-BCAA diets. *P<0.05 vs. LFD, †P<0.05 vs LFD+BCAA, ‡P<0.05 vs HFD+BCAA. Data are shown as mean ± s.e.m.
See also Figure S5.
Comparison of BCAA Catabolism Across Cancers
Given the robust changes of the BCAA catabolic enzymes in HCCs, we examined how they may be similar to, or distinct from, expression changes in other cancers. To characterize expression patterns in the development of various cancers, we performed differential expression analysis on all individual cancer subtypes profiled by TCGA with at least five adjacent normal samples for appropriate comparison (16 cancers in total). Overall, approximately 70% of cancers displayed significant suppression of at least half of the BCAA catabolic pathway (Figure 7A). With exception of the BCATs, enzyme expression was rarely significantly upregulated, leading to a broad net suppression across the profiled cancers (Figure 7B). Oncomine (Rhodes et al., 2004) analysis confirmed that these expression changes were consistent across multiple validated cohorts (Figure S6A). Notably, BCAA catabolic enzyme expression was also strongly associated with tumor progression in some cancers. Similar to HCCs, reduced expression correlated with tumor progression and aggressiveness in cancers of the colon & rectum, stomach, kidney, and adrenal cortex (Figure 7C). Moreover, the expression levels of numerous BCAA catabolic enzymes could be used to robustly predict patient survival among these cancers (Figure S6B).
Figure 7. The Overall Impact of BCAA Tissue Catabolism and Dietary Intake on Cancer Development, Progression, and Mortality.
(A,B) Summary of BCAA catabolic enzyme transcript levels for all cancers profiled by The Cancer Genome Atlas (TCGA) with at least 5 solid normal tissue samples. (A) Heatmap displaying expression in normal and tumor tissues of individual cancers. Cancers are arranged from those with the greatest number of enzymes suppressed (top) to the least (bottom) and genes are arranged from those suppressed in the greatest number of cancers (left) to the least (right). (B) Summary of the BCAA catabolic pathway indicating net expression changes across all tumors.
(C) Summary of BCAA catabolic enzyme transcript levels in tumors of the TCGA datasets COADREAD, STAD, ACC, KIRC, KIRP, and KICH, sorted by stage, grade, local invasion, lymph node invasion, and metastasis.
(D) Quantification of cox proportional hazard ratios (95% confidence intervals), significance (log-rank p-value), robustness, and difference in days of estimated survival for patients across all cancers profiled by TCGA with at least 15 verified deaths with low tumor expression of ACADS, ACADSB, and BCKDHA.
(E,F) Analysis of individuals 50–66 years-old in the NHANES III dataset. (E) Hazard ratios (HR, with 95% confidence intervals) based on BCAA intake, 1adjusted for age, sex, race, total kcal, usual dietary intake, diet change, physical activity, intentional weight loss, waist circumference, smoking, education, and prior diagnosis of cancer, diabetes and cardiovascular disease, 2additionally adjusted for % kcal from other macronutrients. (F) Substitution analysis comparing change in risk when replacing BCAAs with carbohydrate or fat, with the same adjustments as HR2 except total kcal and % kcal from non-BCAA protein. Data are presented as hazard ratio (solid line) with 95% confidence interval (shaded area).
(G) Summary of BCAA catabolism in normal, regenerating, and cancerous liver tissues.
Recent reports have identified enhanced expression of the BCAT enzymes, which catalyze the reversible transamination step, in some cancers (Hattori et al., 2017; Mayers et al., 2016; Raffel et al., 2017). However, opposing metabolic fluxes were observed and conflicting mechanisms were proposed. In some non-small cell lung cancers (NSCLC) and acute myeloid leukemias (AML), enhanced uptake of BCAAs and conversion to α-ketoacids is thought to provide substrates for nucleotide synthesis and maintain α-ketoglutarate homeostasis (Mayers et al., 2016; Raffel et al., 2017). In contrast, chronic myeloid leukemia (CML) displays enhanced production of BCAAs from α-ketoacids, which promotes mTOR activation (Hattori et al., 2017). Why enhanced expression of the same enzyme (BCAT1) leads to opposing net metabolic fluxes in these cancers has not been clarified. As noted, we examined the expression of BCAT1 and BCAT2, but found no consistent changes across the human HCC samples, liver-derived cell lines, and animal tumor models (Figures S1C–E). Moreover, we observed that supplying either BCAAs or α-ketoacids was sufficient to stimulate mTORC1 (Figure S6C), suggesting that these metabolites are readily imported and interconverted. In contrast to effects observed in NSCLC and CML, inhibition of BCAT1 actually enhanced the proliferation of HepG2 cells, an effect lost upon removal of BCAAs from the media (Figures S6D–F). Thus, while examination of the BCATs requires further investigation, our data suggest that the distal irreversible enzymatic steps can also influence the catabolic flux of BCAAs, and indeed may be more important for certain tissues and cancers.
Low Tissue Catabolism and High Dietary Intake of BCAAs is Associated with High Overall Cancer Mortality Risk
Finally, we examined how tissue catabolism and dietary intake of BCAAs may impact overall cancer mortality. In total, TCGA has transcriptionally profiled 33 cancers (i.e. including those without any/sufficient normal tissues for comparison). Therefore, we used the combined expression index of BCKDHA, ACADS, and ACADSB to screen the cancer cohorts with at least 15 verified deaths for differences in clinical outcome. Overall, approximately 60% of cancers had a modest to strong bifurcation in patient survival, with high expression of these three genes associated with better outcome (Figures 7D and S7A). The remaining cancers had minimal difference in survival (Figure S7A). Thus, when collectively analyzed them as a pan-cancer cohort, patients with higher tumor expression of BCKDHA, ACADS, and ACADSB lived significantly longer (1849 days versus 732 days at 70% survival; Figure S7B).
To determine if dietary intake of BCAAs correlated with cancer mortality in humans, we analyzed the National Health and Nutrition Examination Survey (NHANES) III dataset with linked mortality data. In this cohort, we calculated that the BCAAs comprised 17.3% of total protein intake. Given dietary recommendations for protein at 10–35% of total kcal, we set the threshold for low BCAA intake at 1.73% (10% protein/total kcal × 17.3% BCAA/total protein kcal). Few individuals consumed very high protein diets (>35% of total kcal), so we set the threshold for high BCAA intake at 3.89%, which is at the median of recommended protein intake (22.5% protein/total kcal × 17.3%BCAA/total protein kcal). Overall, individuals 50–66 years old in the highest tertile of BCAA intake had a 200% increased risk of death from cancer relative to the lowest tertile, even when adjusting for known confounders, as well as percent kilocalories from fat, carbohydrate, and non-BCAA protein intake (Figures 7E and S7C). When evaluated as continuous substitutive variables, isocalorically replacing 3% of energy from BCAAs with either carbohydrate or fat decreased cancer mortality risk by more than 50% (Figure 7F). While the effect of substituting BCAAs with non-BCAA protein could not be accurately evaluated due to high collinearity between the variables (r=0.975), non-BCAA protein intake was associated with only a modest change in cancer mortality risk (Figure S7D). These associations were not significant in more elderly populations, likely because they have higher basal protein requirements (Figures S7E–G).
Discussion
In summary, utilization of comprehensive transcriptomic and metabolomic analyses of primary HCCs and animal liver cancer models identified an important role for BCAA catabolism in tumor development, progression and growth (Figure 7G). The changes in both enzyme expression and metabolite concentration were consistent in liver cancer across etiological backgrounds from multiple cohorts, positively selected for during disease progression, and significantly associated with patient survival. Animal tumor models and cancer cell lines notably had analogous transcriptomic and metabolomic profiles, while regenerating tissues explicitly did not show similar changes. Taken together, the data provide assurance that the observed suppression of BCAA catabolism is not simply related to generic proliferation, but rather, has some degree of specificity to carcinogenesis. More broadly, combining multi-omic data from the same set of samples helped reveal changes in metabolic fluxes that would be difficult to identify or explain with the data from just one platform, and thus, this experimental approach could potentially provide significant insights when used to analyze other forms of cancer. Of note, the transcriptomic and metabolomic analyses demonstrated that numerous additional metabolites and metabolic pathways were consistently altered in tumors that could be targeted for additional beneficial effects (Figures 1, 2, S1, and S2). Some of these, such as serine/glycine (Maddocks et al., 2017), and proline (Loayza-Puch et al., 2016) metabolism have been characterized in other cancers and warrant further investigation in liver cancer.
Reduced serum BCAAs have been observed in patients with liver cirrhosis and related disease states (Marchesini et al., 2003). We also frequently observed this phenomenon in the animal tumor models analyzed (data not shown). Thus far, it has been assumed that reduced serum BCAAs is a sign of inadequate BCAA intake and/or enhanced BCAA catabolism, leading to recommendations for BCAA supplementation therapy (Marchesini et al., 2005). However, the data presented here suggest low serum BCAAs may in part be a consequence of BCAA accumulation in the liver. In fact, previous reports have observed that liver BCKDH activity is reduced in patients with cirrhosis (Taniguchi et al., 1996). Although the acute beneficial effect of BCAA supplementation on hepatic encephalopathy and clinical parameters has been reported, these results are not consistent, and providing non-BCAA caloric supplements may be just as effective (Charlton, 2006). Considering the data presented here, and the increased risk of liver cancer in cirrhotic and liver disease patients, a careful re-evaluation of this practice should be undertaken. Moreover, BCAA dietary supplements are commercially available and contain 2–8 grams of BCAAs per serving, an additional 100–400% above RDA/AMDR (recommended daily allowance/acceptable macronutrient distribution range) levels. While likely safe for use in individuals with normal liver function, clinicians should be aware if their patients, particularly those with liver diseases, are consuming these products.
Obesity and diabetes are associated with an increased risk of incidence and mortality for a range of cancers (AICR, 2018). This association is particularly strong for liver cancers (Chen et al., 2008), yet the mechanisms involved remain a matter of debate. Previous data have shown that the local expression of tumor-promoting pro-inflammatory cytokines such as TNFα and IL-6 are enhanced after genetic- or diet-induced obesity (Park et al., 2010). While our data are generally consistent with these reports, we have frequently found that the change in expression after HFD feeding to be somewhat minor relative to the increase in pre-tumor tissues versus the normal liver (Figure S5E). As previously reported (Newgard et al., 2009), we observed that a HFD also enhances tissue BCAA accumulation, presumably due to fatty acids competing with BCAAs for access to shared catabolic enzymes (Newgard, 2012). Moreover, multiple groups have reported altered BCAA metabolism in the setting of obesity and diabetes (Newgard et al., 2009; Pedersen et al., 2016; Wang et al., 2011). Thus, the data presented here offer an additional or alternative mechanism by which these metabolic diseases influence the development and progression of certain cancers.
The cellular uptake of BCAAs is largely attributed to the Large-neutral Amino Acid Transporters (LATs) (Wang and Holst, 2015). While there were some changes in expression across the animal tumor models, none of them met our criteria of being upregulated in tumor models but not regenerating tissues (Figure S7H). Moreover, we observed minimal changes in human HCCs relative to adjacent normal liver tissues (Figure S7H), and high tumor expression of the LAT transporters was not associated with significantly worse patient survival (Figure S7I). These transporters also import a wide array of neutral amino acids, and we did not observe a consistent accumulation of other amino acids in tumors. Thus, given a consistent rate of amino acid import, the data suggest that BCAAs are specifically diverted from catabolism due to changes in catabolic enzyme expression. We acknowledge that multiple factors likely influence the intracellular concentration of BCAAs, but our data point towards the suppression of irreversible catabolic enzymes (most notably BCKDHA, ACADS, and ACADSB) as playing a critical role in liver and potentially other cancers.
While BCAAs are believed to have pleiotropic effects through defined (Jang et al., 2016; Mayers et al., 2016) and unknown (Taya et al., 2016) mechanisms, stimulation of mTORC1 is the most potent and extensively-characterized (Laplante and Sabatini, 2012). Although growth factor signaling to mTORC1 is critical, it is not sufficient for activation. Equally important is the nutrient sufficiency signaling to mTORC1, and removal of either input renders mTORC1 inactive (Sancak et al., 2008). Recent comprehensive genetic analyses have revealed that numerous cancers harbor mutations that chronically activate the growth factor arm of mTORC1 (Sanchez-Vega et al., 2018). Interestingly, chronic activation of mTORC1’s growth factor arm plays a particularly central role in the pathogenesis of the cancers displaying the most overt suppression of BCAA catabolism (Figures 7, S6, and S7) (Cancer Genome Atlas, 2012; Cancer Genome Atlas Research, 2013, 2014; Cancer Genome Atlas Research et al., 2016; Zheng et al., 2016). Moreover, like in liver cancer, chronic activation of growth factor signaling to mTORC1 (through the deletion of TSC1 or TSC2) enhances tumorigenesis in cancer models of the kidney (Kobayashi et al., 1999), intestine (Faller et al., 2015), mesothelium (Guo et al., 2014) and other tissues (Hsieh et al., 2012; Liang et al., 2010). Yet it has remained unclear if and how tumors ensure chronic activation of the nutrient sensing arm of mTORC1. The data presented here suggest that tumors may manipulate specific nutrients to achieve this. This mechanism would likely be more favorable for cancer cell growth, as it would provide enhanced pro-growth signaling under most conditions while still allowing for pathway inhibition in cases of extreme starvation.
mTOR plays an important, albeit complex, role in tissue homeostasis and tumorigenesis. In liver cancer, targeting the mTOR pathway with first- or second-generation inhibitors has thus far led to tepid and mixed results (Matter et al., 2014). Although treatments have successfully retarded tumor development and growth in selected animal models and clinical trials (Matter et al., 2014; Menon et al., 2012), broad blockage of the mTOR pathway can also enhance tumorigenesis through the death of normal and regenerating hepatocytes (Umemura et al., 2014). It has also been reported that mTORC1 inhibition can be beneficial to K-ras-driven pancreatic cancers under certain conditions (Palm et al., 2015). Though subsequent work has shown this is due to restoration of the balance between amino acid supply and protein synthesis through the stimulation of macropinocytosis and lysosomal degradation of extracellular proteins (Nofal et al., 2017). These behaviors have been attributed to mutant K-ras, and while nearly all pancreatic cancers have K-ras mutations, only 1% of HCCs harbor these (Ally et al., 2017). In addition, pancreatic cancers have enhanced expression of autophagy/lysosomal genes (Perera et al., 2015), but we found no similar changes in hepatic, gastrointestinal, or renal cancers (data not shown), which have the most overt changes in BCAA catabolism. We also repeatedly found no beneficial effect of mTOR inhibition or extracellular protein supplementation for the liver cancer models analyzed, either in nutrient complete or starved conditions. Thus, cancers of various tissues, particularly those driven by alternate oncogenes, likely handle BCAA intake/catabolism and chronic mTORC1 activation differently. Nonetheless, identification of the mechanisms that specifically regulate mTORC1 in cancer cells may provide targeted and more effective therapeutic results.
Overall, we find that BCAA catabolism is a metabolic pathway potently and robustly altered in certain cancers. Not only can this pathway be examined and utilized for its diagnostic and prognostic value, but dietary and pharmacologic interventions may effectively modulate tumor development and growth while minimally affecting normal and regenerating tissues. In addition, the data presented here and elsewhere (Hattori et al., 2017) raise the intriguing possibility that changes in tumor metabolism may not strictly be used for anabolic purposes, but can also be used to influence established oncogenic signaling pathways.
Limitations of Study
The methods and models used in our study are not without limitations. Current metabolic analytic tools treat metabolic pathways discreetly, whereas in vivo, these pathways are interconnected and constantly in flux. The development of more comprehensive analytic tools will help address the role of individual enzymes in regulating flux within a pathway, as well as the interplay of multiple pathways and their relative contribution to observed phenotypic changes. Moreover, flux studies involving labeled BCAAs and/or related metabolites will be needed to confirm the metabolic changes suggested by the transcriptomic and metabolomic analyses. Repeating in vitro studies in more complex and physiologically relevant models (such as primary hepatocytes and organoids) and in vivo studies in alternate, genetic-based models will also help examine the robustness of the phenotypes observed.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Weiping Han (weiping_han@sbic.a-star.edu.sg).
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Human Subjects and Data
The Singapore General Hospital cohort includes primary tumors and adjacent nontumor tissues that were surgically removed at the National Cancer Centre Singapore between 2008 and 2011. All samples used for analysis were obtained in the context of clinically indicated surgery with informed consent from the patient, in accordance with the Declaration of Helsinki and using protocols approved by the SingHealth Institutional Review Board at the National Cancer Center Singapore. Resected tissues were snap frozen, sectioned, and analyzed by pathologists to score the tumors and identify neoplastic and nonneoplastic regions. RNA-Seq raw count, copy number variation, and mutational data, along with associated clinical information from the TCGA cohorts were downloaded from The Cancer Genome Atlas (cancergenome.nih.gov) and/or UCSC Cancer Genomics Browser (genome-cancer.ucsc.edu). Oncomine data were examined with the filters data type: mRNA, gene rank threshold: all; fold change threshold: 1.5; p-value threshold: 0.05 (oncomine.org). Biopsy immunohistochemical micrographs were downloaded from The Human Protein Atlas (proteinatlas.org). Dietary and mortality data for the NHANES III cohort were downloaded from the Centers for Disease Control and Prevention (cdc.gov/nchs/nhanes).
Animal Studies
All animal studies were approved by the Institutional Animal Care and Use Committee at A*STAR. Animals were fed ad libitum and maintained in a specific pathogen free facility with constant ambient temperature and a 12-hour light cycle. C57BL/6 mice for breeding were obtained from Biological Resource Center, A*STAR.15 day-old pups were injected i.p. with a single 50 mg/kg dose of Diethylnitrosamine (DEN). DEN nontumor and tumor tissues were harvested from injected mice, while normal liver tissues were collected from age-matched mice without DEN injection. ACI rats were obtained from Harlan (Dublin, VA) and used in all rat studies. For the regenerating liver model, 10–12 week-old rats were anesthetized and two-thirds of the liver was removed as previously described (Michalopoulos and DeFrances, 1997). Liver tissue was harvested 24 hours later, at a time when hepatocyte proliferation is at its highest (Michalopoulos and DeFrances, 1997). For the Morris Hepatoma model, 10 week-old rats were anesthetized, 1 million MH3924a cells were injected directly into the liver, and tumors were harvested two weeks later. Control rat liver was harvested from age-matched rats. For rat DEN model, DEN was given to 10 week-old rats via drinking water containing 100 mg/L DEN for up to 4 months, and control rat livers from age-matched rats were analyzed. During sacrifice, animals were anesthetized and blood was collected by cardiac puncture and death was confirmed by cervical dislocation. Livers were resected, measured by electronic calipers and snap frozen in liquid nitrogen. For mice on special diets, animals were randomly assigned to experimental groups upon weaning. Sample sizes were estimated based on prior experience with the DEN model with a power analysis, including a power level of 80% and confidence interval of 95%. Chow diets were obtained from Altromin (#1320, modified), including those with added 0.2% BT2. Custom purified diets were purchased from Research Diets, Inc. (New Brunswick, NJ) and their composition is summarized in Table S1. Diets were given to animals at 12 weeks of age until termination of the study. Body weights were measured weekly and prior to sacrifice lean and fat mass were measured by an EchoMRI Body Composition Analyzer. After sacrifice, tissues were assigned random ID numbers and analyses were performed blinded to experimental group information.
Cell Culture Studies
Cell lines were obtained from ATCC (AML12, HepG2, Hep3B, SNU182, SNU387, SNU398, and SNU449), the Japanese Collection of Research Bioresources (Huh7), and the German Cancer Research Center Tumor Collection (Morris Hepatoma 3924a) and not further verified. Only cell stocks that had tested negative for mycoplasma within the prior 9 months were used. HepG2 cells were maintained in EMEM, and remaining cell lines were maintained in RPMI 1640, with 10% FBS and 1% Pen Strep (Gibco), and grown in a 37°C humidified incubator with 5% CO2. Real-time cell growth measurements were taken on an xCELLigence RTCA SP (Acea) at 15-minute intervals. For studies involving BCAA-reduced/free or AA-free media, amino acid free RPMI (USBiological) was prepared, and supplemented with purified amino acids (Sigma) and/or 10% Dialyzed FBS and 1% Pen Strep (Gibco). Indicated cultures were treated with an additional 3% Albumin (Sigma), 20% FBS (Gibco), Gabapentin (Sigma), or purified amino acids (Sigma) dissolved in media. For transcription factor antagonist studies, cells were seeded in 10cm dishes and treated with GW6471 (Sigma) or vehicle (DMSO) for 48 hours when cells were approximately 80% confluent. For compound treatment growth curves, cells were seeded in quadruplicate at 1,250, 2,500 or 5,000 cells per well and normalized 12 hours later (time 0 hours), immediately prior to changing half of the media to add compounds or vehicle. Proliferation rates were calculated over a 48-hour period beginning 2 hours after addition of BT2 (3,6-dichlorobenzo[b]thiophene-2-carboxylic acid; Matrix Scientific), Rapamycin (Tocris), Torin 1(Tocris), or vehicle (DMSO). For immunoblots, cells were seeded in 10 cm dishes and allowed to grow for at least 24 hours. At approximately 80% confluence the media was changed to include indicated amounts of compound or vehicle and whole cell lysates were harvested 2 hours later. For inducible BCKDHA knockdowns, interfering RNA for target sequences were inserted into the Tet-pLKO-puro vector (Wiederschain et al., 2009) (Addgene plasmid #21915; BCKDHA shRNA1: CCGGTCCTTCTACATGACCAACTATCTC GAGATAGTTGGTCATGTAGAAGGATTTTTG; shRNA2: CCGGGCAGTCACGAAAGAAGGTCATCTCG AGATGACCTTCTTTCGTGACTGCTTTTTG). The resulting constructs, along with lentiviral packaging vectors, were transfected into HEK293T cells with Lipofectamine 2000 (Invitrogen) following manufacturer’s protocol. Supernatants containing lentivirus were collected 24 hours later, passed through 0.2μm filters, and added to recipient cell lines for 24 hours along with 8μg/mL polybrene (Sigma). Cells were then incubated in selection media (1–3μg/mL puromycin, depending on cell line sensitivity) for 5–7 days. For growth curves, cells were maintained in 250μg/mL doxycycline for 5 days, then seeded into wells with (+Dox) or without (-Dox) doxycycline in quadruplicate at 625, 1,250, 2,500 or 5,000 cells per well. Wells were normalized 12 hours later (Day 0) and allowed to grow without media change or addition of more doxycycline. Proliferation rates were calculated over a 7-day period (Day 0 to Day 7). For immunoblots, cells were seeded in 10 or 15 cm dishes, split or received media change every 3–4 days to maintain a doxycycline, Rapamycin, and Torin 1 concentrations, and harvested after 10 days. For CRISPR-Cas9-induced mutagenesis, previously published methods were followed (Sanjana et al., 2014). Briefly, sgRNAs were computationally identified (http://www.genome-engineering.org/crispr) and inserted into the lentiCRISPRv2 vector (Addgene plasmid #52961; BCKDK target the sequence GTCGGCCATCGACGCGGCAG). Lentivirus was produced as detailed above, and Hep3B cells were transduced and underwent with 1μg/mL puromycin selection for 7 days. Single-cell colonies were generated and 80 clones were examined for mutations by sequencing 15–20 PCR products of the target region. LentiCRISPRv2 empty vector-transduced Hep3B cells were used as controls, which behaved similarly to parental Hep3B cells and non-frameshift BCKDK mutant Hep3B clones. For growth curves, 1250 cells from each clone were plated in quadruplicate, normalized 6 hours later, and allowed to grow for 5 days. For immunoblots, cells were seeded into 10cm or 15cm dishes and harvested 10 days later as whole cell lysates, or as purified mitochondrial fractions using an isolation kit (Thermo Fisher). For PPARα knockdowns, target sequences embedded in the lentiviral SMARTvector were obtained from Dharmacon (PPARα shRNA1: AAATGGGTTTATAACTCGT, shRNA2: TCATAAGCTAGCACCCGTG; Nontargeting shRNA: Clone ID VSC11716). Lentivirus was produced as detailed above, and HepG2 cells were transduced and underwent with 1μg/mL puromycin selection for 7 days. Cells were seeded in 10cm dishes and harvested 48 hours later, when the cells were approximately 80% confluent. For ACADS, ACADSB, and BCKDHA overexpression studies, full-length protein-coding regions (CDS) were obtained from Dharmacon (BCKDHA clone ID: 4065221; ACADSB clone ID: PLOHS_100070589) or as a kind gift (ACADS from Dr. Gerard Vockley) and subcloned into a pCMV vector to add a FLAG-tag to the C terminus. The FLAG-tagged CDS was then subcloned into the pLOC lentriviral vector. Lentivirus was produced as detailed above, and Hep3B cells were transduced and underwent with 1μg/mL puromycin selection for 7 days. For Sestrin 1/2/3 knockdowns, interfering RNA for target sequences were inserted into the pLKO-puro vector (Moffat et al., 2006) (Addgene plasmid # 10878; Sestrinl shRNA: TCGTCACTACATTGGAATAAT; Sestrin2 shRNA: GCGGAACCTCAAGGTCTATAT; Sestrin3 shRNA: GCCTTAATGGAAAGGATGAAA). Lentivirus was produced as detailed above, and 293T cells were transduced and underwent with 1μg/mL puromycin selection for 7 days. Knockdowns were confirmed by RT-PCR, with all 3 Sestrins displaying a >90% reduction in mRNA (Chantranupong et al., 2014). RagBS75L, HA-RagCQ99L, Flag-Raptor and Flag-Raptor-Rheb 15 constructs were a gift from David Sabatini via Addgene (plasmid numbers 19303, 19305, 26633, and 26634), and were transfected into HEK-293T cells with Lipofectamine 2000, following manufacturer’s protocol. For all studies, results shown are one of at least three independent experiments.
METHODS DETAIL
Gene Expression Analyses
RNA was extracted using Trizol (Life Technologies) and/or purified on RNAeasy columns (Qiagen), then analyzed for purity using RNA pico chips run on an Agilent 2100 bioanalyzer. Only samples with a RIN > 7 and 28s:18s ratio >1.0 were used in analysis. For the Singapore General Hospital cohort, gene expression was assessed using the HumanHT-12 v3 microarray chip (Illumina) and probe intensities were analyzed using the GenomeStudio software. For TCGA cohorts, raw counts were analyzed with the DESeq2 package in R. Mouse and rat samples were sequenced by Beijing Genomics Institute (BGI, Hong Kong) using the paired-end sequencing method (91 bp) with approximately 40 million reads per sample. For all cohorts, gene lists were analyzed using DAVID (david.ncifcrf.gov) and Ingenuity Pathway Analysis (Qiagen), and heat maps were generated with GenePattern (broadinstitute.org/cancer/software/genepattern). The survMisc package in R and Cutoff Finder (molpath.charite.de/cutoff) were used to identify appropriate cutoff values for splitting patients into high and low expression groups. Combined expression indexes were screened, developed, and analyzed using a method similar to the Steepest Decent (Boutros et al., 2009). Kaplan-Meier survival estimate curves and associated statistics were generated with the Survival package in R. Adjustments in hazard ratios include age at initial pathologic diagnosis, gender (male/female) tumor stage (1–4), tumor grade (1–4), radiation therapy (yes/no/unspecified), pharmaceutical therapy (yes/no/unspecified), additional therapies (additional pharmaceutical therapy, additional radiation therapy, or additional surgery procedure yes/no/unspecified), and tumor subtype (colon adenocarcinoma or rectum adenocarcinoma for colorectal adenocarcinoma; adenocarcinoma diffuse type, intestinal adenocarcinoma tubular type, intestinal adenocarcinoma mucinous type, intestinal adenocarcinoma papillary type, adenocarcinoma signet ring type, adenocarcinoma not otherwise specified, or intestinal adenocarcinoma not otherwise specified for stomach adenocarcinoma). Transcription factor binding sites in the promoters (−2000 to +100) of genes were analyzed using Transfac (geneXplain). RT-PCR reactions were run on an Applied Biosystems StepOnePlus with Power SYBR Green (Life Technologies) and primers listed in Table S2.
Metabolomic Analyses
Amino acids were quantified by HPLC-MS/MS using purified standards (Sigma). Acylcarnitine measurements were made by flow injection tandem mass spectrometry using sample preparation methods described previously (An et al., 2004). The data were acquired using a Waters Acquity™ UPLC system equipped with a TQ (triple quadrupole) detector and a data system controlled by MassLynx 4.1 operating system (Waters, Milford, MA). For tissue BCKDH activity assays, frozen liver samples were pulverized in liquid nitrogen, then 200 mg of tissue was homogenized in 1 mL of ice cold homogenization buffer (30 mM KPi pH7.5, 3 mM EDTA, 5 mM DTT,1 mM α-ketoisovalerate, 3% FBS, 5% Triton X-100,1 μM Leupeptin) using a QIAGEN TissueLyser II set at a frequency of 15/s for 1 minute. Homogenized samples were centrifuged for 10 minutes at 10,000 × g and the supernatant was collected. 50 μL of supernatant was added to 300 μL of assay buffer (50 mM HEPES pH 7.5, 30 mM KPi pH7.5, 0.4 mM CoA, 3 mM NAD+, 5% FBS, 2 mM Thiamine Pyrophosphate, 2 mM MgCl2, 7.8 μM [1-14C] α-ketoisovalerate) in a polystyrene test tube containing a raised 1 M NaOH CO2 trapping system. The tubes were capped and placed in a shaking water bath set at 37°C for 30 min. Tubes were then placed on ice and the reaction mixture was acidified by injection of 100 μl of 70% perchloric acid followed by shaking on an orbital shaker at room temperature for 1 hour. The 14CO2 contained in the 1 M NaOH trap was counted in a liquid scintillation counter. For magnetic resonance spectroscopy studies, approximately 48 mg of [1-13C] 2-ketoisocaproic acid (Sigma #750832), doped with 15 mM trityl-radical (OXO63, GE Healthcare) and 3 μl of gadoterate meglumine (10 mM, Dotarem®, Guerbet), was hyperpolarized in a polarizer, with 60 min of microwave irradiation. The sample was subsequently dissolved in a pressurized and heated alkaline solution, containing 100 mg/L EDTA to yield a solution of 80 mM hyperpolarized sodium [1-13C]2-ketoisocaproate with a polarization of 30%, T1 of 25 seconds and physiological temperature and pH. Rats were positioned in a 9.4 T horizontal bore MR scanner interfaced to an Avance III console (Bruker Biospec), and inserted into a dual-tuned (1H/13C) rat abdominal coil (20 mm diameter). Correct positioning was confirmed by the acquisition of a coronal proton FLASH image (TE/TR, 8.0/100.0 ms; matrix size, 192 × 192; FOV, 50 × 36 mm; slice thickness, 2.0 mm; excitation flip angle, 30°). A respiratory-gated shim was used to reduce the proton linewidth to approximately 230 Hz. Immediately before injection, a respiratory-gated 13CMR pulse-acquire spectroscopy sequence was initiated. 2.0–2.5 mL (0.5 mmol/kg body weight) of hyperpolarized 2-ketoisocaproate was intravenously injected over 10 s into the anesthetized rat. Thirty individual liver spectra were acquired over 1 min after injection (TR, 2 s; excitation flip angle, 25°; sweep width, 8,000 Hz; acquired points, 2,048; frequency centered on the ketoisocaproate resonance). Liver 13C MR spectra were analyzed using the AMARES algorithm as implemented in the jMRUI software package. Spectra were baseline and DC offset-corrected based on the last half of acquired points. To quantify hepatic metabolism, the spectra were summed over the first 30 s upon 2-ketoisocaproate arrival. Metabolite peaks corresponding to [1-13C]2-ketoisocaproate (172.6ppm) and its metabolic derivatives [1-C]leucine (176.8ppm) and [1-13C]bicarbonate (160.8 ppm) were fitted with prior knowledge assuming a Lorentzian line shape, peak frequencies, relative phases, and linewidths. For each animal, tCarbon is defined as the sum of all these three metabolite peaks. The normalized ratios [1-13C]leucine/tCarbon and [1-13C]bicarbonate/tCarbon were computed for statistical analysis.
Immunoblots, Immunocytochemistry, and Immunohistochemistry
Antibodies were obtained from Abcam [Acadsb (ab99951), Bckdk (ab125389 and ab151297), Ki67 (ab15580)], Cell Signaling [BCAT1 (12822), Cleaved Caspase 3 (9664), Cox IV (4844), HA-tag (3724), mTOR (2983), p-Histone H2A.X (9718), S6 (2317), p-S6 (4858), S6K (9202), p-S6K (9234)], Santa Cruz [Bckdha (sc-67200), Gapdh (sc-32233), LAMP-2 (sc-18822)], Bethyl [p-Bckdha (A304–672A)], Sigma [Acads (HPA022271), Flag (F1804), Tubulin (T5168)], and Thermo Fisher [BCAT2 (PA5–21549)]. All samples for Western blot were harvested in RIPA buffer with protease inhibitors (Roche). If mitochondrial fractions were isolated, they were done so using a mitochondrial isolation kit (Thermo), following manufacturer’s protocol. Approximately 10 μg protein samples were run by SDS-PAGE, transferred to PVDF membranes using iBlot2 (Life Technologies), blocked with 5% milk and incubated with primary antibodies in 5% BSA. Membranes were then either incubated with α-mouse/rabbit-HRP secondary antibodies (GE) and developed with ECL prime (GE), or incubated with α-mouse/rabbit-IRDye 680RD/800CW secondary antibodies and imaged on an Odyssey CLx (LI-COR). For immunocytochemistry, cells were grown on glass coverslips in 6-well plates for at least 24 hours before indicated treatments. At time of harvest, cells were fixed with 4% paraformaldehyde, permeabilized with 0.25% Triton X-100, and blocked with Odyssey Blocking Buffer (LI-COR). Cells were then incubated in TBST + 2%BSA with primary antibodies, followed by TBST + 2%BSA with Alexa Fluor 488 or Alexa Fluor 568 secondary antibodies (Thermo). Coverslips were then mounted to glass slides with ProLong Diamond Antifade Mountant with DAPI (Life Technologies) and sealed with nail polish. Confocal images were obtained on a Nikon A1R+si confocal microscope using the same settings for all images. Images were analyzed using Nikon NIS-Elements software package. For immunohistochemistry, liver specimens were fixed with 10% neutral buffered formalin and 70% ethanol and embedded in paraffin. Sections were cut at 7 μm, deparaffinized, subjected to citrate buffer antigen retrieval, and exposed to hydrogen peroxide to quench endogenous peroxidase prior to incubation with primary antibodies. Vectastain ABC kit and ImmPACT DAB (Vector Laboratories) were used for chromagen development, then counterstained with Harris hematoxylin. Images were obtained on a Nikon Ni-E microscope and analyzed using the Nikon NIS-Elements software package.
Human Dietary Analysis
NHANES III (1988–1994) is a US nationally-representative population-based survey of non-institutionalized individuals over 2 years of age (cdc.gov/nchs/nhanes). The study population included 6779 non-pregnant, non-lactating adults with reliable energy intake (400–7000 kcal), ages 50–90 with mortality follow-up from the date of participation (1988–1994) through December 31, 2011. Data from the structured household interview and mobile examination center (MEC) physical examination were included in the present analysis (Parekh et al., 2012). Variables deemed important in the literature were evaluated for potential confounding, and adjusted for in final analyses. Age, sex, race/ethnicity, years of education, cigarette smoking, leisure-time physical activity, dietary intake, dietary behaviors, and doctor-diagnosed medical history were self-reported. Waist circumference was measured by trained personnel to the nearest 0.1cm at the iliac crest. Dummy variables were created for smoking status (current, former, never) based on responses to the questions “Have you smoked at least 100 cigarettes during your entire life?” and “Do you smoke cigarettes now?” Participants were coded as ‘never’ smokers if they had not smoked at least 100 cigarettes and did not currently smoke cigarettes; ‘former’ smokers had smoked at least 100 cigarettes but did not currently smoke, and ‘current’ smokers responded affirmatively to both questions. Three dummy variables were created for race-ethnicity (Non-Hispanic white, Non-Hispanic black, and Mexican-American). Three dummy variables were also created for years of education (less than high school education, high school graduate, and college graduate). Self-reported history (y/n) of doctor diagnosed diabetes, cancer, and cardiovascular disease were included as binary variables in all models. Dietary behaviors including intentional weight loss in the last year (y/n), dietary changes in the last year (y/n), and whether intake on the dietary recall was less than, more than, or comparable to their usual day were included as covariates. Total kilocalories, percent kilocalories from fat, carbohydrate, and non-BCAA protein were computed from the 24-hour recall used to assess dietary intake. Total leisure-time physical activity was estimated by a physical activity questionnaire, which asked participants to report the frequency (over the past 30 days) of engaging in nine activities and up to four additional other activities not queried directly in the survey. Weekly frequencies of each activity were multiplied by a validated intensity rating in metabolic equivalents (MET) and summed for each individual. MET values used in NHANES III were defined by the Compendium of Physical Activities (Pate et al., 1995). Cox Proportional Hazard Models adjusted for covariates were used to evaluate the association between BCAA intake as a percent of total kilocalories and cancer mortality stratified by age group (50–66 and 66 and older). We also estimated the effect of replacing kilocalories from BCAA with carbohydrates and fat by examining the continuous multivariable-adjusted association between BCAA intake and cancer mortality while simultaneously including these macronutrients in the model. Effects of interchanging different macronutrients were estimated by computing the differences between linear coefficients and their corresponding covariance matrix to obtain HRs and 95% CIs (Bernstein et al., 2010). It was not possible to estimate the effect of replacing BCAA with non-BCAA protein due to multicollinearity (r=0.975), but in sensitivity analyses where non-BCAA protein intake was modeled as the primary exposure variable, associations with cancer mortality were attenuated.
QUANTIFICATION AND STATISTICAL ANALYSIS
Differentially expressed genes were quantified with the DESeq2 package in R. Genes with P-values<2.44×10−6 (meeting genome-wide Bonferroni correction criteria) were considered significant. Kaplan-Meier survival estimate curves and associated statistics were generated with the Survival package in R. All analyses performed on the NHANES III dataset were conducted with SAS 9.4 software (SAS Institute). All remaining statistical analyses were performed in Microsoft Excel. Unless otherwise noted, a homoscedastic two-tailed Student’s t-test was performed and considered significant if P<0.05. Standard error of the mean (s.e.m.) are shown for all quantitative data, except when smaller than data point symbols (cell proliferation growth curves) or for clarity (heat maps and dot plots).
DATA AND SOFTWARE AVAILABILITY
Gene expression profiles have been deposited at the Gene Expression Omnibus (GEO) under the accession number GSE75677.
Supplementary Material
Figure S1. BCAA Catabolism Is Suppressed in Hepatocellular Carcinomas and Predicts Patient Survival, Related to Figure 1
(A) Patient characteristics of the cohort recruited at Singapore General Hospital (SGH), including total number, average age (range), and number (percent) of males, females, Hepatitis B+, Hepatitis C+, and indicated racial backgrounds. Additional information on tumor characteristics of the cohort can be found in Figure 1H.
(B) Summary of differential expression analysis from paired HCCs and nontumor liver tissues of the SGH cohort.
(C-E) Immunoblots for BCAT1 and BCAT2 in (C) HCCs and nontumor liver tissues from patients of the SGH cohort, (D) liver-derived cancer cell lines, and (E) normal, tumor, and regenerating liver tissues of animal models.
(F) Representative immunohistochemical micrographs from nontumor liver tissue and HCC biopsies, as profiled by The Human Protein Atlas.
(G) Summary of metabolites that were not significantly different in paired HCCs and nontumor liver tissues from patients of the SGH cohort. Data are shown as mean ± s.e.m.
(H) Summary of BCAA catabolic enzyme transcript levels in HCCs from the SGH and TCGA cohorts, sorted by race/ethnicity, tumor etiology, and extratumoral liver inflammation, as well as the association of these characteristics with tumor aggressiveness.
(I) Quantification of cox proportional hazard ratios (95% confidence intervals), significance (log-rank P-value), robustness, and difference in days of estimated survival for patients of the TCGA-LIHC cohort with low expression of indicated BCAA catabolic enzymes.
Figure S2. Loss of BCAA Catabolism Occurs in Liver Cancers but Not Regenerating Liver Tissues, Related to Figure 2
(A) KEGG pathway analysis of all 1202 genes significantly different in DEN and orthotopic (Morris Hepatoma) tumor models (without exclusion of genes significantly different in regenerating tissues).
(B) KEGG pathway analysis of the 226 genes shared by regenerating tissues, and DEN and orthotopic tumors.
(C) RT-PCR analysis of BCAA catabolic enzymes from rat tumor and regenerating tissues, normalized to normal liver tissues.
(D) RT-PCR analysis of BCAA catabolic enzymes from mouse tumor tissues, normalized to nontumor liver tissue.
(E) Expression summary of all 976 genes identified in the transcriptomic analysis. DEN tumor tissues are compared to DEN nontumor tissues (mouse), and Morris Hepatoma and regenerating liver tissues are compared to normal liver tissues (rat).
(F) Summary of expression changes in the top five KEGG pathways.
(G) Non-targeted metabolomics analysis of rat DEN-induced tumors, Morris hepatoma tumors, and regenerating tissues, normalized to normal liver tissues.
(H) Quantification of immunoblots presented in Figure 2F, normalized to respective normal liver tissue controls.
(I) Representative BCKDK immunohistochemical micrographs from nontumor liver tissue and HCC biopsies, as profiled by The Human Protein Atlas.
(J) Quantification of ex vivo BCKDH complex activity in normal liver tissues and DEN-induced tumors in rats.
(K) Representative coronal FLASH MRI images of DEN-induced liver tumors and normal liver from rats used in the MRS analyses.
*P<0.05, §P<0.01, compared to respective controls. Data are shown as mean ± s.e.m.
Figure S3. Reduced BCAA Catabolic Enzyme Expression Is Associated with Copy Number Variations and Transcription Factor Alterations, Related to Figure 3
(A) Ingenuity Pathway Analysis identifying predicted upstream regulators of all significant, differentially expressed genes of TCGA human liver cancers, animal models (different in tumors but not regenerating tissues), and specifically the BCAA catabolic enzymes.
(B) Summary of transcription factor expression in HCCs from the TCGA-LIHC cohort, sorted by stage, grade, vascular invasion, and local invasion.
(C) Kaplan-Meier survival estimate curves for TCGA-LIHC patients ranked by expression of indicated transcription factors. P-values for log-rank test shown.
(D) Transfac analysis identifying enriched transcription factor sequence motifs in the promoters of all significant, differentially expressed genes of human liver cancers, animal models (different in tumors but not regenerating tissues), and specifically the BCAA catabolic enzymes.
(E) Summary of ENCODE ChIP-seq data identifying enriched transcription factors bound to the promoters of BCAA catabolic enzymes.
(F) RT-PCR analysis of BCAA catabolic enzymes from HepG2 cells expressing shRNAs to PPARα or nontargeting control.
(G) Summary of all non-silent mutations of the TCGA-LIHCin proteins related to the nutrient-sensing arm of Mtorc1.
*P<0.05, compared to respective controls. Data are shown as mean ± s.e.m.
Figure S4. BCAA Catabolism Regulates mTORCl Activity and in vitro Cell Proliferation, Related to Figure 4
(A) Immunoblots of the mTORC1 downstream effector S6K in animal liver tumor and regenerating models.
(B) “mTORCl signaling” enrichment plots from gene set enrichment analysis (GSEA) of the 1405 human HCC and 976 animal tumor model (nor regenerating) data sets.
(C) Real-time proliferation curves, immunoblots detailing knockdown efficiency (BCKDHA) and mTORC1 pathway activity (p-S6KThr389 to total S6K, and p-S6Ser235/236 to total S6 ratios), and intracellular BCAA content of AML12 cells expressing a tet-inducible non-targeting control or BCKDHA shRNAs, in the absence or presence of doxycycline and/or the mTOR inhibitors rapamycin (0.05nM) or Torin 1(0.5nM).
(D) Real-time proliferation curves of Hep3B cells grown in complete or BCAA-free media, with or without 3% albumin, 30% FBS, and/or 100nM Torin 1. Similar results were obtained when using Leucine-free media and/or Rapamycin.
(E) Summary of frame-shift mutations caused by CRISPR-Cas9-mediated insertions and/or deletions in the BCKDK gene of Hep3B clones.
(F) Real-time proliferation curves of the liver cancer cell lines treated with the BCKDK inhibitor BT2, and immunoblots after 2 hours of treatment.
(G-I), Characterization of liver cancer cell lines treated with the mTOR inhibitors Rapamycin or Torin 1. (G) Representative real-time proliferation curves. (H) Calculation of proliferation rates (number of divisions per day).
(I) Immunoblots after 2 hours of treatment.
*P<0.05, compared to respective controls. Data are shown as mean ± s.e.m.
Figure S5. Characterization of Mice on BCAA-Supplemented or -Restricted Diets, Related to Figures 5 and 6
(A) Summary of RT-PCR statistical analyses presented in Figure 5H, comparing tissues of LFD+BCAA, HFD, and HFD+BCAA groups to corresponding tissues of the LFD group.
(B) Quantification of nontumor liver tissue amino acid content of DEN-injected mice fed indicated diets, 5 months post injection, normalized to the LFD group. *P<0.05 vs. LFD. §P<0.05 vs. HFD.
(C) Quantification of tumor and nontumor liver tissue amino acid content of DEN-injected mice fed indicated diets, 8 months post injection, normalized to respective LFD groups. *P<0.05 vs. LFD.
(D) Quantification of tumor and nontumor liver tissue acylcarnitine content from DEN-injected mice fed indicated diets, 8 months post injection, normalized to nontumor tissue of the LFD group. *P<0.05 vs. nontumor LFD, §P<0.05 vs. tumor LFD.
(E) RT-PCR analysis of cytokines and F4/80 (encoded by ADGRE1) in normal liver tissue (from uninjected mice), and nontumor and tumor liver tissues (from DEN-injected mice). Results normalized to normal liver tissue of LFD-fed mice.
(F) Representative histological analyses of livers from DEN-injected mice fed indicated diets, 5 months post injection.
(G) Quantification of immunohistochemical staining related to Figures 5K and S5G as percent area (picosirius red) or percent positive cells (remaining), n ≥ at least two random fields per sample, and at least 3 samples per group.
(H) Characterization of DEN-injected mice fed standard chow diets, or chow diets with 0.02% BT2 added, including quantification of the number of tumors (≥3mm) and size of the largest tumor per mouse.
(I) Summary of RT-PCR statistical analyses presented in Figure 6I, comparing tissues of LFD-lowBCAA and LFD+BCAA groups to corresponding tissues of the LFD group.
(J) Quantification of immunohistochemical staining related to Figure 6G as percent area (picosirius red) or percent positive cells (remaining), n ≥ at least two random fields per sample, and at least 3 samples per group.
Data are shown as mean ± s.e.m.
Figure S6. The Impact of Tissue BCAA Catabolism on Cancer Development and Progression, Related to Figure 7
(A) Oncomine analysis of mRNA expression datasets profiling various cancers compared to nontumor controls. Colors reflect the degree of over- or under-expression, and numbers reflect the quantity of studies included in the summation.
(B) Quantification of cox proportional hazard ratios (95% confidence intervals), significance (log-rank P-value), robustness, and difference in days of estimated survival for patients of the TCGA COADREAD, STAD, ACC, KIRC, and KICP cohorts with low expression of indicated BCAA catabolic enzymes.
(C) Immunoblots for mTOR pathway activity in HepG2 cells cultured in complete media, or cultured in BCAA-free media for 4 hours, then stimulated with indicated concentrations of leucine or α-ketoisocaproate (KIC) for 10 minutes.
(D-F) BCAT1inhibition in HepG2 cells. (D) Real-time proliferation curves of HepG2 cells grown in complete media treated with the BCAT1 inhibitor gabapentin. (E) Immunoblots of HepG2 cells 4 hours after gabapentin treatment. (F) Real-time proliferation curves of HepG2 cells grown in BCAA-free media treated with gabapentin.
Figure S7. The Impact of BCAA Tissue Catabolism and Dietary Intake on Overall Cancer Mortality, Related to Figure 7
(A) Kaplan-Meier survival estimate curves of all cancers profiled by TCGA with at least 15 verified deaths. Within each cancer, individuals were ranked by a combined expression index of ACADS, ACADSB, and BCKDHA, and top and bottom quartiles were compared.
(B) Kaplan-Meier survival estimate curve of the normalized pan-cancer dataset by TCGA. Individuals were ranked by a combined expression index of ACADS, ACADSB, and BCKDHA, and top and bottom quartiles were compared. Cox proportional hazard ratio (HR) for low expression group, and p-value for log-rank test shown.
(C) Population characteristics of NHANES III participants, aged 50–66 years old. Data presented as mean (with standard error).
(D) Change in cancer mortality risk associated with increasing BCAA or non BCAA protein content by 1% kcal in the 50–66 year old cohort.
(E-G) Analysis of cancer mortality risk of individuals >66 years-old. (E) Hazard ratios (with 95% confidence interval) based on BCAA intake, 1adjusted for age, sex, race, total kcal, usual dietary intake, diet change, physical activity, intentional weight loss, waist circumference, smoking, education, and prior diagnosis of cancer, diabetes and cardiovascular disease, 2additionally adjusted for % kcal from other macronutrients. (F) Change in cancer mortality risk associated with increasing BCAA or non-BCAA protein content by 1% kcal. (G) Substitution analysis comparing change in risk when replacing BCAAs with carbohydrate or fat, with the same adjustments as HR2 except total kcal and % kcal from non-BCAA protein. Data are presented as hazard ratio (solid line) with 95% confidence interval (shaded area).
(H) Summary of LAT transporter expression in the animal tumor and regenerating models (mean ± s.e.m.), and human HCC (mean ± s.d.; TCGA-LIHC cohort).
(I) Kaplan-Meier survival estimate curves for patients ranked by tumor LAT transporter expression. P-values for log-rank test shown.
*P<0.05, compared to respective controls.
Table S1. Rodent Diet Composition, Related to STAR Methods
Summary of % of kcal, and source of the fat, carbohydrate, and protein used in the rodent studies. Diets with supplemented BCAAs (+BCAA) included an additional 150% of purified leucine, isoleucine, and valine over baseline levels. Diets with restricted BCAAs (-lowBCAA) lowered leucine, isoleucine, and valine to 50% of baseline levels. *LFD-lowAA and LFD+AA diets were used to control for any possible effects of total protein content in LFD-lowBCAA and LFD+BCAA diets, respectively. Baseline BCAA levels were maintained, while all other amino acids were adjusted proportionally.
Table S2. Real-time PCR Primer Sequences, Related to STAR Methods
Primer sequences used for human, mouse, and rat samples.
Highlights.
BCAA catabolism is lost during the development and progression of multiple cancers
BCAA catabolic enzyme expression predicts tumor aggressiveness and patient survival
Tumors display increased BCAA content and enhanced mTORC1 activation
Dietary BCAAs correlate with tumor burden in mice and cancer mortality in humans
Acknowledgements
This work was supported by A*STAR Biomedical Research Council (WH), the National Institutes of Health NIA grant R01AG045351 (MH), an NIH training grant 5T32-CA059365 to Duke University Cancer Biology Training Program (EM). We thank Prof. TC Südhof for insightful comments and encouragements, M. Grabowska, M.Y.B. Ali, X.Q. Teo, C. Shan, M. Choi, Y. Choy, and L. Xin for technical and experimental support, and everyone involved in TCGA for their efforts.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- AICR (2018). Diet, Nutrition, Physical Activity and Cancer: a Global Perspective. Continuous Update Project Expert Report 2018, W.C.R.F.A.I.f.C. Research., ed. (Washington, DC).
- Ally A, Balasundaram M, Carlsen R, Chuah E, Clarke A, Dhalla N, Holt RA, Jones SJM, Lee D, Ma Y, et al. (2017). Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 169, 1327–1341e1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J, Muoio DM, Shiota M, Fujimoto Y, Cline GW, Shulman GI, Koves TR, Stevens R, Millington D, and Newgard CB (2004). Hepatic expression of malonyl-CoA decarboxylase reverses muscle, liver and whole-animal insulin resistance. Nature medicine 10, 268–274. [DOI] [PubMed] [Google Scholar]
- Bernstein AM, Sun Q, Hu FB, Stampfer MJ, Manson JE, and Willett WC (2010). Major dietary protein sources and risk of coronary heart disease in women. Circulation 122, 876–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros PC, Lau SK, Pintilie M, Liu N, Shepherd FA, Der SD, Tsao MS, Penn LZ, and Jurisica I (2009). Prognostic gene signatures for non-small-cell lung cancer. Proc Natl Acad Sci U S A 106, 2824–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas, N. (2012). Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N. (2013). Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N. (2014). Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N., Linehan WM, Spellman PT, Ricketts CJ, Creighton CJ, Fei SS, Davis C, Wheeler DA, Murray BA, Schmidt L, et al. (2016). Comprehensive Molecular Characterization of Papillary Renal-Cell Carcinoma. N Engl J Med 374, 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantranupong L, Wolfson RL, Orozco JM, Saxton RA, Scaria SM, Bar-Peled L, Spooner E, Isasa M, Gygi SP, and Sabatini DM (2014). The Sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep 9,1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton M (2006). Branched-Chain Amino Acid Enriched Supplements as Therapy for Liver Disease. The Journal of nutrition 136, 295S–298S. [DOI] [PubMed] [Google Scholar]
- Chen CL, Yang HI, Yang WS, Liu CJ, Chen PJ, You SL, Wang LY, Sun CA, Lu SN, Chen DS, et al. (2008). Metabolic factors and risk of hepatocellular carcinoma by chronic hepatitis B/C infection: a follow-up study in Taiwan. Gastroenterology 135, 111–121. [DOI] [PubMed] [Google Scholar]
- Faller WJ, Jackson TJ, Knight JR, Ridgway RA, Jamieson T, Karim SA, Jones C, Radulescu S, Huels DJ, Myant KB, et al. (2015). mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature 517, 497–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Chirieac LR, Bueno R, Pass H, Wu W, Malinowska IA, and Kwiatkowski DJ (2014). Tsc1-Tp53 loss induces mesothelioma in mice, and evidence for this mechanism in human mesothelioma. Oncogene 33, 3151–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori A, Tsunoda M, Konuma T, Kobayashi M, Nagy T, Glushka J, Tayyari F, McSkimming D, Kannan N, Tojo A, et al. (2017). Cancer progression by reprogrammed BCAA metabolism in myeloid leukaemia. Nature 545, 500–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heindryckx F, Colle I, and Van Vlierberghe H (2009). Experimental mouse models for hepatocellular carcinoma research. International journal of experimental pathology 90, 367–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al. (2012). The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485, 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang C, Oh SF, Wada S, Rowe GC, Liu L, Chan MC, Rhee J, Hoshino A, Kim B, Ibrahim A, et al. (2016). A branched-chain amino acid metabolite drives vascular fatty acid transport and causes insulin resistance. Nature medicine 22, 421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenerson HL, Yeh MM, Kazami M, Jiang X, Riehle KJ, McIntyre RL, Park JO, Kwon S, Campbell JS, and Yeung RS (2013). Akt and mTORC1 have different roles during liver tumorigenesis in mice. Gastroenterology 144, 1055–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kladney RD, Cardiff RD, Kwiatkowski DJ, Chiang GG, Weber JD, Arbeit JM, and Lu ZH (2010). Tuberous sclerosis complex 1: an epithelial tumor suppressor essential to prevent spontaneous prostate cancer in aged mice. Cancer Res 70, 8937–8947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Minowa O, Kuno J, Mitani H, Hino O, and Noda T(1999). Renal Carcinogenesis, Hepatic Hemangiomatosis, and Embryonic Lethality Caused by a Germ-Line <em>Tsc2</em> Mutation in Mice. Cancer Research 59, 1206. [PubMed] [Google Scholar]
- Laplante M, and Sabatini DM (2012). mTOR signaling in growth control and disease. Cell 149, 274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P, Leong W, Tan T, Lim M, Han W, and Radda GK (2013). In vivo hyperpolarized carbon-13 magnetic resonance spectroscopy reveals increased pyruvate carboxylase flux in an insulin-resistant mouse model. Hepatology 57, 515–524. [DOI] [PubMed] [Google Scholar]
- Liang MC, Ma J, Chen L, Kozlowski P, Qin W, Li D, Goto J, Shimamura T, Hayes DN, Meyerson M, et al. (2010). TSC1 loss synergizes with KRAS activation in lung cancer development in the mouse and confers rapamycin sensitivity. Oncogene 29,1588–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loayza-Puch F, Rooijers K, Buil LC, Zijlstra J, Oude Vrielink JF, Lopes R, Ugalde AP, van Breugel P, Hofland I, Wesseling J, et al. (2016). Tumour-specific proline vulnerability uncovered by differential ribosome codon reading. Nature 530, 490–494. [DOI] [PubMed] [Google Scholar]
- Lynch CJ, and Adams SH (2014). Branched-chain amino acids in metabolic signalling and insulin resistance. Nature reviews Endocrinology 10, 723–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks ODK, Athineos D, Cheung EC, Lee P, Zhang T, van den Broek NJF, Mackay GM, Labuschagne CF, Gay D, Kruiswijk F, et al. (2017). Modulating the therapeutic response of tumours to dietary serine and glycine starvation. Nature 544, 372–376. [DOI] [PubMed] [Google Scholar]
- Marchesini G, Bianchi G, Merli M, Amodio P, Panella C, Loguercio C, Rossi Fanelli F, and Abbiati R (2003). Nutritional supplementation with branched-chain amino acids in advanced cirrhosis: a double-blind, randomized trial. Gastroenterology 124, 1792–1801. [DOI] [PubMed] [Google Scholar]
- Marchesini G, Marzocchi R, Noia M, and Bianchi G (2005). Branched-Chain Amino Acid Supplementation in Patients with Liver Diseases. The Journal of nutrition 135, 1596S–1601S. [DOI] [PubMed] [Google Scholar]
- Matter MS, Decaens T, Andersen JB, and Thorgeirsson SS (2014). Targeting the mTOR pathway in hepatocellular carcinoma: current state and future trends. Journal of hepatology 60, 855–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayers JR, Torrence ME, Danai LV, Papagiannakopoulos T, Davidson SM, Bauer MR, Lau AN, Ji BW, Dixit PD, Hosios AM, et al. (2016). Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science 353, 1161–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Yecies JL, Zhang HH, Howell JJ, Nicholatos J, Harputlugil E, Bronson RT, Kwiatkowski DJ, and Manning BD (2012). Chronic activation of mTOR complex 1 is sufficient to cause hepatocellular carcinoma in mice. Science signaling 5, ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos GK, and DeFrances MC (1997). Liver Regeneration. Science 276, 60–66. [DOI] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. (2006). A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298. [DOI] [PubMed] [Google Scholar]
- Newgard CB (2012). Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell metabolism 15, 606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, et al. (2009). A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell metabolism 9, 311–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nofal M, Zhang K, Han S, and Rabinowitz JD (2017). mTOR Inhibition Restores Amino Acid Balance in Cells Dependent on Catabolism of Extracellular Protein. Mol Cell 67, 936–946e935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, Park Y, Wright K, Pavlova NN, Tuveson DA, and Thompson CB (2015). The Utilization of Extracellular Proteins as Nutrients Is Suppressed by mTORC1. Cell 162, 259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh N, Lin Y, Craft LL, Vadiveloo M, and Lu-Yao GL (2012). Longitudinal associations of leisure-time physical activity and cancer mortality in the Third National Health and Nutrition Examination Survey (1986–2006). J Obes 2012, 518358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EJ, Lee JH, Yu GY, He G, Ali SR, Holzer RG, Osterreicher CH, Takahashi H, and Karin M (2010). Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell 140, 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pate RR, Pratt M, Blair SN, and et al. (1995). Physical activity and public health: A recommendation from the centers for disease control and prevention and the american college of sports medicine. JAMA 273, 402–407. [DOI] [PubMed] [Google Scholar]
- Pavlova NN, and Thompson CB (2016). The Emerging Hallmarks of Cancer Metabolism. Cell metabolism 23, 27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, and Pearce EJ (2013). Metabolic pathways in immune cell activation and quiescence. Immunity 38, 633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BA, Forslund K, Hildebrand F, Prifti E, Falony G, et al. (2016). Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 535, 376–381. [DOI] [PubMed] [Google Scholar]
- Perera RM, Stoykova S, Nicolay BN, Ross KN, Fitamant J, Boukhali M, Lengrand J, Deshpande V, Selig MK, Ferrone CR, et al. (2015). Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature 524, 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffel S, Falcone M, Kneisel N, Hansson J, Wang W, Lutz C, Bullinger L, Poschet G, Nonnenmacher Y, Barnert A, et al. (2017). BCAT1 restricts alphaKG levels in AML stem cells leading to IDHmut-like DNA hypermethylation. Nature 551, 384–388. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, and Chinnaiyan AM (2004). ONCOMINE: A Cancer Microarray Database and Integrated Data-Mining Platform. Neoplasia (New York, NY) 6,1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, and Sabatini DM (2010). Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, and Sabatini DM (2008). The Rag GTPases Bind Raptor and Mediate Amino Acid Signaling to mTORC1. Science 320, 1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, Dimitriadoy S, Liu DL, Kantheti HS, Saghafinia S, et al. (2018). Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 173, 321–337e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze K, Imbeaud S, Letouze E, Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C, Shinde J, Soysouvanh F, et al. (2015). Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 47, 505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimobayashi M, and Hall MN (2016). Multiple amino acid sensing inputs to mTORC1. Cell Res 26, 7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin AC, Fasshauer M, Filatova N, Grundell LA, Zielinski E, Zhou JY, Scherer T, Lindtner C, White PJ, Lapworth AL, et al. (2014). Brain insulin lowers circulating BCAA levels by inducing hepatic BCAA catabolism. Cell metabolism 20, 898–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K, Nonami T, Nakao A, Harada A, Kurokawa T, Sugiyama S, Fujitsuka N, Shimomura Y, Hutson SM, Harris RA, et al. (1996). The valine catabolic pathway in human liver: Effect of cirrhosis on enzyme activities. Hepatology 24,1395–1398. [DOI] [PubMed] [Google Scholar]
- Taya Y, Ota Y, Wilkinson AC, Kanazawa A, Watarai H, Kasai M, Nakauchi H, and Yamazaki S (2016). Depleting dietary valine permits nonmyeloablative mouse hematopoietic stem cell transplantation. Science. [DOI] [PubMed] [Google Scholar]
- Tso SC, Gui WJ, Wu CY, Chuang JL, Qi X, Skvora KJ, Dork K, Wallace AL, Morlock LK, Lee BH, et al. (2014). Benzothiophene carboxylate derivatives as novel allosteric inhibitors of branched-chain alpha-ketoacid dehydrogenase kinase. The Journal of biological chemistry 289, 20583–20593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. (2015). Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- Umemura A, Park e.j., Taniguchi K, Lee JH, Shalapour S, Valasek MA, Aghajan M, Nakagawa H, Seki E, Hall MN, et al. (2014). Liver damage, inflammation, and enhanced tumorigenesis after persistent mTORC1 inhibition. Cell metabolism 20,133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, and Thompson CB (2009). Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva A, Chiang DY, Newell P, Peix J, Thung S, Alsinet C, Tovar V, Roayaie S, Minguez B, Sole M, et al. (2008). Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology 135, 1972–1983, 1983e1971–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, and Holst J (2015). L-type amino acid transport and cancer: targeting the mTORC1 pathway to inhibit neoplasia. American Journal of Cancer Research 5, 1281–1294. [PMC free article] [PubMed] [Google Scholar]
- Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, et al. (2011). Metabolite profiles and the risk of developing diabetes. Nature medicine 17, 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederschain D, Wee S, Chen L, Loo A, Yang G, Huang A, Chen Y, Caponigro G, Yao YM, Lengauer C, et al. (2009). Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle 8, 498–504. [DOI] [PubMed] [Google Scholar]
- Yang JD, and Roberts LR (2010). Hepatocellular carcinoma: A global view. Nature reviews Gastroenterology & hepatology 7, 448–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Cherniack AD, Dewal N, Moffitt RA, Danilova L, Murray BA, Lerario AM, Else T, Knijnenburg TA,Ciriello G, et al. (2016). Comprehensive Pan-Genomic Characterization of Adrenocortical Carcinoma. Cancer cell 29, 723–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. BCAA Catabolism Is Suppressed in Hepatocellular Carcinomas and Predicts Patient Survival, Related to Figure 1
(A) Patient characteristics of the cohort recruited at Singapore General Hospital (SGH), including total number, average age (range), and number (percent) of males, females, Hepatitis B+, Hepatitis C+, and indicated racial backgrounds. Additional information on tumor characteristics of the cohort can be found in Figure 1H.
(B) Summary of differential expression analysis from paired HCCs and nontumor liver tissues of the SGH cohort.
(C-E) Immunoblots for BCAT1 and BCAT2 in (C) HCCs and nontumor liver tissues from patients of the SGH cohort, (D) liver-derived cancer cell lines, and (E) normal, tumor, and regenerating liver tissues of animal models.
(F) Representative immunohistochemical micrographs from nontumor liver tissue and HCC biopsies, as profiled by The Human Protein Atlas.
(G) Summary of metabolites that were not significantly different in paired HCCs and nontumor liver tissues from patients of the SGH cohort. Data are shown as mean ± s.e.m.
(H) Summary of BCAA catabolic enzyme transcript levels in HCCs from the SGH and TCGA cohorts, sorted by race/ethnicity, tumor etiology, and extratumoral liver inflammation, as well as the association of these characteristics with tumor aggressiveness.
(I) Quantification of cox proportional hazard ratios (95% confidence intervals), significance (log-rank P-value), robustness, and difference in days of estimated survival for patients of the TCGA-LIHC cohort with low expression of indicated BCAA catabolic enzymes.
Figure S2. Loss of BCAA Catabolism Occurs in Liver Cancers but Not Regenerating Liver Tissues, Related to Figure 2
(A) KEGG pathway analysis of all 1202 genes significantly different in DEN and orthotopic (Morris Hepatoma) tumor models (without exclusion of genes significantly different in regenerating tissues).
(B) KEGG pathway analysis of the 226 genes shared by regenerating tissues, and DEN and orthotopic tumors.
(C) RT-PCR analysis of BCAA catabolic enzymes from rat tumor and regenerating tissues, normalized to normal liver tissues.
(D) RT-PCR analysis of BCAA catabolic enzymes from mouse tumor tissues, normalized to nontumor liver tissue.
(E) Expression summary of all 976 genes identified in the transcriptomic analysis. DEN tumor tissues are compared to DEN nontumor tissues (mouse), and Morris Hepatoma and regenerating liver tissues are compared to normal liver tissues (rat).
(F) Summary of expression changes in the top five KEGG pathways.
(G) Non-targeted metabolomics analysis of rat DEN-induced tumors, Morris hepatoma tumors, and regenerating tissues, normalized to normal liver tissues.
(H) Quantification of immunoblots presented in Figure 2F, normalized to respective normal liver tissue controls.
(I) Representative BCKDK immunohistochemical micrographs from nontumor liver tissue and HCC biopsies, as profiled by The Human Protein Atlas.
(J) Quantification of ex vivo BCKDH complex activity in normal liver tissues and DEN-induced tumors in rats.
(K) Representative coronal FLASH MRI images of DEN-induced liver tumors and normal liver from rats used in the MRS analyses.
*P<0.05, §P<0.01, compared to respective controls. Data are shown as mean ± s.e.m.
Figure S3. Reduced BCAA Catabolic Enzyme Expression Is Associated with Copy Number Variations and Transcription Factor Alterations, Related to Figure 3
(A) Ingenuity Pathway Analysis identifying predicted upstream regulators of all significant, differentially expressed genes of TCGA human liver cancers, animal models (different in tumors but not regenerating tissues), and specifically the BCAA catabolic enzymes.
(B) Summary of transcription factor expression in HCCs from the TCGA-LIHC cohort, sorted by stage, grade, vascular invasion, and local invasion.
(C) Kaplan-Meier survival estimate curves for TCGA-LIHC patients ranked by expression of indicated transcription factors. P-values for log-rank test shown.
(D) Transfac analysis identifying enriched transcription factor sequence motifs in the promoters of all significant, differentially expressed genes of human liver cancers, animal models (different in tumors but not regenerating tissues), and specifically the BCAA catabolic enzymes.
(E) Summary of ENCODE ChIP-seq data identifying enriched transcription factors bound to the promoters of BCAA catabolic enzymes.
(F) RT-PCR analysis of BCAA catabolic enzymes from HepG2 cells expressing shRNAs to PPARα or nontargeting control.
(G) Summary of all non-silent mutations of the TCGA-LIHCin proteins related to the nutrient-sensing arm of Mtorc1.
*P<0.05, compared to respective controls. Data are shown as mean ± s.e.m.
Figure S4. BCAA Catabolism Regulates mTORCl Activity and in vitro Cell Proliferation, Related to Figure 4
(A) Immunoblots of the mTORC1 downstream effector S6K in animal liver tumor and regenerating models.
(B) “mTORCl signaling” enrichment plots from gene set enrichment analysis (GSEA) of the 1405 human HCC and 976 animal tumor model (nor regenerating) data sets.
(C) Real-time proliferation curves, immunoblots detailing knockdown efficiency (BCKDHA) and mTORC1 pathway activity (p-S6KThr389 to total S6K, and p-S6Ser235/236 to total S6 ratios), and intracellular BCAA content of AML12 cells expressing a tet-inducible non-targeting control or BCKDHA shRNAs, in the absence or presence of doxycycline and/or the mTOR inhibitors rapamycin (0.05nM) or Torin 1(0.5nM).
(D) Real-time proliferation curves of Hep3B cells grown in complete or BCAA-free media, with or without 3% albumin, 30% FBS, and/or 100nM Torin 1. Similar results were obtained when using Leucine-free media and/or Rapamycin.
(E) Summary of frame-shift mutations caused by CRISPR-Cas9-mediated insertions and/or deletions in the BCKDK gene of Hep3B clones.
(F) Real-time proliferation curves of the liver cancer cell lines treated with the BCKDK inhibitor BT2, and immunoblots after 2 hours of treatment.
(G-I), Characterization of liver cancer cell lines treated with the mTOR inhibitors Rapamycin or Torin 1. (G) Representative real-time proliferation curves. (H) Calculation of proliferation rates (number of divisions per day).
(I) Immunoblots after 2 hours of treatment.
*P<0.05, compared to respective controls. Data are shown as mean ± s.e.m.
Figure S5. Characterization of Mice on BCAA-Supplemented or -Restricted Diets, Related to Figures 5 and 6
(A) Summary of RT-PCR statistical analyses presented in Figure 5H, comparing tissues of LFD+BCAA, HFD, and HFD+BCAA groups to corresponding tissues of the LFD group.
(B) Quantification of nontumor liver tissue amino acid content of DEN-injected mice fed indicated diets, 5 months post injection, normalized to the LFD group. *P<0.05 vs. LFD. §P<0.05 vs. HFD.
(C) Quantification of tumor and nontumor liver tissue amino acid content of DEN-injected mice fed indicated diets, 8 months post injection, normalized to respective LFD groups. *P<0.05 vs. LFD.
(D) Quantification of tumor and nontumor liver tissue acylcarnitine content from DEN-injected mice fed indicated diets, 8 months post injection, normalized to nontumor tissue of the LFD group. *P<0.05 vs. nontumor LFD, §P<0.05 vs. tumor LFD.
(E) RT-PCR analysis of cytokines and F4/80 (encoded by ADGRE1) in normal liver tissue (from uninjected mice), and nontumor and tumor liver tissues (from DEN-injected mice). Results normalized to normal liver tissue of LFD-fed mice.
(F) Representative histological analyses of livers from DEN-injected mice fed indicated diets, 5 months post injection.
(G) Quantification of immunohistochemical staining related to Figures 5K and S5G as percent area (picosirius red) or percent positive cells (remaining), n ≥ at least two random fields per sample, and at least 3 samples per group.
(H) Characterization of DEN-injected mice fed standard chow diets, or chow diets with 0.02% BT2 added, including quantification of the number of tumors (≥3mm) and size of the largest tumor per mouse.
(I) Summary of RT-PCR statistical analyses presented in Figure 6I, comparing tissues of LFD-lowBCAA and LFD+BCAA groups to corresponding tissues of the LFD group.
(J) Quantification of immunohistochemical staining related to Figure 6G as percent area (picosirius red) or percent positive cells (remaining), n ≥ at least two random fields per sample, and at least 3 samples per group.
Data are shown as mean ± s.e.m.
Figure S6. The Impact of Tissue BCAA Catabolism on Cancer Development and Progression, Related to Figure 7
(A) Oncomine analysis of mRNA expression datasets profiling various cancers compared to nontumor controls. Colors reflect the degree of over- or under-expression, and numbers reflect the quantity of studies included in the summation.
(B) Quantification of cox proportional hazard ratios (95% confidence intervals), significance (log-rank P-value), robustness, and difference in days of estimated survival for patients of the TCGA COADREAD, STAD, ACC, KIRC, and KICP cohorts with low expression of indicated BCAA catabolic enzymes.
(C) Immunoblots for mTOR pathway activity in HepG2 cells cultured in complete media, or cultured in BCAA-free media for 4 hours, then stimulated with indicated concentrations of leucine or α-ketoisocaproate (KIC) for 10 minutes.
(D-F) BCAT1inhibition in HepG2 cells. (D) Real-time proliferation curves of HepG2 cells grown in complete media treated with the BCAT1 inhibitor gabapentin. (E) Immunoblots of HepG2 cells 4 hours after gabapentin treatment. (F) Real-time proliferation curves of HepG2 cells grown in BCAA-free media treated with gabapentin.
Figure S7. The Impact of BCAA Tissue Catabolism and Dietary Intake on Overall Cancer Mortality, Related to Figure 7
(A) Kaplan-Meier survival estimate curves of all cancers profiled by TCGA with at least 15 verified deaths. Within each cancer, individuals were ranked by a combined expression index of ACADS, ACADSB, and BCKDHA, and top and bottom quartiles were compared.
(B) Kaplan-Meier survival estimate curve of the normalized pan-cancer dataset by TCGA. Individuals were ranked by a combined expression index of ACADS, ACADSB, and BCKDHA, and top and bottom quartiles were compared. Cox proportional hazard ratio (HR) for low expression group, and p-value for log-rank test shown.
(C) Population characteristics of NHANES III participants, aged 50–66 years old. Data presented as mean (with standard error).
(D) Change in cancer mortality risk associated with increasing BCAA or non BCAA protein content by 1% kcal in the 50–66 year old cohort.
(E-G) Analysis of cancer mortality risk of individuals >66 years-old. (E) Hazard ratios (with 95% confidence interval) based on BCAA intake, 1adjusted for age, sex, race, total kcal, usual dietary intake, diet change, physical activity, intentional weight loss, waist circumference, smoking, education, and prior diagnosis of cancer, diabetes and cardiovascular disease, 2additionally adjusted for % kcal from other macronutrients. (F) Change in cancer mortality risk associated with increasing BCAA or non-BCAA protein content by 1% kcal. (G) Substitution analysis comparing change in risk when replacing BCAAs with carbohydrate or fat, with the same adjustments as HR2 except total kcal and % kcal from non-BCAA protein. Data are presented as hazard ratio (solid line) with 95% confidence interval (shaded area).
(H) Summary of LAT transporter expression in the animal tumor and regenerating models (mean ± s.e.m.), and human HCC (mean ± s.d.; TCGA-LIHC cohort).
(I) Kaplan-Meier survival estimate curves for patients ranked by tumor LAT transporter expression. P-values for log-rank test shown.
*P<0.05, compared to respective controls.
Table S1. Rodent Diet Composition, Related to STAR Methods
Summary of % of kcal, and source of the fat, carbohydrate, and protein used in the rodent studies. Diets with supplemented BCAAs (+BCAA) included an additional 150% of purified leucine, isoleucine, and valine over baseline levels. Diets with restricted BCAAs (-lowBCAA) lowered leucine, isoleucine, and valine to 50% of baseline levels. *LFD-lowAA and LFD+AA diets were used to control for any possible effects of total protein content in LFD-lowBCAA and LFD+BCAA diets, respectively. Baseline BCAA levels were maintained, while all other amino acids were adjusted proportionally.
Table S2. Real-time PCR Primer Sequences, Related to STAR Methods
Primer sequences used for human, mouse, and rat samples.
Data Availability Statement
Gene expression profiles have been deposited at the Gene Expression Omnibus (GEO) under the accession number GSE75677.