Abstract
The generation of thousands of fungal genomes is leading to a better understanding of genes and genomic organization within the kingdom. However, the epigenome, which includes DNA and chromatin modifications, remains poorly investigated in fungi. Large comparative studies in animals and plants have deepened our understanding of epigenomic variation, particularly of the modified base 5-methylcytosine (5mC), but taxonomic sampling of disparate groups is needed to develop unifying explanations for 5mC variation. Here we utilize the largest phylogenetic resolution of 5mC methyltransferases (5mC MTases) and genome evolution to better understand levels and patterns of 5mC across fungi. We show that extant 5mC MTase genotypes are descendent from ancestral maintenance and de novo genotypes, whereas the 5mC MTases DIM-2 and RID are more recently derived, and that 5mC levels are correlated with 5mC MTase genotype and transposon content. Our survey also revealed that fungi lack canonical gene body methylation, which distinguishes fungal epigenomes from certain insect and plant species. However, some fungal species possess independently derived clusters of contiguous 5mC encompassing many genes. In some cases, DNA repair pathways and the N6-methyladenine (6mA) DNA modification negatively coevolved with 5mC pathways, which additionally contributed to interspecific epigenomic variation across fungi.
INTRODUCTION
5mC is a conserved modified base found across all domains of life. Large comparative studies in animals and plants have revealed high levels of variation in relative amount and genomic location of 5mC across these lineages (1-10). However, knowledge of 5mC in fungi is taxonomically limited and dispersed across several independent studies (5, 6, 11-20). Similar to animals and plants, cytosines in repeats and transposons are methylated in some fungi, thus 5mC has been implicated in genome defense. For example, in Neurospora crassa, some duplicated DNA segments are subjected to cytosine-to-thymine mutations by Repeat-Induced Point mutation (RIP), and DNA methylated by the 5mC MTases DIM-2 and RID (9, 21, 22). Additionally, in some filamentous fungi such as Ascobolus immersus, the analogous Methylation Induced Premeiotically (MIP) leads to 5mC of some duplicates (23, 24). Whereas, 5mC in other fungal species maintained by DNA METHYLTRANSFERASE 1 (DNMT1) (6) and/or DNA METHYLTRANSFERASE 5 (DNMT5) (15) has unknown biological roles. Of the limited fungal species sampled to date, 5mC is not present in all and is not restricted to repeats (5, 6, 11-20). The absence/presence of 5mC pathways and genomic patterns of 5mC in fungi have been positively correlated with the evolution of DNA repair mechanisms that correct its mutagenic properties (9) and negatively correlated with 6mA of DNA (25), respectively. However, to thoroughly test hypotheses for 5mC variation, genetic and epigenomic data from a diverse taxonomic sampling of fungal species is needed.
Here we present a comprehensive analysis of 5mC across the fungal tree of life. Using phylogenetic approaches we resolved the evolutionary history of 5mC pathways using the largest sampling of fungi to date: 528 species/strains representing all phyla of Dikarya (Ascomycota: n=167, Basidiomycota: n=125) and ‘early-diverging fungi’ (Blastocladiomycota: n=4, Chytridiomycota: n=32, Cryptomycota: n=1, Microsporidia: n=24, Mucoromycota: n=144, and Zoopagomycota: n=30) (Supplementary Table 1) (11, 12). We then compared the evolution of 5mC pathways to the largest and most taxonomically diverse fungal methylome dataset to date. This dataset included whole-genome bisulfite sequencing (WGBS) (26, 27) from 27 novel and 13 reanalyzed species (6, 14-20), which included both phyla of Dikarya (Ascomycota: n=16, and Basidiomycota: n=14), two phyla of early-diverging fungi (Mucoromycota: n=7, and Zoopagomycota: n=3) (Supplementary Table 2 and http://epigenome.genetics.uga.edu/FungiMethylome/index.html) (28). Methylome analysis revealed extensive variation of 5mC across fungi. Hence, within a phylogenetic framework, we tested hypotheses to better explain this variation. Together, we present an extensive analysis of 5mC across the fungal tree of life.
RESULTS AND DISCUSSION
Evolution of 5mC MTases.
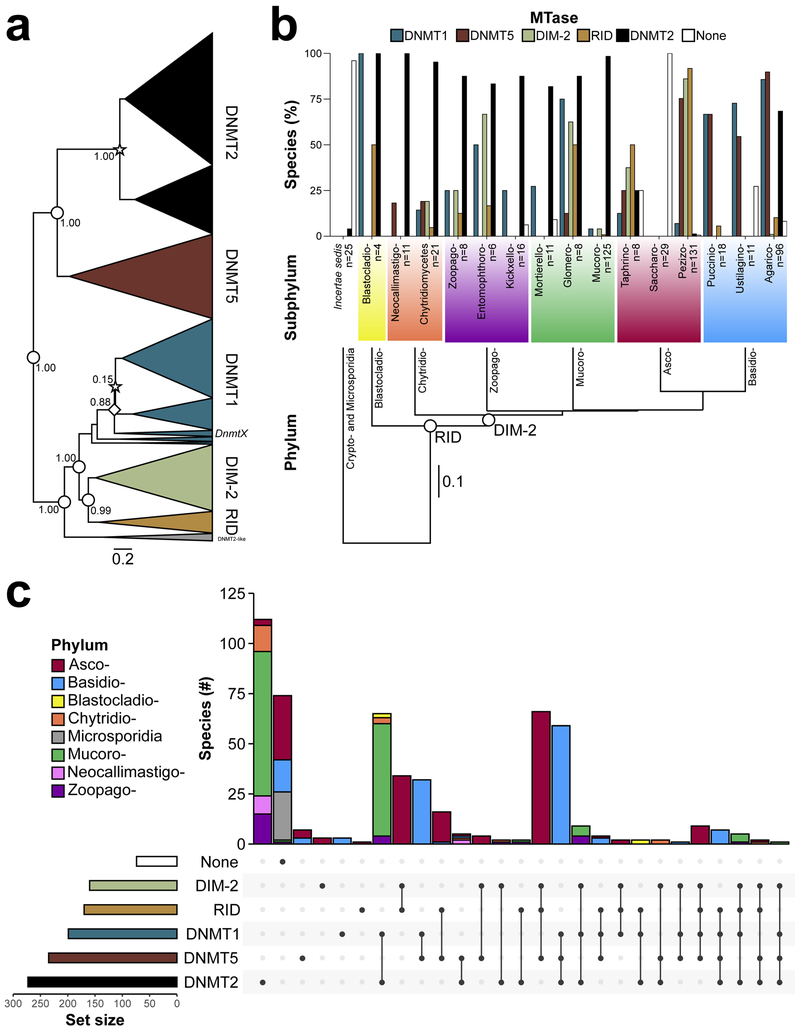
Phylogenetic analysis revealed unique relationships among 5mC MTases in fungi. 5mC MTases group into two monophyletic superclades, which are composed of DNMT5 and DNMT1, DIM-2, and RID (Fig. 1a and Supplementary Fig. 1). Monophyly is also observed for 5mC MTases within the latter superclade (Fig. 1a). However, within DNMT1 relationships are paraphyletic (Fig. 1a). For example, the previously identified ‘DnmtX’ forms a monophyletic clade within the DNMT1 clade (Fig. 1a) (29). Interestingly, the tRNAAsp methyltransferase (tRNA MTase) DNA METHYLTRANSFERASE 2 (DNMT2) (30) is sister to DNMT5 (Fig. 1a). Finally, all fungal species investigated in this study lack the de novo 5mC MTase DNA METHYLTRANSFERASE 3 (DNMT3) (31), which suggests two independent gains in animals and plants or a single loss in the ancestor of all fungi following the divergence from animals.
Figure 1.
Evolution of 5mC MTases across fungi. (a) Phylogenetic relationships of 5mC DNA and tRNA MTases of fungi. Values at selected nodes indicate posterior probability. Nodes with a star specify duplications, and the single node with a diamond specifies the clade containing ‘DnmtX’ (26). Area of the triangle corresponds to the number of taxa. Branch lengths are in units of amino acid substitutions per amino acid site. Phyla abbreviations: Ascomycota (Asco-), Basidiomycota (Basidio-), Blastocladiomycota (Blastocladio-), Chytridiomycota (Chytridio-), Mucoromycota (Mucoro-), and Zoopagomycota (Zoopago-). (b) Proportion of species within subphyla of fungi absent/present for 5mC DNA and tRNA MTases. Empty circles indicate the evolution of a DIM-2 and RID. Branch lengths of the species tree are in substitutions per site. Subphyla abbreviations: Agaricomycotina (Agarico-), Blastocladiomycotina (Blastocladio-), Chytridiomycetes, Entomophthoromycotina (Entomophthoro-), Glomeromycotina (Glomero-), Kickxellomycotina (Kickxelo-), Mortierellomycotina (Mortierello-), Mucoromycotina (Mucoro-), Neocallimastigomycota (Neocallimastigo-), Pezizomycotina (Pezizo-), Pucciniomycotina (Puccinio-), Saccharomycotina (Saccharo-), Taphrinomycotina (Taphrino-), Ustilaginomycotina (Ustilagino-), and Zoopagomycotina (Zoopago-). The number of species within each subphylum investigated is given at the tips. (c) Number of species for the observed combinations of 5mC MTases (5mC MTase genotypes). Phyla abbreviations are identical to a, with the addition of Cryptomycota (Crypto-), and Neocallimastigomycota (Neocallimastigo-).
To better understand 5mC MTase evolution within fungi and across eukaryotes we performed an additional phylogenetic analysis on a subset of DNA methylase domain containing proteins from animals (invertebrates and vertebrates), plants, and fungi with prokaryotic sequences as an outgroup (Supplementary Fig. 2 and Supplementary Table 3). A single prokaryotic clade is sister to a clade containing DNMT1 and the plant ortholog METHYLTRANSFERASE 1 (MET1), the plant specific CHROMOMETHYLASE (CMT), DIM-2 and RID, and a clade containing DNMT3, DNMT5, and the plant specific DOMAINS REARRANGED METHYLTRANSFERASE (DRM) (Supplementary Fig. 2). This relationship suggests a maintenance- and de novo-like 5mC MTase occurred in the ancestor of all eukaryotes (Supplementary Fig. 2). Furthermore, DIM-2 and RID are derived in fungi, with RID evolving prior to DIM-2 rather than jointly as suggested by the fungi exclusive phylogeny (Fig. 1a and Supplementary Fig. 2). The evolution of RID prior to DIM-2 is also supported by taxonomic relationships of subphyla (Fig. 1b). 5mC MTases are evolutionarily old and some have deep ancestry at the base of all eukaryotes and fungi.
Gene duplications and losses have shaped the evolution of 5mC MTases within and across fungi. Species-specific duplications of 5mC (and tRNA) MTases and duplication events in the ancestor of early-diverging fungi and Dikarya are observed for DNMT2 and DNMT1, respectively, and have increased copy number in many fungal species (Fig. 1a and Supplementary Table 1). Conversely, frequent losses of 5mC MTases have occurred during fungal diversification (Fig. 1b). An example of extreme loss is observed in the subphylum Saccharomycotina, with all fungal species investigated being null for all 5mC (and tRNA) MTases (Fig. 1b). Furthermore, losses of all 5mC MTases are observed for Cryptomycota and Microsporidia.
The combination of 5mC MTases (5mC MTase genotype) is diverse in fungi. The highest diversity is observed in the Ascomycota (Fig. 1c, Supplementary Fig. 3 and Supplementary Table 4). The top three most frequent genotypes (disregarding the presence of DNMT2) across all fungi are DNMT1+DNMT5 (n=91), DIM-2+DNMT5+RID (n=88), and DNMT1 (n=68) (Fig. 1c and Supplementary Fig. 4). However, genotypes are not evenly distributed across the fungal phylogeny (Fig. 1c). Basidiomycetes predominantly possess DNMT1+DNMT5, ascomycetes DIM-2+DNMT5+RID or DIM-2+RID, and mucoromycetes DNMT1. Interestingly, the possession of all possible 5mC MTases (DIM-2+DNMT1+DNMT5+RID) is only observed in ascomycetes (Supplementary Table 1). Additionally, we did not observe species with all 5mC MTases and the tRNA MTase DNMT2 (Fig. 1c).
Variation of 5mC across fungi.
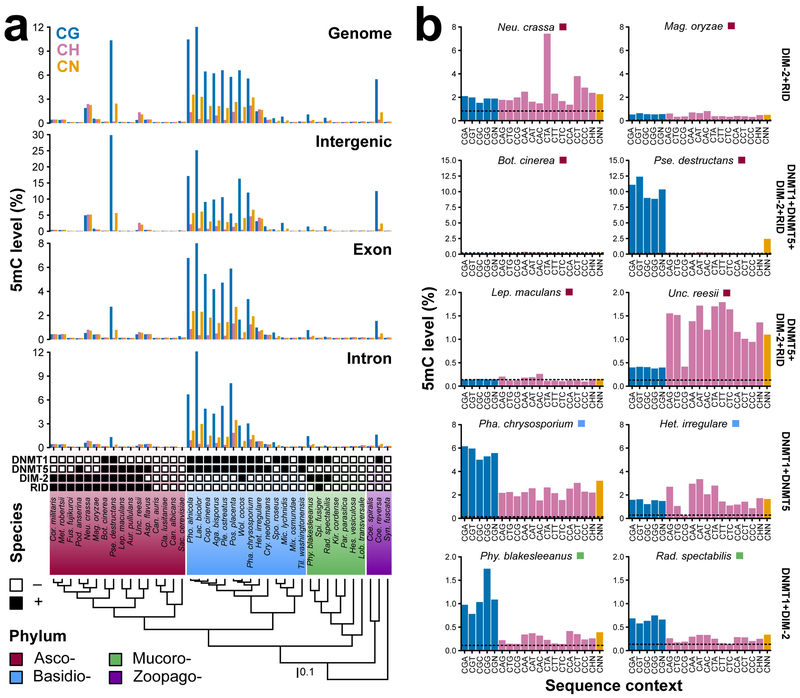
To explore the functional consequences of differential 5mC MTase genotypes we analyzed WGBS data from 40 fungal species (Supplementary Table 2). 5mC levels are on average highest in Basidiomycota compared to other phyla of fungi regardless of genomic location and sequence context (Fig. 2a). Basidiomycota are biased towards the DNMT1+DNMT5 5mC MTase genotype; 5mC MTase genotype is the top predictor of genomic CG methylation levels (Supplementary Fig. 5). Nevertheless, 5mC MTase genotype is not always a predictor of 5mC level. For example, the ascomycetes Botrytis cinerea and Pseudogymnoascus destructans both possess the DIM-2+DNMT1+DNMT5+RID genotype, but the former possesses insignificant levels of 5mC whereas 5mC is abundant in the latter in the tissue-type we sampled (Fig. 2a) Sequence and tissue-specific expression divergence of 5mC MTases between Bot. cinerea and Pse. destructans might explain the discrepancy between 5mC MTase genotype and 5mC levels (Supplementary Fig. 6a). Additionally, some species appear devoid of 5mC when using genome-wide levels of 5mC, however, this does not capture localized, high levels of 5mC that exist in certain species such as in Leptosphaeria maculans ‘brassicae’ (Supplementary Fig. 6b).
Figure 2.
Genome-wide 5mC profiles. (a) Weighted methylation for CG, CH, and CN sites across the genome and various regions of the genome. Empty and filled boxes indicate the absence or presence of 5mC MTases, respectively. Species are ordered based on relationship. Branch lengths of the species tree are in substitutions per site. Phyla abbreviations are identical to Fig. 1a. (b). Weighted 5mC levels at all sequence contexts found across the genome for pairs of fungi with identical 5mC MTase genotypes. Colored boxes beside species names indicate phylum. The dashed line indicates the sodium bisulfite non-conversion rate (estimated background level of methylation).
We observed three major sequence contexts that were targets of fungal 5mC MTases (i) CG, (ii) CH (where H is A, C or T) and (iii) CN (C followed by any nucleotide). Context specificity varies between fungal species with or without similar genotypes suggesting convergent and divergent functions of 5mC MTases (Fig 2b and Supplementary Fig. 7). Convergence is observed between Pse. destructans and the mucoromycete Phycomyces blakesleeanus; 5mC MTase genotypes differ between species, but 5mC is biased towards the CG context with low levels of methylated CH and CN (Fig. 2b). Although, 5mC MTase genotypes are not independent between Pse. destructans and Phy. blakesleeanus and context specificity could be driven by shared DNMT1 and/or DIM-2 (Fig. 2b). An example of divergence is observed in the ascomycetes Neu. crassa and Magnaporthe oryzae, which both possess the DIM-2+RID genotype but preferentially methylate CTA and CAH sites, respectively.
Fungi lack canonical gene-body methylation.
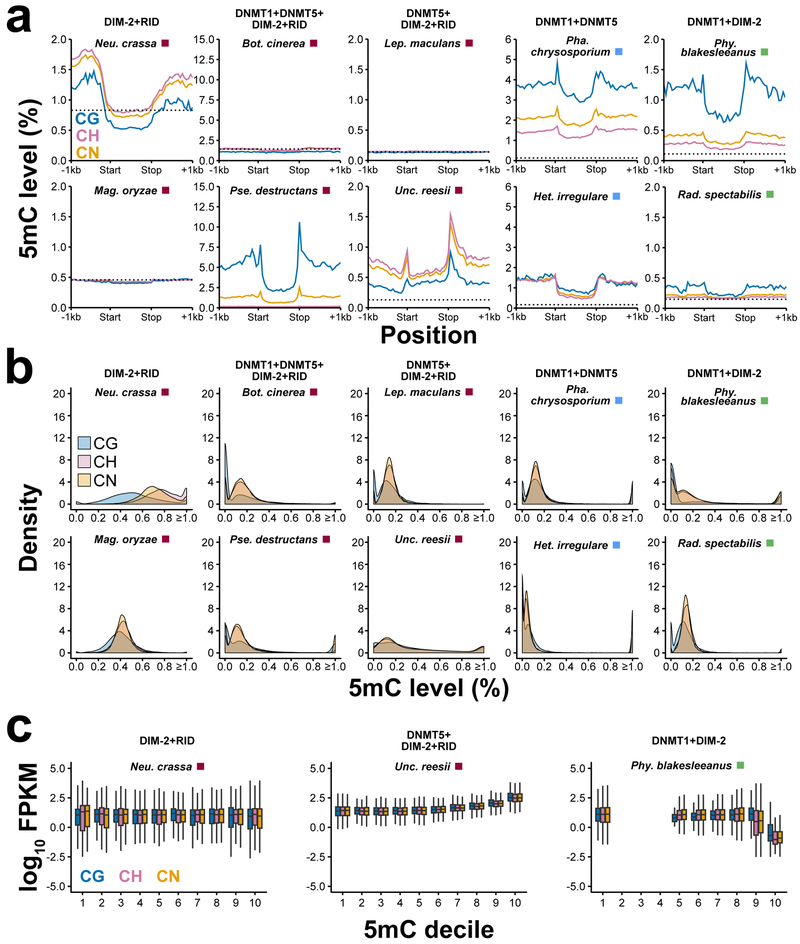
5mC is not uniformly distributed across the genome in fungi. For example, limited 5mC occurs within coding regions (Fig. 3a and Supplementary Fig. 8). This is in contrast to CG DNA methylation enrichment in coding sequences of highly conserved and constitutively expressed genes in some species of insects and angiosperms (i.e., gene body methylation [gbM]) (1, 5, 6, 32, 33). An analogous enrichment has been reported in Unc. reesii at CH contexts (6). Using an enrichment method (1, 34) we confirmed the results in Unc. reesii, and found other species with evidence of enrichment at the CG context (Supplementary Table 5). However, further inspection revealed this 5mC to not be confined to coding regions. Instead, we discovered these genes with 5mC enrichment were localized to long stretches of DNA methylation that spanned genes and intergenic sequences. Furthermore, CG-enriched genes in fungi do not exhibit the same normal-like distribution of CG methylation across the gene body as in plants and certain insect species (Fig. 3a and Supplementary Fig. 9). Genes are typically 5mC-limited (i.e., <1.0%) across fungal species investigated with only a small proportion of genes contributing to the majority of 5mC levels (Fig. 3b). Therefore, these data indicate that the canonical gbM found in other species is absent in fungi.
Figure 3.
5mC profiles of genes. (a) Weighted methylation at CG, CH, and CN sites upstream, within, and downstream of all genes for pairs of fungi with identical 5mC MTase genotypes. The dashed line indicates the sodium bisulfite non-conversion rate (estimated background level of methylation). (b) Distribution of 5mC levels for genes for the same set of ten species in (a). (c) Relationship between gene expression measured as Fragments Per Kilobase of transcript per Million [FPKM] mapped reads and 5mC levels. Empty deciles correspond to missing genic data for those 5mC levels. Boxplot elements: center line, median; upper and lower "hinges", first and third quartiles (the 25th and 75th percentiles), respectively; whiskers, 1.5× interquartile range.
The exact role of genic 5mC in gene expression is debated. In angiosperms, genic CG methylation is associated with constitutively expressed genes (1, 35, 36). However, loss of CG methylation from gene bodies does not lead to steady-state changes to gene expression (37). Non-CG (i.e., CHG and CHH) methylation within genes of plants is typically associated with suppressed gene expression (1). In insects, the association between genic mCG levels and gene expression is negligible (38, 39). The relationship between genic 5mC levels and gene expression fungi is unknown. We tested the association between genic 5mC levels and gene expression in a subset fungal species investigated in this study. Typically, there is no relationship between genic 5mC levels and gene expression (Fig. 3c and Supplementary Fig. 10). However, in 9 of 26 fungal species the most highly 5mC methylated genes have the lowest levels of gene expression (e.g., Het. irregulare, Lac. bicolor, and Pse. destructans). In contrast, one species, Unc. reesii, genic 5mC levels are positively associated with gene expression (Fig. 3c and Supplementary Fig. 10). In most fungi investigated, genic 5mC potentially has no direct effect or suppresses gene expression.
5mC is enriched in repetitive DNA and transposons in fungal genomes.
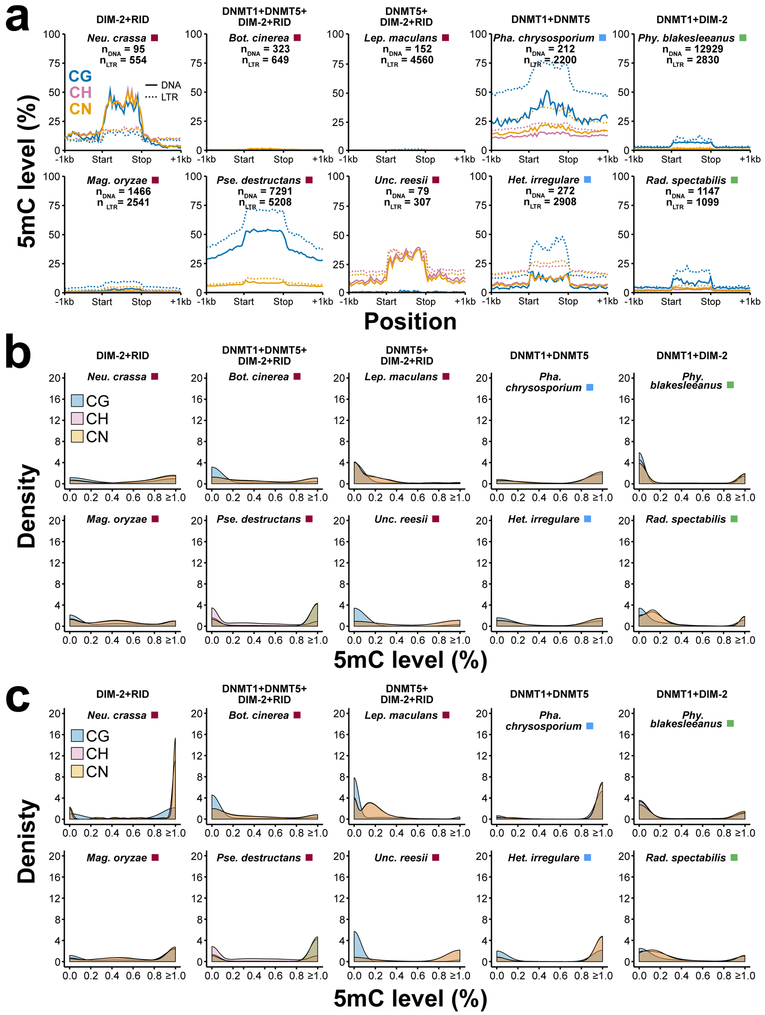
Levels of 5mC are typically highest in repetitive DNA and transposons of animals and plants (5, 6). As in animals and plants, 5mC levels in the few fungal species investigated to date are highest in repetitive DNA and transposons (5, 6). This pattern typically holds true with our increased sampling of fungal diversity for WGBS (Fig. 4a). Similar to genes, transposable elements (TEs) are typically 5mC-limited across fungal species investigated, with only a small proportion of these repeats contributing to the majority of 5mC levels (Fig. 4b and c and Supplementary Fig. 11). Furthermore, as observed for levels of 5mC across the genome and within genes, some species with 5mC DNA MTases have negligible levels of 5mC within repetitive DNA and transposons (Fig. 4a-c). This discrepancy between presence of 5mC DNA MTases and absence of 5mC might reflect developmental, tissue, or cell-type-specific patterns of this modified base.
Figure 4.
5mC profiles of DNA transposons and LTRs. (a) Weighted methylation at CG, CH, and CN sites upstream, within, and downstream of all genes for pairs of fungi with identical 5mC MTase genotypes. The dashed line indicates the sodium bisulfite non-conversion rate (estimated background level of methylation). (b) Distribution of 5mC levels for DNA transposons for the same set of ten species in (a). (c) Distribution of 5mC levels for LTRs for the same set of ten species in (a).
Fungal genomes are punctuated with contiguous regions of 5mC.
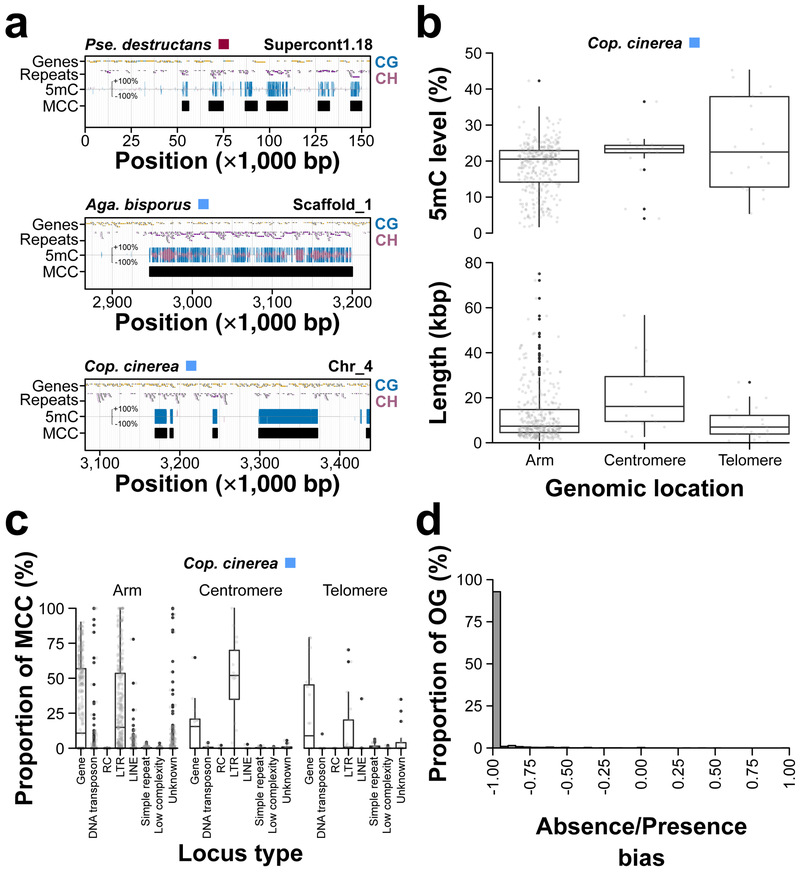
Many fungal species (n=17) possess Methylated Cytosine Clusters (MCCs) (Fig. 5a): long contiguous stretches of highly methylated cytosines (Fig. 4a and Supplementary Fig. 10). MCCs are not taxonomically restricted and are found in fungal species belonging to Ascomycota, Basidiomycota, Mucoromycota, and Zoopagomycota (Supplementary Fig. 12). MCCs are variable between fungal species for proportion of genome, length, and 5mC level. The proportion of genome occupied by MCCs ranged from 0.04% (Phy. blakesleeanus) to 9.90% (Agaricus bisporus) (Supplementary Fig. 12). On average MCCs occupied 3.03% of the genome with a similar level of variation (standard deviation [sd]=3.12%). A large variation in length (base pairs [bp]) of MCCs is also observed: a minimum (min) length of 231 bp (Phanerochaete chrysosporium) and a maximum (max) length of 142,150 bp (Aga. bisporus) with an average (avg) length of 7,517.91 bp and sd of 8,710.77 bp. Variation is less large for 5mC level of MCCs – min=1.44%, max=61.11%, avg=20.55%, and sd=10.20%. MCCs also vary within a species for length, and 5mC level, but some of this variation is dependent on their genomic location (i.e., chromosome arms, centromeres, and telomeres) in at least Coprinopsis cinerea (Fig. 5b). Longer MCCs are on average found within the centromere regions as opposed to arms and telomeres of chromosomes. However, 5mC level of MCCs are identical regardless of their genomic location (Fig. 5b). As expected, repetitive DNA and transposons are found in centromeric and telomeric MCCs, whereas genes are found in MCCs located in chromosome arms (Fig. 5c). However, a similar proportion of Long Terminal Repeats (LTRs) are found in MCCs located in chromosome arms (Fig. 5c). Conservation of MCCs might be driven by the underlying genes, thus genes within MCCs should be orthologous across fungi within these epigenomic features. However, genes within MCCs across fungal species are not orthologous as OrthoGroups (OGs) are not shared among genes found within MCCs (Fig 5d). Furthermore, Gene Ontological (GO) term enrichment suggests genes within MCCs are functionally divergent (Supplementary Fig. 13). Hence, despite MCCs being present in fungal species that are hundreds of millions of years diverged, they appear to be independently derived within each species.
Figure 5.
Methylated Cytosine Clusters (MCCs). (a) Examples of MCCs in Pse. destructans, Agaricus bisporus, and Cop. cinerea. 5mC level is given for both strands of DNA (+ and −). (b) 5mC level and length (kbp) distributions of MCCs located within chromosome arms, centromeres, and telomeres of Cop. cinerea. Boxplot elements: center line, median; upper and lower “hinges”, first and third quartiles (the 25th and 75th percentiles), respectively; whiskers, 1.5× interquartile range; large points, outliers. (c) Genetic content distribution of MCCs located within chromosome arms, centromeres, and telomeres of Cop. cinerea. Boxplot elements: center line, median; upper and lower "hinges", first and third quartiles (the 25th and 75th percentiles), respectively; whiskers, 1.5× interquartile range; large points, outliers. (d) Proportion of orthologous genes absent from or present within MCCs across 17 fungi. Abbreviation: OrthoGroup (OG).
Explanations for interspecific 5mC variation across fungi.
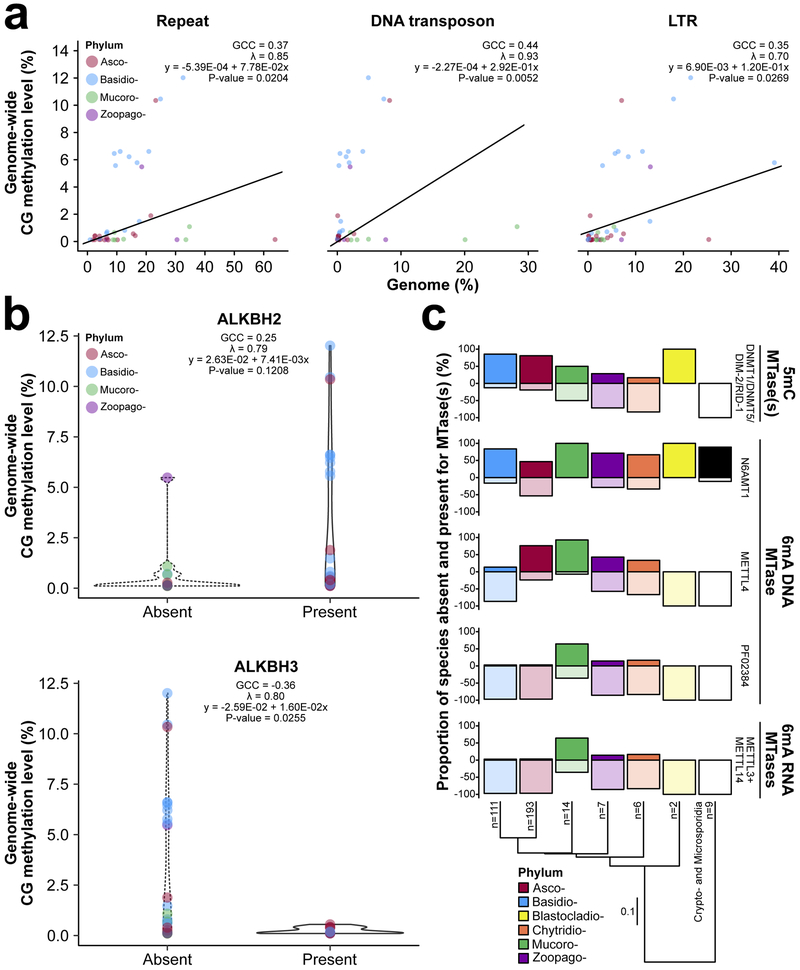
What contributes to interspecific 5mC variation across fungi? Several hypotheses have been tested in animals and plants (1, 5, 9), and our dataset provides a unique opportunity to formally test hypotheses in fungi. One long-standing function for 5mC is its involvement in transcriptional silencing of transposons and other repeats (40). Thus, transposon and repeat content should positively associate with levels of 5mC. We found that repeat content to be a predictor of 5mC in fungi (Supplementary Fig. 5 and 14), and significantly, positively correlates with genome-wide CG methylation levels, with DNA transposon and LTR content as major contributors (Fig. 6a). Overall, repeat content partially explains interspecific 5mC variation between fungal species, and supports a role for 5mC in genome defense.
Figure 6.
Evolutionary relationships of repetitive DNA and transposons, DNA repair pathways and 6mA pathways with 5mC. (a) Correlations between genome-wide CG methylation, and repeat content, DNA transposon, and LTR content adjusted for non-independence of species. The strength of each correlation is given by a ‘generalized correlation coefficient’ (GCC). Lambda (λ) indicates Pagel’s λ and the amount of phylogenetic signal, ranging from 0 (completely independent random walks) to 1 (Brownian motion). (b) Violin plots of genome-wide CG methylation for fungal species absent and present for ALKBH2 and ALKBH3. Correlations between 5mC and DNA repair enzyme are also given. (c) Solid and empty bars correspond to the proportion of species investigated with or without pathways for the establishment and maintenance of 5mC and 6mA of DNA and RNA, respectively. The number of species within each phylum investigated is given at the tips. Branch lengths of the species tree are in substitutions per site. Phyla abbreviations are identical to Fig. 1a.
5mC is mutagenic and causes spontaneous deamination of methylated cytosines to thymines and alkylation damage (41, 42). ALPHA-KETOGLUTARATE DEPENDENT DIOXYGENASE 2 and 3 (ALKBH2 and ALKBH3, respectively) are the only members of the ALKBH family that repair DNA alkylation damage introduced by 5mC (41, 42), and have been observed to associate with DNA MTases in some eukaryotes (8). If ALKBH2 and ALKBH3 coevolved (Supplementary Fig. 15 and Supplementary Table 6) to offset the negative mutational effect of 5mC we would expect these enzymes to coevolve with pathways of 5mC, but negatively correlate with levels of 5mC. Only DNMT5 significantly correlated with ALKBH2 when controlling for non-independence of species (Supplementary Table 7). Levels of 5mC positively correlated with ALKBH2, but not significantly (Fig. 6b and Supplementary Table 9). Whereas, levels of 5mC significantly negatively correlated with ALKBH3 (Fig. 6b and Supplementary Table 8) – i.e., fungi with ALKBH3 tend to have lower levels of 5mC compared to species without ALKBH3. Hence, ALKBH2 and ALKBH3 might be necessary for offsetting the mutational and damaging effect of 5mC. However, counteracting the negative effects of 5mC might be more essential for some fungal species over others. Specifically, those fungal species with high levels of 5mC within coding regions as mutations could disrupt protein function.
The evolutionary relationship between base modifications is poorly understood. A negative relationship between 5mC and 6mA has been reported and proposed to be the consequence of overlapping gene-regulatory functional properties (37). We explored the relationship between 5mC and 6mA by testing for coevolution between the presence 5mC MTases and absence of 6mA DNA and RNA MTases (Supplementary Fig. 16 and Supplementary Tables 9-11). N6AMT1 is responsible for 6mA DNA methylation in humans (43). However, this 6mA DNA MTase is found ubiquitously across fungi and is present in approximately 86.84% and 60.20% of early-diverging fungi and Dikarya (Fig. 6c). Similarly, we observed the presence of a potential 6mA DNA MTase (METHYTRANSFERASE LIKE 4 [METTL4]/ DNA N6-METHYL METHYLTRANSFERASE [DAMT-1]) in many fungal species belonging to the Ascomycota, Chytridiomycota, Mucoromycota, and Zoopagomycota (Fig. 6c). However, METTL4/DAMT-1 was lacking from the majority of Basidiomycota (Fig. 6c). With that said, METTL4/DAMT-1 is present in approximately 47.37% of early-diverging fungi and 53.29% of Dikarya. Other potential 6mA DNA MTases – N-6 DNA Methylase (PF02384) domain containing proteins – were less abundant in the species we sampled (Fig. 6c and Supplementary Table 9). Across fungal species investigated, the coevolution between the presence of 5mC MTases and absence of 6mA MTases was only supported for N6AMT1 (Supplementary Table 7). Specifically, there is a higher rate of losing (9.864) than gaining (1.091) 5mC MTases when N6AMT1 is present. We observed significant positive and negative relationships between 5mC MTases and METTL4/DAMT-1 in Ascomycota and Basidiomycota, respectively (Fig. 6c and Supplementary Table 8). Furthermore, a negative relationship between METTL4/DAMT-1 and the potential 6mA RNA MTases METHYTRANSFERASE LIKE 3 (METTL3) and METHYTRANSFERASE LIKE 14 (METTL14) was observed in both Ascomycota and Basidiomycota. However, this relationship is only significant in the Ascomycota (Fig. 6c and Supplementary Table 7). Resolution of epigenomic conflict that could arise through modified bases with overlapping regulatory functions appears to be more important in some groups of fungi over others.
CONCLUSIONS
Our results identified widespread 5mC variation across the fungal tree of life. 5mC is more associated with DNMT1+DNMT5 MTase genotype, and thus Basidiomycota, than other genotypes and phyla. Unlike animals and plants, fungi lack canonical gbM. Consequently, negligible relationships between genic 5mC levels and gene expression is typically observed. However, like animals and plants, 5mC is present at high levels in repetitive DNA and transposons, which likely reflects a shared mechanism of genome defense. We discovered that 43% of fungal species investigated possess unique methylated clusters (MCCs), potentially reaching up to hundreds of kilobase pairs. We propose that interspecific 5mC variation is the complex combination of 5mC MTase genotype and genome evolution, and in some cases the mitigation of 5mC’s negative effects on sequence changes and distinction of functional roles to other modified bases. However, additional traits might contribute to interspecific 5mC variation in at least fungi (44). Future functional studies controlling for genomic background will elucidate the role of 5mC in fungi. We have added valuable phylogenetic and taxonomic sampling to the field of comparative epigenomics, and unified multiple hypotheses for the explanation of interspecific levels and patterns of 5mC variation.
METHODS
Phylogenetics.
To identify and resolve the relationships of 5mC DNA and tRNA MTases we first identified annotated proteins containing a DNA methylase domain (Pfam domain PF00145) using InterProScan v5.23–62.0 (45) from 528 fungi species/strains. DNA methylase domain containing proteins were also identified from 37 Animalia, 37 Bacteria, and 22 Plantae species (Supplementary Table 3). DNA methylase domains ≥200 amino acids were then extracted from the protein sequences using the coordinates provided by InterProScan v5.23–62.0 (45) and aligned using PASTA v1.6.4 (46). We limited sequence to the DNA methylase domain due to deep divergence times of species, thus sequence divergence, outside of protein functional domains. Phylogenetic relationship among DNA methylase domain containing sequences was estimated using BEAST v2.3.2 (47) with a Blosum62+Γ model of amino acid substitution. A Markov Chain Monte Carlo (MCMC) was ran until stationarity and convergence was reached, and subsequently an appropriate burnin was used prior to summarizing the posterior distribution of tree topologies. A consensus tree was generated using TreeAnnotator v2.3.2, visualized in FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/) and exported for stylization in Affinity Designer v1.5.1 (https://affinity.serif.com/en-us/). 5mC DNA and tRNA MTase clades were assigned based on the placement of previously described methyltransferases in Cryptococcus neoformans var. grubii H99 and Neurospora crassa, best BLASTp hits to Mus musculus, and overall protein domain structure. DNA methylase domain containing sequences not used in the phylogeny were assigned to a clade through best BLASTp hit to sequences assigned to a clade in the phylogeny.
Identical methods described above were used to identify and resolve the relationships of ALKBHs, with minor differences. The number of fungal species included was restricted to those with WGBS. Furthermore, 35 Chordata from Ensembl (http://ensembl.org) (Anolis carolinensis, Astyanax mexicanus, Anas platyrhynchos, Canis familiaris, Choloepus hoffmanni, Ciona intestinalis, Danio rerio, Dasypus novemcinctus, Equus caballus, Echinops telfairi, Gasterosteus aculeatus, Gallus gallus, Gadus morhua, Latimeria chalumnae, Loxodonta africana, Lepisosteus oculatus, Notamacropus eugenii, Myotis lucifugus, Monodelphis domestica, Mus musculus, Ornithorhynchus anatinus, Ovis aries, Oryctolagus cuniculus, Oreochromis niloticus, Oryzias latipes, Homo sapiens, Procavia capensis, Petromyzon marinus, Pelodiscus sinensis, Sorex araneus, Sarcophilus harrisii, Taeniopygia guttata, Takifugu rubripes, Xenopus tropicalis, and Xiphophorus maculatus), and 15 Nematoda from WormBase (http://www.wormbase.org) (Ascaris suum, Brugia malayi, Caenorhabditis briggsae, Caenorhabditis elegans, Dirofilaria immitis, Globodera pallida, Loa loa, Meloidogyne hapla, Meloidogyne incognita, Onchocerca volvulus, Panagrellus redivivus, Pristionchus pacificus, Romanomermis culicivorax, Trichinella spiralis, and Trichuris muris) were included. Annotated proteins containing a 2OG-Fe(II) oxygenase superfamily domain (Pfam domain PF13532) were identified using InterProScan v5.23–62.0 (43). The 2OG-Fe(II) oxygenase superfamily domain was extracted, and subsequently used to estimate phylogenetic relationships of ALKBHs. ALKBH clades were assigned based on the placement of H. sapiens sequences (Supplementary Fig. 15 and Supplementary Table 6).
Identical methods as described for 5mC DNA and tRNA MTases were used to identify potential 6mA DNA and RNA MTases from a set of 342 fungal species with some exceptions. Functional studies on 6mA DNA and RNA MTases are non-existent in fungi, thus we focused on annotated proteins containing the methyltransferase small domain (PF05175), N-6 DNA Methylase domain (PF02384), and the MT–A70 domain (PF05063). Functional work on N6AMT1 – a methyltransferase small domain containing protein – demonstrated this protein as the 6mA DNA MTase in humans (43). Functionally tested and putative 6mA DNA and RNA MTases proteins were identified using InterProScan v5.23–62.0 (45). METTLs were also assigned to specific clades using phylogenetic methods as described for 5mC DNA and tRNA MTases. The location of M. musculus METTL proteins were used to assign clades in Supplementary Fig. 16.
A set of 434 conserved protein coding genes (labeled as JGI_1086) were developed (https://github.com/1KFG/Phylogenomics_HMMs) and searched against proteomes of target species using PHYling (https://github.com/stajichlab/PHYling_unified). Briefly PHYling searches for top hits for each conserved marker using hmmsearch (HMMer v3.1b2; http://hmmer.org, 46) above a minimum e-value threshold (<1.0e-30) in each species proteome. The best hits from each species for each marker are aligned as a multiple alignment using hmmalign (HMMER v3.1.b2) followed by trimmimg with trimal using -automated1 parameter. The trimmed marker alignments were concatenated into a single super alignment and the phylogenetic tree inferred under maximum likelihood (RAxML v8.2.8, 47) using automated bootstrapping which converged after 50 bootstrap replicates (arguments: -f a -m PROTGAMMAAUTO -N autoMRE).
Stochastic character mapping.
A stochastic mutational map was used to estimate the ancestral state at each node, the occurrence and timing of different states, and the timing of changes of 5mC MTase genotypes along the multilocus coalescent tree. Prior to stochastic mutational mapping the multilocus coalescent species tree was converted to a chronogram using the most preferred model of substitution rate variation among branches (relaxed) based on the Akaike information criterion (AIC) using the chronos function in the R package ape v5.0 (50, 61). Stochastic mutational mapping with 1,000 simulations was implemented in the R package phytools v0.6.44 (52). A transition matrix was used allowing for an equal rate of gain and loss of genotypes.
WGBS and methylation analyses.
MethylC-seq libraries for newly sequenced fungal species were prepared according to the protocol described in (27) (Supplementary Table 2). Libraries were single-end 75 or 150 bp sequenced on an Illumina NextSeq500 machine. Unmethylated lambda phage DNA or a mitochondrial genome was used as a control for sodium bisulfite conversion. The non-conversion error rate ranged from 0.30% to 0.11%, with an average rate of 0.16% (sd=0.04%) (Supplementary Table 2). WGBS data was aligned to each species’ respective genome assembly (11, 23, 53-78) using the methylpy pipeline (79). In brief, reads were trimmed of sequencing adapters using Cutadapt (80), and then mapped to both a converted forward strand (cytosines to thymines) and converted reverse strand (guanines to adenines) using bowtie v1.1.1 (81). Reads that mapped to multiple locations, and clonal reads were removed. Mapped sequencing coverage ranged from 4.28× to 51.32×, with an average and standard deviation of 19.01× and 11.36×, respectively (Supplementary Table 2). WGBS data for all newly sequenced species is located on Gene Expression Omnibus (GEO) under accession GSE112636. Previously published WGBS data for Aspergillus flavus (14), Cordyceps militaris (18), Cryptococcus neoformans var. grubii H99 (15), Laccaria bicolor (6), Magnaporthe oryzae (16), Metarhizium robertsii (20), Neurospora crassa (19), Phycomyces blakesleeanus (6), Postia placenta (6), Saccharomyces cerevisiae (17), and Uncinocarpus reesii (6) were downloaded from the Short Read Archive (SRA) using accessions listed in Supplementary Table 2, and aligned identically as described above using the corresponding genome assembly (82-93).
Weighted DNA methylation was calculated for CG, CH, and CN sites by dividing the total number of aligned methylated reads by the total number of methylated plus unmethylated reads (94). For genic and repeat metaplots, the locus body (start to stop codon), 1000 base pairs (bp) upstream, and 1000 bp downstream was divided into 20 windows proportional windows based on sequence length (bp). Weighted DNA methylation was calculated for each window and then plotted in R v3.3.3 (https://www.r-project.org/). CG and CH sequence context enrichment for each gene was determined through a binomial test followed by Benjamini-Hochberg false discovery rate (1, 34). A background methylation level for CG and CH sites was determined from all coding sequence, which was used as a threshold in determining significance with a False Discovery Rate (FDR) correction. Genes were classified as CG- or CH-methylated if they had reads mapping to at least 20 methylated sites with each being sequenced 3× and a q-value ≤0.05 for the context of interest and a q-value >0.05 for the alternative context.
Methylated clusters were identified using a similar method described by Huff and Zilberman (15). First, methylated regions were constructed by defining contiguous runs of cytosines that passed the binomial test from species with 5mC MTases. Methylated clusters were then defined by fusing methylated regions that were ≤1000 bp apart and contained ≥100 methylated cytosines. Methylated cytosines at the CG sequence context were only considered for Asp. flavus, Cry. neoformans var. grubii H99 and Met. robertsii. Methylated clusters were identified in Aga. bisporus, Coe. reversa, Cop. cinerea, Cry. neoformans var. grubii H99, Het. irregulare, Lac. bicolor, Mic. lychnidis A1, Pha. chrysosporium, Pho. alnicola, Phy. blakesleeanus, Ple. ostreatus, Pos. placenta, Pse. destructans, Rad. spectabilis, Spo. roseus, Unc. reesii, and Wol. cocos.
Gene Ontology (GO) term enrichment.
GO term enrichment was performed using Fisher’s exact test implemented in the topGO Bioconductor module in R (95). GO terms were considered significant with a P-value <0.05.
RNA-seq and expression analyses.
RNA-seq libraries for Aga. bisporus (SRR5674591), Asp. flavus (SRR1929577), Aur. pullulans (SRR5145578), Bot. cinerea (SRR5043510, SRR5043508, SRR5040577, SRR5040575, SRR5040545, SRR5040544, SRR5040539, SRR5040538, SRR5040513, SRR5040512, SRR5040511, SRR5040508, SRR5040506, SRR5040505, SRR6924547, SRR6924548, and SRR6924549), Cop. cinerea (ERR364317), Cor. militaris (SRR6252299), Cry. neoformans (SRR6508020), Het. annosum (SRR1797364), Lac. bicolor vSRR1752511), Lep. brassicae (SRR1151407), Mag. oryzae (SRR1015598), Met. robertsii (SRR5282563), Mic. lychnidis (SRR3624826), Neu. crassa (SRR2952639), Pha. chrysosporium (SRR7513038), Pho. alnicola (SRR5501210), Phy. blakesleeanus (SRR5141341), Ple. ostreatus (SRR6986513), Pod. anserina (SRR6974635), Pos. placenta (SRR3929446), Pse. destructans (SRR5770104, SRR5770108, and SRR5770109), Rad. spectabilis (SRR6047893), Spi. fusiger (SRR6053269), Til. washingtonensis (SRR4125823), Unc. reesii (SRR042534), and Wol. cocos (SRR7144104) were downloaded from the SRA. Botrytis cinerea libraries were generated from multiple tissues (ascospores, apothecium disk, apothecium stipes, apothecium primordia, sclerotia, and mycelia).
Raw RNA-seq FASTQ reads were trimmed for adapters and preprocessed to remove low-quality reads using Trimmomatic v0.33 (arguments: TruSeq2-PE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36) (96) prior to mapping. Reads were mapped using HISAT2 v2.1.0 (97) supplied with a reference GTF (General Transfer Format) (arguments: defaults). Following mapping, RNA-Seq alignments were assembled into potential transcripts using StringTie v1.3.3b (97) (arguments: defaults). Mean and standard error of the mean Fragments Per Kilobase of transcript per Million mapped reads (FPKM) was calculated across libraries from the same species and tissue type.
Phylogenetic comparative methods.
Two tests of correlated evolution were conducted: (i) Phylogenetic generalized least squares (PGLS) analysis (98, 99) and (ii) Pagel’s method (100). PGLS is used to test relationships between two (or more) variables while accounting for non-independence of lineages in a phylogeny. The method is a special case of generalized least squares (GLS). PGLS was used to correlate continuous estimates of genome-wide CG methylation to continuous estimates of repeat content and discrete estimates (absence/presence) of ALKBHs. Pagel’s method is similar to PGLS in that it accounts for non-independence of lineages. However, Pagel’s method uses a continuous-time Markov model to simultaneously estimate transition rates in pairs of binary characters on a phylogeny. These rates are then used to test whether a dependent or independent model of evolution is preferred using the likelihood ratio test (LRT). Pagel’s method was used to test for a relationship between the absence/presence of 5mC MTases and the absence/presence of ALKBHs and METTLs, and between the absence/presence of ALKBHs and the absence/presence of DNA methylation. Both tests were implemented in the R package phytools v0.6.44 (52), and the multilocus coalescent species tree was used to account for non-independence of species.
Repeat annotations.
REPET v2.5 (101) was used identify repetitive content and classify conserved and novel repeat elements. These included de novo identification of repeats combined with searches of curated sets from RepBase (102). A set of scripts were developed to run these analyses on the UC Riverside HPCC cluster, (https://github.com/stajichlab/REPET-slurm/). These repeats were classified by matches to RepBase to generate most likely transposable element superfamily categories. The de novo, classified repeats were searched back against each genome to derive a map of repeat element locations for examination of gene and 5mC contexts.
RIP in Neu. crassa mutates C to T at preferentially CA dinucleotides (21). Hence, the frequencies of CA and TA relative to the frequencies of control dinucleotides identify loci that have been subjected to RIP. Specifically, loci with values of the RIP product index (TA / AT) greater than 1.1 and less than 0.9 for the RIP substrate index (CA + TG / AC + GT) have been subjected to RIP. On the other hand, non-mutated loci exhibit values less than 0.8 and greater than 1.1 for RIP product and substrate indices, respectively (103, 104). A composite RIP index (CRI) can be determined by subtracting the substrate index from the product index, thus a positive CRI value implies that the locus has been subjected to RIP (105). We applied these indices to 500 bp non-overlapping windows for the genome assemblies of fungal species listed in Supplementary Table 2. Windows were collapsed into a single locus if neighboring windows on the same molecule exhibited RIP’d or non-mutated index values.
Random forest.
We used an ensemble learning method (Random Forest) analysis to identify variables that best predict levels of genome-wide CG methylation (response variable). Predictor variables tested included both genomic and ecological traits. The number of decision trees was set to 10000, and 5 variables were randomly sampled as candidates at each split. These values were set as they reduce the amount of error. Assessment of the importance of predictors was based on increasing mean squared error (%IncMSE) and the Gini index (%IncNodePurity). Random Forest analysis was implemented in the R package randomForest v4.6.12 (106).
Reporting summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
DATA AVAILABILITY
Genome assemblies and gene annotations are available by following these links listed in Supplementary Table 2. GEO or SRA accessions for RNA-seq and WGBS data generated and used in this study are provided in METHODS.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Elora Demers for DNA from Candida albicans, Clavispora lusitianiae, and Candida auris, Tatiana Giraud for DNA from Microbotryum lychnidis-dioicae A1, Alexander Idnurm for DNA from Sporobolomyces roseus, and Nadia Ponts for DNA from Agaricus bisporus, Botrytis cinerea, Fusarium fujikuroi, Leptosphaeria maculans ‘brassicae’, and Podospora anserina. We also thank Mike Perlin for DNA from Microbotryum lychnidis-dioicae. We thank Nick Rohr and Tina Ethridge for WGBS library preparation for all species sequenced in this study, except Coprinopsis cinerea, Heterobasidion irregulare, and Wolfiporia cocos. We thank Derreck Carter-House and Jericho Ortanez for DNA preparation of Zygomycetes Coemansia spiralis, Hesseltinella vesiculosa, Kirkomyces cordense, Lobosporangium transversale, Parasitella parasitica, Phycomyces blakesleeanus, Radiomyces spectabilis, Spinellus fusiger, Syncephalis fuscata. We thank the following for useful feedback during manuscript preparation: Nick Morffy, and Zack Lewis. We thank the following authors for the use of unpublished genic data: Catherine Aime, Alex Andrianopoulos, Daniele Armaleo, Gerald Bills, Gregory Bonito, Sara Branco, Thomas Bruns, Kathryn Bushley, Ying Chang, In-Geol Choi, Alice Churchill, Luis Corrochano, Christina Cuomo, Alessandro Desirò, Paul Dyer, Javier Franciso, Romina Gazis, John Gladden, Stephen Goodwin, Andrii Gryganskyi, David Hibbett, Derek Johnson, Annegret Kohler, Bjorn Lindahl, François Lutzoni, Jon Magnuson, Jose Maria Barrasa, Francis Martin, Michael Milgroom, Laszlo Nagy, William Nierman, Minou Nowrousian, Donald Nuss, Kerry O'Donnell, Robin Ohm, Chris Pires, Benjamin Schwessinger, Steven Singer, Bernard Slippers, Joseph Spatafora, John Taylor, Adrian Tsang, Sarah Unruh, Kenneth Wolfe, and Larry Zettler. We also thank the Georgia Advanced Computing Resource Center (GACRC) and Georgia Genomics and Bioinformatics Core (GGBC) at the University of Georgia for sequencing and computational resources, respectively. This work was supported by the Office of the Vice President for Research at UGA to RJS and US National Science Foundation grant DEB 1441715 to JES. RJS is a Pew Scholar in the Biomedical Sciences, supported by The Pew Charitable Trusts. Computational analysis on the University of California Riverside High-Performance Computational Cluster were supported by grants from the National Science Foundation DBI-1429826 and National Institutes of Health S10-OD016290. The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02–05CH11231.
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Niederhuth CE & Bewick AJ et al. Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 17, 194 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takuno S, Ran J-H. & Gaut BS. Evolutionary patterns of genic DNA methylation vary across land plants. Nat. Plants 2, 15222 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Bewick AJ, Vogel KJ, Moore AJ & Schmitz RJ Evolution of DNA methylation across insects. Mol. Biol. Evol. 34, 654–665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glastad KG et al. Variation in DNA methylation is not consistently reflected by sociality in Hymenoptera. Genome Biol. Evol. 9, 1687–1698 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng S et al. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. U. S. A. 107, 8689–8694 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zemach A, McDaniel IE, Silva P & Zilberman D Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328, 916–919 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Bewick AJ et al. The evolution of CHROMOMETHYLASES and gene body DNA methylation in plants. Genome Biol. 18, 65 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rošić S, Amouroux R & Requena CE et al. Evolutionary analysis indicates that DNA alkylation damage is a byproduct of cytosine DNA methyltransferase activity. Nat. Genet. Doi: 10.1038/s41588-018-0061-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galagan JE & Selker EU RIP: the evolutionary cost of genome defense. Trends Genet. 20, 417–423 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Gladyshev E & Kleckner N DNA sequence homology induces cytosine-to-thymine mutation by a heterochromatin-related pathway in Neurospora. Nat. Genet. 49, 887–894 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spatafora JW et al. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108, 1028–1046 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stajich JE Fungal genomes and insights into the evolution of the kingdom. Microbiol. Spectr. 5, doi: 10.1128/microbiolspec.FUNK-0055-2016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis ZA et al. Relics of repeat-induced point mutation direct heterochromatin formation in Neurospora crassa. Genome Res. 19, 427–37. (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu S-Y, Lin J-Q & Wu H-L et al. Bisulfite sequencing reveals that Aspergillus flavus holds a hollow in DNA methylation. PLoS One 7, e30349 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huff JT & Zilberman D Dnmt1-independent CG methylation contributes to nucleosome positioning in diverse eukaryotes. Cell 156, 1286–1297 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeon J et al. Genome-wide profiling of DNA methylation provides insights into epigenetic regulation of fungal development in a plant pathogenic fungus, Magnaporthe oryzae. Sci. Rep. 5, 8567 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morselli M et al. In vivo targeting of de novo DNA methylation by histone modifications in yeast and mouse. eLife 4, e06205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang YL et al. Genome-wide analysis of DNA methylation in the sexual stage of the insect pathogenic fungus Cordyceps militaris. Fungal Biol. 119, 1246–1254 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Honda S et al. Dual chromatin recognition by the histone deacetylase complex HCHC is required for proper DNA methylation in Neurospora crassa. Proc. Natl. Acad. Sci. U. S. A. 113, E6135–E6144 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W et al. Differential DNA methylation may contribute to temporal and spatial regulation of gene expression and the development of mycelia and conidia in entomopathogenic fungus Metarhizium robertsii. Fungal Biol. 121, 293–303 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Selker EU Premeiotic instability of repeated sequences in Neurospora crassa. Annu. Rev. Genet. 24, 579–613 (1990). [DOI] [PubMed] [Google Scholar]
- 22.Singer MJ, Marcotte BA, Selker EU DNA methylation associated with repeat-induced point mutation in Neurospora crassa. Mol. Cell Biol. 15, 5586–5597 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhounim L, Rossignol JL, Faugeron G Epimutation of repeated genes in Ascobolus immersus. EMBO J. 11, 4451–4457 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossignol JL & Faugeron G Gene inactivation triggered by recognition between DNA repeats. Experientia 50, 307–317 (1994). [DOI] [PubMed] [Google Scholar]
- 25.Mondo SJ et al. Widespread adenine N6-methylation of active genes in fungi. Nat. Genet. 49, 964–968 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Cokus SJ et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452, 215–219 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urich MA, Nery JR, Lister R, Schmitz RJ, Ecker JR MethylC-seq: Base resolution whole genome bisulfite sequencing library preparation. Nat. Protoc. 10, 475–483 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hofmeister BT & Schmitz RJ Enhanced JBrowse plugins for epigenomics data visualization. BMC Bioinfo. 19, 159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Catania S et al. Epigenetic maintenance of DNA methylation after evolutionary loss of the de novo methyltransferase. Biorxiv doi: 10.1101/149385 (2017). [DOI] [Google Scholar]
- 30.Goll MG et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311, 395–398 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Goll MG, Bestor TH Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74, 481–514 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Stroud H et al. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152, 352–364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bewick AJ & Schmitz RJ Gene body DNA methylation in plants. Curr. Opin. Plant Biol. 36, 103–110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takuno S & Gaut BS Body-methylated genes in Arabidopsis thaliana are functionally important and evolve slowly. Mol. Biol. Evol. 1, 219–227 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Zhang X et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126, 1189–1201 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Zilberman D et al. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39, 61–69 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Bewick AJ et al. On the origin and evolutionary consequences of gene body DNA methylation. Proc. Natl. Acad. Sci. U. S. A. 113, 9111–9116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonasio R et al. Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr. Biol. 22, 1755–1764 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glastad KM, Gokhale K, Liebig J, Goodisman MAD The caste- and sex-specific DNA methylome of the termite Zootermopsis nevadensis. Sci. Rep. 6, 37110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Law JA & Jacobsen SE Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duncan BK & Miller JH Mutagenic deamination of cytosine residues in DNA. Nature 287, 560–561 (1980). [DOI] [PubMed] [Google Scholar]
- 42.Sedgwick B Repairing DNA-methylation damage. Nat. Rev. Mol. Cell Biol. 5, 148–157 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Xiao CL et al. N6-methyladenine DNA modification in the human genome. Mol. Cell. 71, 306–318 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Carlile MJ, Watkinson SC & Gooday GW The Fungi. 2nd edition (Academic Press, London, 2001). [Google Scholar]
- 45.Jones P et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mirarab S et al. PASTA: ultra-large multiple sequence alignment for nucleotide and amino-acid sequences. J. Comput. Biol. 22, 377–386 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bouckaert R et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eddy SR Accelerated Profile HMM Searches. PLoS Comput. Biol. 7, e1002195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stamatakis A RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paradis E, Claude J & Strimmer K APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Popescu A-A, Huber KT & Paradis E ape 3.0: new tools for distance based phylogenetics and evolutionary analysis in R. Bioinformatics 28, 1536–1537 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Revell LJ phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2011). [Google Scholar]
- 53.O’Donnell K, Cigelnik E & Benny GL Phylogenetic relationships among the Harpellales and Kickxellales. Mycologia 90, 624–639 (1998). [Google Scholar]
- 54.O’Donnell K Lutzoni F, Ward TJ & Benny GL Evolutionary relationships among mucoralean fungi (Zygomycota): Evidence for family polyphyly on a large scale. Mycologia 93, 286–296 (2000). [Google Scholar]
- 55.Jones T et al. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 101, 7329–34 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van het Hoog M et al. Assembly of the Candida albicans genome into sixteen supercontigs aligned on the eight chromosomes. Genome Biol. 8, R52 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Espagne E et al. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 9, R77 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Butler G et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459, 657–662 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stajich JE et al. Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus). Proc. Natl. Acad. Sci. U. S. A. 107, 11889–11894 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amselem J et al. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7, e1002230 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rouxel T et al. Effector diversification within compartments of the Leptosphaeria maculans genome affected by Repeat-Induced Point mutations. Nat. Commun. 2, 202 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Floudas D et al. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336, 1715–1719 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Olson A et al. Insight into trade-off between wood decay and parasitism from the genome of a fungal forest pathogen. New Phytol. 194, 1001–1013 (2012). [DOI] [PubMed] [Google Scholar]
- 64.Staats M & van Kan JA Genome update of Botrytis cinerea strains B05.10 and T4. Eukaryot. Cell 11, 1413–1414 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chibucos MC, Crabtree J, Nagaraj S, Chaturvedi S & Chaturvedi V Draft genome sequences of human pathogenic fungus Geomyces pannorum sensu lato and bat white nose syndrome pathogen Geomyces (Pseudogymnoascus) destructans. Genome Announc. 1, e01045–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Muzzey D, Schwartz K, Weissman JS & Sherlock G Assembly of a phased diploid Candida albicans genome facilitates allele-specific measurements and provides a simple model for repeat and indel structure. Genome Biol. 14, R97 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Toome M et al. Genome sequencing provides insight into the reproductive biology, nutritional mode and ploidy of the fern pathogen Mixia osmundae. New Phytol. 202, 554–564 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Walter G et al. DNA barcoding in Mucorales: an inventory of biodiversity. Persoonia 30, 11–47 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wiemann P et al. Deciphering the cryptic genome: genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog. 9, e1003475 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gostincar C et al. Genome sequencing of four Aureobasidium pullulans varieties: biotechnological potential, stress tolerance, and description of new species. BMC Genomics 15, 549 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ohm RA et al. Genomics of wood-degrading fungi. Fungal Genet. Biol. 72, 82–90 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Riley R et al. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc. Natl. Acad. Sci. U. S. A. 111, 9923–9928 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tretter ED et al. An eight-gene molecular phylogeny of the Kickxellomycotina, including the first phylogenetic placement of Asellariales. Mycologia 106, 912–935 (2014). [DOI] [PubMed] [Google Scholar]
- 74.Chang Y et al. Phylogenomic Analyses Indicate that Early Fungi Evolved Digesting Cell Walls of Algal Ancestors of Land Plants. Genome Biol. Evol. 7, 1590–1601 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chatterjee S et al. Draft genome of a commonly misdiagnosed multidrug resistant pathogen Candida auris. BMC Genomics 16, 686 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Perlin MH et al. Sex and parasites: genomic and transcriptomic analysis of Microbotryum lychnidis-dioicae, the biotrophic and plant-castrating anther smut fungus. BMC Genomics. 16, 461 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Drees KP et al. Use of multiple sequencing technologies to produce a high-quality genome of the fungus Pseudogymnoascus destructans, the causative agent of bat white-nose syndrome. Genome Announc. 4, pii: 4/3/e00445–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kijpornyongpan T et al. Broad genomic sampling reveals a smut pathogenic ancestry of the fungal clade Ustilaginomycotina. Mol. Biol. Evol. doi: 10.1093/molbev/msy072 (2018). [DOI] [PubMed] [Google Scholar]
- 79.Schultz MD et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 523, 212–216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martin M & Marcel M Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011). [Google Scholar]
- 81.Langmead B, Trapnell C & Salzberg SL Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goffeau A et al. Life with 6000 genes. Science 274, 563–567 (1996). [DOI] [PubMed] [Google Scholar]
- 83.Galagan JE et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422, 859–868 (2003). [DOI] [PubMed] [Google Scholar]
- 84.Dean RA et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434, 980–986 (2005). [DOI] [PubMed] [Google Scholar]
- 85.Martin F et al. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature 452, 88–92 (2008). [DOI] [PubMed] [Google Scholar]
- 86.Martinez D et al. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc. Natl. Acad. Sci. U. S. A. 106, 1954–1959 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sharpton TJ et al. Comparative genomic analyses of the human fungal pathogens Coccidioides and their relatives. Genome Res. 19, 1722–1731 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gao Q et al. Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet. 7, e1001264 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zheng P et al. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional Chinese medicine. Genome Biol. 12, R116 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Arnaud MB et al. The Aspergillus Genome Database (AspGD): recent developments in comprehensive multispecies curation, comparative genomics and community resources. Nucleic Acids Res. D653–9, doi: 10.1093/nar/gkr875 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hu X et al. Trajectory and genomic determinants of fungal-pathogen speciation and host adaptation. Proc. Natl. Acad. Sci. U. S. A. 111, 16796–16801 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Janbon G et al. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet. 10, e1004261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Corrochano LM et al. Expansion of signal transduction pathways in fungi by extensive genome duplication. Curr. Biol. 26, 1577–1584 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schultz MD, Schmitz RJ & Ecker JR 'Leveling' the playing field for analyses of single-base resolution DNA methylomes. Trends Genet. 28, 583–585 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alexa A & Rahnenfuhrer J topGO: Enrichment Analysis for Gene Ontology. R package version 2.32.0 (2016). [Google Scholar]
- 96.Bolger AM, Lohse M & Usadel B Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics, btu170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pertea M, Kim D, Pertea GM, Leek JT & Salzberg SL Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Grafen A The phylogenetic regression. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 326, 119–157 (1989). [DOI] [PubMed] [Google Scholar]
- 99.Martins EP & Hansen TF Phylogenies and the comparative method: A general approach to incorporating phylogenetic information into the analysis of interspecific data. Am. Nat. 149, 646–667 (1997). [Google Scholar]
- 100.Pagel M Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proc. R. Soc. Lond. B 255, 37–45 (1994). [Google Scholar]
- 101.Flutre T, Duprat E, Feuillet C & Quesneville H Considering transposable element diversification in de novo annotation approaches. PLoS One 6, e16526 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bao W, Kojima KK & Kohany O Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lewis ZA et al. Relics of repeat-induced point mutation direct heterochromatin formation in Neurospora crassa. Genome Res. 19, 427–437 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Margolin BS et al. A methylated Neurospora 5S rRNA pseudogene contains a transposable element inactivated by repeat-induced point mutation. Genet. 149, 1787–1797 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Selker EU et al. The methylated component of the Neurospora crassa genome. Nature 422, 893–897 (2003). [DOI] [PubMed] [Google Scholar]
- 106.Liaw A & Wiener M Classification and regression by randomForest. R News 2, 18–22 (2002). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome assemblies and gene annotations are available by following these links listed in Supplementary Table 2. GEO or SRA accessions for RNA-seq and WGBS data generated and used in this study are provided in METHODS.