There is growing evidence for a link between gastrointestinal bacterial communities and airway disease progression in CF. We demonstrate that infants with CF ≤1 year of age show a distinct stool microbiota versus that of control infants of a comparable age. We detected associations between the gut microbiome and airway exacerbation events in the cohort of infants with CF, and in vitro studies provided one possible mechanism for this observation. These data clarify that current therapeutics do not establish in infants with CF a gastrointestinal microbiota like that in healthy infants, and we suggest that interventions that direct the gastrointestinal microbiota closer to a healthy state may provide systemic benefits to these patients during a critical window of immune programming that might have implications for lifelong health.
KEYWORDS: cystic fibrosis, cytokine, exacerbation, infant, intestine, microbiota, stool
ABSTRACT
Previous work from our group indicated an association between the gastrointestinal microbiota of infants with cystic fibrosis (CF) and airway disease in this population. Here we report that stool microbiota of infants with CF demonstrates an altered but largely unchanging within-individual bacterial diversity (alpha diversity) over the first year of life, in contrast to the infants without CF (control cohort), which showed the expected increase in alpha diversity over the first year. The beta diversity, or between-sample diversity, of these two cohorts was significantly different over the first year of life and was statistically significantly associated with airway exacerbations, confirming our earlier findings. Compared with control infants, infants with CF had reduced levels of Bacteroides, a bacterial genus associated with immune modulation, as early as 6 weeks of life, and this significant reduction of Bacteroides spp. in the cohort with CF persisted over the entire first year of life. Only two other genera were significantly different across the first year of life: Roseburia was significantly reduced and Veillonella was significantly increased. Other genera showed differences between the two cohorts but only at selected time points. In vitro studies demonstrated that exposure of the apical face of polarized intestinal cell lines to Bacteroides species supernatants significantly reduced production of interleukin 8 (IL-8), suggesting a mechanism whereby changes in the intestinal microbiota could impact inflammation in CF. This work further establishes an association between gastrointestinal microbiota, inflammation, and airway disease in infants with CF and presents a potential opportunity for therapeutic interventions beginning in early life.
IMPORTANCE There is growing evidence for a link between gastrointestinal bacterial communities and airway disease progression in CF. We demonstrate that infants with CF ≤1 year of age show a distinct stool microbiota versus that of control infants of a comparable age. We detected associations between the gut microbiome and airway exacerbation events in the cohort of infants with CF, and in vitro studies provided one possible mechanism for this observation. These data clarify that current therapeutics do not establish in infants with CF a gastrointestinal microbiota like that in healthy infants, and we suggest that interventions that direct the gastrointestinal microbiota closer to a healthy state may provide systemic benefits to these patients during a critical window of immune programming that might have implications for lifelong health.
INTRODUCTION
Cystic fibrosis (CF) is an autosomal recessive disease that is known to significantly alter the microenvironments in the airways, intestine, and other organs and is associated with systemic complications such as chronic infections, pancreatic insufficiency, increased systemic inflammation, and ultimately premature morbidity and mortality (1–6). While a great deal of attention has been focused on therapies to treat the chronic airway infections that are often the proximal cause of mortality, it is the gastrointestinal complications that are the cause of significant morbidity for CF patients in early life (7–10). Infants and young children with CF experience difficulty with absorbing nutrients secondary to meconium ileus, pancreatic insufficiency, and small bowel bacterial overgrowth (9, 11–13). Poor weight gain is common, resulting in the need for a high-calorie diet beginning early in life and treatment with pancreatic enzyme replacement therapy (14).
Recent work has suggested an unexpected connection between the gastrointestinal tract microbiota and airway function in young patients with CF. Our group has shown that selected genera (e.g., Enterococcus and Escherichia) colonizing the airway of CF patients tend to first appear in the intestines (15). Furthermore, breastfed infants showed a longer period of pulmonary health and stability prior to their first pulmonary exacerbation, with 20% of breastfed children having no exacerbation over the first 30 months of life, in contrast to exclusively formula-fed infants, all of whom experienced CF exacerbations by 9 months (8). Given the impact of breastfeeding on intestinal microbiota composition (16–18), one possible explanation for these findings in CF is an effect of intestinal bacterial communities on airway disease outcomes. Consistent with this idea, a previous analysis of a subset of subjects from our cohort (aged 0 to 34 months) showed that community composition in the intestine, rather than the airway microbiota, was significantly associated with pulmonary exacerbations prior to age 6 months (8).
In this study, we further probed the relationship between the intestinal microbiota utilizing the Dartmouth Cystic Fibrosis Infant and Children Cohort compared with a subset of infants without CF from a general-population birth cohort, the New Hampshire Birth Cohort Study (NHBCS). Using these two cohorts, we characterize the differences between the intestinal microbiota of patients with CF and the general population over the first year of life, both to understand the differences over time in the microbiota associated with the intestinal tract in these two populations beginning in infancy and to determine if we could gain any mechanistic insight regarding the association between intestinal microbiota and airway disease outcomes through cell culture studies.
RESULTS
Patient population.
We analyzed data from a total of 21 infants with CF and 409 general-population controls (Table 1). The control subjects were selected from a cohort recruited for the New Hampshire Birth Cohort Study from whom at least 2 stool samples were collected during the first year of life at 6 weeks, 4 months, 6 months, 9 months, and/or 12 months (19–21). DNA was extracted from stool samples and analyzed for bacterial composition via amplicon tag sequencing of the 16S rRNA gene as described in Materials and Methods.
TABLE 1.
Summary of the subjects analyzed at each time point
| Subject group | Agea | No. of samples | Feeding modeb (% BF/% formula fed/% mixed) | % with pancreatic insufficiency | Delivery mode (% vaginal) |
|---|---|---|---|---|---|
| Cystic fibrosis patients | 6 wks | 18 | 11/44/44 | 83 | 44 |
| 4 mos | 12 | 8/42/44 | 83 | 67 | |
| 6 mos | 15 | 7/47/47 | 80 | 53 | |
| 9 mos | 14 | 14/57/29 | 79 | 50 | |
| 12 mos | 17 | 12/47/41 | 82 | 53 | |
| Total | 76 | 11/47/42 | 81 | 53 | |
| Control population | 6 wks | 391 | 77/4/19 | NAc | 72 |
| 4 mos | 30 | 77/0/23 | NA | 93 | |
| 6 mos | 28 | 61/0/39 | NA | 93 | |
| 9 mos | 30 | 47/0/53 | NA | 93 | |
| 12 mos | 323 | 30/4/67 | NA | 71 | |
| Total | 802 | 56/3/40 | NA | 74 |
Each time point listed represents a range of samples around that time point as follows: 6 weeks, 2 to 14 weeks; 4 months, 3 to 5 months; 6 months, 5 to 8 months; 9 months, 8 to 11 months; and 12 months, 11 to 16 months.
BF, breastfed. All percentages were rounded to the nearest whole percent. “Mixed” indicates a mixture of BF and formula fed.
NA, not applicable.
The diversity of stool microbial communities is altered in infants with CF in the first year of life.
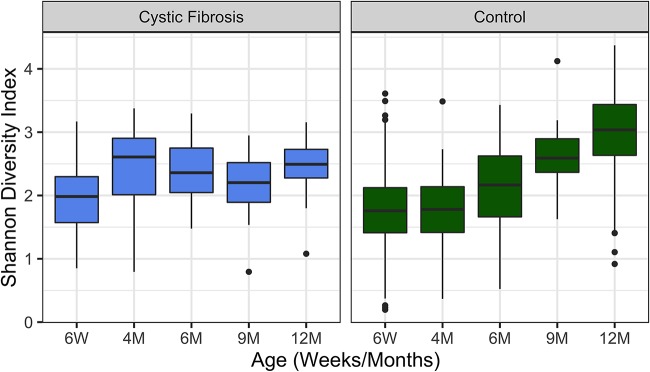
To examine the impact of CF on the bacterial community in individuals at specific time points (alpha diversity), we calculated the Shannon diversity index for infants with and without CF at 6 weeks, 4 months, 6 months, 9 months, and 12 months of age. The alpha diversity of infants with CF did not follow the expected and previously reported (22) pattern of increased diversity over the first year of life observed for the control cohort (Fig. 1). While the difference in alpha diversity between the two cohorts was not significant at 6 weeks of age (Shannon P = 0.18), infants with CF had higher Shannon diversity at 4 months (Shannon P = 0.016) and were surpassed by the control infants by 9 months of age (Shannon P = 0.014). By 12 months of age, infants in the control cohort had significantly higher alpha diversity than infants with CF of the same age (P < 0.001). Within the CF cohort there was only a modest increase in diversity from 6 weeks to 12 months that was not significant by Shannon diversity index after false-discovery rate (FDR) correction (Shannon P = 0.069). This small change in diversity of microbes in the stool for the CF population over the first year of life is in stark contrast to the significant increase we observed in the control cohort (P < 0.0001). We obtained similar findings when analyzing alpha diversity by Shannon diversity index with rarefication or by Fisher’s alpha diversity measures (see Fig. S1 in the supplemental material). These data indicate a significant difference in the community diversity associated with the stool of infants with CF versus the comparison group of control infants as early as 4 months of age.
FIG 1.
Alpha diversity varies between subjects with CF and control infants without CF. Shown are Shannon alpha diversity measures for the cohort of children with CF and the control cohort at the indicated times over the first year of life. The boxes span the first and third quartiles of the data, with the median value indicated as the central horizontal black line. Outliers are indicated as points beyond the span of the whiskers. The difference in alpha diversity between the two cohorts was not significant at 6 weeks of age (Shannon P = 0.18) but was significant at 4 months (Shannon P = 0.016), 9 months (Shannon P = 0.014), and 12 months (P < 0.001) of age. For the CF cohort, there was no significant difference in Shannon diversity index after false-discovery rate (FDR) correction between 6 weeks and 12 months; this difference was significant for the control cohort (P < 0.0001). The Wilcoxon rank sum was used for all comparisons, and FDR correction was applied for multiple-testing correction.
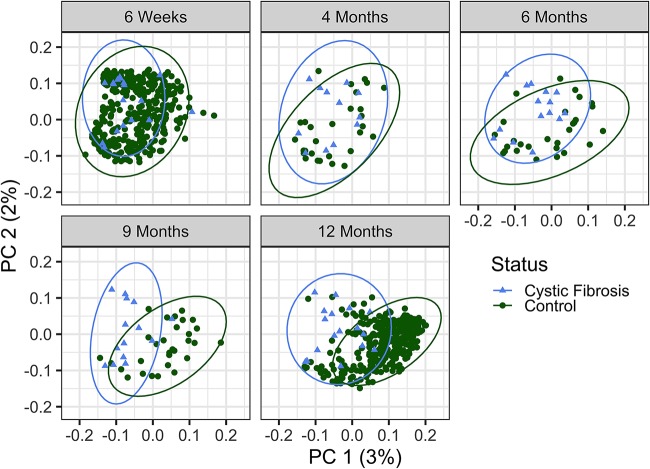
We next analyzed the differences between the samples of the infants with CF in comparison to the control infant group (beta diversity). The gut microbiome of subjects with CF versus that of the control cohort was significantly different across all 5 time points (Fig. 2) (permutational multivariate analysis of variance [PERMANOVA] of generalized UniFrac distances, P = 0.001; PERMANOVA of Bray-Curtis distances, P = 0.001; these data are shown as an aggregated plot with samples from 4 to 14 months in Fig. S2), consistent with the difference in alpha diversity for controls over time in the first year of life. For infants with CF, the similar alpha diversities across the first year are also reflected by the clustering of communities, with no significant difference in community structure across time in the CF cohort (Fig. 2) (PERMANOVA of generalized UniFrac distances, P = 0.397; PERMANOVA of Bray-Curtis distances, P = 0.134).
FIG 2.
Beta diversity of CF and control cohorts over time. The beta diversity of CF samples, as measured by generalized UniFrac distances, is significantly different from that of controls at all time points. Each dot represents a unique sample, and the age and source of the samples are indicated by the color and shape of the dot as depicted in the key. By PERMANOVA, P = 0.015 at 6 weeks, P = 0.004 at 4 months, and P = 0.001 at the remaining time points of 6, 9, and 12 months of age. The percent variability for the total plot region (PC, principal coordinate) is indicated on the x and y axes; plots are subdivided by age for clarity.
Clinical covariates associated with stool microbiota of infants with CF and controls: beta diversity is associated with airway exacerbation.
While feeding mode (exclusively breastfed versus exclusively formula fed or combination fed) was significantly associated with beta diversity in control infants (Fig. S3A) (PERMANOVA of generalized UniFrac distances, P = 0.001; PERMANOVA of Bray-Curtis distances, P = 0.001), this effect did not reach significance among the subjects with CF (PERMANOVA of generalized UniFrac distances, P = 0.081; PERMANOVA of Bray-Curtis distances, P = 0.062). We also noted no significance between these groups for alpha diversity in the CF cohort, while the control cohort did show a significant difference when we compared exclusively breastfed versus exclusively formula-fed infants (Fig. S3B). That is, we found no significant difference between infants with CF who were exclusively breastfed, exclusively formula fed, or combination fed. Similarly, delivery mode has previously been shown to significantly affect the gut microbiota of infants in the New Hampshire Birth Cohort Study (22), but this effect was not observed in the CF cohort (PERMANOVA of generalized UniFrac distances, P = 0.287; PERMANOVA of Bray-Curtis distances, P = 0.121). Additionally, we have noted pancreatic insufficiency as a significant factor in the gut bacterial composition of infants with CF (PERMANOVA of generalized UniFrac distances, P = 0.034; PERMANOVA of Bray-Curtis distances, P = 0.03), but pancreatic insufficiency did not account for the differences observed between infants with CF and controls. That is, when restricting our analysis to only subjects with pancreatic sufficiency, there remained a significant difference between those with CF and controls without CF across all time points (PERMANOVA of generalized UniFrac distances, P = 0.002; PERMANOVA of Bray-Curtis distances, P = 0.001).
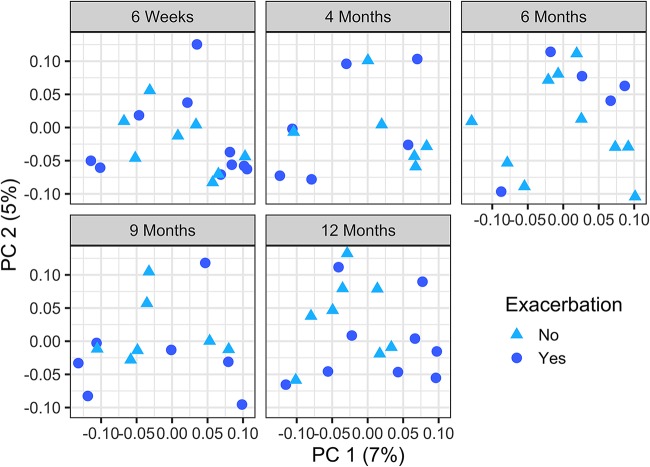
Finally, given that we had observed an association between composition of the stool microbiota and airway exacerbation in CF subjects in a previous study of 13 subjects (0 to 34 months) (8), we examined the correlation between composition of the stool microbiota and airway exacerbation in CF subjects with this larger cohort. As we reported previously, we saw a significant association between bacterial community composition (beta diversity) and airway exacerbation in this CF cohort (Fig. 3) (PERMANOVA of generalized UniFrac distances, P = 0.04; PERMANOVA of Bray-Curtis distances, P = 0.006). These data reinforce the conclusion that there is a correlation between stool bacterial composition and airway disease.
FIG 3.
Beta diversity is associated with airway exacerbation in infants with CF. The principal-coordinate analysis (PCoA) plots depict samples from CF patients at all 5 time points: 6 weeks, 4 months, 6 months, 9 months, and 12 months of age. As indicated in the key, circles indicate that this subject had an exacerbation event in the first year of life, while triangles indicate no such event in the first year. Adjusting for age to account for repeated measures, pulmonary exacerbations in the first year of life were significantly associated with beta diversity, as measured by generalized UniFrac distances (PERMANOVA P = 0.041). The percent variability for the total plot region (PC, principal coordinate) is indicated on the x and y axes; plots are subdivided by age for clarity.
Microbes associated with immune programming are decreased in infants with CF compared to controls.
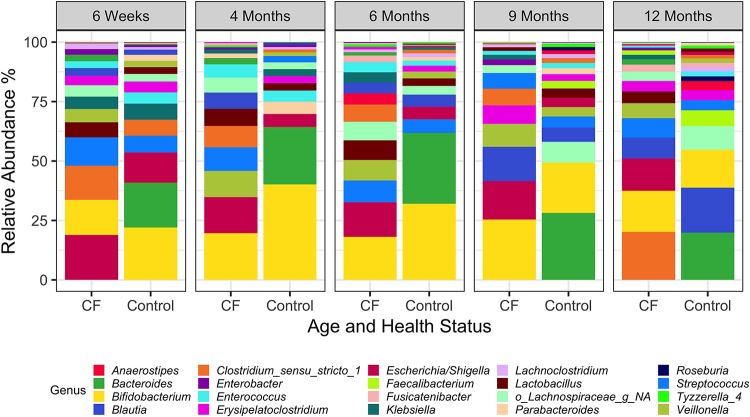
Given the differences in community composition measures between CF and control cohorts in the first year of life, we next examined the relative abundances of the top 20 taxa over the first year of life (Fig. 4 and 5 and Fig. S4). These 20 taxa account for 85% and 88% of the diversity in the CF and control cohorts, respectively.
FIG 4.
Comparison of relative abundances of genera for subjects with CF and the control group over the first year of life. Relative abundance of the top 20 taxa present in stool samples is indicated for the CF and control cohorts. NA, not assigned. The data used to generate this figure are shown in Table S2.
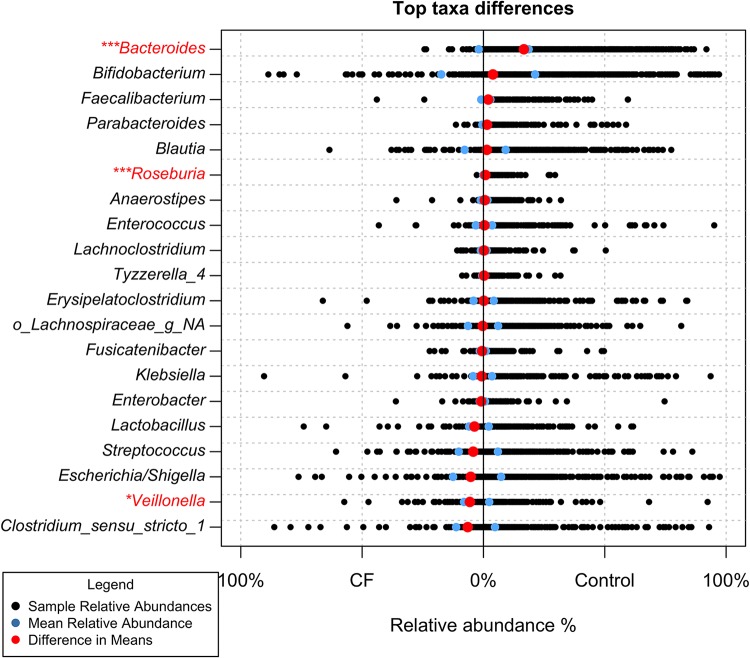
FIG 5.
Relative abundances of genera across the first year of life. Black points represent the relative abundance of each genus in the 76 individual CF samples and 802 individual control samples. The blue points represent the respective mean relative abundance for each genus in the CF and control cohorts. The red points represent the difference in the means (control mean relative abundance − CF mean relative abundance). *, P < 0.05; ***, P < 0.001; significance is marked based on Wilcoxon rank sum P values that were found to be significant after Benjamini-Hochburg multiple-testing correction. Genera significantly different across the first year of life are in red.
Interestingly, we did not observe a major shift in the microbiota between the two cohorts in aggregate or at any particular time point, with the greatest differences observed at 12 months. Indeed, while we observed a marked and significant reduction of the relative abundance of Bacteroides spp. across all time points (average of 1.9% in CF versus 19% in control across all samples [Fig. 5 and 6]), even after correction for the false-discovery rate, in aggregate over the first year of life, the only other taxa significantly different between these populations were Roseburia (0.05% in CF and 0.9% in control) and Veillonella (8% in CF and 2.3% in control [Fig. 5]). A few other selected microbes were significantly less abundant at individual time points, but they were not consistently different over the first year of life (Tables S1 and S2 and Fig. S5).
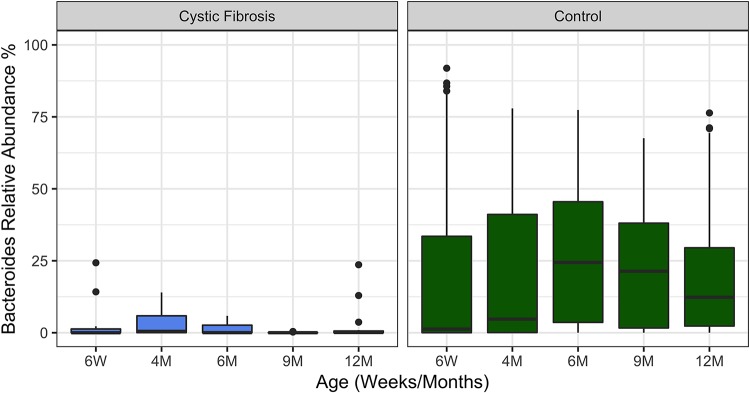
FIG 6.
Relative abundance of Bacteroides in CF and healthy infants over the first year of life. Infants with CF have significantly lower levels of Bacteroides in their stool across all time points in their first year of life (Wilcoxon rank sum P < 0.001). The time points assessed are indicated. The boxes span the first and third quartiles of the data, with the median value indicated in black. Outliers are indicated as points beyond the span of the whiskers. We note that there are similar numbers of outliers in the CF and control cohort, despite the much larger numbers of control patients analyzed, which may be due the low abundance of Bacteroides in the CF cohort making such outliers more easily detected.
Bacteroides has been reported to play an important role in immune programming. Such immune programming microbes, including those in the intestinal tract, impact the formation of a mature immune system in adults (23, 24). Thus, it was unexpected that even for the controls, some individuals had <1% of this genus. To investigate this observation further, the relative abundance of Bacteroides versus Bifidobacterium, a second microbe known to impact immune programming (25–29), was examined on a subject-by-subject basis for both cohorts (Fig. S6). For the control cohort, ∼50% of the 409 subjects with low Bacteroides showed a high relative abundance of Bifidobacterium and vice versa (Fig. S6, control). There was also a group of control subjects with an intermediate level of each microbe. In contrast, most subjects in the CF cohort have a low relative abundance of Bacteroides spp., with only 5 samples containing more than 10% relative abundance of this microbe. Furthermore, only ∼20% of the samples from the CF cohort showed a relative abundance of Bifidobacterium above 50%. Thus, many subjects with CF demonstrated relatively low levels of both of these immune-modulating microbes.
Apical application of Bacteroides cells and secreted products to intestinal epithelial cells reduces IL-8 production.
Our data demonstrate that Bacteroides spp. are significantly reduced in the intestinal microbiota of infants with CF at every time point assessed over the first year of life. Reduction of this immune-modulating microbe is one of the key changes that drive the observed differences in the alpha and beta diversity between the CF and control cohorts. Given the link between intestinal microbiota and airway disease in CF, this observation raises the question of how changes in Bacteroides levels in the intestine might impact the physiology of the airways.
A recent study examining the impact of Bacteroides on intestinal epithelial cells provided a possible clue to the role of Bacteroides in reducing systemic inflammation, including, for example, inflammation in the airways. Some Bacteroides fragilis strains produce enterotoxins that can stimulate the production of interleukin 8 (IL-8). Kim and colleagues (30) showed that exposing the apical face of intestinal epithelial cells to such an enterotoxin resulted in the increased production of IL-8 from the basolateral face of the same cell monolayer. These data indicate that exposing intestinal cells to bacteria on their apical face (i.e., the exposure one would expect from bacteria in the lumen of the intestine) may result in changes in the level of cytokine produced at the basolateral face of the host cells. Such basolaterally secreted cytokines could impact inflammation at a site distant from the intestine. We reasoned that while a toxin on the apical face of the intestinal cell stimulated IL-8 production, a different apical stimulus to the intestinal cells might reduce IL-8 production. Thus, we tested whether apical application of Bacteroides, a bacterium associated with immune modulation, might reduce the production of cytokines from the apical and/or basolateral face of these cells.
To investigate this question, we performed an experiment whereby Caco-2 cells (a human colon cell line derived from an adenocarcinoma) were grown on Transwell filters as polarized monolayers for ∼4 weeks. We used two different Caco-2 cell lines: one line carried loss-of-function mutations in the CFTR gene generated via Cas/CRISPR (designated ΔCFTR), and the other line was wild type (WT) for CFTR but was subjected to mock manipulations used to generate the ΔCFTR line (details provided in Materials and Methods). In initial studies, we exposed a WT Caco-2 cell line to the laboratory strain Bacteroides thetaiotaomicron VPI for 24 h and then assessed a panel of 41 cytokines for changes upon exposure to this microbe, as described in Materials and Methods. Our analysis indicated that IL-8, vascular endothelial growth factor (VEGF), platelet-derived growth factor AA (PDGF-AA), Gro, and IP-10 are reduced by application of Bacteroides. In contrast, interferon α2 (IFN-α2), IL-17α, and IL-1α were unchanged or increased; all other cytokines were below or near the limit of detection of the assay (Table S3). Given that IL-8 is a typical marker for inflammation and that its level was modulated by the addition of B. thetaiotaomicron VPI, IL-8 served as a marker for bacterial cytokine-modulating activity for all subsequent experiments. The basal level of IL-8 produced by the WT and ΔCFTR cell lines was low (∼90 pg/ml of IL-8 on the apical face and ∼20 pg/ml on the basolateral face [data not shown]); thus, we assessed the impact of exposing these lines to tumor necrosis factor alpha (TNF-α) as a means to enhance baseline production of IL-8. Stimulation of the cell lines with 100 ng/ml of TNF-α for 24 h resulted in a marked increase in IL-8 production for both cell lines, and furthermore, we observed significantly more (∼2- to 5-fold) IL-8 production for the ΔCFTR line than for the WT control (unpaired t test, P < 0.05 [Fig. S7]). Thus, in all subsequent experiments we added TNF-α to stimulate IL-8 production and assessed IL-8 production on the ΔCFTR Caco-2 cell line.
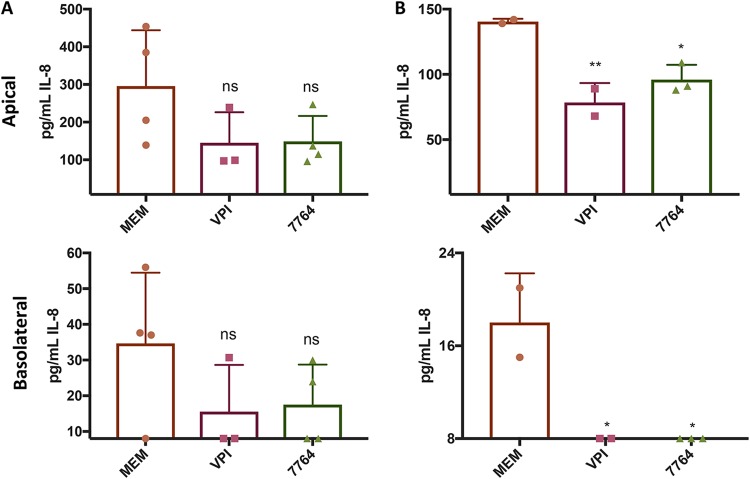
We next tested whether apical application of Bacteroides to these TNF-α-treated cell lines resulted in a reduction in IL-8 levels. To this end, we first applied ∼3.5 × 107 CFU of B. thetaiotaomicron VPI to the apical face of the TNF-α-treated ΔCFTR intestinal cells; B. thetaiotaomicron VPI was not introduced into the basolateral compartment. The IL-8 produced in the apical and basolateral compartments of the Transwells was measured after 24 h of exposure to the bacteria. We found that apically inoculated VPI cells reduced both the apical and basolateral production of IL-8 of the ΔCFTR line (Fig. 7A), although this reduction was not significant. We made a similar observation with a clinical Bacteroides isolate from a patient with CF (strain 7764 [Fig. 7A]).
FIG 7.
Modulation of cytokines by Bacteroides spp. Shown are the apical and basolateral levels of IL-8 for untreated control (MEM) and for the indicated strains: Bacteroides thetaiotaomicron VPI or a clinical Bacteroides isolate (7764). (A) Addition of cells of B. thetaiotaomicron VPI 5482 or the clinical isolate from a CF subject, SMC7764 (7764), to the apical compartment of the Transwell is compared to the untreated control (MEM). Under all conditions, the treatment was supplemented with 100 ng/ml of TNF-α. The filters were incubated anaerobically for 3 h after treatment, and then the added suspension of bacteria was removed and replaced with fresh medium. Filters were returned to anaerobic incubation overnight. Twenty-four hours after initial application of the bacteria, supernatants were collected and IL-8 levels measured via ELISA (PromoKine or Biolegend). To determine CFU, filters were scraped and plated (n = 6); at the end of the experiment there were ∼2 × 109 ± 4 × 108 CFU bacteria/well associated with the apical face of the monolayers, with no significant difference between the strains. (B) Cell-free supernatants (prepared as described in the supplemental material) or medium (MEM) was supplemented with 100 ng/ml of TNF-α and then applied to the apical face of the Caco-2 cells and the filters were incubated anaerobically for 3 h, the supernatant was removed, and then fresh cell-free supernatant supplemented with 100 ng/ml of TNF-α was added and the filters were returned to anaerobic incubation overnight. The exchange of medium at 3 h was performed to mirror the washing step used with cells, as outlined for panel A. Twenty-four hours after initial application of the bacterial supernatants, culture supernatants were collected and IL-8 levels measured via ELISA (PromoKine or Biolegend). For both panels, *, P < 0.05, and **, P < 0.01, by one-way analysis of variance (ANOVA) with Dunnett’s multiple-comparison posttest compared to the untreated (MEM) condition. The pH of the supernatants was ∼8.0 at the start of the experiment and unchanged at ∼8.0 after incubation with the Caco-2 cells.
We next investigated whether secreted products from these strains could also modulate IL-8 production by applying cell-free, sterilized supernatant from the same strains as tested for Fig. 7A to the apical face of the ΔCFTR line. We observed that application of these supernatants to the apical face of the ΔCFTR cell line effectively and significantly reduced IL-8 levels in both the apical and basolateral compartments (Fig. 7B). We tested cells or supernatants from 5 additional clinical isolates of Bacteroides or close relatives; these strains all showed various trends toward reduced IL-8 production, but these trends were not significant (Fig. S8A and B), indicating variability among strains in the ability to modulate IL-8 production in this intestinal cell line.
One limitation of the experimental approach outlined above is the use of Oxyrase to reduce the oxygen levels, as oxygen does limit the growth of the aerotolerant Bacteroides; Oxyrase could potentially interfere with the viability of the ΔCFTR Caco-2 cell line or the IL-8 assay. To mitigate this concern, we prepared supernatant from B. thetaiotaomicron VPI using standard anaerobic culture technique under a headspace of N2. We applied the supernatant from anaerobic bacterial cultures to the ΔCFTR cell line, which was subsequently incubated in a standard incubator with 5% CO2. We found that this anaerobically prepared supernatant significantly reduced IL-8 produced from the apical face of the ΔCFTR Caco-2 cell line (Fig. S8C). Taken together, these data indicate that Bacteroides spp. and close relatives can differentially modulate apical and/or basolateral IL-8 production from an intestinal cell line.
DISCUSSION
In our study, infants with CF showed an intestinal microbiome that is distinct from that of healthy control infants as early as 6 weeks of life, and the composition of the communities remained distinct at 1 year of life. Interestingly, only a small group of genera differed between the cohorts, most significantly, Bacteroides spp. While other studies have also shown a significant reduction in Bacteroides spp. in the stool of patients with CF, this previous work focused on older populations (>1-year-old children, adolescents, or adults) or smaller cohorts and/or reported a larger degree of dysbiosis (31–35), with alterations (i.e., higher or lower relative abundance) in a number of different taxa. Perhaps the larger degree of dysbiosis was observed in these previous studies because older cohorts were evaluated or the studies were performed in cohorts in distinct locations (i.e., South America and other parts of the United States) from the cohort we studied in New England. Some reports also show changes in the abundance of Escherichia coli or other Enterobacteriaceae (32, 35–37), although we did not observe consistent differences in these organisms in our larger cohort over the first year of life. Thus, our work shows that there was a significant reduction of Bacteroides, a critical immune-modulating microbe, in the stool of infants with CF in the first year of life, but there does not appear to be a completely distinct microbiota in the CF cohort compared to that of the controls in this age group. As mentioned above, only two other genera were significantly different across the first year of life (Roseburia was significantly reduced and Veillonella was significantly increased). Other genera showed significant differences but only at selected time points.
Importantly, our previous data (8, 15) and the work presented here indicate a correlation between gut bacterial communities and airway disease, a finding further supported in this larger cohort. Thus, we hypothesized that the inherent intestinal dysbiosis in CF beginning in early life impacts systemic inflammation and immune programming, which likely contributes to airway exacerbation (38–40). Our in vitro data demonstrate that the secreted products of multiple isolates of Bacteroides spp., applied to the apical face of an intestinal cell line defective in CFTR function, can reduce IL-8 secretion from the apical and/or basolateral face of these intestinal cells to various degrees. Our findings in this study are analogous with those in another report showing that apical application of a Bacteroides-derived toxin could alter basolateral production of IL-8. That is, in this previous report (30), it was shown that applying stimulus to the apical face of intestinal cells could alter basolateral cytokine production.
There are several important implications of our findings. First, our data suggest that beneficial strains of Bacteroides can potentially alter systemic signals of inflammation and thereby may impact airway disease through this mechanism. Animal studies will further clarify this link between intestinal microbiota and airway function. Second, while enzyme replacement therapy has clearly had positive transformative impacts on patients with CF (6, 41, 42), the differences we observe in the intestinal microbiota in individuals with CF indicate that they still have underlying defects in intestinal microbiota and suggest a need for further therapeutic interventions. The fact that only a few genera are significantly less abundant over the first year of life in the stool of infants with CF (i.e., Bacteroides and Roseburia) indicates that the alteration(s) in the CF intestinal environment mediated by loss of CFTR function might be relatively specific early in life. Third, our data, in combination with prior work (8, 15), suggest that probiotic therapies or other interventions that shift the intestinal microbiota toward a more health-promoting state may mitigate pulmonary exacerbations. Several systematic reviews of randomized controlled trials of probiotics in groups with CF have shown a positive impact of probiotics on reducing frequency of CF exacerbation and improving gastrointestinal inflammation (43–45); however, mechanisms underlying the observed benefits of altering the intestinal microbiome in CF are not yet clear, and these trials did not supplement the subjects with additional Bacteroides organisms. Given that pulmonary exacerbations are associated with the progressive loss of lung function in this population, resulting in morbidity and ultimately premature mortality, using findings from this and similar studies to reduce the frequency of such clinical episodes beginning in early life would likely assist in maintaining lung function in patients with CF.
MATERIALS AND METHODS
Patient population.
Institutional review board approval was obtained from the Center for the Protection of Human Subjects at Dartmouth College, with yearly renewal of the approval. Eligibility criteria for the CF cohort included diagnosis with CF prenatally or postnatally based upon newborn screening results and subsequent confirmation with sweat chloride and genetic testing. Subjects were eligible if their care for CF would be at clinics affiliated with the Dartmouth-Hitchcock Medical Center in Lebanon, NH, or Manchester, NH. Subjects were excluded if they had non-CF chromosomal anomalies. For the comparison control group without CF, infants were selected from the New Hampshire Birth Cohort Study (NHBCS) if they were delivered at term and provided at least 2 stool samples during their first year of life. For the NHBCS, pregnant women aged 18 to 45 were recruited from prenatal clinics: women were screened for eligibility at an initial prenatal care visit and were enrolled at 24 to 28 weeks’ gestation if they reported using water from a private, unregulated well in their home since their last menstrual period and were not planning a change in residence prior to delivery. Only singleton, live births were included in the cohort.
Analysis of bacterial populations in stool.
Stool samples were analyzed as reported previously (8), with minor modifications. Stool samples were collected from subjects and stored frozen at −80°C until processing. DNA was extracted from stool samples with the Zymo fecal DNA miniprep kit per the manufacturer’s instructions. Illumina targeted sequencing of the V4-V5 hypervariable region of the 16S rRNA gene was performed at the Marine Biological Laboratory in Woods Hole, MA, as we have previously reported (8, 15, 46–49). The 16S rRNA gene sequence reads for subjects with CF and controls were demultiplexed and stored in paired-end forward and reverse read fastq files. Primer sequences were then removed, and all subsequent analyses were performed in R version 3.5.1 (50). All samples were processed through the DADA2 (v1.10.0 [51]) pipeline for quality filtering and trimming, dereplication, and merging of paired-end reads. An amplicon sequence variant (ASV) table was constructed and chimeras were removed. Taxonomy was assigned from the SILVA database (52). The final mean read counts were 116,993.8 (standard deviation [SD], ±50,311.98) in CF patients and 109,920 (SD, ±62,345.95) in controls.
Statistical analyses.
Within-sample alpha diversity was determined using the Shannon diversity index as calculated for each sample in the phyloseq (v1.26.0 [53]) R package function estimate_richness. Significant differences in alpha diversity were determined using Wilcoxon rank sum with FDR correction for multiple testing and subsequently verified with linear regression controlling for age, pancreatic insufficiency, feeding mode, delivery mode, pulmonary exacerbations, and antibiotics use prior to the particular sampling period. For the purposes of this study, antibiotic use prior to sampling is defined as any oral or intravenous antibiotics administered at any time from birth to the time of sample collection. Delivery mode was included in all models that adjusted for antibiotics use to control for confounding, as women who delivered by cesarean section are treated with antibiotics prior to surgery per clinical protocol. The final alpha diversity model selected controlled for age, feeding mode, and delivery mode, as all other covariates were not found to have a significant effect on alpha diversity in the CF cohort. Since Shannon diversity index can be influenced by read depth, we additionally calculated Fisher’s alpha and Shannon diversity index after rarefaction. Differences in the bacterial profiles of stool samples from 21 subjects with CF and 409 comparison infants were analyzed using generalized UniFrac distances from the GUniFrac R package (v1.1 [54]) calculated from a midpoint-rooted phylogenetic tree generated by phangorn (v2.4.0 [55]). Distances were visualized using principal-coordinate analysis (PCoA) plots, and the significance of clustering was determined through permutational multivariate analysis of variance (PERMANOVA) from vegan (v2.5-3 [56]). We additionally confirmed our results using Bray-Curtis distances, which were consistent with the patterns observed using generalized UniFrac distances, as reported in Results. Significant differences in the relative abundance of individual taxa were determined with a Wilcoxon rank sum test, with FDR correction to adjust for multiple testing as indicated, and verified with linear models adjusting for age, feeding mode, antibiotics prior to sampling, delivery mode, pancreatic insufficiency, and pulmonary exacerbations. All code is available at https://github.com/kantosca/CFHCyear1.
Preparation of cell-free supernatants for exposure to intestinal cells.
Bacteroides strains were grown on tryptic soy agar plus 5% sheep’s blood for 48 h in an anaerobic jar. Bacterial cells were scraped off the plate and resuspended in 1 ml of minimal essential medium (MEM), pelleted at 10,000 rpm, and washed once with MEM. These cells were then resuspended in MEM plus 2 mM l-glutamine (MEM–l-Gln) at an optical density at 600 nm (OD600) of 0.1/ml, and 20 μl/ml of Oxyrase was added to the culture. Enough culture was added to overflow a 15-ml Falcon tube, which was then capped tightly and grown on a culture wheel at 37°C for 48 h. Cultures were centrifuged at 5,000 rpm for 10 min, and supernatant was sterile filtered and frozen at −80°C until used. Alternatively, cultures were grown as 25-ml cultures in 200-ml serum vials under a 100% nitrogen headspace using standard anaerobic culture technique. Cell-free supernatants were obtained by removing most cells by centrifugation, followed by passing the supernatants through a 0.22-μm-pore-size filter. Supernatants were prepared aerobically for coculture. Supernatants were diluted 50% in MEM–l-Gln and supplemented with 20 μl/ml of Oxyrase (where indicated) and/or 100 ng/ml of TNF-α (where indicated) before they were exposed to the cell lines. The supernatants prepared from anaerobic culture under a N2 headspace were not supplemented with Oxyrase.
In vitro cell culture studies.
Caco-2 cell lines were grown on Snapwell permeable supports, referred to below and elsewhere as Transwell filters. Cells were cultured in T-75 flasks to 90 to 95% confluency in Gibco Advanced MEM (product number 12492013) supplemented with 2 mM Gibco Glutamax (product number 35050061), Gibco penicillin-streptomycin (50,000 U/500 ml [product number 5140122]), and 10% fetal bovine serum (FBS; Gibco; product number 15140122). Transwell filters were seeded with ∼200,000 cells/filter. All cultures were grown at 37°C with 5% CO2. After 4 weeks, the monolayers were assessed for confluency and polarization by measuring transepithelial resistance as reported previously (57). Briefly, the Transwell filters were mounted in an Ussing chamber (Physiologic Instruments, San Diego, CA) and short circuit current (Isc) was measured by voltage clamping the transepithelial voltage across the monolayers to 0 mV with a voltage clamp (Physiologic Instruments). Amiloride (50 μM) was added to the apical bath solution (5-ml total volume in the apical and basolateral bath solutions) to inhibit Isc attributed to sodium reabsorption, and subsequently Isc was stimulated with forskolin (10 μm), followed by VX-770 (5 μm) to stimulate CFTR-mediated Isc and thiazolidinone (CFTRinh-172, 20 μm; EMD Millipore, Billerica, MA) to inhibit CFTR-mediated Isc. Data were expressed as CFTRinh-172-inhibited Isc in microamps per square centimeter. Data collection and analysis were performed with the Acquire & Analyze data acquisition system (Physiologic Instruments). Monolayers incubated for 2 or 3 weeks did not yield detectable resistance, while cultures at 4 weeks demonstrated measured resistance values of >500 Ω/cm. These 4- to 5-week-old monolayers were washed (both apical and basolateral compartments) twice with fresh MEM (Gibco; product number 51200038), and then 1.5 ml of MEM–l-Gln plus 20 μl/ml of Oxyrase (product number OB0100) was added to the basolateral compartment and 0.25 ml of MEM–l-Gln supplemented with Oxyrase plus bacteria was added to the apical compartment. All experiments included an uninoculated control.
When monolayers were treated with cell-free supernatants, these 4-week-old monolayers were washed (both apical and basolateral compartments) twice with fresh MEM (Gibco product number 51200038), and then 1.5 ml of MEM–l-Gln plus 20 μl/ml Oxyrase was added to the basolateral compartment along with 0.25 ml of MEM–l-Gln diluted 50% with deionized (DI) water and supplementation with 20 μl/ml of Oxyrase (as a control) or cell-free supernatants. Alternatively, when supernatants were generated by anaerobic culture, the monolayers were washed (both apical and basolateral compartments) twice with fresh MEM, and then 1.5 ml of MEM–l-Gln (no Oxyrase) was added to the basolateral compartment along with 0.25 ml of MEM–l-Gln diluted 50% with DI water (as a control) or cell-free supernatants.
Two Caco-2 cell lines were used in this study: one carrying mutations in the CFTR gene generated via Cas-9/CRISPR (designated ΔCFTR) and the other WT for CFTR but was subjected to mock manipulations used to generate the ΔCFTR line. The ΔCFTR line carries frameshifting deletions, and the lines are aneuploid (with most having 3 or 4 copies of chromosome 7 and CFTR). A complete description of the generation of these lines and their characterization is provided elsewhere by Hao and colleagues (58).
Assaying impact of bacteria on intestinal cell production of IL-8.
To prepare the bacterial inoculum for exposure to intestinal cell lines, Bacteroides spp. were grown on tryptic soy agar with 5% sheep blood in an anaerobic atmosphere for 48 h. The bacteria were scraped from the plate, suspend in tissue culture medium, washed once with MEM and the OD600 adjusted to 0.1. At the time of inoculation of the bacteria, TNF-α (product number 8902SC; Cell Signaling Technology, Boston, MA) was added to 100 ng/ml. After 3 h, the apical compartment was washed to remove nonadhered bacteria, and then fresh MEM–l-Gln plus Oxyrase and 100 ng/ml of TNF-α were added to the apical compartment. After an additional 21 h of incubation, supernatants from the apical and basolateral compartments were centrifuged at 1,500 rpm at 4°C for 10 min and stored in 0.5% bovine serum albumin (BSA) at −80°C prior to cytokine quantification.
To determine CFU associated with the apical face of the monolayer of Caco-2 cells, filters were scraped and plated (n = 6); at the end of the experiment there were ∼2 × 109 ± 4 × 108 CFU of B. thetaiotaomicron/well associated with the apical face of the monolayer.
To assess cytokine production, a panel of cytokines produced by the Caco-2 cell line was assayed on plastic after exposure of B. thetaiotaomicron VPI for 24 h. Preparation of the bacterial cells, exposure to the intestinal cells, and preparation of the resulting supernatants from coculturing the bacteria with the intestinal cell line were performed as described in the preceding sections of Materials and Methods. Forty-one cytokines were measured using Millipore human cytokine multiplex kits (EMD Millipore Corporation, Billerica, MA). Calibration curves from recombinant cytokine standards were prepared, and control supernatants from stimulated human peripheral blood mononuclear cells (PBMCs) and dendritic cells were included to determine cytokine recovery. Briefly, wells of 96-well plates were prewet with 100 μl of phosphate-buffered saline (PBS) containing 1% BSA, and then beads together with a standard, sample, spikes, or blank were added in a final volume of 100 μl and incubated at room temperature for 30 min with continuous shaking. Beads were washed three times with 100 μl of PBS containing 1% BSA and 0.05% Tween 20. A cocktail of biotinylated antibodies (50 μl/well) was added to beads for a further 30 min of incubation with continuous shaking. Beads were washed three times, and then streptavidin-phycoerythrin (PE) was added for 10 min. Beads were again washed three times and resuspended in 125 μl of PBS containing 1% BSA and 0.05% Tween 20. The fluorescence intensity of the beads was measured in using the Bio-Plex array reader, and Bio-Plex Manager software with five-parametric-curve fitting was used for data analysis. IL-8 was measured in all subsequent experiments in the apical and basolateral compartments via enzyme-linked immunosorbent assay (ELISA) as per the manufacturer’s instructions (PromoKine and Biolegend).
Isolation and identification of clinical Bacteroides strains.
To isolate clinical Bacteroides strains, stool from patients in our two cohorts was resuspended in phosphate-buffered saline and then dilutions were plated on bile esculin agar to selectively isolate Bacteroides spp. and close relatives after 48 h of anaerobic incubation. Bacteroides spp. and close relatives are identified, in part, by colony blackening due to hydrolysis of the esculin; nonhydrolyzing colonizes are also selected if they grew on this medium and only grew anaerobically. 16S rRNA gene sequencing was performed to confirm the identification of Bacteroides and close relatives and to assign the species, if possible. 16S rRNA gene DNA was amplified directly from strains by colony PCR with standard Taq buffer and reagents with the previously reported (59, 60) universal forward (5′ AGA GTT TGA TCC TGG CTC AG 3′) and reverse (5′ ACG GGC GGT GTG TRC 3′) primers (10 μM each). The annealing temperature was 59°C, with 35 30-s cycles. DNA sequencing was performed on an Applied Biosystems 3730 using both the forward and reverse universal primers.
Accession number(s).
All sequence reads can be found in GenBank with NHBCS reads found under accession number PRJNA296814 (SRP064159), and the CF cohort sequences can be found under accession number PRJNA170783 (SRP014429).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the NIH (grants R37 AI83256-06 and T32HL134598 to G.A.O.; P20GM104416, P20-GM103413, UH3OD023275, NIEHS-P01ES022832/EPA-RD83544201, and 2P42ES007373 to M.R.K. and J.C.M.; P30-DK117469 [NIDDK P30/DartCF]; and T32AI007363), the Cystic Fibrosis Foundation (CFF; grant OTOOLE16G0 to G.A.O., the Harry Shwachman Clinical Investigator Award [MADAN12A0] to J.C.M., grant MADAN18A0 to J.C.M., and the CF-Research Development Program [STANTO07R0]), and the Hearst Foundation (to J.C.M.). We also acknowledge funding from the CFF (DRUMM15R0 and DRUMM15XX1) and NIH (P01 HL128192) to M.L.D. The HPIC and ABBC cores are supported by P30-DK117469.
We thank Andy Goodman for the B. thetaiotaomicron strain and Laurie Comstock for her excellent advice and answering many questions. We thank Roxana Barnaby and the EMIC for assessing transepithelial resistance of the Caco-2 lines and Tom Hampton from the ABBC, and Modupe Coker for consulting on statistics. We also thank Devin Harbin and Michelle Tyler for their assistance.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00274-19.
REFERENCES
- 1.Boucher RC. 2004. New concepts of the pathogenesis of cystic fibrosis lung disease. Eur Respir J 23:146–158. doi: 10.1183/09031936.03.00057003. [DOI] [PubMed] [Google Scholar]
- 2.Lyczak JB, Cannon CL, Pier GB. 2002. Lung infections associated with cystic fibrosis. Clin Microbiol Rev 15:194–222. doi: 10.1128/cmr.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang NN, Van Loon EL, Sheng KT. 1961. The flora of the respiratory tract of patients with cystic fibrosis of the pancreas. J Pediatr 59:512–521. doi: 10.1016/S0022-3476(61)80234-5. [DOI] [PubMed] [Google Scholar]
- 4.Deretic V, Martin DW, Schurr MJ, Mudd MH, Hibler NS, Curcic R, Boucher JC. 1993. Conversion to mucoidy in Pseudomonas aeruginosa. Nat Biotechnol 11:1133–1136. doi: 10.1038/nbt1093-1133. [DOI] [PubMed] [Google Scholar]
- 5.Govan JR, Nelson JW. 1992. Microbiology of lung infection in cystic fibrosis. Br Med Bull 48:912–930. doi: 10.1093/oxfordjournals.bmb.a072585. [DOI] [PubMed] [Google Scholar]
- 6.Castellani C, Assael BM. 2017. Cystic fibrosis: a clinical view. Cell Mol Life Sci 74:129–140. doi: 10.1007/s00018-016-2393-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scanlan PD, Buckling A, Kong W, Wild Y, Lynch SV, Harrison F. 2012. Gut dysbiosis in cystic fibrosis. J Cyst Fibros 11:454–455. doi: 10.1016/j.jcf.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Hoen AG, Li J, Moulton LA, O’Toole GA, Housman ML, Koestler DC, Guill MF, Moore JH, Hibberd PL, Morrison HG, Sogin ML, Karagas MR, Madan JC. 2015. Associations between gut microbial colonization in early life and respiratory outcomes in cystic fibrosis. J Pediatr 167:138–147.e1.3. doi: 10.1016/j.jpeds.2015.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farrelly PJ, Charlesworth C, Lee S, Southern KW, Baillie CT. 2014. Gastrointestinal surgery in cystic fibrosis: a 20-year review. J Pediatr Surg 49:280–283. doi: 10.1016/j.jpedsurg.2013.11.038. [DOI] [PubMed] [Google Scholar]
- 10.Rogers GB, Narkewicz MR, Hoffman LR. 2016. The CF gastrointestinal microbiome: structure and clinical impact. Pediatr Pulmonol 51:S35–S44. doi: 10.1002/ppul.23544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boczar M, Sawicka E, Zybert K. 2015. Meconium ileus in newborns with cystic fibrosis—results of treatment in the group of patients operated on in the years 2000–2014. Dev Period Med 19:32–40. [PubMed] [Google Scholar]
- 12.Bodewes FA, Verkade HJ, Taminiau JA, Borowitz D, Wilschanski M, Working group Cystic Fibrosis Pancreatic Disease of the European Society for Paediatric Gastroenterology Hepatology and Nutrition. 2015. Cystic fibrosis and the role of gastrointestinal outcome measures in the new era of therapeutic CFTR modulation. J Cyst Fibros 14:169–177. doi: 10.1016/j.jcf.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Dorsey J, Gonska T. 2017. Bacterial overgrowth, dysbiosis, inflammation, and dysmotility in the cystic fibrosis intestine. J Cyst Fibros 16(Suppl 2):S14–S23. doi: 10.1016/j.jcf.2017.07.014. [DOI] [PubMed] [Google Scholar]
- 14.Woestenenk JW, van der Ent CK, Houwen RH. 2015. Pancreatic enzyme replacement therapy and coefficient of fat absorption in children and adolescents with cystic fibrosis. J Pediatr Gastroenterol Nutr 61:355–360. doi: 10.1097/MPG.0000000000000784. [DOI] [PubMed] [Google Scholar]
- 15.Madan JC, Koestler DC, Stanton BA, Davidson L, Moulton LA, Housman ML, Moore JH, Guill MF, Morrison HG, Sogin ML, Hampton TH, Karagas MR, Palumbo PE, Foster JA, Hibberd PH, O’Toole GA. 2012. Serial analysis of the gut and respiratory microbiome in CF in infancy: the interaction between intestinal and respiratory tracts and the impact of nutritional exposures. mBio 3:e00251-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chong C, Bloomfield F, O’Sullivan J. 2018. Factors affecting gastrointestinal microbiome development in neonates. Nutrients 10:274. doi: 10.3390/nu10030274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toscano M, De Grandi R, Grossi E, Drago L. 2017. Role of the human breast milk-associated microbiota on the newborns' immune system: a mini review. Front Microbiol 8:2100. doi: 10.3389/fmicb.2017.02100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes S, Mancabelli L, Lugli GA, Rodriguez JM, Bode L, de Vos W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. 2017. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev 81:e00036-17. doi: 10.1128/MMBR.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert-Diamond D, Emond JA, Baker ER, Korrick SA, Karagas MR. 2016. Relation between in utero arsenic exposure and birth outcomes in a cohort of mothers and their newborns from New Hampshire. Environ Health Perspect 124:1299–1307. doi: 10.1289/ehp.1510065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fei DL, Koestler DC, Li Z, Giambelli C, Sanchez-Mejias A, Gosse JA, Marsit CJ, Karagas MR, Robbins DJ. 2013. Association between in utero arsenic exposure, placental gene expression, and infant birth weight: a US birth cohort study. Environ Health 12:58. doi: 10.1186/1476-069X-12-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farzan SF, Korrick S, Li Z, Enelow R, Gandolfi AJ, Madan J, Nadeau K, Karagas MR. 2013. In utero arsenic exposure and infant infection in a United States cohort: a prospective study. Environ Res 126:24–30. doi: 10.1016/j.envres.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madan JC, Hoen AG, Lundgren SN, Farzan SF, Cottingham KL, Morrison HG, Sogin ML, Li H, Moore JH, Karagas MR. 2016. Association of Cesarean delivery and formula supplementation with the intestinal microbiome of 6-week-old infants. JAMA Pediatr 170:212–219. doi: 10.1001/jamapediatrics.2015.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maier E, Anderson RC, Roy NC. 2014. Understanding how commensal obligate anaerobic bacteria regulate immune functions in the large intestine. Nutrients 7:45–73. doi: 10.3390/nu7010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thaiss CA, Levy M, Suez J, Elinav E. 2014. The interplay between the innate immune system and the microbiota. Curr Opin Immunol 26:41–48. doi: 10.1016/j.coi.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 25.Lyons A, O’Mahony D, O’Brien F, MacSharry J, Sheil B, Ceddia M, Russell WM, Forsythe P, Bienenstock J, Kiely B, Shanahan F, O’Mahony L. 2010. Bacterial strain-specific induction of Foxp3+ T regulatory cells is protective in murine allergy models. Clin Exp Allergy 40:811–819. [DOI] [PubMed] [Google Scholar]
- 26.Isolauri E, Sutas Y, Kankaanpaa P, Arvilommi H, Salminen S. 2001. Probiotics: effects on immunity. Am J Clin Nutr 73:444S–450S. doi: 10.1093/ajcn/73.2.444s. [DOI] [PubMed] [Google Scholar]
- 27.Abdulkadir B, Nelson A, Skeath T, Marrs EC, Perry JD, Cummings SP, Embleton ND, Berrington JE, Stewart CJ. 2016. Routine use of probiotics in preterm infants: longitudinal impact on the microbiome and metabolome. Neonatology 109:239–247. doi: 10.1159/000442936. [DOI] [PubMed] [Google Scholar]
- 28.Fanning S, Hall LJ, Cronin M, Zomer A, MacSharry J, Goulding D, Motherway MO, Shanahan F, Nally K, Dougan G, van Sinderen D. 2012. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc Natl Acad Sci U S A 109:2108–2113. doi: 10.1073/pnas.1115621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butel MJ, Suau A, Campeotto F, Magne F, Aires J, Ferraris L, Kalach N, Leroux B, Dupont C. 2007. Conditions of bifidobacterial colonization in preterm infants: a prospective analysis. J Pediatr Gastroenterol Nutr 44:577–582. doi: 10.1097/MPG.0b013e3180406b20. [DOI] [PubMed] [Google Scholar]
- 30.Kim JM, Oh YK, Kim YJ, Oh HB, Cho YJ. 2001. Polarized secretion of CXC chemokines by human intestinal epithelial cells in response to Bacteroides fragilis enterotoxin: NF-kappa B plays a major role in the regulation of IL-8 expression. Clin Exp Immunol 123:421–427. doi: 10.1046/j.1365-2249.2001.01462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enaud R, Hooks KB, Barre A, Barnetche T, Hubert C, Massot M, Bazin T, Clouzeau H, Bui S, Fayon M, Berger P, Lehours P, Bebear C, Nikolski M, Lamireau T, Delhaes L, Schaeverbeke T. 2019. Intestinal inflammation in children with cystic fibrosis is associated with Crohn’s-like microbiota disturbances. J Clin Med 8:645. doi: 10.3390/jcm8050645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Freitas MB, Moreira EAM, Tomio C, Moreno YMF, Daltoe FP, Barbosa E, Ludwig Neto N, Buccigrossi V, Guarino A. 2018. Altered intestinal microbiota composition, antibiotic therapy and intestinal inflammation in children and adolescents with cystic fibrosis. PLoS One 13:e0198457. doi: 10.1371/journal.pone.0198457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miragoli F, Federici S, Ferrari S, Minuti A, Rebecchi A, Bruzzese E, Buccigrossi V, Guarino A, Callegari ML. 2017. Impact of cystic fibrosis disease on archaea and bacteria composition of gut microbiota. FEMS Microbiol Ecol 93:fiw230. doi: 10.1093/femsec/fiw230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flass T, Tong S, Frank DN, Wagner BD, Robertson CE, Kotter CV, Sokol RJ, Zemanick E, Accurso F, Hoffenberg EJ, Narkewicz MR. 2015. Intestinal lesions are associated with altered intestinal microbiome and are more frequent in children and young adults with cystic fibrosis and cirrhosis. PLoS One 10:e0116967. doi: 10.1371/journal.pone.0116967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruzzese E, Callegari ML, Raia V, Viscovo S, Scotto R, Ferrari S, Morelli L, Buccigrossi V, Lo Vecchio A, Ruberto E, Guarino A. 2014. Disrupted intestinal microbiota and intestinal inflammation in children with cystic fibrosis and its restoration with Lactobacillus GG: a randomised clinical trial. PLoS One 9:e87796. doi: 10.1371/journal.pone.0087796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matamouros S, Hayden HS, Hager KR, Brittnacher MJ, Lachance K, Weiss EJ, Pope CE, Imhaus AF, McNally CP, Borenstein E, Hoffman LR, Miller SI. 2018. Adaptation of commensal proliferating Escherichia coli to the intestinal tract of young children with cystic fibrosis. Proc Natl Acad Sci U S A doi: 10.1073/pnas.1714373115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffman LR, Pope CE, Hayden HS, Heltshe S, Levy R, McNamara S, Jacobs MA, Rohmer L, Radey M, Ramsey BW, Brittnacher MJ, Borenstein E, Miller SI. 2014. Escherichia coli dysbiosis correlates with gastrointestinal dysfunction in children with cystic fibrosis. Clin Infect Dis 58:396–399. doi: 10.1093/cid/cit715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Billard L, Le Berre R, Pilorge L, Payan C, Hery-Arnaud G, Vallet S. 2017. Viruses in cystic fibrosis patients’ airways. Crit Rev Microbiol 43:690–708. doi: 10.1080/1040841X.2017.1297763. [DOI] [PubMed] [Google Scholar]
- 39.Smyth A. 2016. Treatment of pulmonary exacerbations in cystic fibrosis—could do better? Paediatr Respir Rev 20(Suppl):6–7. doi: 10.1016/j.prrv.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 40.Pittman JE, Cutting G, Davis SD, Ferkol T, Boucher R. 2014. Cystic fibrosis: NHLBI workshop on the primary prevention of chronic lung diseases. Ann Am Thorac Soc 11(Suppl 3):S161–S168. doi: 10.1513/AnnalsATS.201312-444LD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ozen Alahdab Y, Duman DG. 2016. Pancreatic involvement in cystic fibrosis. Minerva Med 107:427–436. [PubMed] [Google Scholar]
- 42.Schindler T, Michel S, Wilson AW. 2015. Nutrition management of cystic fibrosis in the 21st century. Nutr Clin Pract 30:488–500. doi: 10.1177/0884533615591604. [DOI] [PubMed] [Google Scholar]
- 43.Anderson JL, Miles C, Tierney AC. 2017. Effect of probiotics on respiratory, gastrointestinal and nutritional outcomes in patients with cystic fibrosis: a systematic review. J Cyst Fibros 16:186–197. doi: 10.1016/j.jcf.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 44.Van Biervliet S, Declercq D, Somerset S. 2017. Clinical effects of probiotics in cystic fibrosis patients: a systematic review. Clin Nutr ESPEN 18:37–43. doi: 10.1016/j.clnesp.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 45.Ananthan A, Balasubramanian H, Rao S, Patole S. 2016. Probiotic supplementation in children with cystic fibrosis—a systematic review. Eur J Pediatr 175:1255–1266. doi: 10.1007/s00431-016-2769-8. [DOI] [PubMed] [Google Scholar]
- 46.Filkins LM, Hampton TH, Gifford AH, Gross MJ, Hogan DA, Sogin ML, Morrison HG, Paster BJ, O’Toole GA. 2012. Prevalence of streptococci and increased polymicrobial diversity associated with cystic fibrosis patient stability. J Bacteriol 194:4709–4717. doi: 10.1128/JB.00566-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gifford AH, Alexandru DM, Li Z, Dorman DB, Moulton LA, Price KE, Hampton TH, Sogin ML, Zuckerman JB, Parker HW, Stanton BA, O’Toole GA. 2014. Iron supplementation does not worsen respiratory health or alter the sputum microbiome in cystic fibrosis. J Cyst Fibros 13:311–318. doi: 10.1016/j.jcf.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hampton TH, Green DM, Cutting GR, Morrison HG, Sogin ML, Gifford AH, Stanton BA, O’Toole GA. 2014. The microbiome in pediatric cystic fibrosis patients: the role of shared environment suggests a window of intervention. Microbiome 2:14. doi: 10.1186/2049-2618-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price KE, Hampton TH, Gifford AH, Dolben EL, Hogan DA, Morrison HG, Sogin ML, O’Toole GA. 2013. Unique microbial communities persist in individual cystic fibrosis patients throughout a clinical exacerbation. Microbiome 1:27. doi: 10.1186/2049-2618-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org. [Google Scholar]
- 51.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glockner FO. 2014. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res 42:D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen J, Bittinger K, Charlson ES, Hoffmann C, Lewis J, Wu GD, Collman RG, Bushman FD, Li H. 2012. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics 28:2106–2113. doi: 10.1093/bioinformatics/bts342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schliep KP. 2011. phangorn: phylogenetic analysis in R. Bioinformatics 27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dixon P. 2003. VEGAN, a package of R functions for community ecology. J Veg Sci 14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 57.Kizer NL, Vandorpe D, Lewis B, Bunting B, Russell J, Stanton BA. 1995. Vasopressin and cAMP stimulate electrogenic chloride secretion in an IMCD cell line. Am J Physiol 268:F854–F861. doi: 10.1152/ajprenal.1995.268.5.F854. [DOI] [PubMed] [Google Scholar]
- 58.Hao S, Roesch E, Perez A, Weiner RL, Henderson LC, Cummings L, Consiglio P, Pajka J, Eisenberg A, Yeh L, Cotton CU, Drumm ML. Inactivation of CFTR by CRISPR/Cas9 alters transcriptional regulation of inflammatory pathways and other networks. J Cyst Fibros, in press. doi: 10.1016/j.jcf.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 59.O’Connor RM, Fung JM, Sharp KH, Benner JS, McClung C, Cushing S, Lamkin ER, Fomenkov AI, Henrissat B, Londer YY, Scholz MB, Posfai J, Malfatti S, Tringe SG, Woyke T, Malmstrom RR, Coleman-Derr D, Altamia MA, Dedrick S, Kaluziak ST, Haygood MG, Distel DL. 2014. Gill bacteria enable a novel digestive strategy in a wood-feeding mollusk. Proc Natl Acad Sci U S A 111:E5096–E5104. doi: 10.1073/pnas.1413110111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lane DJ. 1991. 16S/23S sequencing, p 115–175. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.